Published online Nov 7, 2015. doi: 10.3748/wjg.v21.i41.11709
Peer-review started: June 1, 2015
First decision: June 23, 2015
Revised: July 28, 2015
Accepted: September 30, 2015
Article in press: September 30, 2015
Published online: November 7, 2015
Processing time: 158 Days and 18.7 Hours
For two decades Vogelstein’s model has been the paradigm for describing the sequence of molecular changes within protein-coding genes that would lead to overt colorectal cancer (CRC). This model is now too simplistic in the light of recent studies, which have shown that our genome is pervasively transcribed in RNAs other than mRNAs, denominated non-coding RNAs (ncRNAs). The discovery that mutations in genes encoding these RNAs [i.e., microRNAs (miRNAs), long non-coding RNAs, and circular RNAs] are causally involved in cancer phenotypes has profoundly modified our vision of tumour molecular genetics and pathobiology. By exploiting a wide range of different mechanisms, ncRNAs control fundamental cellular processes, such as proliferation, differentiation, migration, angiogenesis and apoptosis: these data have also confirmed their role as oncogenes or tumor suppressors in cancer development and progression. The existence of a sophisticated RNA-based regulatory system, which dictates the correct functioning of protein-coding networks, has relevant biological and biomedical consequences. Different miRNAs involved in neoplastic and degenerative diseases exhibit potential predictive and prognostic properties. Furthermore, the key roles of ncRNAs make them very attractive targets for innovative therapeutic approaches. Several recent reports have shown that ncRNAs can be secreted by cells into the extracellular environment (i.e., blood and other body fluids): this suggests the existence of extracellular signalling mechanisms, which may be exploited by cells in physiology and pathology. In this review, we will summarize the most relevant issues on the involvement of cellular and extracellular ncRNAs in disease. We will then specifically describe their involvement in CRC pathobiology and their translational applications to CRC diagnosis, prognosis and therapy.
Core tip: For many decades the predominant view of molecular functioning of organisms stated that proteins represent the main regulators of genomes and their dysfunctions were the first cause of diseases. This protein-centred view was too simplistic to explain the complexity of cancer. In the last few years many studies have revealed that about 85% of our genome is pervasively transcribed, mainly as non-protein-coding RNAs (ncRNAs). The discovery of countless molecular alterations of ncRNAs related to cancer changed the paradigms of cancer biology. In this review, we report recent advances in the discovery of ncRNAs involved in Colorectal Cancer pathobiologies, and their potential applications in diagnosis, prognosis and therapy.
- Citation: Ragusa M, Barbagallo C, Statello L, Condorelli AG, Battaglia R, Tamburello L, Barbagallo D, Di Pietro C, Purrello M. Non-coding landscapes of colorectal cancer. World J Gastroenterol 2015; 21(41): 11709-11739
- URL: https://www.wjgnet.com/1007-9327/full/v21/i41/11709.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i41.11709
Although Jacob and Monod[1] had suggested in 1961 the centrality of RNA in the flow of genetic information, for many decades the most predominant view remained that proteins represent the main regulatory components of the genome. This “protein-centred” view is indeed simplistic and may be misleading when applied to higher organisms. Recent studies have suggested that there are about 20000 protein-coding genes in the human genome: this is very close to the number of protein-coding genes in C. elegans[2,3]. Such observations suggest that these genes alone are not sufficient to appropriately explain the complexity of higher eukaryotes such as mammals and primates[4,5]. An analogous remark may be made on the model proposed by Fearon and Vogelstein, which describes colorectal cancer (CRC) pathogenesis as a sequence of mutations in protein-coding genes: this model has been the paradigm of CRC pathological evolution and has provided a framework for many other cancer studies[6-8]. However, over the years many observations have shown that this model is not able to recapitulate the complexity and heterogeneity of CRC (in vitro, but especially in vivo)[9,10]. Recent high-throughput studies of the human transcriptome have revealed that about 85%-90% of our genome is dynamically and pervasively transcribed, mostly as non-protein-coding RNAs (ncRNAs)[5,11,12]. In the last decade, many observations have convincingly suggested that ncRNAs significantly contribute to the complex molecular signalling needed to regulate structures and functions in different cells and developmental contexts[13,14]. Accordingly, their dysregulation strongly contributes to the onset and progression of many pathological conditions[15-17]. The discovery of molecular alterations of ncRNAs, related to neoplastic phenotypes, has initiated a shift in the paradigms of cancer biology and has profoundly influenced our understanding of tumour genetics[18-20]. Moreover, several reports have shown that ncRNAs can be secreted by cancer cells into biological fluids, potentially spreading oncogenic signals to other cells: this suggests that cancers may exploit RNA-based, hormone-like mechanisms to advantageously mold their extracellular environment[21,22]. In this review, we will describe recent advances in the discovery of the involvement of ncRNAs in CRC pathobiology, analyzing the contribution of different species of ncRNAs (both cellular and extracellular), their participation to CRC progression and dissemination, and their applications in diagnosis, prognosis and therapy.
It is not precisely known how many ncRNA genes are present in the human genome. ncRNA genes are difficult to identify because of their structural heterogeneity: (1) extreme length variation from 20 nucleotides to > 100 kb; (2) absence of Open Reading Frames (ORFs); (3) no or low evolutionary conservation in many cases; (4) no preferential localization within the genome; and (5) relative tolerance to point mutations[23,24]. The most common (and approximate) classification of ncRNAs is based on their length[25,26]. It divides them into two classes: (1) long non-coding RNAs (lncRNAs), which are longer than 200 nucleotides (nt); and (2) small non-coding RNAs, whose length is equal to or less than 200 nt [i.e., microRNAs (miRNAs), small interfering RNAs (siRNAs), PIWI-interacting RNAs (piRNAs). Other classifications have been proposed to categorize ncRNAs. They can be divided into two classes according to functional features: (1) housekeeping ncRNAs; and (2) regulatory ncRNAs. Housekeeping ncRNAs are constitutively expressed in all cells for their physiological functioning: they include transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), and telomerase RNAs[27,28]. On the other hand, regulatory ncRNAs can be expressed in a cell-specific way, or during defined stages of development and cell differentiation, and finally in response to external stimuli[29,30]. This category comprises miRNAs, siRNAs, lncRNAs (i.e., the RNA molecules more closely involved in cancer biology), and piRNAs[31,32]. It is remarkable that many ncRNAs share features that could allow their assignment to multiple categories, thus eluding systematic classification (for instance: trans-spliced transcripts encompassing huge genomic regions)[33].
This class of ncRNAs includes different types of molecules involved in different steps of RNA synthesis, processing, translation, as well as modulation of transcription initiation [i.e., piRNAs, promoter-associated small RNAs (PASRs)], RNA degradation or protein synthesis block (e.g., miRNAs, siRNAs), RNA maturation (e.g., snoRNAs)[34,35]. Among these RNAs, miRNAs represent the most studied class of ncRNAs: they have also been shown to be tightly associated with neoplastic phenotypes, especially CRC[36,37].
Originally discovered by Victor Ambros in Caenorhabditis elegans, miRNAs are 18-25 nucleotides long, evolutionary conserved, single-stranded RNAs[38]. They are processed from larger precursors through sequential cleavage by two RNase III-like enzymes: Drosha (in the nucleus) and Dicer (in the cytoplasm). By interacting with the protein Ago2, one strand of the resulting duplex can associate with the RNA-induced silencing complex (RISC). In most cases, these miRNAs-RISC complexes target specific mRNA molecules binding to their 3’ untranslated regions (UTRs), which may lead to translational repression or cleavage of the mRNAs[39,40]. The former effect may be due to interference with mRNA cap recognition, inhibition of mRNA interaction with the ribosomal subunit during translation initiation, or increased rate of ribosome drop-off during elongation. Degradation is instead mediated by mRNA decapping and deadenylation[41,42]. Currently, over 2000 human miRNAs have been identified by cloning and sequencing approaches. It is predicted that miRNA genes account for 1%-2% of the human genome and control the expression of at least 50% of all protein-coding genes[43]. MiRNAs regulate fundamental cellular processes, such as cell proliferation, differentiation, migration, angiogenesis and apoptosis: accordingly, they are considered potential oncogenes or tumor suppressors in cancer development and progression[44,45]. The discovery of miRNA dysregulation or mutations, etiopathogenetically related to neoplastic phenotypes, has provided new perspectives for the study of complex gene regulatory networks in CRC and other tumours[46-48].
Long non-coding RNAs are the broadest and most heterogeneous class of non-protein-coding RNAs: their length is greater than 200 nucleotides, frequently reaching up to 100 kb. They include transcripts that may: (1) be located in intergenic regions [long intergenic non-coding RNAs (lincRNAs)][49]; (2) lie within introns of protein coding genes[50]; (3) partially overlap UTRs or promoters of protein coding genes[51,52]; (4) be transcribed from pseudogenes and control the expression of their protein-coding functional paralogs[53]; and (5) be transcribed ultra-conserved regions (tUCRs), which are highly evolutionarily conserved and may be located in intra- or in intergenic regions[54]. Several thousand putative lncRNAs have been already identified and shown to be expressed in a developmental and tissue-specific manner[55,56]. Recent results have convincingly suggested the involvement of lncRNAs in a wide spectrum of biological processes, such as cell-cycle regulation, stemness, differentiation, and apoptosis[57-59]. Unlike miRNAs, which repress gene expression through a common mechanism involving RISC complexes, lncRNAs exhibit a broad range of mechanisms of action through which they are involved in the modulation of epigenetic regulation, alternative splicing, and protein localization and activity. It is probable that these functions are due to the ability of lncRNAs to bind DNA, other RNAs and proteins. LncRNAs can also serve as decoys, which preclude the access to the DNA of regulatory proteins and prevent transcription of specific genes (e.g., lncRNA Gas5, DHFR)[60,61]. Many lncRNAs are associated with polycomb repressive complex-2 (PRC2) or other chromatin-modifying complexes, which modulate epigenetic silencing of target genes (e.g., HOTAIR)[62,63]. LncRNAs can serve as scaffolds to bring two or more proteins into functional complexes (i.e., telomerase RNA TERC)[64], or are required to properly localize protein complexes[65]. A subset of ncRNAs, named Telomere-associated ncRNAs (telomeric repeat-containing RNA, TERRA), negatively regulates telomere length presumably by inhibiting telomerase activity[66]. LncRNAs may be processed into smaller ncRNAs (e.g., MALAT1)[67]; pseudogene transcripts can be processed into siRNAs that regulate protein coding genes through RNA interference (RNAi)[68]. By targeting RNAs through direct sequence complementarity, lncRNAs may also operate as antisense molecules against their targets and modulate alternative splicing events (e.g., antisense of ZEB2), or increase the stability of mRNAs by hiding their miRNA binding sites (e.g., BACEAS)[51,69]. Circular RNAs (circRNAs) belong to an odd, but extremely interesting class of lncRNA molecules, which has been recently described. CircRNAs can act as natural miRNA sponges to lower miRNA levels: accordingly, they perform a critical role modulating the connection between genotype and molecular phenotype[70]. Dysregulation of lncRNAs has been documented for many complex human diseases, including cancer. Dozens of lncRNAs have been reported to have altered expression in neoplasia and to be controlled by specific oncogenic and tumor suppressor pathways[71,72]. These observations strongly suggest that lncRNAs could be added to the list of proto-oncogenes and tumor suppressors, as suspects potentially involved in oncogenesis. Accordingly, they might also be considered as potential biomarkers and targets for novel therapeutic approaches in neoplastic diseases.
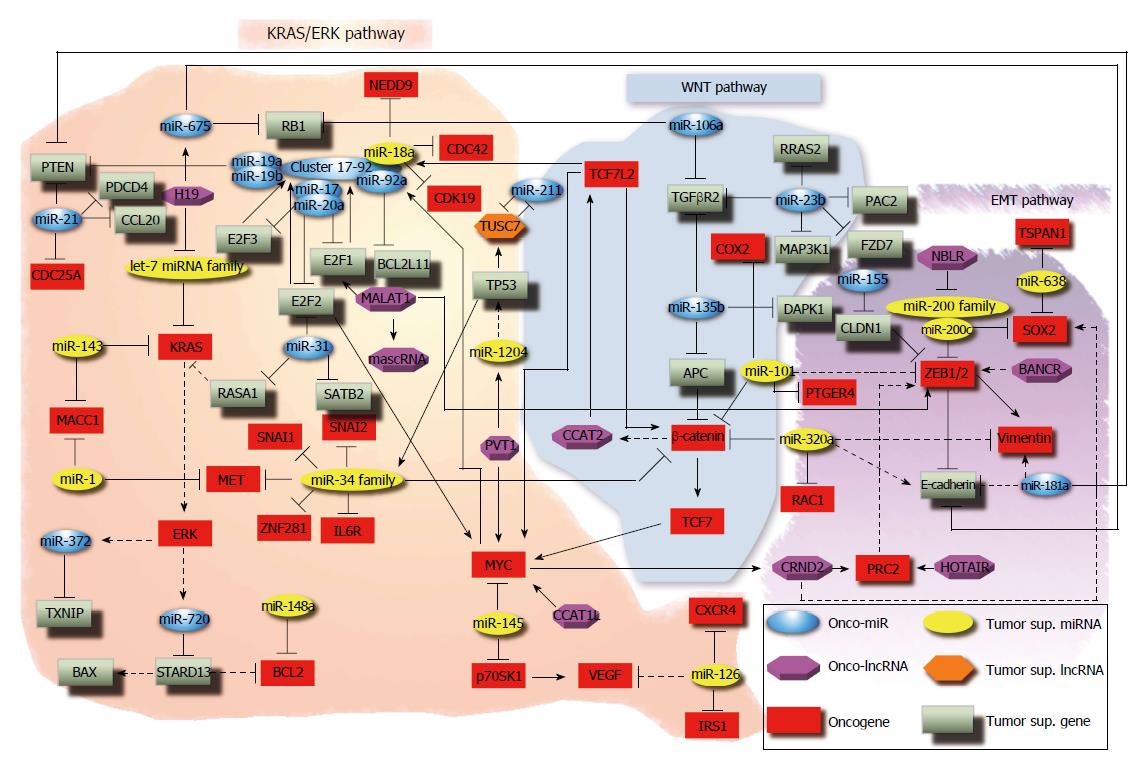
Since the original discovery connecting miRNAs to Chronic Lymphocytic Leukemia[73], researchers have convincingly demonstrated that miRNAs play a critical role in cancer. MiRNA oncogenic activity can be tissue-specific. Altered miRNA expression plays an etiological role in the initiation and progression of colon cancer: global miRNA expression patterns can discriminate between normal tissues and CRC tissues more efficiently than mRNA expression patterns. Furthermore, several investigations have shown the ability of miRNA expression patterns to improve diagnosis of poorly differentiated tumours and predict prognosis in CRC (see next paragraph in this review). In 2003, Michael et al[74] published the first study on miRNAs in CRC: they identified miR-143 and miR-145 as novel dysregulated miRNAs in colon cancer. Since then, the literature on miRNAs in CRC has grown considerably. This paragraph will provide an overview of the etiological connection between the molecular functions of miRNAs and CRC pathobiology. The first discovered CRC-related miRNAs, miR-143 and miR-145, act as tumor suppressor genes and are downregulated in CRC compared with normal colonocytes[74,75]. miR-143 targets KRAS (Kirsten Rat Sarcoma Viral Oncogene Homolog) and MACC1 (metastasis-associated in colon cancer-1), thus playing an important role in the regulation of EGFR (epidermal growth factor receptor) and HGFR (hepatocyte growth factor receptor) signalling[76,77]. Both miR-143 and miR-145 modulate CRC cell proliferation: transfection of DLD-1 and SW480 cells with premiR-143 and premiR-145 reduced cell proliferation[78]. MiR-145 indirectly promotes angiogenesis by binding p70S6K1 mRNA and inhibiting its translation: downregulation of p70S6K1 increases the levels of two proangiogenic factors, HIF1 (hypoxia-inducible factor 1) and VEGF (vascular endothelial growth factor)[79]. MiR-145 also targets the oncogene MYC (v-Myc avian myelocytomatosis viral oncogene homolog), reducing its expression: this explains mR-145-dependent inhibition of cell proliferation in vivo and in vitro[80]. MiR-21 is the most commonly upregulated miRNA in cancer, including CRC[81]. There are many notable downstream effects of elevated miR-21 levels: its genetic locus at 17q23 is amplified in many solid tumours[82,83]; its expression is stimulated by a variety of cancer-associated phenomena, such as inflammation and hypoxia[84-86]. MiR-21 targets various tumor suppressor genes, such as PDCD4 (programmed cell death 4), CCL20 [chemokine (C-C motif) ligand 20], CDC25A (cell division cycle 25 homolog A), PTEN (phosphatase and tensin homolog), thus promoting cell proliferation, invasion/intravasation/metastasis in CRC[87-90].
In addiction to miR-21, many other miRNAs can be induced in cancer cells under hypoxic conditions[91]. In this group of so called “hypoxamir” there is miR-210, which can mediate the hypoxia-induced metastasis of CRC cells[92]. MiR-210 is frequently up-regulated in CRC tissues, 2D, and 3D cultures[92,93]. Its enforced expression in CRC cells promotes the migration and invasion through the repression of its target VMP1 (vacuole membrane protein 1)[92]. Several investigations reported miR-31 upregulation in CRC[94,95]. Overexpression of miR-31 in CRC cells promotes cell proliferation, invasion, and migration in in vivo and in vitro models. Likely, its oncogenic functions are due to targeting of SATB2 (SATB homeobox 2) mRNA, which is followed by SATB2 mRNA and protein downregulation, and by targeting E2F2 (E2F Transcription Factor 2): in turn, E2F2 controls the expression of survivin and other cell cycle genes, such as CCNA2 (Cyclin A2), CDK2 (cyclin-dependent kinase 2), MCM4 (minichromosome maintenance complex component 4), and MYC[96,97]. Notably, members of the E2F family may have a dual function: it has been hypothesized that they may act as oncogenes when they are overexpressed and as tumor suppressors when they are downregulated[96]. MiR-31 also targets RASA1 (RAS p21 GTPase activating protein 1) and inhibits its translation, activating the KRAS signalling pathway[98]. One of the most known oncogenic miRNA clusters in CRC is the miR-17-92 cluster. It consists of six members (miR-17, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92a) with oncogenic functions, all upregulated in CRC[99]. MiR-18a and miR-19 promote angiogenesis by targeting TSP-1 (thrombospondin-1) and CTGF (connective tissue growth factor) mRNAs, respectively[100,101]. Interestingly, different miR-17-92 cluster members can modulate cell proliferation in opposite ways. MiR-19a and miR-19b induce proliferation by acting on PTEN, whereas miR-18a has antiproliferative effects due to its target genes that activate proliferation, such as NEDD9 (neural precursor cell expressed, developmentally down-regulated 9) and CDK19 (cyclin-dependent kinase 19)[102]. MiR-17 and miR-20a target E2F1 (E2F transcription factor 1), whereas miR-20a represses E2F2 and E2F3 (E2F transcription factor 3)[103]. Among miR-17-92 cluster members, only miR-92a has antiapoptotic effects: a negative correlation has been described between miR-92a and BCL2L11 (BCL2-like 11), a pro-apoptotic BCL-2 (B-cell CLL/lymphoma 2) protein family member[101]. Among miR-17-92 cluster members, miR-18a seems to act as a tumor suppressor in CRC: some studies have shown that this miRNA can affect cell proliferation by inhibiting CDC42 (cell division cycle 42)[103], or can promote apoptosis causing hnRNPA1 (heterogeneous nuclear ribonucleoprotein A1) autophagosomal degradation[104]. CDC42 is a small Ras-related GTPase involved in cell cycle progression[105], transendothelial migration through β1 integrin[106], cell motility and cytoskeletal remodelling[107]. The let-7 family is one of the most ancient and conserved group of miRNAs[108]: they act as tumor suppressors in various cancer models, including CRC[109-111]. The let-7 family owes its name to the lethal-7 gene, identified for the first time in C. elegans, where it is involved in development. Later, the same sequence was detected in the genome of Drosophila melanogaster and Homo sapiens, confirming that mature let-7 is highly conserved across animal species. However, the number of members of this family varies in different species: the let-7 family in humans includes 10 mature miRNAs produced from 13 precursor sequences[112]. Several studies have demonstrated that the let-7 family regulates KRAS expression in CRC and other cancer types[111-114]. KRAS is a small monomeric GTPase, involved in signal transduction of stimuli activating proliferation[115]. KRAS mutations or amplifications are frequently detected in CRC patients[116]: they are considered a key step in colorectal carcinogenesis according to the model proposed by Vogelstein[6]. Moreover, KRAS mutation status is a negative predictive factor of the response to anti-EGFR therapy (i.e., Cetuximab)[117]. It has been reported that let-7b and let-7e were downregulated after Cetuximab treatment in a Cetuximab-resistant CRC cell line, suggesting their potential role in the resistance to anti-EGFR therapy[118]. Several reports debated the predictive utility of a let-7 microRNA-binding-site polymorphism in the 3’-UTR of KRAS for CRC outcome, although the results are conflicting[114,119-121]. Together with other let-7 family members, let-7c modulates cell cycle by targeting KRAS, and is also involved in suppressing metastasis via its targets MMP1(matrix metallopeptidase 1) and PBX3 (pre-b-cell leukemia homeobox 3). It has been demonstrated that ectopic expression in Lovo cells or inhibition of let-7c in HT29 cells reduces cell migration and invasion and increases cell motility and invasion, respectively[122]. Overactivation of KRAS signalling could induce the expression of oncogenic miRNAs in CRC. For instance, miR-372 expression is higher in KRAS-mutated CRC samples compared with wild type tumours[123]. miR-372 knockdown decreases cell proliferation and migration and increases apoptosis in CRC cell lines[123]. MiR-372 downregulation results in TXNIP (thioredoxin-interacting protein) overexpression[123]: TXNIP is a tumor suppressor gene involved in apoptosis induction and cell proliferation inhibition[124]. High miR-372 expression is significantly associated with liver metastasis: metastatic CRC samples show higher miR-372 expression than non-metastatic tumours; also, high miR-372 expression is associated with lower 5-year overall survival rate[125]. Similar to miR-372, also miR-720 was found to be more expressed in CRC with mutated KRAS than wild-type KRAS[123,126]. Its overexpression correlates with tumour size, spreading of metastases to distant sites and low 5-year overall survival[126]. MiR-720 knockdown reduces cell proliferation, migration and invasion, and induces apoptosis in CRC cell lines[123,126]. The same miR-720 regulates STARD13 [star-related lipid transfer (START) domain containing 13] expression[126], a GTPase-activating protein (GAP) for Rho and Cdc42. It has been demonstrated that STARD13 knockdown induces upregulation of the antiapoptotic protein BCL2, downregulation of proapoptotic BAX (BCL2-associated X protein), and promotes 3D motility[127]. The miR-200 family is one of the best-known miRNA families in mammals. It consists of five members, which are located at two different loci of chromosome 1 (miR-200a, miR-200b, miR-429) and 12 (miR-200c and miR-141)[128,129]. They are tumor suppressor miRNAs and are significantly involved in inhibition of epithelial-to-mesenchymal transition (EMT), repression of cancer stem cell self-renewal and differentiation, modulation of cell division and apoptosis, and reversal of chemoresistance[128-130]. MiR-200c is particularly interesting in CRC, due to its ability to regulate cell proliferation, invasion, migration, EMT and metastasis. MiR-200c expression is statistically lower in CRC clinical specimens and highly metastatic CRC cell lines. Transfection of CRC cell lines (RKO and SW620) with precursors of miR-200c induced cell proliferation, but reduced invasion and migration. Overexpression of miR-200c in CRC cell lines caused a reduced expression of its target ZEB1/2 (zinc finger E-box binding homeobox 1/2), and resulted in increased E-cadherin and reduced vimentin expression. The associations between miR-200c, its target genes, and EMT markers were validated in primary CRCs and matching liver metastasis tissues[131]. MiR-200c targets SOX2 [SRY (sex determining region Y)-box 2], a pivotal gene required for early development and propagation of undifferentiated embryonic stem cells. Knockdown of miR-200c increased the sphere-forming capacity of CRC cell lines and expression of CRC stem cell markers. MiR-200c suppresses the expression of SOX2, so repressing the activity of the PI3K (phosphoinositide 3-kinase)/AKT (v-Akt murine thymoma viral oncogene homolog 1) pathway[132]. Another tumor suppressor miRNA family controlling EMT in CRC is the miR-34 family. It includes three oncosuppressor members (miR-34a, miR-34b and miR-34c), which are regulated by p53; by promoting mesenchymal-to-epithelial transition (MET) via the inhibition of the EMT-inducing transcription factor SNAI1 (snail family zinc finger-1), they are involved in metastasis suppression. MiR-34a targets involved in CRC invasion and metastasis include IL6R (interleukin 6 receptor), ZNF281 (zinc finger protein 281), MET (MET proto-oncogene, receptor tyrosine kinase), SNAI1, CTNNB1 (β-catenin) and SNAI2 (snail family zinc finger 1)[133-136]. A common event in CRC carcinogenesis is the inactivation of APC (adenomatous polyposis coli), a negative regulator of Wnt signalling pathway through binding to β-catenin, together with Axin and Glycogen Synthase-3, followed by degradation through ubiquitination. Inactivated APC cannot interact with β-catenin, which accumulates in the cytoplasm and then translocates into the nucleus: here β-catenin binds the TCF/LEF (transcription factor/lymphoid enhancer-binding factor) transcription factors, thus leading to MYC and cyclin D transcription and induction of cell proliferation. An miRNA-based regulation of APC has been proposed: miR-135b binds APC mRNA regulating its expression; miR-135b upregulation in CRC cells causes reduced APC protein levels, β-catenin accumulation and Wnt pathway activation[137]. Another study proved that miR-135b also affects apoptosis through its targets TGFβR2 (transforming growth factor β receptor 2) and DAPK1 (death-associated protein kinase 1), both frequently downregulated in CRC[138]. Similar to miR-135b, also miR-135a is frequently upregulated in CRC[139,140]. Acting as an oncomiR, miR-135a promotes cell proliferation, motility and invasion of CRC cells. A target of miR-135a is MTSS1 (metastasis suppressor-1), which is downregulated in CRC, promoting malignant phenotypes in vitro[141]. Interestingly, the oncogenic effects of miR-135b on the Wnt pathway can be counterbalanced by the tumor suppressive action of miR-320a on the same pathway[142-144]. Overexpression of miR-320a in CRC cell lines reduces cell proliferation, blocking cell cycle in G1. Cell cycle arrest is due to miR-320a and β-catenin mRNA interaction, which results in decreased levels of β-catenin protein, together with those of its transcriptional target genes: MYC, cyclin D, survivin[142]. MiR-320a expression inversely correlates with proliferation and migration of CRC cells and is significantly lower in metastatic compared with non-metastatic samples[143,144]. MiR-320a overexpression suppresses migration and invasion by targeting RAC1 (ras-related C3 botulinum toxin substrate-1). It has also been observed that miR-320a overexpression induces vimentin downregulation and E-cadherin upregulation, suggesting that miR-320a is also involved in EMT regulation[144]. Wnt signalling is also controlled by miR-181a. Its upregulation in CRC cell lines induces cell proliferation by means of its WIF1 target (WNT inhibitory factor-1), which is involved in apoptosis promotion, and PTEN, involved in the AKT signalling pathway[145,146]. Moreover, miR-181a overexpression causes downregulation of the epithelial markers E-cadherin and β-catenin and increased expression of vimentin, suggesting that miR-181a also promotes EMT in CRC[145]. MiR-106a acts as an oncogene in CRC, and its upregulation leads to decreased protein levels of RB1 (retinoblastoma protein-1)[81,147]. Active (unphosphorylated) RB1 binds transcription factors E2F and TFDP1 (transcription factor DP-1) at the promoter of the E2F-regulated genes, which are involved in cell cycle progression. The presence of the RB/TFDP1/E2F complex at the promoter inhibits transcription and recruits chromatin-remodelling complexes, inducing gene silencing and blocking cell cycle progression[148]. MiR-106a binds to the 3’UTR of the RB1 mRNA, inhibiting its translation and inducing cell cycle progression in CRC[147]. By targeting TGFβR2, a tumor suppressor commonly inactivated in CRC, miR-106a is also involved in migration and invasion in vitro and in vivo[149,150]. Upregulation of miR-155 in CRC has been shown by several studies[151-153]. It has been demonstrated that this oncomiR promotes migration and invasion by targeting the CLDN1 (claudin 1) protein, a component of tight junctions[152]. In turn, CLDN1 is involved in EMT, causing E-cadherin upregulation through ZEB1 downregulation[154]. Adrenaline-induced miR-155 upregulation modulates cell proliferation and chemoresistance in HT29 CRC cell line. Adrenaline increases miR-155 levels via NFκB (nuclear factor kappa-light-chain-enhancer in activated B cells), inducing cell proliferation and inhibiting cisplatin-induced apoptosis[153]. It has been recently demonstrated that cyclo-oxygenase 2/prostaglandin-endoperoxide synthase 2 (COX2/PTGS2) and prostaglandin E2 (PGE2), a COX2 metabolite, play an important role in colon cancer progression and are potential targets for prevention and therapeutic strategies[155,156]. COX2 is the inducible isoform of the key enzyme in prostaglandin biosynthesis and it has been associated with several malignancies, including CRC[157,158]. Its upregulation promotes cell proliferation in vitro and in vivo[159]. MiR-101 acts as a tumor suppressor in CRC patients and cell lines[158,160]. Among miR-101 targets are COX2 and PTGER4 (prostaglandin E receptor 4), a G protein-coupled cell surface receptor involved in PGE2 signal transduction[158,161]. MiR-101 negatively regulates PTGER4: both miR-101 overexpression and PTGER4 silencing reduce motility and colony formation in vitro[161]. MiR-101 overexpression also promotes cell adhesion and inhibits colonosphere formation, cell growth, invasiveness and survival in hypoxic conditions[160]. MiR-101 repression and Wnt signalling pathway activation show a strong association: miR-101 overexpression reduces β-catenin accumulation in the nucleus and transcriptional activity, leading to increased E-cadherin and decreased ZEB1 mRNA levels; this suggests miR-101 involvement in EMT regulation[160]. MiR-638 downregulation promotes EMT through its target SOX2[162]: it has been demonstrated that SOX2 overexpression induces dedifferentiation and EMT[163]. MiR-638 expression is reduced in CRC tissues and cell lines: its downregulation inversely correlates with tumour progression and predicts poor survival[162,164]. MiR-638 ectopic expression in CRC cell lines results in a reduction of migration, invasion and cell proliferation: this is due to overexpression of its target TSPAN1 (tetraspanin 1)[161,164], which is involved in CRC cell cycle and invasion regulation[165]. MiR-1 levels are reduced in CRC compared with normal tissues: this significantly correlates with MET gene overexpression. Moreover, miR-1 ectopic expression in CRC cell lines impairs MET-induced cell viability, migration and invasion[166]. MiR-23b acts as a tumor suppressor in CRC cell lines by performing a pleiotropic modulation of different cancer-related biological processes. Its ectopic expression strongly inhibits migration and invasion in vitro and primary tumour growth and metastasis in vivo. MiR-23b overexpression reduces cell resistance to anoikis, programmed cell death induced by detachment of anchorage-dependent cells from the extracellular matrix. MiR-23b may also promote mesenchymal-to-epithelial transition: it has been observed that its expression induces E-cadherin upregulation and vimentin reduction. These tumor suppressive effects are due to miR-23b pro-metastatic targets, FZD7 (frizzled class 7 receptor), MAP3K1 (MEKK1, mitogen-activated protein kinase kinase 1, E3 ubiquitin protein ligase), PAK2 [p21 protein (Cdc42/Rac)-activated kinase 2], TGFβR2, RRAS2 [related RAS viral (r-Ras) oncogene homolog 2], and uPA (plasminogen activator, urokinase). Furthermore, miR-23b overexpression inhibits angiogenesis by indirectly suppressing VEGF through FZD7 and MAP3K1[167]. Several papers reported the downregulation of miR-126 in CRC tissues and cell lines[168-170]. Methylation studies suggest that miR-126 is epigenetically silenced by CpG methylation of the promoter of its host gene EGFL7 (EGF-like-domain multiple 7)[171]. Through its target PIK3R2 (phosphoinositide-3-kinase, regulatory subunit 2), a regulatory subunit involved in the PI3K signalling pathway, miR-126 ectopic expression results in cell proliferation impairment[168]. MiR-126 also binds the mRNAs encoding IRS1 (insulin receptor substrate 1) and CXCR4 [chemokine (C-X-C motif) receptor 4], thus regulating AKT and ERK1/2 (mitogen-activated protein kinase 3/1) activation. MiR-126 inhibits cell proliferation, inducing G0/G1 arrest, migration and invasion in CRC cell lines[168-171]. It has been shown that miR-126 ectopic expression in CRC cell lines impairs VEGF secretion in culture medium, suggesting that miR-126 affects angiogenesis[171]. Similarly, DNA hypermethylation of CpG islands seems to cause miR-148a downregulation in various cancers. In CRC cell lines, miR-148a upregulation promotes apoptosis through BCL2 inhibition[172]. Moreover, miR-148a downregulation in CRC correlates with increased tumour size[173]. All data reported in this paragraph (i.e., CRC related miRNAs, their functions and targets) are summarized in Table 1.
| microRNA | Oncogene/tumor suppressor | Process in CRC | Targets | PMID |
| Let-7 | Tumor suppressor | Cell proliferation, migration, invasion, metastasis | KRAS, MMP11, PBX3 | 23167843, 21984339, 16651716 |
| miR-1 | Tumor suppressor | Cell proliferation, invasion, migration | MET | 22179665 |
| miR-16 | Tumor suppressor | Cell proliferation | PTGS2 | 22049153 |
| miR-17-92 cluster | Oncogene | Angiogenesis, proliferation, metastasis | TSP-1, CTGF, PTEN, BCL2L11, E2F1, E2F2, E2F3, TGFBR2, CDKN1A, BIM | 19460962, 16878133, 22308110, 24212931, 21883694 |
| miR-18a | Tumor suppressor | Cell proliferation, migration | CDC42, HNRNPA1 | 25379703, 24166503 |
| miR-21 | Oncogene | Cell proliferation, migration, invasion, metastasis, stemness | PDCD4, CCL20, Cdc25A, TGFBR2, PTEN, RHOB, RASA1 | 22677902, 17968323, 22099878, 19826040, 22120473, 23788041, 23544170, 23174819, 22072622, 21872591, 25663768 |
| miR-23b | Tumor suppressor | Cell migration, invasion, angiogenesis | FZD7, MEKK1, PAK2, TGFBR2, RRAS2, PLAU, VEGF | 22109528 |
| miR-31 | Oncogene | Cell proliferation, invasion, migration | CDKN2B, RASA1 | 21062447, 25202407, 23322774 |
| miR-34 | Tumor suppressor | Migration, invasion, metastasis, EMT | IL6R, ZNF281, MET, SNAIL, CTNNB1, SLUG, ZEB1 | 24642471, 24185900, 23243217, 22134354 |
| miR-101 | Tumor suppressor | Cell proliferation, motility, invasion | EP4, PTGS2 | 22353936, 19133256 |
| miR-103/miR-107 | Oncogene | Invasion, migration, metastasis | DAPK, KLF4 | 22593189 |
| miR-106a | Oncogene | Cell proliferation, migration, invasion, metastasis | RB1, TGFBR2 | 23178825, 22912877 |
| miR-126 | Tumor suppressor | Cell proliferation, migration, invasion, metastasis, angiogenesis | PIK3R2, IRS1, CXCR4, VEGF | 18663744, 24312276, 24189753, 24653631, 23900443 |
| miR-135a | Oncogene | Cell proliferation, migration, invasion | MTSS1 | 23017832 |
| miR-135b | Oncogene | Cell proliferation, invasion | APC, TGFBR2, DAPK1 | 18632633, 24735923 |
| miR-143 | Tumor suppressor | Cell proliferation, metastasis | KRAS, ERK5, MACC1, HK2, IGF1R, DNMT3A | 19137007, 16969504, 22533346, 22691140, 23574723, 19638978 |
| miR-145 | Tumor suppressor | Cell proliferation, angiogenesis | MYC, CDK6, E2F1, CCND2, p70S6K1, PAK4 | 21278451, 15944709, 19843336, 21917858, 22766504 |
| miR-148a | Tumor suppressor | Cell proliferation | BCL2 | 21455217 |
| miR-155 | Oncogene | Cell proliferation, migration, invasion, chemoresistance | CLDN1 | 23588589, 23036199 |
| miR-181a | Oncogene | Cell proliferation, invasion, metastasis, EMT | PTEN, WIF1 | 24685694, 24755295 |
| miR-200c | Tumor suppressor | Cell proliferation, invasion, migration, EMT, metastasis | ZEB1, ETS1, FLT1, CDH1, VIM | 22735571, 22407310 |
| miR-210 | Oncogene | Cell migration, invasion | VMP1 | 24632577 |
| miR-320a | Tumor suppressor | Cell proliferation, migration, invasion, metastasis, EMT | CTNNB1, RAC1, NRP1 | 22459450, 24265291, 22134529 |
| miR-372 | Oncogene | Cell proliferation | TXNIP, LATS2 | 22660396, 22456107 |
| miR-638 | Tumor suppressor | Cell invasion, migration | SOX2, TSPAN1 | 24885288, 25301729 |
| miR-720 | Oncogene | Cell proliferation, invasion, migration | STARD13 | 25286763, 22660396 |
Over the last few years, several papers have reported on the involvement of lncRNAs in CRC genesis and progression through a number of molecular mechanisms. LncRNAs impact on critical CRC signalling pathways by acting both as oncogenes and tumor suppressors through interactions with other regulatory molecules, such as DNA, RNA, and proteins. Although several lncRNAs have been reported to be dysregulated in CRC, which suggests their potential diagnostic/prognostic power (see paragraph “Clinicopathological significance of lncRNAs in CRC”), their molecular mechanisms of action in CRC biology were elucidated only for few of them. One of the most known cancer-related lncRNAs is Metastasis-Associated Lung Adenocarcinoma transcript 1 (MALAT1), located on chromosome 11q13.1 and 8000 nt long[174]. MALAT1 is highly expressed in metastases of various tumours, such as non-small cell lung cancer, hepatocellular carcinoma, and endometrial stromal sarcoma[174-176]. Several intracellular functions of MALAT1 have been proposed. It may have an important role in pre-mRNA metabolism: it is indeed associated with SC35 splicing domains within the nucleus[177]. It is concentrated in nucleoli as a “riboregulator”, controlling expression of its target genes[178]. Moreover, MALAT1 is involved in the regulation of tumor suppressor proteins [e.g., PTB-associated splicing factor (PSF)][179], and has also been found to regulate the activity of E2F1, a pivotal transcription factor for cell cycle progression[180]. In vitro silencing of MALAT1 has been shown to affect bladder cancer cell migration. MALAT1 acts as a negative regulator of EMT-associated ZEB1, ZEB2 and SNAI2, and positive regulator of E-cadherin[181]. Aberrant mitosis, with a large fraction of cells accumulating at the G2/M boundary and increased cell death, result from MALAT1 depletion[182]. Recently, a 3’ end processing mechanism for MALAT1 has been identified: the primary transcription product of the MALAT1 locus is a 6.7 kb nuclear-retained lncRNA and a cytoplasmic 61-nt tRNA-like ncRNA, known as mascRNA (MALAT1-associated small cytoplasmic RNA)[67]. When the MALAT1 RNA fragment containing mascRNA was overexpressed in CRC cells, cell proliferation and invasion were induced. Point mutations of MALAT1 were detected in CRC cell lines and tissues[183]. BRAF-activated non-protein coding RNA (BANCR) seems to be closely associated with V600EBRAF, one of the most frequent mutation types of the BRAF (B-Raf proto-oncogene, serine/threonine kinase) gene in several tumours, including CRC[184]. BANCR is frequently overexpressed in CRC tissues: this overexpression significantly correlates with lymph node metastasis and tumour stage[185]. Enforced expression of BANCR increases cell migration of CRC cell lines, whereas its knockdown inhibits it. BANCR induces EMT by affecting the expression of epithelial and mesenchymal markers through a MEK (mitogen-activated protein kinase kinase 7)/ERK dependent mechanism, thus contributing to CRC migration[185]. Genome wide association studies (GWAS) have identified a set of risk loci, which are linked to susceptibility for different diseases (including CRC) on human chromosome 8q24[186]. Interestingly, this region (about 2 Mb) is within a large protein-coding-gene desert, but several non-coding genes map on it (i.e., CCAT1, CCAT2, PCAT2, and PRNCR1)[187]. The rs6983267 SNP (single nucleotide polymorphism), mapping to the chromosomal region 8q24.21, has been strongly associated with an increased risk of CRC[188]. The genomic region spanning rs6983267 was found to contain DNA enhancer elements, and the allelic variants confer different binding affinity to TCF7L2 [transcription factor 7-like 2 (T-cell specific, HMG-box)], a transcription factor that has a central role in the transcriptional activation of Wnt target genes[189,190]. The SNP status of lncRNA colon cancer associated transcript 2 (CCAT2), which encompasses rs6983267, affects CCAT2 expression: the risk allele G produces more CCAT2 transcripts in CRC[191]. CCAT2 interacts with TCF7L2 and upregulates MYC, miR-17-5p and miR-20a; it also overactivates Wnt signalling[191]. Interestingly, CCAT2 is itself a Wnt downstream target, which suggests the existence of a positive feedback loop[191]. The long isoform of colon cancer associated transcript 1 (CCAT1-L) is upregulated and positively related to tumour stage and progression in CRC[192]. CCAT1-L is located in the nucleus, while the short isoform (CCAT1-S) is found in the cytoplasm. CCAT1-L can interact with a transcriptional enhancer of MYC (MYC-335) by chromatin looping, which in turn interacts with the MYC-promoter[193]. Knockdown of CCAT1-L leads to a decreased level of MYC mRNA, strongly suggesting that this lncRNA may regulate MYC expression in cis[193]. Another lncRNA locus mapping at 8q24 is Plasmacytoma Variant Translocation 1 (PVT1): PVT1 is located downstream of MYC on chromosome 8q24 and produces a wide variety of spliced RNAs, such as a cluster of six miRNAs (i.e., miR-1204, miR-1205, miR-1206, miR-1207-5p, miR-1207-3p, and miR-1208)[194]. PVT1 is upregulated in CRC because of a copy number amplification of chromosome 8q24[195]. Knockdown of PVT1 in CRC cells leads to a significant reduction of proliferation and invasion through activation of TGF-β and apoptotic signalling. Increased PVT1 expression is required for high MYC protein levels in 8q24-amplified CRC cells. PVT1 and MYC protein expression correlate in primary tumours, while ablation of PVT1 from MYC-driven colon cancer line HCT116 diminished its tumorigenic potential[195]. Surprisingly, PVT1 is also a p53-inducible target gene: p53 binds and activates a canonical response element within the vicinity of miR-1204, and induces the endogenous PVT1 transcripts and consequent upregulation of miR-1204[196]. Ectopic expression of miR-1204 leads to increased p53 levels and causes cell death in a partially p53-dependent manner[196]. The most intensively studied lncRNA in different neoplastic diseases is the Hox transcript antisense intergenic RNA (HOTAIR). It is located within the Homeobox C (HOXC) gene cluster on chromosome 12 and is coexpressed with HOXC genes[197]. HOTAIR interacts with PRC2; it functions as a scaffold to assemble PRC2 on the HOXD gene cluster, inducing the trimethylation of histone H3 lysine-27 (H3K27me3) of the HOXD locus[198]. By silencing multiple metastasis suppressor genes (such as HOXD10, PGR, and the protocadherin gene family), HOTAIR epigenetically regulates HOXD expression and promotes metastasis in breast cancer[199]. Expression analysis on CRC reveals a close correlation between HOTAIR expression and members of the PRC2 complex (i.e., SUZ12, EZH2, and H3K27me3): this suggests that HOTAIR expression is associated with epigenetic functions of PRC2, which is involved in maintaining the mesenchymal and undifferentiated status in CRC cells[200]. Similarly to HOTAIR, also the lncRNA CRNDE (colorectal neoplasia differentially expressed) has been shown to be physically and functionally associated to PRC2[201]. Khalil et al[201] showed an overlap in the lists of genes affected by knockdown of CRNDE and PRC2. These data would suggest an involvement of CRNDE in the epigenetic remodelling of chromatin, and specifically in the downregulation of gene expression via targeted histone methylation by the PRC2 complex. The CRNDE promoter is bound by some pluripotency-related transcription factors [i.e., MYC, MYCN (v-Myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog)] and CRNDE knockdown decreases the expression of several pluripotency markers (i.e., SOX2, KLF4, NANOG, and OCT4)[202]. CRNDE expression appears highest in the early stages of mammalian development and progressively decreases thereafter. It is required for the maintenance of pluripotency in mouse embryonic stem cells and is potentially involved in tumorigenesis[202]. The tumor suppressor candidate 7 (TUSC7) is a p53-regulated tumor suppressor, which reduces tumour cell growth both in vitro as well as in vivo in CRC[203]. Specifically, its fourth exon (containing two miR-211 binding sites) is responsible for inhibition of tumour cell growth. In vivo studies confirmed that TUSC7 can bind to miR-211 inducing its downregulation[203]. It has been shown that miR-211 promotes cell growth in CRC cell lines. Accordingly, TUSC7 works as an endogenous miRNA sponge. Interestingly, TUSC7 is a target of miR-211, showing the existence of a reciprocal negative feedback loop between TUSC7 and miR-211[203]. H19 is a paternally imprinted, maternally expressed, oncofetal gene[204]. It is highly expressed from the early stages of embryogenesis to fetal life in many organs, but is nearly completely downregulated postnatally[205]. H19 is upregulated in many cancers, including CRC[206], and acts as the primary miRNA precursor of miR-675 in mammals[207]. RB1 mRNA is a direct target of miR-675: in fact, knockdown of miR-675 increases RB1 expression and at the same time decreases cell growth in CRC. On the contrary, miR-675 gain-of-function causes RB1 downregulation and enhances tumour cell growth. Both TGF-β and hypoxia concomitantly induce H19 and miR-675, together with induction of EMT markers and suppression of E-cadherin protein expression[207]. Interestingly, H19 harbours both canonical and non-canonical binding sites for the let-7 miRNA family, whose critical tumor suppressive role in the development of CRC has already been discussed above. H19 is able to downmodulate let-7 availability by acting as a molecular sponge[208]. Recently, the role of another lncRNA, functionally linked to EMT, has been characterized in CRC[209]. N-BLR is an lncRNA involved in the apoptotic pathway: its inhibition leads to downregulation of XIAP (X-linked inhibitor of apoptosis protein) and subsequent increase of cell death. N-BLR also promotes invasion and migration by modulating vimentin and E-cadherin expression. Intriguingly, N-BLR seems to be regulated by members of the miR-200 family (i.e., miR-141, and miR-200c). As previously mentioned, the miR-200 family has been strongly linked to EMT: indeed, its members target the ZEB1/ZEB2 transcription factors that are repressors of E-cadherin expression. According to the model proposed by Rigoutsos et al[209], the increase of N-BLR expression in CRC samples would attract the available endogenous miR-141/miR-200c and relieve their targeting ZEB1, thereby upregulating its levels. This would lead to a subsequent decrease of E-cadherin; it also would confer a mesenchymal phenotype to the cells, which is correlated with increased invasiveness and migratory potential of CRC cells. All data reported in this paragraph (i.e., CRC related lncRNAs, their functions and targets) are summarized in Table 2.
| lncRNA | Oncogene/tumor suppressor | Biological process | Target | PMID |
| AK123657 | Tumor suppressor | Cell proliferation, invasion | 24809982 | |
| BANCR | Oncogene | Cell migration, EMT | 25013510 | |
| BX648207 | Tumor suppressor | Cell proliferation, invasion | 24809982 | |
| BX649059 | Tumor suppressor | Cell proliferation, invasion | 24809982 | |
| CCAT1-L | Oncogene | MYC expression regulation | MYC-335 | 24662484 |
| CCAT2 | Oncogene | WNT signalling pathway activation | TCF7L2, MYC, miR-17-5p, miR-20a | 23796952 |
| CRNDE | Oncogene | Epigenetic remodelling of chromatin | 19571010 | |
| H19 | Oncogene | miR-675 precursor, cell proliferation, EMT, miRNA sponge | let-7 | 17237358, 19926638 |
| HOTAIR | Oncogene | HOXD expression regulation, metastasis | HOXD10, PGR | 24075995, 20393566 |
| MALAT1 | Oncogene | pre-mRNA metabolism, target gene expression regulator, tumor suppressor protein regulation, cell cycle progression, cell migration, MET | PSF, E2F1 | 17270048, 16878148, 18067128, 22078878, 22722759 |
| mascRNA | Oncogene | Cell proliferation, invasion | 21503572 | |
| N-BLR | Oncogene | Apoptosis, cell migration, invasion | http://dx.doi.org/10.1101/004796 | |
| PVT1 | Oncogene | miRNAs precursor, cell proliferation, invasion | 18194563, 25043044 | |
| TUSC7 | Tumor suppressor | Cell proliferation, miRNA sponge | miR-211 | 23558749 |
Recently, it has been discovered that hundreds of human genes are also expressed in a circular RNA isoform[210]. Initially, these circRNAs were thought to be rare RNA species representing just transcriptional noise. High-throughput sequencing of the RNase R treated, ribosomal-depleted fractions of RNAs showed a ubiquitous expression of circRNAs in human and mouse cells[211]. CircRNAs represent a class of little known post-transcriptional regulators, which compete with other RNAs for binding to miRNAs and RNA binding proteins (RBPs): they may have a role in modulating the local concentration of RBPs and RNAs, as part of the competing endogenous RNA network[211]. Moreover, in contrast with classical ceRNAs, circRNAs have no accessible termini, which makes them resistant to miRNA-mediated RNA degradation or other exonucleolytic activities. ciRS-7 [also termed CDR1as (cerebellar degeneration-related protein 1 antisense)], one of the most studied circRNAs, is a circular miR-7 inhibitor/sponge that binds miR-7, resulting in reduced miR-7 activity and increased levels of miR-7 targets[212]. As miR-7 negatively controls the expression of several oncogenes, impairing miR-7 activity would have an important impact on the cell phenotype. Recently, Bachmayr-Heyda et al[213] found a global reduction of circRNA abundance in CRC cell lines and CRC tissues compared with normal tissues and detected a negative correlation between circRNA expression and proliferation. The authors explained these findings by hypothesizing that the back-splice machinery, responsible for RNA circularization, is dysfunctional in tumoral cells; otherwise, downregulation of circRNAs could be due to increased degradation by oncomiRNAs which are deregulated in CRC[213].
As previously reported, there is overwhelming evidence indicating that post-transcriptional and translational controls, mediated by various miRNAs, exert critical pleiotropic actions on different features of CRC evolution. Based on these premises, tremendous effort was made towards the discovery and characterization of miRNAs as predictive and prognostic biomarkers in CRC. Unsurprisingly, most miRNAs involved in CRC regulation also exhibit potential predictive/prognostic properties (Table 3). MiR-31 is the most frequently mentioned in miRNA-based biomarker discovery studies for CRC patients. MiR-31 is upregulated in colon cancer tissues, compared with adjacent non-neoplastic normal tissues, across all clinical stages; its expression correlates with clinical stages[94,97,214]. It was observed that upregulation of miR-31 in tumour samples is positively associated with advanced tumour-node-metastasis (TNM) stage, presence of lymph node metastasis, and distant metastases[94,97,214]; furthermore, high expression of miR-31 correlates with patients’ short survival[97]. On the other hand, Slaby et al[95] detected upregulation of miR-31 in CRC tissues, but surprisingly found no association with tumour stage. They reported a low expression of miR-31 mainly in poorly differentiated tumours[95]. MiR-21 has been frequently reported as being involved in many neoplasias, including CRC. It has also been demonstrated that it possesses diagnostic and prognostic power. High miR-21 levels correlate with short disease-free survival[215], clinical stage and distant metastases[95]. Analyzing colon and rectal cancer tissues separately, overexpression of miR-21 was an independent prognostic factor of unfavourable recurrence-free survival only for T3-4a colon cancer patients[216]. MiR-21 levels increase, while miR-143 and miR-145 expression decrease, going from well (G1) to poorly (G3) differentiated tumours. Decreased miR-143 and miR-145 expression is also preferentially associated with increased tumour size and localization in the proximal colon[95]. Vickers et al[217] reported that miR-21, miR-135a and miR-335 were upregulated in CRC compared with normal adjacent tissues, in particular in metastatic primary tumours, whereas miR-206 levels inversely correlated with CRC progression. Moreover, let-7a showed elevated expression in metastatic CRC compared with normal mucosa or non-metastatic disease, but only in KRAS-mutated tumours. This prognostic signature of miR-21, miR-135a, miR-206, miR-335, and let-7a, used to detect the presence of metastases, had a specificity of 87% and sensitivity of 76%: these data suggest their application as a prognostic tool in CRC[217]. Díaz et al[218] showed an association between downregulation of miR-126 and age under 50 on CRC diagnosis. miR-126 inversely correlated with metastasis and its expression levels were significantly lower in metastatic CRC than in localized tumours[219]. The same authors also observed a correlation between high miR-106a expression and 5-year disease-free survival and overall survival[218]. Recurrence-free survival of patients with stage II CRC was also independently associated with high expression of miR-320 and miR-498[220]. MiR-181a is upregulated in CRC tissues compared with normal tissues and in liver-metastatic CRC: this suggests its correlation with liver metastasis and also with poor overall survival in EGFR-targeted therapy[145,221]. On the other hand, low miR-181b and let-7g expression correlates with a positive response to 5-fluorouracil-based antimetabolite S-1[222]. Overexpression of miR-15b, miR-181b, miR-191 and miR-200c in CRC, compared to normal colorectal tissues, was reported by Xi et al[223]. Kaplan-Meier survival analysis showed that patients with higher miR-200c expression had shorter survival time compared with patients with lower expression[223]. MiR-372 and miR-720 (both controlled by the KRAS pathway) showed high expression levels, which are significantly associated with CRC tumour size and distant metastases. Metastatic CRC samples showed higher miR-372 and miR-720 expression compared with the non-metastatic samples; moreover, their upregulation was found to be associated with lower 5-year overall survival[125,126]. The presence of metastases in CRC patients is also associated with reduced miR-34a expression, caused by high CpG methylation of miR-34a and miR-34b/c promoters[135]. Expression levels of the oncogenic miR-17-92 cluster and two of its paralogs (miR-106a and miR-106b) are significantly elevated in CRC. Although the authors observed no significant association between deregulation of these miRNAs and clinico-pathological features of patients, high levels of miR-17 were related to reduced overall survival of CRC patients[224]. A promising non invasive approach for CRC screening is to assay stools for molecular biomarkers that mirror the molecular alterations associated with cancer: colon cancer tissues consistently shed cancer cells into stools; accordingly, it would be an ideal substrate to detect specific CRC biomarkers. Several papers showed quantitative changes in the expression of some miRNAs in stools of CRC patients with respect to normal controls. Ahmed et al[225,226], reported 12 upregulated miRNAs (miR-7, miR-17, miR-20a, miR-21, miR-92a, miR-96, miR-106a, miR-134, miR-183, miR-196a, miR-199a-3p and miR-214) and 8 downregulated miRNAs (miR-9, miR-29b, miR-127-5p, miR-138, miR-143, miR-146a, miR-222 and miR-938) in the stools of CRC patients; and these alterations were more pronounced in later carcinoma stages. MiR-21 and -106a upregulation in stools of adenoma and CRC patients was also reported in different papers[227,228]. Other studies on CRC stools suggested miR-18a, miR-31, miR-135b, and miR-221 as potential biomarkers of adenoma and carcinoma[229,230].
| Clinical feature | Oncogenes | Tumor suppressor genes | PMID |
| Fluorouracil based therapy positive response | let-7g, miR-181b, miR-26a-1 (SNP) | 18172508, 20585341 | |
| Metastasis | miR-21, miR-372, miR-720, miR-181a, miR-135a, miR-335 | miR-126, miR-34a, miR-27a (SNP) | 22120473, 24653631, 22456107, 25286763, 23243217, 24755295, 25078482 |
| Poor fluorouracil based therapeutic prognosis | miR-21 | 18230780 | |
| Progression | miR-100 (SNP) | 20585341 | |
| Survival | miR-21, miR-17, miR-181a, miR-181b, miR-372, miR-720, miR-106a, miR-20a, miR-203, miR-423 (SNP), miR-196a-2 (SNP) | miR-200c, miR-320, miR-498, miR-608 (SNP), miR-219-1 (SNP) | 18230780, 18079988, 22065543, 24098024, 18676867, 22456107, 25286763, 22028396, 22661538, 22161766 |
| Tumour size | miR-720 | miR-143, miR-145 | 18196926, 25286763 |
| Undifferentiated phenotype | miR-21 | miR-143, miR-145 | 18196926 |
Several high-throughput profiling and reverse transcription polymerase chain reaction (RT-PCR) studies were published in the last few years that showed the potential diagnostic and prognostic power of lncRNAs in CRC (Table 4). By searching through previously published gene expression microarray data, Hu et al[231] analyzed lncRNA profiles of large cohorts of CRC patients and identified a prognostic six-lncRNA signature. These six lncRNAs (i.e., AK024680, AK026784, AK123657, BX648207, BX649059, and CR622106) significantly correlated with disease-free survival: this signature was able to classify CRC patients into two subgroups: high-risk (shortened survival) and low risk (prolonged overall survival). Moreover, functional experiments demonstrated that repression of lncRNAs AK123657, BX648207 and BX649059 in CRC cell lines increased cell proliferation and invasion[231]. One of the most upregulated lncRNAs associated with CRC is CRNDE[232]. CRNDE exists in different splice variants that may have diagnostic usefulness: the most diagnostically relevant isoform CRNDE-h showed a sensitivity of 95% and a specificity of 96% for distinguishing adenomas from normal tissues and a sensitivity of 80% and a specificity of 96% for carcinoma vs normal tissues[233]. High expression levels of HOTAIR in CRC patients were closely related to poor prognosis[200]. Specifically, Kogo et al[200] divided 100 patients with CRC into two groups: a high HOTAIR expression group and a low expression group: their data show that CRC patients with the highest HOTAIR expression exhibit less differentiated histology, greater tumour size, and higher propensity to liver metastasis than CRC patients with low HOTAIR expression. Similar results were obtained for MALAT1 by Zheng et al[234], who statistically associated expression of MALAT1 to clinico-pathological parameters, disease-free survival, and overall survival. Stage II/III CRC patients with higher expression of MALAT1 showed a significantly higher risk of metastasis after radical surgery and significantly poorer overall survival[234]. 91H (also named LINC01219), an antisense RNA of lncRNA H19, is overexpressed in CRC tumour tissues with respect to adjacent normal tissues. Clinico-pathological factors were compared between CRC patients with high and low expression of 91H. Statistical analysis showed that high expression of 91H was significantly correlated with distant metastases and poorer prognosis[235]. LncRNA PVT1 is a precursor of a number of spliced ncRNAs and of six miRNAs. The biomolecular functions of PVT1 remain elusive, even though several studies were performed to determine them[236,237]. Takahashi et al[238] investigated the clinical significance of PVT1 expression on 164 CRC patients, showing that high PVT1 expression was positively related to the size of lymph node metastasis, venous invasion and poor prognosis. Furthermore, univariate and multivariate analyses showed that PVT1 expression was an independent risk factor for overall survival of CRC patients. NcRAN (non-coding RNA expressed in aggressive neuroblastoma) is an lncRNA whose expression is highly upregulated in neuroblastoma patients with poor prognosis; it could also have an important pathogenetic role in human bladder cancer[239,240]. Qi et al[241] demonstrated that ncRAN expression is significantly reduced in CRC tumour tissues and cell lines, compared with adjacent normal tissues and a normal intestinal mucous cell line. Downregulation of ncRAN was more evident in poorly differentiated or undifferentiated tumours and in CRC with liver metastases. Kaplan-Meier survival analysis showed that CRC patients with lower ncRAN expression had a worse overall survival. Overexpression of PCAT1 (prostate cancer associated transcript 1) was associated with distant metastases and poorer overall survival in CRC patients: in few cases, this was explained by gene copy number variation of the PCAT1 locus[242]. Long intergenic noncoding RNA-p21 (lincRNA-p21) is transcriptionally induced by p53 in CRC cell lines, following apoptosis induction by nutlin-3; it works as a repressor in p53-dependent transcriptional responses. Its expression levels were lower in CRC tumour tissues, when compared with adjacent normal tissues from the same patient. Expression of lincRNA-p21 was higher in rectum cancers with respect to colon cancers. Its overexpression was significantly correlated to higher primary tumour (pT) and vascular invasion[243]. Alaiyan et al[244] screened the expression of CCAT1 in normal colon tissues and in various stages of CRC development (i.e., adenoma, invasive carcinoma, lymph nodes metastases, and distant metastases). By using quantitative RT-PCR (qRT-PCR) and in situ hybridization (ISH), they found: (1) increased CCAT1 expression in colon adenocarcinoma compared with normal tissues; and (2) heterogeneous upregulation across the colon adenoma-carcinoma sequence. CCAT1 upregulation was evident in premalignant conditions and through all disease stages, including advanced metastatic disease: these data suggest its potential role both in transformation and in metastasis. Transcribed ultraconserved regions are a recently discovered class of non-coding RNAs with a high degree of evolutionary conservation: this strongly suggests a critical role in the physiology of mammals and other vertebrates. T-UCRs mainly work as antisense molecules for protein-coding genes and miRNAs; their dysregulation has been reported for different types of tumours[245,246]. Sana et al[247] analyzed the expression in CRC tissues and adjacent normal tissues of t-UCR uc.43, uc.73, uc.134, uc.230, uc.339, uc.388, uc.399, which previously had been found to be associated with CRC. Among these t-UCRs, only uc.73 and uc.388 were significantly downregulated in CRC tissues; uc.73 showed a positive correlation with overall survival of CRC patients. The tumor suppressor lncRNA TUSC7 (LOC285194) is depleted in osteosarcoma, causing abnormal proliferation of osteoblasts; its deletions were associated with poor survival of osteosarcoma patients[248]. Qi et al[249] found that expression levels of TUSC7 significantly decreased also in CRC samples. In addition, this downregulation was correlated with larger tumour size, higher tumour stage, more distant metastases, and poor disease-free survival[249]. By microarray profiling of CRC samples and adjacent normal tissues from non-metastatic and metastatic patients, six aberrantly expressed lncRNAs (i.e., AK097793, ENST00000423943, ENST00000393516, ENST00000428029, RP11-462C24.1, and uc002wvk.2) were identified[250]. Among these, RP11-462C24.1 (also called RPL34-AS1), whose functions are still unknown, exhibited interesting prognostic properties: its expression level decreased as the malignant degree of CRC increased. Furthermore, downregulation of RP11-462C24.1 significantly correlated with more distant metastases and a poor disease-free survival[250]. Some studies on the diagnostic and prognostic power of lncRNAs in CRC focused on their expression in metastatic sites rather than CRC tissues. LncRNA expression profiling in metastatic lymph nodes (MLN), normal lymph nodes (NLN), and tumour tissues from CRC patients showed that 14 lncRNAs were specifically upregulated (e.g., AK021444, ENST00000425785, and AK307796) and 5 lncRNAs were specifically downregulated (e.g., ENST00000465846) in the MLN group compared with the NLN group and tumour tissue group[251]. These data suggest that specific lncRNA dysregulation in MLN may have an important role in facilitating the occurrence of lymph node metastasis in CRC. However, the molecular functions of these lncRNAs are still unknown. The lncRNA highly upregulated in liver cancer (HULC) is usually upregulated in hepatocellular carcinoma and may perturb the circadian rhythm of hepatoma cells[252,253]. HULC is neither expressed in CRC tissues nor in normal colon mucosa; however, it was found to be significantly expressed in CRC with metastases in the liver, but not in lymph nodes[254]. These data suggest that HULC could play a role in the metastatic process of CRC tumours in the liver.
| Clinical feature | Oncogenes | Tumor suppressor genes | PMID |
| Disease-free survival | AK026784, AK024680, MALAT1 | AK123657, CR622106, BX649059, BX648207, TUSC7, RPL34-AS1 | 24809982, 25031737, 23680400, 24908062 |
| Distant metastasis | 91H, PCAT1, CCAT1 | TUSC7, RPL34-AS1 | 25058480, 23640607, 23594791, 23680400, 24908062 |
| Liver metastasis | HOTAIR, HULC | ncRAN | 21862635, 24519959, 19445043 |
| Lymph node metastasis | PVT-1, AK021444, ENST00000425785, AK307796 | ENST00000465846 | 24196785, 25009386 |
| Metastasis risk | MALAT1 | 25031737 | |
| Overall survival | MALAT1, PVT-1, PCAT1 | ncRAN, uc.73 | 25031737, 24196785, 24519959, 23640607, 22328099 |
| Plasma diagnostic marker | CRNDE | 22393467 | |
| Poor prognosis | HOTAIR, 91H, PVT-1 | 21862635, 25058480, 24196785 | |
| Tumour risk | PRNCR1 (SNP) | 24330491 | |
| Tumour size | HOTAIR, PRNCR1 (SNP) | TUSC7 | 21862635, 23680400, 24330491 |
| Tumour stage | CCAT1 | lincRNA-p21, TUSC7, RPL34-AS1 | 24012455, 23594791, 23680400, 24908062 |
| Undifferentiated phenotype | HOTAIR, PRNCR1 (SNP) | ncRAN | 21862635, 24519959, 24330491 |
| Venous invasion | PVT-1 | lincRNA-p21 | 24196785, 24012455 |
One of the most challenging tasks in cancer treatment is to identify patient subpopulations who could benefit from chemotherapy and avoid over-treatment of chemorefractory patients. Discovery of predictive biomarkers would provide information on the likelihood of response to a given therapeutic agent and help to optimize therapy decisions. Much effort was made to find feasible predictive biomarkers in CRC, however, except for KRAS mutations, no clinical study provided markers which have entered into the clinical management of CRC. Recent studies revealed that expression levels of certain miRNAs could be associated with specific therapeutic outcomes in CRC, suggesting that miRNAs may be considered potential predictive biomarkers. Some in vitro studies on CRC cell lines showed that 5-fluorouracil can alter profoundly miRNA expression patterns and miRNAs modulate the expression of key proteins involved in the regulation of cell proliferation, apoptosis and drug response[255,256]. In one of the first studies on CRC patients, miRNA expression was evaluated in cancer biopsies before therapy and two weeks after starting preoperative capecitabine chemoradiotherapy: miR-137 and miR-125b were upregulated two weeks after starting therapy and higher levels of both of them were associated with worse response to the therapy[257]. Hansen et al[258], studied the predictive value of miR-126 in clinical response to capecitabine and oxaliplatin in CRC metastatic patients by in situ hybridization. MiR-126 expression levels were significantly higher in patients responding to therapy with respect to not responding patients: furthermore, its high expression was associated with a more prolonged free survival progression[258]. Overexpression of miR-145 was found in CRC patients who showed a good response to neoadjuvant chemoradiotherapy (5-FU and 50.4 Gy) and tumor regression[259], while miR-17-5p was found increased in chemoresistant patients[260]. Della Vittoria Scarpati et al[261] using microarray expression profiling detected specific miRNA signatures associated with complete response of CRC patients after neoadjuvant chemoradiotherapy. Specifically, they found a set of 13 miRNAs (i.e., 11 upregulated and 2 downregulated miRNAs) strongly associated with good response to chemoradiotherapy. Among them, the upregulation of miR-622 and miR-630 showed 100% sensitivity and specificity in predicting the patients’ response[261]. In a similar study, Svoboda et al[262] compared miRNA profiles of CRC patients classified as sensitive or resistant to neoadjuvant chemoradiotherapy. They identified an overexpression of miR-215, miR-190b, and miR-29b-2* in non-responder patients, while the upregulation of let-7e, miR-196b, miR-450a, miR-450b-5p and miR-99a* was associated with good responders[262]. In another profiling study, Hotchi et al[263] analyzed miRNA expression in rectal cancer patients prior to pre-operative chemoradiotherapy and correlated them to different methods to evaluate the patients’ response. They found different miRNA signatures that discriminated responders from non responders. MiR-223 was the only miRNA common to the different evaluation parameters: its expression was significantly higher in responders compared with non-responders and showed 100% and 78% sensitivity and specificity, respectively, in the prediction of response to pre-operative chemoradiotherapy[263]. In the last few years some evidence on the potential involvement of lncRNAs in molecular mechanisms controlling cancer drug resistance has been reported. Cancer Upregulated Drug Resistant (CUDR) is a lncRNA upregulated in several cancers, including CRC. Enforced overexpression of CUDR induced resistance to doxorubicin and etoposide, as well as drug-induced apoptosis in human squamous carcinoma A431 cells[264]. Lee et al[265], by analyzing the expression of 90 lncRNAs in 5-FU-resistant CRC cell lines, found that SnaR and BACE1AS were significantly downregulated in resistant cell lines; decreased expression of SnaR was responsible for increased cell viability after 5-FU treatment. Recently, colorectal cancer-associated lncRNA (CCAL) was found significantly upregulated in CRC patients with worse response to adjuvant chemotherapy[266]. CCAL regulates CRC progression and multidrug resistance (MDR) through activation of the Wnt pathway by suppressing AP-2α and leading to upregulation of MDR1/P-gp expression[266].
According to the multigenic model of cancer development, combinations of polymorphic genetic variants in susceptibility genes could contribute to CRC risk. Recently, functional polymorphisms in miRNAs or their binding sites in mRNA targets have been discovered in association with pathological phenotypes. A SNP embedded in a miRNA sequence may alter miRNA maturation or its targeting and, accordingly, contribute to the onset and evolution of cancer. One of the first studies showing a prognostic role of miRNA SNPs in CRC reported a significant association of SNP rs7372209 in pri-miR26a-1 to positive chemotherapy response, and SNP rs1834306 in the pri-miR-100 to longer time to progression, suggesting that miRNA polymorphisms could be potential predictors of clinical CRC outcome[267]. Xing et al[268], by screening seven SNPs in 408 CRC patients of a Chinese population, identified two SNPs statistically associated with prognostic features: rs6505162 in pre-miR-423 was correlated to the overall survival and recurrence-free survival, while, rs4919510 in pre-miR-608 was correlated with recurrence-free survival. CRC patients with both SNPs had a 2.84-fold increased risk of recurrence and/or death.These associations were evident only in patients receiving chemotherapy[268]. In another study, performed on 1,097 CRC patients recruited at the University of Texas MD Anderson Cancer Center, rs4919510 in miR-608 associated with increased risk for recurrence and death, and rs213210 in miR-219-1 in association with death of patients with stage III disease were found[269]. Patients carrying both SNPs showed a 5.6-fold increased risk of death. SNP rs11614913 in miR-196a-2 was associated to a significantly increased CRC risk in a Korean population[270]. Association between rs11614913 and CRC susceptibility was also valuated in two different Chinese studies, but the results were conflicting[271,272]. Recently, a study on SNP rs895819 in pre-miR-27a, previously associated with different cancers, was performed to valuate the potential association to CRC susceptibility in a Han Chinese population: GG genotype was significantly associated with risk of CRC and metastasis[273].Data on association to CRC risk of previously mentioned SNPs in miR-196a-2 and miR-27a was not confirmed in a Central-European Caucasian population[274]. The discrepancies in the diagnostic potential of miRNA SNPs in CRC above reported may be due to different molecular pathogenetic mechanisms that differently contribute to cancer or population-specific factors, such as the different genetic backgrounds of the studied population. In addiction to the above-mentioned SNP rs6983267 mapping to a genomic region abundant in lncRNAs, five SNPs in the lncRNA PRNCR1 (prostate cancer associated non-coding RNA 1) were investigated in a Chinese case-control study of 313 cases with CRC and 595 ethnicity-matched controls[275]. The final results of this study were that rs13252298 and rs1456315 are associated with significantly decreased risks of CRC. Tumours of patients expressing rs1456315G were larger than 5 cm. Patients expressing rs7007694C and rs16901946G had a decreased risk of developing poorly differentiated tumours; on the contrary, expression of rs1456315G was found to be associated with an increased risk[275].
Among the most ambitious goals of oncology is the application of fast and non-invasive methods for the identification of specific biomarkers, to be applied to diagnosis and prognosis (i.e., liquid biopsies): these could be useful to discriminate between healthy individuals and cancer patients or as markers of specific responses to therapy. In this regard, blood is the election sample: it contains a high amount of important biomolecules, such as proteins, hormones, metabolites, DNA and different RNAs such as mRNAs and miRNAs. The stoichiometric ratio of its components is highly dynamic and reflects the health status of patients. Blood sampling is fast and easy: serum and plasma have been used for a long time as a source of biomarkers for different conditions. MiRNA profiling is an established method for discriminating cancerous tissues from their healthy counterparts[276]. The discovery, in 2008, of miRNAs in human plasma opened new intriguing perspectives in cancer diagnosis[277,278]. An important requisite of biomarkers is stability in the medium where they are released: miRNAs have been shown to be present in serum in a stable form that prevents their digestion by RNases. Little is known about the origin and features of serum miRNAs, but it is known that they are cell-free molecules: therefore, there have to be other ways to protect them from degradation[277]. Currently, two major release mechanisms of circulating miRNAs have been proposed: (1) active secretion of miRNA-containing shedding microvesicles or exosomes[279]; and (2) active secretion of miRNAs in the form of ribonucleoprotein complexes[280]. Exosomes have been described as a new mechanism of cell-to-cell communication that has similar features to hormonal signalling[281]. Exosomes originate from inward budding of endosomal membranes forming multivesicular bodies, which are secreted into the extracellular environment when late endosomes fuse with the plasma membrane[282]. These nanovesicles are produced by several cell types, both in normal and pathological conditions, and were found in serum and other body fluids[283-286]. Interestingly, exosomes have been shown to carry a set of miRNAs that are specifically targeted to them[281,287-289]. The endosomal sorting complexes required for transport (ESCRT) has been hypothesized to be major regulators of types and amounts of biomolecules sorted into exosomes (i.e., miRNAs, mRNAs, proteins)[290]. The molecular mechanisms involved were uncharacterized until recently. The identification of the role of sumoylated hnRNPA2B1 in transfering miRNAs to exosomes is the first evidence of a possible mechanism involved in the specific miRNA sorting into exosomes[291]. Despite the great interest and growing research in the exosome field, accumulating data suggest that these nanovesicles contain only 10% of circulating miRNAs, and that most of them are associated with RBPs[292]. In 2011, Arroyo et al[293] found that the majority of circulating miRNAs are destabilized by plasma digestion with proteinase K, demonstrating that a protein complex is responsible for their protection in the RNase-enriched plasma environment. Notably, for the miRNAs that are not carried by microvesicles or exosomes, a strong association with Argonaute-2 protein was detected. Another mechanism putatively involved in miRNA preservation in serum is the packaging of circulating miRNAs in high-density lipoproteins (HDL), which in turn can transfer them to recipient cells: this would identify a potential new role for HDL particles in cell communication[294]. Whatever the origin of circulating miRNAs, their expression profiling has proven to have prognostic relevance, as confirmed by several studies. For instance, in 2008 it was shown for the first time that miRNA profiling of circulating exosomes in ovarian cancer patients could potentially be used as a non-invasive diagnostic method of this malignancy. Eight miRNAs (miR-21, miR-141, miR-200a, miR-200c, miR-200b, miR-203, miR-205 and miR-214), known to have a diagnostic value in ovarian cancer tissues, were shown to have the same expression pattern in the exosomes of cancer patients: this was different than that of healthy controls[295]. Moreover, it has been noted that tumour cells possess increased exosome-shedding properties compared with normal cells[295]. In 2009, Rabinowits et al[296] made similar observations in lung cancer patients; in addition, they showed that the serum of lung adenocarcinoma patients is highly enriched in exosomes compared with healthy patients. In 2014, Eichelser et al[297] showed that cell-free miR-101 and miR-373 can be used as breast cancer-specific markers, and that exosomal miR-373, whose expression is increased in Erb-b2 receptor tyrosine kinase 2 (ERBB2/HER2) - negative tumours, has a potential role as a blood-based biomarker for more aggressive tumours. High levels of exosomes in the plasma of CRC patients are secreted by poorly differentiated tumours and are associated with decreased overall survival[298]. Serum levels of exosomal let-7a, miR-21, miR-23a, miR-150, miR-223, miR-1229, and miR-1246 were significantly higher in primary CRC patients at any disease stage than in healthy controls; these miRNAs were significantly downregulated after surgical resection of tumours[299]. Interestingly, exosomes secreted by colon tumour cells play a key role in the recruitment of inflammatory CCR6+CD4+Th17+ into tumour sites and subsequently promote tumour growth[300]. Ragusa et al[289,301] have demonstrated significant alterations of exosomal miRNA cargo in CRC cells following treatment with Cetuximab. The most upregulated miRNAs in exosomes from Caco-2 and HCT-116 cells perform a dual biological function: (1) negative regulation of proliferation or apoptosis induction in cancer cells; and (2) immunosuppression in B- and T- cells. In the last few years, several papers reported the discovery of circulating miRNAs with potential diagnostic or prognostic power in CRC patients (Table 5). The first study on plasma miRNA profiling, performed on 90 CRC patients and 50 healthy controls, showed the significant upregulation of miR-17-3p and miR-92a (both encoded by the miR-17-92 cluster). The authors suggested that elevated plasma levels of miR-92a were not associated with inflammation or other gastrointestinal cancers, but most likely were CRC-related[302]. Upregulation of miR-92a (together with miR-29a) in plasma was also reported by Huang et al[303]. They strengthened these data showing that levels of both miR-29a and miR-92a were significantly reduced in post-operative plasma samples when compared with pre-operative samples. In a different study, the same authors found that the levels of plasma miR-601 and miR-760 were significantly decreased in CRC compared with healthy controls[304]. It is interesting to note that by adding expression data of miR-29a and miR-92a for combined receiver operating characteristic (ROC) curve analysis with miR-760, the resulting AUC (Area Under Curve) increased to 0.943 with 83.3% sensitivity and 93.1% specificity in discriminating CRC from controls: this suggests the additive effect in diagnostic power of these 3 miRNAs[304]. Overexpression of miR-29a was also detected in the serum of CRC patients with liver metastases, but not in CRC patients without metastases: this suggests its potential as a novel non-invasive biomarker for early detection of CRC with liver metastasis[305]. By genome-wide miRNA expression profiling on 196 plasma samples from 123 patients with sporadic CRC and 73 healthy individuals, Giráldez et al[306] found significant upregulation of 6 miRNAs (i.e., miR-15b, miR-18a, miR-19a, miR-19b, miR-29a, and miR-335), suggesting specific expression patterns to be considered as biomarkers for CRC. Upregulation of miR-141 levels was demonstrated in plasma of CRC patients with stage IV with respect to patients with stages I-II and unaffected controls; they were also associated with shorter survival. These expression differences of miR-141 may be related to a differential inflammatory response in CRC patients[307]. Similarly, the expression levels of miR-221 in plasma of CRC patients are significantly higher than in the healthy controls. According to the Kaplan-Meier analysis, upregulation of miR-221 is a significant prognostic factor for poor overall survival in CRC patients: patients with higher miR-221 plasma levels had dramatically lower survival rates than those belonging to the low expression group[308]. Upregulation of miR-155, miR-200c, and miR-210 from serum of CRC patients was reported by Chen et al[309] Expression levels of these miRNAs returned to normal levels in patients with good prognosis after surgery and chemotherapy. Unsurprisingly, several authors reported high levels of miR-21 in plasma and serum of CRC. Upregulation of plasma miR-21 discriminates CRC patients from controls with 90% specificity and sensitivity[310]. MiR-21 levels are significantly elevated in preoperative serum from patients with adenomas and CRCs. High miR-21 expression in serum is statistically associated with tumour size, distant metastases, and poor survival[311]. Recently, Monzo et al[312] showed that miR-21 expression was higher in plasma from blood drawn from the mesenteric veins (MV), near the site of primary CRC than in that drawn from the peripheral veins (PV) of the same CRC patients. Moreover, high levels of miR-21 in MV plasma were associated with shorter disease-free survival of patients. These data suggested that CRC cells release high concentrations of miR-21 in the MV, but that these concentrations are progressively diluted in the peripheral circulatory system. Accordingly, miR-21 dosage from MV would be a stronger prognostic CRC marker than from PV. Recently, Clancy et al[313] analyzed miRNA expression in serum of CRC patients in a literature review on about twenty different published studies: they reported that the 6 circulating miRNAs most frequently found to be dysregulated in CRC are miR-18a-5p, miR-21-5p, miR-29a-5p, miR-92a-5p, miR-143-5p and miR-378-5p. Recently, there has been a notable increase of reports on serum or plasma lncRNAs, potentially useful as biomarkers for different types of tumours. One of the first studies associating circulating lncRNA expression to specific cancers reports the upregulation of lncRNA HULC in blood samples of hepatocellular carcinoma patients[252]. The lncRNA H19 has been found to be highly expressed in the plasma of gastric cancer patients compared with healthy controls, and its levels significantly decrease after tumour resection[314]. Recently, a three lncRNA [urothelial cancer associated 1 (UCA1/CUDR), long stress-induced non-coding transcript 5 (LSINCT-5), phosphatase and tensin homolog pseudogene 1 (PTENP1)] signature in serum was identified as a diagnostic marker for gastric cancers[315]. Tong et al[316] identified the lncRNA POU3F3 (POU class 3 homeobox 3) as a novel serum biomarker for esophageal squamous cell carcinoma, showing that combining POU3F3 expression with plasma levels of the squamous cell carcinoma antigen notably improves efficiency of screening for early detection. Quite surprisingly, only one report has been published to date on the expression of circulating lncRNAs as potential non-invasive diagnostic markers in CRC. The expression level of CRNDE-h transcript in plasma of CRC patients was significantly higher than that of healthy controls, with a sensitivity of 87% and specificity of 93% for diagnosing CRC[233] (Table 5).
| Clinical feature | Oncogene ncRNAs | Tumor suppressor ncRNAs | PMID |
| Diagnostic | miR-17-3p, miR-92a, miR-29a, miR-18a, miR-19a, miR-19b, miR-15b, miR-335, miR-221, exo-let-7a, exo-miR-1229, exo-miR-1246, exo-miR-150, exo-miR-21, exo-miR-223, exo-miR-23a, miR-378, miR-21, CRNDE, miR-155, miR-200c, miR-210 | miR-601, miR-760, miR-143 | 19201770, 19876917, 23267864, 22970209, 20880178, 24705249, 25547322, 22868372, 22393467, 24310813 |
| Metastasis detection | miR-29a, miR-21 | 22018950, 23704278 | |
| Survival | miR-141, miR-221, miR-21 | 21445232, 20880178, 23704278 | |
| Tumour size | miR-21 | 23704278 | |
| Tumour stage | miR-141 | 21445232 |
We have discussed the multiple lines of evidence that pinpoint ncRNAs as key regulators of global gene expression for cancer-related processes. NcRNAs appear to constitute multiple control layers that define the correct functioning of classically known coding layers in cells: they perform this job by modulating chromatin architecture and genome expression (RNA synthesis, stability, processing and translation). The existence of this sophisticated RNA-based regulatory system has important biological and biomedical consequences. Indeed, it could appropriately explain the diversity of phenotypes of mammals, despite their relatively similar proteomes. On the other hand, it is becoming increasingly clear that alterations of these regulatory RNA signals may cause cells to acquire a pathological phenotype, including cancer, as reported for CRC in this review. Both miRNAs and lncRNAs contribute to bring about dysfunctions of critical CRC signalling pathways, such as KRAS/ERK, WNT, and EMT (Figure 1). RNA-based regulation in CRC constitutes a complex, and only partially explored, network of RNA-RNA, RNA-protein interactions that performs a multilayered control on proliferation, invasion, and differentiation in CRC cells. The complexity of this scenario is increased by the presence of several positive and negative regulatory loops. For instance, the miRNA cluster 17-92 represses E2F2, which in turn transcriptionally controls MYC. On the other hand, MYC acts as a transcriptional activator for the cluster 17-92[317]. Moreover, several ncRNAs act as mediators in the cross-talk between different molecular processes in CRC. For instance, MALAT1 induces the expression of E2F2, thus altering MYC signalling and increasing proliferation; it also activates the expression of ZEB1, causing the impairment of epithelial differentiation. Indeed, these apparently different non-coding landscapes converge into a unique RNA network that pervades and controls protein-coding patterns. This key role of ncRNAs makes them very attractive and promising targets for innovative therapeutic approaches. Compared with other strategies, RNA-based therapeutics have several advantages. RNAs are molecules more “druggable” than proteins, because their targeting is mainly based on nucleic acid complementarity, and therefore easy to design and inexpensive to synthesize. RNA-based therapeutic approaches can be divided into two different categories: (1) RNA inhibition therapy, when target RNA is overexpressed; and (2) RNA replacement therapy, when the ncRNA is repressed (this is more feasible for miRNAs). A widely used method for miRNA inhibition is the use of chemically modified oligonucleotides complementary to mature miRNAs (antagomiRs). AntagomiRs quench target miRNAs, which may induce miRISC disruption and miRNA degradation[318]. MiRNA sponges with multiple complementary 3’UTR mRNA sites have been developed to inhibit the activity of miRNA families sharing a common target sequence[319]. Another approach to specifically inhibit miRNA function is to exploit miRNA masks (miR-Masks): these are complementary to the binding sites in the 3’UTR of the target mRNA, so that the miRNA is no longer able to perform its repressive activity[320]. MiRNA replacement therapy is based on the restoration of tumour suppressive miRNAs, expressed at lower levels in cancer. This can be obtained by inserting oligonucleotide mimics, containing the same sequence as the mature endogenous miRNAs, into the cells; alternatively, synthetic pre-miRNA mimics can be used[321,322]. The depletion of oncogenic lncRNAs could be obtained by using similar approaches to those applied to miRNAs: siRNAs and shRNAs (small hairpin RNAs) exhibit great RNA selectivity and knockdown efficiency and utilize a similar silencing molecular mechanism (RISC)[323]. However, in the last few years new appealing methods have been developed, which have the potential to substitute RNAi-based approaches [i.e., antisense oligonucleotides (ASOs), ribozymes and aptamers]. ASOs are single stranded DNAs or RNAs (8-50 nt) with sequence complementary to their target RNAs[324]. When ASOs bind to target RNAs, RNase H1 recognizes the DNA:RNA/RNA:RNA heteroduplexes and catalyzes the cleavage of the RNA molecules[324]. Because ASO-mediated silencing is independent from the RISC machinery, this technology produces less off-targeting effects than siRNAs. As a consequence of their double-stranded structure, siRNAs and shRNAs may trigger an innate immune response and induce high levels of inflammatory cytokines and a toll-like receptor (TLR)-mediated response; such effects are less evident for single-stranded molecules[325]. Ribozymes are naturally synthesized RNA molecules that are able to degrade other RNAs by their intrinsic catalytic functions: their potential application as a therapeutic cancer tool has been already mentioned[326]. Ribozymes bind their targets by complementarity and catalyze cleavage of target RNAs, destabilizing their phosphodiester backbone[326]. An important hurdle to antisense-based lncRNA modulation is the presence of extensive secondary structures along these RNA molecules. Indeed, the effectiveness of siRNAs/shRNAs, ASOs and ribozymes could be seriously impaired by secondary structures in target RNAs[327]. This obstacle could be overcome by aptamers as their targeting mechanism is independent from sequence complementarity[328]. Aptamers are short DNA or RNA oligonucleotides or peptides with a stable three-dimensional structure that specifically bind to their targets (i.e., RNAs, proteins, and small molecules) by fitting the three-dimensional shape of their ligands[329]. Aptamers would be able to recognize distinct RNA structures through tertiary interactions and alter their functions by sequestering them or masking potential binding sites. These RNA-based therapeutic strategies applied to ncRNAs have already shown very promising results in cancer, including CRC. The reintroduction of miR-145 has been demonstrated to perform an antitumoral role in a mouse model of colon cancer[321]. In vivo depletion of miR-21 by ASOs strongly inhibits pancreatic ductal adenocarcinoma tumour growth by inducing cell death[330]. Targeting MALAT1 and HOTAIR with ASOs in mouse xenograft models prevented lung metastasis and inhibited pancreatic tumour growth, respectively[331,332]. However, the main issue related to ncRNA therapeutics is to find a delivery mechanism that would allow their stability in the circulation and efficient tissue-specific uptake, at the same time minimizing off-target side effects[333]. Rapid progress in drug delivery systems has provided optimistic perspectives for advances in this field[334]. Different chemical groups, such as steroids and cholesterol, can be covalently added to miRNAs or ASOs to improve intracellular uptake and extend circulation time[335]. Further delivery mechanisms are the use of adenoviral vectors as carriers of RNA therapeutics, cationic liposomes, or polymer-based nanoparticles, which are able to form micelle-like structures[336]. Recently, an exosomal-based miRNA delivery has been reported to be an effective therapeutic delivery system because these nanoparticles are less toxic and better tolerated by the organism. Moreover, they are naturally used by cells for intercellular communication, and have been proven to protect their molecular cargo in the circulation[337]. For the development of RNA therapeutic strategies, it is interesting to consider that several biological functions of miRNAs and lncRNAs may be partially redundant (Figure 1): accordingly, it may be quite difficult to modulate them by targeting a single ncRNA molecule[338]. This should lead researchers to develop multitargeted RNA therapies in order to increase their impact on oncogenic molecular signalling. For instance, to attenuate KRAS overfunctioning in CRC, exogenous let-7 miRNAs could be introduced to repress KRAS expression; concomitantly, ASOs against lncRNA H19 could be applied to deplete its action as a molecular sponge of the let-7 family. This would strengthen the antiproliferative effects triggered by KRAS repression. Such synergic approaches have already provided very encouraging results by simultaneously administrating miRNA mimics, siRNAs and shRNAs against mRNAs in in vitro and in vivo models[339]. Finally, despite the high number of putatively “druggable” ncRNAs, it is clear that researchers still have to overcome many conceptual and technical challenges to develop effective anticancer strategies that can be applied to patients. However, the results obtained so far are promising and provide optimistic perspectives for the future of anticancer treatments.
Experimental evidence of the potential predictive and prognostic value of cellular and circulating ncRNAs strongly suggests the possibility of exploiting them as useful clinical tools for cancer management. Expression changes of specific ncRNAs can be detected in CRC tissues, utilizing slide-based staining assays: these are standard diagnostic procedures in clinical laboratories. Analysis of plasma or serum ncRNAs could detect tumour biomarkers through PCR at the preoperative stage, indicating their value as early-diagnostic tools. Notwithstanding this, only few reliable diagnostic tests, based on dosage of ncRNAs, have been developed and commercialized to date (e.g., the miRNA-based diagnostic tests of Rosetta Genomics). This slow clinical translatability could be due to different biological and technical factors. Many pre-analytical and analytical factors, including those that are donor/patient-related, can interfere with accurate ncRNA quantification. Due to non homogeneous technical approaches or to real biological heterogeneity of different patient cohorts, expression profiles obtained by various research groups may be different. The real predictive power of these profiles has to be carefully investigated on extended cohorts of patients to understand their actual clinical significance; for instance, data reported on the prognostic value of cellular miR-31 in CRC seem to be very promising, but no univocal results have been obtained to date. Many researchers have compared different protocols for plasma/serum preparation and analyzed the differences between the two biological fluids in terms of amount of circulating miRNAs, but contrasting results have been obtained[277,340,341]. This inconsistency could be explained by the different methods used for the separation of plasma and serum from whole blood (such as different centrifugation protocols): these could lead to different amounts of blood cell contamination[342,343]. However, the effective tumour-specificity of ncRNAs as blood biomarkers remains to be substantiated. Many circulating ncRNAs, whose increased expression had been associated to CRC, could also be related to the presence of other neoplastic diseases. In three independent meta-analysis studies, the diagnostic value of circulating miR-21 in CRC was verified: the authors reported that miR-21 has a moderate potential diagnostic value for CRC[344-346]. However, several papers on different types of carcinoma have also proposed circulating miR-21 as a diagnostic marker of cancer[347-349]. Accordingly, the common upregulation of miR-21 in the blood of patients with different types of cancers makes it a poorly specific diagnostic marker. In fact, Wang et al[350] analyzed the value of circulating miR-21 and reached the conclusion that it might be considered a relevant prognostic biomarker in general carcinomas, but not a sensitive specific diagnostic biomarker. The same observations could also be applied to other CRC-related circulating RNA molecules, commonly found in patients with different tumours or other pathological conditions (e.g., miR-17, miR-18a, miR-20a, miR-29a, and miR-92a)[351-354]. Definitively, all these considerations would suggest that ncRNA-based diagnostic approaches have to be greatly improved. CRC screening should be based on a complex panel of different RNA species (both miRNAs and lncRNAs), rather than on a single or few RNAs. Simultaneous dosage of multiple RNAs having complementary discriminatory and prognostic power could represent a powerful predictive tool. This approach could be improved by analyzing the ratio of expression levels among different ncRNAs (as combined biomarkers), rather than evaluating the concentrations of single RNAs[348,355,356]. Finally, the use of ncRNA-based assays will open new and interesting fields in the screening and monitoring of cancers, including CRC. However, many issues must be addressed before these findings can be translated into a clinically useful screening strategy for CRC patients.
The literature on ncRNAs in CRC has grown considerably over the last decade, confirming their role in this neoplasia. Based on this huge amount of data, there is no doubt that ncRNAs will be important biomarkers in cancer diagnosis and prognosis, as well as therapeutic targets for the treatment of CRC[357]. However, before ncRNAs are routinely applied to clinical settings, it will be critically important to activate large collaborative efforts to fully realize the clinical potential of this approach. Moreover, a deeper knowledge on ncRNA molecular targets, together with new selective methods for RNA delivery to cancer cells, would lead to great benefits for the treatment and management of CRC. NcRNA biomarkers and RNA-based therapies are very promising tools and approaches to supplement current diagnostic/prognostic and therapeutic strategies for CRC.
We wish to thank the Scientific Bureau of the University of Catania for language support.
P- Reviewer: Cecchin E, Cui YY, Mahmood S, Vymetalkova V S- Editor: Yu J L- Editor: A E- Editor: Wang CH
| 1. | Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318-356. [PubMed] |
| 2. | International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931-945. [PubMed] |
| 3. | Ezkurdia I, Juan D, Rodriguez JM, Frankish A, Diekhans M, Harrow J, Vazquez J, Valencia A, Tress ML. Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Hum Mol Genet. 2014;23:5866-5878. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 384] [Cited by in RCA: 339] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 4. | Szymanski M, Barciszewski J. RNA regulation in mammals. Ann N Y Acad Sci. 2006;1067:461-468. [PubMed] |
| 5. | Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11-42. [PubMed] |
| 6. | Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-767. [PubMed] |
| 7. | Gatenby RA, Vincent TL. An evolutionary model of carcinogenesis. Cancer Res. 2003;63:6212-6220. [PubMed] |
| 8. | Arends JW. Molecular interactions in the Vogelstein model of colorectal carcinoma. J Pathol. 2000;190:412-416. [PubMed] |
| 9. | Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108-1113. [PubMed] |
| 10. | Zhou D, Yang L, Zheng L, Ge W, Li D, Zhang Y, Hu X, Gao Z, Xu J, Huang Y. Exome capture sequencing of adenoma reveals genetic alterations in multiple cellular pathways at the early stage of colorectal tumorigenesis. PLoS One. 2013;8:e53310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 11. | Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9:e1003569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 516] [Cited by in RCA: 572] [Article Influence: 47.7] [Reference Citation Analysis (0)] |
| 12. | Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet. 2009;10:833-844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 326] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 13. | Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787-1789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 471] [Cited by in RCA: 458] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 14. | Yazgan O, Krebs JE. Noncoding but nonexpendable: transcriptional regulation by large noncoding RNA in eukaryotes. Biochem Cell Biol. 2007;85:484-496. [PubMed] |
| 15. | Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 760] [Cited by in RCA: 768] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 16. | Liu Y, Ferguson JF, Xue C, Ballantyne RL, Silverman IM, Gosai SJ, Serfecz J, Morley MP, Gregory BD, Li M. Tissue-specific RNA-Seq in human evoked inflammation identifies blood and adipose LincRNA signatures of cardiometabolic diseases. Arterioscler Thromb Vasc Biol. 2014;34:902-912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 17. | Wu P, Zuo X, Deng H, Liu X, Liu L, Ji A. Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res Bull. 2013;97:69-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 291] [Article Influence: 24.3] [Reference Citation Analysis (1)] |
| 18. | Malek E, Jagannathan S, Driscoll JJ. Correlation of long non-coding RNA expression with metastasis, drug resistance and clinical outcome in cancer. Oncotarget. 2014;5:8027-8038. [PubMed] |
| 19. | Raza U, Zhang JD, Sahin O. MicroRNAs: master regulators of drug resistance, stemness, and metastasis. J Mol Med (Berl). 2014;92:321-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 20. | Crea F, Clermont PL, Parolia A, Wang Y, Helgason CD. The non-coding transcriptome as a dynamic regulator of cancer metastasis. Cancer Metastasis Rev. 2014;33:1-16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 78] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 21. | Hwang I. Cell-cell communication via extracellular membrane vesicles and its role in the immune response. Mol Cells. 2013;36:105-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 70] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 22. | Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl). 2013;91:431-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 616] [Cited by in RCA: 731] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 23. | Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15 Spec No 1:R17-R29. [PubMed] |
| 24. | Hüttenhofer A, Vogel J. Experimental approaches to identify non-coding RNAs. Nucleic Acids Res. 2006;34:635-646. [PubMed] |
| 25. | Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3924] [Cited by in RCA: 4442] [Article Influence: 277.6] [Reference Citation Analysis (0)] |
| 26. | Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494-1504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1668] [Cited by in RCA: 1870] [Article Influence: 116.9] [Reference Citation Analysis (0)] |
| 27. | Eddy SR. Non-coding RNA genes and the modern RNA world. Nat Rev Genet. 2001;2:919-929. [PubMed] |
| 28. | Gesteland RF, Cech TR, Atkins JF. The RNA World, 2nd ed. The Nature of Modern RNA Suggests a Prebiotic RNA World. New York: Cold Spring Harbor Laboratory Press 1999; 49-77. [DOI] [Full Text] |
| 29. | Brosnan CA, Voinnet O. The long and the short of noncoding RNAs. Curr Opin Cell Biol. 2009;21:416-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 277] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 30. | Guan D, Zhang W, Zhang W, Liu GH, Belmonte JC. Switching cell fate, ncRNAs coming to play. Cell Death Dis. 2013;4:e464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 31. | Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24:R762-R776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 369] [Cited by in RCA: 377] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 32. | Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26:155-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 357] [Cited by in RCA: 408] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 33. | Horiuchi T, Aigaki T. Alternative trans-splicing: a novel mode of pre-mRNA processing. Biol Cell. 2006;98:135-140. [PubMed] |
| 34. | Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 920] [Cited by in RCA: 958] [Article Influence: 68.4] [Reference Citation Analysis (0)] |
| 35. | Dogini DB, Pascoal VD, Avansini SH, Vieira AS, Pereira TC, Lopes-Cendes I. The new world of RNAs. Genet Mol Biol. 2014;37:285-293. [PubMed] |
| 36. | Bonfrate L, Altomare DF, Di Lena M, Travaglio E, Rotelli MT, De Luca A, Portincasa P. MicroRNA in colorectal cancer: new perspectives for diagnosis, prognosis and treatment. J Gastrointestin Liver Dis. 2013;22:311-320. [PubMed] |
| 37. | Dong Y, Wu WK, Wu CW, Sung JJ, Yu J, Ng SS. MicroRNA dysregulation in colorectal cancer: a clinical perspective. Br J Cancer. 2011;104:893-898. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 94] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 38. | Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-854. [PubMed] |
| 39. | Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642-655. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4017] [Cited by in RCA: 3701] [Article Influence: 231.3] [Reference Citation Analysis (0)] |
| 40. | Wahid F, Shehzad A, Khan T, Kim YY. MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim Biophys Acta. 2010;1803:1231-1243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 533] [Cited by in RCA: 613] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 41. | Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573-1576. [PubMed] |
| 42. | Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. Int J Genomics. 2014;2014:970607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 263] [Cited by in RCA: 388] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 43. | Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597-610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3165] [Cited by in RCA: 3676] [Article Influence: 245.1] [Reference Citation Analysis (0)] |
| 44. | Bueno MJ, Malumbres M. MicroRNAs and the cell cycle. Biochim Biophys Acta. 2011;1812:592-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 323] [Cited by in RCA: 316] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 45. | Ivey KN, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7:36-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 347] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 46. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. [PubMed] |
| 47. | Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-866. [PubMed] |
| 48. | Schee K, Lorenz S, Worren MM, Günther CC, Holden M, Hovig E, Fodstad O, Meza-Zepeda LA, Flatmark K. Deep Sequencing the MicroRNA Transcriptome in Colorectal Cancer. PLoS One. 2013;8:e66165. [PubMed] |
| 49. | Popadin K, Gutierrez-Arcelus M, Dermitzakis ET, Antonarakis SE. Genetic and epigenetic regulation of human lincRNA gene expression. Am J Hum Genet. 2013;93:1015-1026. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 50. | Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, Maracaja-Coutinho V, Verjovski-Almeida S, Machado MC, Reis EM. Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol Cancer. 2011;10:141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 139] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 51. | Beltran M, Puig I, Peña C, García JM, Alvarez AB, Peña R, Bonilla F, de Herreros AG. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756-769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 487] [Cited by in RCA: 519] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 52. | Song X, Wang X, Arai S, Kurokawa R. Promoter-associated noncoding RNA from the CCND1 promoter. Methods Mol Biol. 2012;809:609-622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 53. | Chan WL, Chang JG. Pseudogene-derived endogenous siRNAs and their function. Methods Mol Biol. 2014;1167:227-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 54. | Peng JC, Shen J, Ran ZH. Transcribed ultraconserved region in human cancers. RNA Biol. 2013;10:1771-1777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 57] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 55. | Zhu JG, Shen YH, Liu HL, Liu M, Shen YQ, Kong XQ, Song GX, Qian LM. Long noncoding RNAs expression profile of the developing mouse heart. J Cell Biochem. 2014;115:910-918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 56. | Sun J, Lin Y, Wu J. Long non-coding RNA expression profiling of mouse testis during postnatal development. PLoS One. 2013;8:e75750. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 118] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 57. | Kitagawa M, Kitagawa K, Kotake Y, Niida H, Ohhata T. Cell cycle regulation by long non-coding RNAs. Cell Mol Life Sci. 2013;70:4785-4794. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 215] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 58. | Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011;25:2573-2578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 204] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 59. | Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358-369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1874] [Cited by in RCA: 2166] [Article Influence: 154.7] [Reference Citation Analysis (0)] |
| 60. | Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 720] [Cited by in RCA: 950] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 61. | Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666-670. [PubMed] |
| 63. | Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689-693. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2754] [Cited by in RCA: 2649] [Article Influence: 176.6] [Reference Citation Analysis (0)] |
| 64. | Zappulla DC, Cech TR. RNA as a flexible scaffold for proteins: yeast telomerase and beyond. Cold Spring Harb Symp Quant Biol. 2006;71:217-224. [PubMed] |
| 65. | Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570-1573. [PubMed] |
| 66. | Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. EMBO J. 2009;28:2503-2510. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 200] [Cited by in RCA: 202] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 67. | Wilusz JE, Freier SM, Spector DL. 3’ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919-932. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 600] [Cited by in RCA: 580] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 68. | Chan WL, Yuo CY, Yang WK, Hung SY, Chang YS, Chiu CC, Yeh KT, Huang HD, Chang JG. Transcribed pseudogene ψPPM1K generates endogenous siRNA to suppress oncogenic cell growth in hepatocellular carcinoma. Nucleic Acids Res. 2013;41:3734-3747. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 69. | Liu T, Huang Y, Chen J, Chi H, Yu Z, Wang J, Chen C. Attenuated ability of BACE1 to cleave the amyloid precursor protein via silencing long noncoding RNA BACE1-AS expression. Mol Med Rep. 2014;10:1275-1281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 81] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 70. | Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6253] [Cited by in RCA: 6027] [Article Influence: 502.3] [Reference Citation Analysis (0)] |
| 71. | Luo H, Zhao X, Wan X, Huang S, Wu D. Gene microarray analysis of the lncRNA expression profile in human urothelial carcinoma of the bladder. Int J Clin Exp Med. 2014;7:1244-1254. [PubMed] |
| 72. | Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y, Fang G. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139:437-445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 214] [Cited by in RCA: 228] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 73. | Ganetzky B, Wu CF. Neurogenetics of membrane excitability in Drosophila. Annu Rev Genet. 1986;20:13-44. [PubMed] |
| 74. | Michael MZ, O’ Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882-891. [PubMed] |
| 75. | Arndt GM, Dossey L, Cullen LM, Lai A, Druker R, Eisbacher M, Zhang C, Tran N, Fan H, Retzlaff K. Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer. BMC Cancer. 2009;9:374. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 190] [Cited by in RCA: 207] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 76. | Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, Cai X, Wang K, Wang G, Ba Y. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385-1392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 401] [Cited by in RCA: 450] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 77. | Boardman LA. Overexpression of MACC1 leads to downstream activation of HGF/MET and potentiates metastasis and recurrence of colorectal cancer. Genome Med. 2009;1:36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 43] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 78. | Akao Y, Nakagawa Y, Naoe T. MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncol Rep. 2006;16:845-850. [PubMed] |
| 79. | Xu Q, Liu LZ, Qian X, Chen Q, Jiang Y, Li D, Lai L, Jiang BH. MiR-145 directly targets p70S6K1 in cancer cells to inhibit tumor growth and angiogenesis. Nucleic Acids Res. 2012;40:761-774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 210] [Cited by in RCA: 237] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 80. | Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA. 2009;106:3207-3212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 629] [Cited by in RCA: 700] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 81. | Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257-2261. [PubMed] |
| 82. | Griffin CA, Hawkins AL, Packer RJ, Rorke LB, Emanuel BS. Chromosome abnormalities in pediatric brain tumors. Cancer Res. 1988;48:175-180. [PubMed] |
| 83. | Wu G, Sinclair C, Hinson S, Ingle JN, Roche PC, Couch FJ. Structural analysis of the 17q22-23 amplicon identifies several independent targets of amplification in breast cancer cell lines and tumors. Cancer Res. 2001;61:4951-4955. [PubMed] |
| 84. | Löffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermüller J, Kretzschmar AK, Burger R, Gramatzki M, Blumert C, Bauer K. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330-1333. [PubMed] |
| 85. | Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, Iba H. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol. 2008;378:492-504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 314] [Cited by in RCA: 349] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 86. | Ribas J, Ni X, Haffner M, Wentzel EA, Salmasi AH, Chowdhury WH, Kudrolli TA, Yegnasubramanian S, Luo J, Rodriguez R. miR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 2009;69:7165-7169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 334] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 87. | Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128-2136. [PubMed] |
| 88. | Vicinus B, Rubie C, Faust SK, Frick VO, Ghadjar P, Wagner M, Graeber S, Schilling MK. miR-21 functionally interacts with the 3’UTR of chemokine CCL20 and down-regulates CCL20 expression in miR-21 transfected colorectal cancer cells. Cancer Lett. 2012;316:105-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 89. | Wang P, Zou F, Zhang X, Li H, Dulak A, Tomko RJ, Lazo JS, Wang Z, Zhang L, Yu J. microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res. 2009;69:8157-8165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 265] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 90. | Roy S, Yu Y, Padhye SB, Sarkar FH, Majumdar AP. Difluorinated-curcumin (CDF) restores PTEN expression in colon cancer cells by down-regulating miR-21. PLoS One. 2013;8:e68543. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 117] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 91. | Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859-1867. [PubMed] |
| 92. | Qu A, Du L, Yang Y, Liu H, Li J, Wang L, Liu Y, Dong Z, Zhang X, Jiang X. Hypoxia-inducible MiR-210 is an independent prognostic factor and contributes to metastasis in colorectal cancer. PLoS One. 2014;9:e90952. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 66] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 93. | Ota T, Doi K, Fujimoto T, Tanaka Y, Ogawa M, Matsuzaki H, Kuroki M, Miyamoto S, Shirasawa S, Tsunoda T. KRAS up-regulates the expression of miR-181a, miR-200c and miR-210 in a three-dimensional-specific manner in DLD-1 colorectal cancer cells. Anticancer Res. 2012;32:2271-2275. [PubMed] |
| 94. | Wang CJ, Zhou ZG, Wang L, Yang L, Zhou B, Gu J, Chen HY, Sun XF. Clinicopathological significance of microRNA-31, -143 and -145 expression in colorectal cancer. Dis Markers. 2009;26:27-34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 114] [Reference Citation Analysis (0)] |
| 95. | Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 543] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 96. | Li T, Luo W, Liu K, Lv X, Xi T. miR-31 promotes proliferation of colon cancer cells by targeting E2F2. Biotechnol Lett. 2015;37:523-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 97. | Yang MH, Yu J, Chen N, Wang XY, Liu XY, Wang S, Ding YQ. Elevated microRNA-31 expression regulates colorectal cancer progression by repressing its target gene SATB2. PLoS One. 2013;8:e85353. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 82] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 98. | Sun D, Yu F, Ma Y, Zhao R, Chen X, Zhu J, Zhang CY, Chen J, Zhang J. MicroRNA-31 activates the RAS pathway and functions as an oncogenic MicroRNA in human colorectal cancer by repressing RAS p21 GTPase activating protein 1 (RASA1). J Biol Chem. 2013;288:9508-9518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 156] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 99. | Koga Y, Yasunaga M, Takahashi A, Kuroda J, Moriya Y, Akasu T, Fujita S, Yamamoto S, Baba H, Matsumura Y. MicroRNA expression profiling of exfoliated colonocytes isolated from feces for colorectal cancer screening. Cancer Prev Res (Phila). 2010;3:1435-1442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 164] [Article Influence: 10.9] [Reference Citation Analysis (1)] |
| 100. | Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060-1065. [PubMed] |
| 101. | Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603-1614. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 570] [Cited by in RCA: 692] [Article Influence: 62.9] [Reference Citation Analysis (0)] |
| 102. | Humphreys KJ, Cobiac L, Le Leu RK, Van der Hoek MB, Michael MZ. Histone deacetylase inhibition in colorectal cancer cells reveals competing roles for members of the oncogenic miR-17-92 cluster. Mol Carcinog. 2013;52:459-474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 83] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 103. | Humphreys KJ, McKinnon RA, Michael MZ. miR-18a inhibits CDC42 and plays a tumour suppressor role in colorectal cancer cells. PLoS One. 2014;9:e112288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 75] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 104. | Fujiya M, Konishi H, Mohamed Kamel MK, Ueno N, Inaba Y, Moriichi K, Tanabe H, Ikuta K, Ohtake T, Kohgo Y. microRNA-18a induces apoptosis in colon cancer cells via the autophagolysosomal degradation of oncogenic heterogeneous nuclear ribonucleoprotein A1. Oncogene. 2014;33:4847-4856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 54] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 105. | Stengel KR, Zheng Y. Essential role of Cdc42 in Ras-induced transformation revealed by gene targeting. PLoS One. 2012;7:e37317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 106. | Reymond N, Im JH, Garg R, Vega FM, Borda d’Agua B, Riou P, Cox S, Valderrama F, Muschel RJ, Ridley AJ. Cdc42 promotes transendothelial migration of cancer cells through β1 integrin. J Cell Biol. 2012;199:653-668. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 153] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 107. | Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53-62. [PubMed] |
| 108. | Jérôme T, Laurie P, Louis B, Pierre C. Enjoy the Silence: The Story of let-7 MicroRNA and Cancer. Curr Genomics. 2007;8:229-233. [PubMed] |
| 109. | Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753-3756. [PubMed] |
| 110. | Dahiya N, Sherman-Baust CA, Wang TL, Davidson B, Shih IeM, Zhang Y, Wood W, Becker KG, Morin PJ. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS One. 2008;3:e2436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 249] [Cited by in RCA: 278] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 111. | Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903-906. [PubMed] |
| 112. | Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505-516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 934] [Cited by in RCA: 1100] [Article Influence: 64.7] [Reference Citation Analysis (0)] |
| 113. | Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635-647. [PubMed] |
| 114. | Kjersem JB, Ikdahl T, Guren T, Skovlund E, Sorbye H, Hamfjord J, Pfeiffer P, Glimelius B, Kersten C, Solvang H. Let-7 miRNA-binding site polymorphism in the KRAS 3’UTR; colorectal cancer screening population prevalence and influence on clinical outcome in patients with metastatic colorectal cancer treated with 5-fluorouracil and oxaliplatin +/- cetuximab. BMC Cancer. 2012;12:534. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 115. | Chetty R, Govender D. Gene of the month: KRAS. J Clin Pathol. 2013;66:548-550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 116. | Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449-2460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1274] [Cited by in RCA: 1392] [Article Influence: 87.0] [Reference Citation Analysis (2)] |
| 117. | Mekenkamp LJ, Tol J, Dijkstra JR, de Krijger I, Vink-Börger ME, van Vliet S, Teerenstra S, Kamping E, Verwiel E, Koopman M. Beyond KRAS mutation status: influence of KRAS copy number status and microRNAs on clinical outcome to cetuximab in metastatic colorectal cancer patients. BMC Cancer. 2012;12:292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 118. | Ragusa M, Majorana A, Statello L, Maugeri M, Salito L, Barbagallo D, Guglielmino MR, Duro LR, Angelica R, Caltabiano R. Specific alterations of microRNA transcriptome and global network structure in colorectal carcinoma after cetuximab treatment. Mol Cancer Ther. 2010;9:3396-3409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 84] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 119. | Zhang W, Winder T, Ning Y, Pohl A, Yang D, Kahn M, Lurje G, Labonte MJ, Wilson PM, Gordon MA. A let-7 microRNA-binding site polymorphism in 3’-untranslated region of KRAS gene predicts response in wild-type KRAS patients with metastatic colorectal cancer treated with cetuximab monotherapy. Ann Oncol. 2011;22:104-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 108] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 120. | Smits KM, Paranjape T, Nallur S, Wouters KA, Weijenberg MP, Schouten LJ, van den Brandt PA, Bosman FT, Weidhaas JB, van Engeland M. A let-7 microRNA SNP in the KRAS 3’UTR is prognostic in early-stage colorectal cancer. Clin Cancer Res. 2011;17:7723-7731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 85] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 121. | Langevin SM, Christensen BC. Let-7 microRNA-binding-site polymorphism in the 3’UTR of KRAS and colorectal cancer outcome: a systematic review and meta-analysis. Cancer Med. 2014;3:1385-1395. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 122. | Han HB, Gu J, Zuo HJ, Chen ZG, Zhao W, Li M, Ji DB, Lu YY, Zhang ZQ. Let-7c functions as a metastasis suppressor by targeting MMP11 and PBX3 in colorectal cancer. J Pathol. 2012;226:544-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 121] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 123. | Ragusa M, Statello L, Maugeri M, Majorana A, Barbagallo D, Salito L, Sammito M, Santonocito M, Angelica R, Cavallaro A. Specific alterations of the microRNA transcriptome and global network structure in colorectal cancer after treatment with MAPK/ERK inhibitors. J Mol Med (Berl). 2012;90:1421-1438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 78] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 124. | Sheth SS, Bodnar JS, Ghazalpour A, Thipphavong CK, Tsutsumi S, Tward AD, Demant P, Kodama T, Aburatani H, Lusis AJ. Hepatocellular carcinoma in Txnip-deficient mice. Oncogene. 2006;25:3528-3536. [PubMed] |
| 125. | Yamashita S, Yamamoto H, Mimori K, Nishida N, Takahashi H, Haraguchi N, Tanaka F, Shibata K, Sekimoto M, Ishii H. MicroRNA-372 is associated with poor prognosis in colorectal cancer. Oncology. 2012;82:205-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 126. | Wang X, Kuang Y, Shen X, Zhou H, Chen Y, Han Y, Yuan B, Zhou J, Zhao H, Zhi Q. Evaluation of miR-720 prognostic significance in patients with colorectal cancer. Tumour Biol. 2015;36:719-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 127. | Nasrallah A, Saykali B, Al Dimassi S, Khoury N, Hanna S, El-Sibai M. Effect of StarD13 on colorectal cancer proliferation, motility and invasion. Oncol Rep. 2014;31:505-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 128. | Feng X, Wang Z, Fillmore R, Xi Y. MiR-200, a new star miRNA in human cancer. Cancer Lett. 2014;344:166-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 285] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 129. | Hill L, Browne G, Tulchinsky E. ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer. Int J Cancer. 2013;132:745-754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 206] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 130. | Liu S, Tetzlaff MT, Cui R, Xu X. miR-200c inhibits melanoma progression and drug resistance through down-regulation of BMI-1. Am J Pathol. 2012;181:1823-1835. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 134] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 131. | Hur K, Toiyama Y, Takahashi M, Balaguer F, Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR, Goel A. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2013;62:1315-1326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 390] [Cited by in RCA: 452] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 132. | Lu YX, Yuan L, Xue XL, Zhou M, Liu Y, Zhang C, Li JP, Zheng L, Hong M, Li XN. Regulation of colorectal carcinoma stemness, growth, and metastasis by an miR-200c-Sox2-negative feedback loop mechanism. Clin Cancer Res. 2014;20:2631-2642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 85] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 133. | Rokavec M, Öner MG, Li H, Jackstadt R, Jiang L, Lodygin D, Kaller M, Horst D, Ziegler PK, Schwitalla S. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest. 2014;124:1853-1867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 457] [Cited by in RCA: 625] [Article Influence: 56.8] [Reference Citation Analysis (0)] |
| 134. | Hahn S, Jackstadt R, Siemens H, Hünten S, Hermeking H. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 2013;32:3079-3095. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 146] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 135. | Siemens H, Neumann J, Jackstadt R, Mansmann U, Horst D, Kirchner T, Hermeking H. Detection of miR-34a promoter methylation in combination with elevated expression of c-Met and β-catenin predicts distant metastasis of colon cancer. Clin Cancer Res. 2013;19:710-720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 124] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 136. | Siemens H, Jackstadt R, Hünten S, Kaller M, Menssen A, Götz U, Hermeking H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256-4271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 480] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 137. | Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA, Bolijn A, Meijer GA, Agami R. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795-5802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 376] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 138. | Valeri N, Braconi C, Gasparini P, Murgia C, Lampis A, Paulus-Hock V, Hart JR, Ueno L, Grivennikov SI, Lovat F. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell. 2014;25:469-483. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 240] [Cited by in RCA: 245] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 139. | Xu XM, Qian JC, Deng ZL, Cai Z, Tang T, Wang P, Zhang KH, Cai JP. Expression of miR-21, miR-31, miR-96 and miR-135b is correlated with the clinical parameters of colorectal cancer. Oncol Lett. 2012;4:339-345. [PubMed] |
| 140. | Faltejskova P, Svoboda M, Srutova K, Mlcochova J, Besse A, Nekvindova J, Radova L, Fabian P, Slaba K, Kiss I. Identification and functional screening of microRNAs highly deregulated in colorectal cancer. J Cell Mol Med. 2012;16:2655-2666. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 117] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 141. | Zhou W, Li X, Liu F, Xiao Z, He M, Shen S, Liu S. MiR-135a promotes growth and invasion of colorectal cancer via metastasis suppressor 1 in vitro. Acta Biochim Biophys Sin (Shanghai). 2012;44:838-846. [PubMed] |
| 142. | Sun JY, Huang Y, Li JP, Zhang X, Wang L, Meng YL, Yan B, Bian YQ, Zhao J, Wang WZ. MicroRNA-320a suppresses human colon cancer cell proliferation by directly targeting β-catenin. Biochem Biophys Res Commun. 2012;420:787-792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 128] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 143. | Zhang Y, He X, Liu Y, Ye Y, Zhang H, He P, Zhang Q, Dong L, Liu Y, Dong J. microRNA-320a inhibits tumor invasion by targeting neuropilin 1 and is associated with liver metastasis in colorectal cancer. Oncol Rep. 2012;27:685-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 144. | Zhao H, Dong T, Zhou H, Wang L, Huang A, Feng B, Quan Y, Jin R, Zhang W, Sun J. miR-320a suppresses colorectal cancer progression by targeting Rac1. Carcinogenesis. 2014;35:886-895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 145. | Ji D, Chen Z, Li M, Zhan T, Yao Y, Zhang Z, Xi J, Yan L, Gu J. MicroRNA-181a promotes tumor growth and liver metastasis in colorectal cancer by targeting the tumor suppressor WIF-1. Mol Cancer. 2014;13:86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 127] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 146. | Wei Z, Cui L, Mei Z, Liu M, Zhang D. miR-181a mediates metabolic shift in colon cancer cells via the PTEN/AKT pathway. FEBS Lett. 2014;588:1773-1779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 82] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 147. | Catela Ivkovic T, Aralica G, Cacev T, Loncar B, Kapitanovic S. miR-106a overexpression and pRB downregulation in sporadic colorectal cancer. Exp Mol Pathol. 2013;94:148-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 148. | Chinnam M, Goodrich DW. RB1, development, and cancer. Curr Top Dev Biol. 2011;94:129-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 136] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 149. | Feng B, Dong TT, Wang LL, Zhou HM, Zhao HC, Dong F, Zheng MH. Colorectal cancer migration and invasion initiated by microRNA-106a. PLoS One. 2012;7:e43452. [PubMed] |
| 150. | Muñoz NM, Upton M, Rojas A, Washington MK, Lin L, Chytil A, Sozmen EG, Madison BB, Pozzi A, Moon RT. Transforming growth factor beta receptor type II inactivation induces the malignant transformation of intestinal neoplasms initiated by Apc mutation. Cancer Res. 2006;66:9837-9844. [PubMed] |
| 151. | Wang M, Zhang P, Li Y, Liu G, Zhou B, Zhan L, Zhou Z, Sun X. The quantitative analysis by stem-loop real-time PCR revealed the microRNA-34a, microRNA-155 and microRNA-200c overexpression in human colorectal cancer. Med Oncol. 2012;29:3113-3118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 152. | Zhang GJ, Xiao HX, Tian HP, Liu ZL, Xia SS, Zhou T. Upregulation of microRNA-155 promotes the migration and invasion of colorectal cancer cells through the regulation of claudin-1 expression. Int J Mol Med. 2013;31:1375-1380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 91] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 153. | Pu J, Bai D, Yang X, Lu X, Xu L, Lu J. Adrenaline promotes cell proliferation and increases chemoresistance in colon cancer HT29 cells through induction of miR-155. Biochem Biophys Res Commun. 2012;428:210-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 154. | Singh AB, Sharma A, Smith JJ, Krishnan M, Chen X, Eschrich S, Washington MK, Yeatman TJ, Beauchamp RD, Dhawan P. Claudin-1 up-regulates the repressor ZEB-1 to inhibit E-cadherin expression in colon cancer cells. Gastroenterology. 2011;141:2140-2153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 138] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 155. | Brown JR, DuBois RN. COX-2: a molecular target for colorectal cancer prevention. J Clin Oncol. 2005;23:2840-2855. [PubMed] |
| 156. | Chell S, Kaidi A, Williams AC, Paraskeva C. Mediators of PGE2 synthesis and signalling downstream of COX-2 represent potential targets for the prevention/treatment of colorectal cancer. Biochim Biophys Acta. 2006;1766:104-119. [PubMed] |
| 157. | Misron NA, Looi LM, Nik Mustapha NR. Cyclooxygenase-2 expression in invasive breast carcinomas of no special type and correlation with pathological profiles suggest a role in tumorigenesis rather than cancer progression. Asian Pac J Cancer Prev. 2015;16:1553-1558. [PubMed] |
| 158. | Strillacci A, Griffoni C, Sansone P, Paterini P, Piazzi G, Lazzarini G, Spisni E, Pantaleo MA, Biasco G, Tomasi V. MiR-101 downregulation is involved in cyclooxygenase-2 overexpression in human colon cancer cells. Exp Cell Res. 2009;315:1439-1447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 190] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 159. | Ruan D, So SP. Prostaglandin E2 produced by inducible COX-2 and mPGES-1 promoting cancer cell proliferation in vitro and in vivo. Life Sci. 2014;116:43-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 160. | Strillacci A, Valerii MC, Sansone P, Caggiano C, Sgromo A, Vittori L, Fiorentino M, Poggioli G, Rizzello F, Campieri M. Loss of miR-101 expression promotes Wnt/β-catenin signalling pathway activation and malignancy in colon cancer cells. J Pathol. 2013;229:379-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 95] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 161. | Chandramouli A, Onyeagucha BC, Mercado-Pimentel ME, Stankova L, Shahin NA, LaFleur BJ, Heimark RL, Bhattacharyya AK, Nelson MA. MicroRNA-101 (miR-101) post-transcriptionally regulates the expression of EP4 receptor in colon cancers. Cancer Biol Ther. 2012;13:175-183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 162. | Ma K, Pan X, Fan P, He Y, Gu J, Wang W, Zhang T, Li Z, Luo X. Loss of miR-638 in vitro promotes cell invasion and a mesenchymal-like transition by influencing SOX2 expression in colorectal carcinoma cells. Mol Cancer. 2014;13:118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 163. | Herreros-Villanueva M, Zhang JS, Koenig A, Abel EV, Smyrk TC, Bamlet WR, de Narvajas AA, Gomez TS, Simeone DM, Bujanda L. SOX2 promotes dedifferentiation and imparts stem cell-like features to pancreatic cancer cells. Oncogenesis. 2013;2:e61. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 241] [Cited by in RCA: 262] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 164. | Zhang J, Fei B, Wang Q, Song M, Yin Y, Zhang B, Ni S, Guo W, Bian Z, Quan C. MicroRNA-638 inhibits cell proliferation, invasion and regulates cell cycle by targeting tetraspanin 1 in human colorectal carcinoma. Oncotarget. 2014;5:12083-12096. [PubMed] |
| 165. | Chen L, Yuan D, Zhao R, Li H, Zhu J. Suppression of TSPAN1 by RNA interference inhibits proliferation and invasion of colon cancer cells in vitro. Tumori. 2010;96:744-750. [PubMed] |
| 166. | Migliore C, Martin V, Leoni VP, Restivo A, Atzori L, Petrelli A, Isella C, Zorcolo L, Sarotto I, Casula G. MiR-1 downregulation cooperates with MACC1 in promoting MET overexpression in human colon cancer. Clin Cancer Res. 2012;18:737-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 99] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 167. | Zhang H, Hao Y, Yang J, Zhou Y, Li J, Yin S, Sun C, Ma M, Huang Y, Xi JJ. Genome-wide functional screening of miR-23b as a pleiotropic modulator suppressing cancer metastasis. Nat Commun. 2011;2:554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 164] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 168. | Guo C, Sah JF, Beard L, Willson JK, Markowitz SD, Guda K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer. 2008;47:939-946. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 313] [Cited by in RCA: 329] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 169. | Zhou Y, Feng X, Liu YL, Ye SC, Wang H, Tan WK, Tian T, Qiu YM, Luo HS. Down-regulation of miR-126 is associated with colorectal cancer cells proliferation, migration and invasion by targeting IRS-1 via the AKT and ERK1/2 signaling pathways. PLoS One. 2013;8:e81203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 69] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 170. | Liu Y, Zhou Y, Feng X, An P, Quan X, Wang H, Ye S, Yu C, He Y, Luo H. MicroRNA-126 functions as a tumor suppressor in colorectal cancer cells by targeting CXCR4 via the AKT and ERK1/2 signaling pathways. Int J Oncol. 2014;44:203-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 69] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 171. | Zhang Y, Wang X, Xu B, Wang B, Wang Z, Liang Y, Zhou J, Hu J, Jiang B. Epigenetic silencing of miR-126 contributes to tumor invasion and angiogenesis in colorectal cancer. Oncol Rep. 2013;30:1976-1984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 93] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 172. | Zhang H, Li Y, Huang Q, Ren X, Hu H, Sheng H, Lai M. MiR-148a promotes apoptosis by targeting Bcl-2 in colorectal cancer. Cell Death Differ. 2011;18:1702-1710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 180] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 173. | Chen Y, Song Y, Wang Z, Yue Z, Xu H, Xing C, Liu Z. Altered expression of MiR-148a and MiR-152 in gastrointestinal cancers and its clinical significance. J Gastrointest Surg. 2010;14:1170-1179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 174] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 174. | Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031-8041. [PubMed] |
| 175. | Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810-1816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 478] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 176. | Yamada K, Kano J, Tsunoda H, Yoshikawa H, Okubo C, Ishiyama T, Noguchi M. Phenotypic characterization of endometrial stromal sarcoma of the uterus. Cancer Sci. 2006;97:106-112. [PubMed] |
| 177. | Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. [PubMed] |
| 178. | Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851-858. [PubMed] |
| 179. | Garen A, Song X. Regulatory roles of tumor-suppressor proteins and noncoding RNA in cancer and normal cell functions. Int J Cancer. 2008;122:1687-1689. [PubMed] |
| 180. | Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773-788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 458] [Cited by in RCA: 513] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 181. | Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8:2289-2294. [PubMed] |
| 182. | Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925-938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1521] [Cited by in RCA: 1737] [Article Influence: 115.8] [Reference Citation Analysis (0)] |
| 183. | Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3’ end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39:169-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 183] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 184. | Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949-954. [PubMed] |
| 185. | Guo Q, Zhao Y, Chen J, Hu J, Wang S, Zhang D, Sun Y. BRAF-activated long non-coding RNA contributes to colorectal cancer migration by inducing epithelial-mesenchymal transition. Oncol Lett. 2014;8:869-875. [PubMed] |
| 186. | Grisanzio C, Freedman ML. Chromosome 8q24-Associated Cancers and MYC. Genes Cancer. 2010;1:555-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 87] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 187. | Huppi K, Pitt JJ, Wahlberg BM, Caplen NJ. The 8q24 gene desert: an oasis of non-coding transcriptional activity. Front Genet. 2012;3:69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 121] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 188. | Haiman CA, Le Marchand L, Yamamato J, Stram DO, Sheng X, Kolonel LN, Wu AH, Reich D, Henderson BE. A common genetic risk factor for colorectal and prostate cancer. Nat Genet. 2007;39:954-956. [PubMed] |
| 189. | Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882-884. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 554] [Cited by in RCA: 543] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 190. | Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, Björklund M, Wei G, Yan J, Niittymäki I. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885-890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 404] [Cited by in RCA: 406] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 191. | Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, Nishida N, Gafà R, Song J, Guo Z. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446-1461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 456] [Cited by in RCA: 513] [Article Influence: 42.8] [Reference Citation Analysis (0)] |
| 192. | Nissan A, Stojadinovic A, Mitrani-Rosenbaum S, Halle D, Grinbaum R, Roistacher M, Bochem A, Dayanc BE, Ritter G, Gomceli I. Colon cancer associated transcript-1: a novel RNA expressed in malignant and pre-malignant human tissues. Int J Cancer. 2012;130:1598-1606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 240] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 193. | Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513-531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 457] [Cited by in RCA: 579] [Article Influence: 52.6] [Reference Citation Analysis (0)] |
| 194. | Beck-Engeser GB, Lum AM, Huppi K, Caplen NJ, Wang BB, Wabl M. Pvt1-encoded microRNAs in oncogenesis. Retrovirology. 2008;5:4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 91] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 195. | Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell TC. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 506] [Cited by in RCA: 588] [Article Influence: 53.5] [Reference Citation Analysis (0)] |
| 196. | Barsotti AM, Beckerman R, Laptenko O, Huppi K, Caplen NJ, Prives C. p53-Dependent induction of PVT1 and miR-1204. J Biol Chem. 2012;287:2509-2519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 164] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 197. | Wu Y, Zhang L, Wang Y, Li H, Ren X, Wei F, Yu W, Wang X, Zhang L, Yu J. Long noncoding RNA HOTAIR involvement in cancer. Tumour Biol. 2014;35:9531-9538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 116] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 198. | Li L, Liu B, Wapinski OL, Tsai MC, Qu K, Zhang J, Carlson JC, Lin M, Fang F, Gupta RA. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5:3-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 266] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 199. | Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071-1076. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4202] [Cited by in RCA: 4231] [Article Influence: 282.1] [Reference Citation Analysis (0)] |
| 200. | Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320-6326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 922] [Cited by in RCA: 1039] [Article Influence: 74.2] [Reference Citation Analysis (0)] |
| 201. | Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667-11672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2182] [Cited by in RCA: 2352] [Article Influence: 147.0] [Reference Citation Analysis (0)] |
| 202. | Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295-300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1477] [Cited by in RCA: 1574] [Article Influence: 112.4] [Reference Citation Analysis (0)] |
| 203. | Liu Q, Huang J, Zhou N, Zhang Z, Zhang A, Lu Z, Wu F, Mo YY. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41:4976-4987. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 293] [Cited by in RCA: 334] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 204. | Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A, Galun E. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007;2:e845. [PubMed] |
| 205. | Lustig O, Ariel I, Ilan J, Lev-Lehman E, De-Groot N, Hochberg A. Expression of the imprinted gene H19 in the human fetus. Mol Reprod Dev. 1994;38:239-246. [PubMed] |
| 206. | Cui H, Onyango P, Brandenburg S, Wu Y, Hsieh CL, Feinberg AP. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002;62:6442-6446. [PubMed] |
| 207. | Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, Kwok TT. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 340] [Cited by in RCA: 374] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 208. | Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 783] [Cited by in RCA: 895] [Article Influence: 74.6] [Reference Citation Analysis (0)] |
| 209. | Rigoutsos I, Lee SK, Nam SY, Ivkovic TC, Pichler M, Rossi S, Clark P, Yi J, Ling H, Shimizu M. N-BLR, a primate-specific non-coding transcript, modulates the epithelial-to-mesenchymal transition and leads to colorectal cancer invasion and migration. Available from: http://biorxiv.org/content/early/2014/05/06/004796. |
| 210. | Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1589] [Cited by in RCA: 1959] [Article Influence: 150.7] [Reference Citation Analysis (0)] |
| 211. | Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4631] [Cited by in RCA: 6048] [Article Influence: 504.0] [Reference Citation Analysis (0)] |
| 212. | Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609-5612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 629] [Cited by in RCA: 758] [Article Influence: 63.2] [Reference Citation Analysis (0)] |
| 213. | Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 518] [Cited by in RCA: 609] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 214. | Cekaite L, Rantala JK, Bruun J, Guriby M, Agesen TH, Danielsen SA, Lind GE, Nesbakken A, Kallioniemi O, Lothe RA. MiR-9, -31, and -182 deregulation promote proliferation and tumor cell survival in colon cancer. Neoplasia. 2012;14:868-879. [PubMed] |
| 215. | Nielsen BS, Jørgensen S, Fog JU, Søkilde R, Christensen IJ, Hansen U, Brünner N, Baker A, Møller S, Nielsen HJ. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin Exp Metastasis. 2011;28:27-38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 203] [Cited by in RCA: 226] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 216. | Kang WK, Lee JK, Oh ST, Lee SH, Jung CK. Stromal expression of miR-21 in T3-4a colorectal cancer is an independent predictor of early tumor relapse. BMC Gastroenterol. 2015;15:2. [PubMed] |
| 217. | Vickers MM, Bar J, Gorn-Hondermann I, Yarom N, Daneshmand M, Hanson JE, Addison CL, Asmis TR, Jonker DJ, Maroun J. Stage-dependent differential expression of microRNAs in colorectal cancer: potential role as markers of metastatic disease. Clin Exp Metastasis. 2012;29:123-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 91] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 218. | Díaz R, Silva J, García JM, Lorenzo Y, García V, Peña C, Rodríguez R, Muñoz C, García F, Bonilla F. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosomes Cancer. 2008;47:794-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 183] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 219. | Yin J, Bai Z, Song J, Yang Y, Wang J, Han W, Zhang J, Meng H, Ma X, Yang Y. Differential expression of serum miR-126, miR-141 and miR-21 as novel biomarkers for early detection of liver metastasis in colorectal cancer. Chin J Cancer Res. 2014;26:95-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 39] [Reference Citation Analysis (0)] |
| 220. | Schepeler T, Reinert JT, Ostenfeld MS, Christensen LL, Silahtaroglu AN, Dyrskjøt L, Wiuf C, Sørensen FJ, Kruhøffer M, Laurberg S. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68:6416-6424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 388] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 221. | Pichler M, Winter E, Ress AL, Bauernhofer T, Gerger A, Kiesslich T, Lax S, Samonigg H, Hoefler G. miR-181a is associated with poor clinical outcome in patients with colorectal cancer treated with EGFR inhibitor. J Clin Pathol. 2014;67:198-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 222. | Nakajima G, Hayashi K, Xi Y, Kudo K, Uchida K, Takasaki K, Yamamoto M, Ju J. Non-coding MicroRNAs hsa-let-7g and hsa-miR-181b are Associated with Chemoresponse to S-1 in Colon Cancer. Cancer Genomics Proteomics. 2006;3:317-324. [PubMed] |
| 223. | Xi Y, Formentini A, Chien M, Weir DB, Russo JJ, Ju J, Kornmann M, Ju J. Prognostic Values of microRNAs in Colorectal Cancer. Biomark Insights. 2006;2:113-121. [PubMed] |
| 224. | Yu G, Tang JQ, Tian ML, Li H, Wang X, Wu T, Zhu J, Huang SJ, Wan YL. Prognostic values of the miR-17-92 cluster and its paralogs in colon cancer. J Surg Oncol. 2012;106:232-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 77] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 225. | Ahmed FE, Ahmed NC, Vos PW, Bonnerup C, Atkins JN, Casey M, Nuovo GJ, Naziri W, Wiley JE, Mota H. Diagnostic microRNA markers to screen for sporadic human colon cancer in stool: I. Proof of principle. Cancer Genomics Proteomics. 2013;10:93-113. [PubMed] |
| 226. | Ahmed FE, Jeffries CD, Vos PW, Flake G, Nuovo GJ, Sinar DR, Naziri W, Marcuard SP. Diagnostic microRNA markers for screening sporadic human colon cancer and active ulcerative colitis in stool and tissue. Cancer Genomics Proteomics. 2006;6:281-295. [PubMed] |
| 227. | Link A, Balaguer F, Shen Y, Nagasaka T, Lozano JJ, Boland CR, Goel A. Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers Prev. 2010;19:1766-1774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 240] [Cited by in RCA: 251] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 228. | Koga Y, Yamazaki N, Yamamoto Y, Yamamoto S, Saito N, Kakugawa Y, Otake Y, Matsumoto M, Matsumura Y. Fecal miR-106a is a useful marker for colorectal cancer patients with false-negative results in immunochemical fecal occult blood test. Cancer Epidemiol Biomarkers Prev. 2013;22:1844-1852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 66] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 229. | Yau TO, Wu CW, Dong Y, Tang CM, Ng SS, Chan FK, Sung JJ, Yu J. microRNA-221 and microRNA-18a identification in stool as potential biomarkers for the non-invasive diagnosis of colorectal carcinoma. Br J Cancer. 2014;111:1765-1771. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 101] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 230. | Wu CW, Ng SC, Dong Y, Tian L, Ng SS, Leung WW, Law WT, Yau TO, Chan FK, Sung JJ. Identification of microRNA-135b in stool as a potential noninvasive biomarker for colorectal cancer and adenoma. Clin Cancer Res. 2014;20:2994-3002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 111] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 231. | Hu Y, Chen HY, Yu CY, Xu J, Wang JL, Qian J, Zhang X, Fang JY. A long non-coding RNA signature to improve prognosis prediction of colorectal cancer. Oncotarget. 2014;5:2230-2242. [PubMed] |
| 232. | Nagaraj SH, Reverter A. A Boolean-based systems biology approach to predict novel genes associated with cancer: Application to colorectal cancer. BMC Syst Biol. 2011;5:35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 233. | Graham LD, Pedersen SK, Brown GS, Ho T, Kassir Z, Moynihan AT, Vizgoft EK, Dunne R, Pimlott L, Young GP. Colorectal Neoplasia Differentially Expressed (CRNDE), a Novel Gene with Elevated Expression in Colorectal Adenomas and Adenocarcinomas. Genes Cancer. 2011;2:829-840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 221] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 234. | Zheng HT, Shi DB, Wang YW, Li XX, Xu Y, Tripathi P, Gu WL, Cai GX, Cai SJ. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7:3174-3181. [PubMed] |
| 235. | Deng Q, He B, Gao T, Pan Y, Sun H, Xu Y, Li R, Ying H, Wang F, Liu X. Up-regulation of 91H promotes tumor metastasis and predicts poor prognosis for patients with colorectal cancer. PLoS One. 2014;9:e103022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 71] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 236. | You L, Chang D, Du HZ, Zhao YP. Genome-wide screen identifies PVT1 as a regulator of Gemcitabine sensitivity in human pancreatic cancer cells. Biochem Biophys Res Commun. 2011;407:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 102] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 237. | Alvarez ML, DiStefano JK. Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PLoS One. 2011;6:e18671. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 138] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 238. | Takahashi Y, Sawada G, Kurashige J, Uchi R, Matsumura T, Ueo H, Takano Y, Eguchi H, Sudo T, Sugimachi K. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. Br J Cancer. 2014;110:164-171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 219] [Cited by in RCA: 261] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 239. | Yu M, Ohira M, Li Y, Niizuma H, Oo ML, Zhu Y, Ozaki T, Isogai E, Nakamura Y, Koda T. High expression of ncRAN, a novel non-coding RNA mapped to chromosome 17q25.1, is associated with poor prognosis in neuroblastoma. Int J Oncol. 2009;34:931-938. [PubMed] |
| 240. | Zhu Y, Yu M, Li Z, Kong C, Bi J, Li J, Gao Z, Li Z. ncRAN, a newly identified long noncoding RNA, enhances human bladder tumor growth, invasion, and survival. Urology. 2011;77:510.e1-510.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 67] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 241. | Qi P, Xu MD, Ni SJ, Shen XH, Wei P, Huang D, Tan C, Sheng WQ, Zhou XY, Du X. Down-regulation of ncRAN, a long non-coding RNA, contributes to colorectal cancer cell migration and invasion and predicts poor overall survival for colorectal cancer patients. Mol Carcinog. 2015;54:742-750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 242. | Ge X, Chen Y, Liao X, Liu D, Li F, Ruan H, Jia W. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer. Med Oncol. 2013;30:588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 159] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 243. | Zhai H, Fesler A, Schee K, Fodstad O, Flatmark K, Ju J. Clinical significance of long intergenic noncoding RNA-p21 in colorectal cancer. Clin Colorectal Cancer. 2013;12:261-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 90] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 244. | Alaiyan B, Ilyayev N, Stojadinovic A, Izadjoo M, Roistacher M, Pavlov V, Tzivin V, Halle D, Pan H, Trink B. Differential expression of colon cancer associated transcript1 (CCAT1) along the colonic adenoma-carcinoma sequence. BMC Cancer. 2013;13:196. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 127] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 245. | Braconi C, Valeri N, Kogure T, Gasparini P, Huang N, Nuovo GJ, Terracciano L, Croce CM, Patel T. Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proc Natl Acad Sci USA. 2011;108:786-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 182] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 246. | Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215-229. [PubMed] |
| 247. | Sana J, Hankeova S, Svoboda M, Kiss I, Vyzula R, Slaby O. Expression levels of transcribed ultraconserved regions uc.73 and uc.388 are altered in colorectal cancer. Oncology. 2012;82:114-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 248. | Pasic I, Shlien A, Durbin AD, Stavropoulos DJ, Baskin B, Ray PN, Novokmet A, Malkin D. Recurrent focal copy-number changes and loss of heterozygosity implicate two noncoding RNAs and one tumor suppressor gene at chromosome 3q13.31 in osteosarcoma. Cancer Res. 2010;70:160-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 125] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 249. | Qi P, Xu MD, Ni SJ, Huang D, Wei P, Tan C, Zhou XY, Du X. Low expression of LOC285194 is associated with poor prognosis in colorectal cancer. J Transl Med. 2013;11:122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 120] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 250. | Shi D, Zheng H, Zhuo C, Peng J, Li D, Xu Y, Li X, Cai G, Cai S. Low expression of novel lncRNA RP11-462C24.1 suggests a biomarker of poor prognosis in colorectal cancer. Med Oncol. 2014;31:31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 67] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 251. | Han J, Rong LF, Shi CB, Dong XG, Wang J, Wang BL, Wen H, He ZY. Screening of lymph nodes metastasis associated lncRNAs in colorectal cancer patients. World J Gastroenterol. 2014;20:8139-8150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 23] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 252. | Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330-342. [PubMed] |
| 253. | Cui M, Zheng M, Sun B, Wang Y, Ye L, Zhang X. A long noncoding RNA perturbs the circadian rhythm of hepatoma cells to facilitate hepatocarcinogenesis. Neoplasia. 2015;17:79-88. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 79] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 254. | Matouk IJ, Abbasi I, Hochberg A, Galun E, Dweik H, Akkawi M. Highly upregulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. Eur J Gastroenterol Hepatol. 2009;21:688-692. [PubMed] |
| 255. | Rossi L, Bonmassar E, Faraoni I. Modification of miR gene expression pattern in human colon cancer cells following exposure to 5-fluorouracil in vitro. Pharmacol Res. 2007;56:248-253. [PubMed] |
| 256. | Borralho PM, Kren BT, Castro RE, da Silva IB, Steer CJ, Rodrigues CM. MicroRNA-143 reduces viability and increases sensitivity to 5-fluorouracil in HCT116 human colorectal cancer cells. FEBS J. 2009;276:6689-6700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 165] [Article Influence: 10.3] [Reference Citation Analysis (1)] |
| 257. | Svoboda M, Izakovicova Holla L, Sefr R, Vrtkova I, Kocakova I, Tichy B, Dvorak J. Micro-RNAs miR125b and miR137 are frequently upregulated in response to capecitabine chemoradiotherapy of rectal cancer. Int J Oncol. 2008;33:541-547. [PubMed] |
| 258. | Hansen TF, Sørensen FB, Lindebjerg J, Jakobsen A. The predictive value of microRNA-126 in relation to first line treatment with capecitabine and oxaliplatin in patients with metastatic colorectal cancer. BMC Cancer. 2012;12:83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 259. | Drebber U, Lay M, Wedemeyer I, Vallböhmer D, Bollschweiler E, Brabender J, Mönig SP, Hölscher AH, Dienes HP, Odenthal M. Altered levels of the onco-microRNA 21 and the tumor-supressor microRNAs 143 and 145 in advanced rectal cancer indicate successful neoadjuvant chemoradiotherapy. Int J Oncol. 2011;39:409-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 62] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 260. | Fang L, Li H, Wang L, Hu J, Jin T, Wang J, Yang BB. MicroRNA-17-5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expression. Oncotarget. 2014;5:2974-2987. [PubMed] |
| 261. | Della Vittoria Scarpati G, Falcetta F, Carlomagno C, Ubezio P, Marchini S, De Stefano A, Singh VK, D’Incalci M, De Placido S, Pepe S. A specific miRNA signature correlates with complete pathological response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Int J Radiat Oncol Biol Phys. 2012;83:1113-1119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 133] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 262. | Svoboda M, Sana J, Fabian P, Kocakova I, Gombosova J, Nekvindova J, Radova L, Vyzula R, Slaby O. MicroRNA expression profile associated with response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer patients. Radiat Oncol. 2012;7:195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 103] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 263. | Hotchi M, Shimada M, Kurita N, Iwata T, Sato H, Morimoto S, Yoshikawa K, Higashijima J, Miyatani T. microRNA expression is able to predict response to chemoradiotherapy in rectal cancer. Mol Clin Oncol. 2013;1:137-142. [PubMed] |
| 264. | Tsang WP, Wong TW, Cheung AH, Co CN, Kwok TT. Induction of drug resistance and transformation in human cancer cells by the noncoding RNA CUDR. RNA. 2007;13:890-898. [PubMed] |
| 265. | Lee H, Kim C, Ku JL, Kim W, Yoon SK, Kuh HJ, Lee JH, Nam SW, Lee EK. A long non-coding RNA snaR contributes to 5-fluorouracil resistance in human colon cancer cells. Mol Cells. 2014;37:540-546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 266. | Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P, Yang Z, Liu W, Zhang H, Chen N. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signalling pathway via suppression of activator protein 2α. Gut. 2015;Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 268] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 267. | Boni V, Zarate R, Villa JC, Bandrés E, Gomez MA, Maiello E, Garcia-Foncillas J, Aranda E. Role of primary miRNA polymorphic variants in metastatic colon cancer patients treated with 5-fluorouracil and irinotecan. Pharmacogenomics J. 2011;11:429-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 85] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 268. | Xing J, Wan S, Zhou F, Qu F, Li B, Myers RE, Fu X, Palazzo JP, He X, Chen Z. Genetic polymorphisms in pre-microRNA genes as prognostic markers of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2012;21:217-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 73] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 269. | Lin M, Gu J, Eng C, Ellis LM, Hildebrandt MA, Lin J, Huang M, Calin GA, Wang D, Dubois RN. Genetic polymorphisms in MicroRNA-related genes as predictors of clinical outcomes in colorectal adenocarcinoma patients. Clin Cancer Res. 2012;18:3982-3991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 270. | Min KT, Kim JW, Jeon YJ, Jang MJ, Chong SY, Oh D, Kim NK. Association of the miR-146aC& gt; G, 149C& gt; T, 196a2C& gt; T, and 499A& gt; G polymorphisms with colorectal cancer in the Korean population. Mol Carcinog. 2012;51 Suppl 1:E65-E73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 82] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 271. | Zhan JF, Chen LH, Chen ZX, Yuan YW, Xie GZ, Sun AM, Liu Y. A functional variant in microRNA-196a2 is associated with susceptibility of colorectal cancer in a Chinese population. Arch Med Res. 2011;42:144-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 272. | Chen H, Sun LY, Chen LL, Zheng HQ, Zhang QF. A variant in microRNA-196a2 is not associated with susceptibility to and progression of colorectal cancer in Chinese. Intern Med J. 2012;42:e115-e119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 273. | Wang Z, Sun X, Wang Y, Liu X, Xuan Y, Hu S. Association between miR-27a genetic variants and susceptibility to colorectal cancer. Diagn Pathol. 2014;9:146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 274. | Hezova R, Kovarikova A, Bienertova-Vasku J, Sachlova M, Redova M, Vasku A, Svoboda M, Radova L, Kiss I, Vyzula R. Evaluation of SNPs in miR-196-a2, miR-27a and miR-146a as risk factors of colorectal cancer. World J Gastroenterol. 2012;18:2827-2831. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 83] [Cited by in RCA: 95] [Article Influence: 7.3] [Reference Citation Analysis (1)] |
| 275. | Li L, Sun R, Liang Y, Pan X, Li Z, Bai P, Zeng X, Zhang D, Zhang L, Gao L. Association between polymorphisms in long non-coding RNA PRNCR1 in 8q24 and risk of colorectal cancer. J Exp Clin Cancer Res. 2013;32:104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 75] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 276. | Khoury S, Tran N. Circulating microRNAs: potential biomarkers for common malignancies. Biomark Med. 2015;9:131-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 277. | Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513-10518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5636] [Cited by in RCA: 6313] [Article Influence: 371.4] [Reference Citation Analysis (0)] |
| 278. | Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997-1006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3218] [Cited by in RCA: 3554] [Article Influence: 209.1] [Reference Citation Analysis (0)] |
| 279. | Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1443] [Cited by in RCA: 1559] [Article Influence: 155.9] [Reference Citation Analysis (0)] |
| 280. | Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223-7233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1356] [Cited by in RCA: 1525] [Article Influence: 108.9] [Reference Citation Analysis (0)] |
| 281. | Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-659. [PubMed] |
| 282. | Février B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004;16:415-421. [PubMed] |
| 283. | Théry C, Regnault A, Garin J, Wolfers J, Zitvogel L, Ricciardi-Castagnoli P, Raposo G, Amigorena S. Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73. J Cell Biol. 1999;147:599-610. [PubMed] |
| 284. | Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17:879-887. [PubMed] |
| 285. | Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA. 2004;101:13368-13373. [PubMed] |
| 286. | Andre F, Schartz NE, Movassagh M, Flament C, Pautier P, Morice P, Pomel C, Lhomme C, Escudier B, Le Chevalier T. Malignant effusions and immunogenic tumour-derived exosomes. Lancet. 2002;360:295-305. [PubMed] |
| 287. | Guduric-Fuchs J, O’Connor A, Camp B, O’Neill CL, Medina RJ, Simpson DA. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics. 2012;13:357. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 372] [Cited by in RCA: 436] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 288. | Ragusa M, Barbagallo C, Statello L, Caltabiano R, Russo A, Puzzo L, Avitabile T, Longo A, Toro MD, Barbagallo D. miRNA profiling in vitreous humor, vitreal exosomes and serum from uveal melanoma patients: Pathological and diagnostic implications. Cancer Biol Ther. 2015;16:1387-1396. [PubMed] |
| 289. | Ragusa M, Statello L, Maugeri M, Barbagallo C, Passanisi R, Alhamdani MS, Li Destri G, Cappellani A, Barbagallo D, Scalia M. Highly skewed distribution of miRNAs and proteins between colorectal cancer cells and their exosomes following Cetuximab treatment: biomolecular, genetic and translational implications. Oncoscience. 2014;1:132-157. [PubMed] |
| 290. | Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143-1149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 741] [Cited by in RCA: 789] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 291. | Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1165] [Cited by in RCA: 1515] [Article Influence: 137.7] [Reference Citation Analysis (0)] |
| 292. | Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1009] [Cited by in RCA: 1131] [Article Influence: 80.8] [Reference Citation Analysis (0)] |
| 293. | Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003-5008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2345] [Cited by in RCA: 2642] [Article Influence: 188.7] [Reference Citation Analysis (0)] |
| 294. | Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423-433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2292] [Cited by in RCA: 2188] [Article Influence: 156.3] [Reference Citation Analysis (0)] |
| 295. | Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1730] [Cited by in RCA: 1858] [Article Influence: 109.3] [Reference Citation Analysis (0)] |
| 296. | Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 901] [Cited by in RCA: 1010] [Article Influence: 63.1] [Reference Citation Analysis (0)] |
| 297. | Eichelser C, Stückrath I, Müller V, Milde-Langosch K, Wikman H, Pantel K, Schwarzenbach H. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget. 2014;5:9650-9663. [PubMed] |
| 298. | Silva J, Garcia V, Rodriguez M, Compte M, Cisneros E, Veguillas P, Garcia JM, Dominguez G, Campos-Martin Y, Cuevas J. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosomes Cancer. 2012;51:409-418. [PubMed] |
| 299. | Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9:e92921. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 557] [Cited by in RCA: 647] [Article Influence: 58.8] [Reference Citation Analysis (1)] |
| 300. | Deng ZB, Zhuang X, Ju S, Xiang X, Mu J, Liu Y, Jiang H, Zhang L, Mobley J, McClain C. Exosome-like nanoparticles from intestinal mucosal cells carry prostaglandin E2 and suppress activation of liver NKT cells. J Immunol. 2013;190:3579-3589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 72] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 301. | Ragusa M, Barbagallo D, Purrello M. Exosomes: nanoshuttles to the future of BioMedicine. Cell Cycle. 2015;14:289-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 302. | Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375-1381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 855] [Cited by in RCA: 901] [Article Influence: 56.3] [Reference Citation Analysis (0)] |
| 303. | Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127:118-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 750] [Cited by in RCA: 750] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 304. | Wang Q, Huang Z, Ni S, Xiao X, Xu Q, Wang L, Huang D, Tan C, Sheng W, Du X. Plasma miR-601 and miR-760 are novel biomarkers for the early detection of colorectal cancer. PLoS One. 2012;7:e44398. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 176] [Cited by in RCA: 189] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 305. | Wang LG, Gu J. Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol. 2012;36:e61-e67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 166] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 306. | Giráldez MD, Lozano JJ, Ramírez G, Hijona E, Bujanda L, Castells A, Gironella M. Circulating microRNAs as biomarkers of colorectal cancer: results from a genome-wide profiling and validation study. Clin Gastroenterol Hepatol. 2013;11:681-8.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 139] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 307. | Cheng H, Zhang L, Cogdell DE, Zheng H, Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR, Zhang W. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011;6:e17745. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 303] [Cited by in RCA: 340] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 308. | Pu XX, Huang GL, Guo HQ, Guo CC, Li H, Ye S, Ling S, Jiang L, Tian Y, Lin TY. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol. 2010;25:1674-1680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 243] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 309. | Chen J, Wang W, Zhang Y, Chen Y, Hu T. Predicting distant metastasis and chemoresistance using plasma miRNAs. Med Oncol. 2014;31:799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 57] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 310. | Kanaan Z, Rai SN, Eichenberger MR, Roberts H, Keskey B, Pan J, Galandiuk S. Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann Surg. 2012;256:544-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 227] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 311. | Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105:849-859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 345] [Cited by in RCA: 402] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 312. | Monzo M, Martínez-Rodenas F, Moreno I, Navarro A, Santasusagna S, Macias I, Muñoz C, Tejero R, Hernández R. Differential MIR-21 expression in plasma from mesenteric versus peripheral veins: an observational study of disease-free survival in surgically resected colon cancer patients. Medicine (Baltimore). 2015;94:e145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 313. | Clancy C, Joyce MR, Kerin MJ. The use of circulating microRNAs as diagnostic biomarkers in colorectal cancer. Cancer Biomark. 2015;15:103-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 314. | Arita T, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013;33:3185-3193. [PubMed] |
| 315. | Dong L, Qi P, Xu MD, Ni SJ, Huang D, Xu QH, Weng WW, Tan C, Sheng WQ, Zhou XY. Circulating CUDR, LSINCT-5 and PTENP1 long noncoding RNAs in sera distinguish patients with gastric cancer from healthy controls. Int J Cancer. 2015;Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 133] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 316. | Tong YS, Wang XW, Zhou XL, Liu ZH, Yang TX, Shi WH, Xie HW, Lv J, Wu QQ, Cao XF. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol Cancer. 2015;14:3. [PubMed] |
| 317. | Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135-2143. [PubMed] |
| 318. | Stenvang J, Petri A, Lindow M, Obad S, Kauppinen S. Inhibition of microRNA function by antimiR oligonucleotides. Silence. 2012;3:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 364] [Cited by in RCA: 418] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 319. | Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721-726. [PubMed] |
| 320. | Bohen SP, Kralli A, Yamamoto KR. Hold ‘em and fold ‘em: chaperones and signal transduction. Science. 1995;268:1303-1304. [PubMed] |
| 321. | Ibrahim AF, Weirauch U, Thomas M, Grünweller A, Hartmann RK, Aigner A. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res. 2011;71:5214-5224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 313] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 322. | Terasawa K, Shimizu K, Tsujimoto G. Synthetic Pre-miRNA-Based shRNA as Potent RNAi Triggers. J Nucleic Acids. 2011;2011:131579. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 323. | Rao DD, Vorhies JS, Senzer N, Nemunaitis J. siRNA vs. shRNA: similarities and differences. Adv Drug Deliv Rev. 2009;61:746-759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 367] [Cited by in RCA: 435] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 324. | Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 952] [Cited by in RCA: 1048] [Article Influence: 69.9] [Reference Citation Analysis (0)] |
| 325. | Robbins M, Judge A, MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19:89-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 305] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 326. | Citti L, Rainaldi G. Synthetic hammerhead ribozymes as therapeutic tools to control disease genes. Curr Gene Ther. 2005;5:11-24. [PubMed] |
| 327. | Gredell JA, Berger AK, Walton SP. Impact of target mRNA structure on siRNA silencing efficiency: A large-scale study. Biotechnol Bioeng. 2008;100:744-755. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 45] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 328. | Watrin M, Dausse E, Lebars I, Rayner B, Bugaut A, Toulmé JJ. Aptamers targeting RNA molecules. Methods Mol Biol. 2009;535:79-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 329. | Mascini M, Palchetti I, Tombelli S. Nucleic acid and peptide aptamers: fundamentals and bioanalytical aspects. Angew Chem Int Ed Engl. 2012;51:1316-1332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 255] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 330. | Sicard F, Gayral M, Lulka H, Buscail L, Cordelier P. Targeting miR-21 for the therapy of pancreatic cancer. Mol Ther. 2013;21:986-994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 178] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 331. | Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180-1189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1087] [Cited by in RCA: 1300] [Article Influence: 100.0] [Reference Citation Analysis (0)] |
| 332. | Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616-1625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 631] [Cited by in RCA: 685] [Article Influence: 57.1] [Reference Citation Analysis (0)] |
| 333. | Zhao X, Pan F, Holt CM, Lewis AL, Lu JR. Controlled delivery of antisense oligonucleotides: a brief review of current strategies. Expert Opin Drug Deliv. 2009;6:673-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 66] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 334. | Cho WC. MicroRNAs as therapeutic targets and their potential applications in cancer therapy. Expert Opin Ther Targets. 2012;16:747-759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 56] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 335. | Lorenz C, Hadwiger P, John M, Vornlocher HP, Unverzagt C. Steroid and lipid conjugates of siRNAs to enhance cellular uptake and gene silencing in liver cells. Bioorg Med Chem Lett. 2004;14:4975-4977. [PubMed] |
| 336. | Chen Y, Zhu X, Zhang X, Liu B, Huang L. Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol Ther. 2010;18:1650-1656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 375] [Cited by in RCA: 399] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 337. | Tran TH, Mattheolabakis G, Aldawsari H, Amiji M. Exosomes as nanocarriers for immunotherapy of cancer and inflammatory diseases. Clin Immunol. 2015;160:46-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 140] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 338. | Concepcion CP, Han YC, Mu P, Bonetti C, Yao E, D’Andrea A, Vidigal JA, Maughan WP, Ogrodowski P, Ventura A. Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet. 2012;8:e1002797. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 155] [Cited by in RCA: 168] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 339. | Nishimura M, Jung EJ, Shah MY, Lu C, Spizzo R, Shimizu M, Han HD, Ivan C, Rossi S, Zhang X. Therapeutic synergy between microRNA and siRNA in ovarian cancer treatment. Cancer Discov. 2013;3:1302-1315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 132] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 340. | McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem. 2011;57:833-840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 492] [Cited by in RCA: 521] [Article Influence: 37.2] [Reference Citation Analysis (0)] |
| 341. | Wang K, Yuan Y, Cho JH, McClarty S, Baxter D, Galas DJ. Comparing the MicroRNA spectrum between serum and plasma. PLoS One. 2012;7:e41561. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 441] [Cited by in RCA: 517] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 342. | Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods. 2010;50:298-301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 867] [Cited by in RCA: 932] [Article Influence: 62.1] [Reference Citation Analysis (0)] |
| 343. | Cheng HH, Yi HS, Kim Y, Kroh EM, Chien JW, Eaton KD, Goodman MT, Tait JF, Tewari M, Pritchard CC. Plasma processing conditions substantially influence circulating microRNA biomarker levels. PLoS One. 2013;8:e64795. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 224] [Cited by in RCA: 242] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 344. | Shan L, Ji Q, Cheng G, Xia J, Liu D, Wu C, Zhu B, Ding Y. Diagnostic value of circulating miR-21 for colorectal cancer: a meta-analysis. Cancer Biomark. 2015;15:47-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 345. | Jiang JX, Zhang N, Liu ZM, Wang YY. Detection of microRNA-21 expression as a potential screening biomarker for colorectal cancer: a meta-analysis. Asian Pac J Cancer Prev. 2014;15:7583-7588. [PubMed] |
| 346. | Xu F, Xu L, Wang M, An G, Feng G. The accuracy of circulating microRNA-21 in the diagnosis of colorectal cancer: a systematic review and meta-analysis. Colorectal Dis. 2015;17:O100-O107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 347. | Asaga S, Kuo C, Nguyen T, Terpenning M, Giuliano AE, Hoon DS. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem. 2011;57:84-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 349] [Cited by in RCA: 362] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 348. | Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi K, Fujiwara H. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102:1174-1179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 449] [Cited by in RCA: 509] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 349. | Abue M, Yokoyama M, Shibuya R, Tamai K, Yamaguchi K, Sato I, Tanaka N, Hamada S, Shimosegawa T, Sugamura K. Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer. Int J Oncol. 2015;46:539-547. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 162] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 350. | Wang Y, Gao X, Wei F, Zhang X, Yu J, Zhao H, Sun Q, Yan F, Yan C, Li H. Diagnostic and prognostic value of circulating miR-21 for cancer: a systematic review and meta-analysis. Gene. 2014;533:389-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 124] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 351. | Wang M, Gu H, Wang S, Qian H, Zhu W, Zhang L, Zhao C, Tao Y, Xu W. Circulating miR-17-5p and miR-20a: molecular markers for gastric cancer. Mol Med Rep. 2012;5:1514-1520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 76] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 352. | Zeng X, Xiang J, Wu M, Xiong W, Tang H, Deng M, Li X, Liao Q, Su B, Luo Z. Circulating miR-17, miR-20a, miR-29c, and miR-223 combined as non-invasive biomarkers in nasopharyngeal carcinoma. PLoS One. 2012;7:e46367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 122] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 353. | Shrivastava S, Petrone J, Steele R, Lauer GM, Di Bisceglie AM, Ray RB. Up-regulation of circulating miR-20a is correlated with hepatitis C virus-mediated liver disease progression. Hepatology. 2013;58:863-871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 89] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 354. | Guo F, Tian J, Lin Y, Jin Y, Wang L, Cui M. Serum microRNA-92 expression in patients with ovarian epithelial carcinoma. J Int Med Res. 2013;41:1456-1461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 355. | Kawaguchi T, Komatsu S, Ichikawa D, Morimura R, Tsujiura M, Konishi H, Takeshita H, Nagata H, Arita T, Hirajima S. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. Br J Cancer. 2013;108:361-369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 183] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 356. | Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Morimura R, Nagata H, Kosuga T, Iitaka D, Konishi H, Shiozaki A. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer. 2011;105:104-111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 191] [Cited by in RCA: 206] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 357. | Tokarz P, Blasiak J. The role of microRNA in metastatic colorectal cancer and its significance in cancer prognosis and treatment. Acta Biochim Pol. 2012;59:467-474. [PubMed] |