Published online Mar 6, 2024. doi: 10.12998/wjcc.v12.i7.1227
Peer-review started: November 22, 2023
First decision: December 23, 2023
Revised: February 2, 2024
Accepted: January 29, 2024
Article in press: January 29, 2024
Published online: March 6, 2024
Processing time: 99 Days and 16.6 Hours
Despite being one of the most prevalent sleep disorders, obstructive sleep apnea hypoventilation syndrome (OSAHS) has limited information on its immunologic foundation. The immunological underpinnings of certain major psychiatric di
To investigate the immune cells' association with OSAHS via genetic methods, guiding future clinical research.
A comprehensive two-sample mendelian randomization study was conducted to investigate the causal relationship between immune cell characteristics and OS
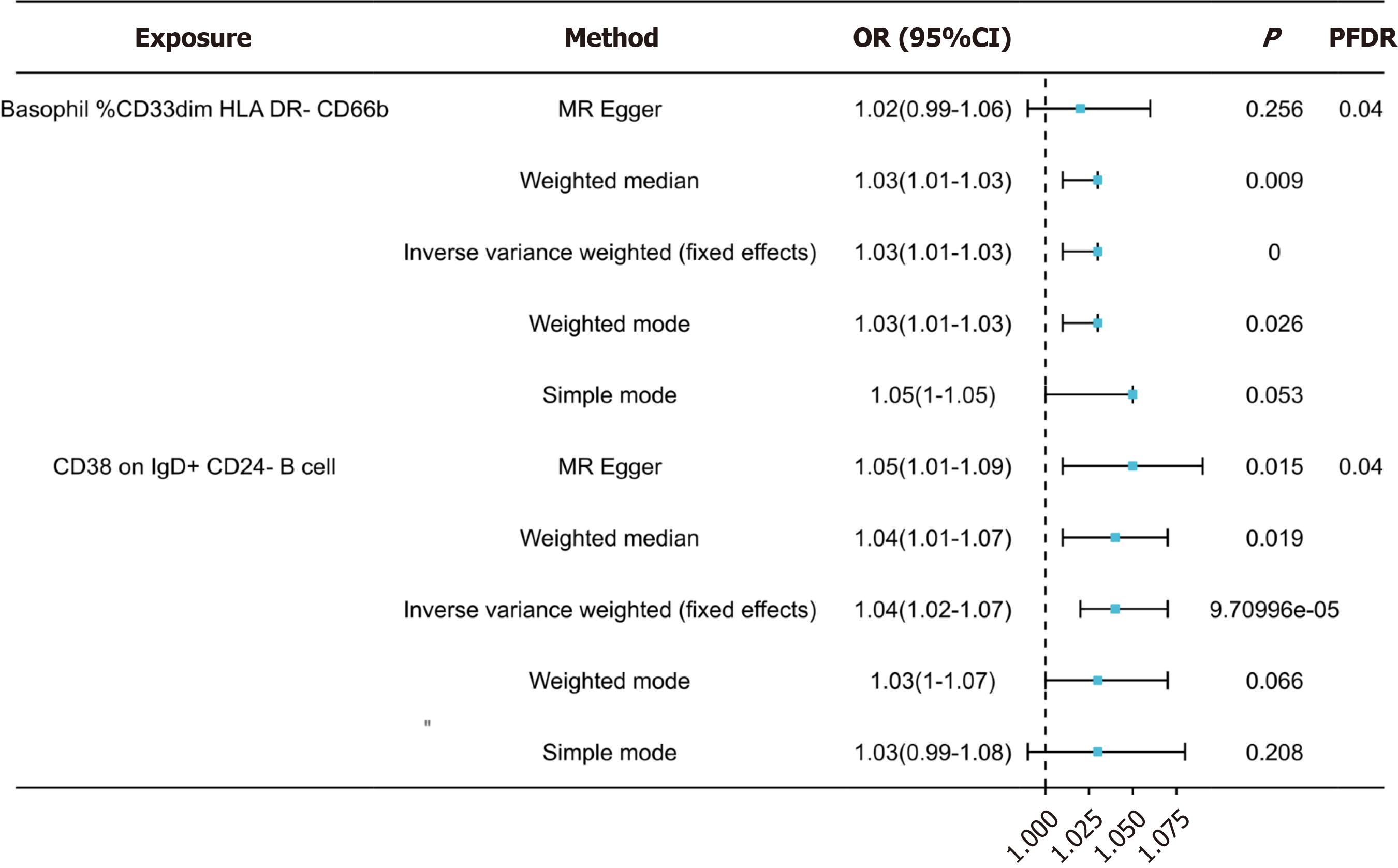
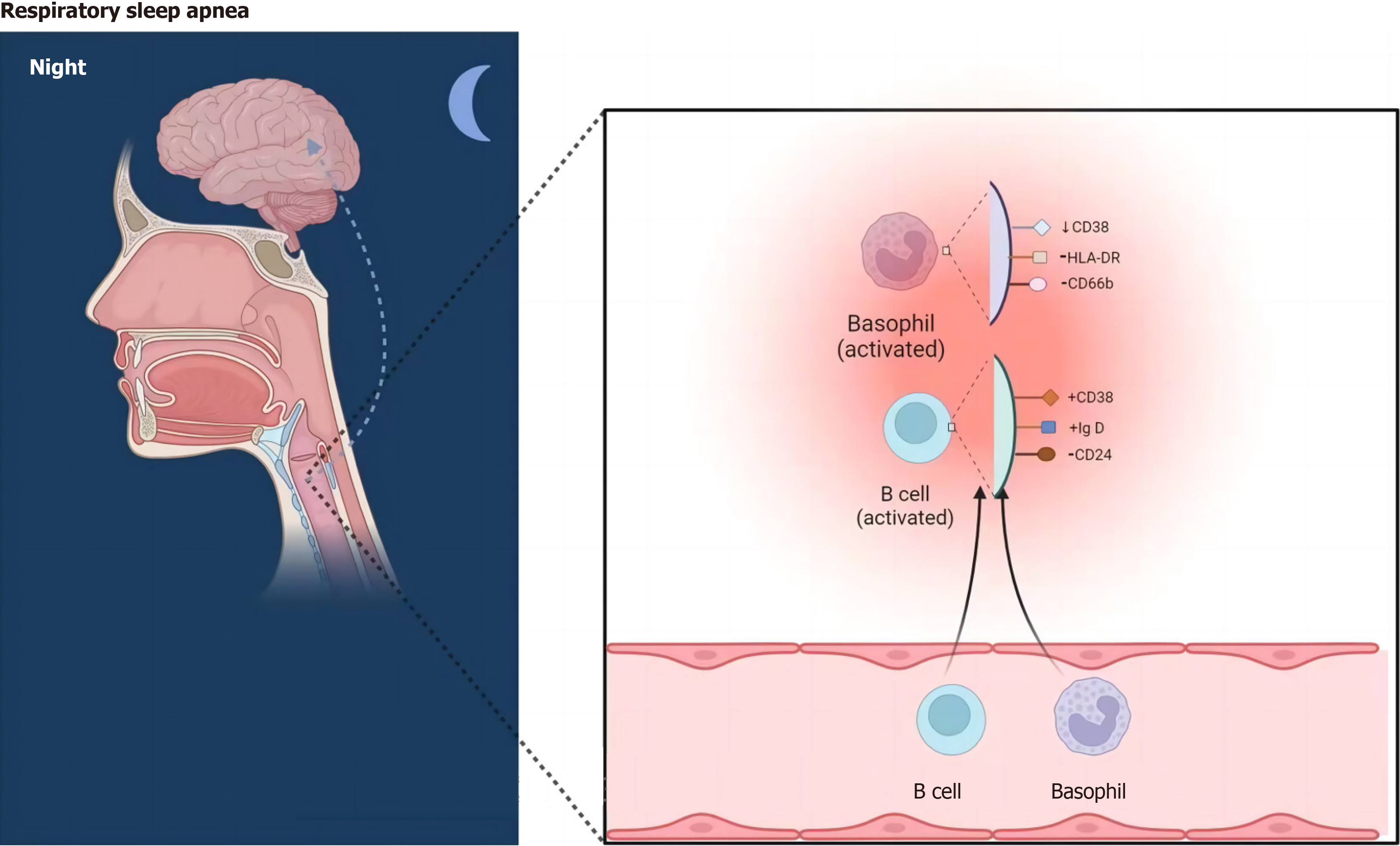
Following false discovery rate (FDR) correction, no statistically significant effect of OSAHS on immunophenotypes was observed. However, two lymphocyte subsets were found to have a significant association with the risk of OSAHS: Basophil %CD33dim HLA DR- CD66b- (OR = 1.03, 95%CI = 1.01-1.03, P < 0.001); CD38 on IgD+ CD24- B cell (OR = 1.04, 95%CI = 1.02-1.04, P = 0.019).
This study shows a strong link between immune cells and OSAHS through a gene approach, thus offering direc
Core Tip: Our comprehensive bidirectional mendelian randomization analysis has revealed causal links between various immunophenotypes and obstructive sleep apnea-hypopnea syndrome (OSAHS), shedding light on the intricate web of relationships between OSAHS and the immune system.
- Citation: Zhao HH, Ma Z, Guan DS. Causal role of immune cells in obstructive sleep apnea hypopnea syndrome: Mendelian randomization study. World J Clin Cases 2024; 12(7): 1227-1234
- URL: https://www.wjgnet.com/2307-8960/full/v12/i7/1227.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i7.1227
Obstructive sleep apnea-hypopnea syndrome (OSAHS) is characterized by an apnea-hypopnea index of 5 or more, ac
Current research is exploring the complicated relationships between OSAHS and the immune system, particularly fo
Mendelian randomization (MR) functions as a statistical method primarily utilized for inferring epidemiological cau
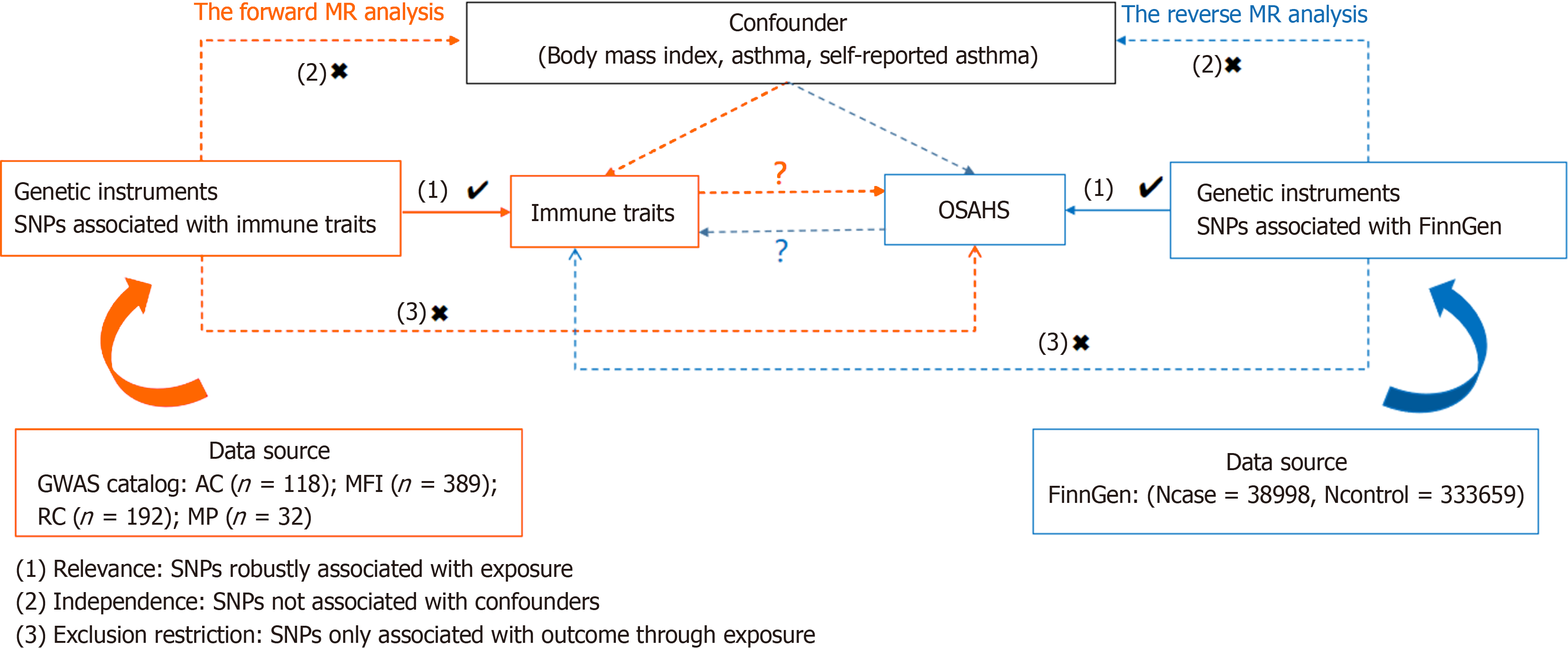
We investigated the causal relationship of the 731 immune cell profiles with OSAHS by two-sample MR analysis. MR requires the use of genetic variation as a proxy variable for risk variation to satisfy three critical hypotheses in causal in
Comprehensive summary statistical data (ranging from accession numbers GCST0001391 to GCST0002121) containing all immunization profiles in the GWAS catalog are readily available[15]. This Genomic Study involved 3757 non-overlapping Europeans analyzed using high-density arrays based on the Sardinian Sequence Reference Panel[16]. The dataset com
FinnGen provided the genome-wide association study (GWAS) summary statistics for OSAHS (https://www.finngen.fi/en). 372657 European people were included in the research (Ncase = 38998, Ncontrol = 333659) for a GWAS. The GWAS identified over 16 million independent single nucleotide polymorphisms (SNPs).
An instrumental variable (IV) extracted from version v1.90 was used to modify SNPs using a distance of 500 kb with a chain disequilibrium (LD) r2 threshold of less than 0.1[17]. Calculation of LD r2 used the 1000 Genomes Project as a re
All studies were analyzed using R version 4.3.1 software (http://www.Rproject.org). In particular, to explore the causal links between the 731 immunophenotypes and OSAHS, a set of analyses were performed using the "Mendelian Randomization" software (version 0.4.3)[18], including median-based weighted analysis[19], pattern-based weighted analysis[20], and inverse variance weighted (IVW) analysis[21]. Instrumental heterogeneity across variables was examined based on Cochran's Q statistic and P value (IV), supported with MR-Egger test, which identifies cross-sectional multidimensiona
Using false discovery rate (FDR) correlation (PFDR < 0.05), we identified two protective immunophenotypes against OSAHS: Basophil %CD33dim HLA DR- CD66b- and CD38 on IgD+ CD24- B cell. In particular, the ratio of basophil %CD33dim HLA DR- CD66b- to the risk of OSAHS was 1.03 (95%CI = 1.01-1.03, PFDR = 0.04, P = 0.256, Supple
In exploring the causal effect of OSAHS on immunophenotypes, we used the IVW approach as the primary analytical method for the two-sample MR analysis. Although adjusted for multiplicity of tests using the FDR method, we did not identify any immunologic features at the 0.05 significance level. However, when loosely thresholding the FDR, we iden
We explored the causal link between 731 immune cell characteristics and OSAHS by leveraging an extensive dataset of publicly available genetic information. This remains the sole MR investigation delving into the causal relationship bet
Our studies have shown that the risk of developing OSAHS increases with the percentage of CD38 in IgD+ CD24-B cells. Altered CD38 expression or increased function of the cyclic ADP ribozyme associated with CD38 in this cell subset has been directly linked to the treatment of a variety of diseases, including cancer, asthma, and neuroimmune diseases[24]. It has been shown that CD38 plays a role in calcium regulation in airway smooth muscle (ASM) and that upregulation of CD38 levels improves Ca2+ responses when airway smooth muscle is exposed to contractile agonists[25,26]. Experimental studies have also shown that CD38 increases airway inflammation and responsiveness by modulating intracellular calcium levels in mouse smooth muscle contractile (ASM) cells. Through a mechanism that is not dependent on CD38, bronchodilators are often used for clinical guidance in the medical management of chronic airway disease[27]. Additionally, in CRS patients with nasal polyps, elevated IgD CSR in mucosa-associated lymphocyte B-cell populations activates mast cells and may promote IgE production and eosinophilic inflammation[28]. Although the exact relationships between these variables are yet unknown, they all have an indirect impact on how OSAHS develops.
Plenty of studies have been done on the connection between basophils and airway inflammation. It has been de
This study utilized a two-sample Mendelian randomization method, and the data were obtained from a sizable ge
In conclusion, our extensive bi-directional MR analyses revealed a causal relationship between various immune pheno
Despite being one of the most prevalent sleep disorders, obstructive sleep apnea hypoventilation syndrome (OSAHS) has limited information on its immunologic foundation. The immunological underpinnings of certain major psychiatric di
In summary, our comprehensive bidirectional mendelian randomization (MR) analysis has revealed causal links between various immunophenotypes and OSAHS, shedding light on the intricate web of relationships between OSAHS and the immune system. Moreover, Reverse causality, other variables, and other unavoidable confounding factors have all been successfully reduced in impact by our study, offering a fresh perspective for researchers to delve into the biological un
This study employed two-sample Mendelian randomization analysis using data from a large genomic research cohort of approximately 372657 individuals, assuring great statistical efficiency. The outcomes of the study were based on genetic instrumental variables, and causal inferences were conducted by various robust Mendelian randomization analysis te
A comprehensive two-sample MR study was conducted to investigate the causal relationship between immune cell characteristics and OSAHS. Summary statistics for each immune cell feature were obtained from the GWAS catalog. Information on 731 immune cell properties, such as morphologic parameters, median fluorescence intensity, absolute cellular, and relative cellular, was compiled using publicly available genetic databases. The results' robustness, heterogeneity, and horizontal pleiotropy were confirmed using extensive sensitivity examination.
After false discovery rate correction, OSAHS had no statistically significant effect on immunophenotypes. However, Two lymphocyte subsets were identified to be significantly associated with OSAHS risk: (OR = 1.03, 95%CI = 1.01-1.03, P = 0.000); CD28+CD4+T cell (OR = 1.04, 95%CI = 1.02-1.04, P = 0.019).
The study has shown the close association between immune cells and OSAHS through genetic methods, thereby offering direction for future clinical research.
This groundbreaking study employs bidirectional MR analysis to unveil crucial immunological links in OSAHS. By establishing causal relationships between diverse immunophenotypes and OSAHS, the research offers a fresh lens for exploring the disorder's biological foundations. Successfully addressing confounding factors, the study presents oppor
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Immunology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Verma V, United States S-Editor: Liu JH L-Editor: A P-Editor: Xu ZH
| 1. | Lee JJ, Sundar KM. Evaluation and Management of Adults with Obstructive Sleep Apnea Syndrome. Lung. 2021;199:87-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 103] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 2. | Akarsu FG, Algin DI, Erdinç OO. Evaluation of comorbid diseases in obstructive sleep apnea syndrome. Rev Assoc Med Bras (1992). 2023;69:421-425. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 3. | Lv R, Liu X, Zhang Y, Dong N, Wang X, He Y, Yue H, Yin Q. Pathophysiological mechanisms and therapeutic approaches in obstructive sleep apnea syndrome. Signal Transduct Target Ther. 2023;8:218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 151] [Reference Citation Analysis (0)] |
| 4. | Antonaglia C, Passuti G. Obstructive sleep apnea syndrome in non-obese patients. Sleep Breath. 2022;26:513-518. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 5. | Singh P, Bonitati A. Obstructive Sleep Apnea Syndrome - A Review for Primary Care Physicians and Pulmonologists. R I Med J (2013). 2021;104:10-13. [PubMed] |
| 6. | Challamel MJ, Beydon N, Coutier L, Launois S, Seailles T, Vecchierini MF, Franco P. [Diagnostic criteria for obstructive sleep apnea syndrome in adolescent]. Rev Mal Respir. 2021;38:829-839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 7. | Kim DK, Lee BC, Park KJ, Son GM. Effect of Obstructive Sleep Apnea on Immunity in Cases of Chronic Rhinosinusitis With Nasal Polyps. Clin Exp Otorhinolaryngol. 2021;14:390-398. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 8. | Wu Y, She L, Huang DH. [A review about changes of immune function in patients with obstructive sleep apnea hypopnea syndrome]. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2022;57:649-655. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 9. | de Azevedo PG, Guimarães MLR, Albuquerque ALB, Alves RB, Gomes Fernandes B, Marques de Melo F, Guimaraes Corrêa Do Carmo Lisboa Cardenas R, Friedman E, De Marco L, Bastos-Rodrigues L. Whole-exome identifies germline variants in families with obstructive sleep apnea syndrome. Front Genet. 2023;14:1137817. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 10. | Wang Q, Yang C, Gelernter J, Zhao H. Pervasive pleiotropy between psychiatric disorders and immune disorders revealed by integrative analysis of multiple GWAS. Hum Genet. 2015;134:1195-1209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 11. | Spiga F, Gibson M, Dawson S, Tilling K, Davey Smith G, Munafò MR, Higgins JPT. Tools for assessing quality and risk of bias in Mendelian randomization studies: a systematic review. Int J Epidemiol. 2023;52:227-249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 39] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 12. | Burgess S, Mason AM, Grant AJ, Slob EAW, Gkatzionis A, Zuber V, Patel A, Tian H, Liu C, Haynes WG, Hovingh GK, Knudsen LB, Whittaker JC, Gill D. Using genetic association data to guide drug discovery and development: Review of methods and applications. Am J Hum Genet. 2023;110:195-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 76] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 13. | Hou R, Ye G, Liu Y, Chen X, Pan M, Zhu F, Fu J, Fu T, Liu Q, Gao Z, Baldwin DS, Tang Z. Effects of SSRIs on peripheral inflammatory cytokines in patients with Generalized Anxiety Disorder. Brain Behav Immun. 2019;81:105-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 14. | Wingo AP, Gibson G. Blood gene expression profiles suggest altered immune function associated with symptoms of generalized anxiety disorder. Brain Behav Immun. 2015;43:184-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 15. | Orrù V, Steri M, Sidore C, Marongiu M, Serra V, Olla S, Sole G, Lai S, Dei M, Mulas A, Virdis F, Piras MG, Lobina M, Pitzalis M, Deidda F, Loizedda A, Onano S, Zoledziewska M, Sawcer S, Devoto M, Gorospe M, Abecasis GR, Floris M, Pala M, Schlessinger D, Fiorillo E, Cucca F. Complex genetic signatures in immune cells underlie autoimmunity and inform therapy. Nat Genet. 2020;52:1036-1045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 616] [Article Influence: 123.2] [Reference Citation Analysis (0)] |
| 16. | Sidore C, Busonero F, Maschio A, Porcu E, Naitza S, Zoledziewska M, Mulas A, Pistis G, Steri M, Danjou F, Kwong A, Ortega Del Vecchyo VD, Chiang CWK, Bragg-Gresham J, Pitzalis M, Nagaraja R, Tarrier B, Brennan C, Uzzau S, Fuchsberger C, Atzeni R, Reinier F, Berutti R, Huang J, Timpson NJ, Toniolo D, Gasparini P, Malerba G, Dedoussis G, Zeggini E, Soranzo N, Jones C, Lyons R, Angius A, Kang HM, Novembre J, Sanna S, Schlessinger D, Cucca F, Abecasis GR. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nat Genet. 2015;47:1272-1281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 212] [Cited by in RCA: 259] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 17. | Vierstra J, Lazar J, Sandstrom R, Halow J, Lee K, Bates D, Diegel M, Dunn D, Neri F, Haugen E, Rynes E, Reynolds A, Nelson J, Johnson A, Frerker M, Buckley M, Kaul R, Meuleman W, Stamatoyannopoulos JA. Global reference mapping of human transcription factor footprints. Nature. 2020;583:729-736. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 226] [Cited by in RCA: 238] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 18. | Wang B, Gao L, Zhang J, Meng X, Xu X, Hou H, Xing W, Wang W, Wang Y. Unravelling the genetic causality of immunoglobulin G N-glycans in ischemic stroke. Glycoconj J. 2023;40:413-420. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 19. | Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40:304-314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4015] [Cited by in RCA: 5686] [Article Influence: 631.8] [Reference Citation Analysis (0)] |
| 20. | Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46:1985-1998. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 643] [Cited by in RCA: 2088] [Article Influence: 298.3] [Reference Citation Analysis (0)] |
| 21. | Patel A, Ye T, Xue H, Lin Z, Xu S, Woolf B, Mason AM, Burgess S. MendelianRandomization v0.9.0: updates to an R package for performing Mendelian randomization analyses using summarized data. Wellcome Open Res. 2023;8:449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 2] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 22. | Cho Y, Haycock PC, Sanderson E, Gaunt TR, Zheng J, Morris AP, Davey Smith G, Hemani G. Exploiting horizontal pleiotropy to search for causal pathways within a Mendelian randomization framework. Nat Commun. 2020;11:1010. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 84] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 23. | Verbanck M, Chen CY, Neale B, Do R. Publisher Correction: Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:1196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 186] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 24. | Deshpande DA, Guedes AGP, Graeff R, Dogan S, Subramanian S, Walseth TF, Kannan MS. CD38/cADPR Signaling Pathway in Airway Disease: Regulatory Mechanisms. Mediators Inflamm. 2018;2018:8942042. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 25. | Deshpande DA, Guedes AGP, Lund FE, Subramanian S, Walseth TF, Kannan MS. CD38 in the pathogenesis of allergic airway disease: Potential therapeutic targets. Pharmacol Ther. 2017;172:116-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 26. | Boslett J, Hemann C, Christofi FL, Zweier JL. Characterization of CD38 in the major cell types of the heart: endothelial cells highly express CD38 with activation by hypoxia-reoxygenation triggering NAD(P)H depletion. Am J Physiol Cell Physiol. 2018;314:C297-C309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 27. | Guedes AG, Dileepan M, Jude JA, Deshpande DA, Walseth TF, Kannan MS. Role of CD38/cADPR signaling in obstructive pulmonary diseases. Curr Opin Pharmacol. 2020;51:29-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 28. | Choi JH, Wang KW, Zhang D, Zhan X, Wang T, Bu CH, Behrendt CL, Zeng M, Wang Y, Misawa T, Li X, Tang M, Scott L, Hildebrand S, Murray AR, Moresco EM, Hooper LV, Beutler B. IgD class switching is initiated by microbiota and limited to mucosa-associated lymphoid tissue in mice. Proc Natl Acad Sci U S A. 2017;114:E1196-E1204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 29. | Motomura Y, Morita H, Moro K, Nakae S, Artis D, Endo TA, Kuroki Y, Ohara O, Koyasu S, Kubo M. Basophil-derived interleukin-4 controls the function of natural helper cells, a member of ILC2s, in lung inflammation. Immunity. 2014;40:758-771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 254] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 30. | Rohn H, Lang C, Schramm S, Heinemann FM, Trilling M, Gäckler A, Witzke O, Horn PA, Rebmann V. Effect of HLA-G5 Immune Checkpoint Molecule on the Expression of ILT-2, CD27, and CD38 in Splenic B cells. J Immunol Res. 2022;2022:4829227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 31. | Ahmad SF, Ansari MA, Nadeem A, Bakheet SA, Al-Ayadhi LY, Alotaibi MR, Alhoshani AR, Al-Hosaini KA, Attia SM. Dysregulation of the expression of HLA-DR, costimulatory molecule, and chemokine receptors on immune cells in children with autism. Int Immunopharmacol. 2018;65:360-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 32. | Chang KH, Wu YR, Chen YC, Fung HC, Chen CM. Association of genetic variants within HLA-DR region with Parkinson's disease in Taiwan. Neurobiol Aging. 2020;87:140.e13-140.e18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 33. | Enz LS, Zeis T, Schmid D, Geier F, van der Meer F, Steiner G, Certa U, Binder TMC, Stadelmann C, Martin R, Schaeren-Wiemers N. Increased HLA-DR expression and cortical demyelination in MS links with HLA-DR15. Neurol Neuroimmunol Neuroinflamm. 2020;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |