Published online Jun 26, 2021. doi: 10.12998/wjcc.v9.i18.4585
Peer-review started: November 16, 2020
First decision: January 17, 2021
Revised: January 26, 2021
Accepted: February 24, 2021
Article in press: February 24, 2021
Published online: June 26, 2021
Processing time: 207 Days and 1.9 Hours
Diffuse large B-cell lymphoma (DLBCL) is a common non-Hodgkin lymphoma. The development of immunotherapy greatly improves the patient prognosis but there are some exceptions. Thus, screening for better biomarkers for prognostic evaluation could contribute to the treatment of DLBCL patients.
To screen the novel mediators involved in the development of DLBCL.
The GSE60 dataset was applied to identify the differentially expressed genes (DEGs) in DLBCL, and the principal components analysis plot was used to determine the quality of the included samples. The protein-protein interactions were analyzed by the STRING tool. The key hub genes were entered into to the GEPIA database to determine their expressions in DLBCL. Furthermore, these hub gene alterations were analyzed in cBioportal. The UALCAN portal was employed to analyze the expression of the hub genes in different stages of DLBCL. The Estimation of Stromal and Immune cells in Malignant Tumor tissues using Expression data Score was conducted to evaluate the correlation between the gene expression and tumor purity. The gene-gene correlation analysis was conducted in the GEPIA. The stromal score analysis was conducted in TIMER to confirm the correlation between the gene expression and infiltrated stromal cells. The correlation between the indicated genes and infiltration level of cancer-associated fibroblasts (CAFs) was also completed in TIMER with two methods, MCP-Counter and Tumor immune dysfunction and exclusion. The correlation between fibronectin (FN1) protein level and secreted protein acidic and cysteine-rich (SPARC) messenger ribonucleic acid expression was confirmed in the cBioportal.
The top 20 DEGs in DLBCL were identified, and the principal components analysis plot confirmed the quality of the significant DEGs. The pairwise correlation coefficient analysis among all samples showed that these DEGs have a certain co-expression pattern. The DEGs were subjected to STRING to identify the hub genes, alpha-2-macroglobulin (A2M), cathepsin B (CTSB), FN1, matrix metallopeptidase 9 (MMP9), and SPARC. The five hub genes were confirmed to be overexpressed in DLBCL. The cBioportal portal detected these five hub genes that had gene alteration, including messenger ribonucleic acid high amplification and missense mutation, and the gene alteration percentages of A2M, FN1, CTSB, MMP9, and SPARC were 5%, 8%, 5%, 2.7%, and 5%, respectively. Furthermore, the five hub genes had a potential positive correlation with tumor stage. The correlation analysis between the five genes and tumor purity confirmed that the five genes were overexpressed in DLBCL and had a positive correlation with the development of DLBCL. More interestingly, the five genes had a significant correlation with the stromal infiltration scores. The correlation analysis between the fives genes and CAFs also showed a significant value, among which the top two genes, FN1 and SPARC, had a remarkable co-expression pattern.
The top DEGs were identified, and the five hub genes were overexpressed in DLBCL. Furthermore, the gene alterations were confirmed and the positive correlation with tumor purity revealed the overexpression of the five genes and close association with the development of DLBCL. More interestingly, the five genes were positively correlated with stromal infiltration, especially in CAFs. The top two genes, FN1 and SPARC, showed a co-expression pattern, which indicates their potential as novel therapeutic targets for DLBCL.
Core Tip: In this project, we identified five genes that were overexpressed in diffuse large B-cell lymphoma (DLBCL), and the five gene signatures were closely associated with the development of DLBCL. More importantly, the five genes were positively correlated with stromal cell infiltration, especially in cancer-associated fibroblasts. Taken together, these genes might be novel therapeutic targets for DLBCL.
- Citation: Nan YY, Zhang WJ, Huang DH, Li QY, Shi Y, Yang T, Liang XP, Xiao CY, Guo BL, Xiang Y. Evaluation of a five-gene signature associated with stromal infiltration for diffuse large B-cell lymphoma. World J Clin Cases 2021; 9(18): 4585-4598
- URL: https://www.wjgnet.com/2307-8960/full/v9/i18/4585.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i18.4585
Diffuse large B-cell lymphoma (DLBCL) is a common type of non-Hodgkin lymphoma, which accounts for about 30% of non-Hodgkin lymphoma patients[1]. DLBCL is a highly malignant tumor; the survival time for those untreated patients is only several months[2]. The current treatments for DLBCL are mainly dependent on the clinical stages. The limited stage contains stages 1 and 2, the treatment guideline for which suggests the combination of systemic chemoimmunotherapy, including rituximab, cyclophosphamide, doxorubicin, vincristine and prednisone (R-CHOP), and the involved-field radiation therapy[3,4]. However, to some degree, involved-field radiation therapy is not applicable to patients with advanced stages. As the first-line treatment for DLBCL, many patients could benefit from R-CHOP. However, there are still 30%-40% of patients expressing resistance and recurrence[5-7]. Therefore, screening for novel targets involved in the development of DLBCL could prove an encouraging work for precision medicine.
Considering the risks of drug resistance and recurrence in the treatment of DLBCL, this study aimed to reveal the significant genes in the development of DLBCL. The discovery of novel molecules could provide further understanding about the regulatory mechanism of DLBCL, finding therapeutic drug targets, and further understanding of the tumor microenvironment in the regulation of drug resistance and tumor recurrence[8,9]. However, we are still unclear about the tumor microenvironment in the development of DLBCL, especially in the regulation of stromal and immune infiltration.
In this study, overexpressed genes in DLBCL were identified, and the key hub genes were confirmed for further analysis. We identified alpha-2-macroglobulin (A2M), fibronectin (FN1), cathepsin B (CTSB), matrix metallopeptidase 9 (MMP9) and secreted protein acidic and cysteine-rich (SPARC) as gene signatures, which were closely associated with the development of DLBCL. More interestingly, we confirmed the significantly positive correlation between the five gene signatures and stromal score in tissue samples of DLBCL. In detail, the five gene signatures could predict the infiltration level of cancer-associated fibroblasts (CAFs). Finally, the top two genes, FN1 and SPARC, showed a co-expression profile in DLBCL, which might be encouraged as novel therapeutic targets for DLBCL.
The GSE60 dataset was employed for the expression profiling assay on the DLBCL samples[10]. The expression profile was obtained from the Gene Expression Omnibus, and treated with log transformation. The Benjamini and Hochberg (false discovery rate) was applied to adjust the P value. The heatmap of differentially expressed genes (DEGs), principal components analysis plot, and correlation coefficient for all samples were produced by the Image GP tool (http://www.ehbio.com/ImageGP/index.
The top 20 DEGs were subjected to analysis through the STRING database (version 11.0)[11], and the multiple protein manners were applied, including the active interaction sources of text mining, experiments, databases, co-expression, neighborhood, gene fusion, and co-occurrence. The minimally required interaction score was set as medium confidence (0.40). The node genes were considered as the hub genes, and the PPI enrichment value expresses statistical difference.
The hub gene expression in tumor cells and normal cells was assessed in the GEPIA database[12]. DLBCL cells from 47 patients were included in the tumor group, and the Genotype-Tissue Expression[13] and normal lymphocytes were included in the normal group (n = 337). The expression profiling was investigated by the log2 (TPM + 1) method.
The genomic alteration analysis of indicated genes was conducted in the cBioportal for cancer genomics (https://www.cbioportal.org/)[14]. The DLBCL (The Cancer Gene Atlas [TCGA], PanCancer Atlas) was selected, and the genomic alternation included missense mutation, amplification, deep deletion, and messenger ribonucleic acid-high. The correlation analysis between the indicated genes and the different stages of DLBCL was conducted in the UALCAN portal in the TCGA database. The patients’ information about the disease stage was obtained from the UALCAN[15].
The correlation analysis between indicated genes and tumor purity and infiltration levels of immune/stromal cells in the tumor tissues was conducted by the Estimation of Stromal and Immune cells in Malignant Tumor tissues using Expression data (ESTIMATE)[16], which is a tool to predict the tumor purity and the infiltration levels of immune or stromal cells in tumor tissues. The ESTIMATE score represents tumor purity, and the stromal score correlates to the presence of stroma in the tumor tissues. For further analysis, CAFs, one kind of stromal cells, were employed for the correlation analysis with the MCP-Counter method[17] and tumor immune dysfunction and exclusion (TIDE)[18].
The data analyses were conducted with GraphPad Prism version 8.0 (GraphPad Software incorporated, La Jolla, CA, United States). The data were shown as the mean ± SD. Student’s t-test was applied to compare the statistical difference between groups. Correlation analysis was performed with the Spearman test. P < 0.05 was considered statistically significant.
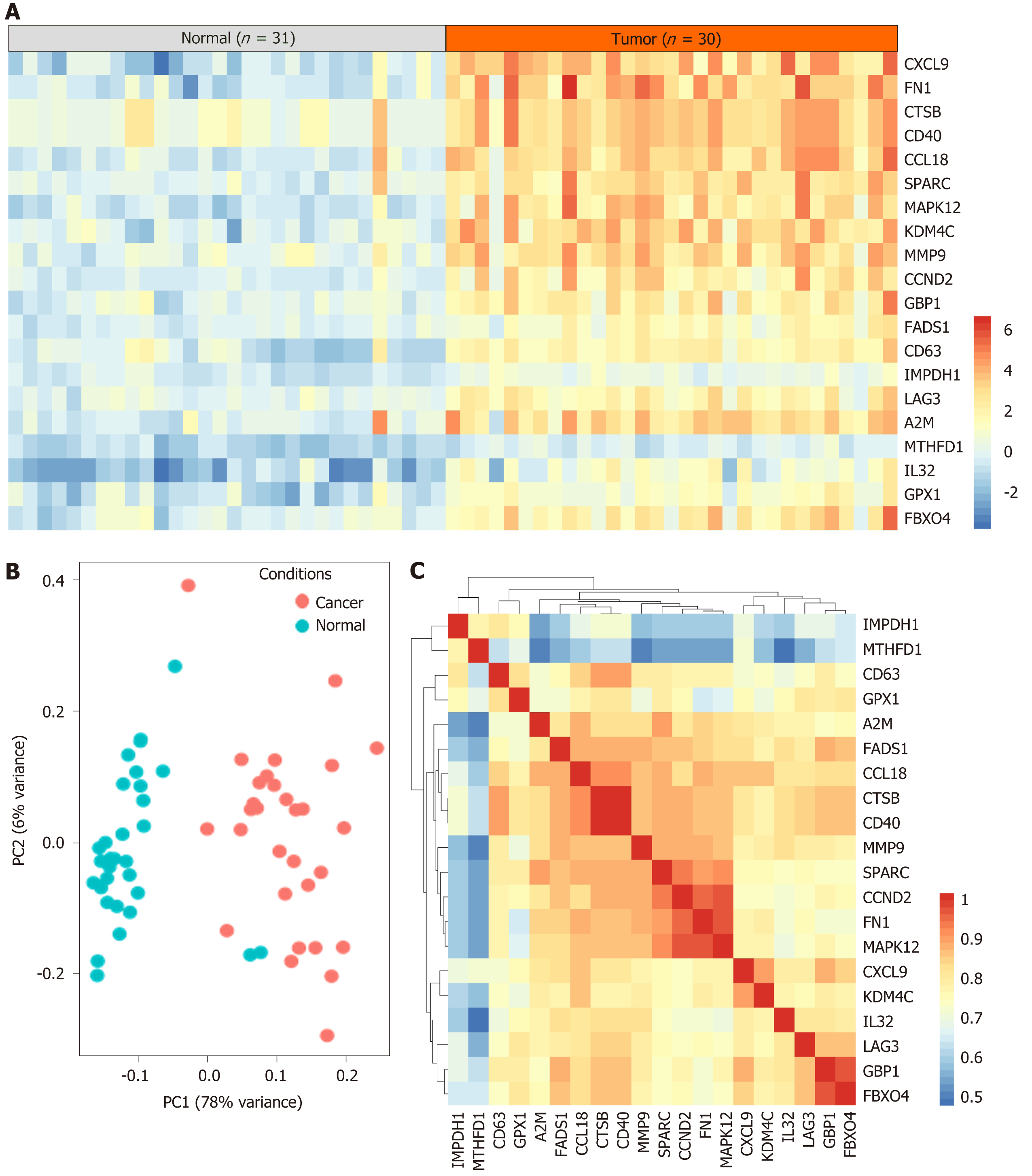
A total of 61 case samples, which involved 31 cases of normal cells and 30 cases of tumor cells were subjected to determine the DEGs. The top 20 overexpressed genes in DLBCL are shown in Figure 1A. Two samples in the normal group were significantly different from the other samples. The principal components analysis on 31 cases of normal cells and 30 cases of DLBCL tumor cells was conducted and shown in Figure 1B, and results were consistent with those shown in Figure 1A. Furthermore, the gene expression pattern of all samples was analyzed by the Spearman correlation matrix for the evaluation of the correlation coefficient, and the results showed that these DEGs have a certain co-expression manner.
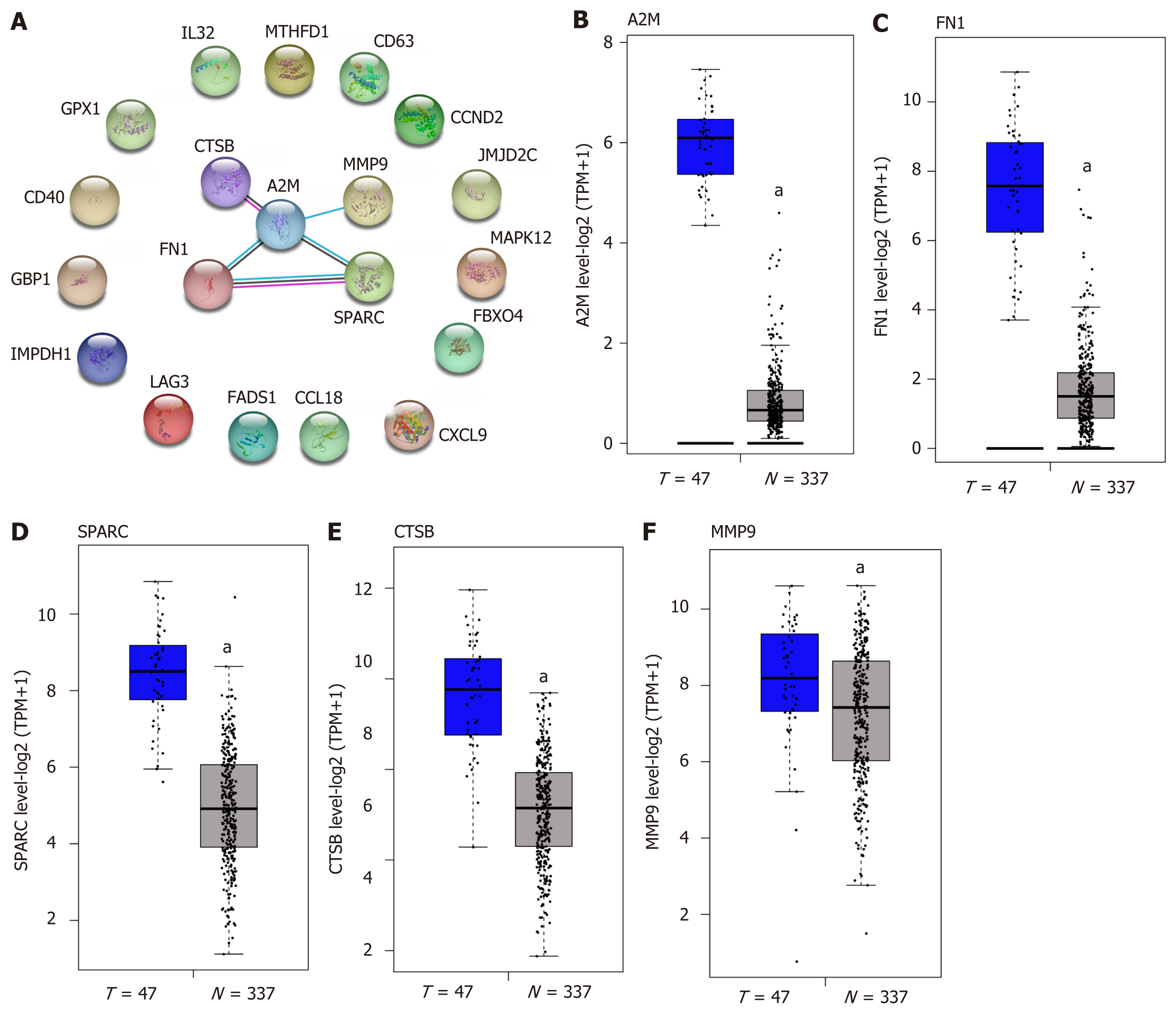
The top 20 DEGs were subjected to STRING for PPI analysis, and screening the hub genes in the regulation of DLBCL. As Figure 2A reveals, A2M, FN1, SPARC, CTSB, and MMP9 were identified as the hub genes. We further analyzed the differential expression of the hub genes in DLBCL in TCGA-DLBCL dataset. The results were similar to those shown in Figure 1 of the GSE60 dataset (Figure 2B-F), and these five hub genes were overexpressed in the DLBCL. Furthermore, the difference of the five genes in germinal center B-like or activated B-like DLBCL was also conducted, as presented in Supplementary Figure 1, and the results showed no significant difference between the two subtypes of DLBCL. Therefore, the five genes were deemed general regulators in DLBCL, and might be important mediators in the regulation of DLBCL.
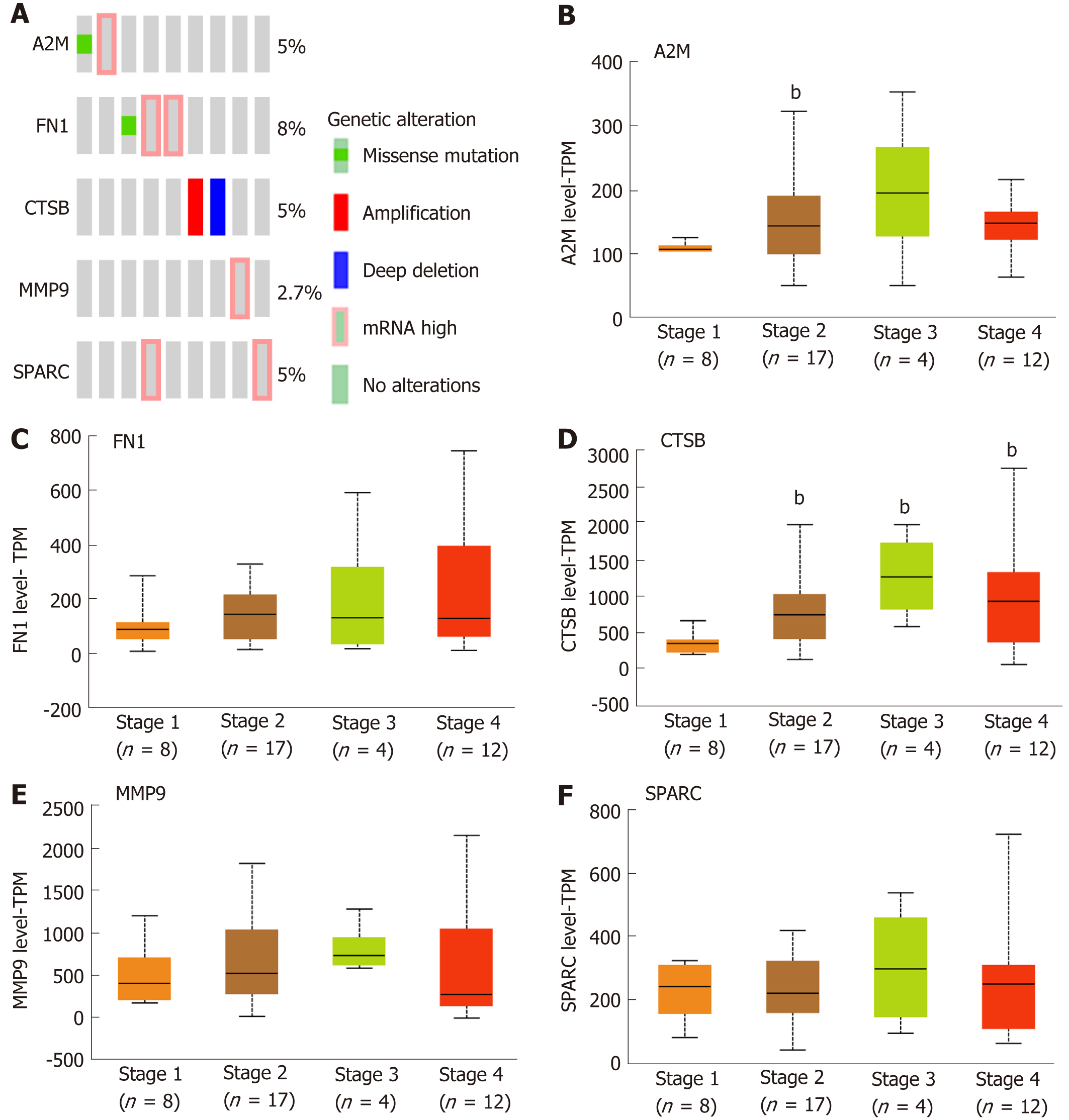
As above mentioned, the five hub genes were identified and confirmed as overexpressed in the DLBCL. Further analysis showed that the percentages of gene alteration among the five hub genes were 5% (A2M), 8% (FN1), 5% (CTSB), 2.7% (MMP9) and 5% (SPARC), respectively (Figure 3A), suggesting the significantly different alteration of the five hub genes, which promotes the overall understanding of the potential association between the hub genes and development of DLBCL. As Figure 3B-F shows, with increased tumor stage, the A2M, FN1, CTSB, MMP9, and SPARC expression becomes higher than that in the early stage (Stage 1). Through the analysis of the five genes associated with the development of DLBCL, we found that the FN1 expression was increased in a stage-dependent manner (Figure 3C). To further analyze the importance of FN1 in the development of DLBCL, and the correlation between FN1 expression and international prognosis index (IPI) score was tested. In detail, the DLBCL samples were divided into three groups according to the IPI score value 0, 1-2, 3-4 respectively, and the result showed that the FN1 expression showed no significant difference in three groups with different IPI scores (Supplementary Figure 2), suggesting that FN1 was an independent factor compared with the common IPI score system.
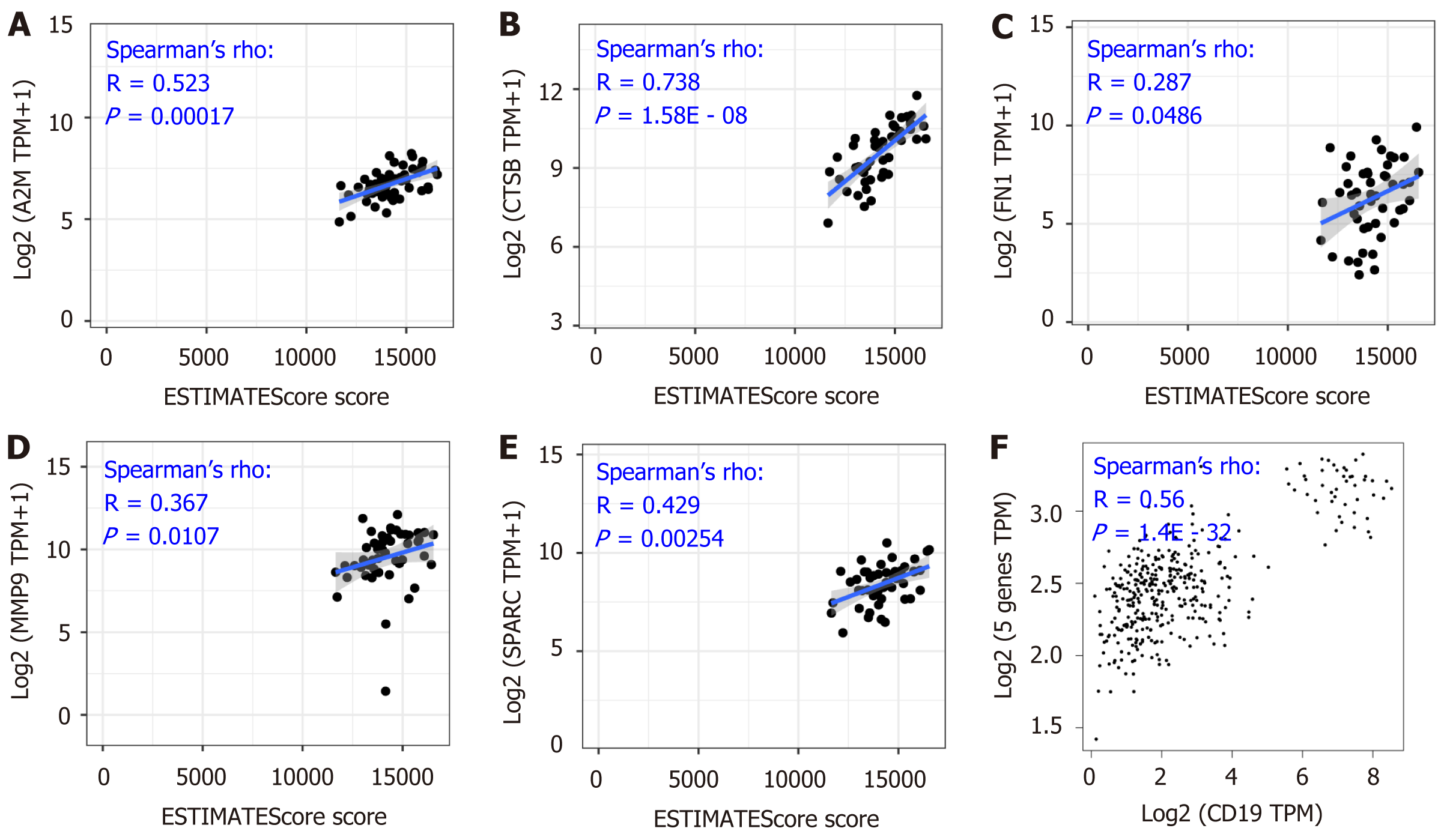
The above results showed the overexpression of these five genes in DLBCL compared with the normal lymphocytes group (Figure 2B-F), and the expression level had a positive correlation with the tumor stage (Figure 3B-F), suggesting the significance of these five genes in the development of DLBCL. To further confirm the findings, tumor purity analysis with the five gene signatures was conducted by the ESTIMATE score. As Figure 4A-E shows, the tumor purity score was positively correlated with the levels of A2M, CTSB, FN1, MMP9, and SPARC gene expression. Furthermore, CD19 is a specific marker for DLBCL; thus, the gene correlation analysis between CD19 and the five gene signatures was conducted and showed a positive correlation between the five gene signatures and the tumor purity (Figure 4F). All of these results showed that the five gene signatures were positively correlated with the tumor purity, suggesting the close association between the five gene signatures and DLBCL development.
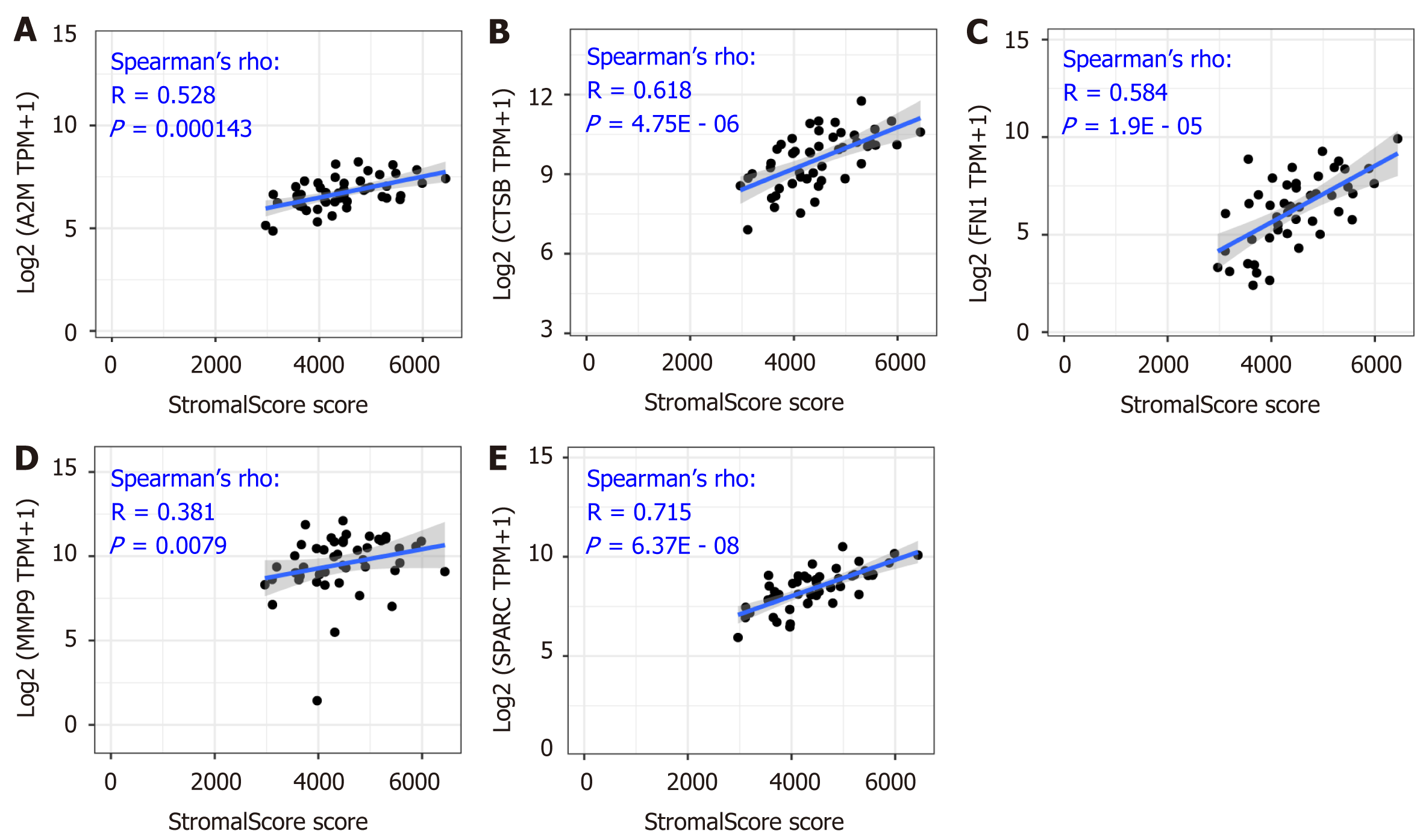
The close correlation between the five hub genes and the tumor purity in the tumor microenvironment was shown above (Figure 4), and the stromal infiltration analysis is an important method to express detailed information regarding the tumor microenvironment. As Figure 5A-E shows, the correlation value between A2M, CTSB, FN1, MMP9, SPARC and the stromal score was 0.528 (P = 0.000143), 0.618 (P = 4.75E - 06), 0.584 (P = 1.9E - 05), 0.381 (P = 0.0079), and 0.715 (P = 6.37E - 08), respectively. The significant positive correlation revealed that the five hub genes might play an important role in the regulation of stromal cells in the tumor microenvironment.
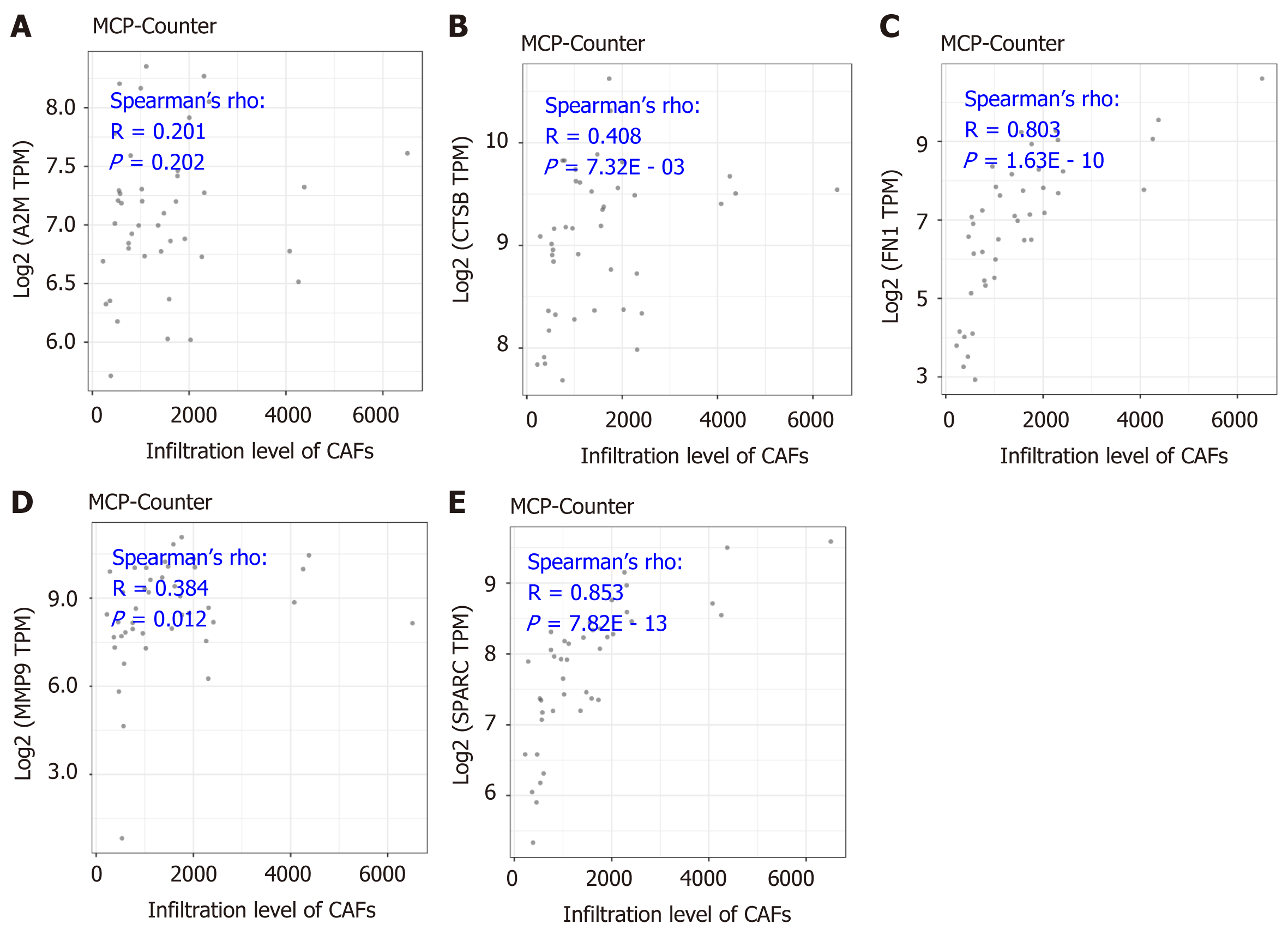
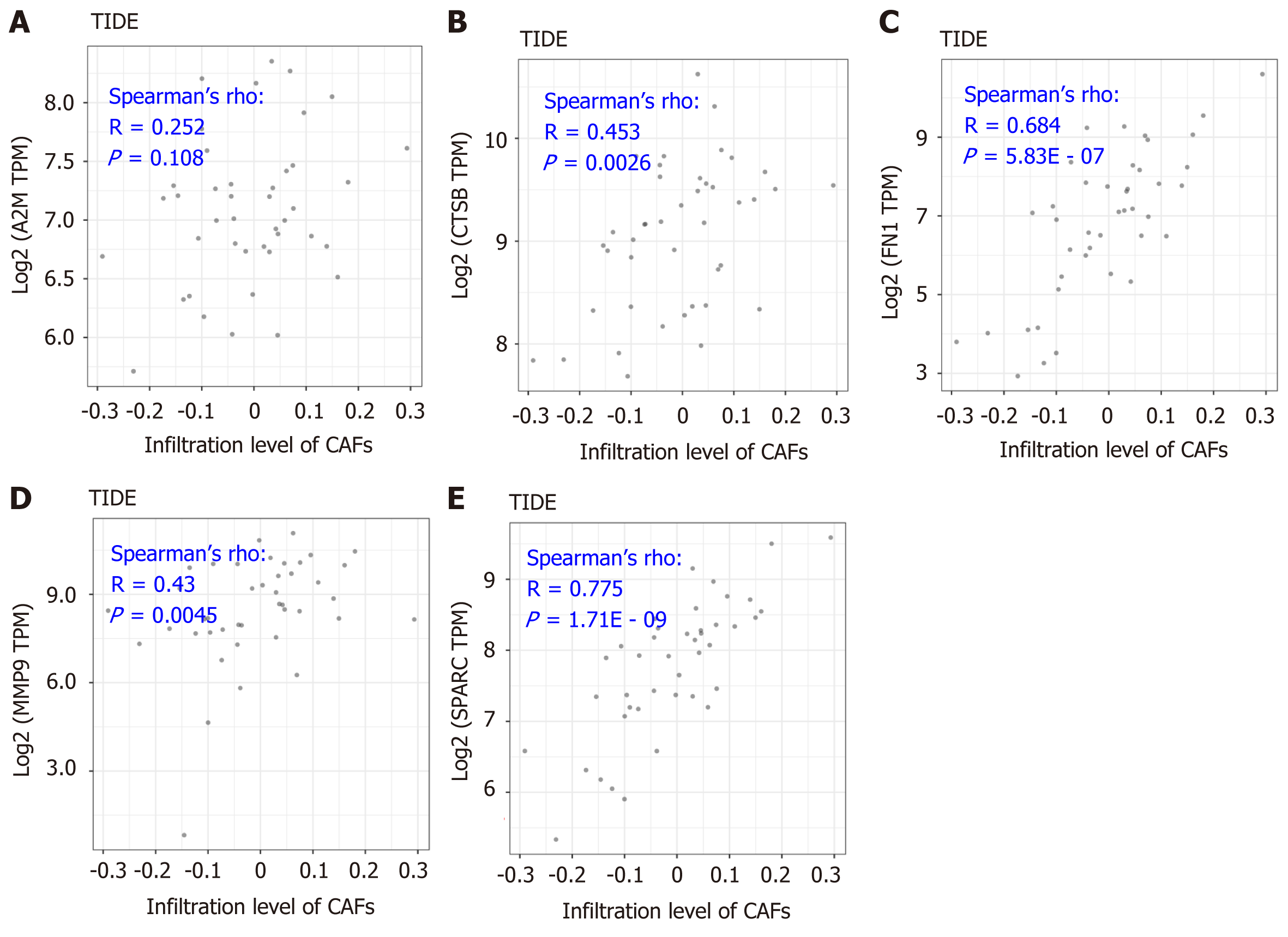
As is indicated in Figure 5, the significantly positive correlation between the five gene signatures and the stromal score suggests a close relationship between the five indicated genes and the tumor microenvironment. To further understand the detailed relationship between the five genes and the stromal infiltration among the tumor microenvironment, and the importance of CAFs in the tumor stromal analysis, we conducted correlation analysis between the five genes’ expression and the CAFs’ infiltration. The results by the MCP-Counter method revealed that the expression of CTSB, FN1, MMP9, and SPARC were positively correlated with the CAFs’ infiltration, having correlation coefficients of 0.201 (P = 0.202), 0.408 (P = 7.32E - 03), 0.803 (P = 1.63E - 10), 0.384 (P = 0.012) and 0.853 (P = 7.82E - 13), respectively (Figure 6B-F). However, A2M showed no significant correlation with the infiltration of CAFs (Figure 6A). More importantly, the results were confirmed with the TIDE portal. The results were consistent with the results from the MCP-Counter method, and the expression of CTSB, FN1, MMP9, and SPARC showed a significant positive correlation with the infiltration levels of CAFs (Figure 7B-E). The uncertain positive correlation between A2M and the degree of CAFs’ infiltration was also confirmed with the TIDE portal. From the above results, we confirmed that the two top genes were FN1 and SPARC. FN1 is a glycoprotein expressing gene, encoding an important component of the extracellular matrix, which could interact with the integrin receptor[19,20]. SPARC is a protein-coding gene rich in cysteine and could be expressed in fibroblasts, osteoblasts, chondrocytes, epithelial cells, and platelets[21]. Previous studies have shown a significant role of the two proteins, FN1 and SPARC, in the regulation of various physiological and pathological processes, including tissue reconstruction, cell migration, and morphogenesis[22-24]. In this study, we identified that the two genes were closely involved in regulation of the DLBCL tumor microenvironment.
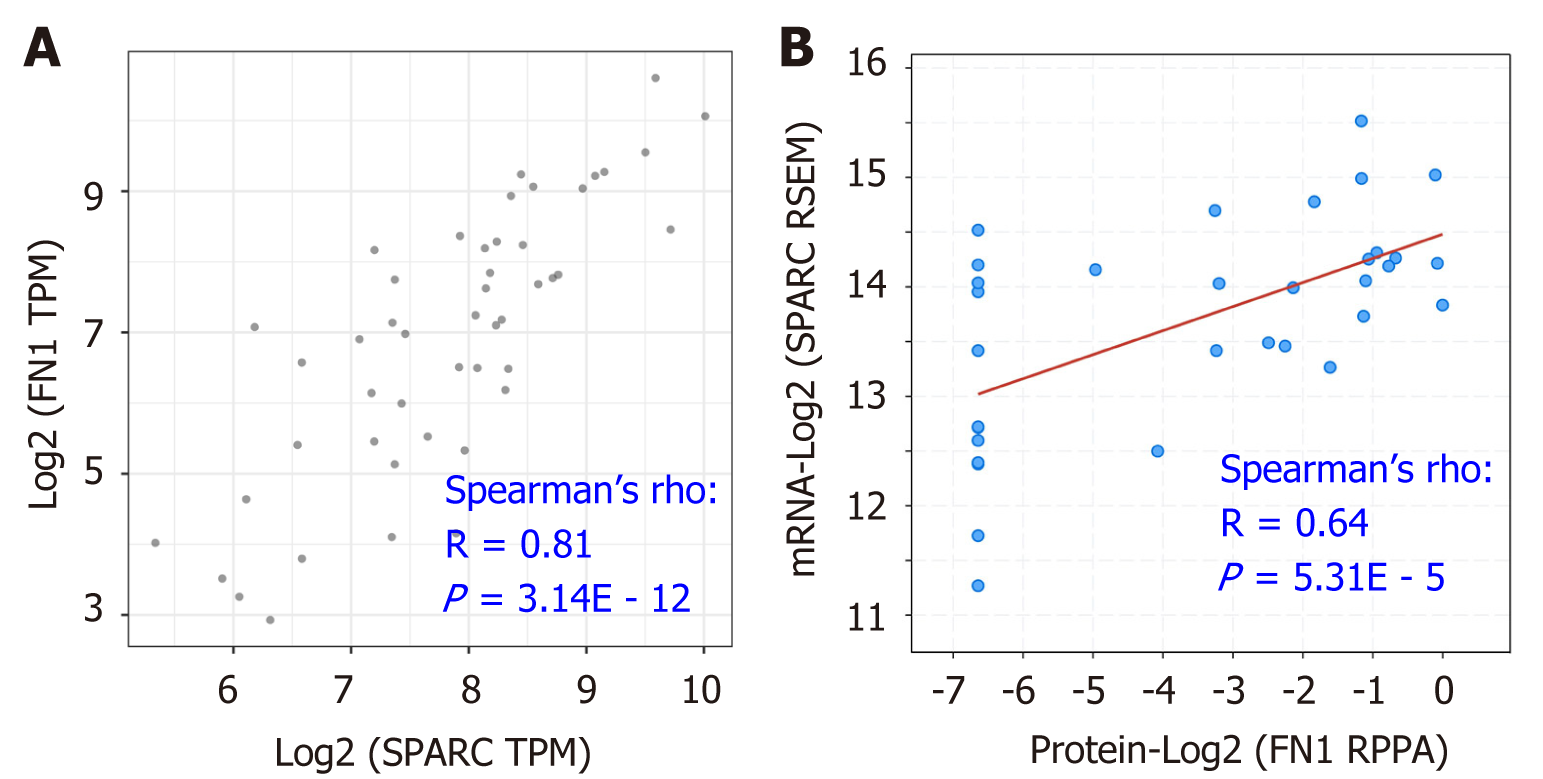
As Figure 6 and 7 show, FN1 and SPARC were the top two molecules correlated with the CAFs’ infiltration, suggesting that the two genes might play a role as pro-oncogenes in the regulation of DLBCL. To further study the potential association between FN1 and SPARC, we conducted the gene expression correlation analysis. As is shown in Figure 8A, FN1 was remarkably correlated with the SPARC expression, with the correlation value of up to 0.81 (P = 3.14E - 12). Besides, similar results were confirmed by the correlation analysis between the FN1 protein level and the SPARC messenger ribonucleic acid level (r = 0.64, P = 5.31E – 5; Figure 8B). These data suggested that FN1 was closely associated with SPARC, and the two genes might be in a co-expression pattern in the DLBCL condition.
DLBCL is a highly heterogeneous tumor; clinical studies showed that DLBCL patients have multiple subtypes and different responses to the treatment of R-CHOP, which is widely recognized by clinicians[5,25]. As a classical example, DLBCL patients with c-Myc gene translocation have poor prognosis after R-CHOP therapy. Therefore, these resistant patients should turn to other treatments, such as chimeric antigen receptor T cells therapy, immunomodulators (immune-inhibitors or immune-agonists), and hematopoietic stem cell transplantation[26-29]. From this aspect, the prognostic prediction analysis based on the patient’s individual gene-mapping information could significantly improve the outcome of precision therapy. However, the identification of prognostic biomarkers for DLBCL has been seldom reported. Though DLBCL is a highly heterogeneous tumor, the stromal infiltration level in DLBCL is unclear. Thus, this study aimed to identify gene signatures for DLBCL and to evaluate the potential significance of these genes in the development of DLBCL.
In this study, we first identified the top 20 overexpressed genes in the DLBCL, and found a correlation coefficient pattern among them (Figure 1), suggesting that they might be involved in the DLBCL and play similar roles in the development of DLBCL. Based on the significant correlation co-efficient pattern of these overexpressed genes, this study was designed to identify the hub genes using the STRING database. With this portal, we analyzed the PPI and obtained five gene signatures (including A2M, CTSB, FN1, MMP9, and SPARC) (Figure 2A). The overexpression of the five genes was confirmed in another DLBCL dataset (Figure 2B-F). Besides, this study identified that these five genes showed certain percentages of genomic alteration, which was 5% (A2M), 8% (FN1), 5% (CTSB), 2.7% (MMP9) and 5% (SPARC), respectively (Figure 3A). The positive correlation between the five genes and disease stages revealed that the gene signature might be involved in the development of DLBCL and might perform as a predictor for the disease progression (Figure 3B-F). The hypothesis was further confirmed by the ESTIMATE analysis, which suggested that the significantly positive correlation between the five genes’ expression and the tumor purity in the DLBCL tumor tissues also reflected the close association between the five gene signatures and DLBCL (Figure 4). More importantly, this study evaluated the correlation between the five genes and stromal score (Figure 5). The results suggested that these five genes were closely involved in the regulation of the extracellular matrix, the most important factor for tumor microenvironment, the abnormal regulation of which plays key roles in tumor progression. Considering the significance of CAFs in the extracellular matrix of some cancers[30,31], we hypothesized that these five genes might be closely associated with CAFs’ infiltration. Figure 6 and 7 show the confirmation of our hypothesis, with the five gene signatures being significantly associated with CAFs’ infiltration. Therefore, based on the inducing effect of CAFs in tumor recurrence and metastasis, the five gene signatures might act as a predictor of CAFs-associated tumor metastasis and recurrence. The detailed analysis revealed that FN1 and SPARC were the most important among the five gene signatures; the remarkable co-expression manner suggested that the two molecules might interact with each other (Figure 8). However, the experimental confirmation of the FN1 and SPARC protein interaction was not included in this study. Moreover, this study was based exclusively on the gene expression level rather than that of the encoded protein. For further study, these points should be attached to much importance, and it could encourage rapid diagnosis for DLBCL patients and provide guidelines for clinical treatment.
Our study found five gene signatures associated with the stromal infiltration, which might provide opportunities to better understand the significance of the tumor microenvironment in DLBCL.
Diffuse large B-cell lymphoma (DLBCL) is a lymphoma with high mortality rates. Even though some therapeutic strategies are applied in clinical practice, the prognoses of DLBCL patients remain unsatisfactory. Therefore, the screening of novel therapeutic targets or prognostic biomarkers could be an important work for DLBCL therapy, which could contribute to the improvement of treatment regimens.
This study aimed to identify the novel biomarkers of DLBCL, and analyze the prognostic value of these biomarkers.
This study addressed the question of the novel biomarkers and potential mechanism involved in the development of DLBCL.
The differentially expressed genes (DEGs) of DLBCL were examined with the GSE60 dataset, and these DEGs were applied to the STRING tool to conduct protein-protein interaction (PPI) analysis. The key hub genes based on PPI analysis were then applied to the GEPIA portal to analyze the expression level in DLBCL. The gene alteration level and the correlation between fibronectin protein level and secreted protein acidic and cysteine-rich messenger ribonucleic acid expression was analyzed in cBioportal. Moreover, the expression level of the hub genes in different stages were investigated in the UALCAN portal. The gene correlation analysis was conducted in GEPIA. The TIMER portal was used to evaluate the correlation between the gene expression and tumor purity, infiltrated stromal cells and infiltrated level of cancer-associated fibroblasts.
The top 20 DEGs in DLBCL were obtained, and the hub genes (A2M, CTSB, FN1, MMP9, and SPARC) were identified based on DEGs through PPI analysis. The five hub genes were overexpressed in DLBCL, and gene alteration was also confirmed in cBioportal, including messenger ribonucleic acid high amplification and missense mutation. Furthermore, the five hub genes had a positive correlation with the tumor stage. Besides, the positive correlation between the five hub genes levels and the tumor purity was also confirmed by the overexpression of the five hub genes in DLBCL. More interestingly, there was a significant correlation between the five hub genes’ expression level and the stromal infiltration score, especially in the correlation analysis with cancer-associated fibroblasts’ infiltration level.
A five hub gene signatures were identified in DLBCL, and the overexpression of these five genes were closely associated with the progression of DLBCL. The mechanism evaluation showed positive correlation between the five genes’ expression levels and infiltrated levels of stromal cells, especially for the cancer-associated fibroblasts. In summary, the five gene signatures have potential values as novel therapeutic targets or biomarkers for DLBCL.
In this project, we identified five gene signatures in DLBCL and that the overexpression of the five genes is closely associated with the disease development, suggesting that the five gene signatures might be novel therapeutic targets for DLBCL, especially in the regulation of cancer-associated fibroblasts. In our subsequent work, the detailed mechanism underlying the regulation of the five genes in the tumor microenvironment will be addressed, which could promote the further understanding of these five gene signatures in DLBCL.
Manuscript source: Unsolicited manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kuo SH S-Editor: Zhang L L-Editor: Filipodia P-Editor: Zhang YL
| 1. | Freedman A, Jacobsen E. Follicular lymphoma: 2020 update on diagnosis and management. Am J Hematol. 2020;95:316-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 151] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 2. | Ferreri AJM, Calimeri T, Ponzoni M, Curnis F, Conte GM, Scarano E, Rrapaj E, De Lorenzo D, Cattaneo D, Fallanca F, Nonis A, Foppoli M, Lopedote P, Citterio G, Politi LS, Sassone M, Angelillo P, Guggiari E, Steffanoni S, Tarantino V, Ciceri F, Bordignon C, Anzalone N, Corti A. Improving the antitumor activity of R-CHOP with NGR-hTNF in primary CNS lymphoma: final results of a phase 2 trial. Blood Adv. 2020;4:3648-3658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 3. | Harker-Murray PD, Pommert L, Barth MJ. Novel Therapies Potentially Available for Pediatric B-Cell Non-Hodgkin Lymphoma. J Natl Compr Canc Netw. 2020;18:1125-1134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 4. | Pettengell R, Długosz-Danecka M, Andorsky D, Belada D, Georgiev P, Quick D, Singer JW, Singh SB, Pallis A, Egorov A, Salles G. Pixantrone plus rituximab vs gemcitabine plus rituximab in patients with relapsed aggressive B-cell non-Hodgkin lymphoma not eligible for stem cell transplantation: a phase 3, randomized, multicentre trial (PIX306). Br J Haematol. 2020;188:240-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 5. | Rushton CK, Arthur SE, Alcaide M, Cheung M, Jiang A, Coyle KM, Cleary KLS, Thomas N, Hilton LK, Michaud N, Daigle S, Davidson J, Bushell K, Yu S, Rys RN, Jain M, Shepherd L, Marra MA, Kuruvilla J, Crump M, Mann K, Assouline S, Connors JM, Steidl C, Cragg MS, Scott DW, Johnson NA, Morin RD. Genetic and evolutionary patterns of treatment resistance in relapsed B-cell lymphoma. Blood Adv. 2020;4:2886-2898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 80] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 6. | Poeschel V, Held G, Ziepert M, Witzens-Harig M, Holte H, Thurner L, Borchmann P, Viardot A, Soekler M, Keller U, Schmidt C, Truemper L, Mahlberg R, Marks R, Hoeffkes HG, Metzner B, Dierlamm J, Frickhofen N, Haenel M, Neubauer A, Kneba M, Merli F, Tucci A, de Nully Brown P, Federico M, Lengfelder E, di Rocco A, Trappe R, Rosenwald A, Berdel C, Maisenhoelder M, Shpilberg O, Amam J, Christofyllakis K, Hartmann F, Murawski N, Stilgenbauer S, Nickelsen M, Wulf G, Glass B, Schmitz N, Altmann B, Loeffler M, Pfreundschuh M; FLYER Trial Investigators; German Lymphoma Alliance. Four vs six cycles of CHOP chemotherapy in combination with six applications of rituximab in patients with aggressive B-cell lymphoma with favourable prognosis (FLYER): a randomised, phase 3, non-inferiority trial. Lancet. 2019;394:2271-2281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 159] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 7. | Smith SD, Till BG, Shadman MS, Lynch RC, Cowan AJ, Wu QV, Voutsinas J, Rasmussen HA, Blue K, Ujjani CS, Shustov A, Cassaday RD, Fromm JR, Gopal AK. Pembrolizumab with R-CHOP in previously untreated diffuse large B-cell lymphoma: potential for biomarker driven therapy. Br J Haematol. 2020;189:1119-1126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 8. | Alame M, Pirel M, Costes-Martineau V, Bauchet L, Fabbro M, Tourneret A, De Oliveira L, Durand L, Roger P, Gonzalez S, Cacheux V, Rigau V, Szablewski V. Characterisation of tumour microenvironment and immune checkpoints in primary central nervous system diffuse large B cell lymphomas. Virchows Arch. 2020;476:891-902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 9. | Ciavarella S, Vegliante MC, Fabbri M, De Summa S, Melle F, Motta G, De Iuliis V, Opinto G, Enjuanes A, Rega S, Gulino A, Agostinelli C, Scattone A, Tommasi S, Mangia A, Mele F, Simone G, Zito AF, Ingravallo G, Vitolo U, Chiappella A, Tarella C, Gianni AM, Rambaldi A, Zinzani PL, Casadei B, Derenzini E, Loseto G, Pileri A, Tabanelli V, Fiori S, Rivas-Delgado A, López-Guillermo A, Venesio T, Sapino A, Campo E, Tripodo C, Guarini A, Pileri SA. Dissection of DLBCL microenvironment provides a gene expression-based predictor of survival applicable to formalin-fixed paraffin-embedded tissue. Ann Oncol. 2018;29:2363-2370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 82] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 10. | Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503-511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7015] [Cited by in RCA: 6303] [Article Influence: 252.1] [Reference Citation Analysis (0)] |
| 11. | Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607-D613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10161] [Cited by in RCA: 11676] [Article Influence: 1946.0] [Reference Citation Analysis (1)] |
| 12. | Tang Z, Li C, Kang B, Gao G, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5550] [Cited by in RCA: 7044] [Article Influence: 880.5] [Reference Citation Analysis (0)] |
| 13. | GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4162] [Cited by in RCA: 3804] [Article Influence: 380.4] [Reference Citation Analysis (0)] |
| 14. | Cline MS, Craft B, Swatloski T, Goldman M, Ma S, Haussler D, Zhu J. Exploring TCGA Pan-Cancer data at the UCSC Cancer Genomics Browser. Sci Rep. 2013;3:2652. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 209] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 15. | Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649-658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2365] [Cited by in RCA: 4194] [Article Influence: 524.3] [Reference Citation Analysis (0)] |
| 16. | Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA, Carter SL, Getz G, Stemke-Hale K, Mills GB, Verhaak RG. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3056] [Cited by in RCA: 6397] [Article Influence: 581.5] [Reference Citation Analysis (0)] |
| 17. | Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, Selves J, Laurent-Puig P, Sautès-Fridman C, Fridman WH, de Reyniès A. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 987] [Cited by in RCA: 2226] [Article Influence: 247.3] [Reference Citation Analysis (0)] |
| 18. | Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, Li Z, Traugh N, Bu X, Li B, Liu J, Freeman GJ, Brown MA, Wucherpfennig KW, Liu XS. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24:1550-1558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1184] [Cited by in RCA: 3359] [Article Influence: 479.9] [Reference Citation Analysis (0)] |
| 19. | Lee J, Park B, Moon B, Park J, Moon H, Kim K, Lee SA, Kim D, Min C, Lee DH, Lee G, Park D. A scaffold for signaling of Tim-4-mediated efferocytosis is formed by fibronectin. Cell Death Differ. 2019;26:1646-1655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 20. | Wang W, He Y, Zhao Q, Zhao X, Li Z. Identification of potential key genes in gastric cancer using bioinformatics analysis. Biomed Rep. 2020;12:178-192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 21. | Bao JM, Dang Q, Lin CJ, Lo UG, Feldkoren B, Dang A, Hernandez E, Li F, Panwar V, Lee CF, Cen JJ, Guan B, Margulis V, Kapur P, Brekken RA, Luo JH, Hsieh JT, Tan WL. SPARC is a key mediator of TGF-β-induced renal cancer metastasis. J Cell Physiol. 2021;236:1926-1938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 22. | Sun Y, Zhang Y, Wu X, Chi P. A Four Gene-Based Risk Score System Associated with Chemoradiotherapy Response and Tumor Recurrence in Rectal Cancer by Co-Expression Network Analysis. Onco Targets Ther. 2020;13:6721-6733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 23. | Munk R, Martindale JL, Yang X, Yang JH, Grammatikakis I, Di Germanio C, Mitchell SJ, de Cabo R, Lehrmann E, Zhang Y, Becker KG, Raz V, Gorospe M, Abdelmohsen K, Panda AC. Loss of miR-451a enhances SPARC production during myogenesis. PLoS One. 2019;14:e0214301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 24. | Wang H, Ning T, Song C, Luo X, Xu S, Zhang X, Deng Z, Ma D, Wu B. Priming integrin α5 promotes human dental pulp stem cells odontogenic differentiation due to extracellular matrix deposition and amplified extracellular matrix-receptor activity. J Cell Physiol. 2019;234:12897-12909. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 25. | Meriranta L, Pasanen A, Alkodsi A, Haukka J, Karjalainen-Lindsberg ML, Leppä S. Molecular background delineates outcome of double protein expressor diffuse large B-cell lymphoma. Blood Adv. 2020;4:3742-3753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 26. | Abdulla M, Guglielmo P, Hollander P, Åström G, Ahlström H, Enblad G, Amini RM. Prognostic impact of abdominal lymph node involvement in diffuse large B-cell lymphoma. Eur J Haematol. 2020;104:207-213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 27. | Karkhanis V, Alinari L, Ozer HG, Chung J, Zhang X, Sif S, Baiocchi RA. Protein arginine methyltransferase 5 represses tumor suppressor miRNAs that down-regulate CYCLIN D1 and c-MYC expression in aggressive B-cell lymphoma. J Biol Chem. 2020;295:1165-1180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 28. | Lekakis LJ, Moskowitz CH. The Role of Autologous Stem Cell Transplantation in the Treatment of Diffuse Large B-cell Lymphoma in the Era of CAR-T Cell Therapy. Hemasphere. 2019;3:e295. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 29. | Kim YR, Yoon SO, Kim SJ, Cheong JW, Chung H, Lee JY, Jang JE, Kim Y, Yang WI, Min YH, Kim JS. Upfront autologous hematopoietic stem cell transplantation for high-risk patients with double-expressor diffuse large B cell lymphoma. Ann Hematol. 2020;99:2149-2157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 30. | Haro M, Orsulic S. A Paradoxical Correlation of Cancer-Associated Fibroblasts With Survival Outcomes in B-Cell Lymphomas and Carcinomas. Front Cell Dev Biol. 2018;6:98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 31. | Wu SZ, Roden DL, Wang C, Holliday H, Harvey K, Cazet AS, Murphy KJ, Pereira B, Al-Eryani G, Bartonicek N, Hou R, Torpy JR, Junankar S, Chan CL, Lam CE, Hui MN, Gluch L, Beith J, Parker A, Robbins E, Segara D, Mak C, Cooper C, Warrier S, Forrest A, Powell J, O'Toole S, Cox TR, Timpson P, Lim E, Liu XS, Swarbrick A. Stromal cell diversity associated with immune evasion in human triple-negative breast cancer. EMBO J. 2020;39:e104063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 278] [Article Influence: 55.6] [Reference Citation Analysis (0)] |