Published online Aug 6, 2020. doi: 10.12998/wjcc.v8.i15.3197
Peer-review started: March 24, 2020
First decision: April 24, 2020
Revised: May 26, 2020
Accepted: July 15, 2020
Article in press: July 15, 2020
Published online: August 6, 2020
Processing time: 134 Days and 23.7 Hours
AMPD2 is a critical enzyme catalyzing smooth muscle energy supply and metabolism; however, its cellular biological function and clinical implication in colorectal cancer (CRC) are largely unknown.
To clarify the role of AMPD2 in CRC and study the pathway and prognostic value of its role.
AMPD2 expression was analyzed by integrated bioinformatics analysis based on TCGA data sets and immunohistochemistry in tissue microarrays, and the correlation between AMPD2 expression and clinicopathological parameters, Notch3 expression, and prognostic features was assessed. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analysis were then performed to investigate the regulatory pathway involved. The effects of AMPD2 expression on CRC cells and Notch3 protein expression were investigated by downregulation and overexpression of AMPD2.
AMPD2 mRNA was significantly overexpressed in tumor tissue when compared with normal tissue in a cohort of the TCGA-COAD data set. Biological function enrichment analysis indicated that the Notch pathway strongly correlated with AMPD2 expression, and that the expression of Notch3 and JAG2 mRNA was positively associated with AMPD2 in CRC tissues. In vitro, AMPD2 overexpression markedly reduced Notch3 protein expression in CRC cells, while knockdown of AMPD2 showed the opposite findings. In addition, protein expression was significantly up-regulated in our CRC cohort as indicated by tissue microarray analysis. High expression of AMPD2 protein correlated with advanced depth of tumor and poor differentiation. Furthermore, high AMPD2 expression in CRC tissues was an indicator of poor outcome for CRC patients.
AMPD2 is commonly overexpressed in CRC, and acts as a metabolism oncogene to induce CRC progression through the Notch signaling pathway. Thus, AMPD2 may be a novel prognostic biomarker for CRC.
Core tip: In this study, we summarized and analyzed the mRNA expression data of TCGA-COAD and obtained the expression features of AMPD2 in cancer tissues and adjacent tissues in patients with colorectal cancer. We constructed cell samples that were not subjected to high-throughput sequencing of RNA-seq, and although the results were closer to clinical reality, in the case of verification experiments, the probability of false negatives was increased. In addition, our cytology validation experiments need to be carried out in more detail, and our colorectal cancer cohort sample was only 201 cases. We expect to add cytological evidence and a large sample of prospective prognostic cohorts in subsequent studies.
- Citation: Gao QZ, Qin Y, Wang WJ, Fei BJ, Han WF, Jin JQ, Gao X. Overexpression of AMPD2 indicates poor prognosis in colorectal cancer patients via the Notch3 signaling pathway. World J Clin Cases 2020; 8(15): 3197-3208
- URL: https://www.wjgnet.com/2307-8960/full/v8/i15/3197.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v8.i15.3197
Colorectal cancer (CRC) is the third most common cancer and the fourth leading cause of cancer-related deaths worldwide, and remains a major health threat in China[1,2]. CRC patients at an advanced stage often have a poor prognosis due to abnormal tumor metabolism and recurrence. Therefore, it is of clinical significance to identify novel biomarkers that control the severity of CRC as well as biomarkers for the prediction of prognosis[3].
Recently, there has been increased interest in studying the metabolic reprogramming in cancer in the hope of developing metabolism-based anticancer therapy[4]. Accordingly, various studies aimed at identifying the metabolic index in blood, body fluids, carcinoma tissue, and gut microbiota have been performed to establish a complete metabolic atlas and explain the possible mechanisms by which these abnormalities are present in gastrocolorectal tumors[5]. In several studies, it has been shown that in addition to primary metabolism, many specific secondary metabolites have an important impact on the development and progression of CRC, thereby suggesting that specific metabolic markers can be used as biomarkers for metabolic abnormalities[6]. These findings suggested that AMPD2 may act as a metabolic tissue enzyme, which is closely related to the type of CRC, and may play an important role in CRC.
AMPD2 is an important enzyme involved in adenosine monophosphate metabolism and plays an important biological function in adenylate metabolism in smooth muscle tissue[7]. In previous studies, it was shown that AMPD2 affected the nephrotic syndrome and hypercholesterolemia[8]. In addition, it has been demonstrated that AMPD2 dramatically affected physiological and pathological processes[9]. Despite recent efforts to understand the activities of AMPD2 in the biological mechanism of cancer, little is known about the role of AMPD2 in CRC and it remains to be elucidated whether AMPD2 expression correlates with any of the clinicopathological features of CRC[10].
In this study, we combined TCGA-COAD data set analyses of both tumor and normal samples to investigate the significance of AMPD2 in CRC. Therefore, in vitro studies were performed to investigate how AMPD2 may affect the CRC-associated regulatory pathway. A total of 201 samples were collected using tissue microarray validation cohort studies to investigate whether AMPD2 could be used as a biomarker for poor CRC prognosis.
In this study, the TCGA dataset was used for analysis of gene expression in CRC tissues and normal tissues. TCGA-COAD RNA-seq data and related clinical data of cancer tissues (478), and normal tissues (41) were downloaded from https://xenabrowser.net/[11]. We continued to match and screen the organization label, and obtained 40 pairs of cancer tissues and paired adjacent normal tissues. The data were updated to November 2018. To assess the prospective functions of the genes influenced by AMPD2, we selected a total of 50 TCGA-COAD specimens with the highest expression of AMPD2 and 50 TCGA-COAD specimens with the lowest expression of AMPD2, and follow-up biologic function prediction analysis was performed in the abnormally high expression and low expression groups. Gene Ontology (GO)[12] and Kyoto Encyclopedia of Genes and Genomes (KEGG)[13] analyses were employed using R software (version 3.6) Bioconductor module. The datasets GSE97023 were downloaded from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/)[14], to analyze RNA-seq baseline data for CRC cell lines.
Human CRC cell lines SW480 and Co115 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in Dulbecco’s Modified Eagle’s Medium (Biowest, France) with 10% fetal bovine serum (Gibco, United States) and 1% penicillin-streptomycin antibiotics (Gibco, United States). Cells were cultured in an incubator at 37°C with 5% CO2. For AMPD2 overexpression, cells were transfected with AMPD2 overexpression plasmid or blank plasmid purchased from Gene Pharma (Shanghai, China). For AMPD2 knockdown, an AMPD2 specific siRNA (siAMPD2) and a negative control siRNA (siNEG) were chemically synthesized by Gene Pharma (Shanghai, China). Transfection of cells with siRNA was performed using Lipofectamine 2000 (Invitrogen, ThermoFisher Scientific, United States) according to the manufacturer’s guidelines.
Cells were harvested and lysed in RIPA buffer, containing proteinase and phosphatase inhibitors. The concentration of proteins in cell lysates was determined by a BCA kit (Applygen Technologies, China) following the manufacturer’s instructions. Equal amounts of cell lysates (20 μg protein/lane) were electrophoresed on SDS-polyacrylamide gels and transferred to nitrocellulose membranes. Membranes were blocked, and then probed overnight with appropriate primary antibodies (AMPD2, 1:1000; Notch3, 1:500; GAPDH, 1:8000) in blocking buffer at 4°C. Bound antibodies were detected with Beyotime horseradish (HRP)-conjugated goat anti-rabbit IgG or anti-mouse IgG (at 1:10000 dilution; Genetech, China) and visualized using a Bio-Rad ChemiDoc XRS imaging system.
A total of 201 CRC patients who underwent CRC surgery between January and August 2014 in the Oncology Department, Affiliated Hospital of Jiangnan University, Wuxi, China, were enrolled in the present study. The patients were followed-up until December 2018. Based on the patient's medical records, formalin-fixed paraffin-embedded (FFPE) tissue specimens from these patients were collected. Clinical and pathological parameters, including patient age, gender, tumor size, histological grade, primary tumor (pT), lymph node (pN) metastasis, pathological staging, neurological invasion, lymphatic invasion, and vascular invasion were analyzed. Patient data and tissue samples collected during our study were approved by the Ethics Committee of Affiliated Hospital of the Jiangnan University (Jiangnan, China). After collecting the paraffin blocks, we fabricated tissue microarrays (TMA) using a Tissue microarray sampler (Quick-Ray; UNITMA, Korea). From each colorectal adenocarcinoma tissue, two representative tumor cores with a diameter of 1.5 mm were obtained. Hematoxylin and eosin (H&E) staining was performed on each tissue to confirm tumor tissue integrity and cell morphology. Cases with insufficient cancer tissue or cases with no adenocarcinoma in the core were not included in the study. To ensure the stability of subsequent TMA studies, two areas were sampled in each tumor tissue and normal epithelial tissue.
Immunohistochemistry was performed using 4 µm sections from FFPE tissue specimens of CRC patients. After incubation in xylene three times for five minutes each time, and subsequent passages in gradient alcohol, the sections were immersed in distilled water for 5 min. Next, an antigen retrieval procedure was performed using a sodium citrate solution for 20 min in boiling water. Sections were incubated with 3% hydrogen peroxide for 5 min to block endogenous peroxidase. Subsequently, the sections were incubated overnight at 4°C with a primary polyclonal antibody directed against AMPD2 (1:200, ProteinTech, China) and Notch3 (1:200, ProteinTech, China). The sections were washed, incubated with the amplification agent and polymerase (reagent A, GTVisionTM III Kit supply, China), then stained with 3,3’-diaminobenzidine (reagent B and C, GTVisionTM III Kit supply, Shanghai, China). Sections were counterstained using hematoxylin. In each section, the staining was scored according to the percentage of positively-stained cells in the entire section (no positive staining or ≤ 5% = 0; 6%-25% positive = 1; 26%-50% positive = 2; 51%-75% positive = 3; and 76%-100% positive = 4). The scoring intensity was determined as previously described: no staining = 0, weak staining = 1, moderate staining = 2, strong staining = 3[15].
Cut-off values for AMPD2 and Notch3 were obtained from the receiver operating characteristic curve (ROC) and the area under curve (AUC). Pearson’s Chi-square tests or Fisher’s exact tests were employed to analyze the association between AMPD2 expression, clinicopathological parameters, and Notch3. Moreover, Kaplan-Meier survival analysis was used to determine the probability of overall survival (OS). Data were analyzed using log-rank tests. Univariate and multivariate survival analyses were performed using the Cox proportional hazard model and OS. Statistical analyses were performed using SPSS for Windows 22.0 software (IBM). P < 0.05 was considered statistically significant.
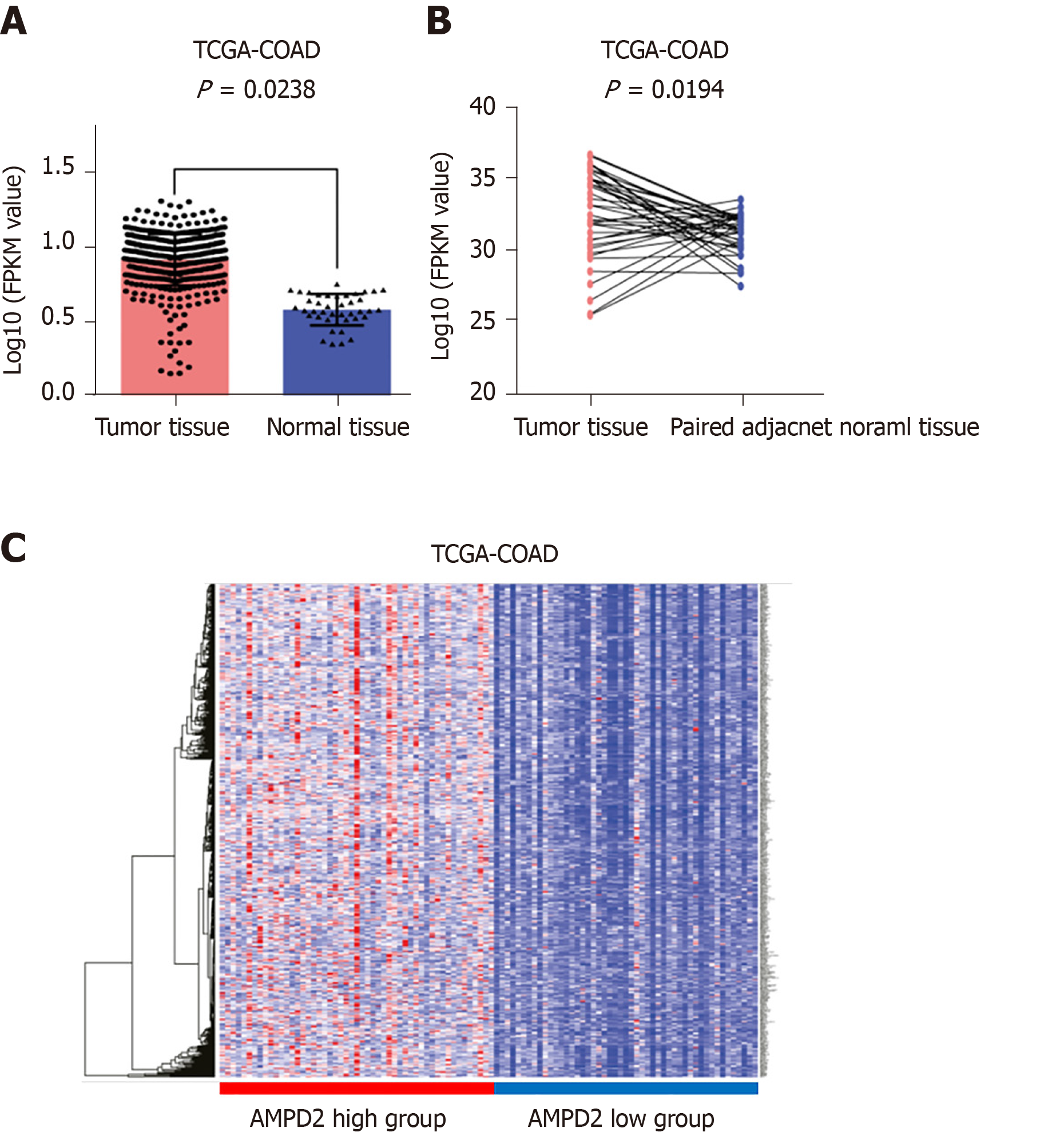
In this study, we evaluated AMPD2 expression in CRC and normal colorectal tissues, and changes in transcriptional levels of AMPD2 were first validated using microarray values in all tumor tissue and normal tissue from the TCGA-COAD data sets. The results showed that AMPD2 mRNA was significantly increased in CRC tissues in the TCGA-COAD data set (P = 0.0238; Figure 1A). Moreover, we demonstrated that an increase in AMPD2 mRNA abundance was observed in paired cancerous tissue when compared with paired adjacent normal tissue from the TCGA data set (Figure 1B). In addition, we used the TCGA-COAD data set to correlate mRNA expression with AMPD2. A total of 50 specimens with abnormally-high expression of AMPD2 and 50 specimens of abnormally-low expression of AMPD2 were selected, and differential gene expression was analyzed. According to the high and low expression of AMPD2, a heat map was drawn as shown in Figure 1C.
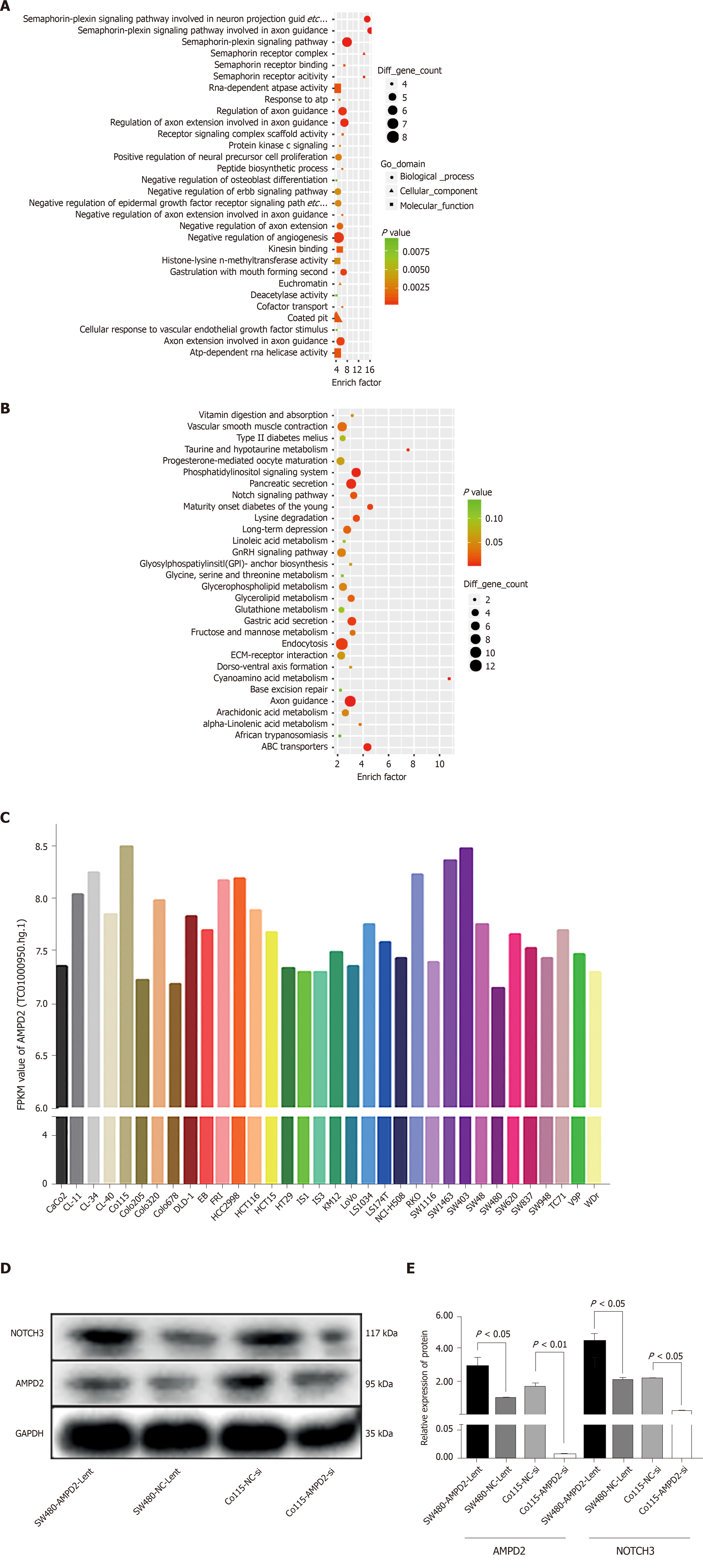
To further investigate the biological function of the differential expression of transcription levels caused by high expression of AMPD2, we performed GO and KEGG enrichment analysis of the genes that were up-regulated in the AMPD2 overexpression group (Figure 2A and B). When compared to the AMPD2 low expression group, the AMPD2 overexpression group showed a positive regulation of neural precursor cell proliferation. In addition, KEGG analysis showed that the Notch pathway was strongly correlated with AMPD2 expression, and the expression of Notch3 and JAG2 mRNA was positively associated with AMPD2 in CRC tissues. These results suggested that high expression of the Notch signaling pathway in CRC patients with AMPD2 up-regulation affected the progression of CRC.
Figure 2C shows AMPD2 (gene probe: TC01000950.hg.1) baseline mRNA expression values in various CRC cell lines. We selected SW480 and Co115 cell lines for subsequent up-regulation and silencing studies of cell biology. Protein expression levels were determined. In brief, AMPD2 and Notch3 protein expression was evaluated in SW480 and Co115 cells. Both AMPD2 and Notch3 were up-regulated at the protein level (Figure 2D and E) in SW480 and Co115 cells with AMPD2-Lent when SW480 and Co115 cells were compared with the AMPD2 negative control. Similarly, both AMPD2 and Notch3 were down-regulated at the protein level (Figure 2D and E) in SW480 and Co115 cells treated with siAMPD2 when compared to SW480 and Co115 cells that were transfected with the negative control.
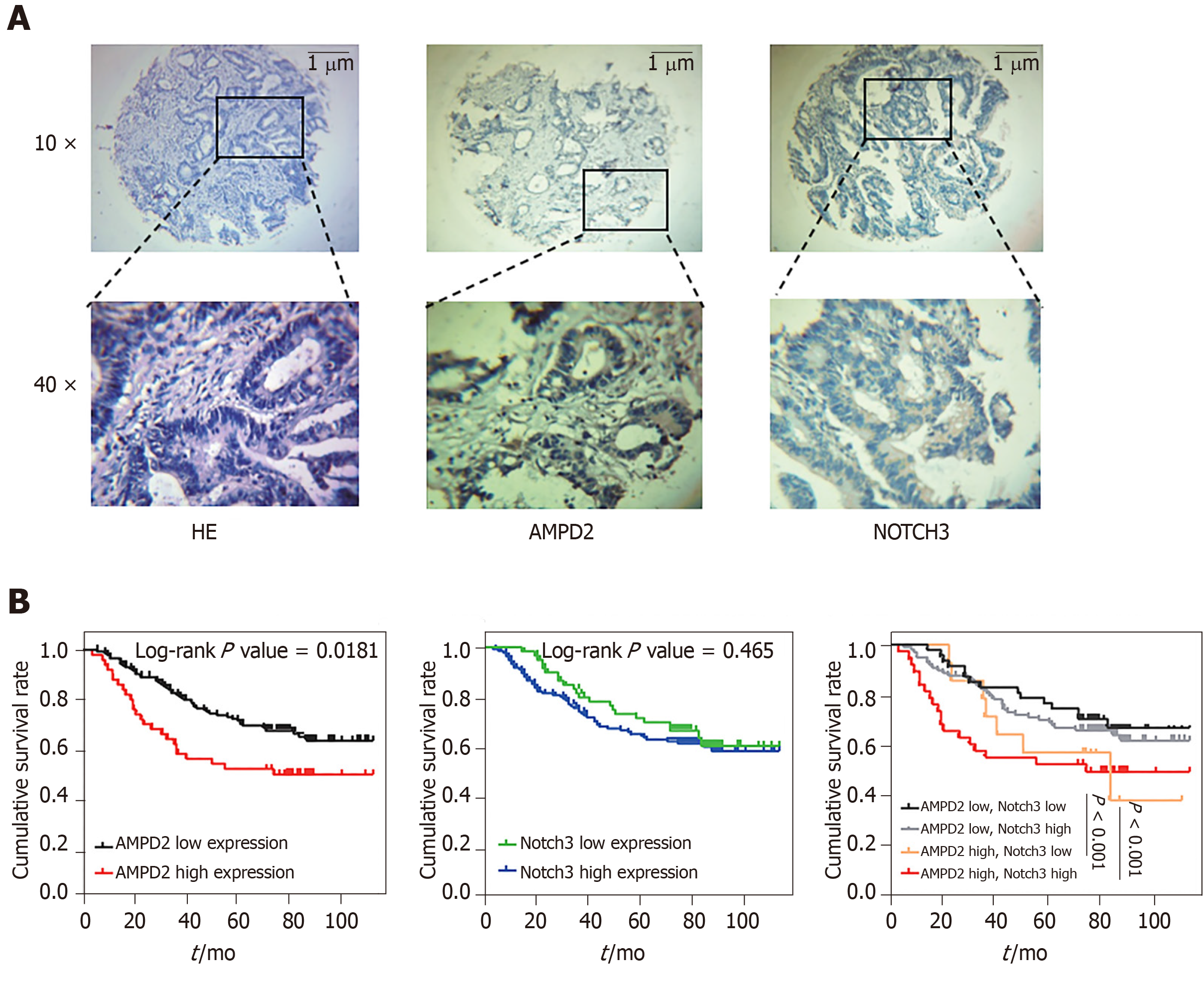
We analyzed the correlation between high expression of AMPD2 and Notch3 and clinicopathological parameters in CRC patients. Clinicopathological and demographic characteristics are presented in Table 1. Immunohistochemical analysis of AMPD2 and Notch3 expression in 201 CRC patients revealed that AMPD2 and Notch3 were predominantly expressed in the cytoplasm of carcinoma cells (Figure 3A). To assess the classification of AMPD2 and Notch3 protein expression, we used the immunohistochemical semi-quantitative score and predicted the ROC curve for survival by AMPD2 and Notch3 expression levels, and calculated the cut-off value in conjunction with the results of the Youden index. Thus, patients were categorized as high expression of AMPD2 if their immunohistochemical scores were > 0.50. As shown in Table 1, overexpression of AMPD2 significantly correlated with advanced depth of the tumor (P = 0.022) and poor differentiation (P = 0.037). As shown in Table 2, univariate Cox analysis for OS indicated a correlation with higher TNM stage (HR = 3.198, 95%CI: 1.915-5.343, P < 0.001) and increased expression of AMPD2 (HR = 1.769, 95%CI: 1.094-2.862, P = 0.020). Furthermore, multivariate regression analysis indicated that a higher TNM stage was associated with a relative risk of death of 3.727 (95%CI: 2.206-6.296, P < 0.001) when compared with that of Stage I-II. In addition, overexpression of AMPD2 was associated with a relative risk of death of 2.133 (95%CI: 1.295-3.513, P = 0.003). Patients with colorectal adenocarcinomas who belonged to the AMPD2 high expression group (n = 61) had a significantly lower cumulative survival rate when compared to patients in the AMPD2 low expression group (n = 140) (log-rank P = 0.0181). Patients with CRC who had high expression of Notch3 (n = 70) had a significantly lower cumulative survival rate when compared to those with low expression of Notch3 (n = 131) (log-rank P = 0.465). Subgroup survival analysis revealed that patients who showed high expression of both AMPD2 and Notch3 had the worst prognosis when compared with the other three subgroups. Patients with low expression of AMPD2 and low expression of Notch3 showed the highest survival when compared to the other subgroups (P < 0.001, Figure 3B).
| Parameters | Total (n = 201) | AMPD2 expression | Chi-square value | P value | |
| High | Low | ||||
| Gender | 0.664 | 0.415 | |||
| Male | 114 | 32 | 82 | ||
| Female | 87 | 20 | 67 | ||
| Age (yr) | 0.004 | 0.953 | |||
| < 60 | 78 | 20 | 58 | ||
| ≥ 60 | 123 | 32 | 91 | ||
| Location | 3.045 | 0.081 | |||
| Colon | 80 | 26 | 54 | ||
| Rectum | 121 | 26 | 95 | ||
| Vascular invasion | 0.502 | 0.479 | |||
| Negative | 165 | 41 | 124 | ||
| Positive | 36 | 11 | 25 | ||
| Neural invasion | 0.780 | 0.337 | |||
| Negative | 170 | 42 | 128 | ||
| Positive | 31 | 10 | 21 | ||
| Depth of tumor | 5.270 | 0.022a | |||
| T1/T2 | 60 | 9 | 51 | ||
| T3/T4 | 141 | 43 | 98 | ||
| Lymph metastasis | 0.048 | 0.826 | |||
| N0 | 94 | 25 | 69 | ||
| N1/N2 | 107 | 27 | 80 | ||
| Differentiation | 3.887 | 0.037a | |||
| Well/mod | 184 | 44 | 140 | ||
| Poor | 17 | 8 | 9 | ||
| TNM stage | 0.487 | 0.485 | |||
| I-II | 96 | 27 | 69 | ||
| III-IV | 105 | 25 | 80 | ||
| Notch3 | 0.389 | 0.553 | |||
| High | 140 | 38 | 102 | ||
| Low | 61 | 14 | 47 | ||
| Clinicopathological Parameters | Univariate | Multivariate | ||
| HR (95%CI) | P value | HR (95%CI) | P value | |
| Gender (female/male) | 1.004 (0.636-1.585) | 0.986 | 0.973 (0.610-1.552) | 0.909 |
| Age (≥ 60/< 60) | 1.060 (0.664-1.693) | 0.806 | 1.066 (0.661-1.721) | 0.793 |
| Location (rectum/colon) | 0.889 (0.561-1.409) | 0.616 | 0.888 (0.553-1.428) | 0.625 |
| Differentiation (poor, moderate/well) | 0.496 (0.249-0.988) | 0.041 | 0.727 (0.450-1.173) | 0.191 |
| TNM stage (III-IV/I-II) | 3.198 (1.915-5.343) | < 0.001a | 3.727 (2.206-6.296) | <0.001a |
| AMPD2 (high/low) | 1.769 (1.094-2.862) | 0.020a | 2.133 (1.295-3.513) | 0.003a |
| Notch3 (high/low) | 1.237 (0.747-2.048) | 0.408 | 1.119 (0.668-1.874) | 0.669 |
Metabolic intermediate and related metabolites are widely present in cells of myometrial and glandular tissues[16]. Their associated metabolic pathways have been reported to be central to cancer growth and metastasis[17]. Thus, upregulation of AMPD2 in the tumor may explain the metabolic shift in adenosine phosphate synthesis in cancer. This evidence from different studies robustly supports that AMPD2 might be the fundamental gene in tumor transformation, resulting in metabolic adaptation in cancer cells[18]. Based on these findings, we suggested that AMPD2 may be a tumor metabolic marker that can be used for the analysis of tumor progression and the prediction of patient prognosis.
In this study, we summarized and analyzed the mRNA expression data of TCGA-COAD, and obtained the expression characteristics of AMPD2 in cancer tissues and adjacent tissues in patients with CRC. A gene function prediction analysis of the differences between high and low AMPD2 expression groups was performed, and the results were enriched in the Notch signaling pathway. Based on these findings, cytological mechanism validation and clinical prognosis-related survival analysis were employed. Our data showed that AMPD2 resulted in CRC progression and poor prognosis in colorectal and CRC tissues. However, our analysis based on gene function enrichment was based on TCGA public database analysis. We constructed cell samples that were not subjected to high-throughput sequencing of RNA-seq. The results were closer to clinical reality; however, when verifying the data, the probability of false negatives was increased. In addition, our cytology validation experiments needed to be carried out in detail, and our CRC cohort sample only included 201 cases. We expect to add cytological evidence and increase the sample size of prospective prognostic cohorts in subsequent studies.
In the present work, we revealed for the first time that overexpression of AMPD2 can promote CRC progression by augmenting tumor proliferation and halting apoptosis, and may serve as a poor prognostic marker for CRC patients. In addition, our data provided evidence that the Notch signaling pathway can affect proline metabolism via AMPD2 in CRC. Taken together, these findings, and our previous report, highlight the potential therapeutic value of targeting AMPD2 as a modulator of proline metabolism in CRC.
As an important gastrointestinal tumor, colorectal cancer (CRC) is often accompanied by abnormal energy metabolism in the case of malignant progression and poor prognosis.
As the abnormal energy metabolism in tumor cells has received considerable attention, we have combined the tissue type and biological characteristics of CRC. With AMPD2 as the entry point for research, a variety of scientific methods can be integrated for research.
Clarify the role of AMPD2 in CRC and study the pathway and prognostic value of its role.
RNAseq data analysis of the TCGA-COAD data set, GO and KEGG analysis. Western blot was used to detect the expression of Notch3-related pathway proteins, as well as clinical pathological samples combined with patient prognosis analysis.
AMPD2 is commonly overexpressed in TCGA CRC tissues, and the function of AMPD2 is associated with the Notch signaling pathway in CRC in vitro. In the CRC cohort, as indicated by tissue microarray analysis, high expression of AMPD2 protein was correlated with advanced depth of tumor and poor differentiation.
The mRNA and protein levels of AMPD2 are significantly highly expressed in CRC tissues. The high expression of AMPD2 has a positive relationship with activation of the Notch3 signaling pathway. In patients with CRC, those with high expression of AMPD2 have a poor prognosis.
In the future, we plan to further study the biological mechanism of AMPD2 stimulation of abnormal lipid metabolism in CRC cells.
The authors thank Yin ZG and Wang ZW of the Department of Pathology, Affiliated Hospital of Jiangnan University for their help in this study.
Manuscript source: Unsolicited manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Mattioli AV, Kopljar M, Sipos F S-Editor: Yan JP L-Editor: Webster JR E-Editor: Xing YX
| 1. | Robertson DJ, Ladabaum U. Opportunities and Challenges in Moving From Current Guidelines to Personalized Colorectal Cancer Screening. Gastroenterology. 2019;156:904-917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 2. | Aymeric L, Donnadieu F, Mulet C, du Merle L, Nigro G, Saffarian A, Bérard M, Poyart C, Robine S, Regnault B, Trieu-Cuot P, Sansonetti PJ, Dramsi S. Colorectal cancer specific conditions promote Streptococcus gallolyticus gut colonization. Proc Natl Acad Sci USA. 2018;115:E283-E291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 97] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 3. | May-Wilson S, Sud A, Law PJ, Palin K, Tuupanen S, Gylfe A, Hänninen UA, Cajuso T, Tanskanen T, Kondelin J, Kaasinen E, Sarin AP, Eriksson JG, Rissanen H, Knekt P, Pukkala E, Jousilahti P, Salomaa V, Ripatti S, Palotie A, Renkonen-Sinisalo L, Lepistö A, Böhm J, Mecklin JP, Al-Tassan NA, Palles C, Farrington SM, Timofeeva MN, Meyer BF, Wakil SM, Campbell H, Smith CG, Idziaszczyk S, Maughan TS, Fisher D, Kerr R, Kerr D, Passarelli MN, Figueiredo JC, Buchanan DD, Win AK, Hopper JL, Jenkins MA, Lindor NM, Newcomb PA, Gallinger S, Conti D, Schumacher F, Casey G, Aaltonen LA, Cheadle JP, Tomlinson IP, Dunlop MG, Houlston RS. Pro-inflammatory fatty acid profile and colorectal cancer risk: A Mendelian randomisation analysis. Eur J Cancer. 2017;84:228-238. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 85] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 4. | Allen J, Sears CL. Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: contributions to colorectal cancer development. Genome Med. 2019;11:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 132] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 5. | Lehr S, Hartwig S, Sell H. Adipokines: a treasure trove for the discovery of biomarkers for metabolic disorders. Proteomics Clin Appl. 2012;6:91-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 242] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 6. | Severino M, Zara F, Rossi A, Striano P. Teaching Neuro Images: Figure of 8: The clue to the diagnosis of AMPD2 pontocerebellar hypoplasia (PCH9). Neurology. 2017;89:e172-e173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 7. | Bhattacharya I, Drägert K, Albert V, Contassot E, Damjanovic M, Hagiwara A, Zimmerli L, Humar R, Hall MN, Battegay EJ, Haas E. Rictor in perivascular adipose tissue controls vascular function by regulating inflammatory molecule expression. Arterioscler Thromb Vasc Biol. 2013;33:2105-2111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 8. | Helmering J, Juan T, Li CM, Chhoa M, Baron W, Gyuris T, Richards WG, Turk JR, Lawrence J, Cosgrove PA, Busby J, Kim KW, Kaufman SA, Cummings C, Carlson G, Véniant MM, Lloyd DJ. A mutation in Ampd2 is associated with nephrotic syndrome and hypercholesterolemia in mice. Lipids Health Dis. 2014;13:167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 9. | Baron B, Fernandez MA, Carignon S, Toledo F, Buttin G, Debatisse M. GNAI3, GNAT2, AMPD2, GSTM are clustered in 120 kb of Chinese hamster chromosome 1q. Mamm Genome. 1996;7:429-432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 10. | Wang F, Liu Y, Jiang J, Qin Y, Ge X, Li J, Qi X, Mao Y. High expression of AMPD2 and obesity are associated with poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2018;11:216-223. [PubMed] |
| 11. | Khalifa AM, Watson-Siriboe A, Shukry SG, Chiu WL, Nelson ME, Geng Y, Fischer-Stenger K, Porter JH, Stewart JK. Thr136Ile polymorphism of human vesicular monoamine transporter-1 (SLC18A1 gene) influences its transport activity in vitro. Neuro Endocrinol Lett. 2012;33:546-551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 12. | Fukuoka S, Shitara K, Noguchi M, Kawazoe A, Kuboki Y, Bando H, Okamoto W, Kojima T, Doi T, Ohtsu A, Yoshino T. Prophylactic Use of Oral Dexamethasone to Alleviate Fatigue During Regorafenib Treatment for Patients With Metastatic Colorectal Cancer. Clin Colorectal Cancer. 2017;16:e39-e44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 13. | Toiyama Y, Ajay Goel. The Diagnostic, Prognostic, and Predictive Potential of MicroRNA Biomarkers in Colorectal Cancer. Curr Colorect Cancer Rep. 2012;8:22-31. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 14. | Bayrer JR, Mukkamala S, Sablin EP, Webb P, Fletterick RJ. Silencing LRH-1 in colon cancer cell lines impairs proliferation and alters gene expression programs. Proc Natl Acad Sci USA. 2015;112:2467-2472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 48] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 15. | Wang F, Wu J, Qiu Z, Ge X, Liu X, Zhang C, Xu W, Wang F, Hua D, Qi X, Mao Y. ACOT1 expression is associated with poor prognosis in gastric adenocarcinoma. Hum Pathol. 2018;77:35-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 16. | Hirschowitz L, Mayall FG, Ganesan R, McCluggage WG. Intravascular adenomyomatosis: expanding the morphologic spectrum of intravascular leiomyomatosis. Am J Surg Pathol. 2013;37:1395-1400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 17. | Szydłowska M, Roszkowska A. Expression patterns of AMP-deaminase isozymes in human hepatocellular carcinoma (HCC). Mol Cell Biochem. 2008;318:1-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 18. | Toledo F, Buttin G, Debatisse M. The origin of chromosome rearrangements at early stages of AMPD2 gene amplification in Chinese hamster cells. Curr Biol. 1993;3:255-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 62] [Article Influence: 3.1] [Reference Citation Analysis (0)] |