Published online Dec 6, 2019. doi: 10.12998/wjcc.v7.i23.3990
Peer-review started: September 27, 2019
First decision: October 24, 2019
Revised: October 31, 2019
Accepted: November 15, 2019
Article in press: November 15, 2019
Published online: December 6, 2019
Processing time: 69 Days and 19.4 Hours
Although it has been shown that arylsulfatases are lost in colorectal cancer (CRC) cell lines, their exact role in the carcinogenesis and behavior of this cancer was not elucidated. No data about the correlation between serum and immunohistochemical (IHC) level of arylsulfatases (ARSA, ARSB) in patients with CRC were published yet.
To evaluate the possible prognostic value of ARSA and/or ARSB in CRC, at circulating and protein levels.
The present study included 45 consecutive patients who were prospectively diagnosed with CRC. For IHC stains (protein expression) ARSA, ARSB and maspin expression were quantified. For these markers, cytoplasmic expression was taken into account. For gene expression study, circulating mRNA was isolated from all patients, before surgery. A group of 45 healthy patients without inflammatory or tumor pathologies was used as control group. Reverse transcription and Taqman Gene Expression Array were used for ARSB gene expression.
The preoperative circulating RNA level of the ARSB gene was significantly decreased in patients with CRC (RQ < 1), compared with the control group (RQ > 1). A more significant decrease (RQ < 0.5) occurred in ulcero-infiltrative maspin-positive adenocarcinomas, with a higher degree of tumor budding, diagnosed in locally advanced stages (pT3/4). ARSA/maspin immunopositivity indicated a higher risk for lymph node metastasis, while triple positivity for maspin/ARSA/ARSB and ARSB gene expression level < 0.5 were indicators of CRC aggressive behavior, independent of lymph node status.
The significant independent negative prognostic factors of CRC are the ulcero-infiltrative aspect, high budding degree, triple positivity for maspin, ARSA and ARSB, and low ARSB gene expression.
Core tip: In this paper we tried to emphasize the role of arylsulfatases (ARSA, ARSB) in colorectal cancer (CRC) beahaviour and possible role of ARSB serum level in follow-up of patients. This is the first study in literature which proved that a low ARSB gene expression in serum (RQ < 1) might be a non-invasive indicator of risk of CRC. Moreover, triple positivity for maspin/ARSA/ARSB and ARSB gene expression level < 0.5 were proved to be indicators of CRC aggressive behavior, independent of lymph node status.
- Citation: Kovacs Z, Jung I, Szalman K, Banias L, Bara TJ, Gurzu S. Interaction of arylsulfatases A and B with maspin: A possible explanation for dysregulation of tumor cell metabolism and invasive potential of colorectal cancer. World J Clin Cases 2019; 7(23): 3990-4003
- URL: https://www.wjgnet.com/2307-8960/full/v7/i23/3990.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v7.i23.3990
Colorectal cancer (CRC) usually affects people over 60 years of age and remains one of the leading causes of cancer mortality in Europe[1]. Although the involvement of several molecular factors in colorectal carcinogenesis and the invasiveness of CRC have been described, these processes are not yet fully understood[2]. In this paper, we sought to characterize the possible role of two arylsulfatases (ARSA and ARSB) in CRC behavior. As the correlation between the immunohistochemical (IHC) and gene expression of ARSB has not yet been examined in CRC, we aimed to formulate a new hypothesis about the molecular background of CRC.
Arylsulfatases are lysosomal enzymes that are able to catalyze the hydrolysis of sulfate esters. A total deficiency of ARSA is known to induce metabolic disorders such as metachromatic leukodystrophy, a lysosomal storage disorder characterized by the accumulation of cerebroside sulfate within lysosomes and the further destruction of the brain's white matter[3,4].
Chondroitin sulfate and dermatan sulfate are the targets of ARSB, which are glycosaminoglycans. The total deficiency of ARSB leads to Maroteaux-Lamy syndrome, a genetic disorder with severe neurological dysfunction[4,5]. The pseudo-deficiency and/or extralysosomal localization of ARSA and/or ARSB is not known to cause serious health problems[4,6].
In one of the recently published reviews of the literature, we showed that fewer than 40 papers concerning the possible role of ARSA/ARSB in tumorigenesis were published in the English literature and indexed in the Medline database prior to 2018[4]. These studies were mainly based on cell lines and took into account malignant melanoma (about one paper), lung cancer (about two papers), urogenital cancer (about 10 papers) and CRC (about 20 papers)[4]. Independently of carcinoma localization, a decreased expression pattern of ARSA and ARSB was described in the above-mentioned papers[7].
In CRC, it seems that ARSB is expressed in normal colonic mucosa and shows a loss of intensity in tumor cells, with a role in carcinoma invasiveness and metastatic capacity[4,7,8]. To our knowledge, no data about the role of ARSA in CRC have yet been published[4].
In this paper, we examined the IHC expression of ARSA and ARSB in 45 CRC specimens, together with circulating ARSB gene expression. All of the obtained data were correlated with clinicopathological parameters, including the degree of budding and overall survival rate[9]. For budding quantification, we used the IHC biomarker maspin, which is a serine protease that is known to be involved in the inhibition of tumor cell proliferation, angiogenesis and apoptosis promotion[9-12]. We chose Maspin for budding assessment, because it was proved to be a specific marker for budding quantification[11,12] and an reliable prognostic and possible predictive marker, for CRC[9-12].
The aim of this study was to examine the unexplored pathological significance of ARSA and ARSB, and their possible interaction with maspin in patients with CRC. ARSA/ARSB IHC expression level was correlated with maspin positivity and ARSB preoperative gene expression level, in order to see the possible positive correlation between ARSA/ARSB and maspin protein level together with ARSB gene expression and tumor behavior and agressivity.
The present study included 45 consecutive cases of patients who were prospectively diagnosed with CRC. The Ethical Approval of Mures County Emergency Hospital and signed informed consent was obtained before surgery.
In all of the patients, surgical resection was performed and blood was taken one day before surgery. Only those patients who survived for at least 20 d after surgery were included. No preoperative radiotherapy was performed before surgery. Cases from the upper rectum also involved the recto-sigmoid junction and no preoperative chemotherapy were administered. The patient follow-up period was between 20 and 509 d.
All of the cases comprised adenocarcinomas without distant metastases (M0). The eighth edition of the American Joint Committee on Cancer and the current World Health Organization classification were used for tumor staging. Tumor buds were quantified under light microscopy using the criteria proposed by Ueno et al[13] in 2012, adapted with the International Tumor Budding Consensus Conference criteria from 2016[14]. To ensure the objectivity of budding assessment, maspin quantification was also conducted, based on the criteria mentioned in the previously published papers[11,12]. Cases were divided into G1 (below five buds/HPF), G2 (five-nine buds) and G3 (over 10 isolated cells or clusters/HPF)[10-12].
ARSA, ARSB and maspin expression were quantified using the IHC markers presented in Table 1. For ARSA, ARSB and maspin, cytoplasmic expression was taken into account. Although maspin can also show nuclear positivity, especially in tumor buds[12], due to the low number of cases, the subcellular expression was not taken into account. Cases were classified as showing low or high expression, based on the percentage of positive cells and the intensity of immunostaining[10-12].
| Marker | Clone | Incubation | pH |
| Arylsulfatase A | Mouse monoclonal (Santa Cruz Biotechnology) | High pH (Dako) | 9.0 |
| Arylsulfatase B | Rabbit policlonal (Abcam) | High pH (Dako) | 9.0 |
| Maspin | Mouse monoclonal (Santa Cruz Biotechnology) | 0.01 mol/L citrate (Novocastra) | 6.0 |
Circulating mRNA was isolated from all 45 patients with CRC (Table 2), from which 2 mL of intravenous blood was taken one day before surgery. For the control group, we used blood from 45 healthy patients without inflammatory or tumor pathologies, in which colonoscopy was conducted for screening purposes.
| Characteristics | Number | ARSA expression | P value | |
| Low | High | |||
| Age, yr | ||||
| ≤ 60 | 19 | 8 | 11 | 0.761 |
| > 60 | 26 | 13 | 13 | |
| Gender | ||||
| Male | 26 | 12 | 14 | > 0.991 |
| Female | 19 | 9 | 10 | |
| Macroscopic aspect | ||||
| Polypoid | 12 | 9 | 3 | 0.041 |
| Ulcero-infiltrative | 33 | 12 | 21 | |
| Microscopic aspect | ||||
| G1 | 14 | 11 | 3 | 0.022 |
| G2 | 24 | 9 | 15 | |
| G3+4 | 7 | 2 | 5 | |
| Localization | ||||
| Proximal | 8 | 5 | 3 | 0.502 |
| Distal | 21 | 10 | 11 | |
| Upper rectum | 16 | 6 | 10 | |
| Lymph node ratio | ||||
| < 0.1 | 34 | 19 | 15 | 0.041 |
| ≥ 0.1 | 11 | 2 | 9 | |
| Lymph node metastasis | ||||
| Absent | 29 | 17 | 12 | 0.031 |
| Present | 16 | 4 | 12 | |
| pT stage | ||||
| ≤ T2 | 15 | 3 | 12 | 0.011 |
| ≥ T3 | 30 | 18 | 12 | |
| Budding grade | ||||
| G1 | 15 | 8 | 7 | 0.812 |
| G2 | 9 | 4 | 5 | |
| G3 | 21 | 9 | 12 | |
| Maspin expression | ||||
| Positive | 22 | 3 | 19 | < 0.00011 |
| Negative | 23 | 18 | 5 | |
| ARSB expression | ||||
| Low | 11 | 5 | 6 | 0.921 |
| High | 34 | 16 | 18 | |
Blood mRNA isolation was performed using a Roche High Pure RNA Isolation kit (Roche Diagnostics, Germany) in line with the user's guide. Reverse transcription was conducted using a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, United States) following the recommended protocol. A Taqman Gene Expression Array was used for Arylsulfatase B gene expression, according to the user's guide (Applied Biosystems, United States), with the following PCR protocol: (1) UNG (uracil N-glycosylase) incubation 50°C, two minutes, one cycle (for preventing carryover contamination); (2) Enzyme activation 95°C, 20 s, one cycle; and (3) Denaturation, 95°C, one second with Anneal/Extend at 60°C, 20 s for 40 cycles.
Statistical analysis was performed using GraphPad Prims8 software. A P value < 0.05 (with a 95% confidence interval) was considered statistically significant, calculated using the χ2 (and Fisher’s exact test and Yate’s continuity correction) test and Kruskal-Wallis, while the Mann-Whitney test was used for ARSB gene expression. The possible correlation between the examined IHC markers and the ARSB gene was assessed using a Venn diagram. Overall survival was evaluated using Kaplan-Meier survival curves.
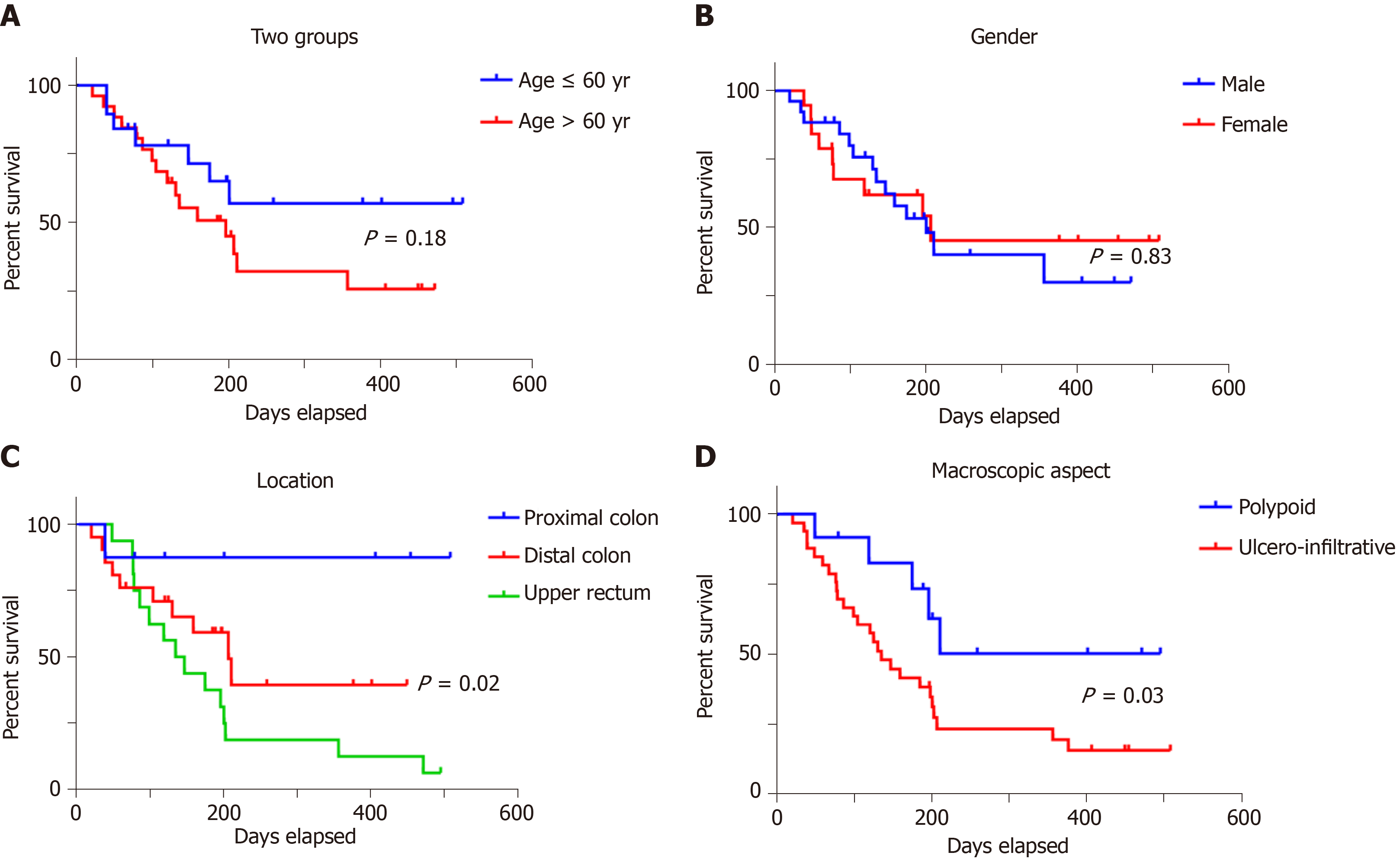
The immunoexpression of the three examined markers (ARSA, ARSB and maspin) was not correlated with the patients’ gender or age (Tables 2-4). Although a slightly increased ARSB expression was seen in aging patients, it was not statistically significant when the age of 60 was taken into account (Table 3). The median age of patients was 57 ± 13.24 years (range 26-86 years), most of whom (n = 26; 57.8%) were diagnosed with CRC over the age of 60.
| Characteristics | Number | ARSB expression | P value | |
| Low | High | |||
| Age, yr | ||||
| ≤ 60 | 19 | 7 | 12 | 0.091 |
| > 60 | 26 | 4 | 22 | |
| Gender | ||||
| Male | 26 | 5 | 21 | 0.341 |
| Female | 19 | 6 | 13 | |
| Macroscopic aspect | ||||
| Polypoid | 12 | 7 | 5 | 0.0031 |
| Ulcero-infiltrative | 33 | 4 | 29 | |
| Microscopic aspect | ||||
| G1 | 14 | 2 | 12 | 0.0072 |
| G2 | 24 | 4 | 20 | |
| G3+4 | 7 | 5 | 2 | |
| Localization | ||||
| Proximal | 8 | 2 | 6 | 0.992 |
| Distal | 21 | 5 | 16 | |
| Upper rectum | 16 | 4 | 12 | |
| Lymph node ratio | ||||
| < 0.1 | 34 | 9 | 25 | 0.701 |
| ≥ 0.1 | 11 | 2 | 9 | |
| Lymph node metastasis | ||||
| Absent | 29 | 9 | 20 | 0.271 |
| Present | 16 | 2 | 14 | |
| pT stage | ||||
| ≤ T2 | 15 | 5 | 10 | 0.461 |
| ≥ T3 | 30 | 6 | 23 | |
| Budding grade | ||||
| G1 | 15 | 3 | 12 | 0.822 |
| G2 | 9 | 2 | 7 | |
| G3 | 21 | 6 | 15 | |
| Maspin expression | ||||
| Positive | 22 | 2 | 20 | 0.03 |
| Negative | 23 | 9 | 14 | |
| Characteristics | Number | Maspin expression | P value | |
| Negative | Positive | |||
| Age, yr | ||||
| ≤ 60 | 19 | 9 | 10 | 0.661 |
| > 60 | 26 | 14 | 12 | |
| Gender | ||||
| Male | 26 | 13 | 13 | 0.861 |
| Female | 19 | 10 | 9 | |
| Macroscopic aspect | ||||
| Polypoid | 12 | 10 | 2 | 0.011 |
| Ulcero-infiltrative | 33 | 13 | 20 | |
| Microscopic aspect | ||||
| G1 | 14 | 11 | 3 | 0.032 |
| G2 | 24 | 10 | 14 | |
| G3+4 | 7 | 2 | 5 | |
| Localization | ||||
| Proximal | 8 | 1 | 7 | < 0.00012 |
| Distal | 21 | 19 | 2 | |
| Upper rectum | 16 | 3 | 13 | |
| Lymph node ratio | ||||
| < 0.1 | 34 | 19 | 15 | 0.261 |
| ≥ 0.1 | 11 | 4 | 7 | |
| Lymph node metastasis | ||||
| Absent | 29 | 19 | 10 | 0.011 |
| Present | 16 | 4 | 12 | |
| pT stage | ||||
| ≤ T2 | 15 | 13 | 2 | 0.0011 |
| ≥ T3 | 30 | 10 | 20 | |
| Budding grade | ||||
| G1 | 15 | 14 | 1 | <0.0011 |
| G2 | 9 | 5 | 4 | |
| G3 | 21 | 4 | 17 | |
ARSA and ARSB expression was not influenced by tumor localization (Tables 2 and 3). Most of the maspin-negative cases (n = 19; 42.22%) involved the distal colon, while the positive cases were located in the upper rectum (Table 4).
Most of the cases (n = 33; 73.33%) showed an ulcero-infiltrative aspect, with high expression of all of the three examined markers. Independently of ARSB expression (Table 3), polypoid tumors were mostly maspin negative (Table 4) and showed low ARSA intensity (Table 2).
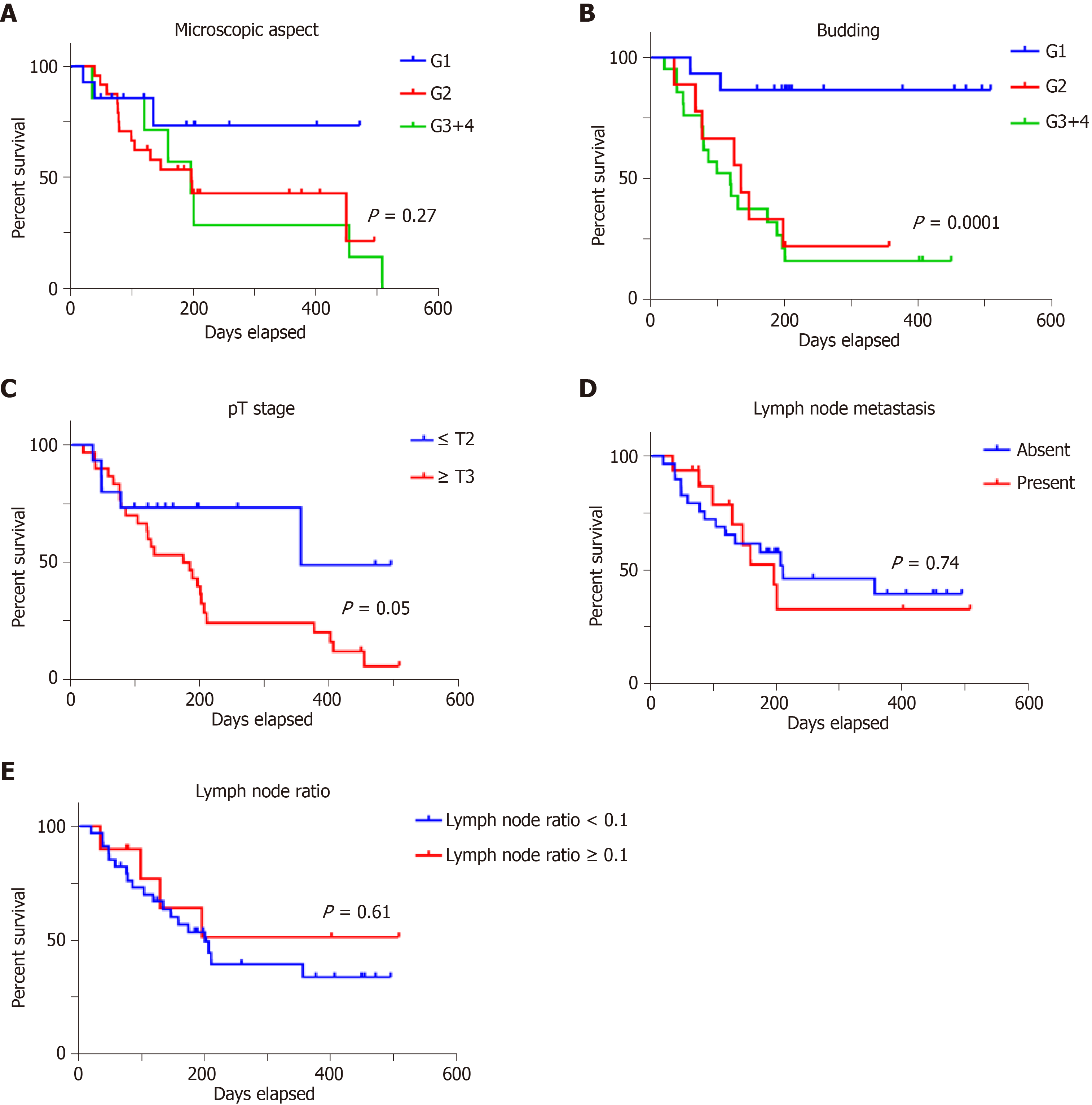
The ARSA and maspin intensity increased in parallel with tumor dedifferentiation, with only G1 cases associated with significantly low ARSA and negative maspin (Tables 2 and 4). ARSB expression was high in G1 and G2 cases and particularly decreased in G3 and G4 specimens (Table 3).
There was predominantly a loss of ARSA expression in cases diagnosed in locally advanced stages (pT3+4), without lymph node metastases, independently of the degree of tumor budding (Table 2). ARSB immune expression was not correlated with the pT stage, pN stage, lymph node ratio or the degree of tumor budding (Table 3). Most of the maspin-negative cases were diagnosed in the early stages (pT1+2) and did not show lymph node metastases or a high degree of budding (Table 4).
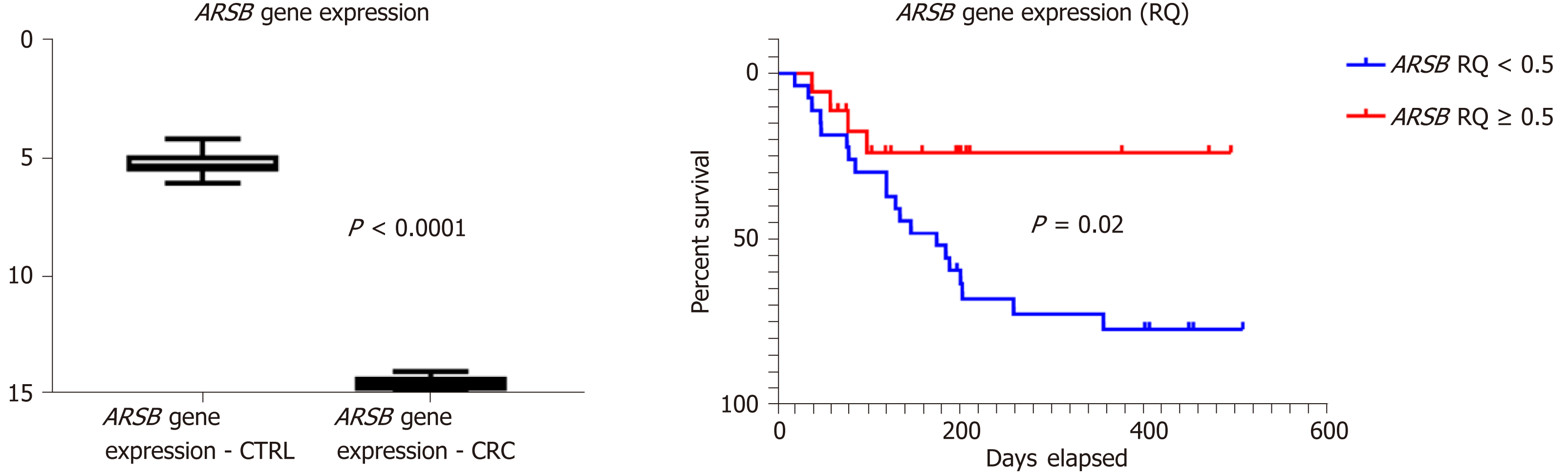
In all of the patients included in the control group (n = 45), without inflammatory or tumor disorders, the circulating ARSB gene expression level was higher than one (ranging from one-five), whereas blood levels higher than one were obtained from the blood of patients with CRC. After normalization with house-keeping, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene expression of ARSB was assessed using Kruskal-Wallis and Mann-Whitney tests[15], with a significantly decreased ARSB gene expression (P < 0.0001) found in patients with CRC (Figure 1).
As a low ARSB gene expression circulating level was found in all of the patients with CRC, after relative quantification (RQ < 1) for statistical assessment (Table 5), the cases were divided into two groups (RQ < 0.5 and RQ ≥ 0.5).
| Characteristics | Number | ARSB gene expression (RQ) | P value | ARSB gene expression (RQ) | P value | |
| RQ < 0.5 | RQ ≥ 0.5 | |||||
| Age, yr | ||||||
| ≤ 60 | 19 | 0.4801 ± 0.2711 | 0.6691 | 10 | 9 | 0.532 |
| > 60 | 26 | 0.5125 ± 0.3261 | 17 | 9 | ||
| Gender | ||||||
| Male | 26 | 0.4938 ± 0.2892 | 0.9591 | 14 | 12 | 0.372 |
| Female | 19 | 0.5058 ± 0.3250 | 13 | 6 | ||
| Macroscopic aspect | ||||||
| Polypoid | 12 | 0.4339 ± 0.2530 | 0.3951 | 4 | 8 | 0.042 |
| Ulcero-infiltrative | 33 | 0.5225 ± 0.3172 | 23 | 10 | ||
| Microscopic aspect | ||||||
| G1 | 14 | 0.4155 ± 0.3034 | 0.2953 | 4 | 10 | 0.014 |
| G2 | 24 | 0.5496 ± 0.2793 | 17 | 7 | ||
| G3+4 | 7 | 0.4915 ± 0.3747 | 6 | 1 | ||
| Localization | ||||||
| Proximal | 8 | 0.3246 ± 0.2332 | 0.1153 | 5 | 3 | 0.064 |
| Distal | 21 | 0.4911 ± 0.3179 | 9 | 12 | ||
| Upper rectum | 16 | 0.5962 ± 0.2824 | 13 | 3 | ||
| Lymph node ratio | ||||||
| < 0.1 | 34 | 0.5057 ± 0.3020 | 0.9261 | 20 | 14 | > 0.992 |
| ≥ 0.1 | 11 | 0.4799 ± 0.3293 | 7 | 4 | ||
| Lymph node metastasis | ||||||
| Absent | 29 | 0.4749 ± 0.2838 | 0.5451 | 17 | 12 | > 0.992 |
| Present | 16 | 0.5422 ± 0.3358 | 10 | 6 | ||
| pT stage | ||||||
| ≤ T2 | 15 | 0.4987 ± 0.3296 | 0.96521 | 3 | 12 | 0.00022 |
| ≥ T3 | 30 | 0.4989 ± 0.2997 | 24 | 6 | ||
| Budding grade | ||||||
| G1 | 15 | 0.5193 ± 0.2963 | 0.97743 | 4 | 11 | 0.0014 |
| G2 | 9 | 0.4835 ± 0.3272 | 5 | 4 | ||
| G3 | 21 | 0.4894 ± 0.3086 | 18 | 3 | ||
| ARSA IHC expression | ||||||
| Low | 21 | 0.5164 ± 0.2751 | 0.75613 | 13 | 8 | > 0.992 |
| High | 24 | 0.4835 ± 0.3275 | 14 | 10 | ||
| ARSB IHC expression | ||||||
| Low | 11 | 0.4849 ± 0.3034 | 0.96131 | 4 | 7 | 0.082 |
| High | 34 | 0.5034 ± 0.3050 | 23 | 11 | ||
| Maspin IHC expression | ||||||
| Positive | 22 | 0.5066 ± 0.2981 | 0.98781 | 18 | 4 | 0.0052 |
| Negative | 23 | 0.4979 ± 0.3175 | 9 | 14 | ||
The preoperative ARSB gene circulating level did not prove to be correlated with patients' age or gender, either in the presence or absence of lymph node metastases (Table 5). ARSB was found to have a significantly lower gene expression profile in ulcero-infiltrative tumors, especially those from the upper rectum, diagnosed in locally advanced stages (pT3+4), which showed a high degree of budding (over 10 buds/HPF) and a high grade of dedifferentiation (Table 5).
No correlation was found between ARSA and the tissue protein level of ARSB (Table 2) or its RNA circulating level (Table 5). The ARSB protein level in surgical specimens was inversely correlated with the preoperative ARSB gene circulating level (Table 5).
A significant direct correlation was observed between maspin and the tissue protein level of both ARSA (Table 2) and ARSB (Table 3). The maspin protein tissue level was inversely correlated with the preoperative ARSB gene circulating level (Table 5).
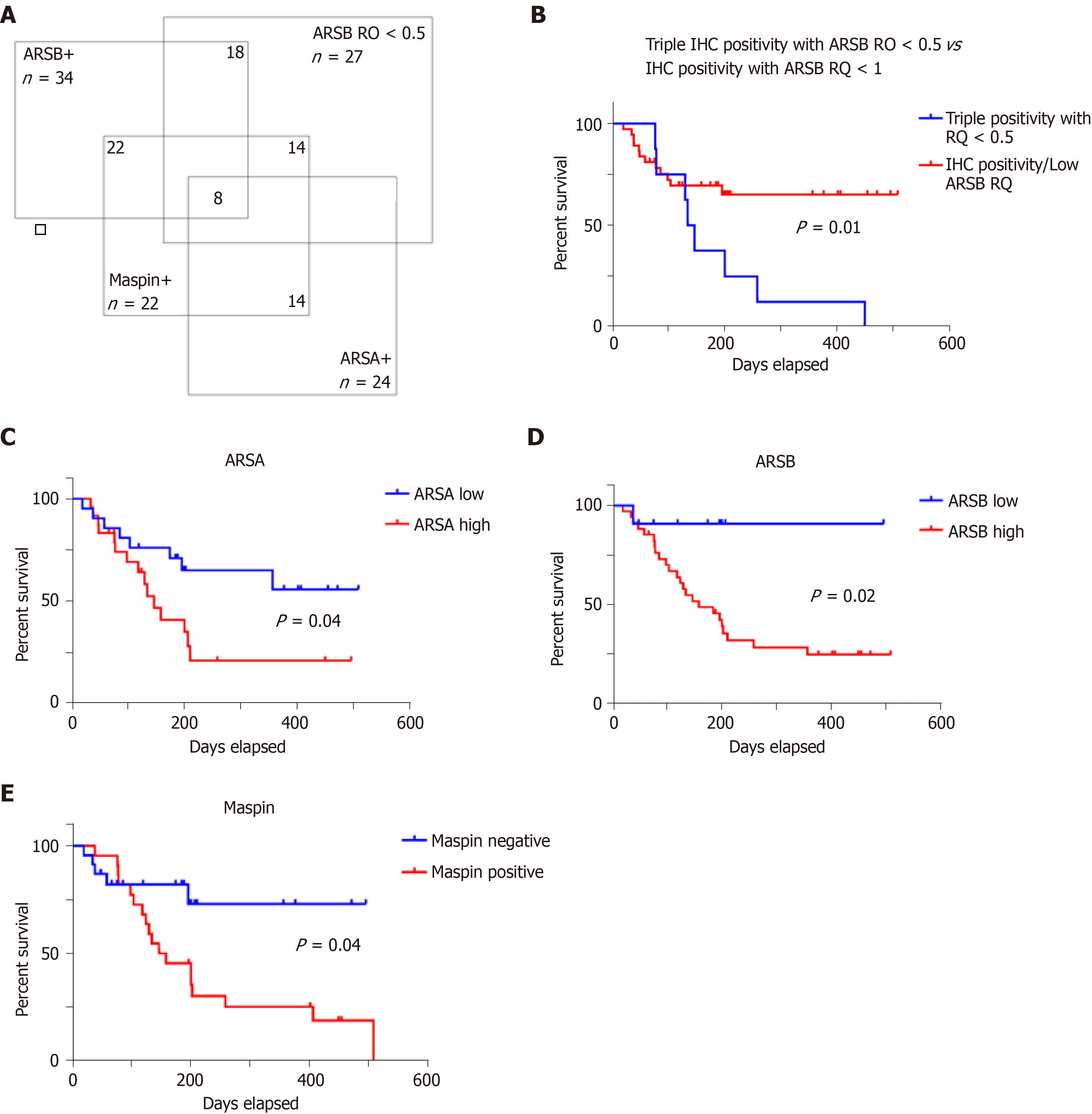
A Venn diagram illustrates that from the 45 cases, 14 (31.11%) showed positivity for maspin, high ARSA and ARSB protein levels and low ARSB gene expression (Figure 2). All of the 14 cases were ulcero-infiltrative carcinomas of the upper rectum, showing a high degree of budding.
Examination of the independent prognostic value of the examined prognostic factors showed no correlation with overall survival for patients' age or gender (Figure 3), or with the tumor grade of differentiation (Figure 4). A longer but not significant overall survival was associated with lymph node status (Figure 4). Ulcero-infiltrative tumors (Figure 3) with a high degree of budding and diagnosed in the T3-4 stage, especially those of lower rectum, presented a shorter overall survival rate (Figure 4). Triple high protein expression of ARSA, ARSB and maspin, correlated with low ARSB gene expression, were indicators of short overall survival (Figure 2).
Multivariate correlation indicates that negative prognostic value was associated with ulcero-infiltrative carcinomas of the lower rectum, with a high degree of budding, triple positivity for ASRSA/ARSB/maspin and low ARSB gene expression.
The present study confirms the possible role of ARSA and ARSB in colorectal carcinogenesis and tumor invasiveness. As no similar studies have previously been published in the Medline database, it is difficult to interpret the obtained results. Accordingly, we took into account the well-known clinicopathological prognostic parameters and maspin, one of the serine proteases intensely examined by our team and used in daily diagnosis for tumor budding quantification[9-12].
As the previous studies proved the unquestionably prognostic role of tumor stages, the present findings show that this parameter remains an independent prognostic factor, but the most strongly statistically independent prognostic value was found for the degree of tumor budding, especially for ulcero-infiltrating tumors, in accordance with other published papers[12-14].
In the early stages (pT1/2N0 cases with low grade budding), maspin was mostly lost in tumor cells compared with normal mucosa. Maspin was found to be positive in advanced stages (pT3/4N1-2 with high grade budding), whereas ARSA was firstly high, then lost in patients with pT3/4N0 carcinomas, and re-expressed in cases with lymph node metastases. These aspects confirm that ARSA is positively maintained in the early stages, as in normal colonic mucosa, functioning as a protector of local invasiveness but not lymph vessel invasion. The ARSA protein level then decreases in parallel with local tumor invasiveness but is re-expressed when tumor cells invade the lymph vessel inva, independently of the degree of tumor budding.
Regarding ARSB, the present study confirms the previously supposed loss of ARSB expression in patients with CRCs, compared with healthy patients[7,8]. For the first time in the literature, we have aimed to prove the possible mechanism of CRC cell ARSB-related aggressiveness. Firstly, we showed that the RNA circulating level of the ARSB gene (RQ < 1) can be used as a screening indicator of CRC, especially in patients with RQ < 0.5. On the one hand, the indicators of local aggressiveness, such as the ulcero-infiltrative aspect, advanced stage (T3,4), a high degree of budding and maspin positivity, were correlated with an ARSB gene circulating level of < 0.5, independently of the presence or absence of lymph node metastases. On the other hand, independently of the tumor stage or other clinicopathological parameters, the ARSB protein level expressed in normal colonic mucosa was maintained in CRC cells in over 75% of cases, especially in ulcero-infiltrative, well-differentiated (G1) adenocarcinomas. An inverse correlation between the circulating and protein level of ARSB was statistically shown. These aspects confirm that in early stages of colorectal carcinogenesis, maspin and ARSB are lost and then re-expressed in tumor cells, as an indicator of aggressiveness.
Although based on a small number of cases, the present study demonstrates that in early stages of carcinogenesis, both ARSA and ARSB protein expression is maintained in tumor cells. Then, the evolution depends on the arylsulfatase that is expressed the most, in correlation with other genetic or environmental factors. A loss of ARSA might indicate a high capacity of tumor cells for local invasiveness and a high risk of local recurrence after surgery. Independently of the depth of infiltration, in cases with high ARSA intensity, there is a higher risk of lymph vessel invasion and lymph node metastases, independently of ARSB expression. Although ARSB is maintained in tumor cells, its circulating level decreases in parallel with the depth of tumor infiltration. Maspin positivity remains an indicator of a high degree of tumor budding, especially for locally advanced carcinomas.
It is also worth mentioning that a triple positivity for ARSA/ARSB/maspin, correlated with an ARSB gene circulating level of < 0.5, is an indicator of a lower survival rate, independently of the other clinicopathological parameters. This association was found in one third of the cases, with a high degree of budding in each case.
In the literature, the ARSB-related potential aggressive behavior of G2/3 ulcero-infiltrative CRC is thought to be induced by either the epithelial-mesenchymal transition of CRC cells or ARSB-mediated hypoxia in colonic epithelial cells, underlining the importance of this enzyme in human CRC[7,8,16-19].
Based on the obtained data and data from literature, it is tempting to emit a hypothesis regarding the possible role of arylsulfatases in carcinogenesis and evolution of CRC, and their possible interaction with maspin. As we have mentioned, till now, it was proved that loss of ARSA/ARSB induce metabolic disorders[3-6]. It was also proved and confirmed in our material that ARSA/ARSB protein level decreases in carcinomas[4]. Based on these facts and our result, we can suppose that, in CRC, the arylsulfatases are firstly lost, possible as result of metabolism imbalance, which is specific for malignant tumors[20]. On the other hand, in parallel with decreased serum expression of ARSB gene, decreased protein levels are seen in tumor cells, in early stages.
In parallel with lymph vessels invasion, when the tumor cells metabolism is accelerated (metabolic reprogramming), compared with normal metabolism, as it was previously described in literature[20], arylsulfatases might be upregulated and induce tumor cells aggressiveness. It was previously proved that metabolic adaptation of tumor cells is realized though aerobic glycolysis, also known as Warburg effect[20], which is mediated by arylsulfatases and is dysregulated in metabolic disorders[4]. As regarding maspin, it was previously proved that its expression is correlated with hypoxic-induced angiogenesis, via VEGF-A (Vascular Endothelial Growth Factor), in several malignant tumors such gastric carcinoma[21], squamous cell carcinoma[22], liposarcoma[23] and CRC[11]. As this showed that, a maspin-arylsulfatases interaction might exist, we can suppose that this interaction evokes the genetically-induced dysregulation of tumor cell metabolism.
Further studies are needed to elucidate the molecular mechanism associated with arylsulfatases in colorectal carcinogenesis and the behavior of this tumor.
Arylsulfatase A and B (ARSA, ARSB) are lysosomal enzymes playing an important role in cellular metabolism. It is described in the literature the genetic condition characterized by a total deficiency of these two enzymes but few aspects are known about their role in carcinogenesis and evolution of colorectal cancer (CRC).
As no data about the correlation of ARSA, ARSB and maspin were published yet, this study is original and represents first step in understanding the arylsulfatases influence upon malignant cells.
This paper aimed to compare the immunohistochemical (IHC) stain and gene expression profile of ARSA and ARSB in CRC. Correlation with maspin and classical clinicopathological parameters was also done.
For IHC study, the expression of ARSA, ARSB and maspin were quantified in the cytoplasm of CRC cells. For gene expression study circulating mRNA, using Roche HighPure RNA kit, was isolated from all patients before surgery. A group of 45 healthy patients without inflammatory or tumor pathologies was used as a control group. Reverse transcription and Taqman Gene Expression Array were used for Arylsulfatase B gene expression. Statistical analysis was performed using GraphPad Prims8 software using the χ2 and Kruskal-Wallis test, while the Mann-Whitney test was used for ARSB gene expression, considering a P-value < 0.05 (with a 95% confidence interval) statistically significant.
The preoperative gene expression level of ARSB was significantly decreased in patients with CRC (RQ < 1), compared with the control group (RQ > 1). A more significant decrease (RQ < 0.5) occurred in ulcero-infiltrative maspin-positive adenocarcinomas, with a higher degree of tumor budding, diagnosed in locally advanced stages (T3/4). The ARSB protein level in surgical specimens was inversely correlated with the preoperative ARSB gene circulating level.
High IHC expression of ARSA and ARSB, correlated with maspin positivity can be used as indicators of prognosis of CRC. Triple positivity of ARSA/ARSB/maspin correlated with an ARSB gene circulating level of <0.5, is an indicator of a lower survival rate, independently of the other clinicopathological parameters.
Based on the high IHC and low gene expression of ARSB, further investigations should be done, to elucidate the precise mechanism of this contradictory protein-gene expression profile.
Manuscript source: Invited manuscript
Specialty type: Medicine, research and experimental
Country of origin: Romania
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Fukushi K, Kanat O, Vynios D S-Editor: Ma YJ L-Editor: A E-Editor: Qi LL
| 1. | Rosso T, Malvezzi M, Bosetti C, Bertuccio P, Negri E, La Vecchia C. Cancer mortality in Europe, 1970-2009: an age, period, and cohort analysis. Eur J Cancer Prev. 2018;27:88-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 2. | Obuch JC, Ahnen DJ. Colorectal Cancer: Genetics is Changing Everything. Gastroenterol Clin North Am. 2016;45:459-476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 3. | Echeverri Olga Y, Salazar Diego A, Rodriguez-Lopez A, Janneth G, Almeciga-Diaz Carlos J, Barrera Luis A. Understanding the Metabolic Consequences of Human Arylsulfatase A Deficiency through a Computational Systems Biology Study. Cent Nerv Syst Agents Med Chem. 2017;17:72-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 4. | Kovacs Z, Jung I, Gurzu S. Arylsulfatases A and B: From normal tissues to malignant tumors. Pathol Res Pract. 2019;215:152516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 5. | Ittiwut C, Boonbuamas S, Srichomthong C, Ittiwut R, Suphapeetiporn K, Shotelersuk V. Novel Mutations, Including a Large Deletion in the ARSB Gene, Causing Mucopolysaccharidosis Type VI. Genet Test Mol Biomarkers. 2017;21:58-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 6. | Prabhu SV, Bhattacharyya S, Guzman-Hartman G, Macias V, Kajdacsy-Balla A, Tobacman JK. Extra-lysosomal localization of arylsulfatase B in human colonic epithelium. J Histochem Cytochem. 2011;59:328-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 7. | Bhattacharyya S, Tobacman JK. Arylsulfatase B regulates colonic epithelial cell migration by effects on MMP9 expression and RhoA activation. Clin Exp Metastasis. 2009;26:535-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 8. | Bhattacharyya S, Feferman L, Tobacman JK. Increased expression of colonic Wnt9A through Sp1-mediated transcriptional effects involving arylsulfatase B, chondroitin 4-sulfate, and galectin-3. J Biol Chem. 2014;289:17564-17575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 9. | Gurzu S, Szentirmay Z, Jung I. Molecular classification of colorectal cancer: a dream that can become a reality. Rom J Morphol Embryol. 2013;54:241-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 10. | Gurzu S, Szentirmay Z, Toth E, Jung I. Possible predictive value of maspin expression in colorectal cancer. Recent Pat Anticancer Drug Discov. 2013;8:183-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 11. | Gurzu S, Szentirmay Z, Popa D, Jung I. Practical value of the new system for Maspin assessment, in colorectal cancer. Neoplasma. 2013;60:373-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 12. | Banias L, Gurzu S, Kovacs Z, Bara T, Bara T, Jung I. Nuclear maspin expression: A biomarker for budding assessment in colorectal cancer specimens. Pathol Res Pract. 2017;213:1227-1230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 13. | Ueno H, Kajiwara Y, Shimazaki H, Shinto E, Hashiguchi Y, Nakanishi K, Maekawa K, Katsurada Y, Nakamura T, Mochizuki H, Yamamoto J, Hase K. New criteria for histologic grading of colorectal cancer. Am J Surg Pathol. 2012;36:193-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 161] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 14. | Lugli A, Kirsch R, Ajioka Y, Bosman F, Cathomas G, Dawson H, El Zimaity H, Fléjou JF, Hansen TP, Hartmann A, Kakar S, Langner C, Nagtegaal I, Puppa G, Riddell R, Ristimäki A, Sheahan K, Smyrk T, Sugihara K, Terris B, Ueno H, Vieth M, Zlobec I, Quirke P. Recommendations for reporting tumor budding in colorectal cancer based on the International Tumor Budding Consensus Conference (ITBCC) 2016. Mod Pathol. 2017;30:1299-1311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 575] [Cited by in RCA: 730] [Article Influence: 91.3] [Reference Citation Analysis (0)] |
| 15. | Marusteri M, Bacarea V. Comparing groups for statistical differences: how to choose the right statistical test? Biochem Med. 2010;20:15-32. [RCA] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 83] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 16. | Vicente CM, Lima MA, Yates EA, Nader HB, Toma L. Enhanced tumorigenic potential of colorectal cancer cells by extracellular sulfatases. Mol Cancer Res. 2015;13:510-523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 17. | Bhattacharyya S, Feferman L, Han X, Ouyang Y, Zhang F, Linhardt RJ, Tobacman JK. Decline in arylsulfatase B expression increases EGFR expression by inhibiting the protein-tyrosine phosphatase SHP2 and activating JNK in prostate cells. J Biol Chem. 2018;293:11076-11087. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 18. | Bhattacharyya S, Tobacman JK. Hypoxia reduces arylsulfatase B activity and silencing arylsulfatase B replicates and mediates the effects of hypoxia. PLoS One. 2012;7:e33250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 19. | Bhattacharyya S, Feferman L, Tobacman JK. Arylsulfatase B regulates versican expression by galectin-3 and AP-1 mediated transcriptional effects. Oncogene. 2014;33:5467-5476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 20. | Romero-Garcia S, Lopez-Gonzalez JS, Báez-Viveros JL, Aguilar-Cazares D, Prado-Garcia H. Tumor cell metabolism: an integral view. Cancer Biol Ther. 2011;12:939-948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 169] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 21. | Gurzu S, Kadar Z, Sugimura H, Orlowska J, Bara T, Bara T, Szederjesi J, Jung I. Maspin-related Orchestration of Aggressiveness of Gastric Cancer. Appl Immunohistochem Mol Morphol. 2016;24:326-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 22. | Ciortea CD, Jung I, Gurzu S, Kövecsi A, Turdean SG, Bara T. Correlation of angiogenesis with other immunohistochemical markers in cutaneous basal and squamous cell carcinomas. Rom J Morphol Embryol. 2015;56:665-670. [PubMed] |
| 23. | Jung I, Gurzu S, Turdean S, Ciortea D, Sahlean DI, Golea M, Bara T. Relationship of endothelial area with VEGF-A, COX-2, maspin, c-KIT, and DOG-1 immunoreactivity in liposarcomas versus non-lipomatous soft tissue tumors. Int J Clin Exp Pathol. 2015;8:1776-1782. [PubMed] |