Published online May 26, 2020. doi: 10.12998/wjcc.v8.i10.1923
Peer-review started: February 24, 2020
First decision: March 24, 2020
Revised: April 2, 2020
Accepted: April 15, 2020
Article in press: April 15, 2020
Published online: May 26, 2020
Processing time: 91 Days and 5.2 Hours
Sensitive, novel, and accurate biomarkers for the detection of physiological changes in type 2 diabetes (T2DM) at an early stage are urgently needed.
To build a multi-parameter diagnostic model for the early detection of T2DM.
MiR-148b, miR-223, miR-130a, and miR-19a levels were detected by real-time polymerase chain reaction in serum of healthy controls, individuals with impaired glucose regulation, and T2DM patients. The diagnostic value of miR-148b, miR-223, miR-130a, and miR-19a, alone or in combination, was analyzed.
The area under the curve (AUC) of miR-223, which had the best diagnostic value for discriminating the impaired glucose regulation and T2DM groups, was 0.84, and the sensitivity and specificity were 73.37% and 81.37%, respectively. The AUC of the four-miRNA signature was 0.90, and the sensitivity and specificity were 78.82% and 88.23%, respectively. In the validation set, the AUC was 0.88, and the sensitivity and specificity were 78.36% and 87.63%, respectively.
In summary, we have built a multi-parameter diagnostic model consisting of miR-148b, miR-223, miR-130a, and miR-19a for the detection of T2DM. It may be a potential tool for the early detection of T2DM.
Core tip: The expression of microRNAs in serum is stable and can be reproducibly detected, and they have the potential to be biomarkers for type 2 diabetes. We built a multi-parameter diagnostic model containing miR-148b, miR-223, miR-130a, and miR-19a. In the validation set, the area under curve of this model for the detection of type 2 diabetes was 0.90, and the sensitivity and specificity were 78.36% and 87.63%, respectively. This diagnostic model may be a novel tool for the detection of type 2 diabetes.
- Citation: Yan LN, Zhang X, Xu F, Fan YY, Ge B, Guo H, Li ZL. Four-microRNA signature for detection of type 2 diabetes. World J Clin Cases 2020; 8(10): 1923-1931
- URL: https://www.wjgnet.com/2307-8960/full/v8/i10/1923.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v8.i10.1923
Type 2 diabetes (T2DM) is a common chronic disease worldwide[1]. It is related to insulin resistance and often leads to serious adverse effects on organs, such as cardiovascular disease, nephropathy, retinopathy, and nerve damage[2]. According to the estimation of the International Diabetes Association, there will be more than 500 million people with diabetes in 2030. In China, which has undergone improvements in quality of life and eating habits, the incidence of T2DM in adults over 18 years of age is 10.4%, and that in individuals over 60 years of age is above 20%. Among diabetes cases in China, T2DM accounts for more than 90.0%[3,4]. Although clinical treatments for T2DM, such as the routine use of glucose-lowering drugs, are mature, the etiology and pathogenesis of T2DM remain unclear. In clinical practice, sensitive, novel, and accurate biomarkers for the detection of physiological changes in T2DM at an early stage are urgently needed.
Currently, the most common diagnostic criteria and classification criteria for T2DM in China are fasting plasma glucose level and 2-h glucose level after a 75 g glucose load, which were recommended in guidelines from the World Health Organization in 1999[5]. The clinical diagnosis of diabetes should be made based on venous plasma glucose levels and not the results of a capillary blood glucose test. After meeting the diagnostic criteria for diabetes, a diagnosis based on etiological evidence is performed. In addition, triglyceride level, total cholesterol level, apolipoprotein level, creatinine level, lack of physical activity, diet, smoking, high waist-to-hip ratio, high blood pressure, and genetic risk factors have been demonstrated to be related to the development of T2DM[6]; however, the biomarkers among these factors are usually detected during metabolic imbalance and cannot be used to assess T2DM susceptibility. With the rapid development of molecular detection methods, single nucleotide polymorphisms, DNA methylation, microRNAs (miRNAs), mRNAs, proteomes, metabolomes, and exosomes have been demonstrated to be potential biomarkers for various cancers[7-9]. MiRNAs are found not only in the cell but also outside the cell, including body fluids, such as serum, saliva, and nasal secretions. Extracellular miRNAs were demonstrated to be stable in serum or plasma due to their presence as protein complexes. Abnormal expression of miRNAs is related to the occurrence and development of many diseases, such as cancer and T2DM[10-15]. MiRNAs in the serum or plasma are stable, can be reproducibly detected, and therefore have the potential as novel biomarkers for a variety of diseases.
Many previous studies have demonstrated that miRNAs are related to the occurrence and development of T2DM. MiR-375 and miR-9 were found to be increased in the peripheral blood of patients with pre-diabetes and T2DM and positively related to blood glucose levels[16]. MiR-192 and miR-194 were also shown to be related to the development of T2DM[17]. By Solexa sequencing, miR-126-3p, miR-28-3p, and miR-486-5p were found to be present only in pre-diabetes[18]. In addition, miR-184, miR-182-5p, and miR-127-3p are involved in insulin biosynthesis and secretion, and miR-125, miR-29, miR-103/107, and miR-15a are involved in insulin resistance. By meta-analysis of reports from the PubMed, ENBASE, and ISI Web of Science databases and three Chinese databases, miR-148b, miR-223, miR-130a, miR-19a, miR-26b, and miR27b were found to be potential biomarkers for T2DM[19].
In the present study, we detected miR-148b, miR-223, miR-130a, and miR-19a levels in the sera of healthy controls, individuals with impaired glucose regulation, and T2DM patients by real-time polymerase chain reaction (PCR). By detecting and analyzing the levels of individual and multi-parameter biomarkers, we aimed to build a multi-parameter diagnostic model for the early detection of T2DM.
All the individuals assessed in our study signed written informed consent forms, and this study was approved by the Ethics Committee of the Third Affiliated Hospital of Inner Mongolia Medical University (Inner Mongolia Baogang Hospital). The diagnosis of T2DM was based on blood glucose levels and clinical symptoms. Criteria for the diagnosis of T2DM were announced by the World Health Organization and International Diabetes Federation in 1999 and approved by the Diabetes Association of the Chinese Medical Association. With typical symptoms, the criterion for a diagnosis of T2DM is a fasting blood glucose level of 7.0 mmol/L or a postprandial blood glucose level ≥ 11.1 mmol/L. Without typical symptoms, when a fasting blood glucose level of 7.0 mmol/L or a postprandial blood glucose level of 11.1 mmol/L is measured, the measurement should be repeated, and those who still exhibit the above values can be diagnosed with T2DM. Only those exhibiting a fasting blood glucose level of 7.0 mmol/L or a postprandial blood glucose level of 11.1 mmol/L and a plasma glucose level of 11.1 mmol/L 2 h after the glucose tolerance test were diagnosed with T2DM. A diagnosis of impaired glucose regulation was based on the following criteria. If the plasma glucose level was between 7.8 and 11.1 mmol/L for 2 h in the glucose tolerance test, glucose tolerance was impaired; if the plasma glucose level was 6.1-7.0 mmol/L, fasting glucose was impaired. Healthy controls were enrolled from a physical examination center with the following criteria: Normal examination, not pregnant, not lactating, no history of chronic disease, no history of long-term medication, normal hematuria, normal blood glucose, normal liver function, normal kidney function, normal electrocardiogram, and normal chest radiograph examination. Sixty-eight healthy controls, 91 individuals with impaired glucose regulation, and 102 T2DM patients were used for diagnostic model validation in our study. Eighty-seven individuals with impaired glucose regulation and 113 T2DM patients were used for diagnostic model training in our study. Age and sex were matched between the three groups.
Fasting blood was collected, and the serum was aliquoted after centrifugation and stored in a refrigerator at -80 °C. Total RNA was isolated from 200 μL of serum using the mirVana PARIS kit (AM1556, Ambion) according to the instructions. The A260/A280 value was calculated to assess the quality of the total RNA. DNase I was used to remove DNA. A TaqMan microRNA RT kit (4366596, Ambion) was used for reverse transcription. The reverse transcription reaction contained 2.08 μL of DEPC-treated water, 0.75 μL of 10 × reverse transcription buffer, 0.095 μL of reverse transcription inhibitor, 0.075 μL of dNTPs, 0.5 μL of reverse transcriptase, 1.5 μL of 20 × miRNA-specific primer, and 2.5 μL of total RNA. The reverse transcription procedure was as follows: 16 °C for 30 min, 42 °C for 30 min, and 85 °C for 5 min. Real-time PCR reactions to detect miR-148b (assay ID: 000471, Thermo Fisher), miR-223 (assay ID: 002098, Thermo Fisher), miR-130a (assay ID: 000454, Thermo Fisher), and miR-19a (assay ID: 000395, Thermo Fisher) included 4.5 μL of water, the PCR product, 5.0 μL of TaqMan Gene Expression Master Mix, and 0.5 μL of 5 × TaqMan microRNA assay buffer. The ABI PRISM 7300 system was used to detect the miRNAs, and the amplification program was 95 °C for 5 min and then 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The internal reference miRNA was U6, and the 2-ΔΔCt method was used to calculate the relative expression of miRNAs.
SPSS 22.0 was used for statistical analyses. The normality test was performed first to assess the data distribution. If the data were normally distributed, the t-test was used. If the data was non-normally distributed, pairwise comparison was performed using the nonparametric Mann-Whitney method. Multi-group comparison was performed using the Kruskal-Wallis method. The diagnostic value of serum miRNAs was evaluated using the receiver operator characteristic curve and the area under curve (AUC). Multi-parameter evaluation of serum miRNAs was carried out by binary logistic regression. AUC comparison of multi-parameter and individual biomarker evaluation was performed using the Z-score test. P < 0.05 indicated statistical significance.
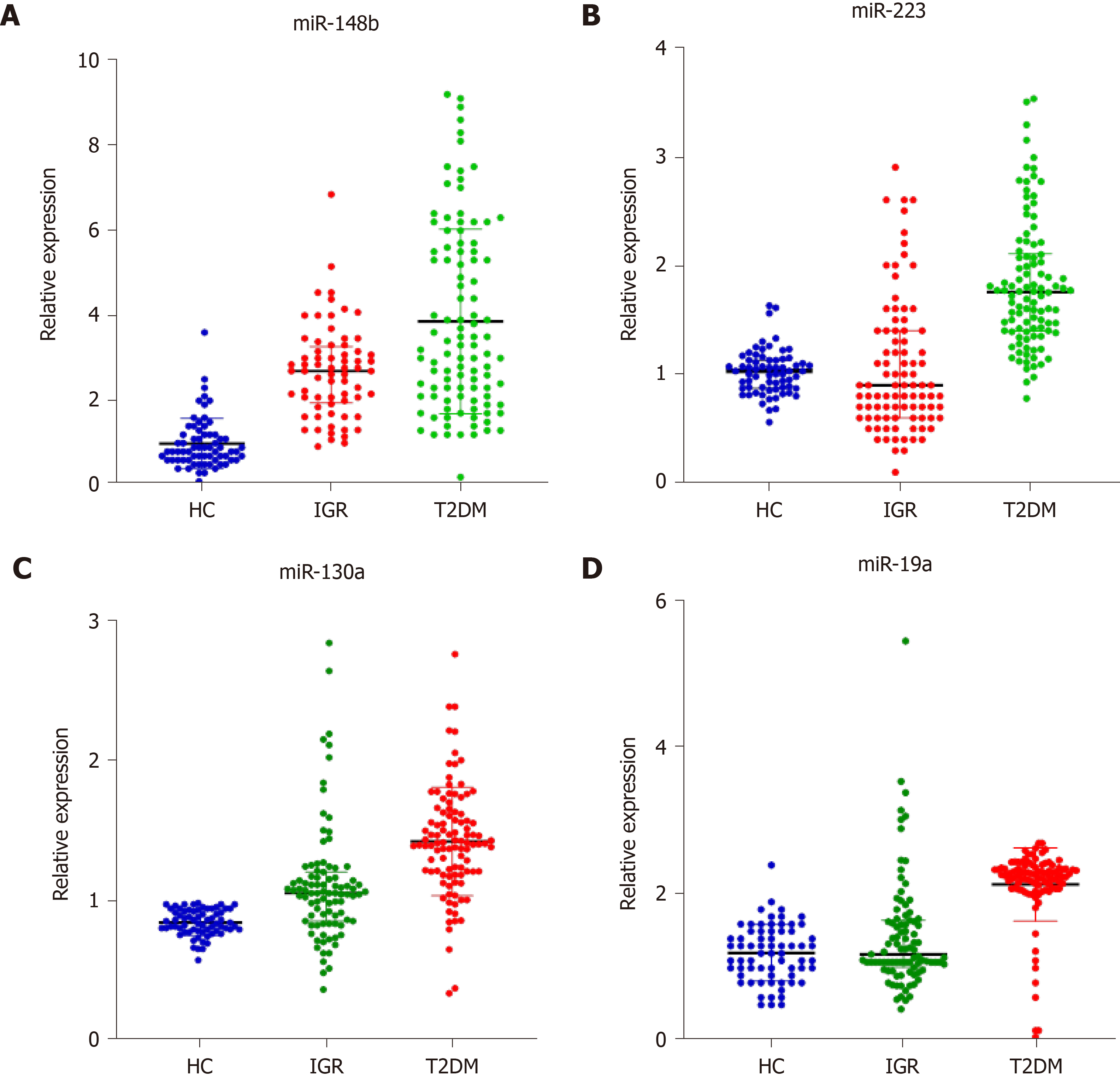
The median (25%, 75%) relative expression levels of miR-148b, miR-223, miR-130a, and miR-19a in the healthy control group, impaired glucose regulation group, and T2DM group are shown. As shown in Figure 1, the relative expression of miR-148b, miR-223, miR-130a, and miR-19a in the three groups was 0.82 (0.62, 1.22), 2.69 (1.94, 3.27), and 3.25 (2.10, 5.50); 1.03 (0.87, 1.13), 0.91 (0.60, 1.41), and 1.76 (1.40, 2.11); 0.84 (0.78, 0.93), 1.05 (0.85, 1.20), and 1.41 (1.20, 1.62); and 1.20 (0.90, 1.50), 1.19 (1.01, 1.65), and 2.27 (2.08, 2.36), respectively. Compared with those in the healthy control group, miR-148b and miR-130a levels in the impaired glucose regulation group were significantly different (P < 0.05). MiR-148b, miR-223, miR-130a, and miR-19a were significantly different in the T2DM group compared with the other groups (P < 0.05). MiR-148b, miR-223, miR-130a, and miR-19a expression was significantly different in the T2DM group compared with the impaired glucose regulation group (P < 0.05).
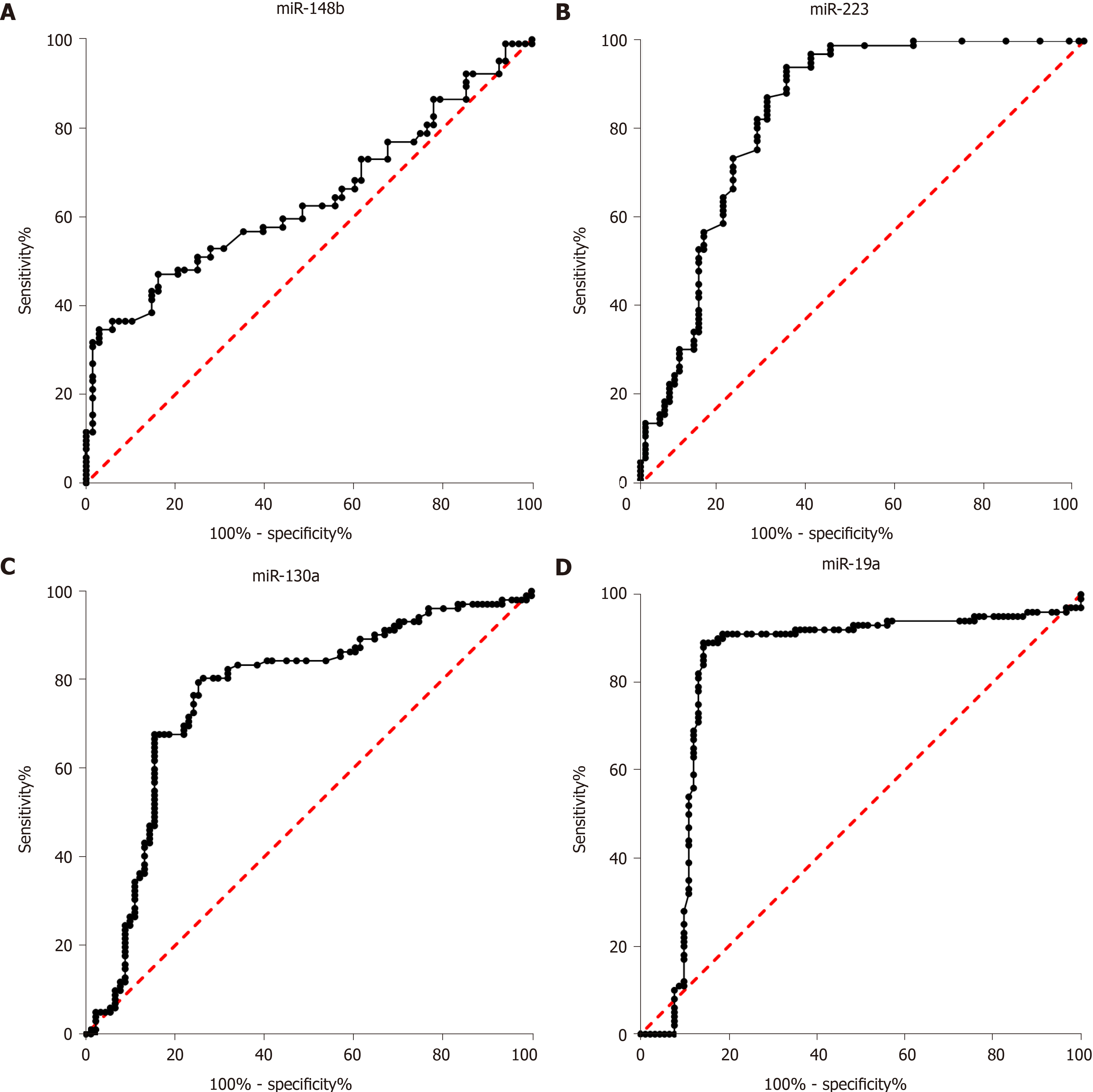
In clinical practice, biomarkers for discriminating between impaired glucose regulation and T2DM are important. In our study, the diagnostic value of miR-148b, miR-223, miR-130a, and miR-19a for discriminating between impaired glucose regulation and T2DM was evaluated. As shown in Figure 2, the AUCs of miR-148b, miR-223, miR-148b, and miR-148b, each alone, for the discrimination between impaired glucose regulation and T2DM were 0.64 (0.56, 0.722), 0.84 (0.77, 0.90), (0.69, 0.84), and 0.83 (0.76, 0.90), respectively (P < 0.05 for all). The four biomarkers showed potential as biomarkers in clinical practice. Among the four miRNA biomarkers, miR-223 had the best diagnostic value, and when the cutoff was 1.32, its sensitivity and specificity were 73.37% and 81.37%, respectively.
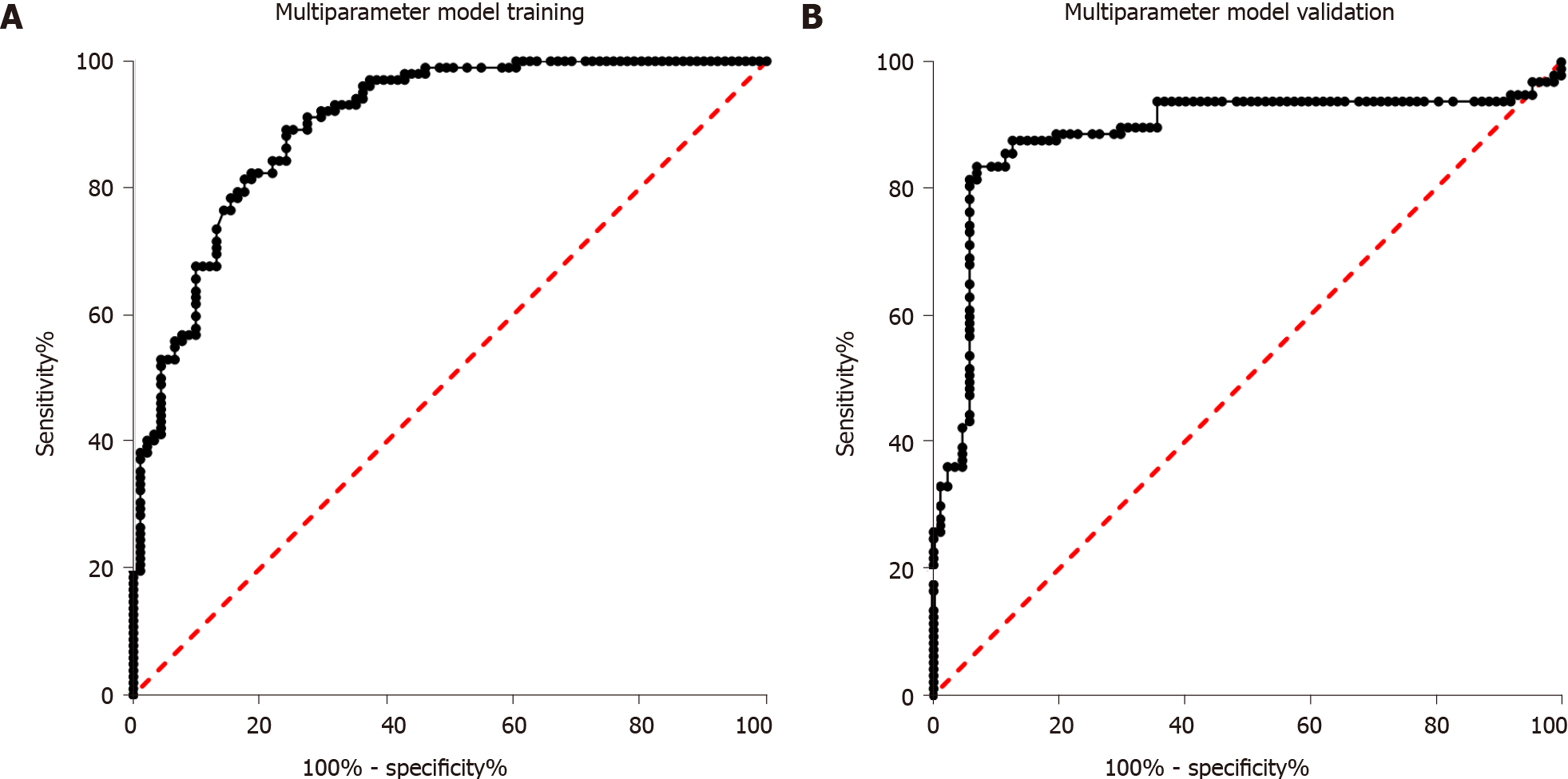
Sixty-eight healthy controls, 91 individuals with impaired glucose regulation, and 102 T2DM patients were used for diagnostic model training. First, the four miRNAs were analyzed alone. Then, binary logistic regression was used for multi-parameter analysis of the four miRNAs. As shown in Figure 3A, the AUC for multi-parameter analysis was 0.90 (0.86, 0.94). Compared with the AUC for the best diagnostic biomarker, miR-223, alone, the AUC for multi-parameter analysis was significantly different (P < 0.05), and the sensitivity and specificity were 78.82% and 88.23%, respectively. After multi-parameter diagnostic model building with miR-148b, miR-223, miR-130a, and miR-19a, 87 individuals with impaired glucose regulation and 113 T2DM patients were independently used for diagnostic model validation. As shown in Figure 3B, the AUC was 0.88 (0.83, 0.94), and the sensitivity and specificity were 78.36% and 87.63%, respectively.
T2DM is a metabolic syndrome characterized mainly by hyperglycemia. Decreased islet β cell function and insulin resistance are the main pathophysiological features in the pathogenesis of T2DM. Studies have demonstrated that miRNA disorders are involved in β cell development, insulin sensitivity, insulin resistance, insulin production, insulin secretion, and insulin signaling pathways and finally lead to the development of T2DM[20]. MiR-338-3p was found to be significantly decreased in T2DM and related to impaired insulin resistance. MiR-15a has been shown to be involved in the pathogenesis of T2DM. Plasma miR-15a is increased in T2DM patients, and miR-15a plays an important role in insulin production. Both miR-1249 and miR-486-5p were significantly decreased in the plasma of patients with T2DM, and these two miRNAs may be diagnostic markers and therapeutic targets for T2DM. MiR-126 was significantly decreased in the sera of T2DM patients and is also considered a potential diagnostic marker for T2DM[21]. We aimed to evaluate these four miRNAs that showed potential diagnostic value for T2DM in the serum to build a multi-parameter diagnostic model.
MiR-148b was reported to be a promising biomarker for T2DM. One target of miR-148b is DNMT1, a very important enzyme for DNA methylation. DNA methylation is involved in regulating β-cell formation. The mechanism by which miR-148b regulates T2DM may be through its regulation of DNA methylation[22]. MiR-148b was also reported to be a promising biomarker for breast cancer[23]. In our study, miR-148b was significantly increased in the T2DM group. This finding is inconsistent with the results of previous studies, possibly because the internal control was inconsistent and the enrolled patients differed. A previous study reported that miR-223 was increased in the islets of both diabetic mice and humans and that miR-223 deficiency significantly suppressed β-cell proliferation and insulin secretion. By inhibiting forkhead box O1 and SRY-box 6 signaling, its overexpression could promote β-cell proliferation and function[24]. MiR-223 was recognized as an important factor for maintaining β-cell mass and metabolic stress. It was also reported to be related to obesity and cardiovascular disease, which are related to T2DM[25,26]. In addition, miR-223 was reported to be associated with positive responses to T2DM treatment. In our study, we also demonstrated that miR-223 has potential diagnostic value for T2DM[27]. MiR-130a was significantly decreased in endothelial progenitor cells from patients with T2DM. Furthermore, miR-130a targets Runx3, which negatively regulates endothelial progenitor cell function. In addition, it also positively regulates the ERK/vascular endothelial growth factor and Akt1 pathways[28] and negatively regulates the c-Jun N-terminal kinases pathway by targeting MAP3K12[29]. MiR-130a plays an important role in endothelial progenitor cell function and is related to T2DM[30]. In addition, it is involved in the proliferation of vascular smooth muscle cells in hypertension, hepatic insulin sensitivity, and liver steatosis[31,32]. In our study, miR-130a also showed potential diagnostic value for T2DM. By targeting tissue factor transcripts, miR-19a reduces tissue factor expression in both endothelial and monocytic cells, and tissue factor was demonstrated to be related to antiphospholipid syndrome and systemic lupus erythematosus[33]. MiR-19a may be involved in tissue factor-mediated thrombogenicity in T2DM. In addition, miR-19a is involved in vascular smooth muscle cell function, insulin resistance, and rapamycin treatment[34-36]. In our study, miR-19a levels in the T2DM group were significantly different, and these results are consistent with those of previous studies.
Although we have built a miRNA diagnostic model for the detection of T2DM, there were still some limitations to our study. First, we evaluated only four miRNAs based on the results of previous meta-analysis, and the other miRNAs were not detected in our study. Second, although we have trained and validated the multi-parameter diagnostic model, the sample size was so small that the results cannot sufficiently reflect the real diagnostic value. Third, we evaluated only the diagnostic value, and the specific mechanism by which the miRNAs are involved in T2DM was not explored.
In summary, we have successfully built a multi-parameter diagnostic model consisting of miR-148b, miR-223, miR-130a, and miR-19a for the detection of T2DM. These miRNAs may be potential biomarkers for the early detection of T2DM.
Sensitive, novel, and accurate biomarkers for the detection of physiological changes in type 2 diabetes (T2DM) at an early stage are urgently needed. MicroRNAs (miRNAs) are found not only in the cell but also outside the cell, including body fluids, such as serum, saliva, and nasal secretions. Extracellular miRNAs were demonstrated to be stable in serum or plasma due to their presence as protein complexes. Abnormal expression of miRNAs is related to the occurrence and development of many diseases, such as cancer and T2DM.
MiRNAs in the serum or plasma are stable, can be reproducibly detected, and therefore have the potential as novel biomarkers for a variety of diseases, include T2DM.
This study aimed to build a multi-parameter diagnostic model for the early detection of T2DM.
MiR-148b, miR-223, miR-130a, and miR-19a levels were detected in serum of healthy controls, individuals with impaired glucose regulation, and T2DM patients. The detection value of these miRNAs, alone and in combination, was analyzed.
The area under the curve of miR-223, which had the best diagnostic value for discriminating the impaired glucose regulation and T2DM groups, was 0.84, and the sensitivity and specificity were 73.37% and 81.37%, respectively. The area under curve of the four-miRNA signature was 0.90, and the sensitivity and specificity were 78.82% and 88.23%, respectively.
In this study, the authors built a multi-parameter diagnostic model containing miR-148b, miR-223, miR-130a, and miR-19a for the detection of T2DM. It may be a potential tool for the early detection of T2DM.
The specific mechanism by which the miRNAs are involved in T2DM can be explored in the future.
Manuscript source: Unsolicited manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Alsina A, Mathur A, Ko E S-Editor: Wang JL L-Editor: Wang TQ E-Editor: Liu MY
| 1. | DeFronzo RA. Pathogenesis of type 2 diabetes mellitus. Med Clin North Am. 2004;88:787-835, ix. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 715] [Cited by in RCA: 743] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 2. | Fletcher B, Gulanick M, Lamendola C. Risk factors for type 2 diabetes mellitus. J Cardiovasc Nurs. 2002;16:17-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 225] [Article Influence: 9.8] [Reference Citation Analysis (1)] |
| 3. | Yang L, Shao J, Bian Y, Wu H, Shi L, Zeng L, Li W, Dong J. Prevalence of type 2 diabetes mellitus among inland residents in China (2000-2014): A meta-analysis. J Diabetes Investig. 2016;7:845-852. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 121] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 4. | Ye Q, Fu JF. Paediatric type 2 diabetes in China-Pandemic, progression, and potential solutions. Pediatr Diabetes. 2018;19:27-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 5. | Grimaldi A, Heurtier A. [Diagnostic criteria for type 2 diabetes]. Rev Prat. 1999;49:16-21. [PubMed] |
| 6. | Tajik S, Mirzababaei A, Ghaedi E, Kord-Varkaneh H, Mirzaei K. Risk of type 2 diabetes in metabolically healthy people in different categories of body mass index: an updated network meta-analysis of prospective cohort studies. J Cardiovasc Thorac Res. 2019;11:254-263. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 7. | Grant AK, Golden L. Technological Advancements in the Management of Type 2 Diabetes. Curr Diab Rep. 2019;19:163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 8. | Sun Q, Qiao J, Zhang S, He S, Shi Y, Yuan Y, Zhang X, Cai Y. Changes in DNA methylation assessed by genomic bisulfite sequencing suggest a role for DNA methylation in cotton fruiting branch development. PeerJ. 2018;6:e4945. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 9. | Yu J, Zhang Y, Liu J, Wang L, Liu P, Yin Z, Guo S, Ma J, Lu Z, Wang T, She Y, Miao Y, Ma L, Chen S, Li Y, Dai S. Proteomic discovery of H2O2 response in roots and functional characterization of PutGLP gene from alkaligrass. Planta. 2018;248:1079-1099. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 10. | Rome S. Are extracellular microRNAs involved in type 2 diabetes and related pathologies? Clin Biochem. 2013;46:937-945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 11. | Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014;15:429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 224] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 12. | Banerjee J, Nema V, Dhas Y, Mishra N. Role of MicroRNAs in Type 2 Diabetes and Associated Vascular Complications. Biochimie. 2017;139:9-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 13. | Sandiford OA, Moore CA, Du J, Boulad M, Gergues M, Eltouky H, Rameshwar P. Human Aging and Cancer: Role of miRNA in Tumor Microenvironment. Adv Exp Med Biol. 2018;1056:137-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 14. | Yaribeygi H, Katsiki N, Behnam B, Iranpanah H, Sahebkar A. MicroRNAs and type 2 diabetes mellitus: Molecular mechanisms and the effect of antidiabetic drug treatment. Metabolism. 2018;87:48-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 57] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 15. | Pordzik J, Jakubik D, Jarosz-Popek J, Wicik Z, Eyileten C, De Rosa S, Indolfi C, Siller-Matula JM, Czajka P, Postula M. Significance of circulating microRNAs in diabetes mellitus type 2 and platelet reactivity: bioinformatic analysis and review. Cardiovasc Diabetol. 2019;18:113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 107] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 16. | Al-Muhtaresh HA, Al-Kafaji G. Evaluation of Two-Diabetes Related microRNAs Suitability as Earlier Blood Biomarkers for Detecting Prediabetes and type 2 Diabetes Mellitus. J Clin Med. 2018;7:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 17. | Jaeger A, Zollinger L, Saely CH, Muendlein A, Evangelakos I, Nasias D, Charizopoulou N, Schofield JD, Othman A, Soran H, Kardassis D, Drexel H, Eckardstein AV. Circulating microRNAs -192 and -194 are associated with the presence and incidence of diabetes mellitus. Sci Rep. 2018;8:14274. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 18. | Matsha TE, Kengne AP, Hector S, Mbu DL, Yako YY, Erasmus RT. MicroRNA profiling and their pathways in South African individuals with prediabetes and newly diagnosed type 2 diabetes mellitus. Oncotarget. 2018;9:30485-30498. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 19. | Liang YZ, Li JJ, Xiao HB, He Y, Zhang L, Yan YX. Identification of stress-related microRNA biomarkers in type 2 diabetes mellitus: A systematic review and meta-analysis. J Diabetes. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 20. | Guay C, Regazzi R. MicroRNAs and the functional β cell mass: For better or worse. Diabetes Metab. 2015;41:369-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 21. | Zhang T, Lv C, Li L, Chen S, Liu S, Wang C, Su B. Plasma miR-126 is a potential biomarker for early prediction of type 2 diabetes mellitus in susceptible individuals. Biomed Res Int. 2013;2013:761617. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 102] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 22. | Azizi M, Teimoori-Toolabi L, Arzanani MK, Azadmanesh K, Fard-Esfahani P, Zeinali S. MicroRNA-148b and microRNA-152 reactivate tumor suppressor genes through suppression of DNA methyltransferase-1 gene in pancreatic cancer cell lines. Cancer Biol Ther. 2014;15:419-427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 23. | Shen J, Hu Q, Schrauder M, Yan L, Wang D, Medico L, Guo Y, Yao S, Zhu Q, Liu B, Qin M, Beckmann MW, Fasching PA, Strick R, Johnson CS, Ambrosone CB, Zhao H, Liu S. Circulating miR-148b and miR-133a as biomarkers for breast cancer detection. Oncotarget. 2014;5:5284-5294. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 24. | Li Y, Deng S, Peng J, Wang X, Essandoh K, Mu X, Peng T, Meng ZX, Fan GC. MicroRNA-223 is essential for maintaining functional β-cell mass during diabetes through inhibiting both FOXO1 and SOX6 pathways. J Biol Chem. 2019;294:10438-10448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 25. | Kilic ID, Dodurga Y, Uludag B, Alihanoglu YI, Yildiz BS, Enli Y, Secme M, Bostancı HE. MicroRNA -143 and -223 in obesity. Gene. 2015;560:140-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 26. | Taïbi F, Metzinger-Le Meuth V, Massy ZA, Metzinger L. miR-223: An inflammatory oncomiR enters the cardiovascular field. Biochim Biophys Acta. 2014;1842:1001-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 136] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 27. | Catanzaro G, Besharat ZM, Chiacchiarini M, Abballe L, Sabato C, Vacca A, Borgiani P, Dotta F, Tesauro M, Po A, Ferretti E. Circulating MicroRNAs in Elderly Type 2 Diabetic Patients. Int J Endocrinol. 2018;2018:6872635. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 28. | Meng S, Cao J, Zhang X, Fan Y, Fang L, Wang C, Lv Z, Fu D, Li Y. Downregulation of microRNA-130a contributes to endothelial progenitor cell dysfunction in diabetic patients via its target Runx3. PLoS One. 2013;8:e68611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 66] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 29. | Ye M, Li D, Yang J, Xie J, Yu F, Ma Y, Zhu X, Zhao J, Lv Z. MicroRNA-130a Targets MAP3K12 to Modulate Diabetic Endothelial Progenitor Cell Function. Cell Physiol Biochem. 2015;36:712-726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 30. | Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111:1217-1226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 364] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 31. | Wu WH, Hu CP, Chen XP, Zhang WF, Li XW, Xiong XM, Li YJ. MicroRNA-130a mediates proliferation of vascular smooth muscle cells in hypertension. Am J Hypertens. 2011;24:1087-1093. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 77] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 32. | Xiao F, Yu J, Liu B, Guo Y, Li K, Deng J, Zhang J, Wang C, Chen S, Du Y, Lu Y, Xiao Y, Zhang Z, Guo F. A novel function of microRNA 130a-3p in hepatic insulin sensitivity and liver steatosis. Diabetes. 2014;63:2631-2642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 65] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 33. | Witkowski M, Tabaraie T, Steffens D, Friebel J, Dörner A, Skurk C, Witkowski M, Stratmann B, Tschoepe D, Landmesser U, Rauch U. MicroRNA-19a contributes to the epigenetic regulation of tissue factor in diabetes. Cardiovasc Diabetol. 2018;17:34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 34. | Sun G, Song H, Wu S. miR19a promotes vascular smooth muscle cell proliferation, migration and invasion through regulation of Ras homolog family member B. Int J Mol Med. 2019;44:1991-2002. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 35. | Chakraborty C, George Priya Doss C, Bandyopadhyay S. miRNAs in insulin resistance and diabetes-associated pancreatic cancer: the 'minute and miracle' molecule moving as a monitor in the 'genomic galaxy'. Curr Drug Targets. 2013;14:1110-1117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 61] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 36. | Belenchia AM, Gavini MP, Toedebusch RG, DeMarco VG, Pulakat L. Comparison of Cardiac miRNA Transcriptomes Induced by Diabetes and Rapamycin Treatment and Identification of a Rapamycin-Associated Cardiac MicroRNA Signature. Oxid Med Cell Longev. 2018;2018:8364608. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |