Published online May 15, 2025. doi: 10.4251/wjgo.v17.i5.105872
Revised: March 27, 2025
Accepted: April 17, 2025
Published online: May 15, 2025
Processing time: 96 Days and 0.1 Hours
Hepatocellular carcinoma (HCC) is a major global contributor to cancer-related mortality, with advanced stages presenting substantial therapeutic challenges. Although targeted immunotherapy shows potential, many patients exhibit poor responses, underscoring the need for predictive tools to optimize treatment strategies. Emerging data indicate that ultrasound features (e.g., tumor stiffness) and serum biomarkers may serve as predictors of treatment outcomes. However, an integrated model for these predictors remains unavailable. This paper introduces a machine learning-based approach that combines ultrasound and serological data to forecast immunotherapy efficacy in patients with advanced HCC.
To develop a non-invasive predictive model for targeted immunotherapy in advanced HCC, incorporating both internal and external validation.
Patients with advanced HCC who received targeted immunotherapy at two medical centers were enrolled and divided into internal training, internal validation, and external validation cohorts. Comprehensive clinical data were gathered. Initially, 13 machine learning algorithms were tested using the internal training cohort. The algorithm yielding the highest area under the curve (AUC) in the internal validation cohort was selected to construct a predictive model, termed the Target Immunotherapy Predictive Model (TIPM). TIPM performance was then compared with that of traditional tumor staging systems (tumor-node-metastasis, Barcelona Clinic Liver Cancer, China Liver Cancer, Hong Kong Liver Cancer, and C-reactive protein and alpha-fetoprotein in immunotherapy).
A total of 306 patients participated in the study, with 143 in the internal training cohort, 62 in the internal validation cohort, and 101 in the external validation cohort. In the internal validation cohort, the random forest model achieved the highest AUC (0.975, 95% confidence interval: 0.924-0.998). The key predictors for TIPM were tumor size, platelet count, tumor stiffness change, and white blood cell count. During external validation, TIPM outperformed conventional models, reaching an AUC of 0.899 (95% confidence interval: 0.840-0.957). Calibration curves demonstrated strong concordance with observed outcomes, while decision curve analysis confirmed TIPM’s enhanced clinical value. Additional metrics, such as the net reclassification index and integrated discrimination improvement, further supported TIPM’s superior predictive accuracy.
TIPM provides a robust tool for predicting targeted immunotherapy efficacy in advanced HCC, facilitating personalized treatment planning.
Core Tip: Predicting treatment response in patients with advanced hepatocellular carcinoma undergoing targeted immunotherapy remains a significant challenge. This study developed and validated the Target Immunotherapy Predictive Model, a novel, non-invasive tool that integrates pre-treatment ultrasound features (tumor diameter, changes in tumor stiffness) and accessible serum biomarkers (platelet count, white blood cell count) using machine learning. The Target Immunotherapy Predictive Model outperformed traditional staging systems in predicting treatment efficacy, providing a promising solution for personalized treatment selection in advanced hepatocellular carcinoma. This approach has the potential to minimize unnecessary treatments, optimize therapeutic strategies, and ultimately improve patient outcomes. A user-friendly web calculator further enhances its clinical applicability.
- Citation: Tu HB, Feng SY, Chen LH, Huang YJ, Zhang JZ, Peng SY, Lin DL, Ye XJ. Integrating ultrasound and serum indicators for evaluating outcomes of targeted immunotherapy in advanced liver cancer. World J Gastrointest Oncol 2025; 17(5): 105872
- URL: https://www.wjgnet.com/1948-5204/full/v17/i5/105872.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i5.105872
Hepatocellular carcinoma (HCC) remains a leading global cause of cancer mortality, characterized by high prevalence and poor prognoses, presenting significant clinical challenges[1]. Recent advancements in immunotherapy have shown considerable potential in the treatment of advanced HCC. Combined therapies that integrate immunotherapy with anti-angiogenic agents have demonstrated superior efficacy compared to monotherapies[2]. Notably, the atezolizumab-bevacizumab regimen has emerged as a first-line treatment, showing improved survival outcomes over sorafenib[3]. Despite these advances, the overall effectiveness of immunotherapy in advanced HCC is still limited[4], with a large proportion of patients failing to achieve a meaningful clinical response. This highlights the critical need for strategies to better identify potential responders and refine treatment approaches[5-7].
Researchers are investigating potential biomarkers to predict treatment response and guide patient selection. Previous studies have explored genetic mutations and noncoding RNAs as predictors of targeted immunotherapy efficacy[8,9]. Wu et al[10] highlighted the accuracy of nuclear medicine in predicting treatment outcomes. However, these approaches are technically complex and costly, limiting their widespread clinical application. Currently, there is a notable absence of predictive models specifically designed to assess the efficacy of targeted immunotherapy in HCC. Conventional staging systems - tumor-node-metastasis (TNM), Barcelona Clinic Liver Cancer (BCLC), China Liver Cancer (CNLC), and Hong Kong Liver Cancer (HKLC) - rely predominantly on clinical and anatomical factors. Although these models provide important prognostic insights, they fail to incorporate more dynamic, biologically relevant biomarkers that could offer a more precise prediction of treatment outcomes. The C-reactive protein (CRP) and alpha-fetoprotein (AFP) in immunotherapy (CRAFITY) score, developed by Scheiner et al[11], has demonstrated good predictive accuracy for immunotherapy outcomes, reinforcing the feasibility of predicting targeted immunotherapy efficacy. Emerging ultrasonographic features, such as tumor stiffness and internal echogenicity, alongside serum markers like AFP, CRP, and white blood cell count (WBC), have shown promise in predicting tumor behavior and postoperative recurrence. Research has de
Machine learning not only identifies linear relationships within data but also uncovers complex nonlinear interactions, which are essential for understanding the intrinsic relationships among diverse indicators. Machine learning’s application in disease prediction and management is well-established. However, its use in predicting the efficacy of targeted immunotherapy for HCC remains underutilized. Thus, leveraging machine learning to predict treatment outcomes in targeted immunotherapy represents a significant and innovative advancement. This study attempted to address this gap by evaluating the predictive potential of integrating ultrasonographic and serological biomarkers with machine learning algorithms to forecast targeted immunotherapy response in advanced HCC. Through a comprehensive methodology that includes model development, validation, and comparison with existing tumor staging systems, the study aimed to provide valuable insights to enhance clinical decision-making and refine therapeutic strategies for advanced HCC.
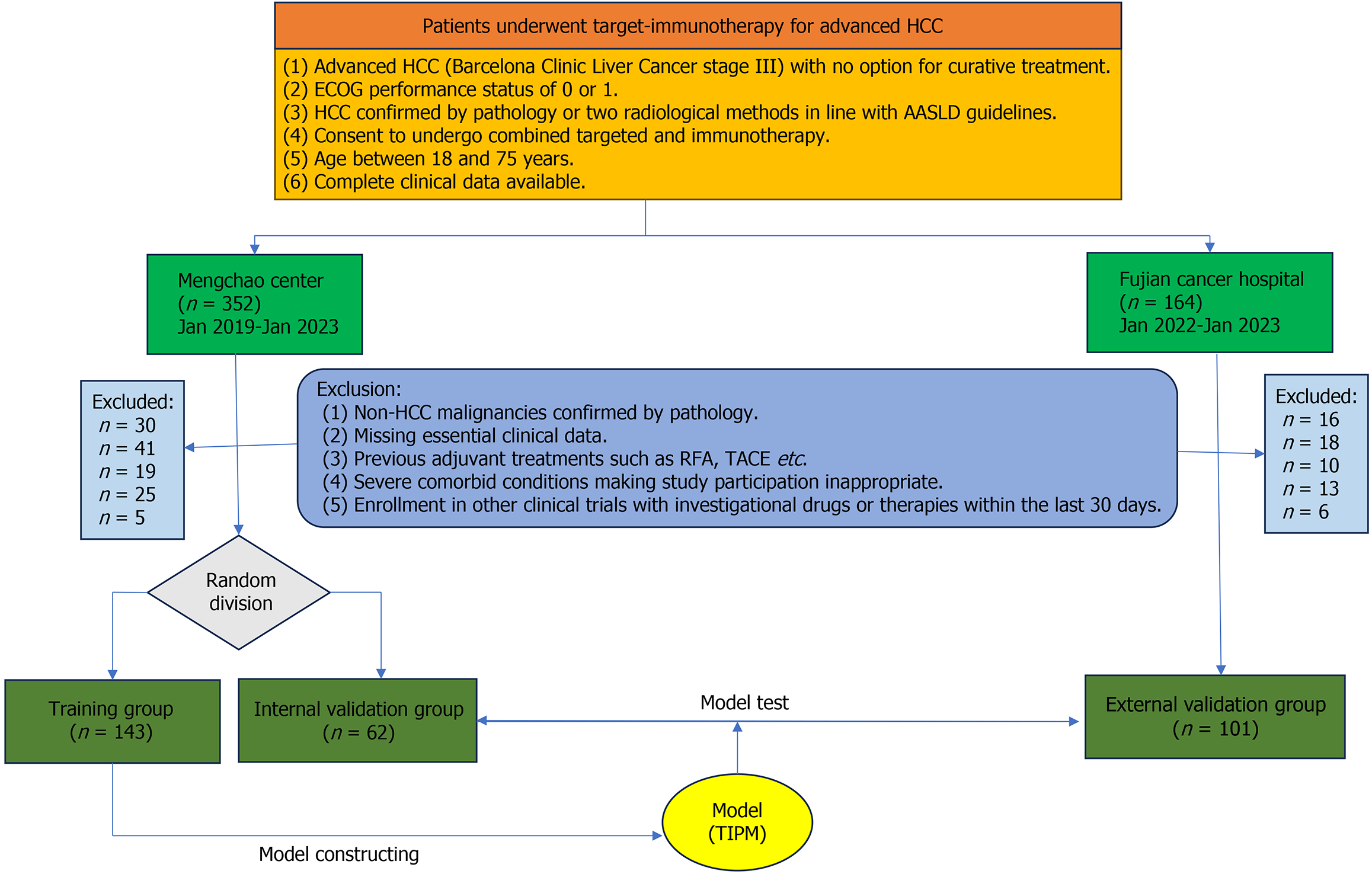
This retrospective analysis evaluated the outcomes of patients with advanced HCC undergoing combined targeted therapy and immunotherapy. Data were collected from Mengchao Hepatobiliary Hospital (2019–2023) and Fujian Province Tumor Hospital (2022–2023). Ethical approval was obtained from the Mengchao Hepatobiliary Hospital Ethics Committee (Approval No. 2021_084_01), and written informed consent was secured from all participants. All procedures adhered to ethical standards and the Declaration of Helsinki. The patient inclusion flow is shown in Figure 1.
The inclusion criteria were: (1) Confirmed diagnosis of advanced HCC or cases with no curative treatment options; (2) Pathological confirmation of HCC or dual radiological assessment following American Association for the Study of Liver Diseases criteria; (3) Agreement to receive combined targeted therapy and immunotherapy; and (4) Availability of comprehensive clinical records.
The exclusion criteria included: (1) Pathologically confirmed non-HCC malignancies; (2) Missing or incomplete clinical records; (3) Prior systemic therapies for HCC, including transarterial chemoembolization or radiofrequency ablation; (4) Severe comorbidities precluding participation; and (5) Participation in other drug trials within the past month.
Treatment response was assessed using 3.0T Siemens magnetic resonance imaging, with scans captured during the arterial, portal, and delayed phases. Monthly imaging data were collected pre- and post-treatment, continuing for up to 26 weeks or until death. Two radiologists independently evaluated the scans according to modified response evaluation criteria in solid tumors criteria[15]. At the 26-week mark, patients were categorized into two groups: Target and immunotherapy effective and noneffective. The target and immunotherapy effective group included patients with complete response, partial response, or stable disease, while the noneffective group consisted of those with progressive disease. Initially, two senior radiologists independently reviewed the imaging findings, and any disagreements were resolved through collaborative consensus.
To ensure data accuracy and reliability, fasting serum samples were meticulously collected in the morning. The biomarker panel analyzed included various biochemical and hematological markers such as albumin, alkaline phosphatase, alanine aminotransferase, platelet count (PLT), and WBC. Quantification of AFP and CRP levels was also performed. The neutrophil-to-lymphocyte ratio was calculated to assess the inflammatory and immune response. These indices collectively provided a comprehensive biochemical and hematological profile, offering valuable insights into the underlying pathophysiology and progression of the disease.
Prior to treatment, essential patient characteristics such as sex, age, height, weight, and body mass index were recorded.
Camrelizumab (Airuika, Shengdiya) was administered intravenously for immunotherapy, while lenvatinib mesylate capsules (Leweima, Weicai) were used for targeted therapy until the patient either voluntarily withdrew, experienced disease progression, or developed severe complications.
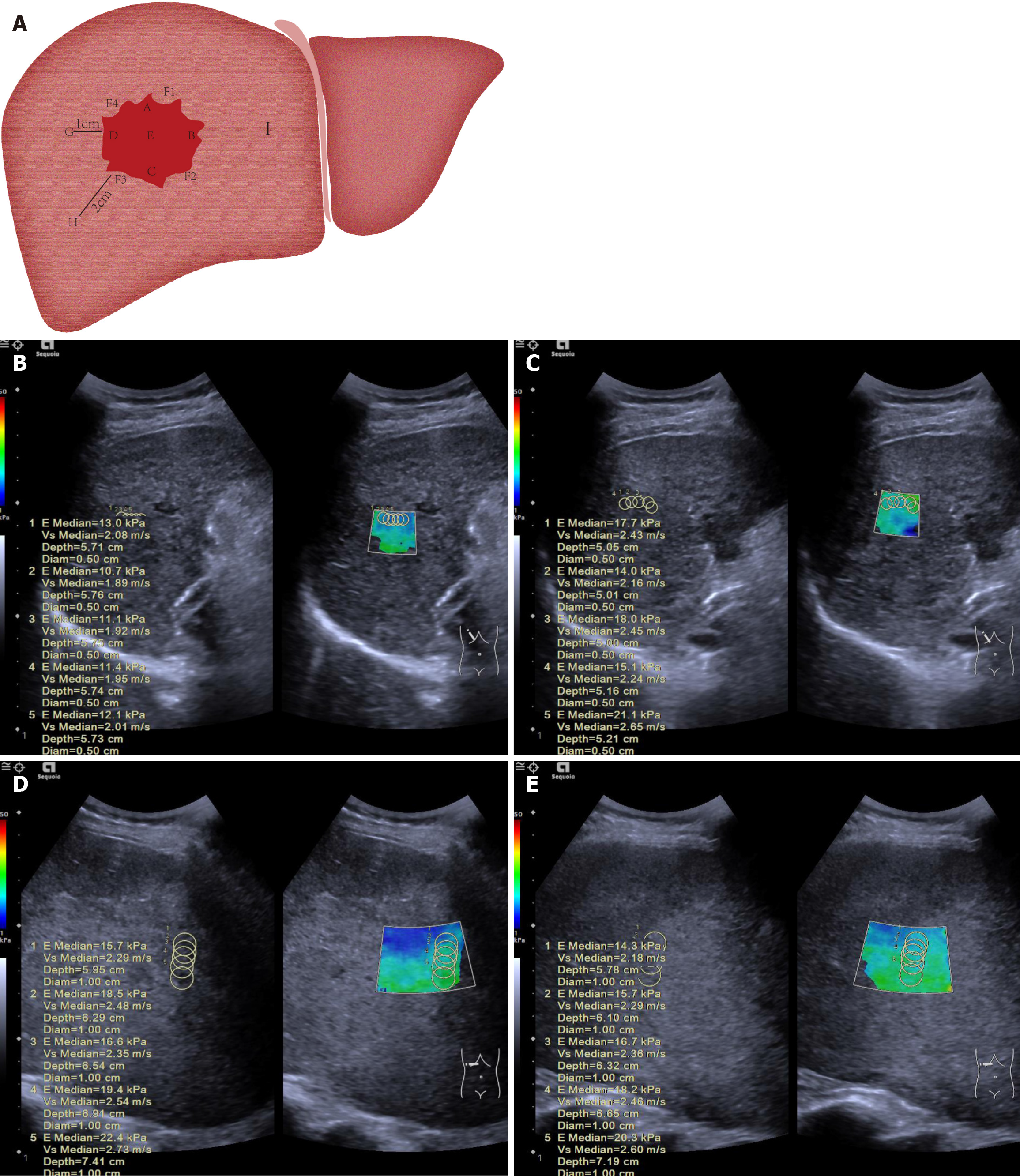
Ultrasound imaging was performed using a Siemens Sequoia system (Germany) with a C3-7 probe (frequency range: 3-7 MHz) at Fujian Province Tumor Hospital and Mengchao Hepatobiliary Hospital. To ensure consistency and accuracy in stiffness measurements, all scans were conducted by experienced radiologists with a minimum of 10 years in liver sonography and elastography. These radiologists underwent standardized training, which included protocol reviews, hands-on practice with phantoms, and supervised patient scanning. All pre-treatment scans were completed within 3 days prior to the initiation of treatment, providing a consistent baseline across all participants. In cases involving multiple lesions, the lesion with the largest diameter was designated as the index lesion. The following characteristics were recorded: Maximum tumor diameter, capsule presence and smoothness, margin clarity, internal echogenicity homogeneity, and the presence of satellite nodules.
The region of interest was segmented into three distinct areas. A 12-point clock-face method was employed to define specific locations within and around the tumor: Point A marked the superior aspect of the tumor (12 o’clock position, closest to the transducer); point B represented the left lateral margin (3 o’clock position); point C indicated the inferior aspect (6 o’clock position, farthest from the transducer); point D denoted the right lateral margin (9 o’clock position); and point E represented the central region. Peritumoral tissue, defined as normal liver tissue adjacent to the tumor capsule, was sampled at four locations: F1 (2 o’clock), F2 (5 o’clock), F3 (8 o’clock), and F4 (11 o’clock). Normal liver tissue was also sampled at increasing distances from the tumor margin: Point G (1 cm), point H (2 cm), and point I (greater than 2 cm). A schematic representation of these measurement points is presented in Figure 2. After selecting the target locations, the patient was instructed to hold his/her breath, and the elastography mode was activated. Five stiffness measurements were taken at each of the aforementioned sites, with the median value recorded as the study parameter. This procedure was performed both pre-treatment and at the sixth week post-treatment. If stiffness increased at five or more of the nine measurement points within the tumor and adjacent peritumoral liver tissue, it was recorded as ΔT > 0; otherwise, ΔT was recorded as < 0. Similarly, if stiffness increased at two or more of the remaining three points, it was recorded as ΔP > 0; otherwise, ΔP was recorded as < 0.
Hepatic nodules exhibiting typical arterial enhancement followed by rapid washout on two or more of the following imaging modalities – contrast-enhanced CT, contrast-enhanced magnetic resonance imaging, or contrast-enhanced ultrasound – or confirmed by histopathology, were classified as HCC.
Various staging systems are used, each assigning points based on tumor characteristics such as size, extent, vascular involvement, and metastatic spread.
TNM staging system: (1) Stage I: 1 point; (2) Stage II: 2 points; (3) Stage III-A: 3 points; (4) Stage III-B: 4 points; (5) Stage III-C: 5 points; and (6) Stage IV: 6 points.
BCLC staging system: (1) Stage 0: 1 point; (2) Stage A: 2 points; (3) Stage B: 3 points; (4) Stage C: 4 points; and (5) Stage D: 5 points.
CNLC staging system: (1) Stage I: 1 point; (2) Stage II: 2 points; (3) Stage III: 3 points; and (4) Stage IV: 4 points.
HKLC staging system: The HKLC staging system is as followed: (1) Stage I: 1 point; (2) Stage II: 2 points; (3) Stage III: 3 points; (4) Stage IV: 4 points; and (5) Stage V: 5 points.
CRAFITY score[11]: (1) AFP level exceeding 100 ng/mL: 1 point; CRP level greater than 1 mg/dL: 1 point; and (2) Patient stratification is performed based on the following categories: 0 points: CRP ≤ 1 mg/dL and AFP ≤ 100 ng/mL; 1 point: Presence of either CRP > 1 mg/dL or AFP > 100 ng/mL. 2 points: Co-occurrence of both CRP > 1 mg/dL and AFP > 100 ng/mL.
Continuous variables in this study are presented as median (25th percentiles, 75th percentiles), and categorical variables as proportions. For normally distributed continuous data with homogeneous variance, analysis of variance was used; otherwise, the Kruskal-Wallis test was applied. Categorical data were compared using the χ2-test or Fisher’s exact test, as appropriate. Thirteen machine learning algorithms were employed, including random forest (RF), gradient boosting, light gradient boosting machine, voting classifier, support vector classifier, logistic regression, extreme gradient boosting, extra trees, k-nearest neighbors, decision tree, naive Bayes, AdaBoost, and Ridge classifier. A random seed was set, and patients from Mengchao Hepatobiliary Hospital were divided into an internal training cohort and an internal validation cohort, while data from Fujian Cancer Hospital formed the external validation cohort. All cohorts underwent standardization prior to model construction to ensure uniformity.
Following model construction with the internal training cohort, model performance was assessed using the internal validation cohort. Predictive performance was evaluated using metrics including positive predictive value, negative predictive value, sensitivity, specificity, accuracy, and area under the receiver operating characteristic curve (AUC-ROC). The algorithm yielding the highest AUC was selected for further analysis. Variables were ranked according to importance, and the relationship between the number of variables and AUC was examined. In adherence to model parsimony, the minimal number of variables required to achieve satisfactory predictive performance was incorporated into the final model. The final model, termed the Targeted Immunotherapy Predictive Model (TIPM), was developed using the selected variables.
TIPM was then compared against established tumor staging systems (TNM, BCLC, CNLC, HKLC, and CRAFITY) within the external validation cohort. AUCs of the different models were compared using DeLong’s test. Calibration curves were plotted, and metrics such as Brier scores, expected calibration error, Hosmer-Lemeshow P values, and maximum calibration error were compared. Decision curve analysis (DCA) was performed, and the integrated discrimination improvement and net reclassification index (NRI) were calculated to assess the clinical benefit of each model. To enhance clinical applicability, an interactive web calculator was developed, incorporating the key variables identified in TIPM. Statistical analyses were performed using R (4.1.1) and Python (3.6.5). All tests were two-sided, with statistical significance set at P < 0.05.
A total of 306 patients participated in this study, including 143 in the internal training cohort, 62 in the internal validation cohort, and 101 in the external validation cohort. Among these, 227 patients (74.2%) exhibited an effective response to targeted immunotherapy, while 79 (25.8%) did not. No significant differences in key indicators were observed across the cohorts, confirming their comparability (Table 1).
| Variable | All patients | Group 1 | Group 2 | Group 3 | Test statistic (F/H/χ²) | P value |
| Diameter | 8.46 (7.09, 9.31) | 8.46 (7.27, 9.32) | 8.43 (7.32, 9.24) | 8.28 (7.01, 9.31) | 1.4 | 0.49 |
| Age | 51.00 (46.00, 57.00) | 50.00 (45.00, 56.00) | 52.00 (47.25, 62.00) | 50.00 (45.00, 55.00) | 4.41 | 0.11 |
| PV | 1.10 (1.00, 1.18) | 1.06 (1.00, 1.15) | 1.10 (1.00, 1.17) | 1.10 (1.00, 1.20) | 2.05 | 0.35 |
| Height | 160.00 (155.00, 162.00) | 159.00 (155.00, 162.00) | 159.50 (155.00, 162.00) | 160.00 (156.00, 162.00) | 2.13 | 0.34 |
| Weight | 57.00 (52.00, 62.00) | 56.50 (52.00, 62.00) | 56.50 (51.25, 62.00) | 57.50 (52.00, 62.00) | 1.11 | 0.57 |
| BMI | 22.46 (21.00, 24.22) | 22.49 (21.04, 24.13) | 22.10 (21.04, 24.17) | 22.43 (21.00, 24.61) | 0.51 | 0.77 |
| WBC | 5.29 (4.56, 6.55) | 5.29 (4.58, 6.68) | 5.42 (4.60, 6.09) | 5.28 (4.56, 6.50) | 0.32 | 0.85 |
| Hemoglobin | 123.00 (110.25, 131.00) | 122.00 (110.50, 131.00) | 126.50 (116.50, 131.75) | 120.00 (110.00, 130.00) | 2.19 | 0.33 |
| PLT | 238.50 (201.25, 280.00) | 241.00 (202.00, 279.00) | 231.50 (191.00, 255.00) | 244.00 (205.00, 284.00) | 2.94 | 0.23 |
| Alanine aminotransferase | 59.00 (34.00, 108.00) | 65.00 (41.00, 123.50) | 50.00 (31.25, 82.00) | 56.00 (33.00, 88.00) | 6.52 | 0.03 |
| Aspartate aminotransferase | 68.00 (41.00, 94.00) | 73.00 (43.00, 105.00) | 57.00 (34.75, 88.25) | 68.00 (40.00, 93.00) | 2.91 | 0.23 |
| Alkaline phosphatase | 235.00 (132.25, 377.50) | 243.00 (150.50, 382.50) | 192.50 (132.25, 300.00) | 248.00 (111.00, 482.00) | 1.9 | 0.38 |
| γ-glutamyl transpeptidase | 241.00 (108.00, 442.75) | 247.00 (125.50, 448.00) | 166.50 (108.00, 373.25) | 204.00 (103.00, 448.00) | 1.78 | 0.41 |
| Albumin | 39.00 (35.00, 42.00) | 38.00 (35.00, 42.00) | 39.00 (35.00, 43.00) | 38.00 (35.00, 41.00) | 1.01 | 0.60 |
| Total bilirubin | 16.75 (11.25, 28.80) | 17.30 (11.70, 31.30) | 14.80 (11.00, 20.10) | 15.60 (11.00, 24.80) | 2.92 | 0.23 |
| Direct bilirubin | 3.65 (2.40, 9.20) | 4.20 (2.40, 13.05) | 3.10 (2.23, 5.47) | 3.70 (2.40, 8.10) | 3.81 | 0.14 |
| Indirect bilirubin | 12.10 (8.60, 17.30) | 12.50 (9.35, 18.00) | 11.70 (8.60, 16.73) | 11.10 (8.30, 17.00) | 1.97 | 0.37 |
| Cholesterol | 5.25 (4.26, 6.90) | 5.26 (4.26, 6.90) | 5.33 (4.30, 6.59) | 5.16 (4.12, 7.50) | 0.06 | 0.96 |
| Prothrombin time | 12.10 (11.60, 12.50) | 12.00 (11.60, 12.50) | 12.20 (11.60, 12.50) | 12.20 (11.70, 12.60) | 0.69 | 0.70 |
| Prothrombin time activity | 100.00 (100.00, 125.00) | 100.00 (100.00, 128.50) | 100.00 (100.00, 112.00) | 100.00 (100.00, 132.00) | 2.09 | 0.35 |
| International normalized ratio | 1.00 (0.94, 1.00) | 1.00 (0.96, 1.00) | 1.00 (0.94, 1.00) | 1.00 (0.94, 1.00) | 0.05 | 0.97 |
| TIN | 79 (25.82) | 40 (27.97) | 11 (17.74) | 28 (27.72) | 2.65 | 0.26 |
| TIE | 227 (74.18) | 103 (72.03) | 51 (82.26) | 73 (72.28) | 2.65 | 0.26 |
| NLR > 3 | 140 (45.75) | 65 (45.45) | 28 (45.16) | 47 (46.53) | 0.04 | 0.98 |
| NLR ≤ 3 | 166 (54.25) | 78 (54.55) | 34 (54.84) | 54 (53.47) | 0.04 | 0.98 |
| ΔT non-increasement | 136 (44.44) | 65 (45.45) | 22 (35.48) | 49 (48.51) | 2.75 | 0.25 |
| ΔT increasement | 170 (55.56) | 78 (54.55) | 40 (64.52) | 52 (51.49) | 2.75 | 0.25 |
| ΔP non-increasement | 245 (80.07) | 114 (79.72) | 51 (82.26) | 80 (79.21) | 0.24 | 0.88 |
| ΔP increasement | 61 (19.93) | 29 (20.28) | 11 (17.74) | 21 (20.79) | 0.24 | 0.88 |
| ECOG (0) | 213 (69.61) | 95 (66.43) | 42 (67.74) | 76 (75.25) | 2.3 | 0.31 |
| ECOG (1) | 93 (30.39) | 48 (33.57) | 20 (32.26) | 25 (24.75) | 2.3 | 0.31 |
| Male | 268 (87.58) | 125 (87.41) | 55 (88.71) | 88 (87.13) | 0.1 | 0.95 |
| Female | 38 (12.42) | 18 (12.59) | 7 (11.29) | 13 (12.87) | 0.1 | 0.95 |
| Uneven echo | 108 (35.29) | 57 (39.86) | 19 (30.65) | 32 (31.68) | 2.47 | 0.29 |
| Homogeneous echo | 198 (64.71) | 86 (60.14) | 43 (69.35) | 69 (68.32) | 2.47 | 0.29 |
| Smooth envelope | 37 (12.09) | 19 (13.29) | 5 (8.06) | 13 (12.87) | 1.2 | 0.54 |
| Rough envelope | 269 (87.91) | 124 (86.71) | 57 (91.94) | 88 (87.13) | 1.2 | 0.54 |
| Drinking history (Yes) | 78 (25.49) | 40 (27.97) | 10 (16.13) | 28 (27.72) | 3.59 | 0.16 |
| Drinking history (No) | 228 (74.51) | 103 (72.03) | 52 (83.87) | 73 (72.28) | 3.59 | 0.16 |
| AFP > 100 ng/mL | 195 (63.73) | 89 (62.24) | 38 (61.29) | 68 (67.33) | 0.86 | 0.64 |
| AFP ≤ 100 ng/mL | 111 (36.27) | 54 (37.76) | 24 (38.71) | 33 (32.67) | 0.86 | 0.64 |
| CRP > 1 mg/dL | 176 (57.52) | 86 (60.14) | 33 (53.23) | 57 (56.44) | 0.92 | 0.63 |
| CRP ≤ 1 mg/dL | 130 (42.48) | 57 (39.86) | 29 (46.77) | 44 (43.56) | 0.92 | 0.63 |
| Rough gallbladder | 213 (69.61) | 95 (66.43) | 42 (67.74) | 76 (75.25) | 2.3 | 0.31 |
| Smooth gallbladder | 93 (30.39) | 48 (33.57) | 20 (32.26) | 25 (24.75) | 2.3 | 0.31 |
| Macrovascular invasion (Yes) | 192 (62.75) | 85 (59.44) | 39 (62.90) | 68 (67.33) | 1.58 | 0.45 |
| Macrovascular invasion (No) | 114 (37.25) | 58 (40.56) | 23 (37.10) | 33 (32.67) | 1.58 | 0.45 |
| Viral infection (Yes) | 269 (87.91) | 124 (86.71) | 59 (95.16) | 86 (85.15) | 3.98 | 0.13 |
| Viral infection (No) | 37 (12.09) | 19 (13.29) | 3 (4.84) | 15 (14.85) | 3.98 | 0.13 |
| TNM (IIIA) | 46 (15.03) | 21 (14.69) | 8 (12.90) | 17 (16.83) | 8.79 | 0.18 |
| TNM (IIIB) | 61 (19.93) | 23 (16.08) | 12 (19.35) | 26 (25.74) | 8.79 | 0.18 |
| TNM (IIIC) | 92 (30.07) | 47 (32.87) | 24 (38.71) | 21 (20.79) | 8.79 | 0.18 |
| TNM (IV) | 107 (34.97) | 52 (36.36) | 18 (29.03) | 37 (36.63) | 8.79 | 0.18 |
| BCLC (B) | 61 (19.93) | 28 (19.58) | 10 (16.13) | 23 (22.77) | 1.67 | 0.79 |
| BCLC (C) | 153 (50.00) | 69 (48.25) | 34 (54.84) | 50 (49.50) | 1.67 | 0.79 |
| BCLC (D) | 92 (30.07) | 46 (32.17) | 18 (29.03) | 28 (27.72) | 1.67 | 0.79 |
| CNLC (II) | 61 (19.93) | 35 (24.48) | 10 (16.13) | 16 (15.84) | 4.75 | 0.31 |
| CNLC (III) | 183 (59.80) | 81 (56.64) | 36 (58.06) | 66 (65.35) | 4.75 | 0.31 |
| CNLC (IV) | 62 (20.26) | 27 (18.88) | 16 (25.81) | 19 (18.81) | 4.75 | 0.31 |
| HKLC (II) | 62 (20.26) | 27 (18.88) | 12 (19.35) | 23 (22.77) | 3.59 | 0.73 |
| HKLC (III) | 92 (30.07) | 43 (30.07) | 15 (24.19) | 34 (33.66) | 3.59 | 0.73 |
| HKLC (IV) | 122 (39.87) | 57 (39.86) | 28 (45.16) | 37 (36.63) | 3.59 | 0.73 |
| HKLC (V) | 30 (9.80) | 16 (11.19) | 7 (11.29) | 7 (6.93) | 3.59 | 0.73 |
| CRAFITY (0) | 48 (15.69) | 23 (16.08) | 12 (19.35) | 13 (12.87) | 1.63 | 0.80 |
| CRAFITY (1) | 145 (47.39) | 65 (45.45) | 29 (46.77) | 51 (50.50) | 1.63 | 0.80 |
| CRAFITY (2) | 113 (36.93) | 55 (38.46) | 21 (33.87) | 37 (36.63) | 1.63 | 0.80 |
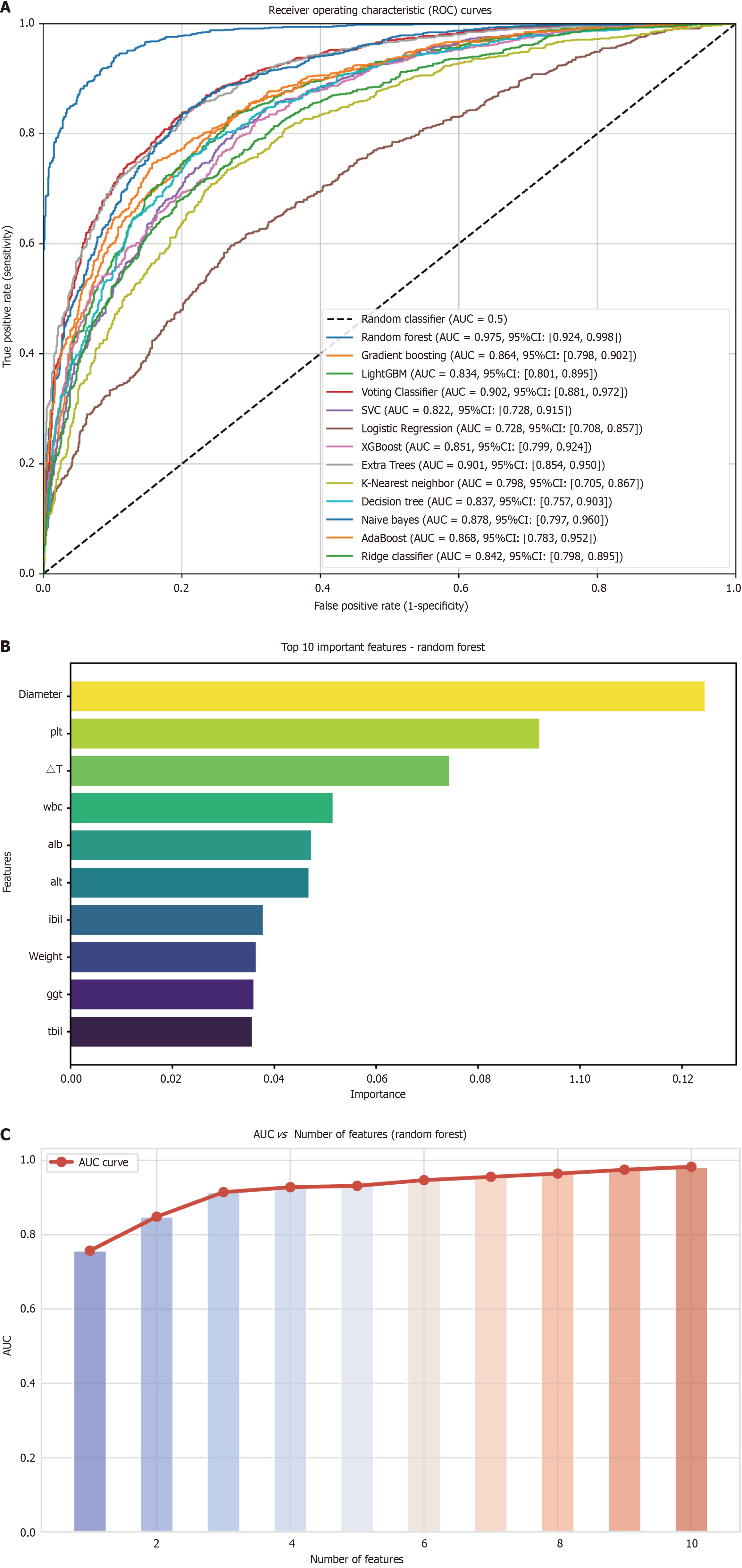
Thirteen machine learning algorithms were utilized to construct predictive models based on data from the internal training cohort. Model performance was evaluated via cross-validation, with metrics such as AUC and other indices calculated for each model. Evaluation results are summarized in Supplementary Table 1. Subsequent validation using the internal validation cohort is presented in Table 2. RF demonstrated the highest AUC among the evaluated models. The corresponding ROC curve is shown in Figure 3A. Based on this outcome, further refinements were incorporated into the model in subsequent phases. To assess the influence of individual variables, Figure 3B displays the importance ranking for each variable in the RF model. The top four variables were: Diameter, PLT, ΔT, and WBC, in order of importance. To optimize clinical utility, the study identified the minimal set of variables for a robust predictive model, as shown in Figure 3C. A high AUC was achieved using just four variables, leading to the final selection of diameter, PLT, ΔT, and WBC for the predictive model, subsequently named TIPM.
| Model | AUC | AUC 95%CI | Accuracy | Sensitivity | Specificity | PPV | NPV |
| Random forest | 0.975 | 0.924-0.998 | 0.951 | 0.727 | 0.981 | 0.957 | 0.944 |
| Gradient boosting | 0.864 | 0.798-0.902 | 0.983 | 0.909 | 0.852 | 0.942 | 0.980 |
| LightGBM | 0.834 | 0.801-0.895 | 0.967 | 0.818 | 0.891 | 0.944 | 0.962 |
| Voting classifier | 0.902 | 0.881-0.972 | 0.983 | 0.909 | 0.948 | 0.879 | 0.980 |
| Support vector classifier | 0.822 | 0.728-0.915 | 0.967 | 0.818 | 0.927 | 0.943 | 0.962 |
| Logistic regression | 0.728 | 0.708-0.857 | 0.967 | 0.909 | 0.980 | 0.909 | 0.980 |
| XGBoost | 0.851 | 0.799-0.924 | 0.983 | 0.909 | 0.893 | 0.915 | 0.980 |
| Extra trees | 0.901 | 0.854-0.95 | 0.935 | 0.727 | 0.980 | 0.888 | 0.943 |
| K-nearest neighbors | 0.798 | 0.705-0.867 | 0.887 | 0.636 | 0.941 | 0.701 | 0.923 |
| Decision tree | 0.837 | 0.757-0.903 | 0.919 | 0.95 | 0.901 | 0.687 | 0.913 |
| Naive Bayes | 0.878 | 0.797-0.960 | 0.838 | 0.818 | 0.843 | 0.529 | 0.955 |
| AdaBoost | 0.868 | 0.783-0.952 | 0.919 | 0.818 | 0.941 | 0.751 | 0.962 |
| Ridge classifier | 0.842 | 0.798-0.895 | 0.967 | 0.909 | 0.980 | 0.909 | 0.980 |
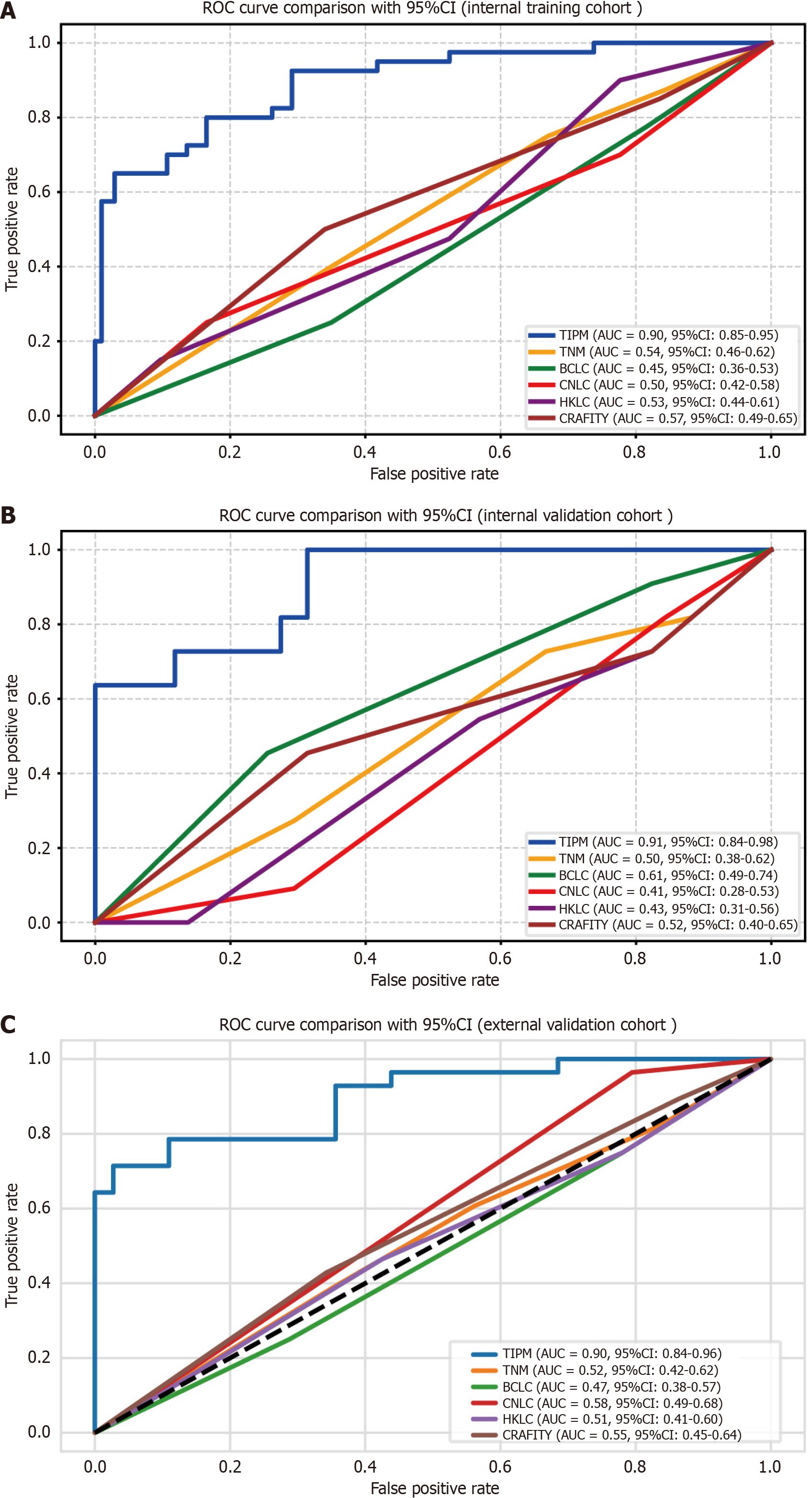
ROC curve analysis: Model validation was conducted through both internal and external assessments, with ROC curves generated for all algorithms. In the internal training cohort, TIPM achieved the highest AUC, as shown in Figure 4A. The AUC for TIPM reached 0.907 in the internal validation cohort (Figure 4B), and in the external validation cohort, TIPM again recorded the peak AUC of 0.899 (Figure 4C). Performance metrics for each cohort are provided in Table 3.
| Model | AUC | AUC 95%CI | Sensitivity | Specificity | PPV | NPV | Accuracy | |
| Internal training cohort | TIPM | 0.895 | 0.845-0.945 | 0.65 | 0.961 | 0.866 | 0.876 | 0.874 |
| TNM | 0.541 | 0.460-0.623 | 0.672 | 0.721 | 0.735 | 0.655 | 0.624 | |
| BCLC | 0.446 | 0.364-0.527 | 0.685 | 0.734 | 0.724 | 0.751 | 0.675 | |
| CNLC | 0.500 | 0.419-0.582 | 0.648 | 0.654 | 0.812 | 0.802 | 0.741 | |
| HKLC | 0.526 | 0.444-0.608 | 0.724 | 0.801 | 0.751 | 0.754 | 0.657 | |
| CRAFITY | 0.571 | 0.490-0.652 | 0.85 | 0.721 | 0.654 | 0.793 | 0.751 | |
| Internal validation cohort | TIPM | 0.907 | 0.835-0.979 | 0.636 | 0.980 | 0.875 | 0.925 | 0.919 |
| TNM | 0.515 | 0.375-0.624 | 0.624 | 0.655 | 0.561 | 0.688 | 0.675 | |
| BCLC | 0.614 | 0.492-0.735 | 0.715 | 0.457 | 0.558 | 0.724 | 0.742 | |
| CNLC | 0.405 | 0.283-0.527 | 0.537 | 0.621 | 0.597 | 0.739 | 0.567 | |
| HKLC | 0.432 | 0.308-0.555 | 0.522 | 0.439 | 0.621 | 0.751 | 0.744 | |
| CRAFITY | 0.524 | 0.400-0.649 | 0.727 | 0.702 | 0.701 | 0.755 | 0.685 | |
| External validation cohort | TIPM | 0.899 | 0.840-0.957 | 0.642 | 0.972 | 0.901 | 0.876 | 0.881 |
| TNM | 0.518 | 0.420-0.615 | 0.577 | 0.715 | 0.544 | 0.674 | 0.655 | |
| BCLC | 0.474 | 0.376-0.571 | 0.622 | 0.702 | 0.658 | 0.587 | 0.724 | |
| CNLC | 0.584 | 0.488-0.680 | 0.478 | 0.658 | 0.728 | 0.653 | 0.726 | |
| HKLC | 0.505 | 0.408-0.603 | 0.685 | 0.653 | 0.648 | 0.724 | 0.658 | |
| CRAFITY | 0.546 | 0.449-0.644 | 0.892 | 0.654 | 0.722 | 0.769 | 0.685 |
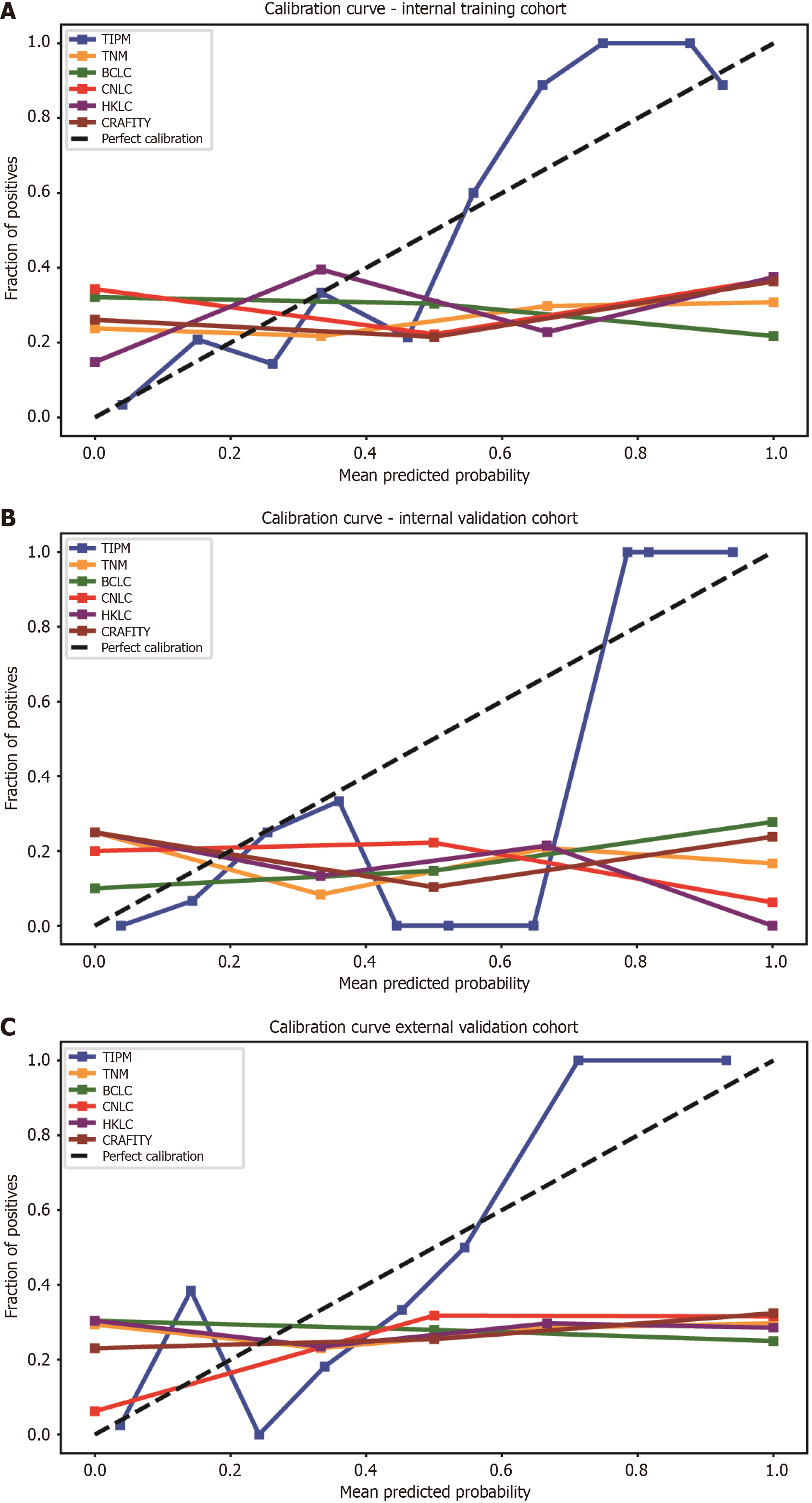
Calibration curve: Calibration curves for all models were plotted across distinct cohorts. In the internal training cohort, TIPM most closely aligned with the ideal calibration curve, while other models tended to overestimate patient risk (Figure 5A). Figure 5B illustrates the calibration curves for each model in the internal validation cohort, with TIPM closely approximating the diagonal. Figure 5C shows the calibration curves for the external validation cohort, where TIPM demonstrated superior calibration compared to other models. Calibration metrics for all models are summarized in Table 4, with TIPM outperforming its counterparts.
| Model | Brier score | HL statistic | HL P value | ECE | MCE | |
| Internal training cohort | TIPM | 0.111 | 3.945 | 0.861 | 0.111 | 0.251 |
| TNM | 0.429 | 25000 | < 0.01 | 0.353 | 0.692 | |
| BCLC | 0.435 | 81000 | < 0.01 | 0.433 | 0.782 | |
| CNLC | 0.344 | 14400 | < 0.01 | 0.416 | 0.629 | |
| HKLC | 0.317 | 16000 | < 0.01 | 0.318 | 0.625 | |
| CRAFITY | 0.400 | 36000 | < 0.01 | 0.393 | 0.636 | |
| Internal validation cohort | TIPM | 0.086 | 5.456 | 0.707 | 0.221 | 0.647 |
| TNM | 0.446 | 40000 | < 0.01 | 0.447 | 0.833 | |
| BCLC | 0.362 | 10000 | < 0.01 | 0.391 | 0.722 | |
| CNLC | 0.419 | 40000 | < 0.01 | 0.471 | 0.937 | |
| HKLC | 0.367 | 90000 | < 0.01 | 0.475 | 0.925 | |
| CRAFITY | 0.423 | 90000 | < 0.01 | 0.469 | 0.761 | |
| External validation cohort | TIPM | 0.098 | 10.11 | 0.256 | 0.102 | 0.287 |
| TNM | 0.427 | 25000 | < 0.01 | 0.541 | 0.702 | |
| BCLC | 0.400 | 49000 | < 0.01 | 0.304 | 0.75 | |
| CNLC | 0.301 | 10000 | < 0.01 | 0.062 | 0.684 | |
| HKLC | 0.309 | 49000 | < 0.01 | 0.548 | 0.714 | |
| CRAFITY | 0.403 | 90000 | < 0.01 | 0.230 | 0.675 |
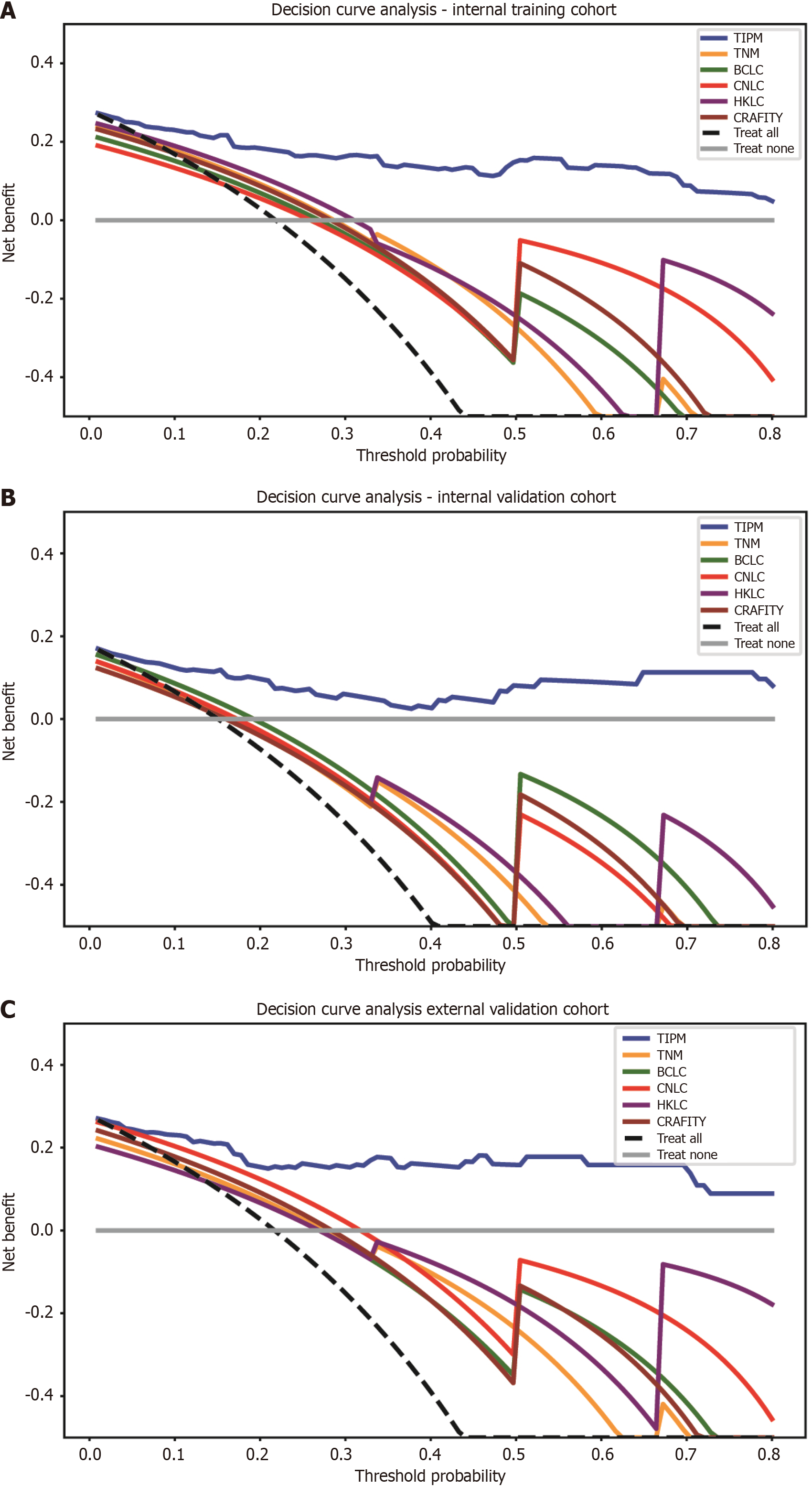
DCA curve: To evaluate clinical net benefit, DCA curves were plotted for all models. DCA (Figure 6) shows that TIPM provides the highest net benefit across a range of threshold probabilities in both the internal and external validation cohorts. Table 5 presents the NRI and integrated discrimination improvement values, further confirming that TIPM delivers the greatest clinical benefit.
| Model | Internal training cohort, NRI | Internal training cohort, IDI | Internal validation cohort, NRI | Internal validation cohort, IDI | External validation cohort, NRI | External validation cohort, IDI |
| TIPM vs TNM | 0.175 | 0.374 | 0.258 | 0.481 | 0.418 | 0.435 |
| TIPM vs BCLC | 0.330 | 0.497 | 0.238 | 0.330 | 0.605 | 0.492 |
| TIPM vs CNLC | 0.387 | 0.423 | 0.440 | 0.587 | 0.534 | 0.355 |
| TIPM vs HKLC | 0.278 | 0.385 | 0.602 | 0.558 | 0.754 | 0.454 |
| TIPM vs CRAFITY | 0.225 | 0.340 | 0.329 | 0.451 | 0.484 | 0.401 |
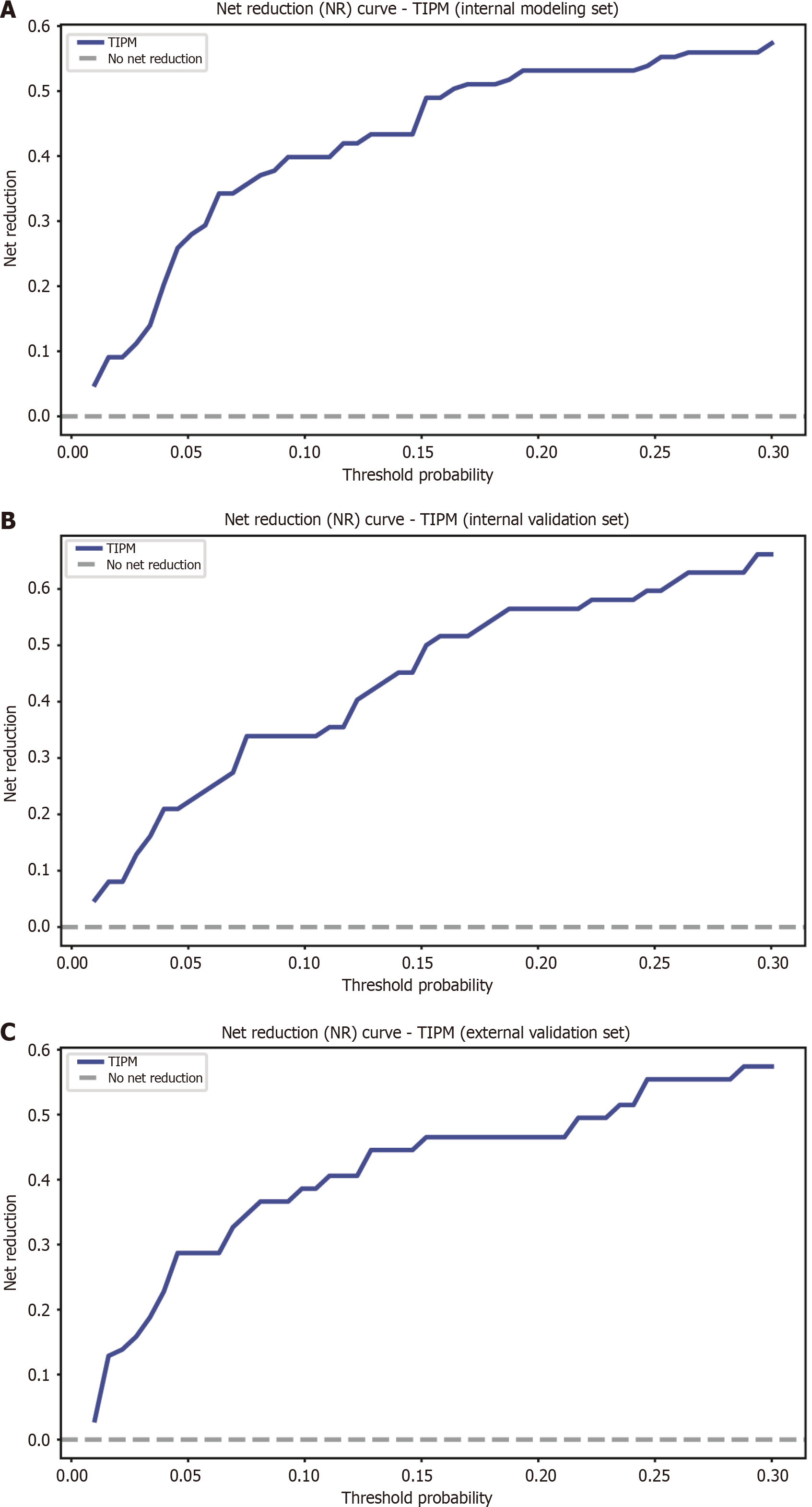
Net reduction curve: Beyond evaluating clinical net benefit, the potential of TIPM to reduce unnecessary interventions was explored. The NRI curves (Figure 7) indicate that TIPM yields a positive net reduction in risk across various threshold probabilities in both the internal and external validation cohorts, suggesting improved risk stratification.
A web-based calculator has been developed to utilize tumor diameter, PLT, tumor stiffness increase, and WBC as input parameters. The tool is accessible via the following URL: http://47.113.207.161. By entering the corresponding values, users can calculate the probability of disease progression, with higher probabilities indicating an increased likelihood of treatment failure. This web-based tool enhances accessibility and convenience, facilitating integration into clinical practice.
This study evaluated 13 machine learning algorithms to predict response to targeted immunotherapy in patients with advanced HCC. Among these, RF exhibited the highest accuracy, leading to the development of TIPM, which incorporates four key variables: Tumor size, PLT, ΔT, and WBC. The model effectively forecasts treatment response, and a user-friendly web-based calculator has been provided for clinical use. TIPM outperformed traditional models, such as TNM, BCLC, CNLC, HKLC, and CRAFITY, as demonstrated by various performance metrics. These findings highlight TIPM’s potential as a valuable tool for predicting treatment efficacy in advanced HCC, particularly for personalized therapy.
TIPM’s superior performance relative to traditional models, such as TNM, BCLC, CNLC, and HKLC, is noteworthy. While these classical models have been long utilized for assessing HCC prognosis, their reliance on staging criteria focused on tumor size, lymph node involvement, and metastasis may not fully predict treatment outcomes, especially in the context of immunotherapy. In contrast, TIPM integrates a set of routinely accessible clinical parameters. While the TNM, BCLC, CNLC, and HKLC staging systems are primarily intended for tumor prognosis prediction, they also play a critical role in guiding treatment decisions. The comparison between TIPM and these staging systems aimed to assess its additional value in predicting the efficacy of immunotherapy and targeted therapies, which are becoming standard in treatment protocols. This comparison highlights the potential of TIPM to refine personalized treatment strategies within the current clinical framework.
The CRAFITY score, developed by Scheiner et al[11], is a simple and effective model for predicting immunotherapy efficacy in patients with HCC based on two biomarkers - AFP and CRP. While valuable, this model is limited by its reliance on only these two factors. TIPM, by contrast, incorporates additional clinical variables such as tumor stiffness and PLT, employing machine learning techniques that capture more complex, non-linear relationships between variables. These differences in model design may explain TIPM’s enhanced performance across all cohorts. While the CRAFITY score remains a useful tool, our findings suggest that integrating a broader set of biomarkers and clinical factors can improve predictive accuracy for immunotherapy. Previous studies have shown that a reduction in AFP following treatment is an indicator of therapeutic efficacy. Although AFP data were initially collected for analysis, it was excluded in later stages due to its correlation with changes in tumor stiffness post-treatment. Furthermore, our study suggests that pre-treatment AFP levels are not predictive of treatment efficacy[14].
This study demonstrates that larger tumor size predicts poorer efficacy of targeted immunotherapy in HCC, aligning with existing prognostic evidence. Zhang et al[16] linked larger tumors to worse survival in HCC without vascular invasion, emphasizing size as a critical factor. Similarly, Li et al[17] identified large tumors as complicating treatment response, suggesting that their size confers resistance to systemic therapies. In contrast, Mohseni et al[18] found tumor size insignificant in advanced HCC under combined targeted immunotherapy, with viable tumor volume serving as a more reliable predictor. This may reflect the greater relevance of tumor viability in predicting therapy response compared to size. Furthermore, Qi et al[19] prioritized radiomic features capturing tumor heterogeneity over diameter, highlighting that larger, more heterogeneous tumors may respond differently to immunotherapy. These inconsistencies may arise from the varying importance of tumor viability in targeted therapies, the broader influence of heterogeneity beyond size (as larger tumors tend to be more variable), or the effect of combined regimens (e.g., anti-angiogenic agents) altering the role of tumor size. Additionally, the response evaluation criteria in solid tumors diameter-based approach may overlook nuanced responses, such as immune infiltration, that volumetric or radiomic methods capture more effectively. Patient characteristics, such as early vs advanced HCC, may also modify the predictive value of tumor size. Thus, while our findings corroborate the prognostic relevance of tumor burden, integrating size with viability and heterogeneity offers a more comprehensive understanding of targeted immunotherapy outcomes in HCC.
PLT and platelet-related biomarkers have emerged as key predictors of immunotherapy outcomes in HCC[20]. Elevated PLTs consistently correlate with poorer prognosis in patients with HCC undergoing immunotherapy, likely due to their role in fostering tumor progression and immune evasion[21,22]. Higher platelet-to-lymphocyte ratios have also been linked to worse prognosis in these patients[21], supporting broader evidence that thrombocytosis and elevated PLR signal poor prognosis across various cancers[23]. Research indicates that PLT promotes immune escape by upregulating programmed death ligand 1 on tumor cells[23], thereby enhancing immune evasion by inhibiting cytotoxic T lymphocytes and diminishing the anti-tumor effect of immune checkpoint inhibitors[24]. Furthermore, preoperative platelet-related biomarkers, such as the alkaline phosphatase-to-PLT ratio, predict poor disease-free survival and overall survival in HCC, further underscoring the prognostic significance of platelets in HCC management[24]. Although PLT and related indices show potential for predicting treatment response, additional research is required to validate their applicability across diverse clinical settings and to explore their integration with other prognostic factors[25]. By synthesizing these insights, our study reinforces the pivotal role of platelets in HCC immunotherapy and establishes a mechanistic foundation for future investigations aimed at optimizing treatment strategies[21].
The interpretation of ΔT as a biomarker of treatment response is highly context-dependent, varying significantly across therapeutic strategies. Our research, alongside that of Qayyum et al[26], demonstrates that increased ΔT may signal a favorable response to targeted immunotherapy or immunotherapy in advanced HCC. This contrasts sharply with neoadjuvant chemotherapy in breast cancer, where decreased stiffness typically correlates with efficacy[27]. These divergent trends reflect fundamentally different underlying mechanisms. In effective immunotherapy, increased stiffness likely results from substantial immune cell infiltration (e.g., cytotoxic T lymphocytes)[26], which enhances cellular density within the tumor. Concurrently, tumor microenvironment (TME) remodeling occurs, involving inflammation, stromal alterations (including collagen deposition), and vascular changes, all contributing to increased stiffness. While tumor cell death is the ultimate goal, the initial immune response and associated inflammatory and repair processes can transiently or persistently elevate stiffness. In contrast, chemotherapy’s direct cytotoxic effects reduce tumor cellularity, often coupled with extracellular matrix alterations that decrease stiffness[27]. Therefore, ΔT’s predictive value is not universal; its interpretation must be context-specific, considering the therapy and its biological effects on the TME. In HCC treated with immunotherapy, increased stiffness may thus be a valuable indicator of a robust and likely beneficial anti-tumor immune response.
Elevated WBC counts often signify systemic inflammation, which is associated with aggressive tumor progression in various cancers. Our findings align with this understanding, suggesting that inflammation, as indicated by WBC count, plays a pivotal role in the TME and immune evasion. Liu et al’s study[28] further corroborates this, identifying similar patterns where lymphocyte count - a key component of WBC count - was linked to treatment efficacy. Specifically, elevated lymphocyte levels were associated with poorer responses to anti-angiogenic therapies combined with programmed death 1 inhibitors in advanced HCC. This suggests that the inflammatory response, as reflected in WBC and lymphocyte counts, may serve as a valuable predictive marker for treatment outcomes. While discrepancies across studies may arise from differences in treatment regimens or immune-modulatory effects, these findings underscore the critical role of immune activity, as indicated by WBC or lymphocyte levels, in HCC prognosis under targeted and immunotherapies. Despite the promising results of this study, several limitations warrant consideration. The retrospective design introduces potential selection bias and other inherent biases. Additionally, although external validation was performed, it was limited to a single external dataset. While a broad range of clinically relevant variables was included, other potential predictors, such as genetic and molecular markers, were not considered. Furthermore, while machine learning models like TIPM show promise, their interpretability remains a challenge, which could hinder their widespread adoption in clinical practice.
Future research should aim to validate TIPM using larger, multicenter samples, particularly through randomized controlled trials. Incorporating additional biomarkers, genetic data, and molecular profiles could further enhance the model’s predictive accuracy and expand its applicability. Integrating TIPM into clinical decision-support systems would facilitate its real-time use, aiding clinicians in making informed treatment decisions. Investigating the combination of TIPM with other therapeutic strategies, such as targeted therapies or combination treatments, may provide valuable insights into optimizing personalized treatment approaches for HCC.
TIPM, which integrates easily accessible clinical, ultrasonographic, and serological data, presents a promising non-invasive method for predicting the efficacy of targeted immunotherapy in advanced HCC. TIPM outperformed traditional staging systems and, with the support of a web-based calculator, has the potential to refine patient selection, personalize treatment plans, and ultimately improve outcomes in this complex disease. Further validation in larger, prospective studies is essential.
Dr. Lin from Fujian Province Cancer Hospital has shared many cases, for which we express our gratitude.
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64684] [Article Influence: 16171.0] [Reference Citation Analysis (177)] |
| 2. | Cappuyns S, Corbett V, Yarchoan M, Finn RS, Llovet JM. Critical Appraisal of Guideline Recommendations on Systemic Therapies for Advanced Hepatocellular Carcinoma: A Review. JAMA Oncol. 2024;10:395-404. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 72] [Article Influence: 72.0] [Reference Citation Analysis (0)] |
| 3. | France NL, Blair HA. Tremelimumab: A Review in Advanced or Unresectable Hepatocellular Carcinoma. Target Oncol. 2024;19:115-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 4. | Sadagopan N, He AR. Recent Progress in Systemic Therapy for Advanced Hepatocellular Carcinoma. Int J Mol Sci. 2024;25:1259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 18] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 5. | Guo Y, Pan Z, Kan X, Li T, Gong B, Li Y, Yang L, Zheng C. Immunotherapy improved the efficacy of TACE or TACE plus MTTs in HCC patients: A meta-analysis. Int Immunopharmacol. 2025;147:114006. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 6. | Foerster F, Gairing SJ, Ilyas SI, Galle PR. Emerging immunotherapy for HCC: A guide for hepatologists. Hepatology. 2022;75:1604-1626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 163] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 7. | Yao J, Zhu X, Wu Z, Wei Q, Cai Y, Zheng Y, Hu X, Hu H, Zhang X, Pan H, Zhong X, Han W. Efficacy and safety of PD-1 inhibitor combined with antiangiogenic therapy for unresectable hepatocellular carcinoma: A multicenter retrospective study. Cancer Med. 2022;11:3612-3622. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 8. | Vianello C, Monti E, Leoni I, Galvani G, Giovannini C, Piscaglia F, Stefanelli C, Gramantieri L, Fornari F. Noncoding RNAs in Hepatocellular Carcinoma: Potential Applications in Combined Therapeutic Strategies and Promising Candidates of Treatment Response. Cancers (Basel). 2024;16:766. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 9. | Jeng LB, Wang J, Teng CF. Predictive Biomarkers of Immune Checkpoint Inhibitor-Based Mono- and Combination Therapies for Hepatocellular Carcinoma. J Cancer. 2024;15:484-493. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 10. | Wu M, Wang Y, Yang Q, Wang X, Yang X, Xing H, Sang X, Li X, Zhao H, Huo L. Comparison of Baseline (68)Ga-FAPI and (18)F-FDG PET/CT for Prediction of Response and Clinical Outcome in Patients with Unresectable Hepatocellular Carcinoma Treated with PD-1 Inhibitor and Lenvatinib. J Nucl Med. 2023;64:1532-1539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 11. | Scheiner B, Pomej K, Kirstein MM, Hucke F, Finkelmeier F, Waidmann O, Himmelsbach V, Schulze K, von Felden J, Fründt TW, Stadler M, Heinzl H, Shmanko K, Spahn S, Radu P, Siebenhüner AR, Mertens JC, Rahbari NN, Kütting F, Waldschmidt DT, Ebert MP, Teufel A, De Dosso S, Pinato DJ, Pressiani T, Meischl T, Balcar L, Müller C, Mandorfer M, Reiberger T, Trauner M, Personeni N, Rimassa L, Bitzer M, Trojan J, Weinmann A, Wege H, Dufour JF, Peck-Radosavljevic M, Vogel A, Pinter M. Prognosis of patients with hepatocellular carcinoma treated with immunotherapy - development and validation of the CRAFITY score. J Hepatol. 2022;76:353-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 188] [Article Influence: 62.7] [Reference Citation Analysis (0)] |
| 12. | Haas Y, Dosch MP, Vogl TJ. Response comparison of PLC and SLC with magnetic resonance elastography after TACE. Sci Rep. 2022;12:8317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Reference Citation Analysis (0)] |
| 13. | Hou S, Hua S, Cui K, Liu F, Ding K, Yuan J. The course and prognostic value of tumor stiffness detected by ultrasound elastography for transarterial chemoembolization of hepatocellular carcinoma. Quant Imaging Med Surg. 2023;13:3962-3972. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 14. | Colloca GA, Venturino A. Radiographic and serologic response in patients with unresectable hepatocellular carcinoma receiving systemic antineoplastic treatments: A trial-level analysis. Cancer. 2024;130:1773-1783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 15. | Yu H, Bai Y, Xie X, Feng Y, Yang Y, Zhu Q. RECIST 1.1 versus mRECIST for assessment of tumour response to molecular targeted therapies and disease outcomes in patients with hepatocellular carcinoma: a systematic review and meta-analysis. BMJ Open. 2022;12:e052294. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 20] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 16. | Zhang Y, Zhang JG, Yu W, Liang L, Wu C, Zhang CW, Xie YM, Huang DS, Shi Y. Prognostic impact of tumor size on isolated hepatocellular carcinoma without vascular invasion may have age variance. Front Surg. 2022;9:988484. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 17. | Li S, Zhou ZF, Long HJ, Yin JX, Wang HZ, Zhao JF. Multimodal combination regimen for a patient with advanced huge hepatocellular carcinoma: a case report. Transl Gastroenterol Hepatol. 2025;10:17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 18. | Mohseni A, Baghdadi A, Madani SP, Shahbazian H, Mirza-Aghazadeh-Attari M, Borhani A, Afyouni S, Zandieh G, Baretti M, Kim AK, Yarchoan M, Kamel IR. Predicting survival of patients with advanced hepatocellular carcinoma receiving combination targeted immunotherapy: an evaluation of volumetric imaging parameters. Abdom Radiol (NY). 2024;49:2595-2605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 19. | Qi L, Zhu Y, Li J, Zhou M, Liu B, Chen J, Shen J. CT radiomics-based biomarkers can predict response to immunotherapy in hepatocellular carcinoma. Sci Rep. 2024;14:20027. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 20. | Dharmapuri S, Özbek U, Lin JY, Sung M, Schwartz M, Branch AD, Ang C. Predictive value of neutrophil to lymphocyte ratio and platelet to lymphocyte ratio in advanced hepatocellular carcinoma patients treated with anti-PD-1 therapy. Cancer Med. 2020;9:4962-4970. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 93] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 21. | Xu H, Yuan Q, Wu Z, Xu Y, Chen J. Integrative transcriptome and single-cell sequencing technology analysis of the potential therapeutic benefits of oleanolic acid in liver injury and liver cancer. Aging (Albany NY). 2023;15:15267-15286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 22. | Zheng J, Cai J, Li H, Zeng K, He L, Fu H, Zhang J, Chen L, Yao J, Zhang Y, Yang Y. Neutrophil to Lymphocyte Ratio and Platelet to Lymphocyte Ratio as Prognostic Predictors for Hepatocellular Carcinoma Patients with Various Treatments: a Meta-Analysis and Systematic Review. Cell Physiol Biochem. 2017;44:967-981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 188] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 23. | Zhang Y, Zhang B, Gong L, Xiong L, Xiao X, Bu C, Liang Z, Li L, Tang B, Lu Y. Preoperative alkaline phosphatase-to-platelet count ratio as a prognostic factor for hepatocellular carcinoma with microvascular invasion. Cancer Med. 2023;12:17545-17558. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 24. | Guo Q, Malloy MW, Roweth HG, McAllister SS, Italiano JE, Battinelli EM. Platelets upregulate tumor cell programmed death ligand 1 in an epidermal growth factor receptor-dependent manner in vitro. Blood Adv. 2022;6:5668-5675. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 25. | Lu YC, Yang YC, Ma D, Wang JQ, Hao FJ, Chen XX, Chen YJ. FOLFOX-HAIC combined with targeted immunotherapy for initially unresectable hepatocellular carcinoma: a real-world study. Front Immunol. 2024;15:1471017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 26. | Qayyum A, Hwang KP, Stafford J, Verma A, Maru DM, Sandesh S, Sun J, Pestana RC, Avritscher R, Hassan MM, Amin H, Rashid A, Wistuba II, Ehman RL, Ma J, Kaseb AO. Immunotherapy response evaluation with magnetic resonance elastography (MRE) in advanced HCC. J Immunother Cancer. 2019;7:329. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 27. | Mahadevan GSV, Chakkalakkoombil SV, Kayal S, Dharanipragada K, Toi PC, Ananthakrishnan R. Evaluation of change in tumor stiffness measured by acoustic radiation force impulse imaging for early prediction of response to neoadjuvant chemotherapy in breast cancer. J Clin Ultrasound. 2022;50:666-674. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 28. | Liu C, Zhu S, Dong Y, Shao J, Liu B, Shen J. The Potential Predictive Biomarkers for Advanced Hepatocellular Carcinoma Treated With Anti-Angiogenic Drugs in Combination With PD-1 Antibody. Front Immunol. 2022;13:930096. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |