Published online Nov 26, 2022. doi: 10.12998/wjcc.v10.i33.12089
Peer-review started: June 18, 2022
First decision: August 6, 2022
Revised: August 18, 2022
Accepted: October 9, 2022
Article in press: October 9, 2022
Published online: November 26, 2022
Processing time: 157 Days and 18.5 Hours
Esophageal cancer is one of the most common malignant tumors of the digestive system, with a 5-year survival rate of 15% to 50%. Cuproptosis, a unique kind of cell death driven by protein lipoylation, is strongly connected to mitochondrial metabolism. The clinical implications of cuproptosis-related genes in esophageal cancer, however, are mainly unknown.
To identify cuprotosis-related genes that are differentially expressed in esophageal cancer and investigate their prognostic significance.
With |log fold change| > 1 and false discovery rate < 0.05 as criteria, the Wilcox test was used to evaluate the differentially expressed genes between 151 tumor tissues and 151 normal esophageal tissues. Cuproptosis-related genes were selected to be linked with prognosis using univariate Cox regression analysis. Genes were separated into high- and low- expression groups based on their cutoff value of gene expression, and the correlation between the two groups and overall survival or progression-free survival was investigated using the log-rank test. The C-index, calibration curve, and receiver operator characteristic (ROC) curve were used to assess a nomogram containing clinicopathological characteristics and cuproptosis-related genes.
Pyruvate dehydrogenase A1 (PDHA1) was found to be highly correlated with prognosis in univariate Cox regression analysis (hazard ratio = 22.96, 95% confidence interval = 3.09-170.73; P = 0.002). According to Kaplan-Meier survival curves, low expression of PDHA1 was associated with a better prognosis (log-rank P = 0.0007). There was no significant correlation between PDHA1 expression and 22 different types of immune cells. Tumor necrosis factor superfamily member 15 (TNFSF15) (P = 3.2 × 10-6; r = 0.37), TNFRSF14 (P = 8.1 × 10-8; r = 0.42), H long terminal repeat-associating 2 (P = 6.0 × 10-8; r = 0.42) and galectin 9 (P = 3.1 × 10-6; r = 0.37) were all found to be considerably greater in the high PDHA1 expression group, according to an analysis of genes related to 47 immunological checkpoints. The low PDHA1 expression group had significantly lower levels of cluster of differentiation 44 (CD44) (P = 0.00028; R = -0.29), TNFRSF18 (P = 1.2 × 10-5; R = -0.35), programmed cell death 1 ligand 2 (P = 0.0032; R = -0.24), CD86 (P = 0.018; R = -0.19), and CD40 (P = 0.0047; R = -0.23), and the differences were statistically significant. We constructed a prognostic nomogram incorporating pathological type, tumor-node-metastasis stage, and PDHA1 expression, and the C-index, calibration curve, and ROC curve revealed that the nomogram’s predictive performance was good.
Cuproptosis-related genes can be used as a prognostic predictor for esophageal cancer patients, providing novel insights into cancer treatment.
Core Tip: Esophageal carcinoma has a poor prognosis and is one of the major causes of cancer-related deaths worldwide. Despite recent advancements in the surgical and pharmacological treatment of esophageal cancer, the prognosis remains poor. Copper toxicity has been linked to the incidence and progression of esophageal cancer in numerous studies. At the gene level, however, the probable biochemical mechanism is unknown. We included 19 cuproptosis-related genes and screened a gene that could successfully predict the prognosis of esophageal cancer by statistical analysis to further elucidate the role of cuproptosis-related genes in impacting the prognosis of esophageal cancer.
- Citation: Xu H, Du QC, Wang XY, Zhou L, Wang J, Ma YY, Liu MY, Yu H. Comprehensive analysis of the relationship between cuproptosis-related genes and esophageal cancer prognosis. World J Clin Cases 2022; 10(33): 12089-12103
- URL: https://www.wjgnet.com/2307-8960/full/v10/i33/12089.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i33.12089
Esophageal cancer is a major global health issue, and its incidence is rapidly increasing[1]. Esophageal cancer is classified into two types: esophageal squamous cell carcinoma (ESCC), which accounts for 90% of all occurrences, and esophageal adenocarcinoma (EAC). In recent years, epidemiological research has revealed that the incidence of EAC has grown 3-4 times, with the proportion increasing[2,3]. Despite significant advances in the diagnosis and treatment of esophageal cancer, the mortality rate varies from 15% to 20%, placing it fourth among all cancer-related deaths[1].
Esophageal cancer is the result of a complex process involving various causes and polygene alterations. Using high-throughput sequencing technologies, a comprehensive mutation catalog was evaluated, and substantial genetic alterations have been discovered in the malignancies. Gene alterations are often linked to aberrant expression and are becoming more essential for the early diagnosis and prognosis of esophageal cancer[4]. Currently, several gene expression products are employed as indicators for esophageal cancer diagnosis and prognosis[5,6]. Somatic mutations in tumor protein p53 (TP53) have been found in more than 83% of ESCCs. Adenocarcinoma and squamous cell carcinoma both have TP53 point mutations[7,8]. In addition, numerous cell cycle-controlling genes are overexpressed in ESCC. For example, cyclin-dependent kinase 4/cyclin-dependent kinase 6 accounted for 23.6%, murine double minute 25.7%, and cyclin D1 46.4%, showing that the above components are implicated in the incidence and development of ESCC[9]. As a result, there is a pressing need to uncover genetic anomalies in esophageal cancer and understand their molecular basis to enhance early diagnosis and minimize esophageal cancer mortality.
Previous studies have reported several types of precisely programmed cell death, including apoptosis, pyroptosis, necrosis, and ferroptosis[10]. Similar to iron, copper is a trace metal in cells that plays an integral role in maintaining protein functions. Excessive copper can cause cytotoxicity, but the exact mechanism is unclear[11]. Tsvetkov et al[12] discovered that the copper carrier elesclomol, which was originally used to treat cancer, kills cells in excess of copper. Elesclomol does not trigger cell death on its own, suggesting that copper toxicity is to blame. Dar et al[13] found that patients with esophageal cancer had considerably higher mean blood copper levels than controls, with a mean copper concentration of 169 g/dL in the cancer group and 149 g/dL in the control group. Therefore, we hypothesized that copper shortage or excess is linked to the occurrence and progression of esophageal cancer. Copper, however, has been linked to the development and progression of esophageal cancer in few studies.
There has not been any research on the role of cuproptosis-related genes in the genesis, progression, and metastasis of esophageal cancer to date. Cuproptosis-related genes and their processes need to be better understood in order to improve the prognosis of malignant tumors and uncover novel treatment targets. Bioinformatics analysis was used to evaluate the expression profile of cuproptosis-related genes and its predictive significance in esophageal cancer in this study.
The study did not include any human participants, data, or tissue, nor did it include any animals. All of the information was gathered from a public database.
The Cancer Genome Atlas (https://portal.gdc.cancer.gov/) provided gene expression data and clinical information for 151 esophageal cancer samples, whereas the Genotype-Tissue Expression database (https://xenabrowser.net/) provided gene expression levels for 151 healthy tissue samples. The "limma" package in R was used to conduct matrix normalization. We selected 19 genes from the scientific literature that have been linked to cuproptosis in prior studies[12].
In 151 patients with esophageal cancer and 151 healthy controls, a total of 55185 genes were acquired. The log2 (x+1) scale was used to standardize all of the expression data. Differentially expressed genes (DEGs) were screened between tumor tissue and normal esophageal tissue using the "limma" package and Wilcox-test, with the conditions of |log fold change [FC]| > 1 and false discovery rate (FDR) < 0.05. Using the "limma" package, we evaluated those genes that were differently expressed between esophageal cancer and healthy control tissues based on 19 cuproptosis-related genes. The FDR less than 0.05 was used as a criterion for further investigation. The researcher employed a univariate Cox regression analysis to find predictive markers linked to overall survival (OS) and progression-free survival (PFS). The optimum cutoff value for a prognostic gene was identified using a receiver operating characteristic (ROC) curve. Patients were classified into high-risk (less than the cutoff) and low-risk (greater than the cutoff) esophageal cancer groups based on gene expression cutoff values.
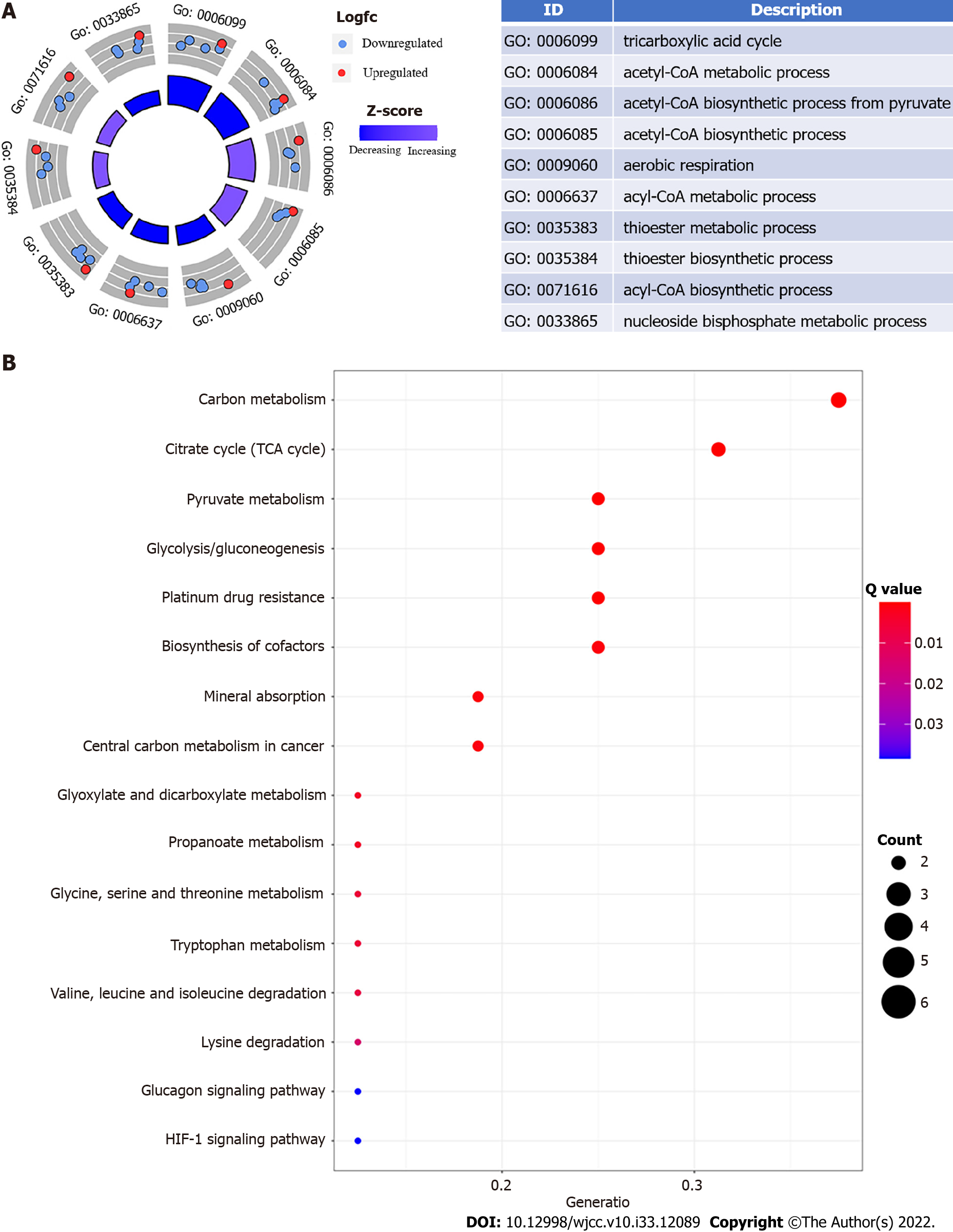
The R package "ClusterProfiler" was used to analyze gene ontology (GO) and pathway enrichment [Kyoto Encyclopedia of Genes and Genomes (KEGG)]. It was deemed substantially enriched when the P value and adjusted P value were both less than 0.05. To identify the functional role of the genes, GO analysis was done on the significantly expressed genes, and the expression levels of the genes were displayed in GO circle plots using the R package "GOplot". To determine tumor-infiltrating immune-cell fractions in esophageal cancer patients, we employed the CIBERSORT algorithm[14]. On samples with a CIBERSORT result of P value less than 0.05, further analysis was performed. Then for reference, we collected 547 gene expression profiles from the CIBERSORT website (http://cibersort.stanford.edu/). The Pearson’s test and the "corrplot" program were used to correlate infiltrated immune cells.
A cuproptosis-related gene and numerous clinicopathological variables were also subjected to univariate and multivariate Cox regression analyses. For predicting the OS, we utilized the R "rms" package to generate a nomogram including pathological categories, American Joint Committee on Cancer (AJCC)-tumor-node-metastasis (TNM) stages, and differential cuproptosis-related genes. The uniformity of the nomogram was assessed using calibration curves to anticipate various OS results. Harrell’s concordance index was used to generate the C-index. The area under the receiver operating characteristic (AUC) curve and ROC were both utilized to evaluate how predictive our nomogram was.
All statistical analyses were performed using R software, version 4.1.0 (http://www.rproject.org/). The Students t-test was used to compare variables such as age at diagnosis, sex, AJCC-TNM stage, mutational status of v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS), v-Raf murine sarcoma viral oncogene homolog B1 genes (BRAF), epidermal growth factor receptor (EGFR), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA, unpaired t-test). The Cox proportional hazards model was used to find genes and clinicopathological variables linked to survival. Based on the optimal cutoff of differential cuproptosis-related genes, the Youden index method determined that esophageal cancer patients should be divided into high- and low-risk groups. For assessing survival in the high- and low-risk groups, Kaplan-Meier survival curves were employed, and log-rank tests were performed to compare survival rates. For two-sided P values, statistical significance was considered as a value less than 0.05.
Our study comprised 151 esophageal cancer patients and 151 healthy controls. Table 1 shows the characteristics of patients with EAC (n = 74) and ESCC (n = 77). All esophageal cancer patients had a median survival duration of 13.4 mo. EAC had a median survival time of 14.8 mo, whereas ESCC had a median survival time of 13 mo.
| Variables | Total, n = 151 | Esophagus cancer, n = 151 | |
| Adenocarcinoma, n = 74 | Squamous cell carcinoma, n = 77 | ||
| Age, yr | |||
| Median | 60 | 68.5 | 57 |
| Interquartile range | (53-72) | (57-77) | (51-63.5) |
| Sex | |||
| Female | 22 (14.6%) | 11 (14.9%) | 11 (14.3%) |
| Male | 129 (85.4%) | 63 (85.1%) | 66 (85.7%) |
| AJCC-TNM Stage | |||
| Ⅰ | 18 (11.9%) | 11 (14.9%) | 7 (9.1%) |
| Ⅱ | 70 (46.4%) | 24 (32.4%) | 46 (59.7%) |
| Ⅲ | 51 (33.8%) | 31 (41.9%) | 20 (26.0%) |
| Ⅳ | 12 (7.9%) | 8 (10.8%) | 4 (5.2%) |
| EGFR status | |||
| Mutant | 5 (3.3%) | 3 (4.1%) | 2 (2.6%) |
| Wild-type | 146 (96.7%) | 71 (95.9%) | 75 (97.4%) |
| KRAS status | |||
| Mutant | 2 (1.3%) | 2 (2.7%) | 0 (0) |
| Wild-type | 149 (98.7%) | 72 (97.3%) | 77 (100%) |
| BRAF status | |||
| Mutant | 1 (0.7%) | 0 (0) | 1 (1.3%) |
| Wild-type | 150 (99.3%) | 74 (100%) | 76 (98.7) |
| PIK3CA status | |||
| Mutant | 14 (9.3%) | 4 (5.4%) | 10 (13.0%) |
| Wild-type | 137 (90.7%) | 70 (94.6%) | 67 (87.0%) |
| OS event | |||
| Event | 58 (38.4%) | 36 (48.6%) | 22 (28.6%) |
| Non-event | 93 (61.6%) | 38 (51.4%) | 55 (71.4%) |
| OS months | |||
| Median | 13.4 | 14.8 | 13.0 |
| Range | (7.8-22.9) | (7.3-27.5) | (11.0-18.6) |
| PFS event | |||
| Event | 73 (48.3%) | 38 (51.4%) | 35 (45.5%) |
| Non-event | 78 (51.7%) | 36 (48.6%) | 42 (54.5%) |
| PFS months | |||
| Median | 10.7 | 10.3 | 10.7 |
| Range | (5.1-18.9) | (5.4-24.5) | (3.7-15.8) |
Table 2 summarizes the relationships between clinical pathological features and these individuals’ OS or PFS. When compared to ESCC, EAC was related to a lower OS (P = 0.011). AJCC-TNM stages III and IV were linked to OS (P = 0.021) and PFS (P = 0.013). When compared to females, males had a lower PFS (P = 0.009). Age, EGFR, BRAF, KRAS, or PIK3CA status, among other clinicopathological variables, were not linked to OS or PFS.
| Parameters | Overall survival | Progression-free survival | ||||
| Non-event | Event | P value | Non-event | Event | P value | |
| Age, yr | 0.758 | 0.628 | ||||
| ≤ 65 | 57 | 37 | 50 | 44 | ||
| > 65 | 36 | 21 | 28 | 29 | ||
| Sex | 0.102 | 0.009 | ||||
| Female | 17 | 5 | 17 | 5 | ||
| Male | 76 | 53 | 61 | 68 | ||
| AJCC-TNM stage | 0.021 | 0.013 | ||||
| I and II | 61 | 27 | 53 | 35 | ||
| III and IV | 32 | 31 | 25 | 38 | ||
| Pathological type | 0.011 | 0.469 | ||||
| Adenocarcinoma | 38 | 36 | 36 | 38 | ||
| SCC | 55 | 22 | 42 | 35 | ||
| EGFR status | 0.596 | |||||
| Wild-type | 3 | 2 | 0.941 | 2 | 3 | |
| Mutant | 90 | 56 | 76 | 70 | ||
| BRAF status | 0.428 | 0.332 | ||||
| Wild-type | 1 | 0 | 1 | 0 | ||
| Mutant | 92 | 58 | 77 | 73 | ||
| KRAS status | 0.734 | 0.962 | ||||
| Wild-type | 1 | 1 | 1 | 1 | ||
| Mutant | 92 | 57 | 77 | 72 | ||
| PIK3CA status | 0.828 | 0.896 | ||||
| Wild-type | 9 | 5 | 7 | 7 | ||
| Mutant | 84 | 53 | 71 | 66 | ||
With |log FC| > 1 and FDR < 0.05, a total of 7055 DEGs were identified in the healthy control group, comprising 3494 upregulated genes and 3561 downregulated genes. Then, using the FDR < 0.05 threshold, gene expression analysis was done to identify cuproptosis-related genes and 18 genes fulfilled our requirements (Supplementary Table 1). In the intersection of two gene sets, there were five genes: high-affinity copper uptake protein 1 (SLC31A1), ferredoxin, lipoyl(octanoyl) transferase 2, pyruvate dehydrogenase A1 (PDHA1), and programmed cell death 6 interacting protein (DEGs and cuproptosis-related genes). Furthermore, using univariate Cox regression analysis, we discovered that one gene (PDHA1) was strongly linked to prognosis [hazard ratio (HR) = 22.96, 95% confidence interval (CI) = 3.09-170.73, P = 0.002] (Table 3).
| Gene name | HR | HR.95L | HR.95H | P value |
| PDHA1 | 22.96 | 3.09 | 170.73 | 0.002 |
| ATP7A | 3.83 | 0.96 | 15.21 | 0.057 |
| CDKN2A | 0.84 | 0.64 | 1.10 | 0.204 |
| PDHB | 1.76 | 0.37 | 8.50 | 0.479 |
| GLS | 1.50 | 0.48 | 4.67 | 0.484 |
| ATP7B | 1.15 | 0.77 | 1.71 | 0.496 |
| FDX1 | 1.52 | 0.41 | 5.59 | 0.527 |
| DLD | 1.82 | 0.27 | 12.11 | 0.534 |
| LIPT2 | 0.87 | 0.46 | 1.64 | 0.662 |
| DLST | 0.58 | 0.05 | 6.70 | 0.662 |
| NFE2L2 | 0.73 | 0.18 | 3.04 | 0.666 |
| DLAT | 1.43 | 0.23 | 8.89 | 0.702 |
| DBT | 0.83 | 0.27 | 2.54 | 0.744 |
| GCSH | 0.87 | 0.32 | 2.36 | 0.777 |
| MTF1 | 0.85 | 0.26 | 2.80 | 0.785 |
| LIAS | 0.86 | 0.25 | 2.93 | 0.806 |
| NLRP3 | 0.91 | 0.44 | 1.90 | 0.809 |
| LIPT1 | 1.14 | 0.29 | 4.41 | 0.853 |
| SLC31A1 | 1.08 | 0.19 | 6.15 | 0.928 |
Using GO enrichment and KEGG pathway analyses, we explored the biological processes, cellular components, and molecular activities of DEGs. Energy metabolism and glycolysis/gluconeogenesis signaling pathways were highly enriched among DEGs with cuproptosis relevance. The phrases "tricarboxylic acid cycle," "acetylCoA metabolic process," and "acetylCoA biosynthetic process from pyruvate" are considerably enriched in Figure 1A. "Carbon metabolism," "tricarboxylic acid cycle," "pyruvate metabolism," "glycolysis/gluconeogenesis," "platinum drug resistance," "biosynthesis of cofactors," "mineral absorption," and "central carbon metabolism in cancer" were the most substantially enriched pathways (Figure 1B).
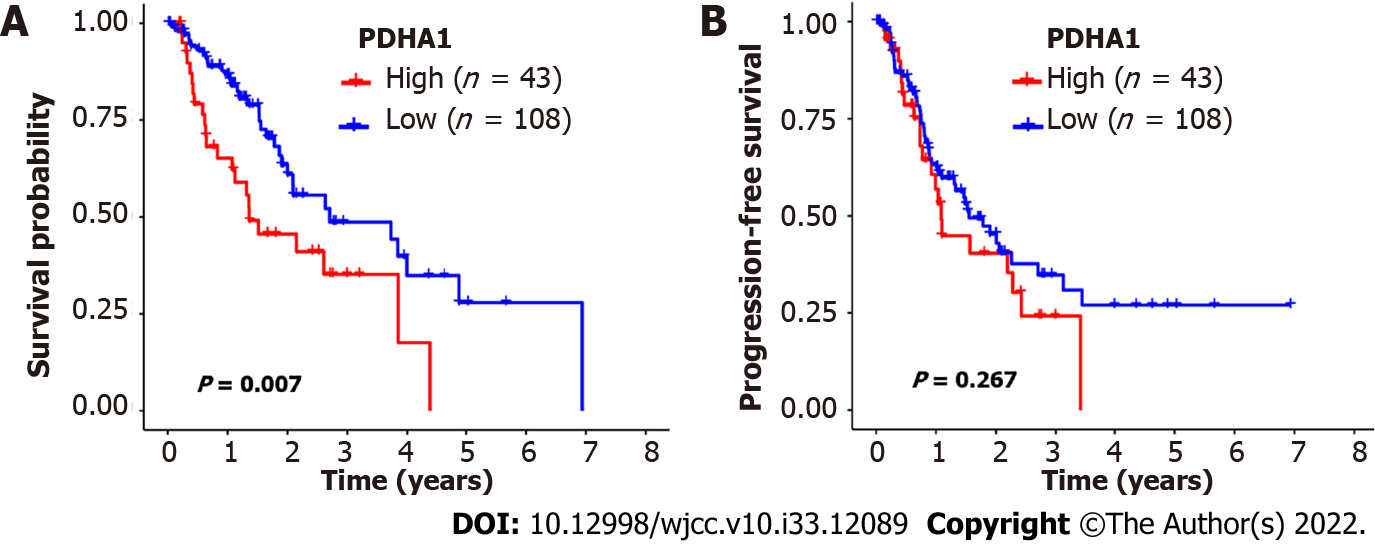
The 151 esophageal cancer patients were divided into two groups based on the cutoff value: Those with high PDHA1 expression and those with low PDHA1 expression. In the analysis of OS, high PDHA1 expression was linked to considerably lower OS rates than low expression (P = 0.007; Figure 2A). However, there was no link between PDHA1 expression and PFS (Figure 2B). Within a multivariate context, we investigated the link between PDHA1 expression and survival result (Table 4). When we controlled for clinical prognostic indicators that were significant (P < 0.05) in univariate Cox regression models, we discovered that overexpression of the PDHA1 gene might predict poor clinical outcomes. The expression of PDHA1 (HR: 1.67, 95 CI: 1.03-2.73; P = 0.0386) and AJCC-TNM stage (HR: 2.30, 95%CI: 1.58-3.35; P < 0.001) were independent risk factors for OS, but not for PFS, according to multivariate Cox regression analysis.
| Variables | Overall survival | Progression-free survival | ||
| HR (95%CI of HR) | P value | HR (95%CI of HR) | P value | |
| PDHA1 low | 1.00 (Reference) | 1.00 (Reference) | ||
| UVA PDHA1 high | 2.34 (1.42-3.87) | 0.0009 | 1.51 (0.95-2.38) | 0.08 |
| MVA PDHA1 high | 1.67 (1.03-2.73) | 0.0386 | ||
| Age ≤ 65 | 1.00 (Reference) | 1.00 (Reference) | ||
| Age > 65 | 0.81 (0.47-1.41) | 0.46 | 0.97 (0.60-1.55) | 0.89 |
| Sex, Female | 1.00 (Reference) | 1.00 (Reference) | ||
| UVA Male | 2.12 (0.84-5.34) | 0.11 | 2.97 (1.19-7.39) | 0.02 |
| MVA Male | 2.27 (0.90- 5.73) | 0.08 | ||
| Pathological types, adenocarcinoma | 1.00 (Reference) | 1.00 (Reference) | ||
| SCC | 0.81 (0.47-1.39) | 0.45 | 1.11 (0.70-1.77) | 0.66 |
| AJCC stage I, II | 1.00 (Reference) | 1.00 (Reference) | ||
| UVA AJCC stage III, IV | 2.53 (1.76-3.64) | 5.86 × 10-7 | 1.98 (1.46-2.67) | 9.48 × 10-6 |
| MVA AJCC stage III, IV | 2.30 (1.58-3.35) | 1.46 × 10-5 | 1.86 (1.37-2.52) | 6.98 × 10-5 |
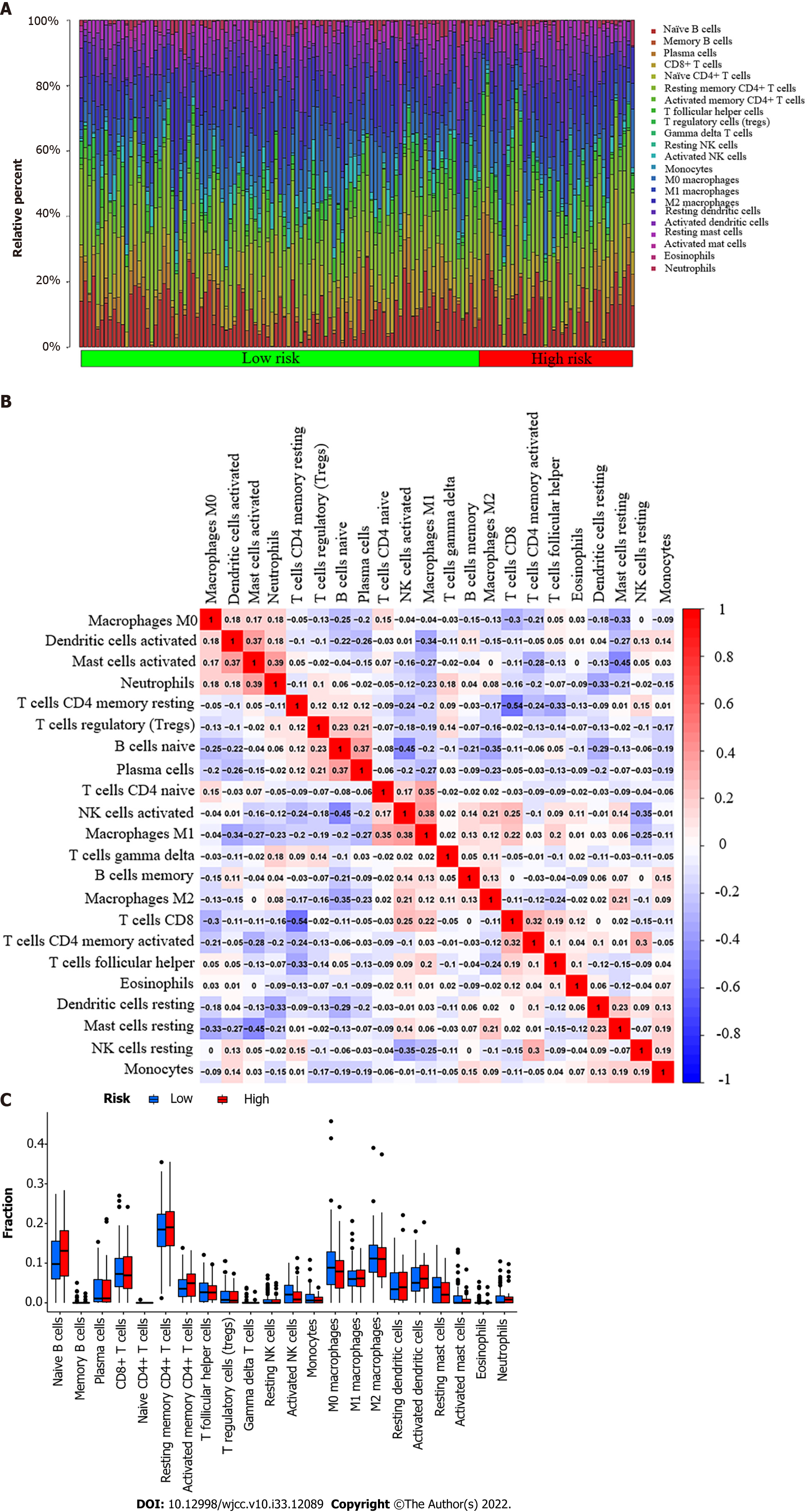
Our study used the CIBERSORT algorithm to examine the ratio of tumor-infiltrating immune cells (TICs) in esophageal cancer to further verify the correlation between PDHA1 and immune cells (Figure 3A). P < 0.05 was used to classify samples as statistically different. The ratio of TICs in the low-risk group was represented by the first 108 of 151 esophageal cancer patients, whereas the ratio of TICs in the high-risk group was represented by the last 43 samples. The correlation between 22 immune cells was represented using a heatmap (Figure 3B). Neutrophils and activated mast cells, macrophages M1 and activated natural killer cells, plasma cells and naive B cells, activated mast cells and activated dendritic cells, and macrophages M1 and naive CD4+ T cells were the top five results with a positive correlation. CD8+ T cells and resting memory CD4+ T cells, on the other hand, were the immune cells that were most negatively associated. Furthermore, the bar graph revealed that the immune infiltration of 22 immune cell types in high and low PDHA1 esophageal cancer patients did not statistically significantly differ (Figure 3C).
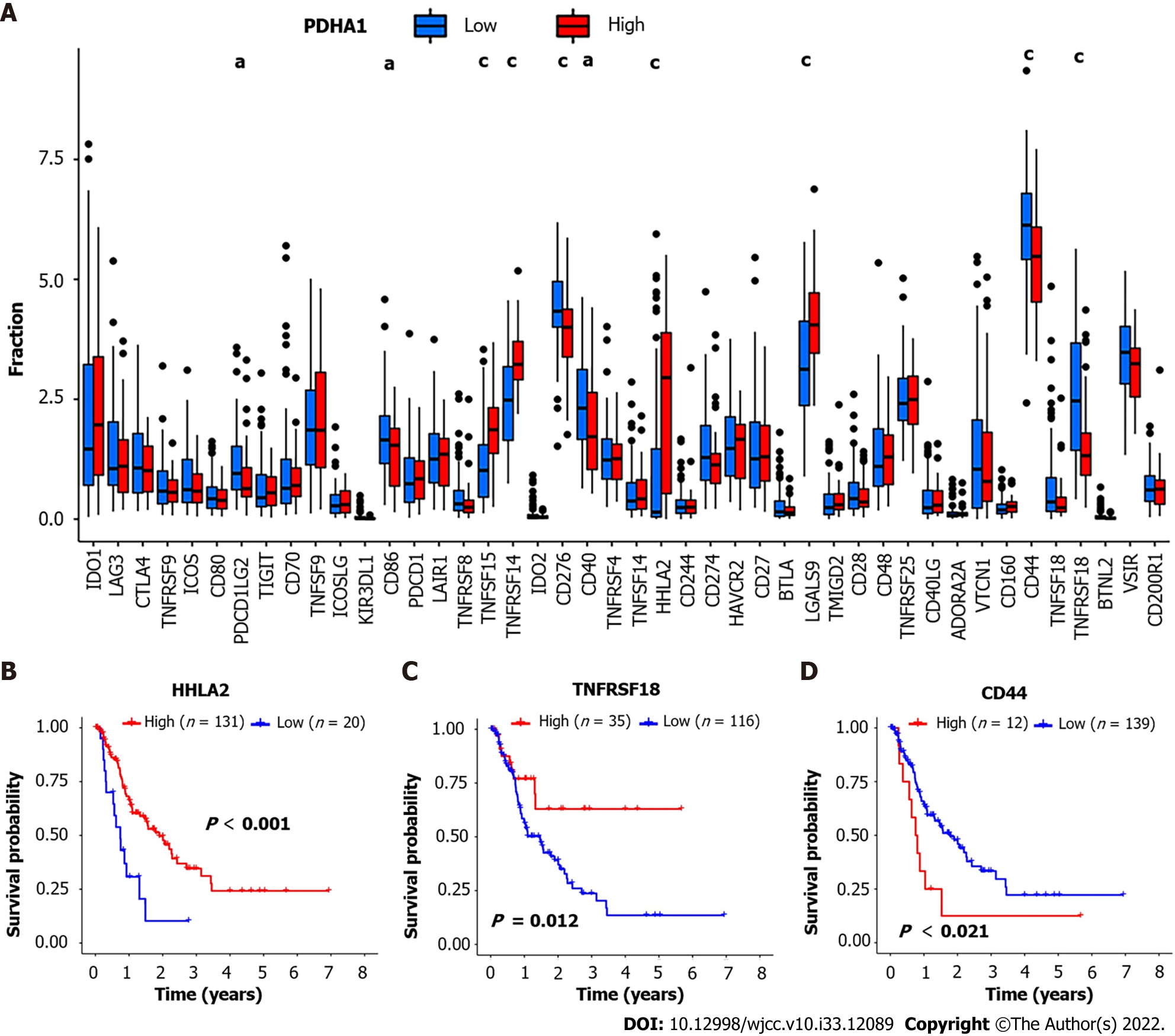
We also investigated the relationships between PDHA1 and 47 genes associated with immunological checkpoints, which have been reported in the literature[15]. The results showed that tumor necrosis factor superfamily member 15 (TNFSF15) (P = 3.2 × 10-6; r = 0.37), TNFRSF14 (P = 8.1 × 10-8; r = 0.42), H long terminal repeat-associating 2 (HHLA2) (P = 6.0 × 10-8; r = 0.42) and galectin 9 (LGALS9) (P = 3.1 × 10-6; r = 0.37) were significantly higher than that of the low expression group of PDHA1. Cluster of differentiation 44 (CD44) (P = 0.00028; R = -0.29), TNFRSF18 (P = 1.2 × 10-5; R = -0.35), programmed cell death 1 ligand 2 (PDCD1LG2) (P = 0.0032; R = -0.24), CD86 (P = 0.018; R = -0.19), and CD40 (P = 0.0047; R = -0.23) were significantly lower than that of the low expression group of PDHA1 (Figure 4A). The expression profiles of 47 immune checkpoint genes involved in cuproptosis were investigated further. In addition, we categorized the above genes into high and low expression groups based on their cutoff values. Kaplan-Meier analysis and log-rank testing were used to assess OS. The findings revealed that low HHLA2, TNFRSF18, and CD44 overexpression was linked to a significantly shorter OS and a worse prognosis (Figure 4B-D).
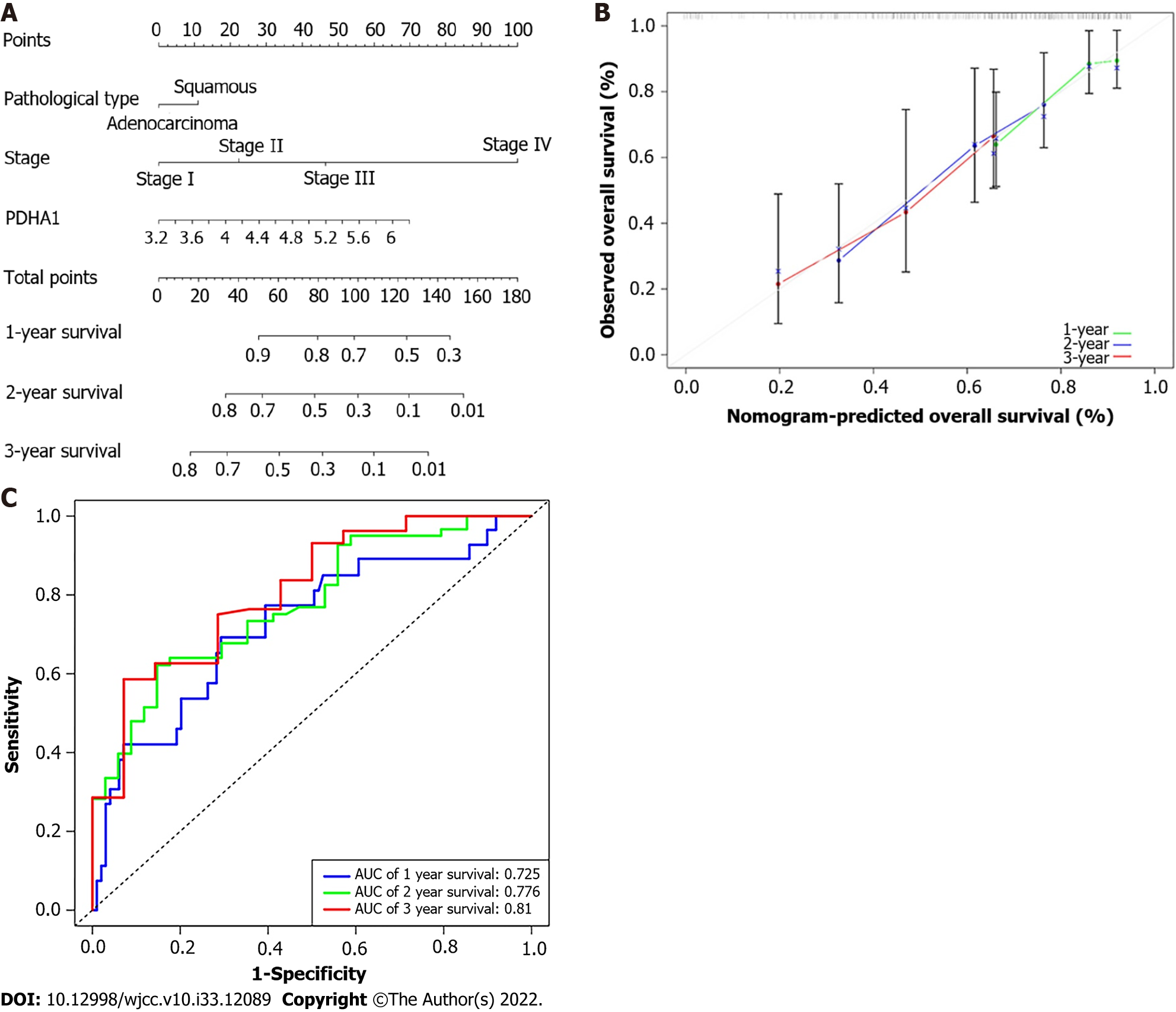
The 1-, 2-, and 3-year OS probability was calculated using a nomogram that included the pathological type, AJCC-TNM stage, and PDHA1 expression (Figure 5A). The actual observed vs anticipated rates of the 1-, 2-, and 3-year OS demonstrated near to the ideal 45° oblique line on the calibration curve (Figure 5B). Furthermore, the AUC of ROC curves for 1-, 2-, and 3-year survival were 0.725, 0.776, 0.619, and 0.810, respectively (Figure 5C). With a C-index of 0.703 for OS, the nomogram showed promising discrimination.
In the present study, we investigated the expression signature of 19 cuproptosis-related genes in esophageal cancer tissues and explored their relationships with OS and PFS. A prognostic nomogram involving gene expression and clinicopathological parameters was constructed for the first time. Functional analysis showed that DEGs were enriched in energy metabolism, especially in pathways related to the tricarboxylic acid cycle. Cuproptosis-related genes were also confirmed to be associated with immune checkpoint genes.
In our study, 151 patients with esophageal cancer and 151 healthy controls were included. In univariate Cox regression analysis, PDHA1 was selected from 19 cuproptosis-related genes to be associated with the prognosis of esophageal cancer. At the same time, PDHA1 expression was also different in tumor tissues and healthy tissues. The results of the present study suggested that the PDHA1 expression was relatively low in cancer tissues, which was consistent with previous studies[16,17]. Li et al[17] also reported that high expression of PDHA1 in ovarian cancer cells was significantly correlated with better OS and PFS. Our results suggested that the high expression of PDHA1 expression was associated with poor OS, but not with PFS.
Li et al[18] showed that PDHA1 knockout inhibited glucose entering the tricarboxylic acid cycle, resulting in the reconnection of glutamine metabolism by increasing glutaminase 1 and glutamate dehydrogenase 1 expression, thus increasing the survival rate of glutamine-dependent cells. Consistent with our results of functional enrichment, these DEGs were significantly enriched in tricarboxylic acid cyclic-related pathways. We hypothesized that PDHA1 gene knockdown or low expression causes mitochondrial malfunction, resulting in aberrant generation of intracellular reactive oxygen species (ROS) and adenosine triphosphate, which is compatible with the cuproptosis mechanism.
TNM staging is linked to OS and PFS in esophageal cancer, whereas pathological types are associated with OS in esophageal cancer, according to our findings. Through univariate and multivariate Cox analyses, Zhang et al[19] also claimed that pathological stage is an independent risk factor for OS and PFS in patients with operable esophageal cancer. Therefore, pathological types and TNM stages were included in the prognostic nomogram. Men and women had different rates of esophageal cancer, with men having nearly twice as many cases as women[20]. We did not include sex in our nomogram because, while our analysis indicated that sex impacted PFS, the OS of esophageal cancer was not connected to sex.
CIBERSORT and ssGSEA techniques were used to analyze the composition of tumor-infiltrating immune cells (TICs) in each sample. Regulatory T cells and CD8+ T cells have been shown to play an important role in anti-tumor immunity in previous studies[21-23]. Studies have demonstrated that PDHA1 mediates metabolic reprogramming in macrophages[24,25]. However, there was no significant difference between the 22 types of immune cells in the high and low PDHA1 expression groups, according to our findings.
Immune checkpoint inhibitors have recently been studied in a variety of cancers, and they provide a novel therapy option[26]. However, no evidence of a link between cuproptosis and immune checkpoint genes has been found. TNFSF15, TNFRSF14, HHLA2, LGALS9, CD44, TNFRSF18, PDCD1LG2, CD86, and CD40 all have a strong relationship with PDHA1 expression in our recent research. The link between immune checkpoint-related genes expression and prognosis in certain esophageal cancer patients is still debated[27,28]. The immune checkpoint-related genes associated with PDHA1 expression were analyzed using the log-rank test, and the results revealed that HHLA2, TNFRSF18, and CD44 were substantially correlated with prognosis.
Our study had a number of limitations. First, the sample size must be increased to analyze EAC and ESCC individually. Second, if the sample size is high enough, the treatment procedures and stages of the esophagus must be unified. Finally, given that prognostic characteristics were generated and analyzed using data from public databases, further biological evidence, in addition to the statistical evidence we present, was required.
This study analyzed the association between cuproprosis-related genes and the prognosis of esophageal cancer in a systematic method. Cuproprosis-related genes, especially PDHA1, can be used as prognostic predictors in esophageal cancer patients, providing additional information on how to treat the disease.
Despite many breakthroughs in treatment, the general prognosis for esophageal cancer, one of the least responsive malignancies to cancer therapy, remains poor. As a result, identifying biomarkers and understanding the molecular mechanisms of esophageal cancer were critical for improving patient outcomes.
A nomogram for predicting the prognosis of esophageal cancer would be developed by evaluating cuproprosis-related genes features and their correlation with prognosis in order to predict the prognosis of esophageal cancer.
Considering cuproprosis-related genes expression was linked to patient prognosis, we intended to develop a nomogram to predict prognosis based on cuproprosis-related genes characteristics and evaluate its prediction performance.
Cuproprosis-related genes were found to be linked with esophageal cancer prognosis using univariate COX regression analysis on 151 esophageal cancer samples. The C-index, calibration curve, and receiver operator characteristic (ROC) curve were used to evaluate the prediction ability of a prognostic nomogram created by combining clinicopathological variables and cuproprosis-related genes.
Univariate COX regression analysis of 19 Cuproprosis-related genes revealed that the expression of pyruvate dehydrogenase A1 (PDHA1) was associated with the prognosis of esophageal cancer. The low PDHA1 expression group had a better prognosis of esophageal cancer, according to the log-rank test. There was no statistical correlation between PDHA1 expression and 22 immune cells; however there was a correlation between PDHA1 expression and several immune checkpoint genes. The C-index, calibration curve, and ROC curve were used to confirm the predictive ability of the esophageal cancer prognostic nomogram, which was developed by combining pathological type, tumor-node-metastasis stage, and PDHA1 expression.
Cuproprosis-related genes are correlated to esophageal cancer prognosis, and a deep understanding of its molecular mechanism might contribute to novel cancer treatments in the future.
To enhance the overall survival of esophageal cancer patients, researchers must investigate cuproprosis biomarkers and anticipate possible therapy targets.
We appreciate the analytical data provided by The Cancer Genome Atlas database (https://cancergenome.nih.gov/) and Genotype-Tissue Expression (https://xenabrowser.net/).
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Surgery
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Endo S, Japan; Mohamed SY, Egypt S-Editor: Chen YL L-Editor: Filipodia P-Editor: Yu HG
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64631] [Article Influence: 16157.8] [Reference Citation Analysis (176)] |
| 2. | Murphy CC, Yang YC, Shaheen NJ, Hofstetter WL, Sandler RS. An age-period-cohort analysis of obesity and incident esophageal adenocarcinoma among white males. Dis Esophagus. 2017;30:1-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 3. | Iwaya Y, Shimamura Y, Goda K, Rodríguez de Santiago E, Coneys JG, Mosko JD, Kandel G, Kortan P, May G, Marcon N, Teshima C. Clinical characteristics of young patients with early Barrett's neoplasia. World J Gastroenterol. 2019;25:3069-3078. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 4] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 4. | Xu Y, Wang N, Liu R, Lv H, Li Z, Zhang F, Gai C, Tian Z. Epigenetic Study of Esophageal Carcinoma Based on Methylation, Gene Integration and Weighted Correlation Network Analysis. Onco Targets Ther. 2021;14:3133-3149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 5. | Jørgensen JT, Mollerup J, Yang H, Go N, Nielsen KB. MET deletion is a frequent event in gastric/gastroesophageal junction/esophageal cancer: a cross-sectional analysis of gene status and signal distribution in 1,580 patients. Ann Transl Med. 2021;9:225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 6. | Tokunaga M, Okimoto K, Akizue N, Ishikawa K, Hirotsu Y, Amemiya K, Ota M, Matsusaka K, Nishimura M, Matsushita K, Ishikawa T, Nagashima A, Shiratori W, Kaneko T, Oura H, Kanayama K, Ohta Y, Taida T, Saito K, Matsumura T, Chiba T, Mochizuki H, Arai M, Kato J, Ikeda JI, Omata M, Kato N. Genetic profiles of Barrett's esophagus and esophageal adenocarcinoma in Japanese patients. Sci Rep. 2021;11:17671. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Reference Citation Analysis (0)] |
| 7. | Zhang H, Huang Z, Song Y, Yang Z, Shi Q, Wang K, Zhang Z, Liu Z, Cui X, Li F. The TP53-Related Signature Predicts Immune Cell Infiltration, Therapeutic Response, and Prognosis in Patients With Esophageal Carcinoma. Front Genet. 2021;12:607238. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 8. | Liu T, Yao Q, Jin H. Plasma Circulating Tumor DNA Sequencing Predicts Minimal Residual Disease in Resectable Esophageal Squamous Cell Carcinoma. Front Oncol. 2021;11:616209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 9. | Torres-Ayuso P, Brognard J. Combing the Cancer Genome for Novel Kinase Drivers and New Therapeutic Targets. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 10. | Bock FJ, Tait SWG. Mitochondria as multifaceted regulators of cell death. Nat Rev Mol Cell Biol. 2020;21:85-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1162] [Cited by in RCA: 1587] [Article Influence: 317.4] [Reference Citation Analysis (0)] |
| 11. | Tsvetkov P, Detappe A, Cai K, Keys HR, Brune Z, Ying W, Thiru P, Reidy M, Kugener G, Rossen J, Kocak M, Kory N, Tsherniak A, Santagata S, Whitesell L, Ghobrial IM, Markley JL, Lindquist S, Golub TR. Mitochondrial metabolism promotes adaptation to proteotoxic stress. Nat Chem Biol. 2019;15:681-689. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 385] [Article Influence: 64.2] [Reference Citation Analysis (0)] |
| 12. | Tsvetkov P, Coy S, Petrova B, Dreishpoon M, Verma A, Abdusamad M, Rossen J, Joesch-Cohen L, Humeidi R, Spangler RD, Eaton JK, Frenkel E, Kocak M, Corsello SM, Lutsenko S, Kanarek N, Santagata S, Golub TR. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375:1254-1261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 2489] [Article Influence: 829.7] [Reference Citation Analysis (1)] |
| 13. | Dar NA, Mir MM, Salam I, Malik MA, Gulzar GM, Yatoo GN, Ahmad A, Shah A. Association between copper excess, zinc deficiency, and TP53 mutations in esophageal squamous cell carcinoma from Kashmir Valley, India--a high risk area. Nutr Cancer. 2008;60:585-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 14. | Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453-457. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4763] [Cited by in RCA: 8913] [Article Influence: 891.3] [Reference Citation Analysis (0)] |
| 15. | Wang S, Zhang X, Leng S, Xu Q, Sheng Z, Zhang Y, Yu J, Feng Q, Hou M, Peng J, Hu X. Immune Checkpoint-Related Gene Polymorphisms Are Associated With Primary Immune Thrombocytopenia. Front Immunol. 2020;11:615941. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 16. | Song L, Liu D, Zhang X, Zhu X, Lu X, Huang J, Yang L, Wu Y. Low expression of PDHA1 predicts poor prognosis in gastric cancer. Pathol Res Pract. 2019;215:478-482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 17. | Li Y, Huang R, Li X, Yu D, Zhang M, Wen J, Goscinski MA, Trope CG, Nesland JM, Suo Z. Decreased expression of pyruvate dehydrogenase A1 predicts an unfavorable prognosis in ovarian carcinoma. Am J Cancer Res. 2016;6:2076-2087. [PubMed] |
| 18. | Li Y, Li X, Zhong Y, Ji Y, Yu D, Zhang M, Wen JG, Zhang H, Goscinski MA, Nesland JM, Suo Z. PDHA1 gene knockout in prostate cancer cells results in metabolic reprogramming towards greater glutamine dependence. Oncotarget. 2016;7:53837-53852. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 19. | Zhang S, Tan Y, Cai X, Luo K, Wu Z, Lu J. Preoperative weight loss is associated with poorer prognosis in operable esophageal cancer patients: A single-center retrospective analysis of a large cohort of Chinese patients. J Cancer. 2020;11:1994-1999. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 20. | Wang C, Wang P, Liu JC, Zhao ZA, Guo R, Li Y, Liu YS, Li SG, Zhao ZG. Interaction of Estradiol and Endoplasmic Reticulum Stress in the Development of Esophageal Carcinoma. Front Endocrinol (Lausanne). 2020;11:410. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 21. | de Ruiter EJ, Ooft ML, Devriese LA, Willems SM. The prognostic role of tumor infiltrating T-lymphocytes in squamous cell carcinoma of the head and neck: A systematic review and meta-analysis. Oncoimmunology. 2017;6:e1356148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 245] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 22. | Saloura V, Izumchenko E, Zuo Z, Bao R, Korzinkin M, Ozerov I, Zhavoronkov A, Sidransky D, Bedi A, Hoque MO, Koeppen H, Keck MK, Khattri A, London N, Kotlov N, Fatima A, Vougiouklakis T, Nakamura Y, Lingen M, Agrawal N, Savage PA, Kron S, Kline J, Kowanetz M, Seiwert TY. Immune profiles in primary squamous cell carcinoma of the head and neck. Oral Oncol. 2019;96:77-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 23. | Balermpas P, Michel Y, Wagenblast J, Seitz O, Weiss C, Rödel F, Rödel C, Fokas E. Tumour-infiltrating lymphocytes predict response to definitive chemoradiotherapy in head and neck cancer. Br J Cancer. 2014;110:501-509. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 212] [Cited by in RCA: 245] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 24. | Semba H, Takeda N, Isagawa T, Sugiura Y, Honda K, Wake M, Miyazawa H, Yamaguchi Y, Miura M, Jenkins DM, Choi H, Kim JW, Asagiri M, Cowburn AS, Abe H, Soma K, Koyama K, Katoh M, Sayama K, Goda N, Johnson RS, Manabe I, Nagai R, Komuro I. HIF-1α-PDK1 axis-induced active glycolysis plays an essential role in macrophage migratory capacity. Nat Commun. 2016;7:11635. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 259] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 25. | Tan Z, Xie N, Cui H, Moellering DR, Abraham E, Thannickal VJ, Liu G. Pyruvate dehydrogenase kinase 1 participates in macrophage polarization via regulating glucose metabolism. J Immunol. 2015;194:6082-6089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 254] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 26. | Tanaka T, Nakamura J, Noshiro H. Promising immunotherapies for esophageal cancer. Expert Opin Biol Ther. 2017;17:723-733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 27. | Dhupar R, Van Der Kraak L, Pennathur A, Schuchert MJ, Nason KS, Luketich JD, Lotze MT. Targeting Immune Checkpoints in Esophageal Cancer: A High Mutational Load Tumor. Ann Thorac Surg. 2017;103:1340-1349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 28. | Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, Barron DA, Zehir A, Jordan EJ, Omuro A, Kaley TJ, Kendall SM, Motzer RJ, Hakimi AA, Voss MH, Russo P, Rosenberg J, Iyer G, Bochner BH, Bajorin DF, Al-Ahmadie HA, Chaft JE, Rudin CM, Riely GJ, Baxi S, Ho AL, Wong RJ, Pfister DG, Wolchok JD, Barker CA, Gutin PH, Brennan CW, Tabar V, Mellinghoff IK, DeAngelis LM, Ariyan CE, Lee N, Tap WD, Gounder MM, D'Angelo SP, Saltz L, Stadler ZK, Scher HI, Baselga J, Razavi P, Klebanoff CA, Yaeger R, Segal NH, Ku GY, DeMatteo RP, Ladanyi M, Rizvi NA, Berger MF, Riaz N, Solit DB, Chan TA, Morris LGT. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2239] [Cited by in RCA: 2800] [Article Influence: 466.7] [Reference Citation Analysis (0)] |