Published online Jan 14, 2017. doi: 10.3748/wjg.v23.i2.224
- This article has been retracted.
- Retraction in: World J Gastroenterol. Jul 7, 2019; 25(25): 3281-3282 See also: Errata, Retraction, Duplicate Publication and Comment Policy
Peer-review started: August 3, 2016
First decision: September 21, 2016
Revised: October 5, 2016
Accepted: November 2, 2016
Article in press: November 2, 2016
Published online: January 14, 2017
Processing time: 162 Days and 20.1 Hours
To prepare a Gpm6a/ReelinGFPCreERT2 construct with a rapid and reliable strategy using a bacterial artificial chromosome (BAC).
Gpm6a and Reelin BACs were purified and transformed into SW102 E. coli by electroporation. The GFPCreERT2 fragment was prepared from a shuttle vector and transformed into SW102 E. coli carrying a BAC. Homologous recombination was induced in SW102 E. coli. Recombinant clones were screened and confirmed by PCR and restriction enzyme digestion. Recombinant clones were transformed into SW102 E. coli to remove the kanamycin unit.
A complete BAC was successfully transformed into SW102 E. coli by electroporation because BAC purified from SW102 E. coli showed the same pattern as the original BAC with BamHI digestion. The GFPCreERT2 fragment was deemed to have been prepared successfully because we obtained the same size fragment as expected. Homologous recombination was induced, and GFPCreERT2 was deemed to have been inserted into the correct site of the BAC because we found the band change was the same as the expected pattern after restriction enzyme digestion. The kanamycin unit was deemed to have been removed successfully because we obtained different sizes of bands that were consistent with the results expected by PCR with different primers.
The construct of Gpm6aGFPCreERT2 or ReelinGFPCreERT2 was prepared successfully, which will establish a foundation for tracing the hepatic stellate cell lineage and studying its function.
Core tip: Until now, there have been few specific mouse lines that allowed recombination for tracing hepatic mesothelial cells or hepatic stellate cells. Here, we describe a rapid and reliable strategy for construct preparation using a bacterial artificial chromosome. This study prepared a Gpm6a/ReelinGFPCreERT2 construct for the first time, which is the first step for the preparation of a Gpm6aGFPCreERT2 or ReelinGFPCreERT2 mouse line.
- Citation: Shi HB, Lou JL, Shi HL, Ren F, Chen Y, Duan ZP. Construction of Gpm6a/ReelinGFPCreERT2 by BAC recombination using a specific gene in hepatic mesothelial or stellate cells. World J Gastroenterol 2017; 23(2): 224-231
- URL: https://www.wjgnet.com/1007-9327/full/v23/i2/224.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i2.224
Excessive extracellular matrix (ECM) of the liver results in cirrhosis, which is an end stage liver disease with high mortality and for which efficacious medical treatments are not currently available, except for liver transplantation. Hepatic stellate cell (HSC) activation is considered a major mechanism in the formation of fibrosis and cirrhosis. However, fundamental questions concerning the cell fate regulation of HSCs remain largely underexplored. A recent study reported that hepatic mesothelial cells are the potential precursors for HSCs in the development of liver disease and can transdifferentiate into myofibroblast cells in mouse liver fibrosis[1-3].
Until now, there have been few specific mouse lines that cause recombination (Cre recombinase, Cre) for tracing hepatic mesothelial cells or HSCs. The Wt1CreERT2 mice are useful to trace hepatic mesothelial cells, but the labeling efficiency and specificity is low[4-6]. Specific genes have been identified in hepatic mesothelial cells and HSCs by microarray[7,8]. We plan to develop Cre mouse lines with specific markers for the study of HSCs or hepatic mesothelial cells. Based on a previous study, glycoprotein M6a (Gpm6a) has been identified as a specific surface marker of hepatic mesothelial cells. It covers the surface of the liver and migrates from the surface into the center[4]. Reelin is an extracellular matrix glycoprotein, which is a specific HSC marker in the mouse liver and has similar amounts in resting and activated HSCs[9].
The Gpm6aGFPCreERT2 or ReelinGFPCreERT2 mouse line will express a fusion protein of green fluorescent protein (GFP), Cre, and estrogen receptor induced by tamoxifen (ERT2) under the control of the Gpm6a or Reelin promoter. GFP is used to track the labeled protein as a marker. Cre recombinase is used to delete a segment of DNA flanked by LoxP sites (flox). The ERT2 system is used to activate Cre activity by tamoxifen treatment[10,11].
Cre mouse lines are very useful tools that can generate knockout mice through the cross breeding of Cre and flox mouse lines. A mouse line is needed to trace HSC lineages and study their function through the knockout of specific genes in specific cells. It is, therefore, necessary to prepare a Gpm6aGFPCreERT2 or ReelinGFPCreERT2 mouse line. In this study, we investigated the preparation and identification of a Gpm6a/ReelinGFPCreERT2 construct, which is the first step for the preparation of a Gpm6aGFPCreERT2 or ReelinGFPCreERT2 mouse line.
Gpm6a Bacterial artificial chromosome (BAC) (RP23-410D17) and Reelin BAC (RP23-143M9) were purchased from the BACPAC Resource Center (BPRC) located at the Children’s Hospital Oakland Research Institute in Oakland, California, United States. The female (C57BL/6J) mouse BAC library was made from kidney and brain DNA cloned into the pBACe3.6 vector at the EcoRI site and transformed into DH10B E. coli. The reporter gene GFPCreERT2 is located in the shuttle vector that was developed by the Biomed Company. The 5arm and 3arm were inserted into both sites of the reporter gene. The arm sequences were amplified according to the BAC template by polymerase chain reaction (PCR).
DH10B E. coli were streaked onto LB plates with 25 mg/mL chloramphenicol (Cmr; Sigma, St Louis, MO, United States) at 32 °C. A single colony was picked and cultured in 25 mL of LB medium with Cmr. BAC DNA was purified with a large construct kit (Qiagen, Hilden, Germany). BAC DNA was digested with the restriction enzyme BamHI (New England Biolabs, Ipswich, MA, United States).
SW102 E. coli (NCI, Frederick, MD, United States) was streaked onto plates with 50 mg/mL tetracycline (Tc; Sigma) at 32 °C. A single colony was picked and inoculated in LB medium with Tc and incubated for 4-6 h. After placing on ice, competent cells were made from SW102 E. coli. The cells were washed with ice-cold water, and 5 mL of BAC DNA was added to 85 mL of competent cells. A Gene Pulser Xcell (Bio-Rad, Hercules, CA, United States) was used for electroporation at 1.75 KV for 25 mF (time constant: 4.5-5.0). Next, 1 mL of LB was added and incubated at 32 °C for 1 h. Following plating onto LB plates with Cmr, the BAC was purified using a Qiagen kit. BAC DNA was confirmed with the restriction enzyme BamHI.
SW102 E. coli were streaked onto 50 mg/mL kanamycin (Kam; Sigma) plates at 32 °C. A single colony was picked and inoculated in LB medium with Kam and incubated for 4-6 h. The shuttle vector was purified with an Endofree plasmid maxi kit (Qiagen). The vector was digested with NotI and FseI (New England Biolabs), and electrophoresis was performed with agarose gels (Takara, Osaka, Japan). The large fragment of GFPCreERT2 was extracted from the gel with a quick gel extraction kit (Qiagen).
SW102 E. coli carrying BAC was streaked onto LB plates with 50 mg/mL Tc and 25 mg/mL Cmr. SW102 E. coli carrying BAC were induced, and competent cells were made as described above. A total of 200 ng of the fragment of GFPCreERT2 was added to 85 mL of competent cells, which were then electroporated at 1.75 KV for 25 mF (time constant: 4.5-5.0). A total of 0.6 mL of LB was added and cultured at 32 °C for 1 h. The sample was then plated onto LB plates with 25 mg/mL Cmr and 12.5 mg/mL Kam. The recombinant BAC DNA was confirmed by PCR with platinum Taq DNA Polymerase (Invitrogen, Carlsbad, CA, United States) and restriction enzyme digestion.
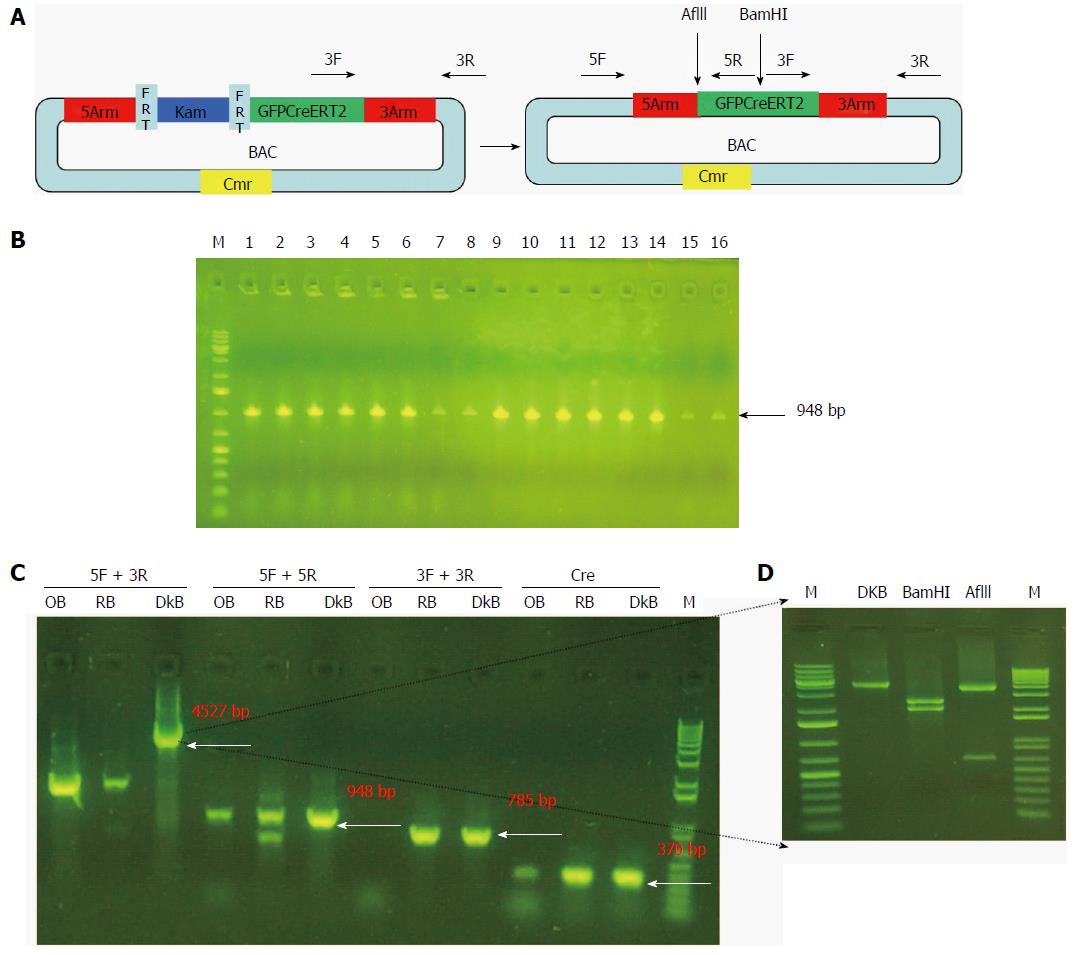
SW105 E. coli (NCI) was streaked without antibiotics. A single colony was picked and competent cells were made. Recombinant BAC DNA was transformed into competent cells with electroporation and inoculated in LB medium with L-arabinose (Sigma) for removal of the Kam unit. They were then plated onto LB plates with Cmr only. We confirmed that no colonies were on the LB plate with Cmr and Kam. Sixteen colonies were picked and checked by PCR using the 5arm primer (Table 1). One colony was confirmed by PCR using different primers (Table 1). The PCR product was digested with BamHI and AflII (New England Biolabs) for further confirmation.
| Forward | Reverse | |
| Gpm6a 3arm | CGG TAC CTT TCA TGT TTT CAT GGT TGT CA | AGG TAC CGG CCG GCC ATG ACA GCA AAC ACT GCC TCT A |
| Gpm6a 5arm | ACC CAA TCT CCC TTT CAG | TGA ACT TGT GGC TTT AGA TC |
| Gpm6a Cre | ACC TGA AGA TGT TCG CGA TTA TCT | ACC GTC AGT ACG TGA GAT ATC TT |
| Reelin 3arm | AGG TAC CAC GGC ATC CCT ACG GCG C | AGG TAC CGG CCG GCC ACA GCC GCT CTG TTT CTT GAG G |
| Reelin 5arm | ACC CAA TCT CCC TTT CAG | TGA ACT TGT GGC TTT ACG TC |
| Reelin Cre | ACT TAA GCT CGT TCG CGC AGC G | AGT CGA CGC CGC CGC GCT CCG T |
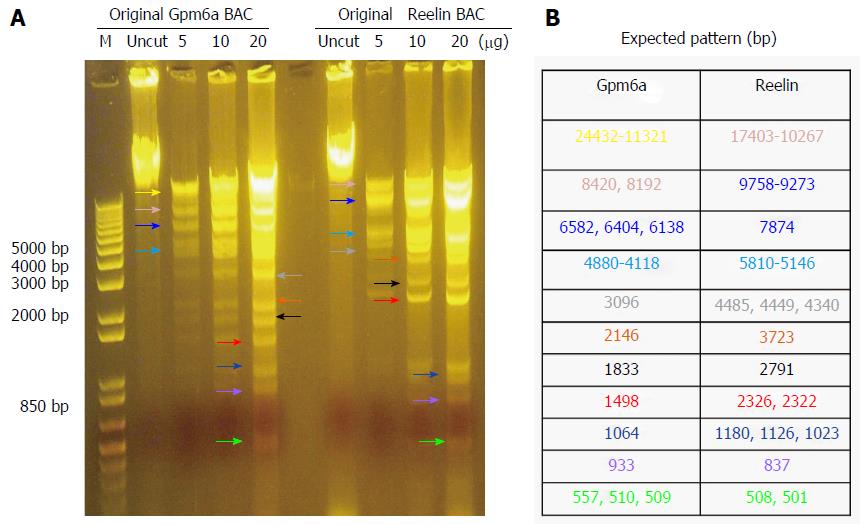
As mentioned previously, we purchased BAC from BPRC and confirmed the BAC sequence. From Figure 1, it can be observed that, after digestion with BamHI, Gpm6a BAC had 11 bands, which was the expected pattern. For Reelin BAC, the same results were obtained. Therefore, we confirmed that we received the BAC clone.
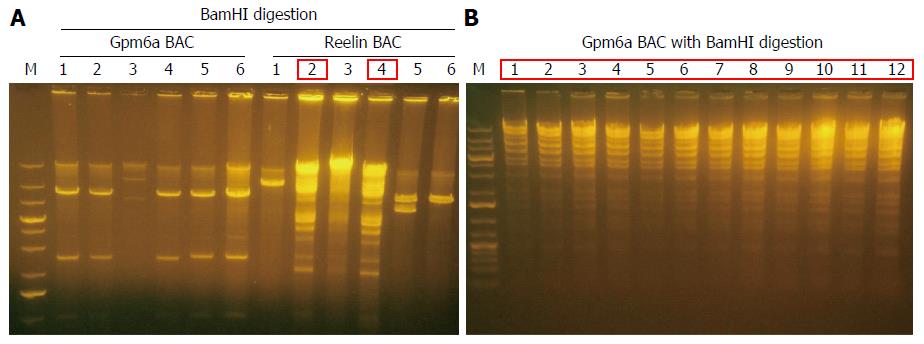
BAC DNA is so long that it is impossible to transform BAC into SW102 E. coli by chemical transduction[12]. We transformed purified BAC into SW102 E. coli by electroporation. After electroporation, we purified BAC from SW102 E. coli and identified BAC with restriction enzyme BamHI digestion. For Gpm6a BAC, after BamHI digestion, selected clones did not show the same digestion pattern with original BAC, as shown in Figure 2, which suggested the original BAC DNA may have been fragmented during the purification step. For Reelin BAC, we obtained two positive clones that showed the same pattern as the original BAC after BamHI digestion. We improved the purification methods, and we finally succeeded in transforming the complete Gpm6a BAC into SW102 E. coli (Figure 2B).
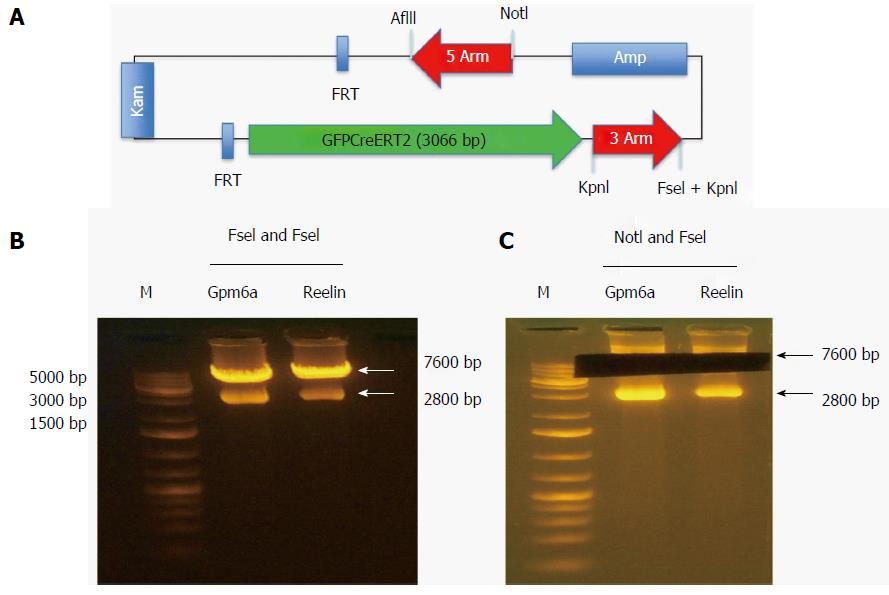
The reporter gene GFPCreERT2 was in the shuttle vector that contained some genes encoding the 5arm, flp recombinase target (FRT), Kam, FRT, GFPCreERT2, 3arm and ampicillin. We used NotI and FseI to digest the shuttle vector and obtained the fragment of GFPCreERT2 flanked by the 5arm and 3arm. As shown in Figure 3, after digestion with NotI and FseI, we obtained two bands; one was approximately 7600 bp, and the other was approximately 2800 bp. We recovered the long band containing GFPCreERT2 from the agarose gels for transformation.
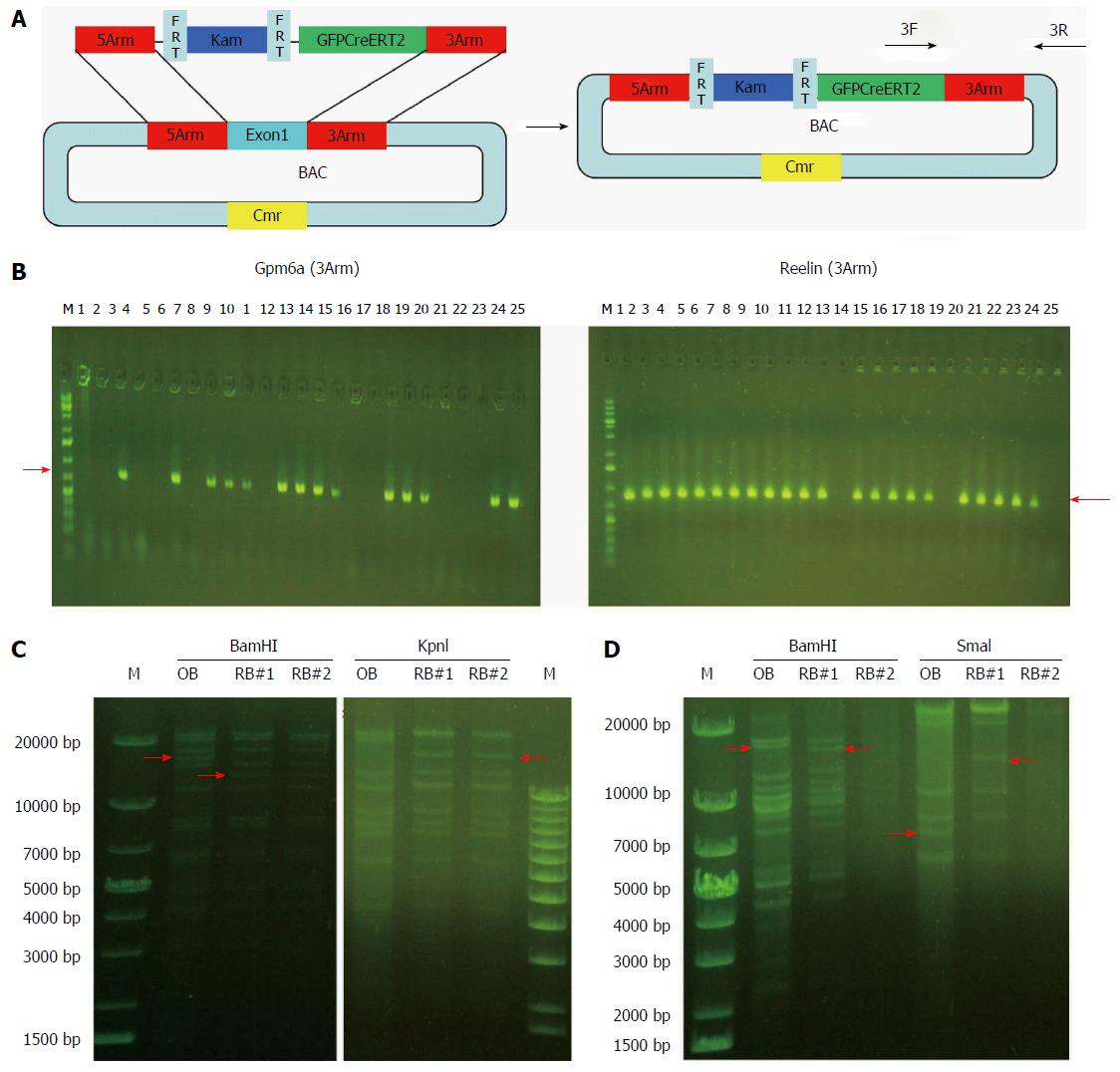
We induced the SW102 E. coli carrying BAC to become competent cells and transformed the fragment of GFPCreERT2 into the SW102 E. coli carrying the BAC. After homologous recombination between BAC and GFPCreERT2, GFPCreERT2 replaced the first exon in the BAC vector. Then, we purified the recombinant BAC for screening and confirmation. As shown in Figure 4B, we obtained 14 positive clones for Gpm6a (14/25, 56%), as evidenced by PCR screening. For Reelin, we obtained 22 positive clones (22/25, 88%). We further confirmed the positive clones by restriction enzyme digestion. For Gpm6a, we found that the band change was the same as the expected pattern after BamHI or KpnI digestion (Figure 4C). Reelin had the same results (Figure 4D). The results suggested that GFPCreERT2 was inserted into the correct site in the BAC vector and that we acquired the recombinant BAC.
Finally, we removed the Kam unit that could interfere with the expression of the reporter gene. We purified the recombinant BAC carrying GFPCreERT2 and transformed it into SW105 E. coli. The Kam unit was removed by homologous recombination. As shown in Figure 5B, we obtained 16 positive clones for Gpm6a (16/16, 100%), as evidenced by PCR screening. We further confirmed the positive clones by PCR with different primers. We obtained different sizes of bands that were consistent with what was expected (Figure 5C). We digested the PCR product of 4527 bp with BamHI or AflII, and the bands obtained were the same as expected (Figure 5D). This suggested that the Kam unit was removed and that we obtained BAC carrying GFPCreERT2 without Kam.
First of all, we needed to choose a vector to prepare the Gpm6a/ReelinGFPCreERT2 construct. As a conventional vector, its advantage was that it is easy to handle, but it had low expression levels of the reporter genes and low specificity because the DNA size is approximately 3-5 kb and contains only a promoter. For the BAC vector, its advantages include relatively high efficiency and relatively high specificity, but BAC DNA is fragile and can break easily because BAC DNA is approximately 200 kb and contains all the regulatory sequences upstream and downstream of the encoding sequence[13,14]. Thus, we chose a BAC vector to prepare the constructs of Gpm6aGFPCreERT2 and ReelinGFPCreERT2.
Second, how to insert reporter genes into BAC DNA was a question. In 200 kb of BAC DNA, there would be 49 recognition sites for 6 nucleotide cutters such as EcoRI and BamHI, so it was impossible to use restriction enzyme digestion and ligation. We, therefore, used homologous DNA recombination for gene insertion[15-18]. In BAC DNA, we designed the primers upstream or downstream of the first exon and obtained the 5arm and 3arm fragment by PCR amplification. In the shuttle vector, the 5arm fragment was inserted upstream of the reporter gene, and the 3arm fragment was inserted downstream of the reporter gene. In SW102 E. coli, the reporter gene that is GFPCreERT2 replaced the first exon in the BAC vector by homologous recombination. The expression of GFPCreERT2 is regulated by the Gpm6a or Reelin promoter.
Finally, we removed the Kam unit by homologous recombination of the FRT using SW105 E. coli. The FRT is similar to LoxP, which is used to delete a segment of DNA flanked by LoxP sites[19]. The FRT cassette was excised with high frequency, which was close to 100%[20]. Then, we obtained the constructs of Gpm6aGFPCreERT2 and ReelinGFPCreERT2 successfully. After microinjection, we can develop the mouse line with Gpm6aGFPCreERT2 and ReelinGFPCreERT2.
Homologous recombination was performed with modified E. coli bacteria strains: SW102 and SW105. SW102 E. coli carries genes such as exo, bet and gam. Exo encodes 5’-3’ exonuclease, and bet encodes the overhang binding protein, which enables annealing and recombination with complementary DNA; gam encodes the inhibitor of E. coli exonuclease to protect the introduced DNA. SW105 E. coli carries the L-arabinose-inducible flp gene, which encodes recombinase, allowing DNA modification without restriction enzymes and DNA ligase[21].
Cre-Lox recombination is a site-specific recombinase technology that is widely used to carry out deletions, insertions, translocations and inversions in the DNA of cells. It allows the DNA modification to be targeted to a specific cell type or to be triggered by a specific external stimulus[22,23]. For a mouse line with Gpm6aGFPCreERT2 and ReelinGFPCreERT2, once the specific genes, such as Gpm6a or Reelin, begin to express, the GFPCreERT2 will express in the specific cell. Using GFP as a marker, we can trace the specific cells. Using the CreERT2 system, we can knock out the specific gene in specific cells, which is a conditional knockout[24].
In the next stage, we will label hepatic mesothelial and HSCs in the Gpm6a/ReelinGFPCreERT2 transgenic mouse to trace the lineage of HSCs. We will also make a conditional TGFβ-knockout mouse through cross breeding of the TGFflox/flox mouse and the Gpm6a/ReelinGFPCreERT2 mouse to explore the function of HSCs during development.
Hepatic stellate cell (HSC) activation is considered a major mechanism in the formation of fibrosis and cirrhosis. However, fundamental questions concerning the cell fate regulation of HSCs remain largely underexplored. A recent study reported that hepatic mesothelial cells are the potential precursors of HSCs in the development of the liver and can transdifferentiate into myofibroblast cells in mouse liver fibrosis. Until now, there have been few specific mouse lines that could cause recombination (Cre) for tracing hepatic mesothelial cells or HSCs. Here, we describe a rapid and reliable strategy for construct preparation using the bacterial artificial chromosome (BAC).
A recent study reported that hepatic mesothelial cells are the potential precursors for HSCs in the development of the liver and can transdifferentiate into myofibroblast cells in mouse liver fibrosis. Glycoprotein M6a (Gpm6a) has been identified as a specific surface marker of hepatic mesothelial cells, which cover the surface of the liver and migrate from the surface into the center. Reelin is an extracellular matrix glycoprotein, which is a specific HSC marker in the mouse liver and has similar amounts in resting and activated HSCs.
Until now, there have been few specific mouse lines that could Cre for tracing hepatic mesothelial cells or HSCs. Here, the authors describe a rapid and reliable strategy for construct preparation using a BAC. This study prepared a Gpm6a/ReelinGFPCreERT2 construct for the first time, which is the first step in the preparation of a Gpm6aGFPCreERT2 or ReelinGFPCreERT2 mouse line.
In this study, a construct of Gpm6aGFPCreERT2 or ReelinGFPCreERT2 was prepared successfully, which will establish the foundation for tracing the HSC lineage and studying its function.
Cre-Lox recombination is a site-specific recombinase technology widely used to create deletions, insertions, translocations and inversions in the DNA of cells. It allows the DNA modification to be targeted to a specific cell type or triggered by a specific external stimulus. In a mouse line with Gpm6aGFPCreERT2 and ReelinGFPCreERT2, once the specific genes such as Gpm6a or Reelin begin to express, the GFPCreERT2 will express in the specific cell. Using GFP as a marker, we can track the specific cells. Using the CreERT2 system, we can knock out the specific gene in a specific cell, which is a conditional knockout.
This is a well-designed study in which the author prepared a construct of Gpm6aGFPCreERT2 or ReelinGFPCreERT2 successfully, which is the first step for the preparation of a Gpm6aGFPCreERT2 or ReelinGFPCreERT2 mouse line. Cre mouse lines are very useful tools that can generate knockout mice through the cross breeding of Cre and flox mouse lines.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Chan WK S- Editor: Qi Y L- Editor: Filipodia E- Editor: Wang CH
| 1. | Lua I, Asahina K. The Role of Mesothelial Cells in Liver Development, Injury, and Regeneration. Gut Liver. 2016;10:166-176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 2. | Lua I, James D, Wang J, Wang KS, Asahina K. Mesodermal mesenchymal cells give rise to myofibroblasts, but not epithelial cells, in mouse liver injury. Hepatology. 2014;60:311-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 3. | Li Y, Lua I, French SW, Asahina K. Role of TGF-β signaling in differentiation of mesothelial cells to vitamin A-poor hepatic stellate cells in liver fibrosis. Am J Physiol Gastrointest Liver Physiol. 2016;310:G262-G272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 4. | Vicente-Steijn R, Scherptong RW, Kruithof BP, Duim SN, Goumans MJ, Wisse LJ, Zhou B, Pu WT, Poelmann RE, Schalij MJ. Regional differences in WT-1 and Tcf21 expression during ventricular development: implications for myocardial compaction. PLoS One. 2015;10:e0136025. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 5. | Li Y, Wang J, Asahina K. Mesothelial cells give rise to hepatic stellate cells and myofibroblasts via mesothelial-mesenchymal transition in liver injury. Proc Natl Acad Sci USA. 2013;110:2324-2329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 165] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 6. | Asahina K, Zhou B, Pu WT, Tsukamoto H. Septum transversum-derived mesothelium gives rise to hepatic stellate cells and perivascular mesenchymal cells in developing mouse liver. Hepatology. 2011;53:983-995. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 265] [Cited by in RCA: 239] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 7. | Lua I, Li Y, Zagory JA, Wang KS, French SW, Sévigny J, Asahina K. Characterization of hepatic stellate cells, portal fibroblasts, and mesothelial cells in normal and fibrotic livers. J Hepatol. 2016;64:1137-1146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 120] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 8. | Mansy SS, Nosseir MM, Zoheiry MA, Hassanein MH, Guda MF, Othman MM, AbuTalab H. Value of reelin for assessing hepatic fibrogenesis in a group of Egyptian HCV infected patients. Clin Chem Lab Med. 2014;52:1319-1328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 9. | Kobold D, Grundmann A, Piscaglia F, Eisenbach C, Neubauer K, Steffgen J, Ramadori G, Knittel T. Expression of reelin in hepatic stellate cells and during hepatic tissue repair: a novel marker for the differentiation of HSC from other liver myofibroblasts. J Hepatol. 2002;36:607-613. [PubMed] |
| 10. | Lizen B, Claus M, Jeannotte L, Rijli FM, Gofflot F. Perinatal induction of Cre recombination with tamoxifen. Transgenic Res. 2015;24:1065-1077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 11. | Miwa H, Era T. Generation and characterization of PDGFRα-GFPCreERT2 knock-In mouse line. Genesis. 2015;53:329-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 12. | Cui B, Smooker PM, Rouch DA, Deighton MA. Enhancing DNA electro-transformation efficiency on a clinical Staphylococcus capitis isolate. J Microbiol Methods. 2015;109:25-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 13. | Hasse S, Hyman AA, Sarov M. TransgeneOmics--A transgenic platform for protein localization based function exploration. Methods. 2016;96:69-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 14. | Hall RN, Meers J, Fowler E, Mahony T. Back to BAC: the use of infectious clone technologies for viral mutagenesis. Viruses. 2012;4:211-235. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 15. | Zhou F, Li Q, Wong SW, Gao SJ. Autoexcision of bacterial artificial chromosome facilitated by terminal repeat-mediated homologous recombination: a novel approach for generating traceless genetic mutants of herpesviruses. J Virol. 2010;84:2871-2880. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 16. | Narayanan K, Williamson R, Zhang Y, Stewart AF, Ioannou PA. Efficient and precise engineering of a 200 kb beta-globin human/bacterial artificial chromosome in E. coli DH10B using an inducible homologous recombination system. Gene Ther. 1999;6:442-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 119] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 17. | Yang XW, Model P, Heintz N. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat Biotechnol. 1997;15:859-865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 403] [Cited by in RCA: 423] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 18. | Wang S, Zhao Y, Leiby M, Zhu J. A new positive/negative selection scheme for precise BAC recombineering. Mol Biotechnol. 2009;42:110-116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 73] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 19. | Shah R, Li F, Voziyanova E, Voziyanov Y. Target-specific variants of Flp recombinase mediate genome engineering reactions in mammalian cells. FEBS J. 2015;282:3323-3333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 20. | Fan HF, Cheng YS, Ma CH, Jayaram M. Single molecule TPM analysis of the catalytic pentad mutants of Cre and Flp site-specific recombinases: contributions of the pentad residues to the pre-chemical steps of recombination. Nucleic Acids Res. 2015;43:3237-3255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 21. | Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 950] [Cited by in RCA: 1016] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 22. | Hubbard EJ. FLP/FRT and Cre/lox recombination technology in C. elegans. Methods. 2014;68:417-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 23. | Yarmolinsky M, Hoess R. The Legacy of Nat Sternberg: The Genesis of Cre-lox Technology. Annu Rev Virol. 2015;2:25-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 24. | Hu MW, Wang ZB, Schatten H, Sun QY. New understandings on folliculogenesis/oogenesis regulation in mouse as revealed by conditional knockout. J Genet Genomics. 2012;39:61-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |