Published online Feb 6, 2023. doi: 10.12998/wjcc.v11.i4.874
Peer-review started: August 17, 2022
First decision: November 11, 2022
Revised: December 21, 2022
Accepted: January 12, 2023
Article in press: January 12, 2023
Published online: February 6, 2023
Processing time: 172 Days and 17.3 Hours
Copy number variation (CNV) has become widely recognized in recent years due to the extensive use of gene screening in developmental disorders and epilepsy research. 1q21.1 microduplication syndrome is a rare CNV disease that can manifest as multiple congenital developmental disorders, autism spectrum disorders, congenital malformations, and congenital heart defects with genetic heterogeneity.
We reported a pediatric patient with 1q21.1 microduplication syndrome, and carried out a literature review to determine the correlation between 1q21.1 microduplication and its phenotypes. We summarized the patient’s medical history and clinical symptoms, and extracted genomic DNA from the patient, her parents, elder brother, and sister. The patient was an 8-mo-old girl who was hospitalized for recurrent convulsions over a 2-mo period. Whole exon seq
Whole exon sequencing combined with quantitative polymerase chain reaction can provide an accurate molecular diagnosis in children with 1q21.1 microduplication syndrome, which is of great significance for genetic counseling and early intervention.
Core Tip: We reported an 8-mo-old girl with 1q21.1 microduplication syndrome, and review the literature to determine the correlation between 1q21.1 microduplication and its phenotypes. Whole exon sequencing and whole genome low-depth sequencing (Copy number variation -seq) were performed on the patient and her family members. This case shows that whole exon sequencing combined with quantitative polymerase chain reaction can provide an accurate molecular diagnosis in children with 1q21.1 microduplication syndrome, which is important for genetic counseling and early intervention in the patients.
- Citation: Huang TT, Xu HF, Wang SY, Lin WX, Tung YH, Khan KU, Zhang HH, Guo H, Zheng G, Zhang G. Identification of 1q21.1 microduplication in a family: A case report. World J Clin Cases 2023; 11(4): 874-882
- URL: https://www.wjgnet.com/2307-8960/full/v11/i4/874.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i4.874
Copy number variation (CNV) has become widely recognized in recent years due to the extensive use of gene screening in developmental disorders and epilepsy research[1]. In this study, we performed molecular genetic analysis on a child admitted to the hospital in October 2019 with unexplained seizures and found approximately 1.58 Mb of duplication in the 1q21.1-1q21.2 (CHR1:145883867-147465312 *3) region. Previous literature reported that epilepsy was associated with CNV of region 1q21.1, but it is mainly seen in microdeletion syndrome and rarely in microduplication syndrome[2,3]. The existing reports show that 1q21.1 microduplication syndrome leads to a phenotype that includes developmental delays, autism spectrum disorders, congenital malformations, and congenital heart defects (typically, tetralogy of Fallot) along with characteristic facial dysmorphic signs[4-6]. Here, we describe a 1q21.1 microduplication in a patient with a 1.58 Mb duplication. The patient experienced seizures, developmental delay, and congenital heart disease in the form of ventricular septal defect. In addition, we summarized and analyzed the clinical characteristics and CNV regions of children with 1q21.1 microduplication syndrome in order to further illustrate the correlation between the duplication region and phenotype.
The patient was an 8-mo-old girl who was hospitalized for recurrent convulsions over a 2-mo period.
She presented with repeated convulsions after admission to the hospital that manifested as upturned eyes, followed by involuntary blinking, cyanosis of the lips and hands and confusion. This was not considered a tonic seizure, and she had no limb shaking. The convulsions occurred approximately 10 times a day, each lasting about 30 s; some episodes were self-relieving. The patient did not present with fever before the convulsions and there was no history of birth asphyxia or trauma. The convulsions were relieved after treatment with sodium valproate. The patient also suffered from developmental retardation as she could not lift her head in the prone position until 6 mo old, and was unable to sit independently at admission (age: 8 mo). Her gross and fine motor skills were poor, but her language and speech skills seemed to be fine. The patient had clear consciousness. Her weight was in the normal range, and she did not show macrocephaly or short stature.
Ventricular septal defect (VSD) was found when she was 4 mo old, and she underwent open-heart repair of the VSD.
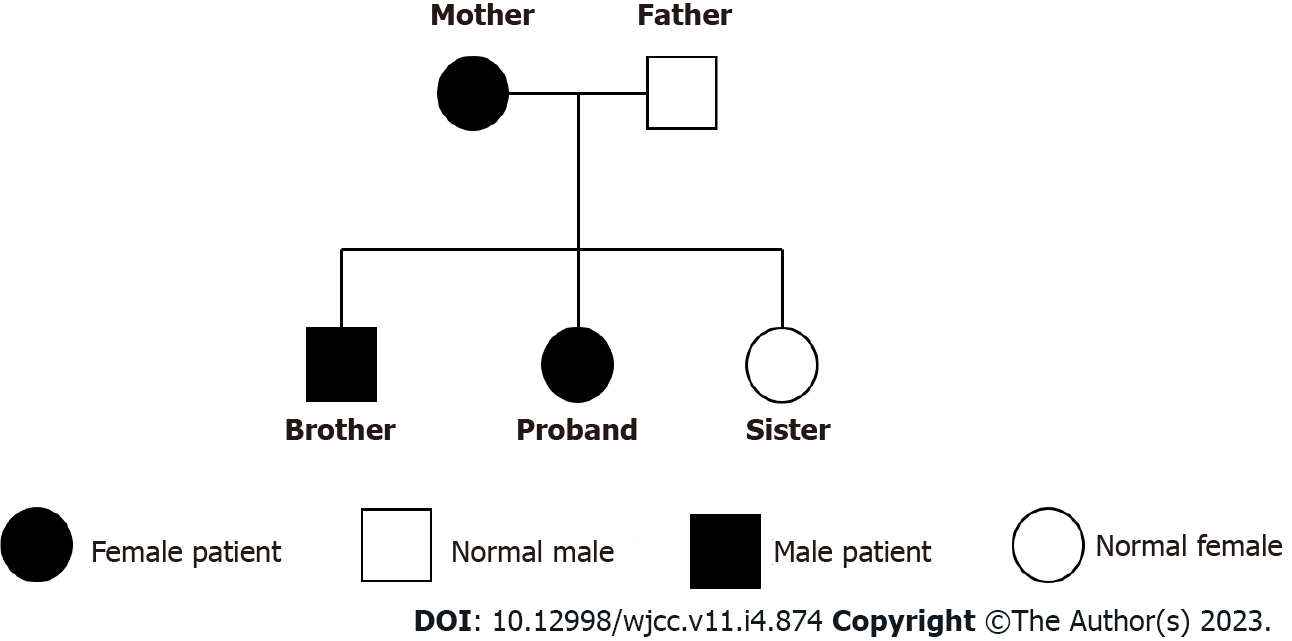
She delivered naturally at term. There was no hypoxia or suffocation or intraventricular hemorrhage during birth. Her parents denied any family history of developmental disorders, mental disorders, or genetic disorders. Both parents were in good health and are unrelated. The mother claimed that she developed a cold in the first 3 mo of the pregnancy. The mother denied any possible exposure to toxic substances such as alcohol, drugs, or other harmful environmental factors during the pregnancy and perinatal period. The child has a 5-year-old sister in good health and a 2.5-year-old brother who has language developmental delay and a two-time history of febrile convulsions. The patient's brother had no hypoxia or asphyxia or intraventricular hemorrhage at birth, has poor gross and fine motor skills, and he also appeared to have poor speech and language skills. He did not present with macrocephaly or short stature.
On physical examination, there was no remarkable dysmorphism in the patient’s appearance. A 6-cm-long surgical scar was observed on the anterior chest. She had normal limb muscle strength and muscle tone, and her physiological reflexes were normal, with no evidence of any pathological signs.
Routine blood, urine, and stool tests showed no abnormalities. Her thyroid hormone and growth hormone profiles were normal.
Electrocardiography showed sinus rhythm, left atrial burden, and T-wave changes in partial leads (V5). Chest radiography revealed increased pulmonary vascular markings, enlarged heart shadow, right upper mediastinal widening, and a thymus. Her video electroencephalography (VEEG) detected sharp slow wave release in the mid-line CZ (central midline point) and PZ (parietal midline point) zone during the sleep course. Cranial magnetic resonance imaging (MRI) showed left lateral ventricle enlargement and dysgenesis of the corpus callosum.
Genomic DNA extraction: Briefly, 2 mL peripheral blood was drawn from the patient, her parents, and siblings, and sent to Kaiumph Medical Diagnosis (Beijing, China) for molecular karyotyping. We used a blood genomic DNA extraction kit (QIAamp DNA Blood Mini Kit, Qiagen) to extract genomic DNA.
Chromosome microarray: We first extracted DNA from peripheral blood and then processed the DNA samples by CMA using whole-genome bacterial artificial chromosome (BAC) microarrays, in accordance with the protocol. Data analysis was performed using algorithm fixed settings. The genomic coverage resolution of this microarray platform is up to 1 Mb. There were a minimum of 3 consecutive BAC clones used for microarray probes to determine copy number variation. We compared the detected CNVs with the known CNVs in the public database, and also classified detected CNVs as “Clinically Significant,” “Likely Benign,” or as “Variants Of Uncertain Clinical Significance (VOUS).” The result of the chromosome microarray test was negative.
Whole exome sequencing: Genomic DNA was quantified using Nanodrop 2000 (Thermal Fisher Scientific, United States). We used the NEXT flex Rapid DNA-Seq Kit to construct a genomic DNA library, used xGen Exome Research Panel v2 (IDT, United States) to capture the constructed libraries, and then used Novaseq 6000 (Illumina, United States) to sequence for 10–12 GB. We screened for variants based on minor allele frequencies in the normal population and performed variant function and prediction by Mutation Tester, Polyphen2, and SIFT. The mutant pathogenicity was assessed in accordance with guidelines of the American College of Medical Genetics and Genomics. We carried out analysis of CNV of patient’s total exome detection results based on Cap CNV (capture copy number variation) analysis which was published in previous articles[1]. The results of gene sequencing and analysis with seizure-related genes such as AAAS, AARS, AASS, and ABAT, and another 1254 genes failed to show any pathogenic variation at the exon region.
Cap CNV analysis: We calculated the depth of each exon using bedtools-2.16.2 coverage for the same batch of samples. The sequencing quantity ratio of the sample and the batch mean was calculated to obtain the corrected depth in order to correct the bias in the sequencing data quantity. The depth ratio is used to determine whether a potential CNV exists in an exonic area, obtained from the corrected sample depth over the batch mean depth. A potential deletion is judged by a depth ratio of less than 0.7, and a potential duplication is judged by a depth ratio of greater than 1.3. The criterion for judging the low-quality area is that the rate of abnormal depth ratio in this area exceeds 20% in this batch, and then it is filtered.
The criteria for screening candidate CNV regions are as follows: (1) It is required that at least two consecutive exons have abnormal depth ratio for OMIM disease-related gene exon regions; and (2) It is required that at least ten consecutive exons have abnormal depth ratio for non OMIM disease-related gene exon regions. Then the regions of candidate CNV were annotated with Decipher and DGV.
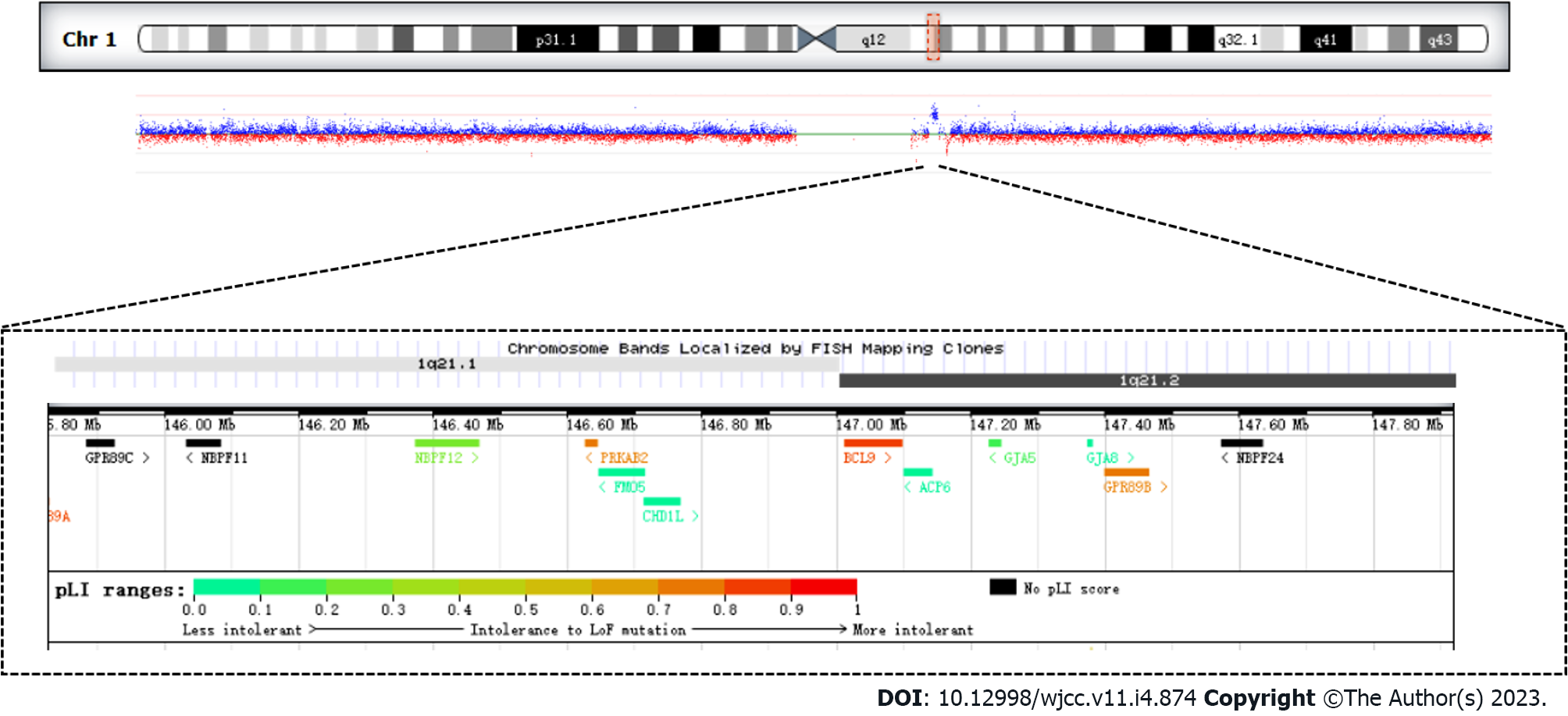
The analysis of CNVs based on the depth of the exon captured sequence showed that there was a 1.58 Mb duplication of heterozygosity in the 1q21.1 region, which included the following 11 genes GJA5, FMO5, CHD1L, PRKAB2, BCL9, BNBPF11, BNBPF12, BNBPF24, GJA8, GPR89B, GPR89C (Figure 1, Table 1).
| Gene | Phenotype | Inheritance | Gene function |
| GJA5 | Atrial Fibrillation, Familial, 11; Atrial Standstill 1; Tetralogy of Fallot; Chromosome 1q21.1 deletion syndrome, 1.35-MB | AD | The GJA5 gene encodes gap junction protein 40 (CX40), a cardiac gap junction protein expressed in the right ventricular outflow tract, which plays a key role in cell adhesion and intercellular communication |
| FMO5 | Duodenal Atresia; Jacobsen Syndrome; Mowat-Wilson Syndrome | AD | The gene acts as Baeyer-Villiger monooxygenase on a broad range of substrates. Catalyzes the insertion of an oxygen atom into a carbon-carbon bond adjacent to a carbonyl, which converts ketones to esters |
| CHD1L | Postcholecystectomy Syndrome; Prostate Calculus; Chromosome 1q21.1 Duplication Syndrome | AD | DNA helicase which plays a role in chromatin-remodeling following DNA damage, targeted to sites of DNA damage through interaction with poly (ADP-ribose) and functions to regulate chromatin during DNA repair. Able to catalyze nucleosome sliding in an ATP-dependent manner. Helicase activity is strongly stimulated upon poly (ADP-ribose)-binding |
| NBPF12 | Amelogenesis Imperfecta, Type Ia; Neuroblastoma; Microcephaly; Autism | Unknown | No data available for molecular function |
| NBPF11 | Neuroblastoma; Duodenal Atresia | Unknown | Predicted to be located in the cytoplasm. No data available for molecular function |
| NBPF24 | Also known asNBPF11 | Unknown | No data available for molecular function |
| PRKAB2 | Chromosome 1q21.1 Duplication Syndrome; Type 2 Diabetes Mellitus | Unknown | Non-catalytic subunit of AMP-activated protein kinase (AMPK), an energy sensor protein kinase that plays a key role in regulating cellular energy metabolism |
| BCL9 | Chromosome 1q21.1 Duplication Syndrome; Lymphoma; Leukemia; Retinitis Pigmentosa | Unknown | Involved in signal transduction through the Wnt pathway. Promotes beta-catenin's transcriptional activity. Gene coding for a large proline-rich protein with two transcripts, expressed in all tissues and a third expressed only in thymus, spleen, small intestine, involved in translocation t(1;14) and t(1;22) |
| GJA8 | Chromosome 1q21.1 Duplication Syndrome; Cataract 1, Multiple Types; Cataract Microcornea Syndrome; Early-Onset Sutural Cataract | AD | Structural component of eye lens gap junctions. Gap junctions are dodecameric channels that connect the cytoplasm of adjoining cells. They are formed by the docking of two hexameric hemichannels, one from each cell membrane. Small molecules and ions diffuse from one cell to a neighboring cell via the central pore |
| GPR89B | Thrombocytopenia-Absent Radius Syndrome; Hypothyroidism, Congenital, Nongoitrous, 1 | Unknown | Voltage dependent anion channel required for acidification and functions of the Golgi apparatus that may function in counter-ion conductance. Plays a role in lymphocyte development, probably by acting as a RABL3 effector in hematopoietic cells |
| GPR89C | Also known as, GPR89B | Unknown | The function of the gene is the same as GPR89B |
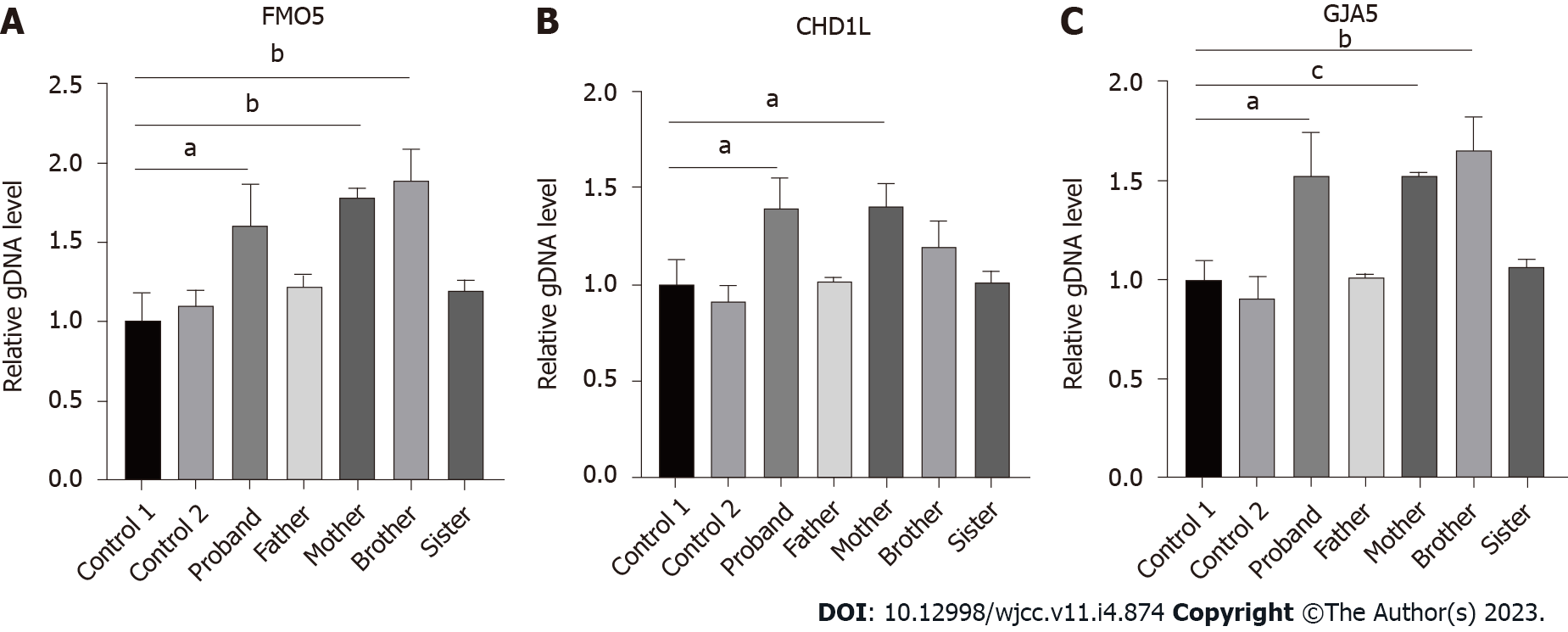
SYBR green fluorescent quantitative polymerase chain reaction: Primers for fluorescent quantitative polymerase chain reaction (qPCR) were designed for FMO5, CHD1L, GJA5, and ABL1 genes in the microduplication region. ABL1, as a housekeeping gene, is used as an internal reference. The relative quantitative values (RQ values) of FMO5 (forward, GAGCCCCATCCCACTTTCC; reverse, CCAACGCCATACCATTCAGG), CHD1L (forward, CCTCCTCAAGACAGCTGGTG; reverse, GCCCACCAGATCCTGATTCC) and GJA5 (forward, CAGAGCCCCGGACCTCTTT; reverse, TCCCCATCTCCCACATTCG) genes were calculated with ABL1 (forward, CTAAAGGTGAAAGCTCCG; reverse, GACTGTTGACTGGCGTGAT) as the internal reference gene by an ABI QuantStudioTM 6 Flex fluorescence quantitative analyzer for statistical analysis. Fluorescence qPCR was performed to determine the expression of related genes in duplicate regions of the genomic DNA from the patient, her parents, and normal controls. We purchased the PCR reaction reagents from Cowin Biosciences Company (Beijing, China), and the primer sequences were synthesized by Shanghai Sangon Biotech Company (Shanghai, China). We randomly selected exons from the FMO5, CHD1L, and GJA5 genes in the 1.58 Mb heterozygous duplication region, which was detected by CNV analysis, according to the results of whole exome sequencing (WES) to design the fluorescent quantitative PCR primers with the ABL as the internal reference gene. The results showed that the patient and her mother were carriers of a heterozygous duplication in FMO5, CHD1L, and GJA5 genes (the patient’s RQ was approximately 1.5, and the mother’s RQ was between 1.4 and 1.8). The father, however, did not show microduplication of FMO5, CHD1L, and GJA5 genes (RQ: approximately 1), which indicated that the mutation originated from the mother (Figure 2).
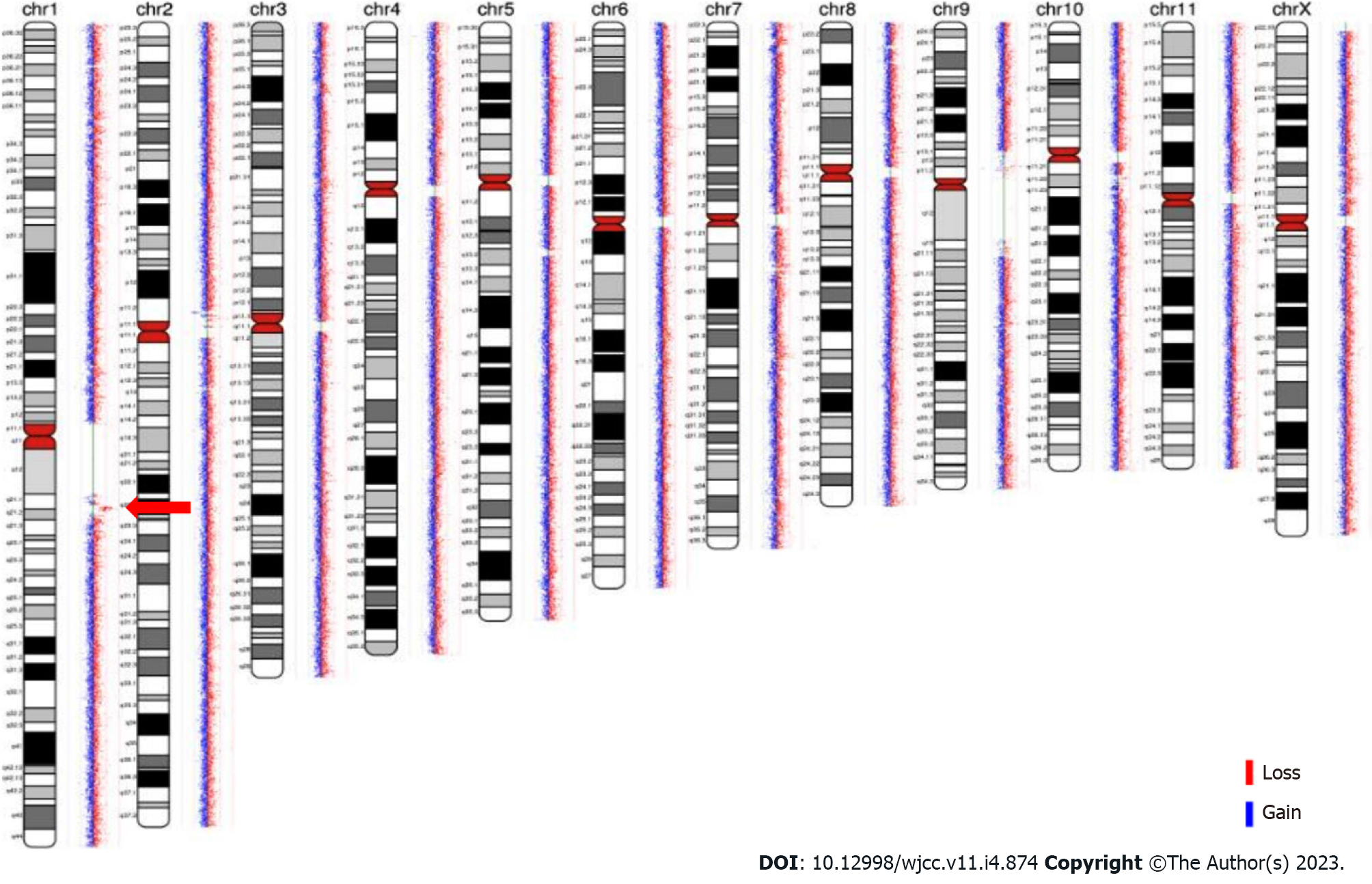
Familial analysis of low-depth whole genome sequencing: Low-depth whole genome sequencing detected a duplication of 2.096 Mb of heterozygosity in chromosome 1q (CHR1:145828373-147924436). The region has not yet been reported in the Decipher database. Low-depth whole genome sequencing, which was performed on the DNA of the patient’s mother and brother, also revealed a 2.096 Mb duplication of heterozygosity in the CHR1:145828373-147924436 region (as shown in Figure 3). These results indicated that the duplicate fragment originated from the mother and was also carried by the brother.
The patient’s family tree: The patient, her brother and mother had a duplication of heterozygosity in the 1q21.1 region, her father and sister did not (Figure 4).
The patient was diagnosed with 1q21.1 microduplication syndrome.
During the hospital stay, the patient had no epileptic seizures. The convulsions were relieved after treatment with sodium valproate.
Detailed history-taking and follow-up examinations showed that the patient and her brother are currently lagging behind in motor development and have language developmental delay and are likely on the autistic spectrum.
The 1q21.1 region is considered the most genetically unstable fragment due to multiple low-copy duplication, and one of the largest regions in the human genome that carries repeated duplication, thereby making it prone to CNV[2,4]. CNVs in the region are associated with developmental disorders, feeding problems, intellectual disability (ID), developmental delays (DD), congenital heart defects, behavioral problems (including autistic disorder and attention deficit hyperactivity disorder), and various other congenital deformities. There are 20-40 genes in the 1q21.1 region. The CNV of 1q21.1 can present as two main types: The one containing only the distal end region of 1q21.1 is called type I (approximately 1.8 Mb), and the one extending proximally to encompass the thrombocytopenia absent radius syndrome region is called type 2 (approximately 2.7 Mb)[4,5].
1q21.1 microduplication syndrome is a rare chromosome 1 mutation. It can be an autosomal dominant inheritance or a novel mutation. There is no significant difference in the sex ratio. Microduplication of 1q21.1 is observed in approximately 0.03% adults, and the frequency in live births is estimated to be 1/6309. It presents with variable and partially explicit phenotypes; the common clinical manifestations are multiple congenital developmental disorders, which include DD, autism spectrum disorders, congenital malformations, and congenital heart defects, wherein tetralogy of Fallot is the most common. It can also be seen in normal individuals[4,6-8]. In 2017, Busè et al[9] expanded the phenotype of the 1q21.1 syndrome and suggested that the 1q21.1 microduplication syndrome showed special facial features such as macrocephaly or relative macrocephaly, prominent forehead, and widening of the eye distance. The relationship between triangular head deformity and 1q21.1 microduplication syndrome was also mentioned[9]. Some researchers believed that it is a predisposing locus instead of a clinically distinct syndrome, given its incomplete dominance and variable phenotype[10]. With the development of genome microarray and other methods for detecting CNVs, the improved accuracy of CNVs detection, and the widespread use of gene screening in the study of DD and epilepsy, an increasing number of cases of 1q21.1 microduplication have been reported.
1q21.1 microduplication has been linked to a range of neurodevelopmental disorders, including autism spectrum disorders, ID, and even epilepsy, although there is no known gene for neurological disorders in this region[11]. The UCSC Genome Browser lists two genes located in the distal microduplication region, CHD1L and PRKAB2, which may be associated with epilepsy[2]. The CHD1L gene is also a candidate gene for attention deficit hyperactivity disorder and autism spectrum disorders, and the repetition of CHD1L is implicated in delayed language development[10]. The CHD1L gene, which is associated with a variety of cancers, encodes a helicase responsible for DNA repair. It comes from the same family as CHD2, which is related to epileptic encephalopathy and various generalized epilepsy syndromes[2]. PRKAB2 acts as a regulator of cellular responses to numerous stimuli, encodes the β2 subunits of AMP-activated protein kinase, and appears to play an important role in brain function[12]. With regard to the previously mentioned macrocephaly, it has been reported that it may be related to DUF1220 in this region, because the deletion of DUF1220 was found to be associated with microcephaly in 1q21.1 microdeletion syndrome. HYDIN2, in region 1q21.1, is a gene involved in the DUF1220 protein domain, and a homolog of the HYDIN gene in region 16q22.2. HYDIN, expressed only in the brain, is associated with hydrocephalus, so it may affect head circumference[13-15]. Regarding congenital heart defect, the GJA5 gene is believed to play an important role in the heart phenotype in the 1q21.1 region. It encodes gap junction protein 40 (CX40), a cardiac gap junction protein expressed through the right ventricular outflow tract, which plays a key role in intercellular communication and cellular adhesion and makes GJA5 gene a major candidate for congenital heart defect phenotype in this region[5,16-18].
At present, the sample size of 1q21.1 microduplication syndrome is limited, and the reported phenotypes are different. Using "1q21.1 microduplication syndrome" as the key word, we searched the biomedical literature database (PubMed) for nearly 10 years of literature, and we counted the 1q21.1 duplication regions and the genes contained in the region (Supplementary Table 1). Therefore, we still need more research to clarify the connection between genotype and specific phenotype of 1q21.1 microduplication syndrome. In our case, the result of the patient's gene analysis detected microduplication which contained both the GJA5 and CHD1L genes in the 1q21.1 region. This might explain her seizures and congenital heart disease. In the familial analysis, her mother and brother both carried microduplication of the GJA5 and CHD1L genes. However, her brother, who had a heterozygous duplication of about 2.096 Mb in the CHR1:145826755-147924436 region, showed symptoms presenting with language DD and febrile convulsions, while her mother showed no abnormalities. VEEG and cranial MRI of the patient’s mother did not show any abnormalities, and the language ability and reading ability of her mother were normal as assessed by the simple Wechsler Adult Intelligence Scale-Revised in China. In 2015, Judith and Verhagen et al[17] reported a case of 1q21.1 microduplication in a family of 11 people over three generations. Their patient suffered from severe learning difficulties and was diagnosed with chronic depression and anxiety at the age of 18. All carriers in her family showed hypertelorism and other facial dysmorphism but no congenital heart defect. At the first pregnancy of the proband, ultrasound examination at 20 wk of gestation revealed excessive fetal growth and a complex congenital heart defect, the pregnancy was terminated on the advice of her obstetrician. With these two reports of the same genotype but different phenotypes, we could speculate that the same genetic changes may result in different phenotypes. The phenotypes of our patient and her relatives still require long-term follow-up, for any new potential phenotypes.
The development of the nervous system is a complex process involving nearly 70% gene expression, which may explain why different CNVs have common clinical features. However, there is no explanation as to why the same position and CNV would result in a different phenotype[19]. Genes are epigenetically inherited and subject to environmental modifications; genetic alterations may make CNV carriers more susceptible to environmental influences, which can lead to different phenotypes, depending on the severity of the environmental impact[20,21]. Furthermore, the variable expressivity and incomplete penetrance suggest that the influence of CNV is modified by other genetic loci or environmental factors[22]. Xu et al[23] suggested that women have a higher tolerance for path
1q21.1 microduplication syndrome is a rare CNV disease. This finding has extended our knowledge of the clinical manifestations of 1q21.1 microduplication syndrome and enhanced our understanding of CNV. However, further research is required to clarify the connection between genotype and specific phenotype of 1q21.1 microduplication syndrome. WES combined with qPCR can provide an accurate molecular diagnosis for children carrying this genetic mutation, which is of great significance for genetic counseling and early intervention.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Neurosciences
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Dokponou YCH, Morocco S-Editor: Liu JH L-Editor: A P-Editor: Liu JH
| 1. | Tung Y, Lu H, Lin W, Huang T, Kim S, Hu G, Zhang G, Zheng G. Case Report: Identification of a de novo Microdeletion 1q44 in a Patient With Seizures and Developmental Delay. Front Genet. 2021;12:648351. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 2. | Gourari I, Schubert R, Prasad A. 1q21.1 Duplication syndrome and epilepsy: Case report and review. Neurol Genet. 2018;4:e219. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 3. | Seo HS, Jung YJ, Kim JH, Lee HH, Park CH. The Effect of Metformin on Prognosis in Patients With Locally Advanced Gastric Cancer Associated With Type 2 Diabetes Mellitus. Am J Clin Oncol. 2019;42:909-917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (1)] |
| 4. | Dolcetti A, Silversides CK, Marshall CR, Lionel AC, Stavropoulos DJ, Scherer SW, Bassett AS. 1q21.1 Microduplication expression in adults. Genet Med. 2013;15:282-289. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 5. | Sun G, Tan Z, Fan L, Wang J, Yang Y, Zhang W. 1q21.1 microduplication in a patient with mental impairment and congenital heart defect. Mol Med Rep. 2015;12:5655-5658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 6. | Xavier J, Zhou B, Bilan F, Zhang X, Gilbert-Dussardier B, Viaux-Savelon S, Pattni R, Ho SS, Cohen D, Levinson DF, Urban AE, Laurent-Levinson C. 1q21.1 microduplication: large verbal-nonverbal performance discrepancy and ddPCR assays of HYDIN/HYDIN2 copy number. NPJ Genom Med. 2018;3:24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 7. | Gillentine MA, Lupo PJ, Stankiewicz P, Schaaf CP. An estimation of the prevalence of genomic disorders using chromosomal microarray data. J Hum Genet. 2018;63:795-801. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 8. | Veerapandiyan A, Oh D, Kornitzer J. Distal 1q21.1 and proximal 1q21.2 microduplication in a child with attention-deficit hyperactivity disorder. Acta Neurol Belg. 2019;119:289-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 9. | Busè M, Cuttaia HC, Palazzo D, Mazara MV, Lauricella SA, Malacarne M, Pierluigi M, Cavani S, Piccione M. Expanding the phenotype of reciprocal 1q21.1 deletions and duplications: a case series. Ital J Pediatr. 2017;43:61. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 10. | Benítez-Burraco A, Barcos-Martínez M, Espejo-Portero I, Fernández-Urquiza M, Torres-Ruiz R, Rodríguez-Perales S, Jiménez-Romero MS. Narrowing the Genetic Causes of Language Dysfunction in the 1q21.1 Microduplication Syndrome. Front Pediatr. 2018;6:163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 11. | Dougherty ML, Nuttle X, Penn O, Nelson BJ, Huddleston J, Baker C, Harshman L, Duyzend MH, Ventura M, Antonacci F, Sandstrom R, Dennis MY, Eichler EE. The birth of a human-specific neural gene by incomplete duplication and gene fusion. Genome Biol. 2017;18:49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 12. | Harvard C, Strong E, Mercier E, Colnaghi R, Alcantara D, Chow E, Martell S, Tyson C, Hrynchak M, McGillivray B, Hamilton S, Marles S, Mhanni A, Dawson AJ, Pavlidis P, Qiao Y, Holden JJ, Lewis SM, O'Driscoll M, Rajcan-Separovic E. Understanding the impact of 1q21.1 copy number variant. Orphanet J Rare Dis. 2011;6:54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 13. | Dumas LJ, O'Bleness MS, Davis JM, Dickens CM, Anderson N, Keeney JG, Jackson J, Sikela M, Raznahan A, Giedd J, Rapoport J, Nagamani SS, Erez A, Brunetti-Pierri N, Sugalski R, Lupski JR, Fingerlin T, Cheung SW, Sikela JM. DUF1220-domain copy number implicated in human brain-size pathology and evolution. Am J Hum Genet. 2012;91:444-454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 90] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 14. | Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, Sahoo T, Lalani SR, Graham B, Lee B, Shinawi M, Shen J, Kang SH, Pursley A, Lotze T, Kennedy G, Lansky-Shafer S, Weaver C, Roeder ER, Grebe TA, Arnold GL, Hutchison T, Reimschisel T, Amato S, Geragthy MT, Innis JW, Obersztyn E, Nowakowska B, Rosengren SS, Bader PI, Grange DK, Naqvi S, Garnica AD, Bernes SM, Fong CT, Summers A, Walters WD, Lupski JR, Stankiewicz P, Cheung SW, Patel A. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466-1471. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 490] [Cited by in RCA: 443] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 15. | O'Bleness M, Searles VB, Varki A, Gagneux P, Sikela JM. Evolution of genetic and genomic features unique to the human lineage. Nat Rev Genet. 2012;13:853-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 107] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 16. | Soemedi R, Topf A, Wilson IJ, Darlay R, Rahman T, Glen E, Hall D, Huang N, Bentham J, Bhattacharya S, Cosgrove C, Brook JD, Granados-Riveron J, Setchfield K, Bu'lock F, Thornborough C, Devriendt K, Breckpot J, Hofbeck M, Lathrop M, Rauch A, Blue GM, Winlaw DS, Hurles M, Santibanez-Koref M, Cordell HJ, Goodship JA, Keavney BD. Phenotype-specific effect of chromosome 1q21.1 rearrangements and GJA5 duplications in 2436 congenital heart disease patients and 6760 controls. Hum Mol Genet. 2012;21:1513-1520. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 95] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 17. | Verhagen JM, de Leeuw N, Papatsonis DN, Grijseels EW, de Krijger RR, Wessels MW. Phenotypic Variability Associated with a Large Recurrent 1q21.1 Microduplication in a Three-Generation Family. Mol Syndromol. 2015;6:71-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 18. | Wang HD, Liu L, Wu D, Li T, Cui CY, Zhang LZ, Wang CZ. Clinical and molecular cytogenetic analyses of four families with 1q21.1 microdeletion or microduplication. J Gene Med. 2017;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 19. | Deshpande A, Weiss LA. Recurrent reciprocal copy number variants: Roles and rules in neurodevelopmental disorders. Dev Neurobiol. 2018;78:519-530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 20. | Rosenfeld JA, Coe BP, Eichler EE, Cuckle H, Shaffer LG. Estimates of penetrance for recurrent pathogenic copy-number variations. Genet Med. 2013;15:478-481. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 209] [Cited by in RCA: 270] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 21. | Qiao Y, Badduke C, Tang F, Cowieson D, Martell S, Lewis SME, Peñaherrera MS, Robinson WP, Volchuk A, Rajcan-Separovic E. Whole exome sequencing of families with 1q21.1 microdeletion or microduplication. Am J Med Genet A. 2017;173:1782-1791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 22. | Maher BS, Riley BP, Kendler KS. Psychiatric genetics gets a boost. Nat Genet. 2008;40:1042-1044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 21] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 23. | Xu M, Ji Y, Zhang T, Jiang X, Fan Y, Geng J, Li F. Clinical Application of Chromosome Microarray Analysis in Han Chinese Children with Neurodevelopmental Disorders. Neurosci Bull. 2018;34:981-991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |