Published online May 15, 2025. doi: 10.4251/wjgo.v17.i5.102417
Revised: February 3, 2025
Accepted: February 27, 2025
Published online: May 15, 2025
Processing time: 209 Days and 23.8 Hours
Colorectal cancer (CRC) is the second most prevalent cause of cancer-related mortality and is increasing in younger individuals. Chemotherapy, a crucial adjuvant systemic therapy for CRC management, often leads to resistance through poorly characterized underlying molecular mechanisms. The long noncoding RNA SNHG5 is highly expressed in CRC and promotes tumor proliferation and invasion, prompting us to hypothesize that SNHG5 may play a crucial role in the chemotherapeutic agent 5-fluorouracil (5-Fu) resistance in CRC.
To identify the function and mechanism of SNHG5 in 5-Fu resistance in CRC.
Quantitative real-time polymerase chain reaction was performed to examine the expression of SNHG5 in CRC tissues from 22 5-Fu-sensitive patients and 14 5-Fu-resistant patients and in CRC cells and 5-Fu-resistant CRC cells. Cell viability and apoptosis were assessed in SNHG5-overexpressing CRC cells and SNHG5-knockdown 5-Fu-resistant CRC cells. SNHG5 function in 5-Fu resistance in CRC was further analyzed using a xenograft mouse model. SNHG5 interactions with microRNAs were predicted by bioinformatics analysis. Luciferase reporter and RNA immunoprecipitation assays were performed to verify the binding between SNHG5 and miR-26b. Rescue experiments were performed to validate the functional interaction between SNHG5 and the miR-26b/p-glyco
SNHG5 expression was upregulated in 5-Fu-resistant CRC tissues and 5-Fu-resistant CRC cells. In vitro functional experiments demonstrated that SNHG5 overexpression significantly reduced cell apoptosis and enhanced cell viability, whereas SNHG5 knockdown in 5-Fu-resistant CRC cells increased cell apoptosis and decreased cell viability upon 5-Fu treatment. In a xenograft mouse model, we confirmed that SNHG5 overexpression led to a reduction in 5-Fu sensitivity in CRC in vivo. Mechanistically, SNHG5 acted as a molecular sponge for miR-26b. Rescue experiments validated that SNHG5 conferred 5-Fu resistance in CRC by regulating the miR-26b/Pgp axis.
SNHG5/miR-26b/Pgp regulates CRC chemosensitivity, providing potential therapeutic targets for the treatment of 5-Fu-resistant CRC.
Core Tip: This study confirmed that the expression of the long noncoding RNA SNHG5 was upregulated in 5-fluorouracil (5-Fu)-resistant colorectal cancer (CRC) tissues and 5-Fu-resistant CRC cells. Functional experiments confirmed that SNHG5 overexpression reduced 5-Fu sensitivity in CRC in vitro and in vivo. Mechanistically, SNHG5 acted as a molecular sponge for miR-26b. In addition, SNHG5 conferred 5-Fu resistance in CRC by regulating the miR-26b/p-glycoprotein axis. Our results elucidate potential mechanisms of noncoding RNAs in CRC chemoresistance and provide new strategies for CRC chemotherapy.
- Citation: Wang B, Zhou Q, Cheng CE, Gu YJ, Jiang TW, Qiu JM, Wei GN, Feng YD, Ren LH, Shi RH. Long noncoding RNA SNHG5 promotes 5-fluorouracil resistance in colorectal cancer by regulating miR-26b/p-glycoprotein axis. World J Gastrointest Oncol 2025; 17(5): 102417
- URL: https://www.wjgnet.com/1948-5204/full/v17/i5/102417.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i5.102417
Colorectal cancer (CRC) is a common cancer that ranks third in incidence among human cancers and is the second leading cause of cancer-related mortality[1]. Moreover, the incidence rate of CRC in younger patients has increased over the last several decades[2]. The prognosis of CRC patients has gradually improved with the adoption of several clinical techniques, including surgery, radiation, and chemotherapy[3,4]. 5-fluorouracil (5-Fu), a first-line chemotherapeutic agent, has been widely used for CRC treatment[5]. However, resistance to 5-Fu is a critical limitation to its clinical appli
Long noncoding RNAs (lncRNAs) are classified as noncoding RNAs (ncRNAs) that are more than 200 nucleotides in length and have no protein-coding capacity. Numerous studies have shown that lncRNAs participate in 5-Fu resistance by modifying signaling pathways and regulating gene expression. For example, recent research revealed that the lncRNA NEAT1 promoted 5-Fu resistance by remodeling chromatin, which increased the acetylation levels of histones and c-Myc and promoted the expression of aldehyde dehydrogenases 1 and c-Myc[8]. The lncRNA HAND2-AS1 was shown to inhibit 5-Fu resistance by modulating the miR-20a/PDCD4 axis in CRC[9]. Additionally, the lncRNA CRNDE was shown to attenuate chemoresistance via SRSF6-regulated alternative splicing of PICALM in gastric cancer[10].
The lncRNA SNHG5 is highly expressed in a variety of tumor tissues, including CRC, and promotes tumor proliferation and invasion through multiple pathways[11-13]. However, the function and mechanism of SNHG5 in 5-Fu resis
In this study, we attempted to identify the function and mechanism of SNHG5 in 5-Fu resistance in CRC. We demon
We obtained clinical colorectal tissues, including tumor tissues (n = 36) and adjacent normal tissues (n = 16), from individuals at Changshu No. 2 People’s Hospital. The clinical samples were stored in liquid nitrogen after surgical removal. The patients received postoperative adjuvant chemotherapy based on 5-Fu followed by chest X-ray, computed tomography and/or gastrointestinal endoscopy to monitor for tumor progression. Clinicopathological characteristics of the 36 CRC patients were recorded (Table 1) and the response to 5-Fu was evaluated based on the response evaluation criteria in solid tumors (version 1.1). 5-Fu resistance was defined as disease progression within 6 months after primary chemotherapy or during primary chemotherapy, whereas 5-Fu sensitivity was defined as progression beyond 6 months or no progression[14,15]. The experimental procedures involving clinical samples were approved by the Institutional Review Board of Changshu No. 2 People’s Hospital (No. 2020-KY-008).
| Characteristics | 5-Fu-sensitive group (n = 22) | 5-Fu-resistant group (n = 14) | P value |
| Age (years) | |||
| < 60 | 3 | 2 | 1.000 |
| ≥ 60 | 19 | 12 | |
| Sex | |||
| Male | 14 | 9 | 1.000 |
| Female | 8 | 5 | |
| Tumor size | |||
| < 5 cm | 17 | 9 | 0.462 |
| ≥ 5 cm | 5 | 5 | |
| Location | |||
| Colon | 14 | 6 | 0.307 |
| Rectum | 8 | 8 | |
| Differentiation | |||
| Well/moderate | 15 | 10 | 1.000 |
| Poor | 7 | 4 | |
| Tumor depth | |||
| T1 + T2 | 3 | 2 | 1.000 |
| T3 + T4 | 19 | 12 | |
| AJCC stage | |||
| II | 10 | 4 | 0.485 |
| III | 12 | 10 | |
| SNHG5 expression | |||
| Low | 17 | 5 | 0.018a |
| High | 5 | 9 |
CRC cell lines (LOVO, HT-29, HCT-8, and SW116), human normal colorectal epithelial cells (FHCs), and HEK-293T cells were obtained from American type culture collection (MD, United States), and 5-Fu-resistant cells (LOVO/5-Fu, HT-29/5-Fu) were obtained as described in our previous study[16]. CRC cell lines were cultured in Dulbecco’s modified Eagle’s medium (Gibco, Thermo Fisher Scientific, United States) supplemented with penicillin (100 U/mL) and streptomycin (100 μg/mL). Resistance cells were cultured as described previously[16].
LncRNA SNHG5 small interfering RNA (siRNA), control siRNA, miR-26b mimics and miR-control (miR-NC) mimics were obtained from RiboBio (Guangzhou, Guangdong Province, China). The pcDNA3.1-SNHG5 vector (SNHG5), pcDNA3.1-Pgp vector (Pgp) and control vectors were purchased from Biovector Co., Ltd. (Beijing, China).
SNHG5 sequences with predicted miR-26b binding sites [wild-type (WT): 5’-GTGCCACGAGGTTTACTTGAC-3’] and the corresponding mutant (Mut) sequences (Mut: 5’-GTGCCACGAGGTAATGAACTC-3’) were cloned and inserted into the pmirGLO vector (Promega, Madison, WI, United States). Before the luciferase reporter assay, HEK-293T cells were seeded and cultured in 24-well plates. The plasmids encoding SNHG5-WT or SNHG5-Mut (500 ng) were transfected with miR-NC or miR-26b mimics (100 nM) into HEK-293T cells. After 24 hours, the cells were lysed with 1 × passive lysis buffer for 15 minutes, and a dual-luciferase reporter assay system was used to measure luciferase activity (E1910, Promega, Madison, WI, United States).
Total RNA was extracted using TRIzol reagent (15596026, Invitrogen, Carlsbad, CA, United States). RNA was isolated from the cytoplasm and nucleus of CRC cells using a cytoplasmic and nuclear RNA purification kit (Cat. 21000, 37400, Norgen, Thorold, Canada). Complementary DNA (cDNA) for lncRNA analysis was synthesized using a lnRcute lncRNA cDNA kit (KR202-02, TIANGEN, Beijing, China), and cDNA for miRNA analysis was synthesized using a HiScript cDNA synthesis kit (R323-01, Vazyme, Nanjing, China). Beta-actin (β-actin) and U6 were used as controls for the mRNA and miRNA assays, respectively. Quantitative polymerase chain reaction (qPCR) was performed using ChamQ SYBR qPCR master mix (high ROX premixed) (Q341-02, Vazyme, Nanjing, Jiangsu Province, China) according to the manufacturer’s instructions. The sequences of primers used were as follows: SNHG5 (forward) 5’-TACTGGCTGCGCACTTCG-3’; SNHG5 (reverse) 5’-TACCCTGCACAAACCCGAAA-3’; miR-26b (forward) 5’-ACACTCCAGCTGGGTTCAAGTAATTCAGG-3’; miR-26b (reverse) 5’- CTCAACTGGTGTCGTGGA-3’; β-actin (forward) 5’-CATGTACGTTGCTATCCAGGC-3’; β-actin (reverse) 5’-CTCCTTAATGTCACGCACGAT-3’; U6 (forward) 5’-CTCGCTTCGGCAGCACA-3’; and U6 (reverse) 5’-AACGCTTCACGAATTTGCGT-3’.
LOVO and HT-29 cells transfected with miR-26b were scraped from culture plates, resuspended in ice-cold PBS, and then lysed in RNA immunoprecipitation (RIP) lysis buffer [25 mmol/L tris potential of hydrogen (pH) = 7.4, 150 mmol/L potassium chloride, 5 mmol/L ethylene diamine tetraacetic acid, 0.5 mmol/L dithiothreitol, 0.5% nonaethylene glycol octylphenyl ether, 100 U/mL RNAase inhibitor SUPERase, and protease inhibitors] (Abcam, United States). The lysates were incubated with a human anti-argonaute 2 (Ago2) antibody (67934-1-Ig, Proteintech, IL, United States) overnight [a human anti-IgG antibody (10284-1-AP, Proteintech, IL, United States) was used as a control] and then incubated with protein A/G beads for one hour. After the beads were washed twice, RNA was isolated with TRIzol reagent and analyzed via quantitative real-time polymerase chain reaction (qRT-PCR).
SNHG5-expressing (SNHG5) and scrambled control, SNHG5-short hairpin RNA (shRNA) (shSNHG5) and negative control (shNC) lentiviruses were purchased from GenePharma (Shanghai, China). After infection, stable cells were selected with puromycin (2 μg/mL; Roche, United States) for 2 weeks. Stable cells were identified via qRT-PCR and pooled for subsequent analysis.
SNHG5-overexpressing CRC cells and SNHG5-knockdown 5-Fu-resistant cells were cultured in 96-well plates (5000 cells/well). After 24 hours, the cells were treated with different concentrations of 5-Fu for 48 hours, and then, cell viability was analyzed using 10% cell counting kit 8 (CCK-8) (Dojindo, Kumamoto, Japan). Stable cells were treated with 2.5 μg/mL 5-Fu for 48 hours. Flow cytometry was performed after annexin-V fluorescein 5-isothiocyanate/propidium iodide staining (A211-01, Vazyme, Nanjing, Jiangsu Province, China) according to the manufacturer’s instructions.
Total protein was isolated from cells using cell lysis buffer [20 mmol/L tris (pH = 7.5), 150 mmol/L sodium chloride, 1% Triton X-100, and phenyl methane sulfonyl fluoride]. Then, 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels were used to separate the proteins. Primary antibodies against Pgp (1:1000; ab261736, Abcam, United States), Bax (1:1000; 50599-2-Ig, Proteintech, IL, United States), Bcl-2 (1:1000; 26593-1-AP, Proteintech, IL, United States) and β-actin (1:1000; 4967S, Cell Signaling Technology, United States) were used.
Stable cells [SNHG5-overexpressing LOVO (LOVO/SNHG5), control LOVO (LOVO/Con), SNHG5-knockdown LOVO/5-Fu (LOVO/5-Fu/shSNHG5), and control LOVO/5-Fu (LOVO/5-Fu/shNC) cells] (5 × 106 cells per mouse) were injected subcutaneously into 4-week-old BALB/c-nude mice. Approximately 7 days later, 30 mg/kg 5-Fu in saline buffer was administered by intraperitoneal injection. The mice were treated with a cyclic regimen composed of three subsequent daily injections followed by two days of recovery. Tumors were measured every two days. After two weeks, the tumors were isolated from the mice and weighed prior to subsequent analyses. Tumor volume was calculated using the following formula: V = 1/2 a2 × b, where a is tumor length and b is tumor width. Immunohistochemistry (IHC) was performed using an anti-Ki-67 antibody (ab15580, Abcam, United States) to analyze the proliferation of tumors in different groups. IHC experiments were performed in accordance with the manufacturer’s instructions (Genetech, China, GK500705). The Ki-67 positive area (%) was calculated by Image J software (NIH, United States). The animal experiments were approved by the Animal Care and Welfare Committee of Southeast University (No. 20200721007).
Continuous data are expressed as the mean ± SD. The Fisher’s exact test was used to analyze categorical variables. For continuous variables, Student’s t test was used to analyze differences between two groups, and one-way analysis of variance was used to determine significant differences among three or more groups. Correlations were analyzed via the Pearson correlation coefficient. Data were considered statistically significant when the P value was < 0.05.
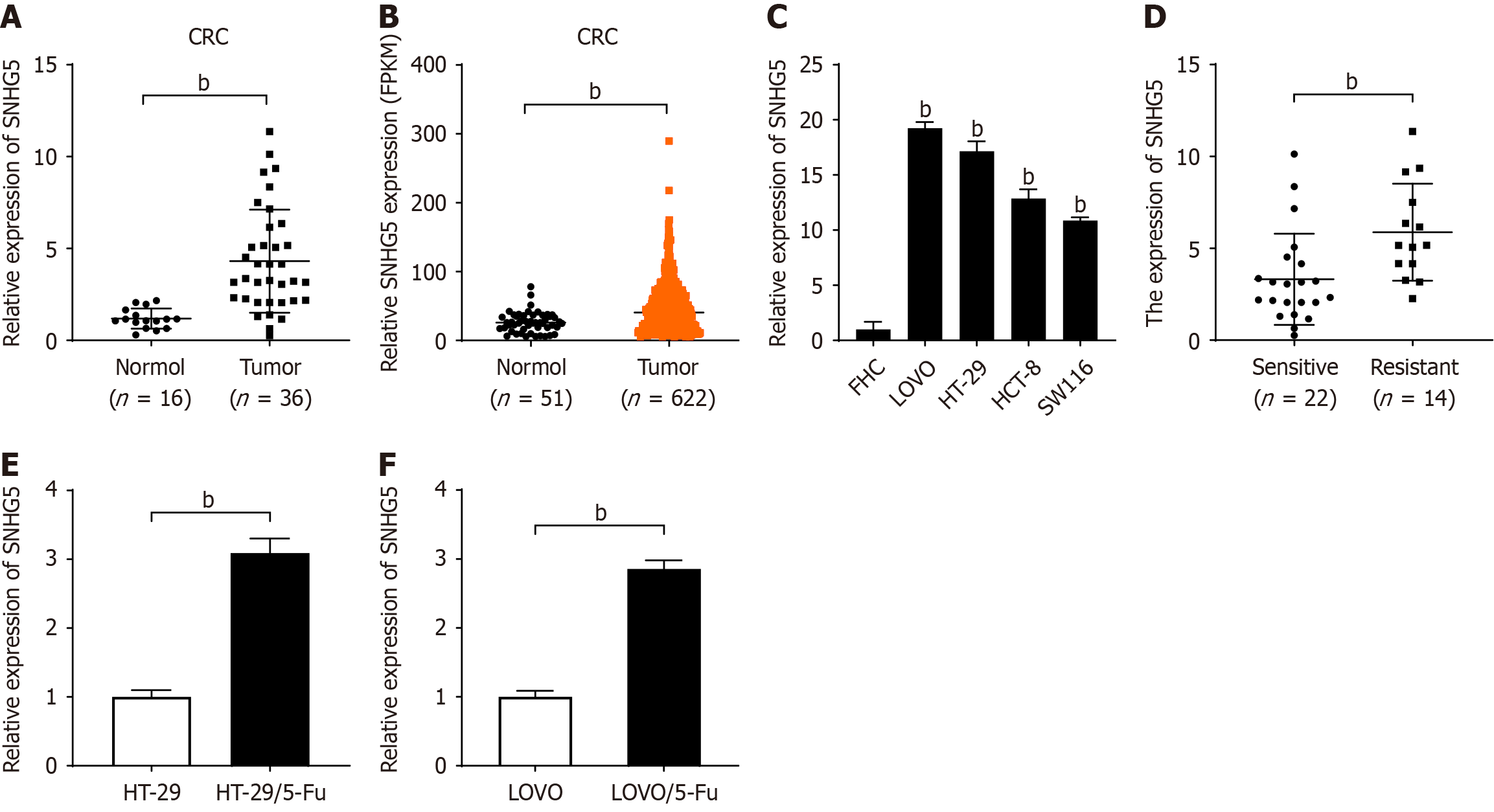
We first explored the expression levels of SNHG5 in CRC tissues (n = 36) and normal colorectal tissues (n = 16) via qRT-PCR. Our results revealed that the level of SNHG5 in CRC tissues was nearly 3.64-fold greater than that in normal tissues (P < 0.01; Figure 1A). We subsequently examined the expression of SNHG5 in CRC by analyzing data from the Cancer Genome Atlas database. We compared data from 622 CRC tissues and 51 normal colorectal tissues and found that the expression level of SNHG5 was 1.77-fold higher in CRC tissues than in normal colorectal tissues (P < 0.01; Figure 1B). We measured the expression levels of SNHG5 in the LOVO, HT-29, HCT-8 and SW116 CRC cell lines and in a normal colorectal epithelial cell line, i.e., FHC cells. We found that SNHG5 expression was higher in the CRC cell lines than in the FHC cell line (P < 0.01; Figure 1C). These results indicated that SNHG5 was highly expressed in CRC.
Next, we divided CRC patients into a 5-Fu-sensitive group (n = 22) and a 5-Fu-resistant group (n = 14) and found that SNHG5 expression was significantly higher in the tumor tissues of the 5-Fu-resistant group than in those of the 5-Fu-sensitive group (1.77-fold, P < 0.01; Figure 1D). The association between chemoresponse and SNHG5 expression was also assessed by analyzing 36 samples (Table 1). The results revealed a significant correlation between 5-Fu resistance and high expression levels of SNHG5 in CRC (P < 0.05). Next, we examined SNHG5 expression in CRC cell lines and con
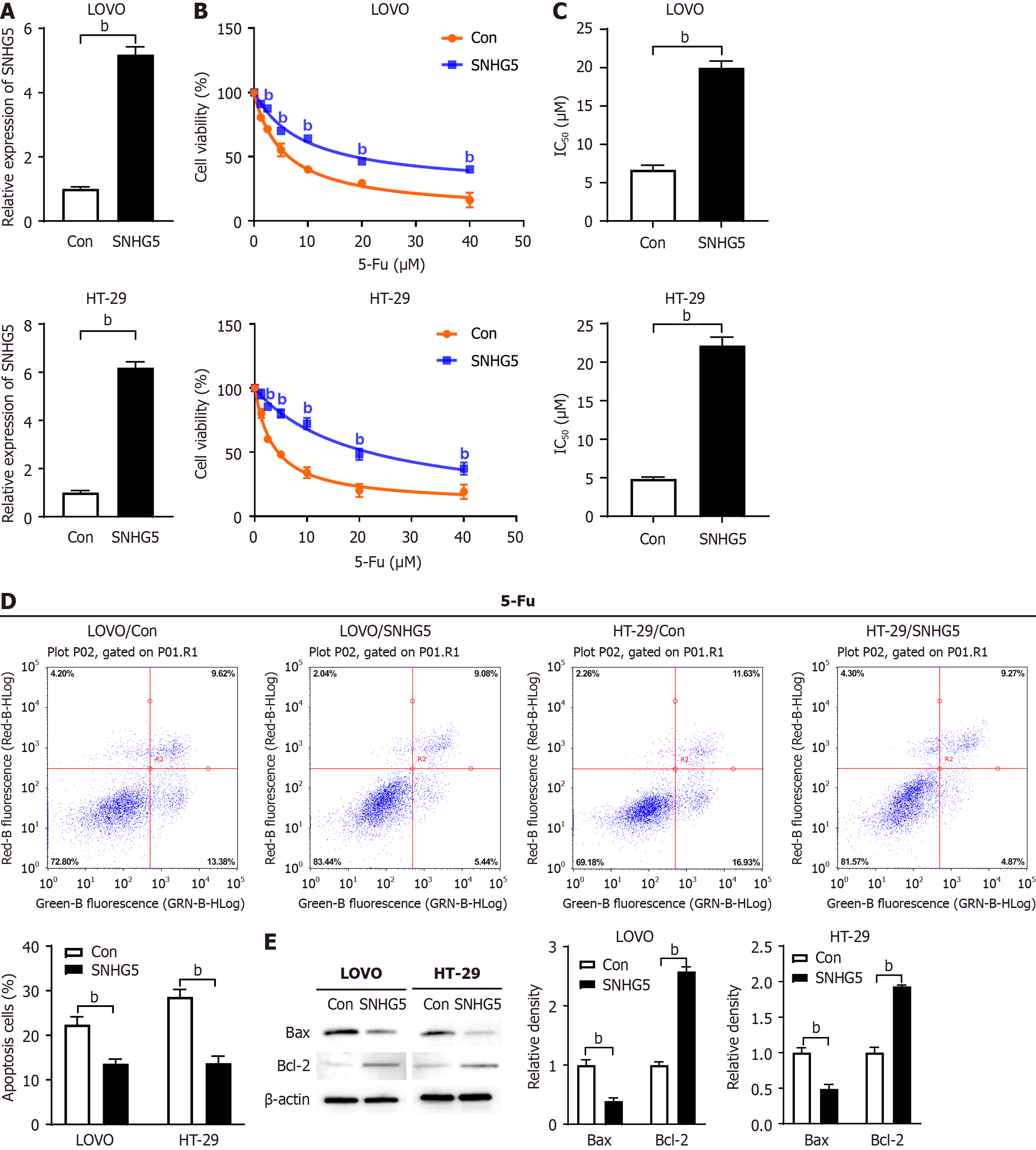
To investigate the effect of SNHG5 on the chemosensitivity of CRC cells to 5-Fu, we first transfected an SNHG5-expre
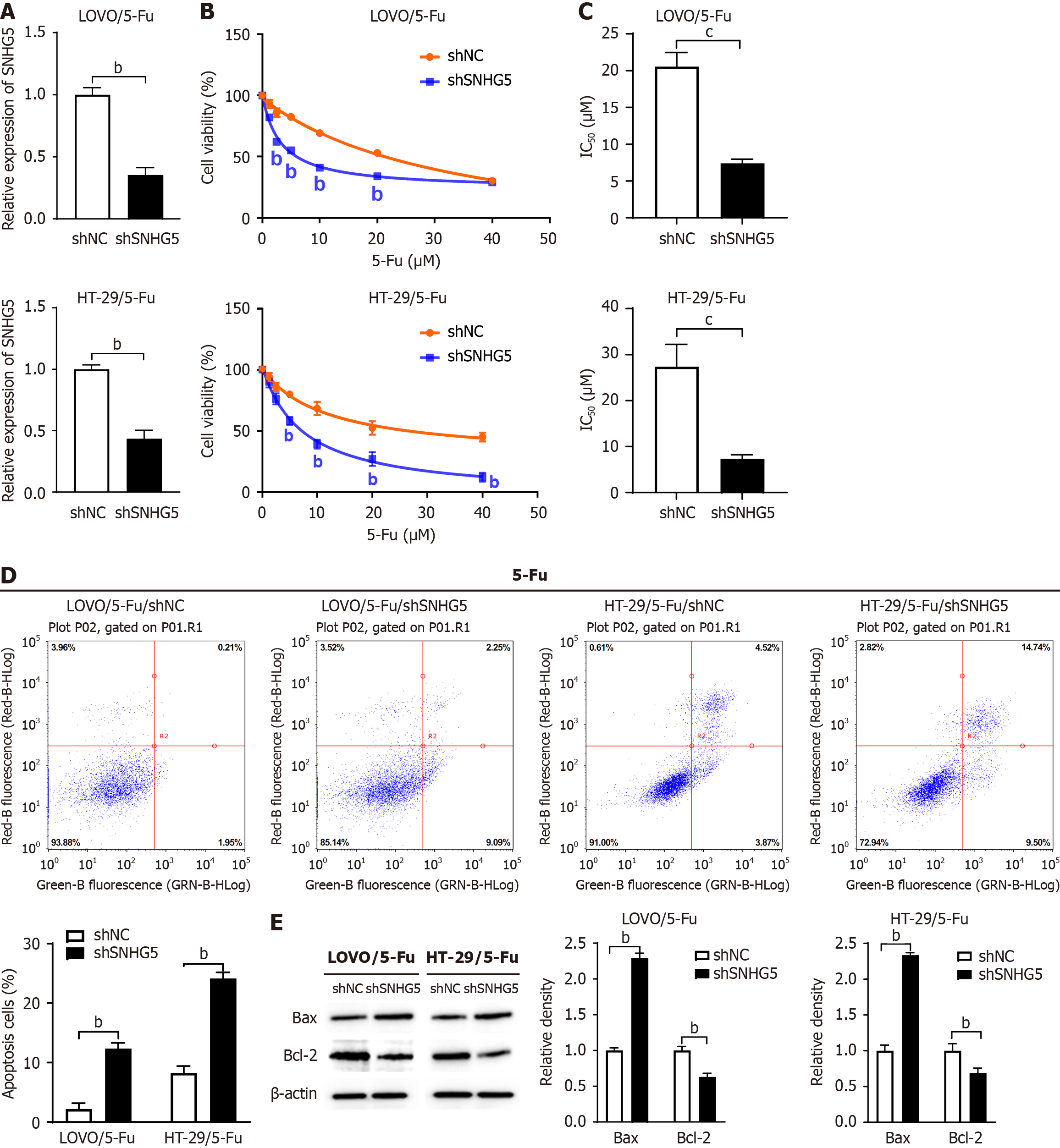
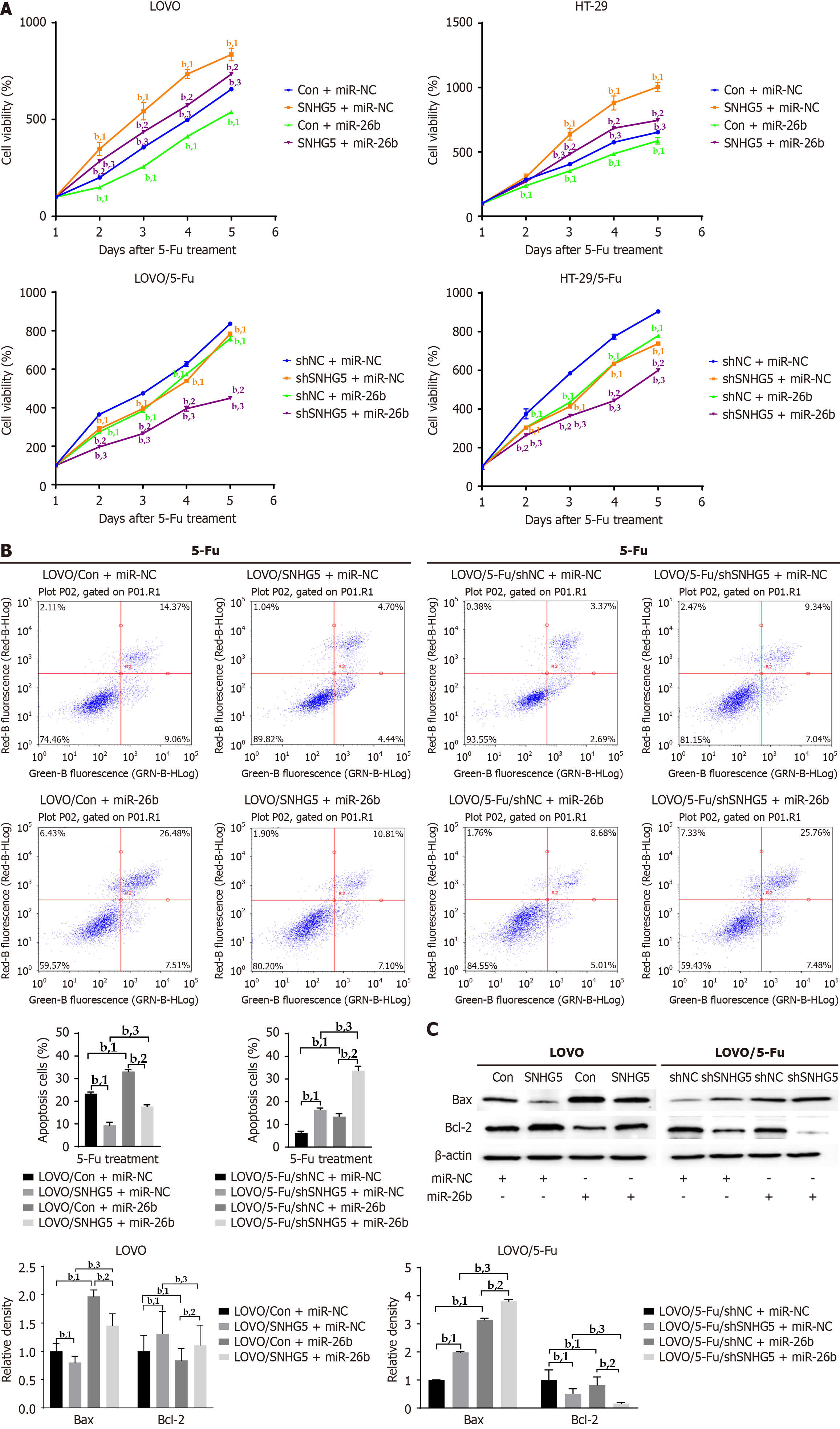
Next, we investigated the effect of SNHG5 knockdown on 5-Fu sensitivity via the use of an SNHG5-shRNA lentivirus (shSNHG5) (Figure 3A). CCK-8 assay results revealed that the knockdown of SNHG5 significantly decreased the 5-Fu resistance of 5-Fu-resistant CRC cells (Figure 3B). We measured the IC50 and found that SNHG5-knockdown cells were more sensitive to 5-Fu than control cells (7.40 ± 0.57 μM vs 20.56 ± 1.92 μM for LOVO, P < 0.001; 7.35 ± 0.89 μM vs 27.37 ± 4.85 μM for HT-29, P < 0.001) (Figure 3C). In contrast to SNHG5 overexpression, SNHG5 knockdown increased cell apoptosis and Bax levels and decreased Bcl-2 levels in CRC cells (Figure 3D and E).
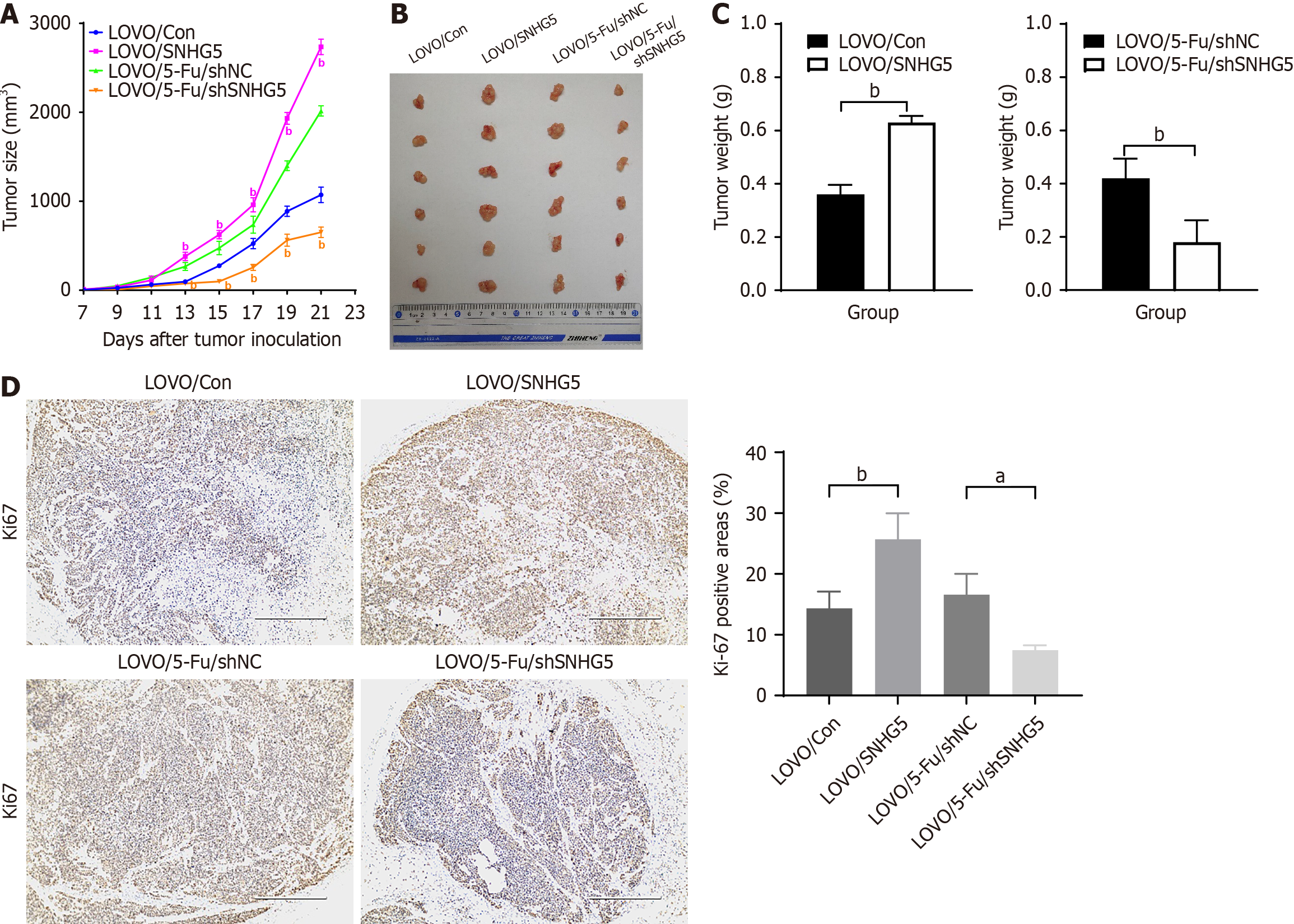
To evaluate whether SNHG5 promotes 5-Fu resistance in CRC in vivo, LOVO/SNHG5 cells, LOVO/Con cells, LOVO/5-Fu/shSNHG5 cells and LOVO/5-Fu/shNC cells were injected subcutaneously into the flanks of 4-week-old nude mice. When the tumor diameter reached 5 mm, 5-Fu was administered by intraperitoneal injection. The results revealed that the tumors derived from LOVO/SNHG5 cells in 5-Fu-treated mice had greater mean volumes, exhibited faster growth, and were heavier than those derived from LOVO/Con cells. In contrast, the tumors derived from LOVO/5-Fu/shSNHG5 cells presented smaller mean volumes, exhibited slower growth, and were lighter than those derived from the LOVO/5-Fu/shNC cells (Figure 4A-C). The proliferation marker Ki-67 was used to test tumorigenesis ability in vivo. Our results revealed that the LOVO/SNHG5 group had a greater Ki-67-positive than the LOVO/Con group did; however, the LOVO/5-Fu/shSNHG5 group presented a lower positive rate than the LOVO/5-Fu/shNC group did (Figure 4D). Taken together, these data indicated that SNHG5 promoted CRC resistance to 5-Fu in vivo.
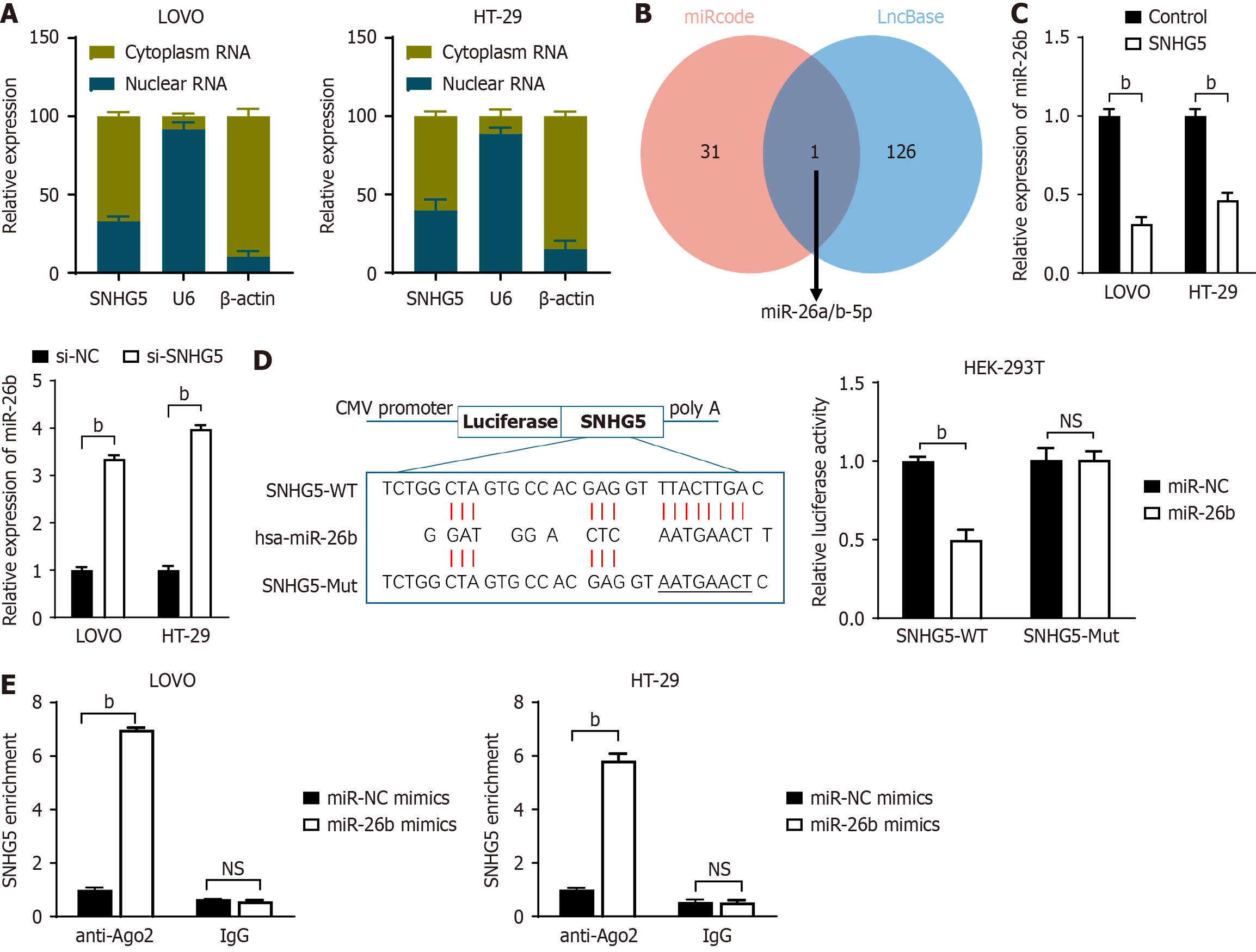
To identify the mechanism by which SNHG5 promotes 5-Fu resistance, we first examined, via qRT-PCR analyses, the cellular location of SNHG5 and found that SNHG5 was enriched in both the cytoplasmic and nuclear fractions of LOVO and HT-29 cells (Figure 5A). Multiple studies have shown that lncRNAs can function as competing endogenous RNAs (ceRNAs) by binding to miRNAs[17,18]. We subsequently searched for miRNAs that have complementary base pairing with SNHG5 via the online software programs miRcode (http://www.miRcode.org) and DIANA-LNCBase (https://diana.e-ce.uth.gr/Lncbasev3). We previously reported that miR-26b, which has been found to suppress 5-Fu resistance by inhibiting Pgp in CRC[16], may form complementary base pairs with SNHG5 (Figure 5B).
To confirm the interaction between SNHG5 and miR-26b, we transfected LOVO and HT-29 cells with an SNHG5-overexpression vector or siRNA and found that miR-26b expression was significantly downregulated or upregulated, respectively (Figure 5C). We subsequently constructed SNHG5 Luciferase reporter plasmids containing WT (SNHG5-WT) and Mut-binding sites (SNHG5-Mut) for miR-26b and transfected them into HEK-293T cells together with a miR-26b mimic (miR-26b) or a control mimic (miR-NC). The miR-26b mimic reduced the luciferase activity of the WT but not the Mut plasmid (Figure 5D). We then conducted anti-Ago2 RIP using LOVO and HT-29 cells transiently overexpressing miR-26b. Endogenous SNHG5 pulled down by Ago2 was significantly enriched in miR-26b-transfected cells (Figure 5E). These data suggested that miR-26b was a miRNA binding partner of SNHG5.
To clarify whether miR-26b is involved in SNHG5-mediated 5-Fu resistance in CRC cells, we increased miR-26b expression with a miR-26b mimic in SNHG5-overexpressing CRC cells and in SNHG5-knockdown 5-Fu-resistant CRC cells. Cell viability analyses revealed that SNHG5 promoted CRC cell proliferation and that this effect was inhibited by the miR-26 mimic. Similarly, the miR-26b mimic inhibited the proliferation of SNHG5-knockdown 5-Fu-resistant CRC cells (Figure 6A). Consistently, the miR-26b mimic increased 5-Fu-induced apoptosis and Bax expression and decreased Bcl-2 expression in SNHG5-overexpressing CRC cells and in SNHG5-knockdown 5-Fu-resistant CRC cells (Figure 6B and C).
Taken together, these data confirmed that SNHG5 promoted CRC cell resistance to 5-Fu by binding to miR-26b.
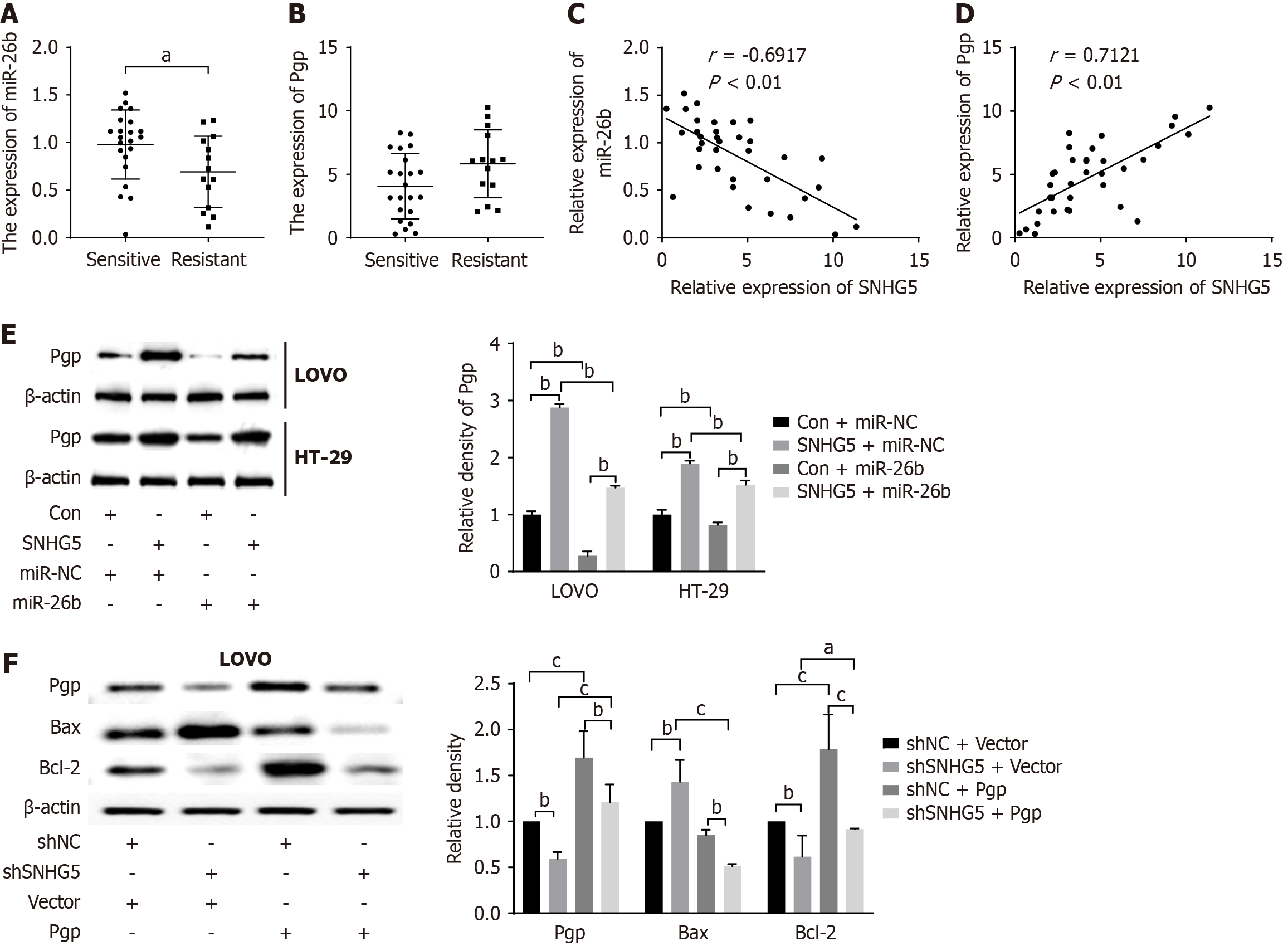
Since our previous study showed that miR-26b suppressed 5-Fu resistance by targeting Pgp[16], we investigated whether SNHG5 regulated the miR-26b/Pgp axis. We first evaluated the expression levels of miR-26b and Pgp in 36 CRC tissues. The results indicated that miR-26 expression was downregulated whereas Pgp expression was upregulated in tumor tissues of 5-Fu-resistant group compared with 5-Fu-sensitive group (Figure 7A and B). Next, we measured the correlation of SNHG5 with miR-26b and Pgp in CRC tissues. The results revealed that SNHG5 was negatively correlated with miR-26b (Figure 7C) and positively correlated with Pgp, the target of miR-26b (Figure 7D). These data indicated that SNHG5 played a vital role in CRC chemoresistance. The Western blot results revealed that SNHG5 increased the expression of Pgp; however, the miR-26b mimic reversed that effect (Figure 7E). The knockdown of SNHG5 suppressed Pgp and Bcl-2 expression and increased Bax expression, whereas Pgp overexpression partially restored the function of SNHG5 (Figure 7F). Collectively, the results demonstrated that SNHG5 served as a miR-26b sponge and increased the expression of Pgp, the target of miR-26b in CRC cells, indicating that SNHG5 conferred 5-Fu resistance in CRC at least partly by regulating the miR-26b/Pgp axis.
As a common clinical therapy, chemotherapy, especially 5-Fu-based chemotherapy, is still the main therapeutic option for patients with CRC at an advanced stage or at high risk of recurrence[19]. Increasing evidence has demonstrated that the phenomenon of drug resistance may become a major obstacle to successful outcomes during clinical adjuvant chemo
A review of recent literature indicates that SNHG5 plays a vital role in human cancers[23]. Chi et al[24] reported that SNHG5 facilitated the proliferation of breast cancer cells by sponging the miR-154-5p/proliferating cell nuclear antigen axis. Zhang et al[11] reported that in CRC, SNHG5 affected cell proliferation, metastasis, and migration by regulating miR-132-3p/CREB5. Damas et al[25] reported that SNHG5 promoted CRC cell survival by counteracting STAU1-mediated mRNA destabilization. However, the functions of SNHG5 in promoting 5-Fu resistance in CRC are currently unclear. In the present study, we found that higher expression levels of SNHG5 were associated with 5-Fu resistance in CRC. Our data demonstrated that SNHG5 overexpression significantly reduced cell apoptosis and enhanced cell viability, whereas SNHG5 knockdown in 5-Fu-resistant CRC cells increased cell apoptosis and decreased cell viability upon 5-Fu treatment. In a xenograft mouse model, we observed that 5-Fu, in combination with shSNHG5, inhibited the growth of 5-Fu-resistant CRC cells, thus confirming that SNHG5 promoted the resistance of CRC cells to 5-FU both in vitro and in vivo.
Multiple studies have shown that SNHG5 regulates chemoresistance in malignancies via various mechanisms, including ceRNAs. Wang et al[26] confirmed that SNHG5 regulated the resistance of acute myeloid leukemia to chemo
The adenosine triphosphate (ATP) binding cassette (ABC) superfamily, a group of membrane transporter proteins, includes 7 subfamilies, ABC-A to ABC-G, in humans and can function as ATP-dependent efflux pumps of antitumor drugs[28]. Pgp, also called ABCB1, is the first identified ABC transporter and contribute to chemotherapy failure in various cancer cells, including CRC cells[29]. Pgp can reduce the accumulation of intracellular antitumor drugs through the export of drugs, leading to multidrug resistance in CRC[30]. The substrates of Pgp include commonly utilized chemotherapeutic agents, which subsequently results in intrinsic resistance in CRC cells. Furthermore, exposure to these chemotherapeutic drugs induces the upregulation of Pgp in CRC cells, leading to acquired resistance[31]. Wang et al[29] reported that Pgp was highly expressed in CRC tissues and correlated with poor prognosis in CRC patients. We found that the expression of Pgp was higher in 5-Fu-resistant CRC tissues than in 5-Fu-sensitive tissues. However, the difference between the two groups was not statistically significant due to the limited sample size. The regulatory networks of Pgp remain unclear, and the potential regulatory mechanisms of ncRNAs have gained much attention[32]. Shi et al[33] reported that miR-29a overexpression restored the sensitivity of doxorubicin (DOX)-resistant CRC cells to DOX treatment via the downregulation of Pgp. Our previous data confirmed the tumor suppressive function of miR-26b in increasing the sensitivity of CRC cells to 5-Fu by negatively regulating Pgp expression[16]. As a further extension, the present study revealed that SNHG5 served as a miR-26b sponge and increased the expression of Pgp in CRC cells. These findings verified the regulatory role of the lncRNA SNHG5/miR-26b/Pgp axis in 5-Fu resistance in CRC and clarified the under
This study also has several limitations. Firstly, our study focused on the regulation of the SNHG5/miR-26b/Pgp axis, which might be a subset of a vast regulatory network in 5-Fu resistance in CRC, and other factors affecting 5-Fu resistance should be explored further. Secondly, the findings were investigated based on in vitro and xenograft mouse models, which might not recapitulate the complexity of CRC. With advances in gene editing techniques, transgenic animal models are applied to validate molecular functions, which are more consistent with the clinical pathogenic mechanism. It is valuable to utilize transgenic animal models with overexpression or specific knockout of SNHG5 to validate the role of SNHG5 in the chemoresistance of CRC in subsequent studies. Additionally, we detected the expression of SNHG5/miR-26b/Pgp in CRC tissues with a limited number of samples. It is necessary to further validate the findings in a large sample size cohort and detect the expression of SNHG5/miR-26b/Pgp in serum to explore novel non-invasive biomarkers for CRC chemotherapy.
Overall, in our study, we confirmed that the lncRNA SNHG5 promoted drug resistance in 5-Fu-resistant CRC. SNHG5 suppressed the sensitivity of CRC cells to 5-Fu treatment and facilitated the growth of cancer cells via miR-26b/Pgp. Our results contribute to elucidating the potential mechanisms of ncRNAs in the chemoresistance of CRC and provide new strategies for CRC chemotherapy.
The authors acknowledge Zhou GQ, Shi ZL, Guo J, Lu FY, Zhu JL and Wei W for their valuable assistance.
| 1. | Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5690] [Cited by in RCA: 8152] [Article Influence: 8152.0] [Reference Citation Analysis (2)] |
| 2. | Siegel RL, Wagle NS, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2023. CA Cancer J Clin. 2023;73:233-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1563] [Reference Citation Analysis (3)] |
| 3. | Smith HG, Nilsson PJ, Shogan BD, Harji D, Gambacorta MA, Romano A, Brandl A, Qvortrup C. Neoadjuvant treatment of colorectal cancer: comprehensive review. BJS Open. 2024;8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 19] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 4. | Shin AE, Giancotti FG, Rustgi AK. Metastatic colorectal cancer: mechanisms and emerging therapeutics. Trends Pharmacol Sci. 2023;44:222-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 287] [Reference Citation Analysis (0)] |
| 5. | Xu R, Du A, Deng X, Du W, Zhang K, Li J, Lu Y, Wei X, Yang Q, Tang H. tsRNA-GlyGCC promotes colorectal cancer progression and 5-FU resistance by regulating SPIB. J Exp Clin Cancer Res. 2024;43:230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 6. | Chen L, Sun K, Qin W, Huang B, Wu C, Chen J, Lai Q, Wang X, Zhou R, Li A, Liu S, Zhang Y. LIMK1 m(6)A-RNA methylation recognized by YTHDC2 induces 5-FU chemoresistance in colorectal cancer via endoplasmic reticulum stress and stress granule formation. Cancer Lett. 2023;576:216420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 7. | Song M, Yang J, Di M, Hong Y, Pan Q, Du Y, Xiang T, Liu J, Tang Y, Wang Q, Li Y, He J, Chen H, Zhao J, Weng D, Zhang Y, Xia JC. Alarmin IL-33 orchestrates antitumoral T cell responses to enhance sensitivity to 5-fluorouracil in colorectal cancer. Theranostics. 2023;13:1649-1668. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 8. | Zhu Y, Hu H, Yuan Z, Zhang Q, Xiong H, Hu Z, Wu H, Huang R, Wang G, Tang Q. LncRNA NEAT1 remodels chromatin to promote the 5-Fu resistance by maintaining colorectal cancer stemness. Cell Death Dis. 2020;11:962. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 9. | Jiang Z, Li L, Hou Z, Liu W, Wang H, Zhou T, Li Y, Chen S. LncRNA HAND2-AS1 inhibits 5-fluorouracil resistance by modulating miR-20a/PDCD4 axis in colorectal cancer. Cell Signal. 2020;66:109483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 47] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 10. | Zhang F, Wang H, Yu J, Yao X, Yang S, Li W, Xu L, Zhao L. LncRNA CRNDE attenuates chemoresistance in gastric cancer via SRSF6-regulated alternative splicing of PICALM. Mol Cancer. 2021;20:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 141] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 11. | Zhang M, Li Y, Wang H, Yu W, Lin S, Guo J. LncRNA SNHG5 affects cell proliferation, metastasis and migration of colorectal cancer through regulating miR-132-3p/CREB5. Cancer Biol Ther. 2019;20:524-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 12. | Li Y, Hu J, Guo D, Ma W, Zhang X, Zhang Z, Lu G, He S. LncRNA SNHG5 promotes the proliferation and cancer stem cell-like properties of HCC by regulating UPF1 and Wnt-signaling pathway. Cancer Gene Ther. 2022;29:1373-1383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 42] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 13. | Huang SL, Huang ZC, Zhang CJ, Xie J, Lei SS, Wu YQ, Fan PZ. LncRNA SNHG5 promotes the glycolysis and proliferation of breast cancer cell through regulating BACH1 via targeting miR-299. Breast Cancer. 2022;29:65-76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 14. | Yue Y, Zhang Q, Wang X, Sun Z. STAT3 regulates 5-Fu resistance in human colorectal cancer cells by promoting Mcl-1-dependent cytoprotective autophagy. Cancer Sci. 2023;114:2293-2305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 15. | Liu S, Lin H, Wang D, Li Q, Luo H, Li G, Chen X, Li Y, Chen P, Zhai B, Wang W, Zhang R, Chen B, Zhang M, Han X, Li Q, Chen L, Liu Y, Chen X, Li G, Xiang Y, Duan T, Feng J, Lou J, Huang X, Zhang Q, Pan T, Yan L, Jin T, Zhang W, Zhuo L, Sun Y, Xie T, Sui X. PCDH17 increases the sensitivity of colorectal cancer to 5-fluorouracil treatment by inducing apoptosis and autophagic cell death. Signal Transduct Target Ther. 2019;4:53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 16. | Wang B, Lu FY, Shi RH, Feng YD, Zhao XD, Lu ZP, Xiao L, Zhou GQ, Qiu JM, Cheng CE. MiR-26b regulates 5-FU-resistance in human colorectal cancer via down-regulation of Pgp. Am J Cancer Res. 2018;8:2518-2527. [PubMed] |
| 17. | Lin X, Zhuang S, Chen X, Du J, Zhong L, Ding J, Wang L, Yi J, Hu G, Tang G, Luo X, Liu W, Ye F. lncRNA ITGB8-AS1 functions as a ceRNA to promote colorectal cancer growth and migration through integrin-mediated focal adhesion signaling. Mol Ther. 2022;30:688-702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 103] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 18. | Wang M, Niu X, Wang M, Zheng P, Liu X, Cao Z, Zhang C. Long non-coding RNA RP11-197K6.1 as ceRNA promotes colorectal cancer progression via miR-135a-5p/DLX5 axis. J Transl Med. 2024;22:469. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 19. | Yang Y, Zhang J, Zhang W, Wang Y, Zhai Y, Li Y, Li W, Chang J, Zhao X, Huang M, Geng Q, Yang Y, Gong Z, Yu N, Shen W, Li Q, Huang S, Guo W. A liquid biopsy signature of circulating extracellular vesicles-derived RNAs predicts response to first line chemotherapy in patients with metastatic colorectal cancer. Mol Cancer. 2023;22:199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 20. | Guo Y, Wang M, Zou Y, Jin L, Zhao Z, Liu Q, Wang S, Li J. Mechanisms of chemotherapeutic resistance and the application of targeted nanoparticles for enhanced chemotherapy in colorectal cancer. J Nanobiotechnology. 2022;20:371. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 41] [Reference Citation Analysis (0)] |
| 21. | Bian Z, Yang F, Xu P, Gao G, Yang C, Cao Y, Yao S, Wang X, Yin Y, Fei B, Huang Z. LINC01852 inhibits the tumorigenesis and chemoresistance in colorectal cancer by suppressing SRSF5-mediated alternative splicing of PKM. Mol Cancer. 2024;23:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 22. | Dong S, Liang S, Cheng Z, Zhang X, Luo L, Li L, Zhang W, Li S, Xu Q, Zhong M, Zhu J, Zhang G, Hu S. ROS/PI3K/Akt and Wnt/β-catenin signalings activate HIF-1α-induced metabolic reprogramming to impart 5-fluorouracil resistance in colorectal cancer. J Exp Clin Cancer Res. 2022;41:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 263] [Article Influence: 87.7] [Reference Citation Analysis (0)] |
| 23. | Han W, Shi J, Cao J, Dong B, Guan W. Latest Advances of Long Non-Coding RNA SNHG5 in Human Cancers. Onco Targets Ther. 2020;13:6393-6403. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 24. | Chi JR, Yu ZH, Liu BW, Zhang D, Ge J, Yu Y, Cao XC. SNHG5 Promotes Breast Cancer Proliferation by Sponging the miR-154-5p/PCNA Axis. Mol Ther Nucleic Acids. 2019;17:138-149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 25. | Damas ND, Marcatti M, Côme C, Christensen LL, Nielsen MM, Baumgartner R, Gylling HM, Maglieri G, Rundsten CF, Seemann SE, Rapin N, Thézenas S, Vang S, Ørntoft T, Andersen CL, Pedersen JS, Lund AH. SNHG5 promotes colorectal cancer cell survival by counteracting STAU1-mediated mRNA destabilization. Nat Commun. 2016;7:13875. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 172] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 26. | Wang D, Zeng T, Lin Z, Yan L, Wang F, Tang L, Wang L, Tang D, Chen P, Yang M. Long non-coding RNA SNHG5 regulates chemotherapy resistance through the miR-32/DNAJB9 axis in acute myeloid leukemia. Biomed Pharmacother. 2020;123:109802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 27. | Li M, Zhang YY, Shang J, Xu YD. LncRNA SNHG5 promotes cisplatin resistance in gastric cancer via inhibiting cell apoptosis. Eur Rev Med Pharmacol Sci. 2019;23:4185-4191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 28. | Bharathiraja P, Yadav P, Sajid A, Ambudkar SV, Prasad NR. Natural medicinal compounds target signal transduction pathways to overcome ABC drug efflux transporter-mediated multidrug resistance in cancer. Drug Resist Updat. 2023;71:101004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 35] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 29. | Wang H, Li JM, Wei W, Yang R, Chen D, Ma XD, Jiang GM, Wang BL. Regulation of ATP-binding cassette subfamily B member 1 by Snail contributes to chemoresistance in colorectal cancer. Cancer Sci. 2020;111:84-97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 30. | Sancha SAR, Szemerédi N, Spengler G, Ferreira MU. Lycorine Carbamate Derivatives for Reversing P-glycoprotein-Mediated Multidrug Resistance in Human Colon Adenocarcinoma Cells. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 31. | Lei ZN, Albadari N, Teng QX, Rahman H, Wang JQ, Wu Z, Ma D, Ambudkar SV, Wurpel JND, Pan Y, Li W, Chen ZS. ABCB1-dependent collateral sensitivity of multidrug-resistant colorectal cancer cells to the survivin inhibitor MX106-4C. Drug Resist Updat. 2024;73:101065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 32. | Wang Y, Wang Y, Qin Z, Cai S, Yu L, Hu H, Zeng S. The role of non-coding RNAs in ABC transporters regulation and their clinical implications of multidrug resistance in cancer. Expert Opin Drug Metab Toxicol. 2021;17:291-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 96] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 33. | Shi X, Valizadeh A, Mir SM, Asemi Z, Karimian A, Majidina M, Safa A, Yosefi B. miRNA-29a reverses P-glycoprotein-mediated drug resistance and inhibits proliferation via up-regulation of PTEN in colon cancer cells. Eur J Pharmacol. 2020;880:173138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 47] [Article Influence: 9.4] [Reference Citation Analysis (0)] |