Published online Aug 12, 2013. doi: 10.5501/wjv.v2.i3.110
Revised: May 31, 2013
Accepted: May 31, 2013
Published online: August 12, 2013
Processing time: 211 Days and 12.5 Hours
Nuclear domain 10 (ND10) are spherical bodies distributed throughout the nucleoplasm and measuring around 0.2-1.0 μm. First observed under an electron microscope, they were originally described as dense bodies found in the nucleus. They are known by a number of other names, including Promyelocytic Leukemia bodies (PML bodies), Kremer bodies, and PML oncogenic domains. ND10 are frequently associated with Cajal bodies and cleavage bodies. It has been suggested that they play a role in regulating gene transcription. ND10 were originally characterized using human autoantisera, which recognizes Speckled Protein of 100 kDa, from patients with primary biliary cirrhosis. At the immunohistochemical level, ND10 appear as nuclear punctate structures, with 10 indicating the approximate number of dots per nucleus observed. ND10 do not colocalize with kinetochores, centromeres, sites of mRNA processing, or chromosomes. Resistance of ND10 antigens to nuclease digestion and salt extraction suggest that ND10 are associated with the nuclear matrix. They are often identified by immunofluorescent assay using specific antibodies against PML, Death domain-associated protein, nuclear dot protein (NDP55), and so on. The role of ND10 has long been the subject of investigation, with the specific connection of ND10 and viral infection having been a particular focus for almost 20 years. This review summarizes the relationship of ND10 and viral infection. Some future study directions are also discussed.
Core tip: We, for the first time, discussed the function of nuclear domain 10 (ND10) as a nuclear structure. Although the ND10 components, especially Promyelocytic Leukemia bodies, Speckled Protein of 100 kDa and death domain-associated protein, have been widely investigated for their roles in viral gene expression and viral replication, individual virus interacts with ND10 differentially as we summarized up in this review. This review is expected to guide readers especially virologists and cell biologists to understanding the interaction of ND10 with viruses.
- Citation: Rivera-Molina YA, Martínez FP, Tang Q. Nuclear domain 10 of the viral aspect. World J Virol 2013; 2(3): 110-122
- URL: https://www.wjgnet.com/2220-3249/full/v2/i3/110.htm
- DOI: https://dx.doi.org/10.5501/wjv.v2.i3.110
Mammalian cells contain differentially functional compartments called organelles, which are separated from the cytoplasm by a lipid bilayer membrane. The nucleus is an extremely dynamic organelle and highly organized compartment with multiple functions (reviewed in Dundr et al[1], Dundr et al[2], and Zhao et al[3]) the nucleoplasm consists of soluble and insoluble materials that keep the genomic structure intact and host the complicated process of gene transcription. Some insoluble and soluble materials congregate together to form shaped structures such as nuclear domain 10 (ND10 can also refer to nuclear dot 10)[4]. When analyzed by indirect immunofluorescence microscopy, many nuclear proteins are seen to localize in distinct structures with punctate staining patterns[5,6]. Nuclear structures, such as speckles, paraspeckles, nucleoli, Cajal bodies, polycomb bodies, and ND10, are formed primarily by protein-protein, protein-RNA, or protein-DNA interactions[1]. Each nuclear body has a matrix protein that is essential for the formation of the specific nuclear body. ND10 are subnuclear structures that gather many different SUMOylated nuclear proteins (such as Daxx and SP100). The formation of ND10 depends on Promyelocytic Leukemia (PML) protein. Past observations confirm that PML knockout cells lack ND10 and that transfecting exogenous PML into PML knockout cells results in the restoration of ND10[7,8]. Most DNA viruses replicate DNA and transcribe genes in the nucleus after their genomic DNA enters the nucleus by facilitated transport through the nuclear pore complex[9]. Once inside the nucleus, viral genomes distribute randomly, but it appears that only those at ND10 replicate and transcribe predominantly[10-13], suggesting specifically that the environment at ND10 is particularly advantageous for the virus. However, the ND10 proteins [such as PML, Speckled Protein of 100 kDa (SP100), and Daxx] are interferon-upregulated and have repressive effects on viral replication[14-25]. Moreover, most DNA viruses encode an immediate-early protein that induces the dispersion of ND10[10,26-29], and in the absence of these viral proteins, replication is severely retarded[13,29,30]. These findings have led to the hypothesis that ND10 are also part of nuclear defense mechanism[4]. At this point, the effects of ND10 on viral replication remain to be settled.
There are five hallmark events in the history of studying ND10. First, a French paper in 1960 described an unknown nuclear structure in rabbit cells with the Papillomavirus as an electron-dense body[31]. It was the earliest description of the nuclear structure, but it left everything unexplained, other than providing that observation. Second, there was not any other information that could lead to a deeper investigation of these nuclear structures until they were first identified (by immunofluorescence analysis, using specific antibodies that were later revealed to be against SP100 and NDP55) as ND10 in 1991 by Ascoli et al[32]. SP100 was later proved to be essential for the formation of ND10[33]; NDP55 has not been characterized so far. Ascoli et al[32] investigated the structure in different types of cells. A combination of immunofluorescence analysis and electronic microscopy confirmed that ND10 are the structures that were previously observed in 1960. Third, in the process of investigating the function of ND10, ND10 were found to be related to herpes simplex virus type 1 (HSV-1) infection[26,34] in 1993 and 1994; these studies (by Maul GG, the Wistar Institute, the United States of America, and Everett RD, the MRC Virology Unit, Glasgow, United Kingdom) awakened the interest of virologists with regard to the interaction of ND10 and many different viruses. The interactions of ND10 and a variety of viruses will be discussed in this review. Fourth, it was determined that PML knockout mice lack ND10, which provided direct evidence that supported the hypothesis that the protein PML is essential to the formation of ND10. It was confirmed, as well, by the experimental results that demonstrate that the transfection of PML into PML-/- cells restores ND10[26,35]. PML-/- mice live normally, which further obscured the function of PML, though later studies were able to determine that PML-/- have a greater tendency to develop cancer than do their PML+/+ counterparts[36]. Fifth, ND10 components were identified. Even though more than 60 nuclear proteins have been shown to be more-or-less related to ND10[37,38], three components are thought to be the primary ND10 proteins (called the prototype proteins of ND10): PML, Daxx, and SP100. Other important events relating to the study of ND10 will be discussed in the following sections.
The molecular mechanism of the biogenesis of ND10 was a complete mystery until PML was identified as forming the matrix of ND10. PML is a tumor-suppressor protein that in both humans and mice is encoded by the PML gene. This gene was found to be involved in translocation with the retinoic acid receptor alpha (RARalpha) gene, causing acute promyelocytic leukemia (APL) (see the review by de Thé et al[39]). The protein encoded by this gene was therefore named after PML. PML is also called tripartite motif (TRIM) 19 because it is a member of the TRIM family[40]. The TRIM motif includes three zinc-binding domains, a RING, two B-boxes, and a coiled-coil region. Phosphorylation is required for the high SUMOylation of PML; SUMOylated PML localizes to ND10, where it functions as a transcription factor and tumor suppressor[41]. Its expression is cell-cycle related; therefore ND10 morphology and number in the nucleus are dependent on the cell cycle[42]. It regulates the p53 response to oncogenic signals, which might explain how the translocation of PML with RARalpha causes APL. Right after its identification, ND10 were shown to be important in cell differentiation and cell growth; this was first indicated in studies of promyelocytes from patients suffering from APL[43-45]. In the promyelocytes from these patients, ND10 cannot be detected. When cells are treated with retinoic acid (RA) or with arsenic trioxide, ND10 are restored and the APL phenotype is reversed and the patients can be cured with these agents (reviewed by Melnick and Licht[46]). PML has about 11 isoforms that are caused by the extensive alternative splicing of this gene. PML isoforms vary in the protein’s central and C-terminal regions; all variants encode the same N-terminus[47]. Some isoforms of PML are cytoplasmic, but most of the isoforms are nuclear proteins important for ND10 formation.
SP100 was first identified by immunofluorescence using autoantibodies from patients with primary biliary cirrhosis, and its cDNA was then isolated and cloned; it was found to encode a human nuclear antigen distributing in the nucleus as speckles[48]. SP100 is a single-copy gene sited in human chromosome 2q37 and, like PML, it is IFN upregulated. The SP100 gene needs to be spliced and gives rise to a number of speckled protein of 100 kDa (SP100) isoforms: SP100A, -B, -C, and -HMG[48-52]. The four SP100 isoforms share a homologous 476 N-terminal amino acid, but differ in their C-terminal part. The most abundant isoform is SP100A, which has 480 amino acids and migrates to 100 kDa on SDS-PAGE[51]. SP100A most likely does not bind to DNA alone because it lacks all other domains of SP100B, -C, and -HMG. It may be recruited to DNA via association with DNA-binding proteins such as hHMG2/DSP1[53], the B-cell-specific transactivator Bright[54], or ETS-1[55]. SP100B contains a SAND domain (SAND stands for SP100, AIRE, NucP41/75, and DEAF1), SP100HMG contains a SAND domain and an HMG box, and newly described SP100C contains SAND, PHD, and Bromo domains[52,56]. SP100 is one of the prototypical proteins of ND10, and it colocalizes with Daxx and PML in ND10. SP100B, -C, and -HMG isoforms contain SAND, PHD, Bromo, and HMG domains and are highly SUMOylated. All the domains are suggestive of a role in chromatin-mediated gene regulation. The three minor isoforms contain a SAND domain that binds to DNA and is required if SP100 is to have transcriptional regulating activity.
Upon its discovery, death domain-associated protein (Daxx beta) was found to be a protein of the classical death receptor[57]. It was found to bind specifically to the Fas death domain via its C-terminal portion. Overexpression of Daxx enhances Fas-mediated apoptosis through activating the Jun N-terminal kinase (JNK) pathway. It was later found that Daxx interacted with CENP-C, one of the few known intrinsic proteins of the human centromere[58]. CENP-C is thought to play structural as well as regulatory roles crucial to proper chromosome segregation and mitotic progression. The interaction between CENP-C and Daxx was then confirmed by an immunofluorescence assay that found the colocalization of these two proteins at discrete spots in the nuclei of some interphase cells[58]. The other Daxx-binding proteins include the transcription factor Pax3[59] and DNA methyltransferase I[60]. They both are related to centromeres such as CENP-C and are not related to ND10. Therefore, Daxx is a protein of centromere. However, Ishov et al [7] found that PML recruited Daxx to ND10. Interestingly, in PML-/- cells, Daxx totally stays in the centromere. Therefore, Daxx might travel from centromere to ND10 or from ND10 to centromere. Ishov et al [42] also found that Daxx and the SWI/SNF protein ATRX are both associated with two intranuclear domains: ND10 and heterochromatin. The accumulation of ATRX at ND10 was mediated by its interaction with the N-terminus of Daxx. Although ATRX was present in heterochromatin during the entire cell cycle, Daxx was actively recruited to this domain at the end of the S-phase. Daxx functions as an adapter for ATRX accumulation at ND10[42]. Daxx can be highly SUMOylated, and SUMOylation was found to be crucial for targeting Daxx to PODs and for the transrepression of several SUMOylated transcription factors, including the glucocorticoid receptors (GR)[61]. Recently, two variants of Daxx were identified. The two novel variants of Daxx were termed Daxx- and Daxx-γ, and these variants are generated by alternative splicing. They have a truncated regulatory C terminus, and Daxx- and Daxx-γ show markedly decreased affinities to PML and have a different nuclear distribution[62].
In summary, all three of the prototypical proteins (PML, SP100, and Daxx) of ND10 share some similar characteristics: (1) They colocalize with ND10. Their colocalization in ND10 depends on PML[7,8], and SP100 can also affect ND10 formation[33]. These proteins are prototypical components of ND10; (2) They can be up regulated by interferon, which provided the first evidence to support the hypothesis that ND10 are defensive against viral infection[4]; (3) The prototypical proteins of ND10 are all highly SUMOylated, SUMOylation is important for the formation of ND10, Daxx function, and the interaction of the three proteins; (4) They are all cancer gene repressors. Although PML-/- mice can still live normally, they are shown to have a higher chance of developing cancer[63]; (5) All three genes produce different isoforms via alternative splicing; and (6) They are all viral replication inhibitors, which will be discussed in the review below.
ND10 came to the forefront because it was found that t(15; 17) translocation causes the fusion of PML and RARA (generating PML/RARA) and the dysfunction of both PML and RARA (consequently resulting in APL). The oncogenic PML/RARA protein disrupts ND10 in a reversible manner upon being treated with retinoic acid and/or arsenic, either of which treatment can cure the patients with APL[64-68]. ND10 number and size are regulated in several cellular responses: viral infection[69], DNA-damage, transformation[70-72], and oxidative stress[73,74]. The transcriptions of PML, SP100, and Daxx are dramatically enhanced by interferons. However, PML-/- mice develop normally and live well without the formation of ND10, demonstrating that ND10 are not required for most basic biological functions. Nevertheless, recent data have implicated PML in the control of cellular senescence and stem cell self-renewal, extending the fields of the investigation of PML function[75,76].
ND10 studies have been so intense in recent years that novel information about these structures is being uncovered continuously; however, the function of PML bodies is still not fully understood. Three models have been proposed: the Depot or Sequestration model; the Hotspot model, and a site of specific nuclear activities. These models are described in the following paragraphs.
The nuclear domains are proposed to be aggregations of excess nucleoplasmic protein[77]. This model suggests that the ND10 components in the nucleoplasm have a dynamic nature, that is, they move from ND10 to the functional sites where they are needed. In other words, the aggregated proteins in ND10 are sequestered. This sequestration is evidenced by the fact that the PML partners in ND10 vary significantly between individual partners and levels of PML expression, as well as SUMOylation. A well-studied sequestered component in ND10 is Daxx, a potent repressor that forms partitions between SUMOylated proteins, including PML and many transcription factors. Sequestration of Daxx by ND10-associated, SUMOylated PML releases transcriptional repression by DNA-bound SUMOylated transcription factors[61,78-80].
This model proposes that ND10 are the sites of the post-translational modification and the degradation of proteins. It is supported by the facts that SUMO-1 molecules aggregate in ND10 and ND10 might be the hot sites for SUMOylation, that the acetylation and phosphorylation of p53 at PML bodies enhance the activity of p53[16,81,82], and that the 19S and 20S proteasome subunits localize at some PML bodies[83].
Proposes ND10 to be sites of specific nuclear activities, such as transcriptional regulation and DNA replication. This model is supported by the detection of nascent RNA around ND10[84], the association of ND10 with regions of high transcriptional activity[85], and the non-random nature of PML body assembly (based on the conservation of their size and position) following dissociation and re-formation as a result of cellular stress[86].
Human herpesviruses are divided into three subfamilies: alpha, beta, and gamma. The alpha subfamily includes Herpes Simplex Virus 1 and 2 (HSV-1 and HSV-2) and the Varicella Zoster Virus (VZV). The beta subfamily has cytomegalovirus (CMV) and human herpesvirus 6 and 7 (HHV-6 and HHV-7). Kaposi’s sarcoma-associated herpes virus (KSHV) and the Epstein-Bar virus (EBV) are in the gamma subfamily. These viruses are characterized by their practice of setting up latency in the host after primary infection. After entering to the nucleus through nuclear pores, these DNA viruses replicate their DNA and transcribe their genes inside the nucleus, preferably at ND10[4]. Therefore, the interaction of ND10 and herpesviruses occurs at the very early stage of infection.
The first virus found to be connected to ND10 was herpes simplex (HSV)-1. In 1993, Maul et al[28] were the first to discover that Vmw110 (ICP0-infected cell protein 0) localizes to ND10. Interestingly, they also showed that the C-terminal portion of ICP0, when linked to a heterologous protein, disrupts the normal distribution of PML. These observations presented the first link between processes involved in the control of cell growth and viral infection and latency. Later, Maul and Everett[11,26,34,87] systemically collaborated on the investigation of the interaction of ND10 and HSV-1, which collaboration typically combined the views from a cell biologist (Maul) and a virologist (Everett) on the direction to revelation of the phenomenal interaction of viral molecules and ND10. This, according to the authors’ opinion, could be the most important contribution to the ND10 field.
It is now known that ICP0 disrupts ND10 through mediating the loss of the SUMO-1-modified forms of PML and the subsequent proteasome-mediated degradation of the PML protein[14-15,88-90]. The results were consistent with the finding that PML residue lysine 160 is the SUMOylation site and the mutation of this residue makes PML resistant to degradation by ICP0[91]. ND10 function might not be so critical for HSV-1 lytic infection because ICP0-deleted HSV-1 can replicate well, especially at a high multiplicity of infection (MOI).
It was visualized that both parental and replicated HSV-1 amplicon genomes were in association with ND10 in live cells[92]. It is likely that the genomes situated at ND10 preferentially form viral replication compartments. Tang et al[12] further figured out that there exist minimal viral DNA sequences and viral proteins that are essential and sufficient for the replication of DNA and the transcription of RNA at ND10 by the virus. For HSV-1 we found that a specific viral DNA sequence, OriS, and the viral immediate-early proteins ICP4 and ICP27 are sufficient for a reporter gene placed in cis at the OriS sequence to transcribe RNA at ND10[12]. HSV-1 DNA replication results in formation of compartments in the nucleus; it has been shown that some, but not all, PML isoforms are recruited to the replication compartments[93]. Viral DNA replication compartments also contain many other viral and cellular proteins that have different functions, many of which are required for DNA replication, DNA repair, and DNA stabilization[94]. However, the function of ND10 proteins in the DNA replication compartments is not fully understood.
HSV-1 with deleted ICP0 has an obvious defect in viral gene expression and plaque formation in limited-passage human fibroblasts (though not in mouse fibroblast cells)[95,96]. This suggests both that ND10 have defensive effect on HSV-1 infection and that ICP0 can abolish the defensive effect of ND10 in human fibroblasts. ICP0 is a RING finger E3 ubiquitin ligase that induces the degradation of PML. Depletion of PML from human fibroblasts increases ICP0-null mutant HSV-1 gene expression but not to wild-type levels[96]. Another major ND10 protein, SP100, has a similar effect on ICP0-deleted HSV-1 gene expression[96]. It has been shown that all four SP100 isoforms stabilize ND10 and protect PML from ICP0-based hydrolysis[18]. Depletion of either all PML isoforms or all SP100 isoforms reduces the other constituent ND10 protein, suggesting that different ND10 proteins use different mechanisms to inhibit virus infection at the immediate-early stage of HSV-1 infection[18]. Simultaneous depletion of both PML and SP100 proteins complements the mutant virus to a greater degree, implying that PML and SP100 could have additive or synergistic effects on viral replication[96].
HSV-1 ICP0 might be important for the activation of lytic infection and the countering of the cell-mediated repression of viral gene expression by HSV-1. This repression is defended by preexisting cellular proteins, and those proteins function as intrinsic antiviral resistance or intrinsic defense. PML and SP100, as we discussed above, are two of the core components of ND10 and contribute to intrinsic resistance. But how about other ND10 proteins, such as, ATRX and Daxx? ATRX and Daxx are known to comprise components of a repressive chromatin-remodeling complex. It has been shown that the infection of ICP0-deleted HSV-1 (not wild-type HSV-1) can replicate at a greater level in both ATRX- and hDaxx-depleted cells than it can in normal cells[97], suggesting that ATRX and hDaxx act as a complex to play intrinsic antiviral resistance to HSV-1 infection, which is counteracted by ICP0.
Cytomegalovirus (CMV) infection differs from that of HSV-1 in host range and replication. HCMV can infect only human cells productively and causes diseases in humans only, and it replicates slowly in cell culture. HCMV is similar to HSV-1 in many ways: (1) setting up latency after primary infection in host; (2) sequential viral gene expression; and (3) viral DNA replication at ND10, preferentially. Following the studies of ND10 and HSV-1 interaction, many ND10 components have been demonstrated to have a repressive effect on CMV gene expression and viral replication (reviewed by Saffert and Kalejta[98]). The first ND10 protein investigated for its role in HCMV gene expression and viral replication was Daxx. In that study, Daxx was found to interact functionally with HCMV tegument protein pp71[16]. The Stamminger group[99] also investigated PML to see whether PML could have any effects on viral gene expression or on viral replication. After comparing HCMV replication in PML-kd or hDaxx-kd cells with that in normal cells, they revealed that immediate-early (IE) gene expression increased to a similar extent, regardless of whether PML or Daxx was depleted[98]. Their experimental results suggest that PML and Daxx might function using different mechanisms to suppress HCMV replication; double-knockdown cells depleted of both PML and hDaxx support the additive enhancement of HCMV infection in the replication efficacy of HCMV compared to that of single-knockdown cells[99]. Finally, they also found that the infection of SP100 knockdown (kd) cells with HCMV resulted in a significantly increased plaque-forming ability[99,100].
Like HSV-1, HCMV infection can also disrupt ND10, but the mechanisms of dispersing ND10 might be different. HSV-1 ICP0 induces the loss of the SUMO-1-modified forms of PML and the proteasome-mediated degradation of the PML protein[14,15,88-90]. However, in CMV-infected cells, PML is not degraded[13,101]. For cytomegaloviruses (including MCMV and HCMV), IE1 has been identified to disperse ND10 by an as yet unknown mechanism, but it is not able to degrade PML[27,101-104]. HCMV IE1’s induction of PML deSUMOylation, reported by Lee et al[101], needs to be investigated for MCMV IE1.
Species-specificity is one of the major characteristics of cytomegaloviruses (CMVs) and is the primary reason for the lack of a mouse model for the direct infection of human CMV (HCMV). It has been determined that CMV cross-species infections are blocked at the post-entry level by intrinsic cellular defense mechanisms[105,106], but few details are known. We discovered that ND10 of human cells is not disrupted by murine CMV (MCMV) and that the ND10 of mouse cells is not disrupted by HCMV[107], although the ND10-disrupting protein, immediate-early protein 1 (IE1), also colocalize with ND10 in cross-species infections[107]. In addition, we found that the UL131-repaired HCMV strain AD169 (vDW215-BADrUL131) can infect mouse cells to produce immediate-early (IE) and early (E) proteins but that neither DNA replication nor viral particles are detectable in mouse cells. Unrepaired AD169 can express only IE1 in mouse cells. In both HCMV-infected mouse cells and MCMV-infected human cells, the knocking-down of ND10 components (PML, Daxx, and SP100) resulted in significantly increased viral-protein production. Our observations provide evidence to support our hypothesis that ND10 and ND10 components might be important defensive factors against CMV cross-species infection.
The relationship of Epstein-Barr Virus (EBV) or Kaposi’s sarcoma-associated herpesvirus (KSHV) with ND10 has been less investigated than has that of HSV-1 or CMV (with ND10). The first study of the interaction of EBV and ND10 also came from the Maul group. Bell et al[108] studied the effect of the EBV on ND10, and its (EBV’s) spatial distribution in the nucleus of cells during latency and lytic reactivation. In EBV, latently-infected Burkitt’s lymphoma, lymphoblastoid, and D98/HR1 cells, ND10 were intact. Fluorescent in situ hybridization (FISH) revealed no association between viral episomes and ND10 during latency, implying that the maintenance replication of EBV, which depends on host cell proliferation, occurs independently of ND10. Upon lytic activation, ND10 become dispersed in cells expressing lytic proteins. Thus, latency does not require or induce the interaction of EBV and ND10 for transcription or replication, whereas lytic replication triggers the dispersion of ND10 proteins and occurs in close association with PML aggregates. The required movement of chromosome-attached latent EBV episomes to ND10 after reactivation from latency might include the physical release of chromosome-bound episomes. Only episomes that come in contact with ND10 after such a release might be able to begin the process of lytic replication[108]. The dispersion of ND10 by EBV in lytic infection might be through molecular and functional interactions between the EBV BZLF1 protein and the PML[109].
There are many fewer functional studies of ND10 proteins in EBV infection or reactivation than there are of those proteins in HSV-1 or CMV. So far, SP100 appears to be an effective ND10 protein that is related to EBV gene expression and viral reactivation. The EBV EBNA-LP protein is a potent gene-specific coactivator of the viral transcriptional activator, EBNA2. Ling et al[17] found that EBNA-LP interacts with ND10 protein SP100 and displaces SP100 and heterochromatin protein 1alpha (HP1alpha) from ND10. Their experimental results suggest that SP100 is a major mediator of EBNA-LP coactivation[17]. Recently, Tsai et al[110] showed that the EBV major tegument protein BNRF1 interacts with host-cell ND10 proteins and promotes viral early gene activation. Specifically, they demonstrated that BNRF1 interacts with the Daxx at ND10 and interferes with the formation of the Daxx-ATRX chromatin remodeling complex. Furthermore, the knockdown of Daxx and ATRX induces the reactivation of EBV from latency in infected lymphoblastoid cell lines, suggesting that Daxx and ATRX play a role in the regulation of viral gene expression and viral replication.
KSHV interacts with ND10 at the very early stage after reactivation. Although EBV and KSHV are so similar in many aspects that they are classified into the gama-herpesviral subfamily, they are different in many other characteristics. For example, KSHV might not be able to disrupt ND10, even though that particular claim is arguable. Wu et al[111] first studied the interaction of ND10 and KSHV and found that the KSHV protein, K8, interacted with PML; nevertheless, they clearly demonstrated that KSHV infection (latent or lytic) cannot disrupt ND10[111,112]. Our unpublished data also support their conclusion that ND10 are not dispersed by KSHV infection. However, this has been recently challenged by other studies[111,112]. In one such study, Marcos-Villar et al[113] stated that the KSHV protein LANA2 increased the levels of SUMOylated PML and induced the disruption of ND10 by a proteasome-mediated mechanism. They also reported that ND10 disruption needs both the integrity of a SUMO interaction motif (SIM) in LANA2 and the lysine 160 from PML. Moreover, they showed that the depletion of LANA2 in PEL cells led to an increase in the PML levels[111,112]. Arguably, KSHV’s dispersion of ND10 was not clearly shown in the published pictures. Interestingly, the authors didn’t cite the paper by Wu et al[111] that is intimately related to the subject.
As for the molecular and functional interaction of KSHV and ND10 proteins, only a few publications have been presented. First, Murakami et al[114] reported that Daxx is a LANA-binding protein and that interaction made LANA inhibit the repressive effect of Daxx on VEGF expression. Their results suggest that LANA contributes to the high expression of the vascular endothelial growth factor (VEGF) receptors in KS lesions by interfering with the interaction of Daxx and Ets-1[114]. Other studies showed the existence of an interaction between PML and KSHV proteins (including K8 and LANA2)[111,115]. The biological significance of this interaction is still unclear.
Adenovirus (Adv) is another virus that interacts with ND10. It was found that Adv infection changed the morphology of ND10 from being spherical punctate structures to being fibrous ones. This morphological change is caused by the molecular interaction of the Adv protein, E4 ORF3, and PML[116]. The other Adv protein found to interact with PML was E1A, which is an oncoprotein[116]. This study suggests that PML in ND10 might be involved in the cancerous consequence of Adv infection. More recently, a study by Hoppe et al[117] showed the PML isoform interacting directly and specifically with Adv E4 Orf3 in vitro and in vivo. Moreover, Hoppe et al[117] reconstructed ND10 in PML-null cells by inducing the transient transfection of different PML isoforms. They observed that only those ND10 formed from PML isoform II were morphologically changed by E4 Orf3. Their data suggest that the interaction of E4 Orf3 and PML isoform II is required for ND10 rearrangement[117].
The E4 ORF3 protein is required for Adv DNA replication when the cells are in the interferon (IFN)-induced antiviral state. ND10 prone proteins are all IFN-upregulated. This may reflect the fact that PML, Daxx, and SP100 are encoded by an interferon-stimulated gene. If so, can the interaction of E4 ORF3 and ND10 have any effect on Adv replication or viral gene expression? Ullman et al[118] demonstrate that the interaction of E4 ORF3 and ND10 antagonizes an innate antiviral response mediated by both PML and Daxx. Depleting any one of these proteins makes it possible to restore the replicative capacity of the virus using the E4 ORF3 protein deleted in the IFN-induced antiviral state. The interaction of Adv and ND10 has been also investigated with respect to SP100. Obviously, SP100 SUMOylation is also affected by E4 ORF3, which in part contributes to the morphological change of ND10[89]. We think that it is critical to investigate whether E4 ORF3’s interaction with ND10 plays a role in the oncogenesis of Adv. The interaction of E1A (an oncoprotein of Adv) and ND10 might be more important in the field of ND10 and viruses.
Using indirect immunofluorescence in combination with fluorescence in situ hybridization, Swindle et al[119] found that human papillomavirus (HPV) DNA replication is targeted to host nuclear domains that are active during the late S phase, when such domains are limited in number. It was also observed that E1 and E2 partially or completely colocalize with ND10. The observation suggests that HPV DNA amplification might be partially coupled to virion assembly[119]. Interestingly, Florin et al[120] showed that the minor capsid protein L2 of HPV interacted with ND10-associated proteins. They observed that (1) the PML was unaffected by L2; (2) SP100 was released from ND10 upon L2 expression; and (3) In contrast to SP100, Daxx was recruited to ND10 by L2 expression. These studies suggested that ND10 might be involved in HPV capsidation.
Simian virus 40 (SV40) is a small DNA virus. Like other DNA viruses, SV40 starts transcription and replication adjacent to ND10. In an early study, we identified a specific viral DNA sequence and its binding protein that determine the location of these synthetic activities at such restricted nuclear sites[121]. A beta-galactosidase gene was introduced into an expression vector that contains partial and overlapping SV40 sequences. Transcripts derived from control plasmids were found throughout the nucleus and at highly concentrated sites but not at ND10. SV40 genomic segments supported ND10-associated transcription only when the origin and the coding sequence for the large T antigen were present. When the large T-antigen coding sequence was eliminated but the T antigen was constitutively expressed in COS-7 cells, the viral origin was sufficient to localize transcription and replication to ND10. Large T antigen expressed from plasmids without the viral core origin did not bind or localize to ND10. Blocking of DNA replication prevented the accumulation of transcripts at ND10, indicating that only sites with replicating templates accumulated transcripts. Transcription at ND10 did not enhance total protein synthesis of plasmid transcripts. These findings suggest that viral transcription at ND10 may only be a consequence of viral genomes directed to ND10 for replication. Although plasmid transcription can take place anywhere in the nucleus, T-antigen-directed replication is apparently restricted to ND10[121].
The first RNA virus studied for its interaction with ND10 was the lymphocytic choriomeningitis virus (LCMV), a single strand RNA virus, but interestingly, the interaction occurs in the cytoplasm. In cells infected with LCMV, the viral zinc-finger (Z) protein forms large bodies primarily in the cytoplasm. Z protein can redistribute PML from the nucleus to the cytoplasm, and PML and Z protein colocalize in the cytoplasm[35,122]. The similar function of Z protein was also found in other viruses of Arenaviridae[35,122]. The interaction of PML and Z proteins may influence certain unique characteristics of arenavirus infection.
Another RNA virus is hepatitis delta virus (HDV). HDV is a single-stranded RNA virus and has only one coding region producing the hepatitis delta antigen (HDAg). HDAg is expressed in two isoforms, small (S-HDAg) and large (L-HDAg). S-HDAg is required for the replication of HDV, while L-HDAg inhibits viral replication and is required for the envelopment of the HDV genomic RNA by hepatitis B virus proteins[123]. Bell et al[124] found that over half of the L-HDAg domains were localized beside ND10. At later times, ND10-associated proteins such as PML were found in larger HDAg complexes, in which PML was found chiefly in the rims of the spheres. Other ND10 components (SP100, Daxx, and NDP55) were found in the centers of the spheres. HDV genomic RNA was distributed more uniformly throughout the nucleus, but nascent viral RNA colocalizes with L-HDAg and the transcriptional repressor PML. These results suggest that this RNA virus, like DNA viruses, can alter the distribution of ND10-associated proteins and preferably transcribe mRNA at ND10. It is not clear whether the ND10-associated proteins (PML) play a role in the regulation of HDV RNA synthesis.
As for human immunodeficiency virus (HIV), the results have been controversial. Bell et al[125] reported that no significant relationship was observed between ND10 or any of the following: HIV-1 DNA, transcription foci, and integrated DNA. Their results showed that HIV-1 did not modify ND10 at early or late times of infection[125]. However, Turelli et al[126] reported that incoming retroviral preintegration complexes trigger the exporting-mediated cytoplasmic export of PML. They further described how the HIV genome associates with PML before nuclear migration. Further experiments are needed to reveal the detailed interaction of HIV and ND10.
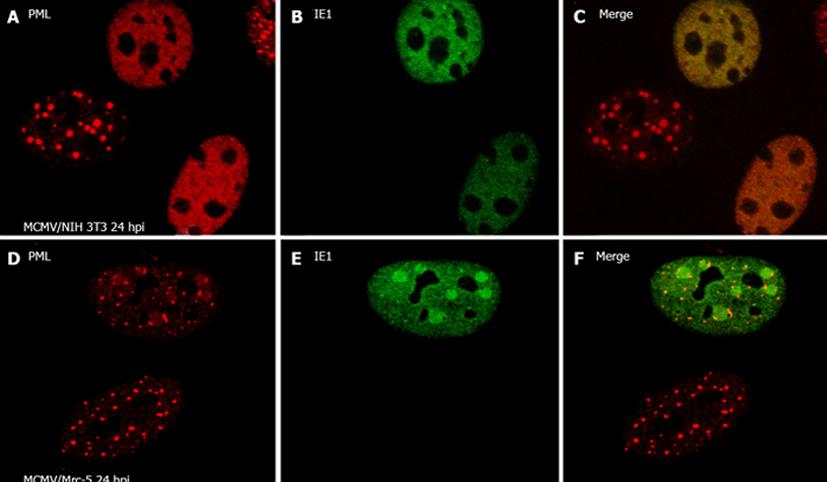
During viral infection, viruses and ND10 interact differently. The modification of ND10 structure can include (1) an increase in the size and number of ND10 per nucleus by double strand RNA viruses because their infection can induce IFN; (2) a change to the shape of ND10; (3) a decrease in the size or the number (of ND10) per nucleus; and (4) a total dispersal of ND10. Only CMV, EBV, and HSV have been clearly shown to disperse ND10. Here we take the MCMV infection as an example to show the real dispersing of ND10 (Figure 1). As we stated above, CMV infection is species-specific. Interestingly, we discovered that murine CMV (MCMV) infection in human cells cannot disperse ND10[107], suggesting the involvement of ND10 in species-specificity. We performed comparative IFA to analyze the ability of IE1 to disperse ND10 in cross-species-infected cells as opposed to in natively infected (mouse) cells. We infected wt-MCMV into both Mrc-5 cells and NIH3T3 cells for 24 h. Cells were fixed and permeabilized and stained with anti-PML to show ND10 (red, Figure 1A, D) and with anti-IE1 to show the distribution of IE1 (green, Figure 1B, E). As can be seen in the MCMV-infected mouse cells, IE1 was diffusely distributed in the nucleus at 24 hpi. Interestingly, the IE1 of MCMV formed domains (Figure 1E) in human cells and lost the ability to disperse ND10, their distribution being different from that found in MCMV-infected mouse cells (Figure 1A-C). There is no standard for judging the level of viral effect on ND10 structure because ND10 number or size can vary in different cell cycles. Therefore, one has to be careful to make conclusions of dispersing or disrupting ND10 by any viral infection or transient transfection.
A great deal of progress regarding the interaction of ND10 and viruses has been made in the past decades. A lot of questions are still left behind us, which makes the future direction of studies in the ND10-viruses field: (1) ND10 structure and ND10 protein are clearly related to cancer development (at least to some types of cancers). Therefore, the interaction of tumor viruses and ND10 should be the future focus of research in this field; (2) ND10 aggregate a lot of nuclear proteins that have different functions; we already know that SUMOylation is important for the formation of ND10. Are there any other nuclear functions needed for ND10 formation? Why do so many nuclear proteins meet in this place? (3) ND10 have been shown to be positioned beside SC35; SC35 is also related to transcribed RNA. What is the functional connection between ND10 and RNA? and (4) HIV DNA locates at SC35, not at ND10. HIV DNA is replicated and not integrated DNA (leftover). Given the fact that ND10 are located next to SC35, is it possible that they have any role with regard to HIV DNA?
ACKNOWLEDGEMENTS
This review is dedicated to the memory of Gerd Maul who was named the Father of ND10 in the international herpesvirus workshop. Dr. Maul was a talented, energetic, and creative cell biologist from the Wistar Institute (Philadelphia, PA) who passed away in August 2010, after devoting over 20 years of his life to ND10.
We apologize to those friends and colleagues whose primary work could not be cited because of space restrictions.
We thank Bob Ritchie of the Ponce School of Medicine and Health Sciences/RCMI Publications Office for his help with manuscript preparation.
P- Reviewers Berardinis PD, Devaux CA, Laassri M, Kawasaki H S- Editor Wen LL L- Editor A E- Editor Lu YJ
| 1. | Dundr M, Misteli T. Functional architecture in the cell nucleus. Biochem J. 2001;356:297-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 171] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 2. | Dundr M, Misteli T. Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000711. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 232] [Cited by in RCA: 212] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 3. | Zhao R, Bodnar MS, Spector DL. Nuclear neighborhoods and gene expression. Curr Opin Genet Dev. 2009;19:172-179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 131] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 4. | Maul GG. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays. 1998;20:660-667. [PubMed] |
| 5. | Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 684] [Cited by in RCA: 672] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 6. | Spector DL. SnapShot: Cellular bodies. Cell. 2006;127:1071. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 133] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 7. | Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF, Maul GG. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147:221-234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 669] [Cited by in RCA: 673] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 8. | Wang ZG, Ruggero D, Ronchetti S, Zhong S, Gaboli M, Rivi R, Pandolfi PP. PML is essential for multiple apoptotic pathways. Nat Genet. 1998;20:266-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 316] [Cited by in RCA: 348] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 9. | Trotman LC, Mosberger N, Fornerod M, Stidwill RP, Greber UF. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat Cell Biol. 2001;3:1092-1100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 228] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 10. | Ishov AM, Maul GG. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815-826. [PubMed] |
| 11. | Maul GG, Ishov AM, Everett RD. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 222] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 12. | Tang Q, Li L, Ishov AM, Revol V, Epstein AL, Maul GG. Determination of minimum herpes simplex virus type 1 components necessary to localize transcriptionally active DNA to ND10. J Virol. 2003;77:5821-5828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 13. | Tang Q, Maul GG. Mouse cytomegalovirus immediate-early protein 1 binds with host cell repressors to relieve suppressive effects on viral transcription and replication during lytic infection. J Virol. 2003;77:1357-1367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 93] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 14. | Chelbi-Alix MK, de Thé H. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene. 1999;18:935-941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 267] [Cited by in RCA: 276] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 15. | Gu H, Roizman B. The degradation of promyelocytic leukemia and Sp100 proteins by herpes simplex virus 1 is mediated by the ubiquitin-conjugating enzyme UbcH5a. Proc Natl Acad Sci USA. 2003;100:8963-8968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 110] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 16. | Hofmann H, Sindre H, Stamminger T. Functional interaction between the pp71 protein of human cytomegalovirus and the PML-interacting protein human Daxx. J Virol. 2002;76:5769-5783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 136] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 17. | Ling PD, Peng RS, Nakajima A, Yu JH, Tan J, Moses SM, Yang WH, Zhao B, Kieff E, Bloch KD. Mediation of Epstein-Barr virus EBNA-LP transcriptional coactivation by Sp100. EMBO J. 2005;24:3565-3575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 77] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 18. | Negorev DG, Vladimirova OV, Maul GG. Differential functions of interferon-upregulated Sp100 isoforms: herpes simplex virus type 1 promoter-based immediate-early gene suppression and PML protection from ICP0-mediated degradation. J Virol. 2009;83:5168-5180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 44] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 19. | Puvion-Dutilleul F, Venturini L, Guillemin MC, de Thé H, Puvion E. Sequestration of PML and Sp100 proteins in an intranuclear viral structure during herpes simplex virus type 1 infection. Exp Cell Res. 1995;221:448-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 20. | Roberts S, Hillman ML, Knight GL, Gallimore PH. The ND10 component promyelocytic leukemia protein relocates to human papillomavirus type 1 E4 intranuclear inclusion bodies in cultured keratinocytes and in warts. J Virol. 2003;77:673-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 21. | Saffert RT, Kalejta RF. Human cytomegalovirus gene expression is silenced by Daxx-mediated intrinsic immune defense in model latent infections established in vitro. J Virol. 2007;81:9109-9120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 126] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 22. | Saffert RT, Kalejta RF. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J Virol. 2006;80:3863-3871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 244] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 23. | Tavalai N, Papior P, Rechter S, Leis M, Stamminger T. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J Virol. 2006;80:8006-8018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 188] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 24. | Tavalai N, Stamminger T. New insights into the role of the subnuclear structure ND10 for viral infection. Biochim Biophys Acta. 2008;1783:2207-2221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 148] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 25. | Woodhall DL, Groves IJ, Reeves MB, Wilkinson G, Sinclair JH. Human Daxx-mediated repression of human cytomegalovirus gene expression correlates with a repressive chromatin structure around the major immediate early promoter. J Biol Chem. 2006;281:37652-37660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 133] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 26. | Everett RD, Maul GG. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062-5069. [PubMed] |
| 27. | Korioth F, Maul GG, Plachter B, Stamminger T, Frey J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res. 1996;229:155-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 190] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 28. | Maul GG, Guldner HH, Spivack JG. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J Gen Virol. 1993;74:2679-2690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 266] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 29. | Mocarski ES, Kemble GW, Lyle JM, Greaves RF. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc Natl Acad Sci USA. 1996;93:11321-11326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 180] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 30. | Stow ND, Stow EC. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J Gen Virol. 1986;67:2571-2585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 350] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 31. | de THE M, BERNHARD W. [Examination by electron microscope of the VX2 tumor of the domestic rabbit derived from the Shope papilloma]. Bull Assoc Fr Etud Cancer. 1960;47:570-584. [PubMed] |
| 32. | Ascoli CA, Maul GG. Identification of a novel nuclear domain. J Cell Biol. 1991;112:785-795. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 213] [Cited by in RCA: 239] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 33. | Negorev DG, Vladimirova OV, Kossenkov AV, Nikonova EV, Demarest RM, Capobianco AJ, Showe MK, Rauscher FJ, Showe LC, Maul GG. Sp100 as a potent tumor suppressor: accelerated senescence and rapid malignant transformation of human fibroblasts through modulation of an embryonic stem cell program. Cancer Res. 2010;70:9991-10001. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 34. | Maul GG, Everett RD. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223-1233. [PubMed] [DOI] [Full Text] |
| 35. | Borden KL, Campbelldwyer EJ, Carlile GW, Djavani M, Salvato MS. Two RING finger proteins, the oncoprotein PML and the arenavirus Z protein, colocalize with the nuclear fraction of the ribosomal P proteins. J Virol. 1998;72:3819-3826. [PubMed] |
| 36. | Bernardi R, Papa A, Pandolfi PP. Regulation of apoptosis by PML and the PML-NBs. Oncogene. 2008;27:6299-6312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 124] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 37. | Ching RW, Dellaire G, Eskiw CH, Bazett-Jones DP. PML bodies: a meeting place for genomic loci? J Cell Sci. 2005;118:847-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 107] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 38. | Dellaire G, Bazett-Jones DP. PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. Bioessays. 2004;26:963-977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 316] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 39. | de Thé H, Le Bras M, Lallemand-Breitenbach V. The cell biology of disease: Acute promyelocytic leukemia, arsenic, and PML bodies. J Cell Biol. 2012;198:11-21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 167] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 40. | Jensen K, Shiels C, Freemont PS. PML protein isoforms and the RBCC/TRIM motif. Oncogene. 2001;20:7223-7233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 360] [Cited by in RCA: 368] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 41. | Lallemand-Breitenbach V, Zhu J, Puvion F, Koken M, Honoré N, Doubeikovsky A, Duprez E, Pandolfi PP, Puvion E, Freemont P. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J Exp Med. 2001;193:1361-1371. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 393] [Cited by in RCA: 404] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 42. | Ishov AM, Vladimirova OV, Maul GG. Heterochromatin and ND10 are cell-cycle regulated and phosphorylation-dependent alternate nuclear sites of the transcription repressor Daxx and SWI/SNF protein ATRX. J Cell Sci. 2004;117:3807-3820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 121] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 43. | de Thé H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15; 17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1024] [Cited by in RCA: 1019] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 44. | Goddard AD, Borrow J, Freemont PS, Solomon E. Characterization of a zinc finger gene disrupted by the t(15; 17) in acute promyelocytic leukemia. Science. 1991;254:1371-1374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 356] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 45. | Kakizuka A, Miller WH, Umesono K, Warrell RP, Frankel SR, Murty VV, Dmitrovsky E, Evans RM. Chromosomal translocation t(15; 17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1081] [Cited by in RCA: 1084] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 46. | Melnick A, Licht JD. Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93:3167-3215. [PubMed] |
| 47. | Geng Y, Monajembashi S, Shao A, Cui D, He W, Chen Z, Hemmerich P, Tang J. Contribution of the C-terminal regions of promyelocytic leukemia protein (PML) isoforms II and V to PML nuclear body formation. J Biol Chem. 2012;287:30729-30742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 48. | Szostecki C, Guldner HH, Netter HJ, Will H. Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J Immunol. 1990;145:4338-4347. [PubMed] |
| 49. | Andreoni M, Faircloth M, Vugler L, Britt WJ. A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J Virol Methods. 1989;23:157-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 145] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 50. | Dent AL, Yewdell J, Puvion-Dutilleul F, Koken MH, de The H, Staudt LM. LYSP100-associated nuclear domains (LANDs): description of a new class of subnuclear structures and their relationship to PML nuclear bodies. Blood. 1996;88:1423-1426. [PubMed] |
| 51. | Guldner HH, Szostecki C, Schröder P, Matschl U, Jensen K, Lüders C, Will H, Sternsdorf T. Splice variants of the nuclear dot-associated Sp100 protein contain homologies to HMG-1 and a human nuclear phosphoprotein-box motif. J Cell Sci. 1999;112:733-747. [PubMed] |
| 52. | Seeler JS, Marchio A, Losson R, Desterro JM, Hay RT, Chambon P, Dejean A. Common properties of nuclear body protein SP100 and TIF1alpha chromatin factor: role of SUMO modification. Mol Cell Biol. 2001;21:3314-3324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 102] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 53. | Lehming N, Le Saux A, Schüller J, Ptashne M. Chromatin components as part of a putative transcriptional repressing complex. Proc Natl Acad Sci USA. 1998;95:7322-7326. [PubMed] |
| 54. | Zong RT, Das C, Tucker PW. Regulation of matrix attachment region-dependent, lymphocyte-restricted transcription through differential localization within promyelocytic leukemia nuclear bodies. EMBO J. 2000;19:4123-4133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 47] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 55. | Wasylyk C, Schlumberger SE, Criqui-Filipe P, Wasylyk B. Sp100 interacts with ETS-1 and stimulates its transcriptional activity. Mol Cell Biol. 2002;22:2687-2702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 52] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 56. | Grötzinger T, Jensen K, Guldner HH, Sternsdorf T, Szostecki C, Schwab M, Savelyeva L, Reich B, Will H. A highly amplified mouse gene is homologous to the human interferon-responsive Sp100 gene encoding an autoantigen associated with nuclear dots. Mol Cell Biol. 1996;16:1150-1156. [PubMed] |
| 57. | Yang X, Khosravi-Far R, Chang HY, Baltimore D. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell. 1997;89:1067-1076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 704] [Cited by in RCA: 720] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 58. | Pluta AF, Earnshaw WC, Goldberg IG. Interphase-specific association of intrinsic centromere protein CENP-C with HDaxx, a death domain-binding protein implicated in Fas-mediated cell death. J Cell Sci. 1998;111:2029-2041. [PubMed] |
| 59. | Hollenbach AD, Sublett JE, McPherson CJ, Grosveld G. The Pax3-FKHR oncoprotein is unresponsive to the Pax3-associated repressor hDaxx. EMBO J. 1999;18:3702-3711. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 176] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 60. | Michaelson JS, Bader D, Kuo F, Kozak C, Leder P. Loss of Daxx, a promiscuously interacting protein, results in extensive apoptosis in early mouse development. Genes Dev. 1999;13:1918-1923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 189] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 61. | Lin DY, Huang YS, Jeng JC, Kuo HY, Chang CC, Chao TT, Ho CC, Chen YC, Lin TP, Fang HI. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol Cell. 2006;24:341-354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 345] [Cited by in RCA: 337] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 62. | Wethkamp N, Hanenberg H, Funke S, Suschek CV, Wetzel W, Heikaus S, Grinstein E, Ramp U, Engers R, Gabbert HE. Daxx-beta and Daxx-gamma, two novel splice variants of the transcriptional co-repressor Daxx. J Biol Chem. 2011;286:19576-19588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 63. | Salomoni P, Guernah I, Pandolfi PP. The PML-nuclear body associated protein Daxx regulates the cellular response to CD40. Cell Death Differ. 2006;13:672-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 64. | Daniel MT, Koken M, Romagné O, Barbey S, Bazarbachi A, Stadler M, Guillemin MC, Degos L, Chomienne C, de Thé H. PML protein expression in hematopoietic and acute promyelocytic leukemia cells. Blood. 1993;82:1858-1867. [PubMed] |
| 65. | Dyck JA, Maul GG, Miller WH, Chen JD, Kakizuka A, Evans RM. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 596] [Cited by in RCA: 603] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 66. | Koken MH, Puvion-Dutilleul F, Guillemin MC, Viron A, Linares-Cruz G, Stuurman N, de Jong L, Szostecki C, Calvo F, Chomienne C. The t(15; 17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO J. 1994;13:1073-1083. [PubMed] |
| 67. | Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell. 1994;76:345-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 500] [Cited by in RCA: 505] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 68. | Zhu J, Koken MH, Quignon F, Chelbi-Alix MK, Degos L, Wang ZY, Chen Z, de Thé H. Arsenic-induced PML targeting onto nuclear bodies: implications for the treatment of acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1997;94:3978-3983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 349] [Cited by in RCA: 335] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 69. | Everett RD. Interactions between DNA viruses, ND10 and the DNA damage response. Cell Microbiol. 2006;8:365-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 111] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 70. | Gurrieri C, Capodieci P, Bernardi R, Scaglioni PP, Nafa K, Rush LJ, Verbel DA, Cordon-Cardo C, Pandolfi PP. Loss of the tumor suppressor PML in human cancers of multiple histologic origins. J Natl Cancer Inst. 2004;96:269-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 259] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 71. | Koken MH, Linares-Cruz G, Quignon F, Viron A, Chelbi-Alix MK, Sobczak-Thépot J, Juhlin L, Degos L, Calvo F, de Thé H. The PML growth-suppressor has an altered expression in human oncogenesis. Oncogene. 1995;10:1315-1324. [PubMed] |
| 72. | Terris B, Baldin V, Dubois S, Degott C, Flejou JF, Hénin D, Dejean A. PML nuclear bodies are general targets for inflammation and cell proliferation. Cancer Res. 1995;55:1590-1597. [PubMed] |
| 73. | Villagra NT, Navascues J, Casafont I, Val-Bernal JF, Lafarga M, Berciano MT. The PML-nuclear inclusion of human supraoptic neurons: a new compartment with SUMO-1- and ubiquitin-proteasome-associated domains. Neurobiol Dis. 2006;21:181-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 74. | Yamada M, Sato T, Shimohata T, Hayashi S, Igarashi S, Tsuji S, Takahashi H. Interaction between neuronal intranuclear inclusions and promyelocytic leukemia protein nuclear and coiled bodies in CAG repeat diseases. Am J Pathol. 2001;159:1785-1795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 56] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 75. | Ito K, Bernardi R, Morotti A, Matsuoka S, Saglio G, Ikeda Y, Rosenblatt J, Avigan DE, Teruya-Feldstein J, Pandolfi PP. PML targeting eradicates quiescent leukaemia-initiating cells. Nature. 2008;453:1072-1078. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 457] [Cited by in RCA: 438] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 76. | Salomoni P, Pandolfi PP. The role of PML in tumor suppression. Cell. 2002;108:165-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 411] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 77. | Negorev D, Maul GG. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene. 2001;20:7234-7242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 213] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 78. | Lehembre F, Müller S, Pandolfi PP, Dejean A. Regulation of Pax3 transcriptional activity by SUMO-1-modified PML. Oncogene. 2001;20:1-9. [PubMed] |
| 79. | Li H, Leo C, Zhu J, Wu X, O’Neil J, Park EJ, Chen JD. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol Cell Biol. 2000;20:1784-1796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 294] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 80. | Lin DY, Lai MZ, Ann DK, Shih HM. Promyelocytic leukemia protein (PML) functions as a glucocorticoid receptor co-activator by sequestering Daxx to the PML oncogenic domains (PODs) to enhance its transactivation potential. J Biol Chem. 2003;278:15958-15965. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 65] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 81. | D’Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, Gostissa M, Coen S, Marchetti A, Del Sal G. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol. 2002;4:11-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 525] [Cited by in RCA: 544] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 82. | Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi PP. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature. 2000;406:207-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 450] [Cited by in RCA: 537] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 83. | Lafarga M, Berciano MT, Pena E, Mayo I, Castaño JG, Bohmann D, Rodrigues JP, Tavanez JP, Carmo-Fonseca M. Clastosome: a subtype of nuclear body enriched in 19S and 20S proteasomes, ubiquitin, and protein substrates of proteasome. Mol Biol Cell. 2002;13:2771-2782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 109] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 84. | Boisvert FM, Hendzel MJ, Bazett-Jones DP. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J Cell Biol. 2000;148:283-292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 206] [Cited by in RCA: 214] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 85. | Wang J, Shiels C, Sasieni P, Wu PJ, Islam SA, Freemont PS, Sheer D. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J Cell Biol. 2004;164:515-526. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 177] [Cited by in RCA: 164] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 86. | Eskiw CH, Dellaire G, Mymryk JS, Bazett-Jones DP. Size, position and dynamic behavior of PML nuclear bodies following cell stress as a paradigm for supramolecular trafficking and assembly. J Cell Sci. 2003;116:4455-4466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 110] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 87. | Everett RD, Maul GG, Orr A, Elliott M. The cellular RING finger protein PML is not a functional counterpart of the herpes simplex virus type 1 RING finger protein Vmw110. J Gen Virol. 1995;76:791-798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 88. | Everett RD, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581-6591. [PubMed] |
| 89. | Müller S, Dejean A. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J Virol. 1999;73:5137-5143. [PubMed] |
| 90. | Parkinson J, Everett RD. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J Virol. 2000;74:10006-10017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 146] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 91. | Boutell C, Orr A, Everett RD. PML residue lysine 160 is required for the degradation of PML induced by herpes simplex virus type 1 regulatory protein ICP0. J Virol. 2003;77:8686-8694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 85] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 92. | Sourvinos G, Everett RD. Visualization of parental HSV-1 genomes and replication compartments in association with ND10 in live infected cells. EMBO J. 2002;21:4989-4997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 87] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 93. | Burkham J, Coen DM, Hwang CB, Weller SK. Interactions of herpes simplex virus type 1 with ND10 and recruitment of PML to replication compartments. J Virol. 2001;75:2353-2367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 56] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 94. | Wilkinson DE, Weller SK. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J Virol. 2004;78:4783-4796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 152] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 95. | Chee AV, Lopez P, Pandolfi PP, Roizman B. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J Virol. 2003;77:7101-7105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 155] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 96. | Everett RD, Parada C, Gripon P, Sirma H, Orr A. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J Virol. 2008;82:2661-2672. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 175] [Cited by in RCA: 177] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 97. | Lukashchuk V, Everett RD. Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J Virol. 2010;84:4026-4040. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 148] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 98. | Saffert RT, Kalejta RF. Promyelocytic leukemia-nuclear body proteins: herpesvirus enemies, accomplices, or both? Future Virol. 2008;3:265-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 43] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 99. | Tavalai N, Papior P, Rechter S, Stamminger T. Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J Virol. 2008;82:126-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 107] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 100. | Adler M, Tavalai N, Müller R, Stamminger T. Human cytomegalovirus immediate-early gene expression is restricted by the nuclear domain 10 component Sp100. J Gen Virol. 2011;92:1532-1538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 101. | Lee HR, Kim DJ, Lee JM, Choi CY, Ahn BY, Hayward GS, Ahn JH. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J Virol. 2004;78:6527-6542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 120] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 102. | Ahn JH, Hayward GS. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599-4613. [PubMed] |
| 103. | Kang H, Kim ET, Lee HR, Park JJ, Go YY, Choi CY, Ahn JH. Inhibition of SUMO-independent PML oligomerization by the human cytomegalovirus IE1 protein. J Gen Virol. 2006;87:2181-2190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 104. | Kelly C, Van Driel R, Wilkinson GW. Disruption of PML-associated nuclear bodies during human cytomegalovirus infection. J Gen Virol. 1995;76:2887-2893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 81] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 105. | Lafemina RL, Hayward GS. Differences in cell-type-specific blocks to immediate early gene expression and DNA replication of human, simian and murine cytomegalovirus. J Gen Virol. 1988;69:355-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 89] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 106. | Tang Q, Maul GG. Mouse cytomegalovirus crosses the species barrier with help from a few human cytomegalovirus proteins. J Virol. 2006;80:7510-7521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 107. | Cosme RC, Martínez FP, Tang Q. Functional interaction of nuclear domain 10 and its components with cytomegalovirus after infections: cross-species host cells versus native cells. PLoS One. 2011;6:e19187. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 108. | Bell P, Lieberman PM, Maul GG. Lytic but not latent replication of epstein-barr virus is associated with PML and induces sequential release of nuclear domain 10 proteins. J Virol. 2000;74:11800-11810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 86] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 109. | Bowling BL, Adamson AL. Functional interactions between the Epstein-Barr virus BZLF1 protein and the promyelocytic leukemia protein. Virus Res. 2006;117:244-253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 110. | Tsai K, Thikmyanova N, Wojcechowskyj JA, Delecluse HJ, Lieberman PM. EBV tegument protein BNRF1 disrupts DAXX-ATRX to activate viral early gene transcription. PLoS Pathog. 2011;7:e1002376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 108] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 111. | Wu FY, Ahn JH, Alcendor DJ, Jang WJ, Xiao J, Hayward SD, Hayward GS. Origin-independent assembly of Kaposi’s sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J Virol. 2001;75:1487-1506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 137] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 112. | Katano H, Ogawa-Goto K, Hasegawa H, Kurata T, Sata T. Human-herpesvirus-8-encoded K8 protein colocalizes with the promyelocytic leukemia protein (PML) bodies and recruits p53 to the PML bodies. Virology. 2001;286:446-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 113. | Marcos-Villar L, Campagna M, Lopitz-Otsoa F, Gallego P, González-Santamaría J, González D, Rodriguez MS, Rivas C. Covalent modification by SUMO is required for efficient disruption of PML oncogenic domains by Kaposi’s sarcoma-associated herpesvirus latent protein LANA2. J Gen Virol. 2011;92:188-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 114. | Murakami Y, Yamagoe S, Noguchi K, Takebe Y, Takahashi N, Uehara Y, Fukazawa H. Ets-1-dependent expression of vascular endothelial growth factor receptors is activated by latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus through interaction with Daxx. J Biol Chem. 2006;281:28113-28121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 47] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 115. | Marcos-Villar L, Lopitz-Otsoa F, Gallego P, Muñoz-Fontela C, González-Santamaría J, Campagna M, Shou-Jiang G, Rodriguez MS, Rivas C. Kaposi’s sarcoma-associated herpesvirus protein LANA2 disrupts PML oncogenic domains and inhibits PML-mediated transcriptional repression of the survivin gene. J Virol. 2009;83:8849-8858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 66] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 116. | Carvalho T, Seeler JS, Ohman K, Jordan P, Pettersson U, Akusjärvi G, Carmo-Fonseca M, Dejean A. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J Cell Biol. 1995;131:45-56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 213] [Cited by in RCA: 220] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 117. | Hoppe A, Beech SJ, Dimmock J, Leppard KN. Interaction of the adenovirus type 5 E4 Orf3 protein with promyelocytic leukemia protein isoform II is required for ND10 disruption. J Virol. 2006;80:3042-3049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 57] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 118. | Ullman AJ, Hearing P. Cellular proteins PML and Daxx mediate an innate antiviral defense antagonized by the adenovirus E4 ORF3 protein. J Virol. 2008;82:7325-7335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 75] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 119. | Swindle CS, Zou N, Van Tine BA, Shaw GM, Engler JA, Chow LT. Human papillomavirus DNA replication compartments in a transient DNA replication system. J Virol. 1999;73:1001-1009. [PubMed] |
| 120. | Florin L, Schäfer F, Sotlar K, Streeck RE, Sapp M. Reorganization of nuclear domain 10 induced by papillomavirus capsid protein l2. Virology. 2002;295:97-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 84] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 121. | Tang Q, Bell P, Tegtmeyer P, Maul GG. Replication but not transcription of simian virus 40 DNA is dependent on nuclear domain 10. J Virol. 2000;74:9694-9700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 68] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 122. | Borden KL, Campbell Dwyer EJ, Salvato MS. An arenavirus RING (zinc-binding) protein binds the oncoprotein promyelocyte leukemia protein (PML) and relocates PML nuclear bodies to the cytoplasm. J Virol. 1998;72:758-766. [PubMed] |
| 123. | Chen R, Linnstaedt SD, Casey JL. RNA editing and its control in hepatitis delta virus replication. Viruses. 2010;2:131-146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 124. | Bell P, Brazas R, Ganem D, Maul GG. Hepatitis delta virus replication generates complexes of large hepatitis delta antigen and antigenomic RNA that affiliate with and alter nuclear domain 10. J Virol. 2000;74:5329-5336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 125. | Bell P, Montaner LJ, Maul GG. Accumulation and intranuclear distribution of unintegrated human immunodeficiency virus type 1 DNA. J Virol. 2001;75:7683-7691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 54] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 126. | Turelli P, Doucas V, Craig E, Mangeat B, Klages N, Evans R, Kalpana G, Trono D. Cytoplasmic recruitment of INI1 and PML on incoming HIV preintegration complexes: interference with early steps of viral replication. Mol Cell. 2001;7:1245-1254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 180] [Article Influence: 7.5] [Reference Citation Analysis (0)] |