Published online Jul 21, 2022. doi: 10.3748/wjg.v28.i27.3435
Peer-review started: February 7, 2022
First decision: April 5, 2022
Revised: April 16, 2022
Accepted: June 3, 2022
Article in press: June 3, 2022
Published online: July 21, 2022
Processing time: 161 Days and 6.1 Hours
Hepatocellular carcinoma (HCC) exhibits high invasiveness and mortality rates, and the molecular mechanisms of HCC have gained increasing research interest. The abnormal DNA damage response has long been recognized as one of the important factors for tumor occurrence and development. Recent studies have shown the potential of the protein RING finger and WD repeat domain 3 (RFWD3) that positively regulates p53 stability in response to DNA damage as a therapeutic target in cancers.
To investigate the relationship between HCC and RFWD3 in vitro and in vivo and explored the underlying molecular signalling transduction pathways.
RFWD3 gene expression was analyzed in HCC tissues and adjacent normal tissues. Lentivirus was used to stably knockdown RFWD3 expression in HCC cell lines. After verifying the silencing efficiency, Celigo/cell cycle/apoptosis and MTT assays were used to evaluate cell proliferation and apoptosis. Subsequently, cell migration and invasion were assessed by wound healing and transwell assays. In addition, transduced cells were implanted subcutaneously and injected into the tail vein of nude mice to observe tumor growth and metastasis. Next, we used lentiviral-mediated rescue of RFWD3 shRNA to verify the phenotype. Finally, the microarray, ingenuity pathway analysis, and western blot analysis were used to analyze the regulatory network underlying HCC.
Compared with adjacent tissues, RFWD3 expression levels were significantly higher in clinical HCC tissues and correlated with tumor size and TNM stage (P < 0.05), which indicated a poor prognosis state. RFWD3 silencing in BEL-7404 and HCC-LM3 cells increased apoptosis, decreased growth, and inhibited the migration in shRNAi cells compared with those in shCtrl cells (P < 0.05). Furthermore, the in vitro results were supported by the findings of the in vivo experiments with the reduction of tumor cell invasion and migration. Moreover, the rescue of RFWD3 shRNAi resulted in the resumption of invasion and metastasis in HCC cell lines. Finally, gene expression profiling and subsequent experimental verification revealed that RFWD3 might influence the proliferation and metastasis of HCC via the Wnt/β-catenin signalling pathway.
We provide evidence for the expression and function of RFWD3 in HCC. RFWD3 affects the prognosis, proliferation, invasion, and metastasis of HCC by regulating the Wnt/β-catenin signalling pathway.
Core Tip: The relationship between RING finger and WD repeat domain 3 (RFWD3) and tumorigenesis process has been reported occasionally, but the relationship between RFWD3 and hepatocellular carcinoma (HCC) is still unclear. This research reports the significant role of RFWD3 in HCC development by using bioinformatics databases, clinical samples, cell phenotypes, in vivo experiments, and microarray analyses. It provides evidence that RFWD3 expression might affect the tumorigenesis process of HCC by regulating the Wnt/β-catenin signalling pathways, laying a basic foundation for future relevant studies.
- Citation: Liang RP, Zhang XX, Zhao J, Lu QW, Zhu RT, Wang WJ, Li J, Bo K, Zhang CX, Sun YL. RING finger and WD repeat domain 3 regulates proliferation and metastasis through the Wnt/β-catenin signalling pathways in hepatocellular carcinoma. World J Gastroenterol 2022; 28(27): 3435-3454
- URL: https://www.wjgnet.com/1007-9327/full/v28/i27/3435.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i27.3435
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors and is highly invasive with a high mortality rate[1]. Reportedly, the incidence of HCC has increased rapidly by approximately 3%–4% per year in the United States[2]. Despite advances in medical research, early diagnosis of HCC remains difficult, and most patients are diagnosed with middle-or late-stage disease[3-5]. Moreover, even though some progress has been made in the clinical diagnosis and treatment of HCC, the long-term survival after treatment for patients diagnosed with early-stage HCC is low[6]. With the development of biomedicine, more basic studies have been carried out on HCC, but the underlying mechanism remains unclear, warranting promising interventions[7,8].
Multiple signal transduction pathways have been demonstrated to be involved in cellular responses to stress. For example, the ubiquitin-proteasome system (UPS) plays an important role in DNA damage repair, transcription regulation, cell cycle arrest, and apoptosis[9-11]. Several studies have focused on the role of the protein RING finger and WD repeat domain 3 (RFWD3) in the ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related pathways of DNA damage response[12-16]. RFWD3 contains a RING finger domain and displays in vitro E3 ubiquitin ligase activity. It comprises an SQ-rich region in the N-terminus, a coiled-coil domain, and a WD40 domain in the C-terminus. In addition, sequence comparison revealed that the residues flanking serine 46 and serine 63 of RFWD3 are conserved[17]. According to the current literature, RFWD3 accumulates at stalled replication forks as part of the DNA damage response[18,19]. At these sites, RFWD3 co-localizes with replication protein A (RPA) and binds via its WD40 domain at the C-terminus to RPA2[20,21]. It functions in replication checkpoint signalling upon replication stress and promotes replication fork restart after homologous recombination (HR)[22,23]. RFWD3 is regarded as a tumor suppressor gene, and recent functional and mechanistic studies have indicated that it maintains the stability of p53[24]. Feeney et al[25] reported that it facilitates RPA-mediated DNA damage response[25]. Furthermore, RFWD3 is required for the proliferation of non-small-cell lung cancer, gastric carcinoma, and colorectal cancer cells and may have an important role[26-28]. However, the relationship between HCC and RFWD3 has not been explored.
In a preliminary experiment using a small number of HCC samples, our team found that RFWD3 expression was higher in HCC tissues than in adjacent non-cancerous tissues[29]. To rule out the effects of small sample numbers, the results were then verified using the open-source database. For this purpose, we chose RNA SEQ data (FPKM normalized) of liver cancer from The Cancer Genome Atlas (TCGA) database directly downloaded from the UCSC Xena database and further transformed it into log2 (TPM + 1) format. The results were concordant with our preliminary findings. Furthermore, the immunohistochemical analysis demonstrated that RFWD3 expression was associated with the clinical TNM stage (and with survival, according to the database). Therefore, in this study, we tested the hypothesis that RFWD3 plays a significant role in HCC development.
To test this hypothesis, in this study, we investigate the relationship between HCC and RFWD3 in vitro using human HCC cell lines and in vivo using HCC xenograft models in nude mice. Further, we unravel the underlying molecular signalling transduction pathways in HCC. The findings of this study provide evidence for the first time that RFWD3 plays a significant role in HCC cell proliferation and may serve as a potential therapeutic target for HCC.
Sixty tissue sample pairs (HCC and adjacent normal liver tissues) were obtained from clinical patients who underwent surgical HCC resection at the First Affiliated Hospital of Zhengzhou University (Zhengzhou, China). Before the operation, all patients were informed that the resected tissues could be used for research, and consent was obtained from each patient. After resection, the tissues were snap-frozen immediately and stored at −80 °C. All the experimental procedures were approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University (Approval No. 10[2017]).
TCGA liver HCC cohort included 374 cancer tissues and 50 adjacent normal tissues. According to the expression level of RFWD3 gene, we used the best cut-off value to divide the HCC patients into high and low expression groups by the Surv function of the survminer package. Then, the Kaplan-Meier method and log-rank test were used to evaluate the differences in overall survival and recurrence-free survival between the two groups. All data processing, statistical analyses, and graphical presentations (of this section) were carried out using R 4.0.5 software.
HCC tissue microarray data of 60 HCC and adjacent tissues, including some basic clinical data shown in Table 1, were obtained from BiochipTech (Zhengzhou, China). Immunohistochemistry (IHC) was performed using a 1:100 dilution of a primary antibody against RFWD3 (Multi Sciences, Hangzhou, China), and all immunohistochemical procedures were performed according to a common standard protocol. First, the expression levels of RFWD3 between HCC and adjacent tissues were compared with a histochemical score (H-score). Then, a scoring system that combined the staining intensity (SI: 0, negative; 1, weak; 2, moderate; 3, strong) and the percentage of positive cells (PP: 0, < 5%; 1, 6%–25%; 2, 26%–50%; 3, 51%–75%; 4, > 75%) was used to compare the correlation of clinical data in patients with RFWD3 expression (SI × PP > 4 was considered high expression; SI × PP ≤ 4 was considered low expression).
The human HCC cell lines BEL-7404 and HCC-LM3, maintained in our laboratory, were cultured (BEL-7404: 10% FBS, 90% RPMI-1640; HCC-LM3: 10% FBS, 90% DMEM; all purchased from Gibco, United States) according to the American type culture collection protocol. All cells were cultured in 50 ml/L CO2 at 37°C in a moderately humid environment. The RFWD3 siRNA sequence (5-CAGAGAATGATGGCAACAT-3) was synthesized and cloned into the AgeI/EcoRI sites of the lentivirus-based GV493-puromycin vector (shRNAi). The negative control lentiviral vector (5-TTCTCCGAACGTGTCACGT-3) was used as a control (shCtrl). The cells were divided into two groups (shCtrl and shRNAi), seeded in six-well plates, and allowed to grow until the logarithmic phase and less than 70% confluency. After verifying the transfection efficiency, the cells were harvested for subsequent in vivo and in vitro experiments. Additionally, the RFWD3-reverse mutation sequence (5-CTGAGAACGACGGAAATAT-3) was synthesized and cloned into the BamHI/AgeI sites of the lentivirus-based GV557-neomycin vector (KD-OE). The virus without anti-Smad served as negative controls (KD-OE-NC) for subsequent phenotypic validation. The GV493-puromycin and GV557-neomycin vectors were obtained from GeneChem (Shanghai, China).
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was used to detect the mRNA expression level of RFWD3 using the following primers: Forward: 5’-AGAGTATGGAGGGGTTGTTGAC-3’ and reverse: 5’-TAGAAGCAGAAGGTAGCAGAGG-3’. Total RNA was extracted from tissues and cells using TRIzol reagent (Thermo Fisher Scientific, United States) and reverse transcribed into cDNA using a Prime-Script RT Reagent Kit with gDNA Eraser (Takara Bio, Japan). Following a common standard protocol, qRT-PCR was performed in a total volume of 10 μL on a 7900-HT Fast Real-time PCR System (Applied Biosystems Inc., United States) to evaluate RFWD3 expression in 60 pairs of HCC tissues and determine the RFWD3 knockdown efficiency in cells transduced with shCtrl and shRNAi.
To determine the RFWD3 virus transfection efficiency in cells transduced with shCtrl, shRNAi, KD-OE, and KD-OE-NC, the levels of RFWD3 protein expression were assessed by western blot (WB). Briefly, the total protein was extracted by incubating the cells in RIPA lysis buffer (Thermo Fisher Scientific, United States) supplemented with a protease inhibitor cocktail (Thermo Fisher Scientific, United States) for 1 h on ice, followed by sonication (Shanghai Experimental Instruments, China) for three 5 s cycles on ice. After centrifugation (14000 ×g, 4 °C, 15 min), the supernatant was collected for protein concentration measurement using a bicinchoninic acid protein assay kit (Pierce Biotechnology, United States). Denatured protein samples (30 μg/10 μL) were separated by SDS-PAGE (Invitrogen, United States) and transferred to polyvinylidene fluoride membranes (Bio-Rad, United States). Afterward, the membranes were blocked with 5% non-fat milk (BD Biosciences, United States) dissolved in 30 mL Tris-buffered saline with Tween-20 (TBST) for 2 h at common room temperature. The membranes were then incubated overnight at 4 °C with primary antibodies against GAPDH (1:1000; Abcam, United States) and RFWD3 (1:1000; Multi Sciences, Hangzhou, China). After three 5 min washes in TBST with agitation, the membranes were incubated for 1 h at room temperature with the appropriate secondary horseradish peroxidase-linked antibodies, including goat anti-rabbit IgG for GAPDH (1:2000; Abcam, United States) and goat anti-rabbit IgG for RFWD3 (1:2000; Multi Sciences, Hangzhou, China). An ECL kit (Pierce Biotechnology, United States) was used to visualize the protein bands.
A Celigo imaging cytometer (Celigo; Nexcelom Bioscience, United States) and MTT assays were used to detect the number of cells at each time point, and the growth status of the cells was analyzed. For the Celigo assay, cells in the logarithmic phase were digested with trypsin (Sangon Biotech, Shanghai, China), seeded in 96-well plates (2000 cells/well in 100 μL medium) with three wells per condition, and cultured in a 37 °C incubator with 50 mL/L CO2. From the first day to the fifth day after seeding, the plate was subjected to high-throughput screening every day. GFP fluorescence was measured in each scan to generate a five days cell proliferation curve. For the MTT assay, two groups of cells were seeded in 96-well plates (2000 cells/well) with three wells per condition and incubated at 37 °C for six consecutive days. Cell viability was analyzed every 24 h by washing the cells twice with PBS and adding MTT solution (5 mg/mL) to each well. After 4 h of incubation, the supernatant in each well was removed, and 100 μL DMSO was added to solubilize the formazan crystals. After 10 min, the optical density (OD) was measured at an excitation wavelength of 490 nm and an emission wavelength of 570 nm using a microplate reader (VarioskanFlash; Thermo Fisher Scientific, United States).
Cell cycle progression was examined using propidium iodide (PI; Sigma, United States) and RNase A (Thermo Fisher, United States). Cells in the logarithmic growth phase in six-well culture plates were digested with trypsin (Sangon Biotech, Shanghai, China) and diluted with the culture medium. Three replicate tubes were analyzed for both the shCtrl and shRNAi groups (1 × 106 cells/tube). The cells were centrifuged at 1300 rpm for 5 min, and the supernatant was discarded. The cell pellet was washed with pre-cooled D-Hanks equilibrium solution (4 °C with pH = 7.2–7.4), and the harvested cells were fixed with 70% cold ethanol at 4 °C overnight. Afterward, the cells were washed twice with 4 °C D-Hanks solution and incubated with 10 mg/mL RNase at 37 °C. Cell cycle progression was monitored by staining the nuclei with PI. The fluorescence of DNA-bound PI in cells was measured using flow cytometry (FCM; FC500 flow cytometer; Cytomics, United States). Finally, ModFit LT software (Windows version 4.0; Verity Software House, United States) was used to analyze the cell populations in different phases of the cell cycle.
Cell apoptosis was assayed using an Annexin V-APC/PI apoptosis detection kit (Biosciences, Thermo Fisher, United States). Cells in the logarithmic phase were digested with trypsin (Sangon Biotech, Shanghai, China) and diluted with the culture medium. Three replicate tubes each were analyzed for the shCtrl, shRNAi, KD-OE, and KD-OE-NC groups of each cell line (1 × 104 cells/tube). The cells were centrifuged at 1300 rpm for 5 min, and the supernatant was discarded. The cell pellet was washed with a pre-cooled D-Hanks equilibrium solution (4 °C with pH = 7.2–7.4). The cells were then washed with 1 × binding buffer, centrifuged at 1300 rpm for 3 min, and resuspended in 200 μL 1 × binding buffer. Next, 10 μL Annexin V-APC staining solution was added, and the samples were incubated for 10–15 min at room temperature in the dark. Finally, apoptosis was detected in the samples.
Cell migration was assayed using wound healing and transwell assays. For the wound-healing assay, cells were plated in 6-well plates and scratched with a sterile pipette tip. The cells were then imaged using a microscope (Olympus, Japan) to observe migration after 12 and 24 h. For the invasion and migration assays, the upper surfaces of transwell chambers (Costar, Cambridge, MA, United States) were uncoated (for the migration experiments) or coated with Matrigel (1:4, BD, United States) (for the invasion experiments). Cells were then added to the upper chambers containing 200 μL serum-free medium (BEL-7404: RPMI-1640; HCC-LM3: DMEM) per well, and 500 μL medium (with 10% FBS) was added to the lower chambers. After 48 h, the chambers were cleaned, stained with hematoxylin, transferred to glass slides after the operation, and then observed under a microscope (Olympus, Japan). Six fields per chamber were used to quantify the invaded cell numbers with 100 × and 200 × magnifications.
All animal experimental protocols were conducted following the ARRIVE (Animal Research: Reporting In Vivo Experiments) guidelines (https://www.nc3rs.org.uk/arrive-guidelines) and performed following protocols approved by the Research Ethics Committee of the First Affiliated Hospital of Zhengzhou University (Approval No. 10[2017]). Nude mice were purchased from Shanghai B&K Universal Group Limited (certificate number: SCXK 2013-0016; Shanghai, China). For the proliferation assay, 5 × 106 HCC-LM3 cells stably transduced with the lentiviral pSC-1-GFP vector carrying shCtrl or shRNAi were suspended in PBS and injected subcutaneously into the flank region of 6-week-old male athymic nude mice (six mice in each group). For the metastasis assay, 2 × 106 HCC-LM3 cells transduced as described above were injected into the tail vein of nude mice. The animals were acclimatized to laboratory conditions (23 °C, 12 h/12 h light/dark, 50% humidity, ad libitum access to food and water). After four weeks, the mice were euthanized by barbiturate overdose (intravenous injection, 150 mg/kg pentobarbital sodium) for tissue collection and photographed.
To elucidate the molecular mechanisms involved in the association between RFWD3 and HCC, we performed microarray-based gene expression analysis to compare gene expression between the shCtrl and shRNAi groups. We then used ingenuity pathway analysis (IPA) to explore the molecular signal transduction pathways involved in HCC. All procedures were performed according to a common protocol, and assistance was provided by the Shanghai Biotechnology Corporation (SBC, Shanghai, China).
Statistical analyses and related statistical illustrations were performed using SPSS (version 21.0; IBM, Armonk, NY, United States) and GraphPad Prism software (ver. 5.0 for Windows; GraphPad Software Inc., United States). The Pearson chi-square test was used to determine the correlation between clinical characteristics and immunohistochemical staining. Other results are expressed as mean ± SD. A t-test was used to compare the two groups, and a P value of < 0.05 was considered statistically significant.
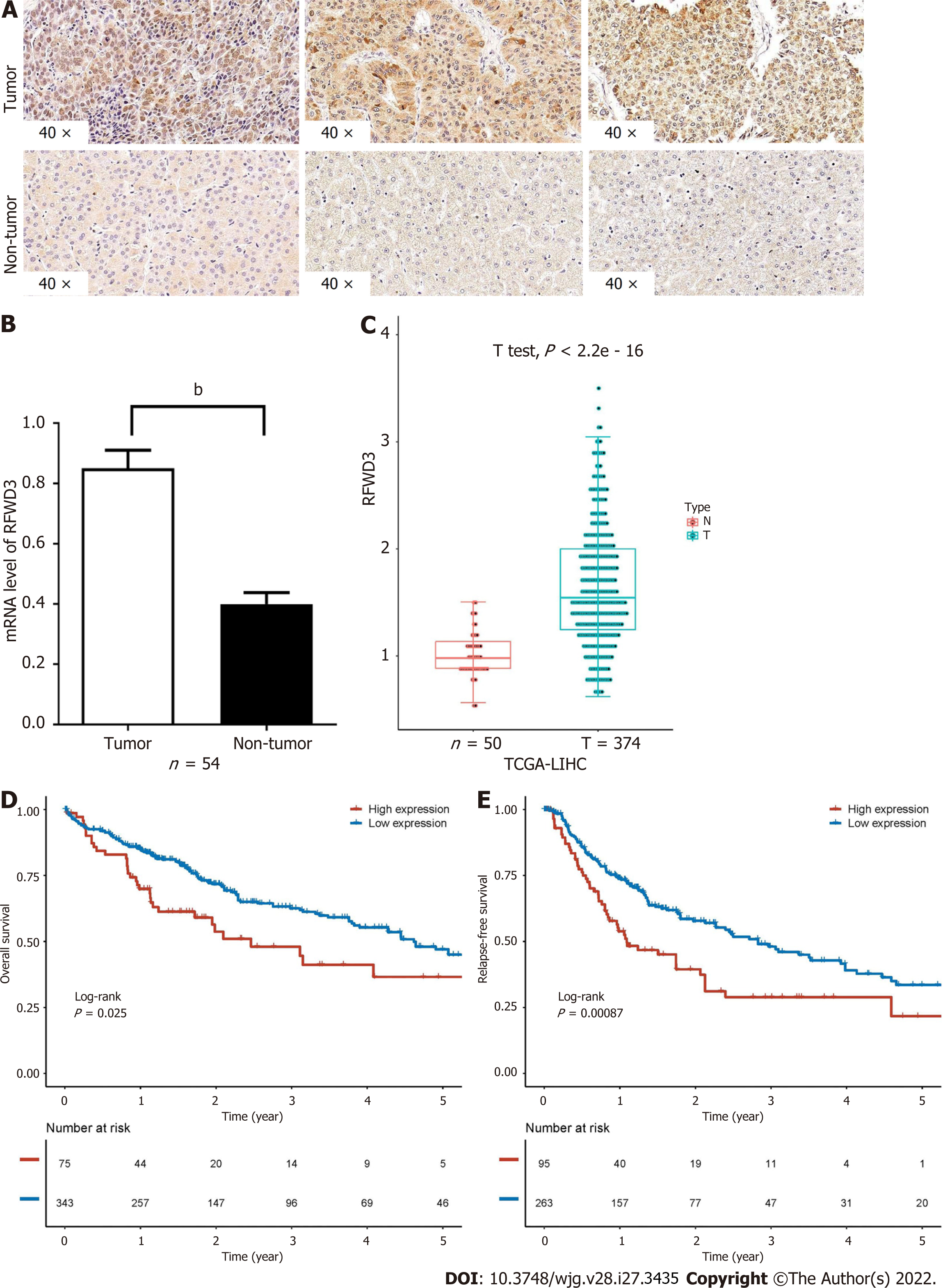
RFWD3 expression was examined in HCC tissues and matched adjacent normal tissues. IHC analysis of 60 tissue pairs revealed that HCC tissues exhibited a higher H-score than non-tumor tissues (Figure 1A)[30]. This result was supported by the qRT-PCR analysis of 54 tissue sample pairs (some outlier value pairs were excluded) (Figure 1B). These results were concordant with the RFWD3 expression data obtained by analyzing the bioinformatics database (Figure 1C). In addition, the bioinformatics database showed that higher RFWD3 expression represents a worse prognosis in HCC (Figure 1D and E). These data indicate that high RFWD3 expression might predict poor HCC development and prognosis (Table 1).
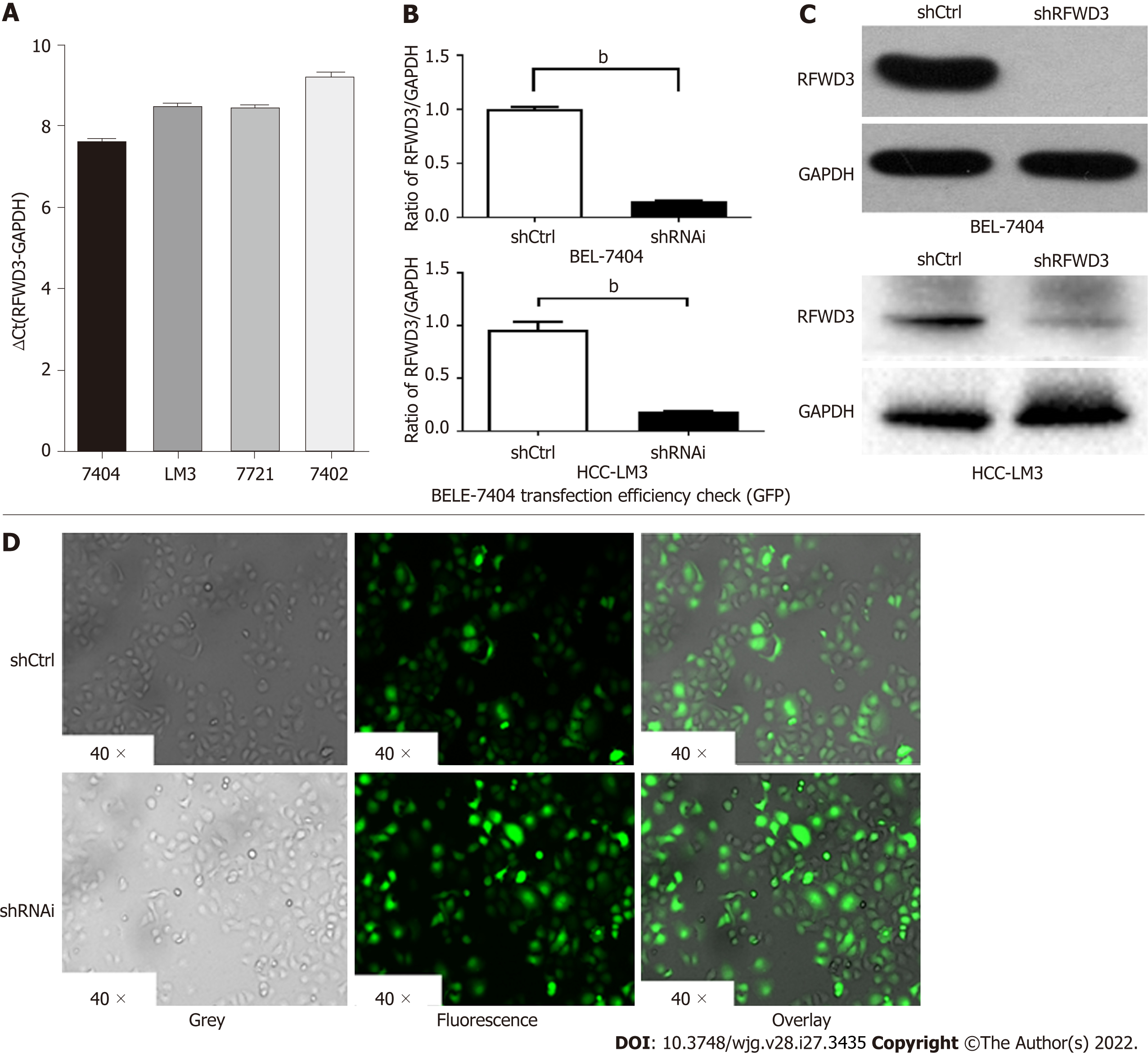
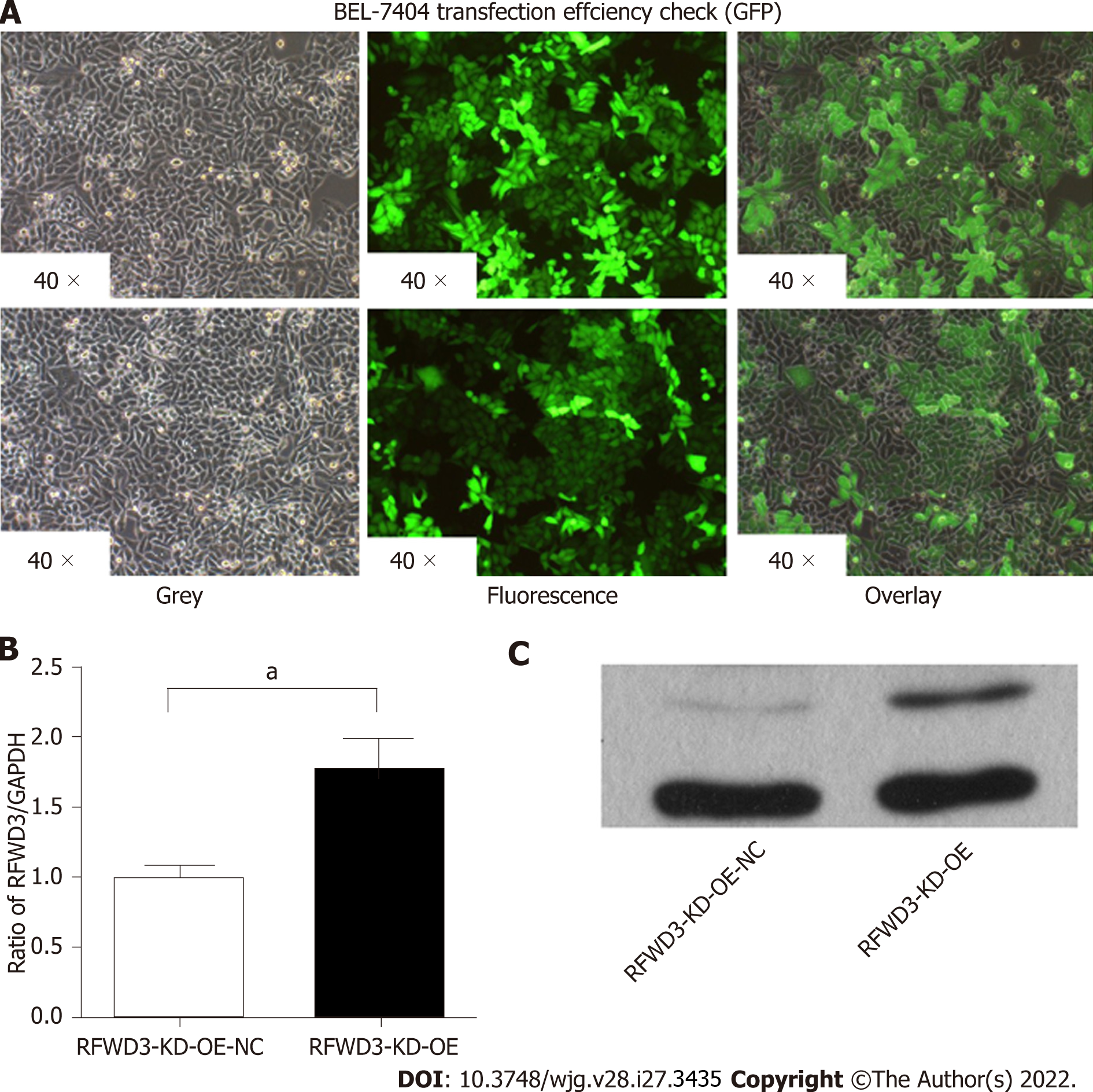
After verifying the high expression of RFWD3 in HCC tissues, we selected HCC cell lines for further in vitro experiments. BEL-7404, HCC-LM3, SMMC-7721, and BEL-7402 cells were evaluated for the stable expression of RFWD3. Based on qRT-PCR and cell growth analyses, we chose BEL-7404 and HCC-LM3 cells for lentiviral-mediated knockdown of RFWD3 expression (Figure 2A). After lentiviral transduction, compared with the shCtrl group, the shRNAi group showed obvious knockdown of RFWD3 (qRT-PCR: BEL-7404: P < 0.01; HCC-LM3: P < 0.01) (Figure 2B). These results were subsequently confirmed with WB analysis and GFP fluorescence imaging (Figure 2C and D).
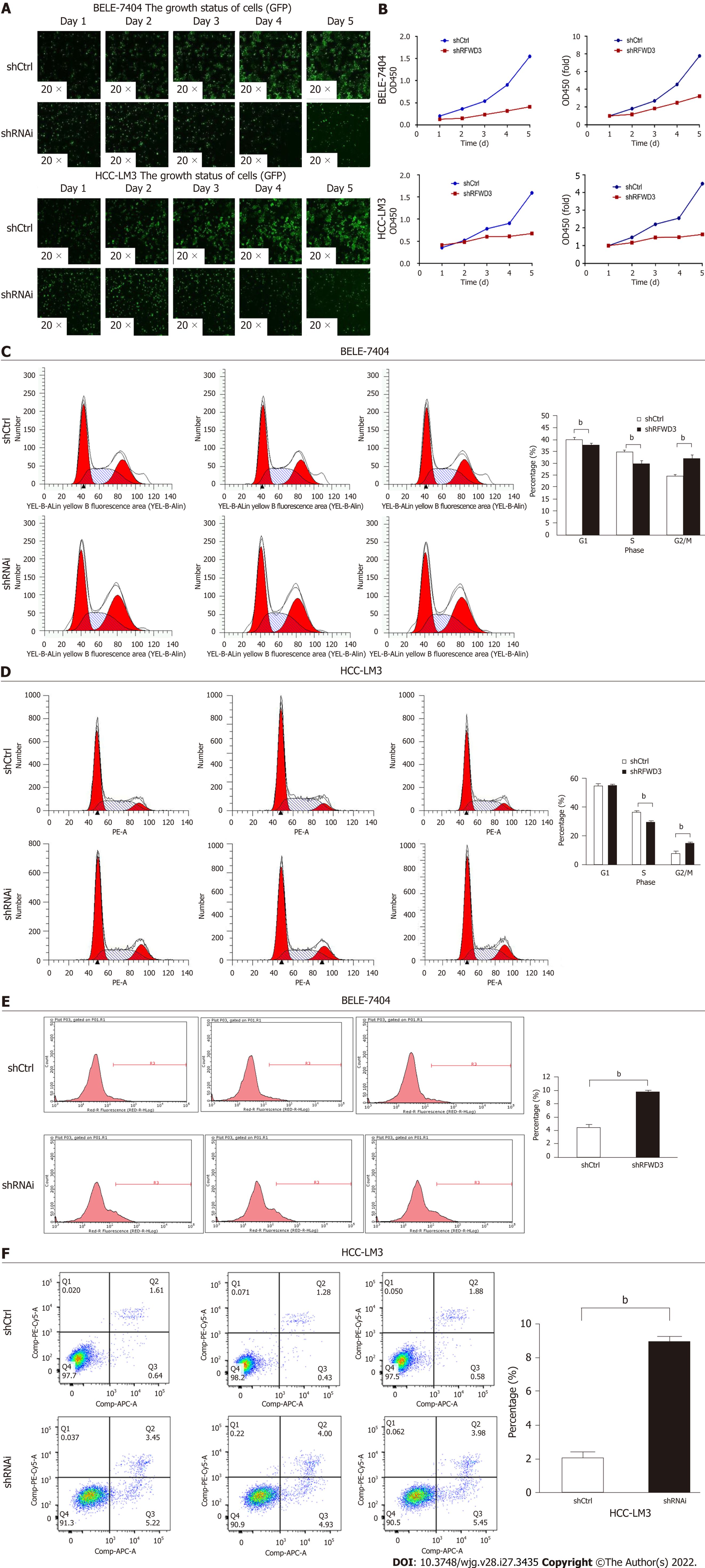
A high-content screening assay (Celigo) was performed to monitor cell growth for five days. The total cell count and growth rate were lower in the shRNAi group than in the shCtrl group (Figure 3A). To further verify these results, we used an MTT assay. After five days of incubation in the same environment, consistent with the Celigo results, the OD value was significantly lower in the shRNAi group than that in the shCtrl group (Figure 3B). Taken together, these results indicate that RFWD3 knockdown significantly reduced the proliferation of HCC cells.
To determine whether RFWD3 affects cell proliferation by regulating the cell cycle, we used PI staining to evaluate cell cycle progression. In the shCtrl and shRNAi groups, the cell cycle distributions were as follows (presented as the mean value of three replicates). For BEL-7404 shCtrl cells, 40.09% were in G0/G1 phase, 35.01% were in the S phase, and 24.9% were in the G2/M phase, while for BEL-7404 shRNAi cells, 37.82% were in G0/G1 phase, 30.03% were in S phase, and 32.15% were in G2/M phase. For HCC-LM3 shCtrl cells, 55.07% were in G0/G1 phase, 36.98% were in the S phase, and 7.95% were in the G2/M phase, while for HCC-LM3 shRNAi cells, 54.98% were in G0/G1 phase, 29.84% were in S phase, and 15.18% were in G2/M phase. Compared with shCtrl cells, shRNAi cells showed a reduced percentage of cells in G0/G1 (BEL-7404: P < 0.01) and S phase (BEL-7404: P < 0.01; HCC-LM3: P < 0.01), and the fraction of cells in the G2/M phase was significantly increased (BEL-7404: P < 0.01; HCC-LM3: P < 0.01). These results indicated that RFWD3 knockdown blocked cell cycle progression through the G2/M phase (Figure 3C and D).
To determine whether RFWD3 affects cell proliferation by inducing apoptosis, an Annexin V-APC/PI apoptosis detection kit (AV/PI) was used to detect apoptosis in the two groups of cells by FCM. The apoptosis ratios were (presented as the mean value of three replicates) 4.47 ± 0.3748 for BEL-7404 shCtrl cells, 9.77 ± 0.1142 for BEL-7404 shRNAi cells, 2.14 ± 0.2234 for HCC-LM3 shCtrl cells, and 9.01 ± 0.2230 for HCC-LM3 shRNAi cells. Compared with shCtrl cells, shRNAi cells had a higher apoptosis rate (BEL-7404: P < 0.01; HCC-LM3: P < 0.01). This result demonstrated that RFWD3 knockdown increased the apoptosis ratio, thus affecting the proliferation of HCC cells (Figure 3E and F).
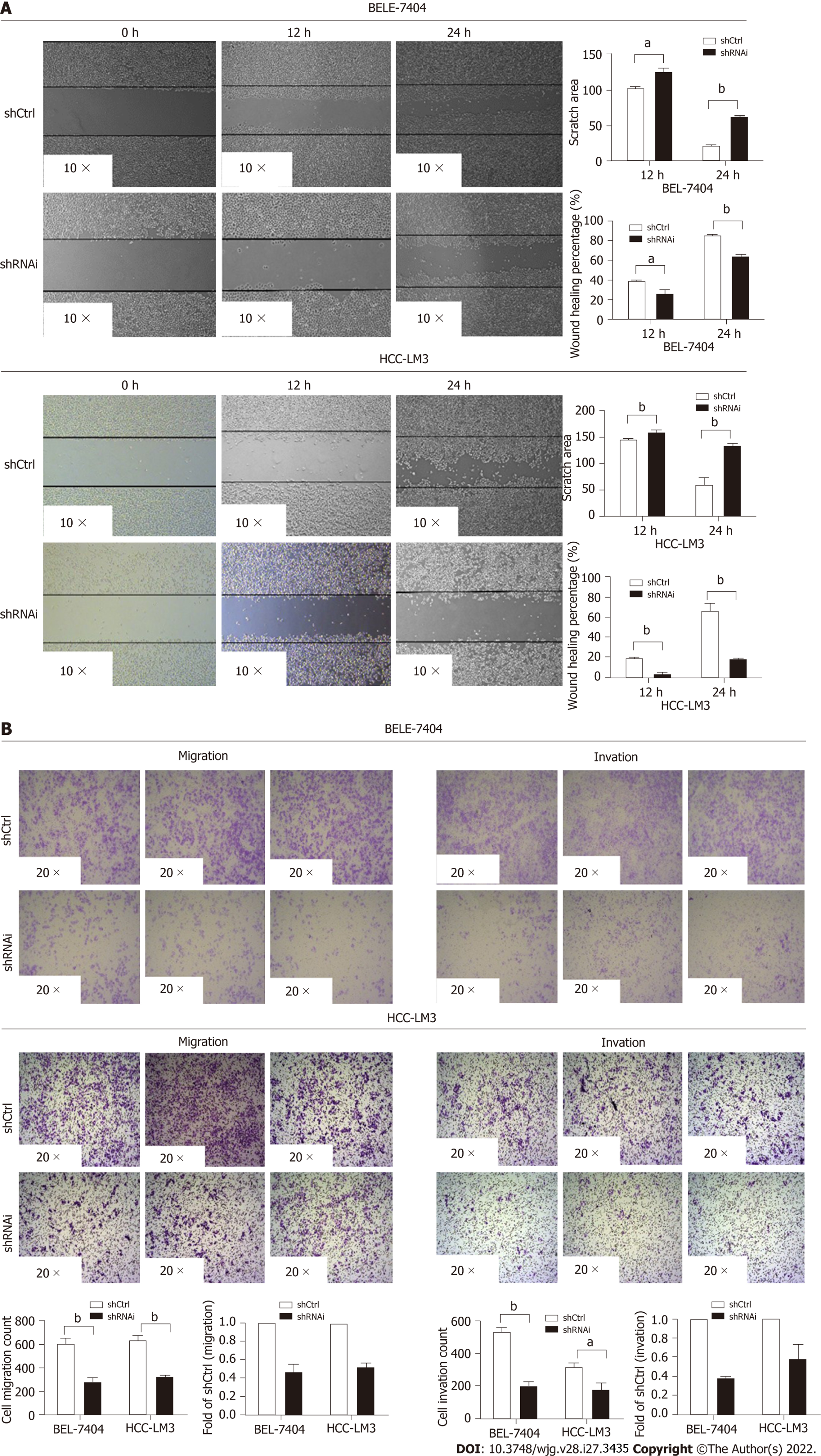
Wound healing and transwell assays were used to evaluate HCC cell invasion and migration. In the wound healing assay, the shCtrl groups exhibited better healing ability (P < 0.05). After 12 h, the wound area was 103.40 ± 1.32 in BEL-7404 shCtrl cells, 125.90 ± 4.46 in BEL-7404 shRNAi cells, 145.40 ± 0.84 in HCC-LM3 shCtrl cells and 159.50 ± 2.21 in HCC-LM3 shRNAi cells. Over time, the difference in the size of the wound area between the two groups increased. After 24 h, the wound area was 22.97 ± 0.22 in BEL-7404 shCtrl cells, 60.94 ± 2.41 in BEL shRNAi cells, 60.22 ± 12.68 in HCC-LM3 shCtrl cells and 134.40 ± 0.84 in HCC-LM3 shRNAi cells (Figure 4A). We next examined cell migration and invasion with transwell assays. The migration of both BEL-7404 and HCC-LM3 shRNAi cells was attenuated compared with shCtrl cells (P < 0.05). Similar results were observed in the invasion assay (Figure 4B). These data revealed that RFWD3 knockdown reduced HCC cell invasion and migration.
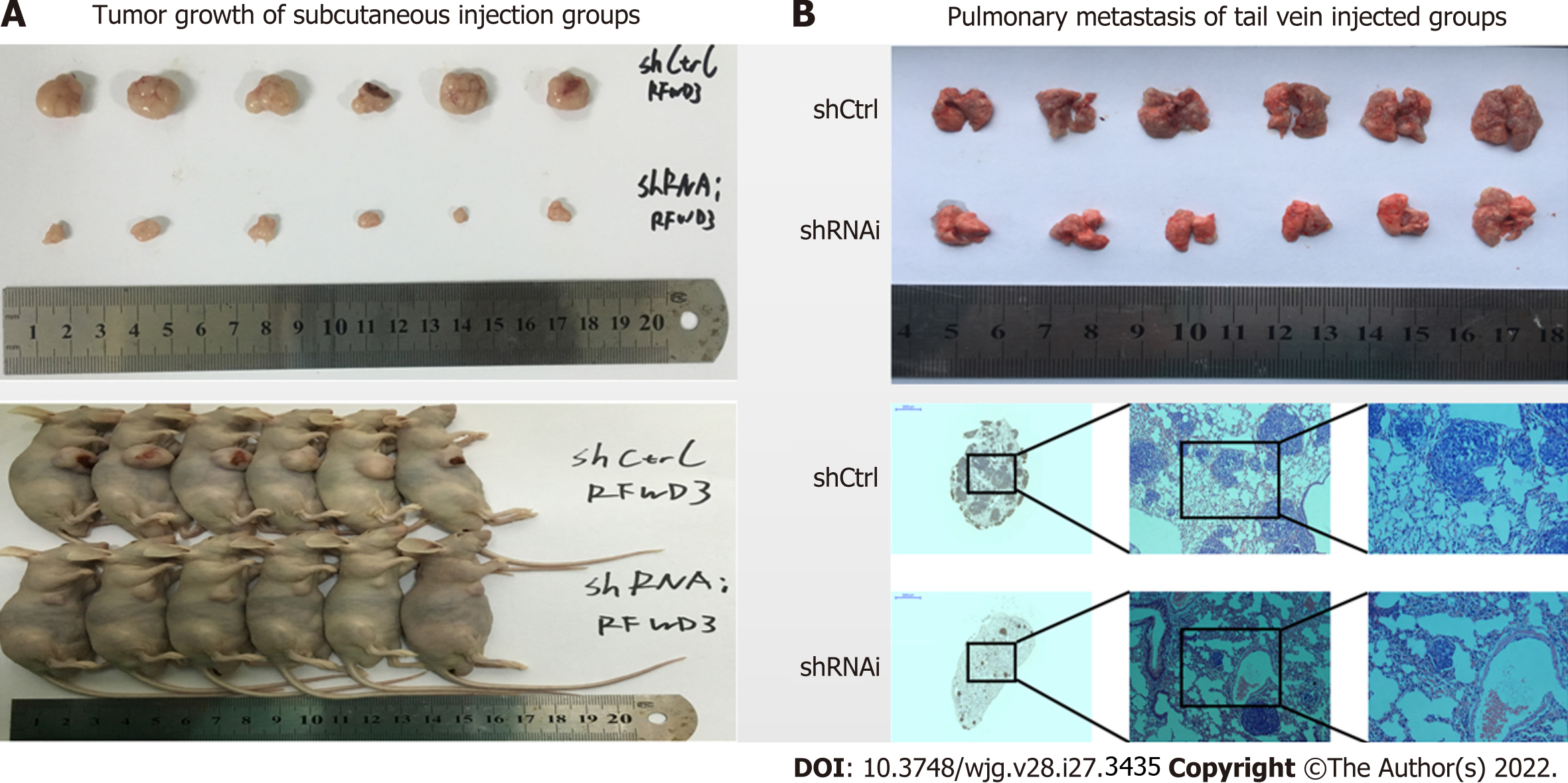
We established an HCC xenograft model in nude mice to verify the results in vivo. HCC-LM3 shCtrl and shRNAi cells were injected subcutaneously (Figure 5A) and into the tail vein (Figure 5B) of nude mice (six mice in each group). After four weeks, the mice were euthanized by cervical dislocation. The tumors (subcutaneous injection groups) and lung tissues (tail vein injection groups) were removed for imaging analysis. The growth and invasion ability of tumors was significantly lower in mice injected with shRNAi cells than in mice injected with shCtrl cells. These results show that silencing RFWD3 effectively reduced HCC tumor proliferation, invasion, and metastasis in vivo.
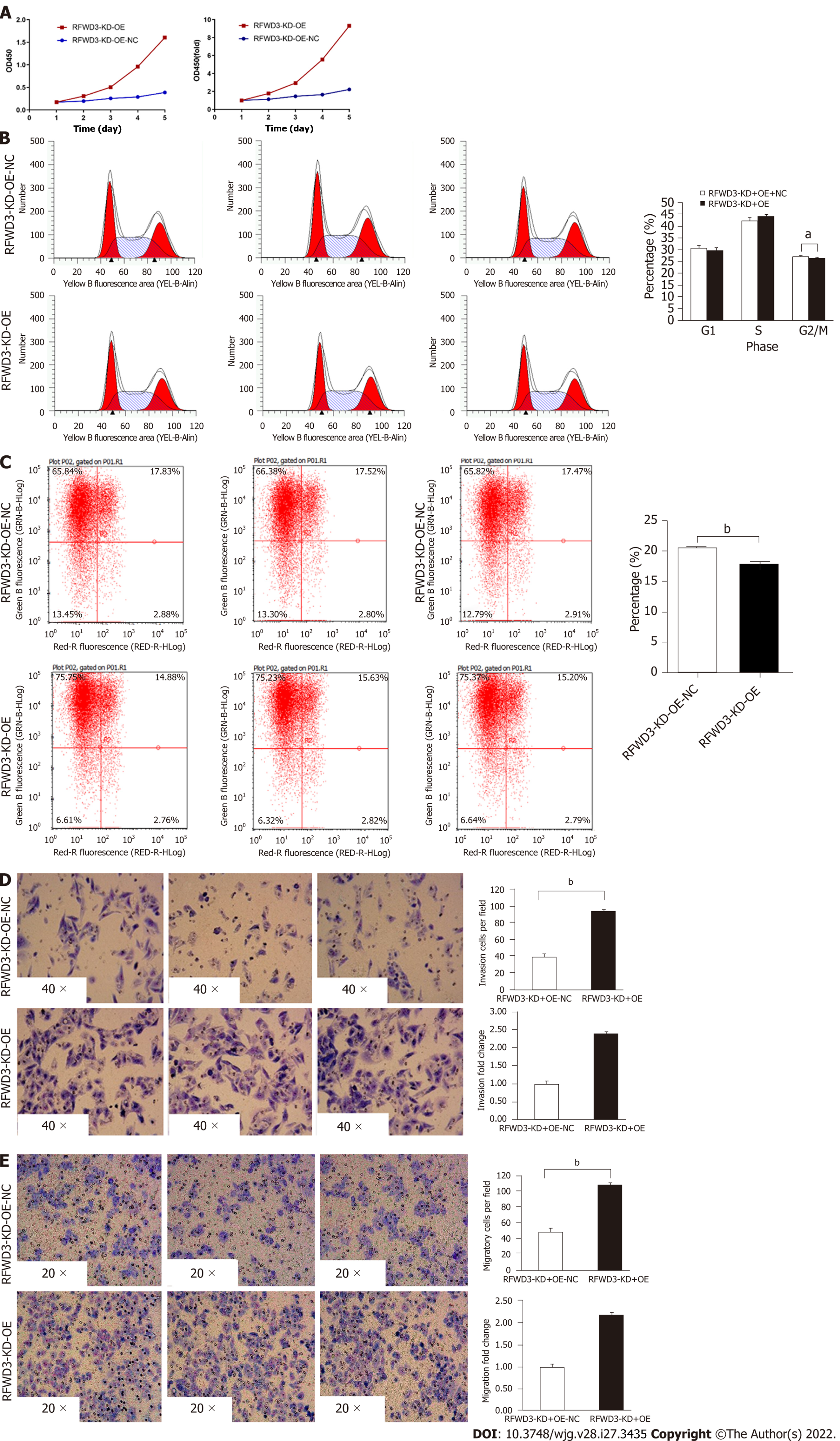
To rule out off-target effects and confirm the biological function of RFWD3 shRNAi, a rescue experiment was performed with RFWD3-KD-OE in HCC cells. After transfection with an LV-RFWD3 lentiviral vector, we used GFP fluorescence imaging to confirm transduction efficiency (Figure 6A). Compared with the RFWD3-KD-OE-NC group, the RFWD3-KD-OE group showed upregulation of RFWD3 in qRT-PCR (Figure 6B). WB analysis also confirmed the lentivirus transduction efficiency (Figure 6C).
In the MTT assay, the OD value of the RFWD3-KD-OE group was higher than that of the RFWD3-KD-OE-NC group. These results indicate that RFWD3-KD-OE rescued the proliferation of shRNAi BEL-7404 cells (Figure 7A). Furthermore, no difference was observed between the G1 and S phases in the two groups of cell cycle progression assays. However, in the G2/M phase, the fraction of RFWD3-KD-OE-NC cells was increased (the average value was 30.51% in RFWD3-KD-OE-NC cells, 29.74% in RFWD3-KD-OE cells, P < 0.05); however, the gap in data was diminished compared with previous BEL-7404 RNAi groups (24.9% in shCtrl cells, 32.15% in shRNAi cells, P < 0.05) (Figure 7B). In the subsequent apoptosis experiments, BEL-7404 RFWD3-KD-OE-NC cells had a higher apoptosis rate (the average value was 20.47% in RFWD3-KD-OE-NC cells, 18.03% in RFWD3-KD-OE cells, P < 0.01) (Figure 7C). This result demonstrated that RFWD3-KD-OE reduced the apoptosis ratio, thus affecting the proliferation of shRNAi BEL-7404 cells. In transwell assays, the migration of BEL-7404 RFWD3-KD-OE cells was higher than that of RFWD3-KD-OE-NC cells (P < 0.01) (Figure 7D). Similar results were observed in the invasion assay (P < 0.01; Figure 7E). These data revealed that RFWD3-KD-OE rescued BEL-7404 shRNAi cell invasion and migration.
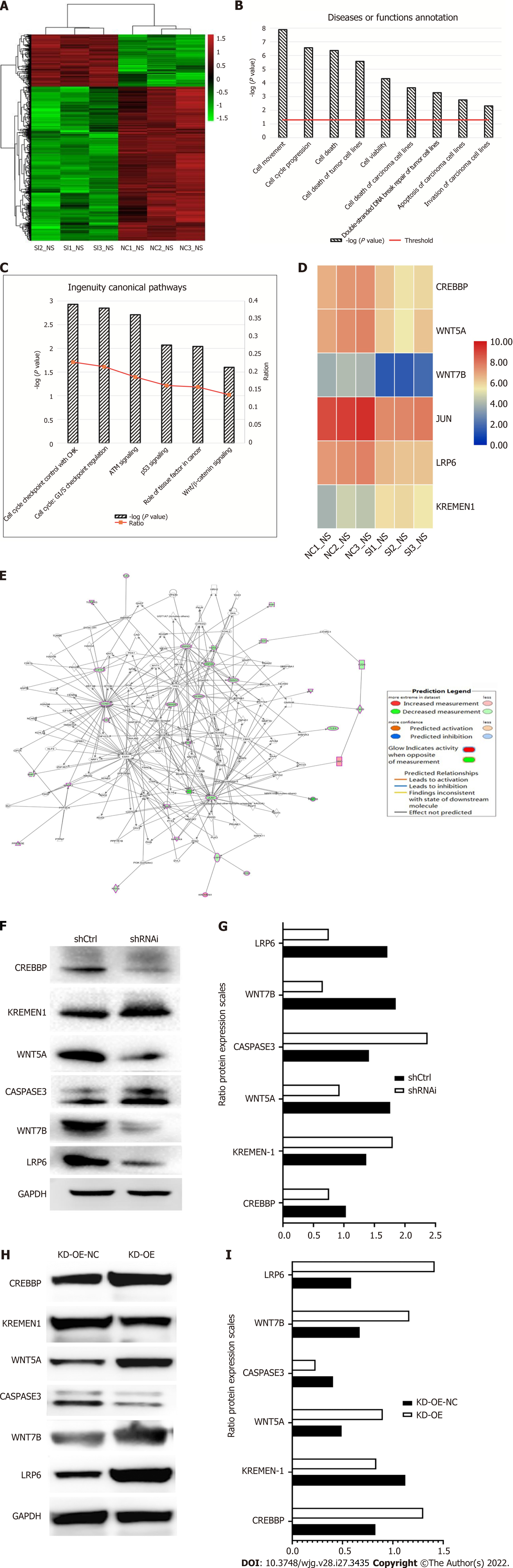
To further validate the results of the in vitro and in vivo experiments, we performed microarray analysis to explore the molecular mechanism underlying the differential RFWD3 expression between shCtrl and shRNAi HCC cells. A total of 454 genes (Figure 8A) exhibited differential mRNA expression between the two groups (> 2-fold, P < 0.05). We then analyzed the disease and function to determine the enriched gene sets. Genes involved in cell death, viability and survival, cell movement and invasion, cellular assembly and organization, DNA replication, recombination and repair, and cellular function and maintenance were significantly enriched (P < 0.05, Figure 8B). Classical pathway analysis revealed that, in addition to the previously reported p53 and cell cycle checkpoint control pathways, the Wnt/β-catenin signalling pathway was altered following RFWD3 silencing (P < 0.05, Figure 8C and D). Next, we used IPA to analyze the knowledge-based interactome associated with the regulation of these pathways (Figure 8E). Some genes involved in tumorigenesis were shown to be regulated by RFWD3, including CREBBP, KREMEN-1, LRP6, JUN, WNT5A, and WNT7B. Finally, we tested the protein expression between shCtrl and shRNAi and RFWD3-KD-OE vs RFWD3-KD-OE-NC groups and found that the expression of CREBBP, WNT5A, JUN, WNT7B, and LRP6 were decreased, and the expression of KREMEN-1 was increased in the shRNAi and RFWD3-KD-OE-NC groups than that in shCtrl and RFWD3-KD-OE groups (Figure 8F-I). These results indicate that the Wnt/β-catenin signalling pathway is involved in the RFWD3-mediated promotion of HCC tumorigenesis.
In this study, we demonstrated reduced growth and proliferation of HCC cells in the shRNAi group. Subsequent evaluation of cell cycle progression revealed disrupted cell cycle progression at the G2/M phase, whereas the FCM analysis showed a higher rate of apoptosis in the shRNAi group than in the ShCtrl group. These results confirmed that RFWD3 silencing caused both cell cycle arrest at the G2/M phase and apoptosis. Furthermore, the in vitro experiments revealed that shCtrl cells had better healing abilities than shRNAi cells. In the subsequent transwell experiments, we also found that the shRNAi cells had about half the metastatic capacity of the shCtrl cells. These results were validated in vivo using mice models, which showed that the tumor volumes in mice injected subcutaneously with shRNAi cells were significantly smaller than those in mice injected with shCtrl cells. Furthermore, the pulmonary metastasis group revealed that lung metastasis was significantly reduced in mice injected with shRNAi cells than in those injected with shCtrl cells.
To rule out off-target effects with RFWD3-shRNAi, we performed a rescue experiment. After verifying the transfection efficiency and screening of stable cell lines, we carried out phenotype verification. Compared with the control group, the cell growth state was restored in the RFWD3-KD-OE group. It also showed a higher OD value in the MTT assay, decreased fraction of G2/M phase, and diminution gap of data compared with RFWD3-KD-OE-NC group. Furthermore, in the apoptosis experiments, the RFWD3-KD-OE group had a lower apoptosis rate than the RFWD3-KD-OE-NC group. The same results were observed in the transwell assays, and the migration and invasion abilities of the RFWD3-KD-OE group were recovered. These data revealed that RFWD3-KD-OE resumed BEL-7404 shRNAi cell invasion and metastasis and proved that the previous RFWD3-shRNAi experimental results were not due to off-target effects.
Subsequently, we proved that RFWD3 silencing plays a positive role in inhibiting the proliferation, invasion, and metastasis of HCC. Previous studies have reported that the essential role of RFWD3 in HR makes it important for cell survival, as it promotes the repair of DNA damage. Therefore, the increased apoptosis could be attributed to the unrepaired DNA damage upon RFWD3 loss. On the contrary, the migration and metastasis phenotypes could be explained by decreased viability due to increased RFWD3 function during DNA repair. In other words, this condition may be explained by a common theory: “There are more firefighters on the fire scene than ordinary people.” However, the questions like whether the increased expression of RFWD3 in HCC is a compensatory mechanism to deal with increased genomic instability or whether RFWD3 is regulated by a Janus-faced regulator that plays other roles in HCC, consequently promoting the progression of HCC remained unanswered.
To answer these questions, we explored the mechanism underlying HCC and identified the molecular signalling transduction pathways using microarray-based gene expression analyses and IPA. The raw data (Supplementary materials 1) were verified with the uniformity, integrity and reliability (Supplementary materials 2). Gene expression profiling revealed that RFWD3 silencing enriched the expression of genes involved in cellular function and maintenance, cell death, viability, movement, invasion and survival, and DNA replication, recombination, and repair (Supplementary materials 3). In terms of the gene pathways involved, in addition to the p53 and cell replication cycle pathways that were previously reported in the literature, we found that the Wnt/β-catenin signalling pathway also had a significant impact. Further examination of the knowledge-based interactome associated with the regulation of Wnt/β-catenin via IPA analysis demonstrated that CREMEN1, JUN, CREBBP, WNT5A, WNT7B, and LRP6, which have been implicated in the proliferation and metastasis of various cancers, were differentially regulated by RFWD3 (> 2-fold, P < 0.05).
Reportedly, Wnt/β-catenin signalling plays an important role in embryonic liver development and regeneration and preserves liver health. An aberrant Wnt signalling activity has been implicated in several human cancers[31,32]. In normal tissues, only a small amount of β-catenin is found in the cytoplasm and is quickly degraded by the UPS. It is bound by a destruction complex containing GSK3B, AXIN, and APC and then ubiquitinated by β-transducin repeat-containing protein and subsequently degraded by the proteasome[33]. In the traditional Wnt/β-catenin pathway in tumors, the destruction complex is disassembled by the phosphorylation of LRP6 and GSK3β, thereby preventing β-catenin degradation[34]. The accumulated β-catenin is transferred from the cytoplasm to the nucleus, where it competes with Groucho to form the β-catenin-TCF/LEF transcriptional complex. Finally, the Wnt signalling pathway is activated to regulate the transcription of target genes such as cyclin D1, c-myc, and MMP9 under the synergistic effect of auxiliary factors, thus regulating tumor progression[35]. KREMEN-1 is a transmembrane protein that contains a kringle domain. Studies have shown that it is a high-affinity receptor for DKK. KREMEN-1, DKK, and LRP5/6 form a ternary complex that inhibits Wnt/β-catenin signalling, thus inducing head defects in Xenopus embryos[36,37]. Furthermore, KREMEN-1 expression is prevalent in adult mammals but is often lacking in tumors. It has been shown that Wnt/β-catenin signalling maintains the low differentiation state of stem cells and undifferentiated cells, but Dickkopf-KREMEN blocks Wnt/β-catenin signalling causing cells to differentiate[38].
Collectively, the findings demonstrated that factors involved in Wnt/β-catenin signallings, such as CREBBP, WNT5A, JUN, WNT7B, and LRP6, were downregulated by RFWD3 silencing, and the expression of KREMEN-1 was increased, indicating that RFWD3 might influence the progression of HCC via the Wnt/β-catenin signalling pathway. Based on these findings, it can be inferred that RFWD3 affects the Wnt/β-catenin signalling pathway and therefore affects HCC tumorigenesis. However, the detailed regulatory mechanism is still unclear and should be explored in future studies.
In this study, we validated the expression and function of RFWD3 in HCC and demonstrated that RFWD3 expression might affect the proliferation and metastasis of HCC by regulating the Wnt/β-catenin signalling pathway. The findings suggest that RFWD3 could be a potential therapeutic target for HCC; however, it warrants further investigations to ascertain these possibilities.
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors with a high mortality rate. The molecular biology research of HCC can reveal the potential mechanism and provide direction for its comprehensive treatment.
The relationship between RING finger and WD repeat domain 3 (RFWD3) and the tumorigenesis process has been reported occasionally; however, the relationship between RFWD3 and HCC is still unclear.
We aimed to investigate the relationship between HCC and RFWD3 and explore the underlying molecular signalling transduction pathways.
We analyzed RFWD3 expression in HCC tissues and evaluated cell phenotypes such as proliferation, apoptosis, migration, and invasion using Lentivirus mediated RFWD3 knockdown. In addition, in vivo experiments were performed to observe tumor growth and metastasis. Next, the phenotype was verified using the lentiviral-mediated rescue of RFWD3 shRNA. Finally, we unraveled the regulatory network underlying HCC using microarray, bioinformatics, and western blot analyses.
RFWD3 expression levels were correlated with tumor size and TNM stage in clinical samples. RFWD3 silencing increased apoptosis, decreased growth, and inhibited migration in HCC cell lines and nude mice, which were resumed with RFWD3 shRNAi rescue. Subsequent experiments revealed that RFWD3 might influence the proliferation and metastasis of HCC via the Wnt/β-catenin signalling pathway.
This study shows that RFWD3 affects the Wnt/β-catenin signalling pathway and therefore affects HCC tumorigenesis.
The findings suggest that RFWD3 could be a potential therapeutic target for HCC. However, it warrants further investigations to ascertain these inferences.
We would like to thank Wen-Feng Song, Hao Chen, Liang-Jie Hong, Dan-Jing Guo, Rong Su, Hao-Hao Chen and Han-Wen Zhang for their technical assistance.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gupta T, India; Limaiem F, Tunisia A-Editor: Morozov S, Russia S-Editor: Fan JR L-Editor: A P-Editor: Zhang YL
| 1. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13213] [Article Influence: 1468.1] [Reference Citation Analysis (3)] |
| 2. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12667] [Cited by in RCA: 15314] [Article Influence: 3062.8] [Reference Citation Analysis (4)] |
| 3. | Li J, Qiu X, Guo W, Yan B, Zhang S. Prospective analysis of tiopronin in prevention of sorafenib and antiviral therapy inducing liver toxicity in advanced hepatitis B virus-related hepatocellular carcinoma. Med Oncol. 2015;32:238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 4. | Tsai CL, Hsu FM, Cheng JC. How to Improve Therapeutic Ratio in Radiotherapy of HCC. Liver Cancer. 2016;5:210-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.0] [Reference Citation Analysis (2)] |
| 5. | Kim KI, Chung HK, Park JH, Lee YJ, Kang JH. Alpha-fetoprotein-targeted reporter gene expression imaging in hepatocellular carcinoma. World J Gastroenterol. 2016;22:6127-6134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 17] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 6. | Trojan J, Zangos S, Schnitzbauer AA. Diagnostics and Treatment of Hepatocellular Carcinoma in 2016: Standards and Developments. Visc Med. 2016;32:116-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 7. | Tang ZY. Hepatocellular carcinoma--cause, treatment and metastasis. World J Gastroenterol. 2001;7:445-454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 296] [Cited by in RCA: 303] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 8. | Hu L, Lau SH, Tzang CH, Wen JM, Wang W, Xie D, Huang M, Wang Y, Wu MC, Huang JF, Zeng WF, Sham JS, Yang M, Guan XY. Association of Vimentin overexpression and hepatocellular carcinoma metastasis. Oncogene. 2004;23:298-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 182] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 9. | Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739-745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1222] [Cited by in RCA: 1284] [Article Influence: 75.5] [Reference Citation Analysis (0)] |
| 10. | Turnbull AP, Ioannidis S, Krajewski WW, Pinto-Fernandez A, Heride C, Martin ACL, Tonkin LM, Townsend EC, Buker SM, Lancia DR, Caravella JA, Toms AV, Charlton TM, Lahdenranta J, Wilker E, Follows BC, Evans NJ, Stead L, Alli C, Zarayskiy VV, Talbot AC, Buckmelter AJ, Wang M, McKinnon CL, Saab F, McGouran JF, Century H, Gersch M, Pittman MS, Marshall CG, Raynham TM, Simcox M, Stewart LMD, McLoughlin SB, Escobedo JA, Bair KW, Dinsmore CJ, Hammonds TR, Kim S, Urbé S, Clague MJ, Kessler BM, Komander D. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature. 2017;550:481-486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 357] [Article Influence: 44.6] [Reference Citation Analysis (0)] |
| 11. | Hou CC, Yang WX. New insights to the ubiquitin-proteasome pathway (UPP) mechanism during spermatogenesis. Mol Biol Rep. 2013;40:3213-3230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 66] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 12. | Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1898] [Cited by in RCA: 1972] [Article Influence: 89.6] [Reference Citation Analysis (0)] |
| 13. | Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616-627. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1417] [Cited by in RCA: 1352] [Article Influence: 79.5] [Reference Citation Analysis (0)] |
| 14. | Mu JJ, Wang Y, Luo H, Leng M, Zhang J, Yang T, Besusso D, Jung SY, Qin J. A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related (ATR) substrates identifies the ubiquitin-proteasome system as a regulator for DNA damage checkpoints. J Biol Chem. 2007;282:17330-17334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 143] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 15. | Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160-1166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2234] [Cited by in RCA: 2400] [Article Influence: 133.3] [Reference Citation Analysis (0)] |
| 16. | Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci U S A. 2007;104:10364-10369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 363] [Cited by in RCA: 343] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 17. | Fu X, Yucer N, Liu S, Li M, Yi P, Mu JJ, Yang T, Chu J, Jung SY, O'Malley BW, Gu W, Qin J, Wang Y. RFWD3-Mdm2 ubiquitin ligase complex positively regulates p53 stability in response to DNA damage. Proc Natl Acad Sci U S A. 2010;107:4579-4584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 63] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 18. | Elia AE, Wang DC, Willis NA, Boardman AP, Hajdu I, Adeyemi RO, Lowry E, Gygi SP, Scully R, Elledge SJ. RFWD3-Dependent Ubiquitination of RPA Regulates Repair at Stalled Replication Forks. Mol Cell. 2015;60:280-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 105] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 19. | Zhang W, Sidhu SS. Development of inhibitors in the ubiquitination cascade. FEBS Lett. 2014;588:356-367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 20. | Knies K, Inano S, Ramírez MJ, Ishiai M, Surrallés J, Takata M, Schindler D. Biallelic mutations in the ubiquitin ligase RFWD3 cause Fanconi anemia. J Clin Invest. 2017;127:3013-3027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 156] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 21. | Wolberger C. Mechanisms for regulating deubiquitinating enzymes. Protein Sci. 2014;23:344-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 22. | Gong Z, Chen J. E3 Ligase RFWD3 participates in replication checkpoint control. J Biol Chem. 2011;286:22308-22313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 23. | Sokratous K, Hadjisavvas A, Diamandis EP, Kyriacou K. The role of ubiquitin-binding domains in human pathophysiology. Crit Rev Clin Lab Sci. 2014;51:280-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 24. | Liu S, Chu J, Yucer N, Leng M, Wang SY, Chen BP, Hittelman WN, Wang Y. RING finger and WD repeat domain 3 (RFWD3) associates with replication protein A (RPA) and facilitates RPA-mediated DNA damage response. J Biol Chem. 2011;286:22314-22322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 63] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 25. | Feeney L, Muñoz IM, Lachaud C, Toth R, Appleton PL, Schindler D, Rouse J. RPA-Mediated Recruitment of the E3 Ligase RFWD3 Is Vital for Interstrand Crosslink Repair and Human Health. Mol Cell. 2017;66:610-621.e4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 59] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 26. | Zhang Y, Zhao X, Zhou Y, Wang M, Zhou G. Identification of an E3 Ligase-encoding gene RFWD3 in non-small cell lung cancer. Front Med. 2020;14:318-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 27. | Jia J, Yang Y, Yan T, Chen T, Lu G. Down-regulation of RFWD3 inhibits cancer cells proliferation and migration in gastric carcinoma. Gen Physiol Biophys. 2020;39:363-371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 28. | Xu F, Xiao Z, Fan L, Ruan G, Cheng Y, Tian Y, Chen M, Chen D, Wei Y. RFWD3 Participates in the Occurrence and Development of Colorectal Cancer via E2F1 Transcriptional Regulation of BIRC5. Front Cell Dev Biol. 2021;9:675356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 29. | Wang WJ, Jin WW, Pan J, Wang LB, Zhu RT, Li J, Zhang CX, Liang RP. The expression of ring finger and WD repeat domain 3 in hepatocellular carcinoma and its clinical relevance. J Hepatopancreat Sur. 2019;31:741-745. [DOI] [Full Text] |
| 30. | Rutgers M, van Pelt MJ, Dhert WJ, Creemers LB, Saris DB. Evaluation of histological scoring systems for tissue-engineered, repaired and osteoarthritic cartilage. Osteoarthritis Cartilage. 2010;18:12-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 176] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 31. | McLin VA, Rankin SA, Zorn AM. Repression of Wnt/beta-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development. 2007;134:2207-2217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 307] [Cited by in RCA: 259] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 32. | Brafman D, Willert K. Wnt/β-catenin signaling during early vertebrate neural development. Dev Neurobiol. 2017;77:1239-1259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 65] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 33. | Fatima S, Luk JM, Poon RT, Lee NP. Dysregulated expression of dickkopfs for potential detection of hepatocellular carcinoma. Expert Rev Mol Diagn. 2014;14:535-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 34. | Murillo-Garzón V, Kypta R. WNT signalling in prostate cancer. Nat Rev Urol. 2017;14:683-696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 261] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 35. | Lee HC, Kim M, Wands JR. Wnt/Frizzled signaling in hepatocellular carcinoma. Front Biosci. 2006;11:1901-1915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 121] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 36. | Nakamura T, Aoki S, Kitajima K, Takahashi T, Matsumoto K, Nakamura T. Molecular cloning and characterization of Kremen, a novel kringle-containing transmembrane protein. Biochim Biophys Acta. 2001;1518:63-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 40] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 37. | Mishra SK, Funair L, Cressley A, Gittes GK, Burns RC. High-affinity Dkk1 receptor Kremen1 is internalized by clathrin-mediated endocytosis. PLoS One. 2012;7:e52190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 38. | Nakamura T, Nakamura T, Matsumoto K. The functions and possible significance of Kremen as the gatekeeper of Wnt signalling in development and pathology. J Cell Mol Med. 2008;12:391-408. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 2.7] [Reference Citation Analysis (0)] |