Published online Jul 7, 2019. doi: 10.3748/wjg.v25.i25.3218
Peer-review started: January 25, 2019
First decision: February 26, 2019
Revised: May 13, 2019
Accepted: May 31, 2019
Article in press: June 1, 2019
Published online: July 7, 2019
Processing time: 162 Days and 21.5 Hours
Several studies have demonstrated a correlation between esophageal cancer (EC) and perturbed urinary metabolomic profiles, but none has described the correlation between urine metabolite profiles and those of the tumor and adjacent esophageal mucosa in the same patient.
To investigate how urinary metabolic phenotypes were linked to the changes in the biochemical landscape of esophageal tumors.
Nuclear magnetic resonance-based metabolomics were applied to esophageal tumor tissues and adjacent normal mucosal tissues alongside patient-matched urine samples.
Analysis revealed that specific metabolite changes overlapped across both metrics, including glucose, glutamate, citrate, glycine, creatinine and taurine, indicating that the networks for metabolic pathway perturbations in EC, potentially involved in but not limited to disruption of fatty acid metabolism, glucose and glycolytic metabolism, tricarboxylic acid cycle and glutaminolysis. Additionally, changes in most urinary biomarkers correlated with changes in biomarker candidates in EC tissues, implying enhanced energy production for rapid cell proliferation.
Overall, these associations provide evidence for distinct metabolic signatures and pathway disturbances between the tumor tissues and urine of EC patients, and changes in urinary metabolic signature could reflect reprogramming of the aforementioned metabolic pathways in EC tissues. Further investigation is needed to validate these initial findings using larger samples and to establish the underlying mechanism of EC progression.
Core tip: Our research provides evidence for distinct metabolic signatures and metabolic pathway disturbances between the tumor tissues and urine of esophageal cancer patients, and changes in the urinary metabolic signature could reflect reprogramming of aforementioned metabolic pathways in esophageal tumor tissues.
- Citation: Liang JH, Lin Y, Ouyang T, Tang W, Huang Y, Ye W, Zhao JY, Wang ZN, Ma CC. Nuclear magnetic resonance-based metabolomics and metabolic pathway networks from patient-matched esophageal carcinoma, adjacent noncancerous tissues and urine. World J Gastroenterol 2019; 25(25): 3218-3230
- URL: https://www.wjgnet.com/1007-9327/full/v25/i25/3218.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i25.3218
Esophageal cancer (EC) is the eighth most common type of malignant tumor and the sixth leading cause of cancer-related death worldwide[1]. Identifying cancer-related biomarkers of EC is essential for its diagnosis and therapeutic intervention in the early stage, which in turn will significantly increase patient survival. While there are a few diagnostic/screening techniques, such as upper gastrointestinal endoscopy, endo-scopy-based balloon cytology, and serum carcinoembryonic antigen (commonly known as CEA) test, high-throughput and sensitive molecular tools are required to elucidate specific disease biomarkers for optimal disease management. Metabolomics, in which the global changes of small molecular weight metabolites in a given biological specimen are investigated, has considerable potential to identify useful biomarkers, thereby stratifying subjects into disease or nondiseased categories[2,3]. Proton nuclear magnetic resonance (1H-NMR) spectroscopy is a well-established, robust, noninvasive and reproducible method for quantifying metabolic profiles[4], and it has several advantages over other analytical techniques, such as liquid ch-romatography mass spectroscopy and gas chromatography mass spectroscopy, including nondestructive analysis of samples, minimal sample preparation, and the ability to detect multiple metabolites within a single experiment[5-8]. NMR data com-bined with multivariate statistical analysis, such as orthogonal partial least squares discriminant analysis (OPLS-DA), which can be utilized for disease cla-ssification (through the use of score plots) and biomarker detection (through the use of loading plots), allow for the identification of potential biomarkers in biological specimens and improve the ability to identify specific metabolic pathways and networks associated with the disease process.
Urine is a biological fluid commonly used by NMR-based metabolomics[9,10] because it is easily collected in large volumes with noninvasive procedures and may provide substantial diagnostic information for many cancer types[7,11-13]. Biomarkers in urine may be derived from cell apoptosis, glomerular filtration of blood plasma, cell slo-ughing, epithelial cell secretion of exosomes, and other processes[14]. Several NMR spectroscopy-based metabolomic studies have reported that metabolite compositions of urine samples from EC patients differ from those of healthy controls (HCs)[15,16]. However, little is known about the systemic mechanistic link between esophageal tissues and urine, owing to the invasiveness of tissue sampling and sensitivity of the urinary metabolome to factors such as environment, food and genetic composition.
The aim of this research was to profile parallel metabolites of EC tissues and ad-jacent noncancerous tissues alongside urine samples from the same patients, to investigate how urinary metabolomic phenotypes associate with tumor tissue, and to identify specific disturbed metabolic pathways in esophageal tissues and urine. Such information would be vital to bridge the gap between the systemic metabolic characteristics of EC tissues and the corresponding samples of urine and may help advance the utility of urinary-based metabolites as relevant indicators of EC tissue microenvironment.
This study was approved by the Ethics Committee of Shantou University Medical College. Written informed consent was obtained from each subject prior to par-ticipation. Early-morning midstream urine samples were collected preoperatively from 41 EC patients and 40 HCs between August 2015 and October 2016 at the Second Affiliated Hospital of Shantou University Medical College. EC patients were diagnosed by microscopy, biopsy, or surgical resection, and the disease stage was determined according to the American Joint Committee on Cancer (AJCC) staging system for esophageal tumors: Stage I/II, 15 patients; stage III, 11 patients; stage IV, 15 patients. Each urine sample was mixed with 50 μL sodium azide preservative and stored at −80 °C until further analysis. Patient-matched EC tissues and their cor-responding adjacent noncancerous tissues (~5 cm away from the tumor margin) were collected from 20 EC patients and immediately stored at −80 °C until NMR analysis. Exclusion criteria for all participants included use of antibiotics, nonsteroidal anti-inflammatory drugs, statins, or probiotics within 2 mo of study participation. Additional exclusion criteria for EC patients included chemotherapy or radiotherapy prior to surgery. The demographic and clinical characteristics of the EC patients and con-trols are summarized in Table 1.
| EC | HC | χ2 | P value | |
| Subjects, n | 41 | 40 | ||
| Age (median, range), yr | 60, 39-77 | 59, 28-78 | ||
| Sex | 6.77 | 0.12 | ||
| Male | 31 | 19 | ||
| Female | 10 | 21 | ||
| Cancer stage | ||||
| Stage I/II | 15 | - | ||
| Stage III | 11 | - | ||
| Stage IV | 15 | - | ||
| CEA, ng/mL | ||||
| Positive | 2 | - | ||
| Negative | 31 | - | ||
| Not measured | 8 | - | ||
| CA 19-9, U/mL | ||||
| Positive | 2 | - | ||
| Negative | 18 | - | ||
| Not measured | 21 | - | ||
| Location | ||||
| Cervical | 1 | - | ||
| Upper thoracic | 5 | - | ||
| Middle thoracic | 24 | - | ||
| Lower thoracic | 11 | - | ||
| Symptoms | ||||
| Dysphagia | 40 | - | ||
| Gastroesophageal reflux | 27 | - |
Frozen urine samples were thawed at room temperature and mixed to suspend any settled precipitate. After adding 300 μL PBS/D2O buffer (0.1 M, pH 7.4) to 600 μL of each urine sample, the mixture was homogenized by vortexing for 60 s and then centrifuged at 8000 rpm for 10 min. Subsequently, a volume of 500 μL of the sup-ernatant was transferred into an Eppendorf vial, to which 50 μL of a stock solution of sodium (3-trimethylsilyl)-2, 2, 3, 3-tetradeuteriopropionate (TSP)/D2O was added, using a chemical shift reference (0.0 ppm) for spectral alignment. Finally, the resulting mixture was centrifuged at 10000 rpm for 5 min, and 500 μL of the prepared sample was transferred into a 5-mm high-resolution NMR tube for 1H-NMR analysis.
The frozen tissue samples weighed ~300 mg and were thawed and cut into small pieces at room temperature. After adding 1.8 mL mixture containing 0.6 mL distilled water (2 mL/g tissue) and 1.2 mL methanol (4 mL/g tissue), the resulting mixture was homogenized at 16000 rpm for 80 s. After homogenization, the mixture was added to chloroform (4 mL/g tissue) and distilled water (4 mL/g tissue) and vortexed for 60 s. The suspension was then left on ice for 15 min and centrifuged at 2000 rpm for 5 min to facilitate separation of the liquid layers. The resulting supernatant was evaporated under a stream of nitrogen and then dried under vacuum for a minimum of 18 h. Subsequently, the lyophilized powder was redissolved with 550 μL PBS/D2O buffer (0.1 mol/L, pH 7.4), to which 50 μL of a stock solution of TSP/D2O was added. After homogenization and centrifugation at 10000 rpm for 5 min, 500 μL supernatant was transferred into a 5-mm high-resolution NMR tube for 1H-NMR analysis.
All samples were detected by a Bruker AVII 400 MHz NMR spectrometer (Bruker Biospin, Germany) operating at a 1H frequency of 400.13 MHz. Magnetic field homogeneity was optimized by gradient or manual shimming prior to acquisition. The temperature was maintained at 298 K and lock performed on the D2O signal. 1H NMR spectra of urine samples were obtained using a 1D nuclear Overhauser enhancement spectroscopy pulse sequence [RD-90°-t1-90°-tm-90°- ACQ], with the following acquisition parameters: Recycle delay (RD) 1.5 s; t1 3 µs; mixing time, tm 100 ms; 90° pulse width 7.3 μs; number of scans (NS) 256; number of points, TD 32768; spectral width (SW) 8012.82 Hz; acquisition time (AQ) 2.04 s. Water suppression was achieved by irradiation of the water peak during RD and tm. Esophageal tissue 1H NMR spectra were recorded using a standard (1D) Carr–Purcell–Meiboom–Gill pulse sequence with the following acquisition parameters: number of dummy scans 4; RD 70 ms; 90° pulse width 10 μs; NS 64; TD 65536; SW 8012 Hz; AQ 4.09 s.
All free induction decays from 1D 1H-NMR of the tissues and urine samples were multiplied by a 0.3 Hz exponential line broadening prior to Fourier transformation. 1H-NMR spectra were then corrected for phase and baseline distortion and calibrated to TSP at 0.0 ppm by using MestReNova software (version 8.1.0, Mestrelab Research, Santiagode Compostella, Spain). To reduce the complexity of the NMR data, the spectral range from 0.8 to 9.0 ppm was segmented into buckets with the equal width of 0.004 ppm. The region of 5.5–4.5 ppm was discarded to eliminate imperfect water suppression. Each bucket was internally normalized to the total integral of the spectrum prior to pattern recognition analysis, to eliminate the dilution or bulk mass differences among samples due to the different sample weight.
To maximize class discrimination between EC patient and HC urine samples, as well as between EC tissues and their corresponding noncancerous tissues, multivariate data analysis was applied to the 1H-NMR spectral data according to previously published and accepted methods[6,7]. The normalized NMR spectral data sets were unit variance scaled and analyzed using the SIMCA-P+ program (version 14.1, Umetrics AB; Umea, Sweden). OPLS-DA was applied to the analysis of 1H-NMR spectral data to optimize the separation between experimental groups. The model quality was evaluated with the R2Y and Q2 values, reflecting the explained fraction of variance and the model predictability. R2Y scores ranged between 0 and 1 and Q2 scores between negative and 1, where a R2Y score of 1 demonstrated that the model explained 100% of variance, and a Q2 score closer to 1 indicated higher reliability of the prediction in the cross-validation procedure. Validation of the OPLS-DA model was also performed by means of a permutation test (400 times). The R2Y in the permutated plot described how well the data fitted with the derived model, whereas Q2 described the predictive ability of the derived model (Q2 > 0.5 considered as good and Q2 > 0.9 as excellent). To further evaluate the predictive power of the robust OPLS-DA model, 80% of samples were applied to construct a model, which was used to predict the remaining 20% of samples. The variable importance in the projection (VIP) values of all peaks from OPLS-DA models was taken as a coefficient for peak selection. The VIP value was represented by a unitless number, where higher values suggested greater dis-criminatory power of the metabolite. Those variables with VIP > 1 were considered potential biomarker candidates for group discrimination.
The relative concentrations of those metabolites with VIP > 1 were calculated by inte-grating the signals in the spectra. Statistical significance was assessed using the Mann–Whitney U test, and P < 0.05 was considered statistically significant. The relative concentrations of those metabolites with VIP > 1 were calculated by integrating the signals in the spectra, and data are presented as the mean fold di-fference in EC metabolite abundance compared to controls. To further evaluate the diagnostic power of the potential biomarkers whose levels significantly differed between experimental groups, receiver operating characteristic (ROC) analysis in SPSS version 16.0 was performed, and the optimal area under the ROC curve (AUROC), specificity and sensitivity of the metabolites were calculated, where AUROC > 0.8 indicated excellent diagnostic ability. Pearson correlation analysis was used to assess the association of biomarker candidates between urine and tumor tissues of EC patients with an OPLS-DA model using a correlation coefficient cut-off of |r| and correlation significance of P < 0.05. Correlation coefficients ranged from 1.0 (maximum positive correlation) to −1.0 (maximum anticorrelation), with a value of 0 representing no correlation. Correspondingly, |r| > 0.44 (calculated for the sample size of 20) was considered to be a statistically significant relationship between the two metabolites.
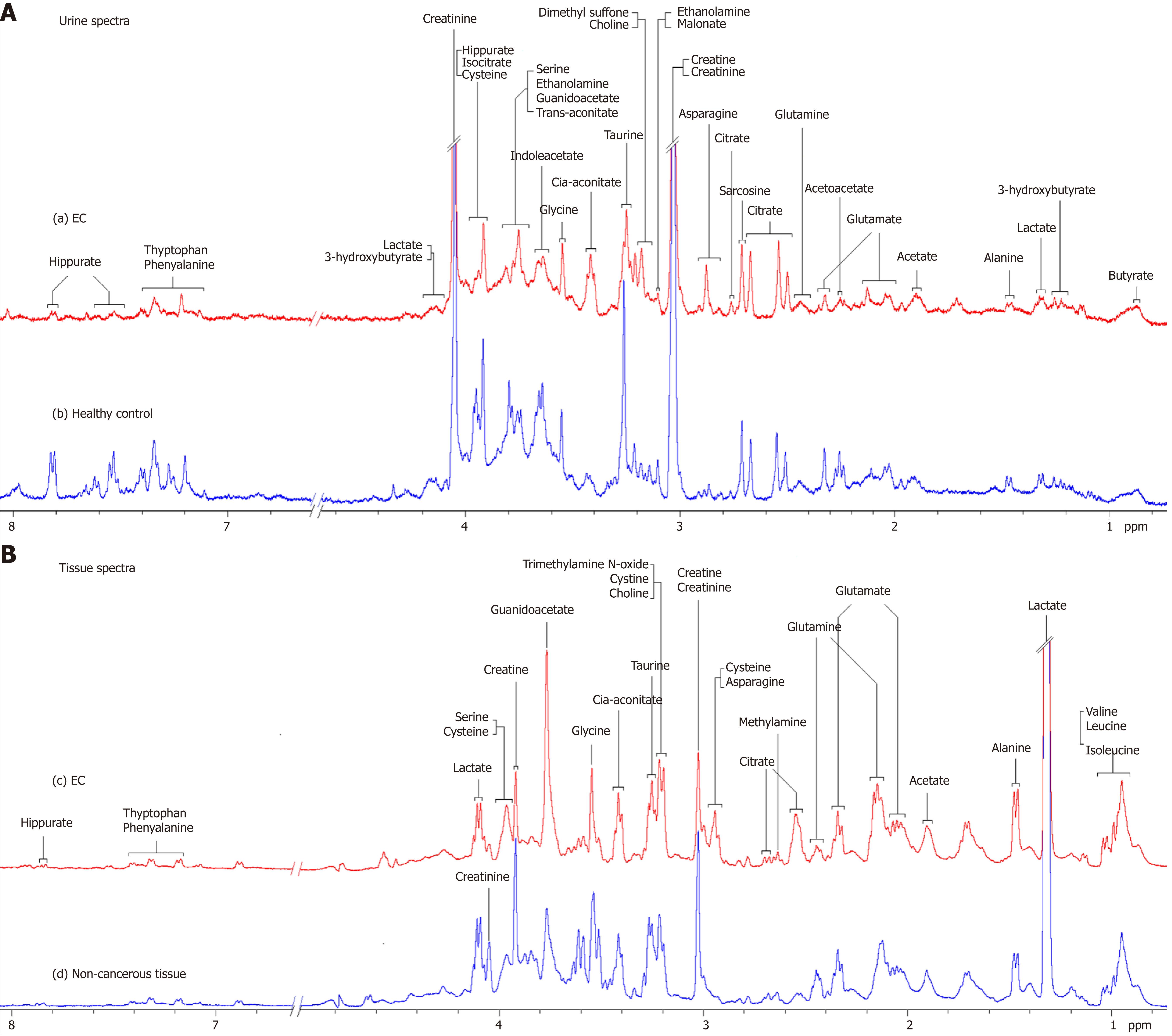
Representative 1D 1H-NMR spectra of urine specimens acquired from EC patients, HCs and patient-matched esophageal tissue extracts are shown in Figure 1A and B. The standard 1D spectrum gave an overview of all metabolites. The major metabolites in the spectra were identified according to previous studies[17,18] and the Human Metabolome Database (http://www.hmdb.ca/). In all urine and esophageal tissue spectra, the aliphatic region at 0.8–4.2 ppm included numerous signals from the following water-soluble metabolites: Glutamate, glutamine, acetoacetate, citrate, cisaconitate, choline, creatine, creatinine and glycine, which are known to be involved in many biochemical processes, especially in energy metabolism.
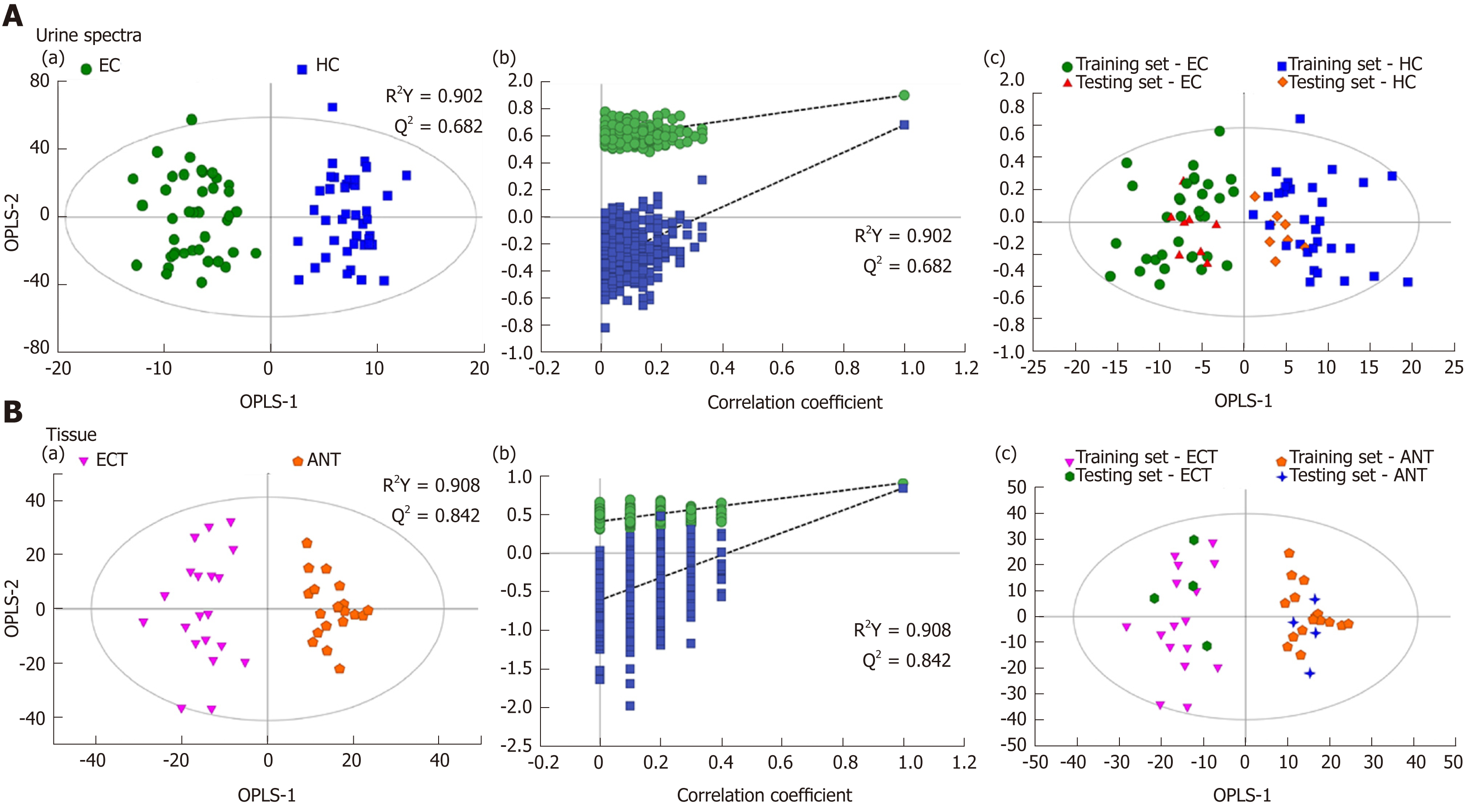
Good discrimination between EC patients and HCs was achieved by an OPLS-DA scores plot generated from 1H-NMR spectra of urine specimens (Figure 2A-a). The predictive ability of the model was calculated by internal validation (R2Y = 0.902, Q2 = 0.682, CV-ANONA P < 0.01), suggesting that the model possessed a satisfactory fit with good predictive power. In order to evaluate the robustness of the OPLS-DA model described above, a random permutation test was performed 400 times to further evaluate the robustness of this model, as exhibited by the steep R2 and Q2 regression lines and difference between R2 and Q2 (R2Y = 0.886 and Q2 = 0.660), indicating that this was an excellent model suitable for data analysis (Figure 2A-b). To further assess the predictive ability of the model for unknown samples, we randomly selected 80% of urine samples (“training set”, 33 EC patients and 32 HCs) to construct an OPLS-DA model, which was then used to predict the remaining 20% of samples (“testing set”, 8 EC patients and 8 HCs). As can be seen in Figure 2A-c, the testing sets of EC patients and HCs were correctly located in their corresponding region of the training sets. Urine metabolites that met the following conditions were considered potential metabolite biomarkers for EC detection: Levels of metabolites with VIP > 1 and the presence of a significant difference (P < 0.01) between metabolite levels of EC patients and HCs according to the Mann–Whitney U test. A total of ten urine meta-bolites were found to be significantly changed in EC patients compared to HCs (Table 2), including higher levels of acetoacetate, cis-aconitate, citrate and glutamate, together with lower amounts of glycine, taurine, creatinine, ethanolamine, glucose and hippurate.
| Metabolite | Chemicalshift, ppm | Fold difference | AUC (95%CI) | Related metabolic pathways | |
| EC / HC | P value | ||||
| Acetoacetate | 2.25–2.28 | 1.55 ↑ | < 0.001 | 0.745 (0.636–0.836) | Fatty acid metabolism, TCA cycle |
| Glutamate | 2.04–2.06 | 1.22 ↑ | 0.002 | 0.702 (0.590–0.798) | Glutaminolysis, TCA cycle |
| Cis-aconitate | 3.40–3.46 | 1.30 ↑ | 0.001 | 0.719 (0.608–0.813) | TCA cycle, glyoxylate and dicarboxylate metabolism |
| Citrate | 2.48–2.54 2.64–2.66 | 1.27 ↑ | 0.002 | 0.643 (0.528–0.746) | TCA cycle |
| Hippurate | 7.51–7.56 7.60–7.65 7.78–7.84 | 0.45 ↓ | 0.002 | 0.702 (0.591–0.799) | Gut microflora metabolism |
| Ethanola-mine | 3.08–3.16 | 0.86 ↓ | 0.001 | 0.723 (0.613–0.817) | Fatty acid metabolism |
| Glycine | 3.54–3.55 | 0.85 ↓ | 0.003 | 0.633 (0.518–0.737) | Amino acid metabolism |
| Creatinine | 3.03–3.06 4.05–4.10 | 0.95 ↓ | < 0.001 | 0.790 (0.685–0.872) | Urea metabolism, Creatinine metabolism |
| Taurine | 3.26–3.28 | 0.53 ↓ | < 0.001 | 0.763 (0.655–0.850) | Amino acid metabolism |
| Glucose | 3.73–3.80 | 0.89 ↓ | 0.007 | 0.694 (0.581–0.792) | Glycolysis; TCA cycle |
Tissue profiles from EC tumor tissues and their corresponding adjacent noncancerous tissues were clearly separated using the OPLS-DA scores plot (Figure 2B-a). Model parameters of the 400 times permutation test generated R2Y = 0.908 and Q2 = 0.842 (Figure 2B-b). Moreover, the OPLS-DA model showed good performance for predicting the unknown samples (Figure 2B-c). Using these criteria (VIP > 1 and P < 0.05), the 15 metabolites listed in Table 3 were found to be significantly changed in EC tissues compared to their corresponding noncancerous tissues, including elevated levels of valine, leucine, alanine, acetate, citrate, succinate, choline and glutamate, as well as depleted levels of creatinine and creatine, glycine, threonine, taurine, glucose, and glutamine.
| Metabolite | Chemicalshift, ppm | Fold difference | AUC (95%CI) | Related metabolic pathways | |
| ECT / ANT | P value | ||||
| Valine | 0.96–0.99 1.02–1.04 | 1.63 ↑ | < 0.001 | 0.988 (0.889-1.000) | Amino acid metabolism |
| Leucine | 0.93–0.95 | 1.32 ↑ | < 0.001 | 0.852 (0.705-0.944) | Amino acid metabolism |
| Glutamate | 1.97–2.08 2.32–2.39 | 1.30 ↑ | < 0.001 | 0.930 (0.803-0.986) | Glutaminolysis, TCA cycle |
| Acetate | 1.88–1.93 | 1.33 ↑ | 0.001 | 0.790 (0.632-0.902) | SCFA metabolism |
| Alanine | 1.43–1.48 | 1.44 ↑ | < 0.001 | 0.920 (0.789-0.982) | Amino acid metabolism, Gluconeogenesis |
| Choline | 3.19–3.22 | 1.59 ↑ | < 0.001 | 0.980 (0.876-1.000) | Choline metabolism, Lipid metabolism |
| Succinate | 2.39–2.40 | 2.12 ↑ | 0.009 | 0.738 (0.575-0.864) | TCA cycle |
| Citrate | 2.68–2.71 | 1.42 ↑ | < 0.001 | 0.942 (0.820-0.991) | TCA cycle |
| Glucose | 3.37–3.44 3.50–3.52 | 0.72 ↓ | < 0.001 | 0.928 (0.800-0.985) | Glycolysis, TCA cycle |
| Creatinine | 3.02–3.03 4.04–4.06 | 0.64 ↓ | < 0.001 | 0.963 (0.849-0.997) | Urea metabolism, Creatine metabolism |
| Glycine | 3.52–3.55 | 0.75 ↓ | < 0.001 | 0.850 (0.702-0.943) | Amino acid metabolism |
| Threonine | 3.58–3.62 | 0.60 ↓ | < 0.001 | 0.933 (0.806-0.987) | Amino acid metabolism |
| Creatine | 3.90–3.94 | 0.78 ↓ | < 0.001 | 0.917 (0.786-0.981) | Urea metabolism, Creatine metabolism |
| Glutamine | 2.42–2.48 | 0.77 ↓ | < 0.001 | 0.895 (0.757-0.969) | Glutaminolysis, TCA cycle |
| Taurine | 3.24–3.28 3.33–3.34 | 0.78 ↓ | < 0.001 | 0.878 (0.735-0.960) | Amino acid metabolism |
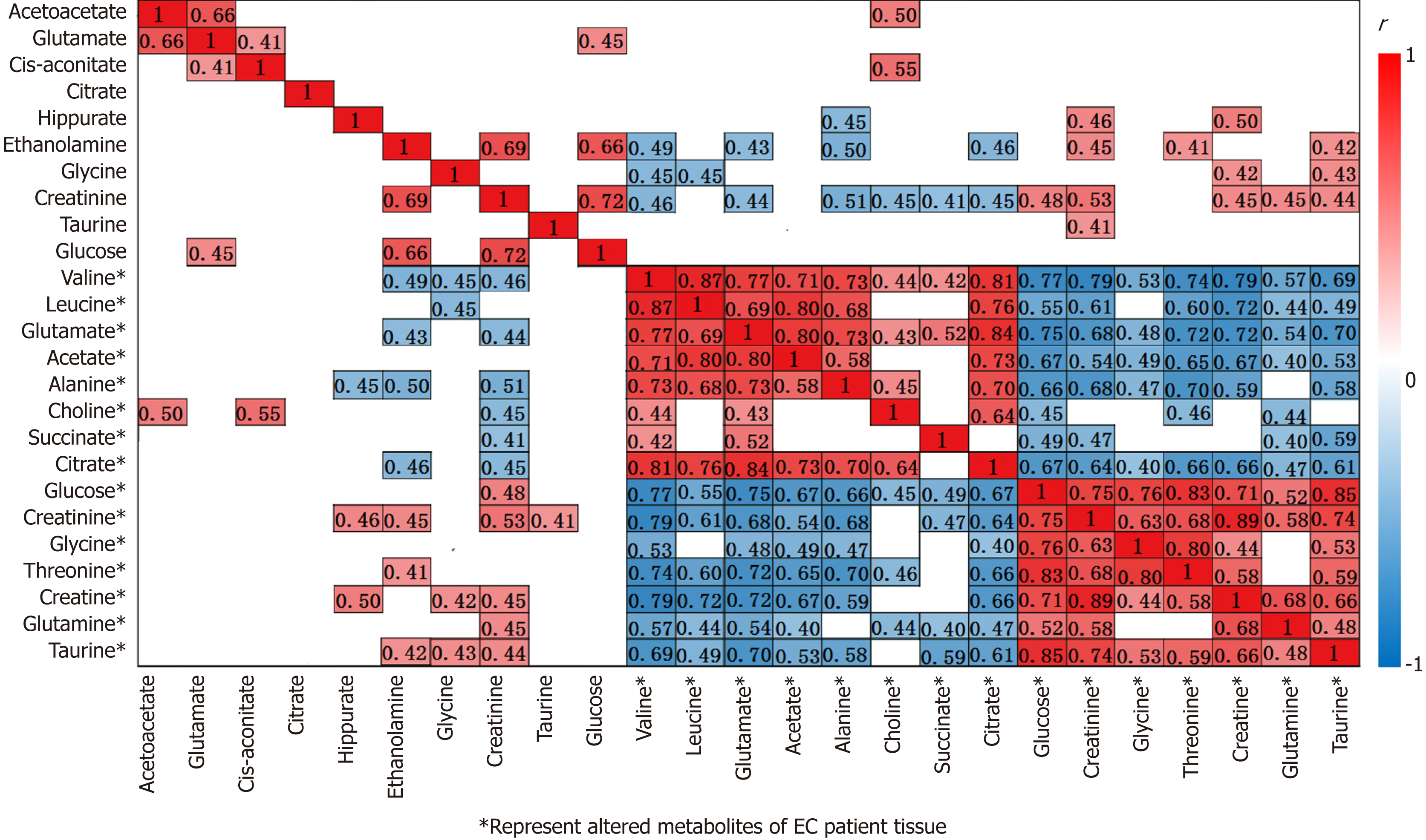
Our parallel investigations revealed a few distinct and overlapping discriminatory metabolites between cancer tissues and urine of EC patients, including decreased levels of glucose, glycine, creatinine and taurine, together with increased levels of glutamate and citrate, as compared to their respective controls. Metabolic profiling associations between potential urine and tissue biomarkers were further analyzed, plotted as correlation heat maps color-coded by the strength of Spearman correlation coefficients (Figure 3). Changes in most potential urinary biomarkers (except for glutamate, citrate and glucose) were found to be correlated with changes in most biomarker candidates in EC tissues (except for glycine and acetate) (|r| > 0.44, P < 0.05).
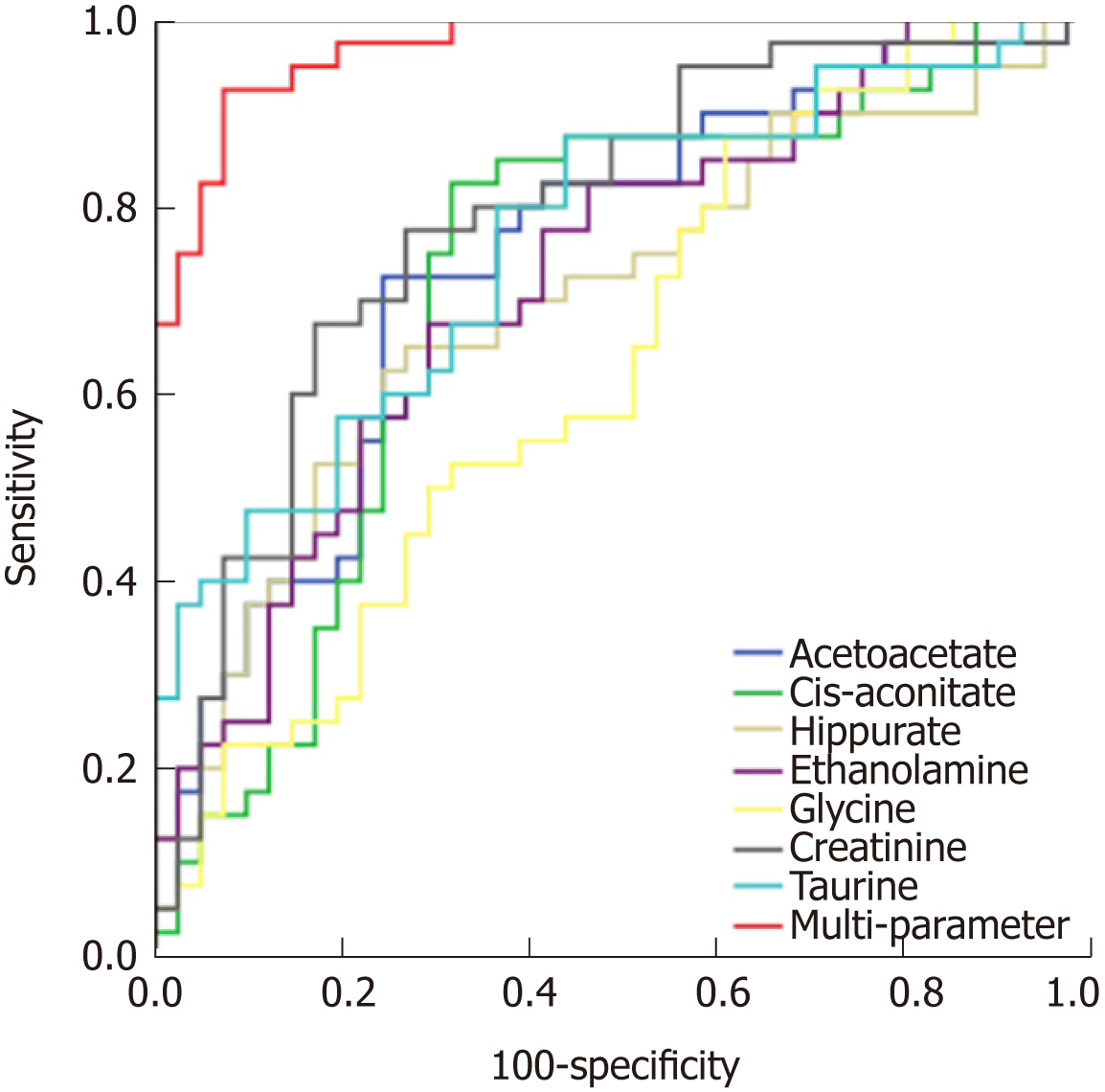
Given that urinary glutamate, citrate and glucose in EC patients did not show cor-relation with esophageal tissue biomarkers, a panel of urinary metabolite markers composed of acetoacetate, cis-aconitate, hippurate, ethanolamine, glycine, creatinine and taurine was selected to compare their diagnostic performance for distinguishing EC patients from HCs. ROC analysis of the comparison of single urinary biomarkers and their combination showed that combined metabolites of the aforementioned metabolites had better diagnostic performance than any single metabolite alone in discriminating EC patients from HCs, with sensitivity, specificity and area under the curve values of 92.68%, 92.50% and 0.971, respectively (Figure 4).
The current study performed parallel investigations of EC tissues and adjacent noncancerous mucosal tissues alongside patient-matched urine samples to investigate how changes of the urine metabolome were linked to the changes of EC tissues metabolic phenotypes. Our study showed significant metabolic alterations in both urine and tumor tissues of EC patients compared to their respective HCs. Correlative analysis of the altered metabolites across both matrices revealed a few distinct and overlapping discriminatory metabolites, such as glucose, glycine, creatinine and taurine (decreased levels) together with glutamate and citrate (increased amounts), suggesting that EC is associated with the following dysregulated metabolic pathway perturbations, including but not limited to fatty acid metabolism, glucose and glycolytic activity, tricarboxylic acid (TCA) cycle and glutaminolysis. Metabolic profiling correlations between esophageal tissues and urine showed that most potential urine biomarkers were correlated with most of the discriminating meta-bolites in EC tissues, indicating that changes in urine metabolic signature could reflect reprogramming of metabolic pathways in tumor tissues, highlighting the significance of the distinct urinary metabolic profiles as potential novel and nonin-vasive indicators for EC detection.
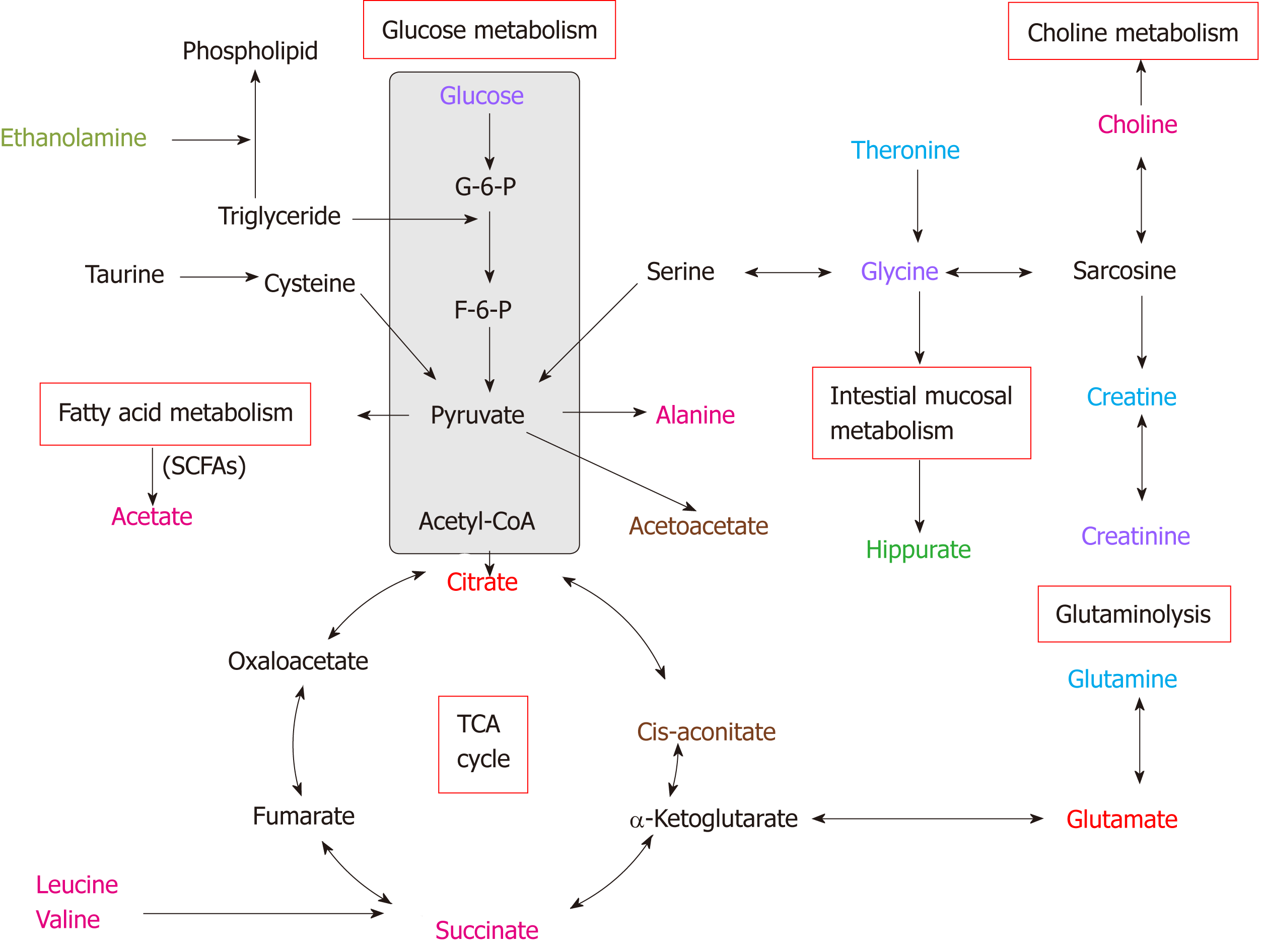
Our NMR-based metabolomic findings identified distinct disturbances occurring in both tissues and urine of EC patients compared to their respective controls (Figure 5). Tumor-microenvironment cooperation may occur in cancer cells, which exhibit a high rate of anabolic metabolism, by which they take up large amounts of nutrients to fuel the TCA cycle and oxidative phosphorylation. Therefore, in order to meet the increased demands of proliferation, tumor cells display changes in energy metabolism and nutrient uptake pathways[19]. In general, tumor tissue has depleted glucose and increased citrate and succinate, the TCA intermediates, reflecting high TCA cycle activity to maintain tumor promotion[8,20]. Reduced glucose and increased citrate levels were evident in EC patient urine, further indicating enhanced glycolysis under hy-poxic conditions required for rapid cancer cell proliferation[21,22]. Acetate, a source for lipid and myelin synthesis[23], is derived from acetyl-CoA via the deacetylation of N-acetylaspartate. The observed elevation of acetate in EC tissues might result from an increase in fatty acid metabolism, and this observation is supported by elevation of alanine derived from metabolism of pyruvate, indicating activation of glycolysis to provide higher energy needs[24]. Glutamine is also regarded as important for energy production in proliferating cells, and it donates nitrogen for nucleotide synthesis, resulting in the formation of glutamate (glutaminolysis). The latter can be converted to α-ketoglutarate to increase transit through the TCA cycle, providing sustainable energy required for rapid cell proliferation[25]. Therefore, increased glutamate along with depleted glutamine in EC tissue could suggest changes in glutaminolytic activity for EC development. The increased glutamate in EC urine observed in this study could also suggest a high energy demand in proliferating cells due to augmented glutaminolysis[26]. Increased leucine and valine in tissues and increased acetoacetate in urine were in agreement with the ingested nutrients that fuel the TCA cycle to support cell proliferation. Similarly, cell growth and proliferation need amino acids to generate proteins required for cancer cell synthesis[27], therefore leading to decreased levels of glycine and threonine in EC pa-tients. Depleted creatine/creatinine levels in tumor tissues have been related to altered energy transfer processes and may reflect increased activity of creatine kinase, which has been previously reported to be lower in lung tumors compared to normal adjacent tissues[28]. Besides, creatinine levels in EC patient urine samples were significantly decreased compared to HCs, which has also been reported to be lower in urine samples from colorectal cancer patients[7]. The observed depletion of taurine in both tissues and urine of EC patients suggest a disruption in taurine metabolism and diffusion of gut microbes associated with EC tumors[29]. Ethanolamine is an important fatty acid for cellular membranes[30], and its decreased levels in EC urine could suggest increased consumption for biosynthesis of cellular membranes and indicate activation of fatty acid metabolism. The observed depletion of hippurate in EC urine was likely the result of gut microbiome per-turbation associated with EC tumorigenesis. Hippurate is metabolized from benzoic acid, which is metabolized from the dietary polyphenol 3-hydroxyphenyl propionic acid by the gut microflora[31]. Choline is one of the major cell membrane phos-pholipids[32] and is overexpressed and highly active in tumor tissues and cell lines. We observed increased choline levels in EC tissues, which was consistent with previous reports[7], indicating that choline could be a viable tissue biomarker as-sociated with tumor promotion. However, this biomarker of EC tumor tissue was not completely detectable at the end of metabolism of urine biomarkers in this study.
In addition to specific metabolite differences between tumor tissues and urine in EC patients and HCs, we also evaluated the relationships between the metabolic net-works in both tissues and urine. Decreased metabolite levels of glucose, glycine, creatinine and taurine, as well as increased citrate and glutamate in EC tissues, were also detectable in the urine of EC patients. These distinct and overlapping metabolites may reflect tumor cell shedding and represent metabolic pathway aberrations across both matrices. This potentially reveals linkages to disturbances of fatty acid me-tabolism, glucose and glycolytic activity, TCA cycle and glutaminolysis associated with tumor proliferation. Correlative analysis of metabolic profiling between EC tissues and urine showed that changes in most potential urinary biomarkers were correlated with changes in most biomarker candidates in EC tissues, implying enhanced energy production required for rapid cell proliferation. Creatinine was found to be the most sensitive predictor of EC in urine metabolite, with an AUC of 0.790. Overall, these associations provide evidence of distinct metabolic signatures and pathway disturbances across both matrices, and changes in urinary metabolic signature could reflect the EC tissue microenvironment.
In conclusion, our parallel investigations of EC patients through 1H-NMR meta-bolomics revealed a great number of altered metabolites and metabolic pathway networks in EC patient urine and tumor tissues compared with HCs. We identified a few overlapping discriminatory metabolites across both matrices, derived from fatty acid metabolism (taurine and glycine), as well as metabolites (e.g., glucose, glutamate, citrate and creatinine) involved in glucose and glycolytic metabolism, the TCA cycle and glutaminolysis. Correlative analysis of metabolic profiling across tumor tissues and urine in EC patients showed that changes in most potential urinary biomarkers were correlated with changes in most candidate biomarkers in EC tissues, implying enhanced energy production required for rapid cell proliferation. In summary, these associations provide clear evidence of different metabolic signatures and metabolic pathway disturbances between EC tissues and urine, and changes in urinary meta-bolic signatures could reflect the EC tissue microenvironment. Our study highlighted the significance of the distinct urinary metabolic profile as a potential noninvasive indicator of EC detection. Further investigation is needed to validate these initial findings using larger samples and to establish the mechanism underlying EC progression.
A large number of studies have revealed changes of urinary metabolites between esophageal cancer (EC) and healthy controls (HCs), and some studies have demonstrated a correlation between EC and perturbed urinary metabolomic profiles.
However, none of the previous studies has described the correlation between urine metabolite profiles and those of the tumor and adjacent colonic mucosa in the same patient. Our study revealed a significant number of altered metabolites and metabolic pathway networks in EC patient urine and tumor tissues compared with HCs.
Our work is the first parallel investigation of esophageal tumor tissues and adjacent normal mucosal tissues alongside patient-matched urine samples to investigate how urinary metabolic phenotypes were linked to changes in the biochemical landscape of esophageal tumors.
All samples were detected by a Bruker AVII 400 MHz nuclear magnetic resonance spectrometer, and all spectral data were applied to pattern recognition analysis and cross-validation by SIMCA-P software. Then, statistical significance was assessed using the Mann-Whitney U test and receiver operating characteristic analysis to calculate biomarker metabolites. Finally, we employed Pearson Correlation Analysis to assess the associations of biomarker candidates between urine and tumor tissues of EC patients.
Our study revealed metabolite changes that overlapped across both metrics, including glucose, glutamate, citrate, glycine, creatinine and taurine, indicating the networks for metabolic pathway perturbations in EC. Additionally, changes in most urinary biomarkers were correlated with changes in biomarker candidates in EC tissues.
Our research is the first parallel investigation to investigate how urinary metabolic phenotypes were linked to the changes in the biochemical landscape of esophageal tumors. Our study showed significant metabolic alterations in both urine and tumor tissues of EC patients compared to their respective HCs. Our research revealed a few distinct and overlapping discri-minatory metabolites, suggesting that EC is associated with the following dysregulated metabolic pathway perturbations. Furthermore, the metabolic profiling correlations between esophageal tissues and urine showed that most urine potential biomarkers were correlated with most of the discriminating metabolites in EC tissues, indicating that changes in the urine metabolic signature could reflect reprogramming of metabolic pathways in tumor tissue, high-lighting the significance of the distinct urinary metabolic profiles as potential novel and noni-nvasive indicators for EC detection.
With experiences in our study, we realized that many metabolites have associations in samples of cancer patients. In our same group, we are now investigating the serum samples of EC pa-tients to see whether the same pattern of serum levels of amino acids can be found in EC patients.
The authors thank Ju-Rong Yang for kindly providing us with the NMR experimental setting and Dr. Hong-Jun Luo for handling of tissue samples.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Hashimoto N, Luyer MDP S-Editor: Cui LJ L-Editor: Filipodia E-Editor: Ma YJ
| 1. | Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20108] [Cited by in RCA: 20513] [Article Influence: 2051.3] [Reference Citation Analysis (20)] |
| 2. | Lindon JC, Nicholson JK, Holmes E, Everett JR. Metabonomics: metabolic processes studied by NMR spectroscopy of biofluids. Magnetic Resonance An Ed J. 2015;12:289-320. [DOI] [Full Text] |
| 3. | Griffin JL, Atherton H, Shockcor J, Atzori L. Metabolomics as a tool for cardiac research. Nat Rev Cardiol. 2011;8:630-643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 199] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 4. | Lin Y, Stephenson MC, Xin L, Napolitano A, Morris PG. Investigating the metabolic changes due to visual stimulation using functional proton magnetic resonance spectroscopy at 7 T. J Cereb Blood Flow Metab. 2012;32:1484-1495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 131] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 5. | Duarte IF, Gil AM. Metabolic signatures of cancer unveiled by NMR spectroscopy of human biofluids. Prog Nucl Magn Reson Spectrosc. 2012;62:51-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 6. | Lin Y, Ma C, Liu C, Wang Z, Yang J, Liu X, Shen Z, Wu R. NMR-based fecal metabolomics fingerprinting as predictors of earlier diagnosis in patients with colorectal cancer. Oncotarget. 2016;7:29454-29464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 97] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 7. | Wang Z, Lin Y, Liang J, Huang Y, Ma C, Liu X, Yang J. NMR-based metabolomic techniques identify potential urinary biomarkers for early colorectal cancer detection. Oncotarget. 2017;8:105819-105831. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 8. | Lin Y, Ma C, Bezabeh T, Wang Z, Liang J, Huang Y, Zhao J, Liu X, Ye W, Tang W, Ouyang T, Wu R. 1 H NMR-based metabolomics reveal overlapping discriminatory metabolites and metabolic pathway disturbances between colorectal tumor tissues and fecal samples. Int J Cancer. 2019;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 9. | Nicholson JK, Lindon JC. Systems biology: Metabonomics. Nature. 2008;455:1054-1056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1368] [Cited by in RCA: 1383] [Article Influence: 81.4] [Reference Citation Analysis (0)] |
| 10. | Rocha CM, Carrola J, Barros AS, Gil AM, Goodfellow BJ, Carreira IM, Bernardo J, Gomes A, Sousa V, Carvalho L, Duarte IF. Metabolic signatures of lung cancer in biofluids: NMR-based metabonomics of blood plasma. J Proteome Res. 2011;10:4314-4324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 122] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 11. | Theodorescu D, Wittke S, Ross MM, Walden M, Conaway M, Just I, Mischak H, Frierson HF. Discovery and validation of new protein biomarkers for urothelial cancer: a prospective analysis. Lancet Oncol. 2006;7:230-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 336] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 12. | M'Koma AE, Blum DL, Norris JL, Koyama T, Billheimer D, Motley S, Ghiassi M, Ferdowsi N, Bhowmick I, Chang SS, Fowke JH, Caprioli RM, Bhowmick NA. Detection of pre-neoplastic and neoplastic prostate disease by MALDI profiling of urine. Biochem Biophys Res Commun. 2007;353:829-834. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 66] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 13. | Theodorescu D, Schiffer E, Bauer HW, Douwes F, Eichhorn F, Polley R, Schmidt T, Schöfer W, Zürbig P, Good DM, Coon JJ, Mischak H. Discovery and validation of urinary biomarkers for prostate cancer. Proteomics Clin Appl. 2008;2:556-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 121] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 14. | Pisitkun T, Johnstone R, Knepper MA. Discovery of urinary biomarkers. Mol Cell Proteomics. 2006;5:1760-1771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 298] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 15. | Hasim A, Ma H, Mamtimin B, Abudula A, Niyaz M, Zhang LW, Anwer J, Sheyhidin I. Revealing the metabonomic variation of EC using ¹H-NMR spectroscopy and its association with the clinicopathological characteristics. Mol Biol Rep. 2012;39:8955-8964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 16. | Davis VW, Schiller DE, Eurich D, Sawyer MB. Urinary metabolomic signature of esophageal cancer and Barrett's esophagus. World J Surg Oncol. 2012;10:271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 61] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 17. | Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, Scalbert A. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801-D807. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2109] [Cited by in RCA: 2188] [Article Influence: 168.3] [Reference Citation Analysis (0)] |
| 18. | Bouatra S, Aziat F, Mandal R, Guo AC, Wilson MR, Knox C, Bjorndahl TC, Krishnamurthy R, Saleem F, Liu P, Dame ZT, Poelzer J, Huynh J, Yallou FS, Psychogios N, Dong E, Bogumil R, Roehring C, Wishart DS. The human urine metabolome. PLoS One. 2013;8:e73076. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1023] [Cited by in RCA: 1011] [Article Influence: 84.3] [Reference Citation Analysis (0)] |
| 19. | Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 720] [Cited by in RCA: 786] [Article Influence: 49.1] [Reference Citation Analysis (0)] |
| 20. | Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1460] [Cited by in RCA: 1592] [Article Influence: 79.6] [Reference Citation Analysis (0)] |
| 21. | Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029-1033. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12491] [Cited by in RCA: 11812] [Article Influence: 738.3] [Reference Citation Analysis (0)] |
| 22. | Echeverry G, Hortin GL, Rai AJ. Introduction to urinalysis: historical perspectives and clinical application. Methods Mol Biol. 2010;641:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 50] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 23. | Phelps TJ, Suflita JM, Little B. Carbon dioxide corrosion and acetate: a hypothesis on the influence of microorganisms. Corrosion Houston Tx. 2012;64:854-859. [DOI] [Full Text] |
| 24. | Chen JQ, Russo J. Dysregulation of glucose transport, glycolysis, TCA cycle and glutaminolysis by oncogenes and tumor suppressors in cancer cells. Biochim Biophys Acta. 2012;1826:370-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 159] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 25. | Curthoys NP, Watford M. Regulation of glutaminase activity and glutamine metabolism. Annu Rev Nutr. 1995;15:133-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 483] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 26. | Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nature Reviews Cancer. 2011;11:85. |
| 27. | Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13:572-583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1038] [Cited by in RCA: 1210] [Article Influence: 100.8] [Reference Citation Analysis (0)] |
| 28. | Joseph J, Cardesa A, Carreras J. Creatine kinase activity and isoenzymes in lung, colon and liver carcinomas. Br J Cancer. 1997;76:600-605. [PubMed] |
| 29. | van Stijn MF, Vermeulen MA, Siroen MP, Wong LN, van den Tol MP, Ligthart-Melis GC, Houdijk AP, van Leeuwen PA. Human taurine metabolism: fluxes and fractional extraction rates of the gut, liver, and kidneys. Metabolism. 2012;61:1036-1040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 30. | Amelio I, Cutruzzolá F, Antonov A, Agostini M, Melino G. Serine and glycine metabolism in cancer. Trends Biochem Sci. 2014;39:191-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 572] [Cited by in RCA: 794] [Article Influence: 72.2] [Reference Citation Analysis (0)] |
| 31. | Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3:431-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 682] [Cited by in RCA: 655] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 32. | Pun WK, Chow SP, Fang D, Cheng CL, Leong JC, Ng C. Post-traumatic oedema of the foot after tibial fracture. Injury. 1989;20:232-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 105] [Article Influence: 3.0] [Reference Citation Analysis (0)] |