Published online Feb 21, 2016. doi: 10.3748/wjg.v22.i7.2284
Peer-review started: July 30, 2015
First decision: September 9, 2015
Revised: September 29, 2015
Accepted: December 19, 2015
Article in press: December 19, 2015
Published online: February 21, 2016
Processing time: 186 Days and 23.3 Hours
Two major types of cancer occur in the esophagus: squamous cell carcinoma, which is associated with chronic smoking and alcohol consumption, and adenocarcinoma, which typically arises in gastric reflux-associated Barrett’s esophagus. Although there is increasing incidence of esophageal adenocarcinoma in Western counties, esophageal squamous cell carcinoma (ESCC) accounts for most esophageal malignancies in East Asia, including China and Japan. Technological advances allowing for massively parallel, high-throughput next-generation sequencing (NGS) of DNA have enabled comprehensive characterization of somatic mutations in large numbers of tumor samples. Recently, several studies were published in which whole exome or whole genome sequencing was performed in ESCC tumors and compared with matched normal DNA. Mutations were validated in several genes, including in TP53, CDKN2A, FAT1, NOTCH1, PIK3CA, KMT2D and NFE2L2, which had been previously implicated in ESCC. Several new recurrent alterations have also been identified in ESCC. Combining the clinicopathological characteristics of patients with information obtained from NGS studies may lead to the development of effective diagnostic and therapeutic approaches for ESCC. As this research becomes more prominent, it is important that gastroenterologist become familiar with the various NGS technologies and the results generated using these methods. In the present study, we describe recent research approaches using NGS in ESCC.
Core tip: Because targeted therapies have not been implemented in the treatment of esophageal squamous cell carcinoma (ESCC) to date, defining the genetic landscape of ESCC would facilitate the use of targeted therapies. Improvements in molecular profiling technologies have provided new insight into the basic molecular events during carcinogenesis as well as the mechanisms of anti-cancer drug resistance. Our invited review offers a current overview of the somatic genetic alterations in ESCC, emphasizing the recent results of large-scale sequencing efforts using next-generation sequencing technology.
- Citation: Sasaki Y, Tamura M, Koyama R, Nakagaki T, Adachi Y, Tokino T. Genomic characterization of esophageal squamous cell carcinoma: Insights from next-generation sequencing. World J Gastroenterol 2016; 22(7): 2284-2293
- URL: https://www.wjgnet.com/1007-9327/full/v22/i7/2284.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i7.2284
Cancer of the esophagus is the eighth leading causes of cancer-related mortality worldwide[1]. It is one of the most deadly gastrointestinal tumors, with a 5-year survival rate of 20%-30% after curative surgery[2]. Two major types of cancer occur in the esophagus, including squamous cell carcinoma and adenocarcinoma, but their epidemiological features differ considerably. The incidence of esophageal cancer varies greatly by geographic location. Esophageal squamous cell carcinoma (ESCC) has a predilection for black and Asian populations and more than 70% of esophageal cancers worldwide are squamous cell carcinomas[3-5]. ESCC is considered an environmental malignancy attributable to chronic smoking and alcohol consumption[6]. In contrast, esophageal adenocarcinoma (EAC) predominantly affects white populations, which typically arises in a premalignant condition called Barrett’s esophagus[7-10]. The changing epidemiology of esophageal cancer, with a dramatic increase in EAC and decrease in ESCC in Western countries indicates that differences exist between the two types of esophageal cancer[2].
Despite recent advances in imaging and surgical techniques, as well as the intensification of treatment with the increased use of chemoradiation, the survival rates for esophageal cancers have remained largely unchanged for several decades[6]. We have observed many patients with ESCC in whom local tumor recurrence or distant metastases occurred during an early disease stage and within a short period after surgery. Therefore, the molecular carcinogenesis and metastatic process of esophageal cancer must be clarified. Understanding tumor biology offers the potential for individualizing treatment and developing targeted therapies to increase cure rates and minimize morbidities. This review provides a current overview of the genomic and molecular characterization of ESCC, emphasizing recent results of large-scale sequencing efforts using next-generation sequencing (NGS) technology.
Since unique mutations have been observed in individual human cancer samples, the identification and characterization of molecular alterations underlying individual cancer patients is a critical step in the development of more effective, personalized therapies. For example, NGS technologies have revolutionized cancer genomics research by providing a comprehensive method for detecting somatic cancer genome alterations, such as point mutations, insertions, deletions, and copy number variations[11,12]. NGS has also revolutionized the field of genomics and improved our understanding of cancer biology. Advances have been achieved in the sequencing of tumor DNA; matched normal DNA was used to filter out germline variants to identify cancer-specific changes. High incidences of activating mutations in ESCCs amenable to drug targeting have also been identified. Investigators have also identified several critical genes and pathways important in the tumorigenesis of ESCC using this technology. This wealth of information undoubtedly improves our understanding of ESCC biology and provides clear targets for drug targeting to guide future personalized medicine.
NGS technologies have several advantages over classical Sanger sequencing, such as the ability to generate large quantities of DNA sequence information in a single run for detecting genetic mosaicism in depth[13]. However, routine usage of these technologies has several limitations, such as high cost, long processing time, and sample scalability. Three NGS platforms are now widely applied in cancer genome studies, including short-read technologies (< 400 bp) from Illumina (Genome analyzer/MiSeq/HiSeq/NextSeq; San Diego, CA, United States) and Thermo Fisher (SOLiD/Ion Torrent, Waltham, MA, United States) as well as a relative long-read technology (< 700 bp) from Roche (GS FLX, Basel, Switzerland). NGS platforms differ in performance metrics such as read length, accuracy, and output. The next next-generation (third-generation) sequencing system from Pacific Biosciences is also available (PacBio RS, Menlo Park, CA, United States), which can sequence a single molecule of DNA without polymerase chain reaction (PCR) amplification[14]. The average read length is 1500 bp, which is longer than that of any NGS technology, although the throughput of PacBio RS is lower than that of the second-generation sequencer. A brief summary of the technical features of these NGS platforms is shown in Table 1.
| 454 GS FLX | GAIIx | MiSeq | HiSeq 2500 | SOLiD 5500 | Ion PGM | Ion Proton | PacBio RS | |
| (318 chip) | ||||||||
| Reads per run | 1 M | 150 M | 50 M | 6 G | 1.4 G | 5.5 M | 60 M | 50 K |
| Read length (bp) | 700 | 2 × 150 | 2 × 150 | 2 × 100 | 2 × 50 | 400 | 200 | 250-10000 |
| Output per run | 700 Mb | 90 Gb | 15 Gb | 600 Gb | 120 Gb | 2 Gb | 12 Gb | 200 Mb |
| Run time | 24 h | 14 d | 55 h | 10 d | 7 d | 5 h | 3 h | 2 h |
| Cost/Mb1 | $10.00 | $0.15 | $0.50 | $0.05 | $0.10 | $1.00 | $0.08 | $2.00 |
| Advantage | Long read length | Widly used | Widly used | High-throughput, widly used | High-throughput, accuracy | Fast, flexible chip | High-throughput, fast | Long read length, fast |
| Disadvantage | Long hand-on time, low output | Long run time | Long run time | Long run time | Long run time, short read length | High error rate (homopolymer) | High error rate (homopolymer) | High error rate |
The NGS market is dominated by Illumina, which occupies the largest market share at 70% (http://www.marketsandmarkets.com). Illumina platforms are based on bridge amplification to clonally amplify the fragments, which are then sequenced using sequencing-by-synthesis chemistry[15]. Sequencing capabilities include both single-end sequencing and paired-end sequencing. The HiSeq 2000/2500 set the standard for high-throughput massively parallel sequencing. The original output was 200 Gb per run, which was improved to 600 Gb per run and can be finished in 10 d. The MiSeq was then released as a lower-throughput fast-turnaround instrument for use in smaller laboratories. Recently, Illumina developed the HiSeq X Ten Sequencing System, a very high-throughput and high-speed sequencing platform that enables sequencing for less than $1000 per genome at 30 × coverage[16].
After the human genome project, the first commercial NGS platform 454 pyrosequencer was developed by 454 Life Sciences Corp in 2005. The platform was purchased by Roche in 2007. Roche 454 platforms use emulsion PCR, and is based on pyrosequencing technology relying on the detection of pyrophosphate released during nucleotide incorporation[17]. The 454 GS-FLX system produces one million 700-bp sequences within 24 h. However, this platform has a significantly lower output compared to other NGS platforms. Additionally, the cost per base is also significantly higher compared with short-read technologies. The GS Junior is a benchtop version of this platform.
Similarly to Roche 454, the SOLiD sequencer relies on emulsion PCR and sequencing by ligation to small beads. Although the reads obtained from the SOLiD 5500 Genetic Analyzer system are only 50-75 bp in length, its system accuracy of 99.99% ranks first among all NGS platforms[18]. Recently, Ion Torrent sequencing technology based on semiconductor sequencing[19] has been released. Ion Torrent platforms use a high-density array of micro-machined wells, each containing a different DNA template. Beneath the wells lies an ion-sensitive layer, which is placed on a proprietary ion sensor to detect changes in pH resulting from incorporation of nucleotides in the new strand of DNA. The compact Ion Personal Genome Analyzer has three different chips, each designed for a specific purpose, including ranging from sequencing small genomes (314 chip, 550 K reads) and targeted gene sequencing (316 chip, 3 M reads) to chromatin immunoprecipitation-sequencing (ChIP-seq) (318 chip, 5.5 M reads). The desktop-type Ion Proton allows for larger chips with higher densities needed for the human exome and whole genome sequencing. The outstanding advantage of Ion Torrent is its speed: it takes 3-5 h from the start of sequencing until completion; however, this method has high error rate in homopolymer regions[20].
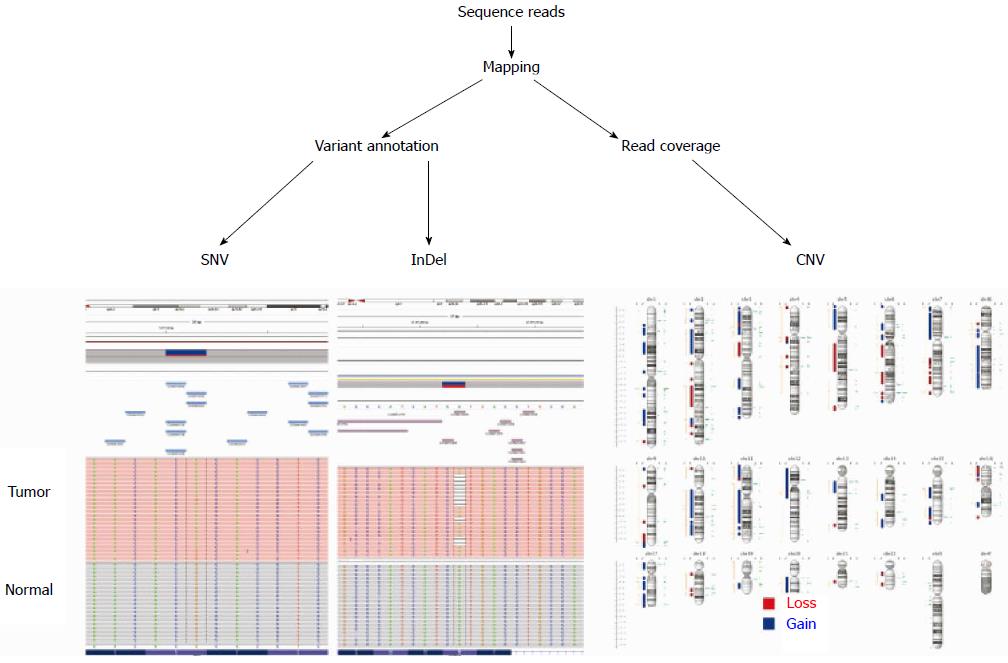
NGS is increasingly used in many areas of cancer research and in the clinical setting. Depending on the purpose, NGS is applied in cancer genome studies, including whole genome, whole exome, targeted gene sequencing, RNA sequencing, and ChIP-Seq[11,12,21-23]. For variant identification by resequencing target regions, whole genomes, or whole exons, it is key to sequence both the tumor and non-malignant tissues of an individual. There are 3-5 million inherited sequence variants per human genome. Consequently, most sequence variants identified in a cancer genome are inherited polymorphisms and are not somatic mutations[24]. Thus, comparing a tumor genome to its paired normal genome is required to efficiently identify somatic sequence variants (Figure 1). In addition to CGH and SNP arrays, NGS techniques can be used to detect copy number variations[12,25]. Targeted sequencing is a variation of re-sequencing where only a small subset of the genome is sequenced, such as a set of genes or particular sequences under interest. Although this approach will not detect most structural variants, such as chromosomal translocations, targeted gene sequencing represents a cost- and resource-efficient approach for identifying somatic mutations in cancer genomes[26,27].
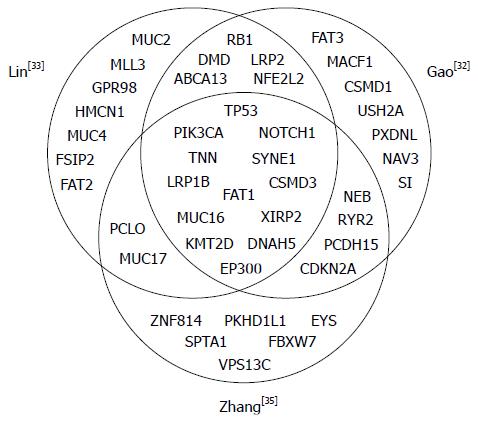
Targeted therapies have been successfully used for the treatment of in certain human solid tumors, including lung adenocarcinoma, colorectal cancer, stomach cancer, breast cancer and renal cell carcinoma as well as hematologic malignancies, but have not been implemented in the treatment of ESCC[28-30]. Therefore, defining the genetic landscape of ESCCs would facilitate the use of targeted therapies. Agrawal and colleagues published the first exome-sequencing study of esophageal cancer, sequencing 12 ESCCs and 11 EACs as well as matched non-neoplastic tissues from subjects in the United States[31], and a handful of NGS studies in ESCC have been published over the last four years (summarized in Table 2)[31-35]. Genetic aberrations identified within these studies, including gene mutation, gene rearrangement, and gene amplification/deletion, increased the understanding of constitutive activation of oncogenes, or loss of function of tumor suppressors. These comprehensive studies have demonstrated recurrent mutations in several genes in ESCC, most notably TP53, NOTCH1, PIK3CA and FAT1 as well as copy-number alterations in CCND1 and CDKN2A (Table 3). Figure 2 shows a Venn diagram of the most significantly mutated genes identified in the three whole genome and whole exome sequencing studies of relatively large cohorts for ESCCs. The Broad Institute (Cambridge, MA, United States) project[36] used this method to examine 149 EAC tumors, and confirmed recurrent driver mutations in TP53, CDKN2A, SMAD4, ARID1A, and PIK3CA. Previously unidentified mutations in SPG20, TLR4, ELMO1, and DOCK2 were also found, and a possible role for the RAC1 GTPase pathway was identified. The genomic landscape of EAC differed from that of ESCC, highlighting the different therapeutic strategies needed to treat esophageal cancers. In this review, we highlight the current knowledge regarding molecular targets, clinical trials of targeted agents, and druggable aberrations in ESCCs.
| Study | Method | Sample number | Number of non-silent mutations/tumor | Additional analyses | Platform |
| Agrawal et al[31] | WES | 12 WES | 83 | - | GA IIx |
| Song et al[34] | WGS and WES | 17 WGS, 71 WES | 61 | 123 CGH | HiSeq 2000 |
| Lin et al[33] | WES and TS | 20 WES, 119 TS | 59 | 4 RNA-seq, 59 CGH, | HiSeq 2000 |
| 125 SNP-array | |||||
| Gao et al[32] | WES | 113 WES | 82 | - | HiSeq 2000 |
| Zhang et al[35] | WGS and WES | 14 WGS, 90 WES | 104 | - | HiSeq 2000 |
| Gene symbol | TP53 | NOTCH1 | PIK3CA | CDKN2A | CCND1 | FAT1 |
| Chromosomal location | 17p13.1 | 9q34.3 | 3q26.3 | 9p21.3 | 11q13 | 4q35.2 |
| Alteration frequency (%) | ||||||
| Agrawal et all[31] | 92 (M) | 33 (M) | 0 (M) | 8 (M) | NA | 8 (M) |
| Song et al[34] | 83 (M) | 9 (M) | 5 (M) | 5 (M) | 46 (G) | 5 (M) |
| 1 (L) | 41 (G) | 44 (L) | ||||
| Lin et al[33] | 60 (M) | 8 (M) | 7 (M) | 3 (M) | 46 (G) | 12 (M) |
| 10 (G) | 33 (L) | |||||
| Gao et al[32] | 93 (M) | 14 (M) | 9 (M) | 8 (M) | 33 (G) | 11 (M) |
| 2 (G) | 12 (L) | |||||
| Zhang et al[35] | 88 (M) | 21 (M) | 17 (M) | 8 (M) | 64 (G) | 15 (M) |
| 64 (L) |
TP53 is one of the most thoroughly studied tumor suppressor genes in human cancer. Genetic mutations in TP53 are present in more than 50% of human cancers, leading to dysregulation of its downstream targets[37,38]. NGS studies have confirmed that TP53 is the most commonly mutated gene in ESCC. The first whole exome sequencing study found that ESCCs contained an average of 83 mutations per tumor, and that the most frequent mutations in ESCC occurred in TP53 (92% of the 12 cases sequenced), NOTCH1 (33%), NOTCH3 (25%), and FBXW7 (17%)[31]. The p53 protein is activated by a variety of cell stresses, such as DNA damage, oncogene activation, spindle damage, and hypoxia. Activated p53 transactivates a number of target genes, many of which are involved in DNA repair, cell cycle arrest, and apoptosis[39-41]. TP53 alterations have been identified as early events in the carcinogenesis of ESCC and have been associated with disease progression and a poor outcome[42-44]. Therapies targeting TP53 loss of function are currently being examined in clinical trials, and several studies suggest that patients harboring TP53 alterations will respond better to angiogenesis inhibitors[45]. The efficacy of intra-tumor injection of p53 adenovirus (Advexin, Introgen Therapeutics Inc., Austin, TX, United States) has been confirmed in Japanese ESCC patients[46]. Additionally, a TP53 adenoviral-based treatment (Gendicine, Shenshen Sibiono Genetech, Shenzhen, China) for patients with squamous cell carcinoma of the head and neck has recently been approved for use in China[47]. Two other p53 family members, p63 and p73, also induce cell cycle arrest and apoptosis and play an important role in development and differentiation[48]. Dominant negative forms lacking the N-terminal transactivation domain (DNp63 and DNp73) are overexpressed in some types of cancers[49]. In esophageal cancer, DNp63 is overexpressed in ESCC but not in EAC, making p63 a useful marker of squamous cell cancer[50,51]. Additionally, at least 30% of head and neck squamous cell carcinomas harbor mutations in genes regulating squamous differentiation, including p63[52].
NOTCH1 is the second most commonly mutated gene in ESCC, with a mutation rate of 8%-33%[31-35]. The Notch signaling pathway is thought to play important roles in regulating normal cell differentiation in a context-dependent manner[53]. The Notch pathway has also been implicated in human carcinogenesis as both an oncogene and a tumor suppressor[54]. The oncogenic activity of this pathway has been observed in a number of hematopoietic cancers[55]. When we characterized the distribution of NOTCH1 somatic mutations obtained from the two studies[32,35], most NOTCH1 mutations observed in ESCC affect the epidermal growth factor (EGF)-like ligand-binding domain (56%, 30 of 54) and are thought to lead to a loss of function. Inactivating mutations in these regions of the gene have also been observed in cutaneous, lung, head, and neck squamous cell carcinomas[56-58]. Thus, the idea that the same gene can function in completely opposite manners in different cell types is important for understanding cell signaling pathways. In addition to NOTCH1, mutations in the NOTCH2 and NOTCH3 genes were detected in ESCC[31,32]. Interestingly, Agrawal et al[31] identified inactivating mutations of NOTCH1 in 21% of ESCC but not in EAC, suggesting tumor-suppressive roles of Notch signaling in squamous cell carcinomas. Notch pathway disruption also results from FBXW7 mutations, which were identified in 5%-17% of ESCC specimens, because FBXW7 forms part of the ubiquitin ligase complex that mediates NOTCH1 degradation[59].
KRAS is one of the most frequently mutated oncogenes in human cancer[60]. In ESCC, KRAS mutations are generally rare[61], although the incidence of KRAS mutations in Chinese patients with ESCC was relatively high, with a mutation rate of 12%[62]. Receptor tyrosine kinases (RTKs) of the EGFR family are involved in development and progression of epithelial tumors and thus represent therapeutic targets for inhibition by tyrosine kinase inhibitors or humanized monoclonal antibodies[63-65]. Upstream RTKs, EGFR, ERBB2, ERBB4, and MET, as well as G-protein-coupled receptors activate phosphoinositide-3-kinase (PI3K) after binding of growth factor ligands[63]. The PI3K pathway plays a key role in regulating multiple cellular events, including cell growth, proliferation, cell cycle progression, and survival[66,67]. PIK3CA is the second most commonly mutated gene, occurring frequently (< 20%) in most cancer types[60]. Overexpression of EGFR has been described in ESCC; most ESCC tumors show increased activity in the absence of somatic mutations[68-70]. In addition to EGFR amplification, this pathways displayed genetic alterations in 78.6% of cases, including FGFR1, ERBB2, RAF1, AKT1, SOS1, SOS2, and PIK3CA mutations and amplifications[33]. Moreover, EGFR transactivation via ectodomain shedding of EGFR ligands plays a role in inflammation as well as tumor growth and metastasis[71,72]. A recent report demonstrated that targeting the sheddase activity of ADAM17, which is responsible for the release of multiple EGFR ligands, decreased head and neck squamous cell carcinoma cell viability and motility through blocking of the EGFR pathway[73]. Since RTKs are well-characterized druggable proteins, targeting components in this pathway may represent valuable investigational avenues for clinical trials in patients with ESCCs. Of interest, KRAS, a frequently aberrant gene in non-squamous tumors that leads to resistance to PI3K pathway inhibitors, was found to be aberrant significantly less frequent in ESCCs. Recent clinical studies have demonstrated that anti-EGFR monoclonal antibody (cetuximab) in combination with irradiation yielded encouraging survival and local control in ESCC patients[74].
The cell cycle regulation pathway is one of the most perturbed pathways in ESCC. Mutations have been observed in the cell-cycle regulatory pathway genes TP53 (88%), CDKN2A (8%), and RB1 (2%)[35]. In addition, ESCC tumors show amplification of CCND1, which encodes for cyclin D1, and deletion of CDKN2A/B, which encodes for p16 and p14. Cyclin D1 is responsible for inducing the G1/S phase transition and is located at 11q13[31-35]. Gains in the chromosomal region 11q13 are some of the most prominent genetic alterations in squamous cell carcinomas and are associated with poor prognosis and metastasis[28,75,76]. Other G1/S transition control molecules, CDK4, CDK6, E2F1, and MDM2, are also amplified in ESCC. The p16 tumor suppressor can inhibit the formation of the CDK4/6 and cyclin D1 complex and plays a role in the oncogene-induced senescence of cells. Gain of CCND1 and/or loss of CDK2NA events occurred in over 70% of ESCC samples[33-35]. Flavopiridol, the first cyclin-dependent kinase inhibitor examined in human clinical trials, was reported to be a targeting drug for ESCC and head and neck squamous cell carcinoma patients[77]. NFE2L2, a frequently mutated gene in ESCC, encodes a sequence-specific transcriptional factor that upregulates genes associated with oxidative stress. Activating missense mutations in the NFE2L2 gene result in accumulation of the NFE2L2 protein and promote aberrant activation of downstream genes that confer resistance to oxidative stress and induce metabolic transformation in cancer cells[78].
Altered genes in the Wnt pathway were also frequently found in ESCC, including mutations in CTNNB1 and SFRP4, and mutations and amplifications of AXIN inhibitors, DAAM2, DVL3, LRP5 and LRP6[34]. In addition, loss of FAT1, by either somatic mutation or deletions, promotes tumorigenesis through activation of Wnt signaling[79].
Dysregulation of proteins involved in chromatin regulation can affect the genome-wide control of gene expression and play key roles in DNA repair and genome maintenance. Mutations in a number of genes involved in histone modifications have been identified in many cancer types[60]. Inactivating missense mutations in several chromatin-remodeling genes, including EP300, CREBBP, and BAP1, in ESCC samples. Moreover, truncating mutations were observed in the chromatin-remodeling genes KMT2D, KMT2C, and KDM6A[35]. Approximately 30% of ESCC tumors contained at least one chromatin remodeling gene alteration.
Recently, inactivating mutations in the Hippo pathway regulator (AJUBA, FAT1, FAT2, FAT3, and FAT4) were observed in ESCC[32]. Hippo signaling cross-talks with commonly mutated cancer genes such as KRAS, PIK3CA, CTNNB1, or FBXW7[80,81].
The identification and characterization of molecular alterations in individual cancer patients is a critical step towards the development of more effective personalized therapies. NGS technologies have revolutionized cancer genomics research by providing a comprehensive method of detecting somatic DNA modifications. ESCC is the major histological type of esophageal cancer in East Asian countries and is one of the most aggressive malignant tumors. Recent studies using NGS have revealed that ESCC is characterized by specific somatic DNA modifications such as exonic mutations, copy-number alterations, and genomic rearrangements. The most common mutation in ESCC is TP53. Pathway assessment has shown that somatic aberrations within ESCC genomes are mainly involved in several important pathways, including cell cycle regulation and the Notch, RTK-MAPK-PI3K, and Wnt pathways. We expect that many new discoveries will increase our understanding of the molecular mechanisms of ESCC for targeted therapies.
P- Reviewer: Chen XL, Komatsu S, Slomiany BL S- Editor: Gong ZM L- Editor: A E- Editor: Liu XM
| 1. | Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893-2917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11128] [Cited by in RCA: 11836] [Article Influence: 845.4] [Reference Citation Analysis (4)] |
| 2. | Holmes RS, Vaughan TL. Epidemiology and pathogenesis of esophageal cancer. Semin Radiat Oncol. 2007;17:2-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 367] [Cited by in RCA: 387] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 3. | Cook MB. Non-acid reflux: the missing link between gastric atrophy and esophageal squamous cell carcinoma? Am J Gastroenterol. 2011;106:1930-1932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 4. | Dawsey SM, Lewin KJ, Liu FS, Wang GQ, Shen Q. Esophageal morphology from Linxian, China. Squamous histologic findings in 754 patients. Cancer. 1994;73:2027-2037. [PubMed] |
| 5. | Dawsey SM, Lewin KJ, Wang GQ, Liu FS, Nieberg RK, Yu Y, Li JY, Blot WJ, Li B, Taylor PR. Squamous esophageal histology and subsequent risk of squamous cell carcinoma of the esophagus. A prospective follow-up study from Linxian, China. Cancer. 1994;74:1686-1692. [PubMed] |
| 6. | Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19:5598-5606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 736] [Cited by in RCA: 747] [Article Influence: 62.3] [Reference Citation Analysis (8)] |
| 7. | Huang Q, Fang DC, Yu CG, Zhang J, Chen MH. Barrett’s esophagus-related diseases remain uncommon in China. J Dig Dis. 2011;12:420-427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 8. | Kountourakis P, Papademetriou K, Ardavanis A, Papamichael D. Barrett’s esophagus: treatment or observation of a major precursor factor of esophageal cancer? J BUON. 2011;16:425-430. [PubMed] |
| 9. | Hongo M, Nagasaki Y, Shoji T. Epidemiology of esophageal cancer: Orient to Occident. Effects of chronology, geography and ethnicity. J Gastroenterol Hepatol. 2009;24:729-735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 270] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 10. | Shaheen N, Ransohoff DF. Gastroesophageal reflux, barrett esophagus, and esophageal cancer: scientific review. JAMA. 2002;287:1972-1981. [PubMed] |
| 11. | Dong H, Wang S. Exploring the cancer genome in the era of next-generation sequencing. Front Med. 2012;6:48-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 12. | Tran B, Dancey JE, Kamel-Reid S, McPherson JD, Bedard PL, Brown AM, Zhang T, Shaw P, Onetto N, Stein L. Cancer genomics: technology, discovery, and translation. J Clin Oncol. 2012;30:647-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 152] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 13. | Suzuki S, Ono N, Furusawa C, Ying BW, Yomo T. Comparison of sequence reads obtained from three next-generation sequencing platforms. PLoS One. 2011;6:e19534. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 67] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 14. | Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2551] [Cited by in RCA: 2371] [Article Influence: 139.5] [Reference Citation Analysis (0)] |
| 15. | Ross JS, Cronin M. Whole cancer genome sequencing by next-generation methods. Am J Clin Pathol. 2011;136:527-539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 121] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 16. | Wang Q, Lu Q, Zhao H. A review of study designs and statistical methods for genomic epidemiology studies using next generation sequencing. Front Genet. 2015;6:149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 17. | Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5541] [Cited by in RCA: 5070] [Article Influence: 253.5] [Reference Citation Analysis (0)] |
| 18. | Daniels MG, Bowman RV, Yang IA, Govindan R, Fong KM. An emerging place for lung cancer genomics in 2013. J Thorac Dis. 2013;5 Suppl 5:S491-S497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 19. | Pourmand N, Karhanek M, Persson HH, Webb CD, Lee TH, Zahradníková A, Davis RW. Direct electrical detection of DNA synthesis. Proc Natl Acad Sci USA. 2006;103:6466-6470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 48] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 20. | Boland JF, Chung CC, Roberson D, Mitchell J, Zhang X, Im KM, He J, Chanock SJ, Yeager M, Dean M. The new sequencer on the block: comparison of Life Technology’s Proton sequencer to an Illumina HiSeq for whole-exome sequencing. Hum Genet. 2013;132:1153-1163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 60] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 21. | Pant S, Weiner R, Marton MJ. Navigating the rapids: the development of regulated next-generation sequencing-based clinical trial assays and companion diagnostics. Front Oncol. 2014;4:78. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 22. | Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012;13:341. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1386] [Cited by in RCA: 1269] [Article Influence: 97.6] [Reference Citation Analysis (1)] |
| 23. | Thomas F, Desmedt C, Aftimos P, Awada A. Impact of tumor sequencing on the use of anticancer drugs. Curr Opin Oncol. 2014;26:347-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 24. | Jones S, Anagnostou V, Lytle K, Parpart-Li S, Nesselbush M, Riley DR, Shukla M, Chesnick B, Kadan M, Papp E. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med. 2015;7:283ra53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 327] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 25. | Wang H, Nettleton D, Ying K. Copy number variation detection using next generation sequencing read counts. BMC Bioinformatics. 2014;15:109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 26. | Singh RR, Patel KP, Routbort MJ, Aldape K, Lu X, Manekia J, Abraham R, Reddy NG, Barkoh BA, Veliyathu J. Clinical massively parallel next-generation sequencing analysis of 409 cancer-related genes for mutations and copy number variations in solid tumours. Br J Cancer. 2014;111:2014-2023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 27. | Nguyen-Dumont T, Pope BJ, Hammet F, Mahmoodi M, Tsimiklis H, Southey MC, Park DJ. Cross-platform compatibility of Hi-Plex, a streamlined approach for targeted massively parallel sequencing. Anal Biochem. 2013;442:127-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 28. | Chen J, Kwong DL, Cao T, Hu Q, Zhang L, Ming X, Chen J, Fu L, Guan X. Esophageal squamous cell carcinoma (ESCC): advance in genomics and molecular genetics. Dis Esophagus. 2015;28:84-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 29. | Shang L, Wang M. Molecular alterations and clinical relevance in esophageal squamous cell carcinoma. Front Med. 2013;7:401-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 30. | Zhang WZ, Chen JZ, Li DR, Chen ZJ, Guo H, Zhuang TT, Li DS, Zhou MZ, Chen CZ. Simultaneous modulated accelerated radiation therapy for esophageal cancer: a feasibility study. World J Gastroenterol. 2014;20:13973-13980. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 18] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (1)] |
| 31. | Agrawal N, Jiao Y, Bettegowda C, Hutfless SM, Wang Y, David S, Cheng Y, Twaddell WS, Latt NL, Shin EJ. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov. 2012;2:899-905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 290] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 32. | Gao YB, Chen ZL, Li JG, Hu XD, Shi XJ, Sun ZM, Zhang F, Zhao ZR, Li ZT, Liu ZY. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet. 2014;46:1097-1102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 557] [Cited by in RCA: 553] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 33. | Lin DC, Hao JJ, Nagata Y, Xu L, Shang L, Meng X, Sato Y, Okuno Y, Varela AM, Ding LW. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet. 2014;46:467-473. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 497] [Cited by in RCA: 504] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 34. | Song Y, Li L, Ou Y, Gao Z, Li E, Li X, Zhang W, Wang J, Xu L, Zhou Y. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 721] [Cited by in RCA: 855] [Article Influence: 77.7] [Reference Citation Analysis (0)] |
| 35. | Zhang L, Zhou Y, Cheng C, Cui H, Cheng L, Kong P, Wang J, Li Y, Chen W, Song B. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am J Hum Genet. 2015;96:597-611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 225] [Cited by in RCA: 276] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 36. | Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C, Bandla S, Imamura Y, Schumacher SE, Shefler E. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45:478-486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 633] [Cited by in RCA: 604] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 37. | Soussi T, Wiman KG. TP53: an oncogene in disguise. Cell Death Differ. 2015;22:1239-1249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 170] [Cited by in RCA: 228] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 38. | Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546-1558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5906] [Cited by in RCA: 5601] [Article Influence: 466.8] [Reference Citation Analysis (0)] |
| 39. | Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5100] [Cited by in RCA: 5117] [Article Influence: 204.7] [Reference Citation Analysis (0)] |
| 40. | el-Deiry WS. Regulation of p53 downstream genes. Semin Cancer Biol. 1998;8:345-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 588] [Cited by in RCA: 575] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 41. | Tokino T, Nakamura Y. The role of p53-target genes in human cancer. Crit Rev Oncol Hematol. 2000;33:1-6. [PubMed] |
| 42. | Egashira A, Morita M, Yoshida R, Saeki H, Oki E, Sadanaga N, Kakeji Y, Tsujitani S, Maehara Y. Loss of p53 in esophageal squamous cell carcinoma and the correlation with survival: analyses of gene mutations, protein expression, and loss of heterozygosity in Japanese patients. J Surg Oncol. 2011;104:169-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 43. | Mandard AM, Hainaut P, Hollstein M. Genetic steps in the development of squamous cell carcinoma of the esophagus. Mutat Res. 2000;462:335-342. [PubMed] |
| 44. | Makino T, Yamasaki M, Miyata H, Yoshioka S, Takiguchi S, Fujiwara Y, Nakajima K, Nishida T, Mori M, Doki Y. p53 Mutation status predicts pathological response to chemoradiotherapy in locally advanced esophageal cancer. Ann Surg Oncol. 2010;17:804-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 46] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 45. | Schwaederlé M, Lazar V, Validire P, Hansson J, Lacroix L, Soria JC, Pawitan Y, Kurzrock R. VEGF-A Expression Correlates with TP53 Mutations in Non-Small Cell Lung Cancer: Implications for Antiangiogenesis Therapy. Cancer Res. 2015;75:1187-1190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 99] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 46. | Shimada H, Matsubara H, Shiratori T, Shimizu T, Miyazaki S, Okazumi S, Nabeya Y, Shuto K, Hayashi H, Tanizawa T. Phase I/II adenoviral p53 gene therapy for chemoradiation resistant advanced esophageal squamous cell carcinoma. Cancer Sci. 2006;97:554-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 66] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 47. | Nemunaitis J, Nemunaitis J. Head and neck cancer: response to p53-based therapeutics. Head Neck. 2011;33:131-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 48. | Irwin MS, Kaelin WG. p53 family update: p73 and p63 develop their own identities. Cell Growth Differ. 2001;12:337-349. [PubMed] |
| 49. | Orzol P, Holcakova J, Nekulova M, Nenutil R, Vojtesek B, Coates PJ. The diverse oncogenic and tumour suppressor roles of p63 and p73 in cancer: a review by cancer site. Histol Histopathol. 2015;30:503-521. [PubMed] |
| 50. | Thépot A, Hautefeuille A, Cros MP, Abedi-Ardekani B, Pétré A, Damour O, Krutovskikh V, Hainaut P. Intraepithelial p63-dependent expression of distinct components of cell adhesion complexes in normal esophageal mucosa and squamous cell carcinoma. Int J Cancer. 2010;127:2051-2062. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 51. | Bergholz J, Xiao ZX. Role of p63 in Development, Tumorigenesis and Cancer Progression. Cancer Microenviron. 2012;5:311-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 52. | Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157-1160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2034] [Cited by in RCA: 1970] [Article Influence: 140.7] [Reference Citation Analysis (0)] |
| 53. | Bolós V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocr Rev. 2007;28:339-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 397] [Cited by in RCA: 398] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 54. | Strizzi L, Hardy KM, Seftor EA, Costa FF, Kirschmann DA, Seftor RE, Postovit LM, Hendrix MJ. Development and cancer: at the crossroads of Nodal and Notch signaling. Cancer Res. 2009;69:7131-7134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 68] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 55. | Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2075] [Cited by in RCA: 2131] [Article Influence: 101.5] [Reference Citation Analysis (0)] |
| 56. | South AP, Purdie KJ, Watt SA, Haldenby S, den Breems NY, Dimon M, Arron ST, Kluk MJ, Aster JC, McHugh A. NOTCH1 mutations occur early during cutaneous squamous cell carcinogenesis. J Invest Dermatol. 2014;134:2630-2638. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 233] [Cited by in RCA: 266] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 57. | Westhoff B, Colaluca IN, D’Ario G, Donzelli M, Tosoni D, Volorio S, Pelosi G, Spaggiari L, Mazzarol G, Viale G. Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci USA. 2009;106:22293-22298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 317] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 58. | Sun W, Gaykalova DA, Ochs MF, Mambo E, Arnaoutakis D, Liu Y, Loyo M, Agrawal N, Howard J, Li R. Activation of the NOTCH pathway in head and neck cancer. Cancer Res. 2014;74:1091-1104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 167] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 59. | Wang L, Ye X, Liu Y, Wei W, Wang Z. Aberrant regulation of FBW7 in cancer. Oncotarget. 2014;5:2000-2015. [PubMed] |
| 60. | Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333-339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2931] [Cited by in RCA: 3365] [Article Influence: 280.4] [Reference Citation Analysis (0)] |
| 61. | Shigaki H, Baba Y, Watanabe M, Miyake K, Murata A, Iwagami S, Ishimoto T, Iwatsuki M, Yoshida N, Baba H. KRAS and BRAF mutations in 203 esophageal squamous cell carcinomas: pyrosequencing technology and literature review. Ann Surg Oncol. 2013;20 Suppl 3:S485-S491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 62. | Liu QW, Fu JH, Luo KJ, Yang HX, Wang JY, Hu Y, Yang H, Bella E. Identification of EGFR and KRAS mutations in Chinese patients with esophageal squamous cell carcinoma. Dis Esophagus. 2011;24:374-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 63. | Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4920] [Cited by in RCA: 5111] [Article Influence: 213.0] [Reference Citation Analysis (1)] |
| 64. | Imai K, Takaoka A. Comparing antibody and small-molecule therapies for cancer. Nat Rev Cancer. 2006;6:714-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 513] [Cited by in RCA: 554] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 65. | Wakeling AE, Guy SP, Woodburn JR, Ashton SE, Curry BJ, Barker AJ, Gibson KH. ZD1839 (Iressa): an orally active inhibitor of epidermal growth factor signaling with potential for cancer therapy. Cancer Res. 2002;62:5749-5754. [PubMed] |
| 66. | Samuels Y, Ericson K. Oncogenic PI3K and its role in cancer. Curr Opin Oncol. 2006;18:77-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 375] [Cited by in RCA: 392] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 67. | Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497-5510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1330] [Cited by in RCA: 1448] [Article Influence: 85.2] [Reference Citation Analysis (0)] |
| 68. | Hanawa M, Suzuki S, Dobashi Y, Yamane T, Kono K, Enomoto N, Ooi A. EGFR protein overexpression and gene amplification in squamous cell carcinomas of the esophagus. Int J Cancer. 2006;118:1173-1180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 233] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 69. | Kato H, Arao T, Matsumoto K, Fujita Y, Kimura H, Hayashi H, Nishiki K, Iwama M, Shiraishi O, Yasuda A. Gene amplification of EGFR, HER2, FGFR2 and MET in esophageal squamous cell carcinoma. Int J Oncol. 2013;42:1151-1158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 70. | Wang CY, Deng JY, Cai XW, Fu XL, Li Y, Zhou XY, Wu XH, Hu XC, Fan M, Xiang JQ. High EGFR and low p-Akt expression is associated with better outcome after nimotuzumab-containing treatment in esophageal cancer patients: preliminary clinical result and testable hypothesis. Oncotarget. 2015;6:18674-18682. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 71. | Xu P, Derynck R. Direct activation of TACE-mediated ectodomain shedding by p38 MAP kinase regulates EGF receptor-dependent cell proliferation. Mol Cell. 2010;37:551-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 213] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 72. | Slomiany BL, Slomiany A. Role of epidermal growth factor receptor transactivation in the amplification of Helicobacter pylori-elicited induction in gastric mucosal expression of cyclooxygenase-2 and inducible nitric oxide synthase. OA Inflammation. 2013;1:1-8. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 73. | Huang Y, Benaich N, Tape C, Kwok HF, Murphy G. Targeting the sheddase activity of ADAM17 by an anti-ADAM17 antibody D1(A12) inhibits head and neck squamous cell carcinoma cell proliferation and motility via blockage of bradykinin induced HERs transactivation. Int J Biol Sci. 2014;10:702-714. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 74. | Chen Y, Wu X, Bu S, He C, Wang W, Liu J, Guo W, Tan B, Wang Y, Wang J. Promising outcomes of definitive chemoradiation and cetuximab for patients with esophageal squamous cell carcinoma. Cancer Sci. 2012;103:1979-1984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 75. | Lam AK. Molecular biology of esophageal squamous cell carcinoma. Crit Rev Oncol Hematol. 2000;33:71-90. [PubMed] |
| 76. | Okumura H, Uchikado Y, Setoyama T, Matsumoto M, Owaki T, Ishigami S, Natsugoe S. Biomarkers for predicting the response of esophageal squamous cell carcinoma to neoadjuvant chemoradiation therapy. Surg Today. 2014;44:421-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 77. | Yan W, Wistuba II, Emmert-Buck MR, Erickson HS. Squamous Cell Carcinoma - Similarities and Differences among Anatomical Sites. Am J Cancer Res. 2011;1:275-300. [PubMed] |
| 78. | Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, Hirohashi S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci USA. 2008;105:13568-13573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 529] [Cited by in RCA: 605] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 79. | Morris LG, Kaufman AM, Gong Y, Ramaswami D, Walsh LA, Turcan Ş, Eng S, Kannan K, Zou Y, Peng L. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat Genet. 2013;45:253-261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 234] [Cited by in RCA: 279] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 80. | Mohamed AD, Tremblay AM, Murray GI, Wackerhage H. The Hippo signal transduction pathway in soft tissue sarcomas. Biochim Biophys Acta. 2015;1856:121-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 81. | Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15:73-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 867] [Cited by in RCA: 928] [Article Influence: 92.8] [Reference Citation Analysis (0)] |