Published online Apr 19, 2024. doi: 10.5498/wjp.v14.i4.563
Peer-review started: October 11, 2023
First decision: December 26, 2023
Revised: January 9, 2024
Accepted: February 28, 2024
Article in press: February 28, 2024
Published online: April 19, 2024
Processing time: 188 Days and 23.7 Hours
Alzheimer’s disease (AD) is a neurodegenerative condition characterized by oxi
To investigate potential Tan-IIA neuroprotective effects in AD and to elucidate their underlying mechanisms.
Hematoxylin and eosin staining was utilized to analyze structural brain tissue morphology. To assess changes in oxidative stress and neuroinflammation, we performed enzyme-linked immunosorbent assay and western blotting. Addi
In vivo, Tan-IIA treatment improved neuronal morphology and attenuated oxidative stress and neuroinflammation in the brain tissue of AD mice. In vitro experiments showed that Tan-IIA dose-dependently ameliorated the amyloid-beta 1-42-induced reduction of neural stem cell viability, apoptosis, oxidative stress, and neuroinflammation. In this process, the lncRNA NEAT1 - a potential therapeutic target - is highly expressed in AD mice and downregulated via Tan-IIA treatment. Mechanistically, NEAT1 promotes the transcription and translation of Rab22a via miR-291a-3p, which activates nuclear factor kappa-B (NF-κB) signaling, leading to activation of the pro-apoptotic B-cell lymphoma 2-associated X protein and inhibition of the anti-apoptotic B-cell lymphoma 2 protein, which exacerbates AD. Tan-IIA intervention effectively blocked this process by inhibiting the NEAT1/miR-291a-3p/Rab22a axis and NF-κB signaling.
This study demonstrates that Tan-IIA exerts neuroprotective effects in AD by modulating the NEAT1/miR-291a-3p/Rab22a/NF-κB signaling pathway, serving as a foundation for the development of innovative approaches for AD therapy.
Core Tip: Tanshinone IIA (Tan-IIA), a compound isolated from Salvia miltiorrhiza, demonstrates neuroprotective effects against Alzheimer’s disease (AD). This study reveals that Tan-IIA improves neuronal health, reduces oxidative stress and neuroinflammation, and promotes neural stem cell viability. Importantly, it targets the nuclear-enriched abundant transcript 1/microRNA-291a-3p/member RAS oncogene family Rab22a/nuclear factor kappa-B pathway, offering a potential therapeutic avenue for AD.
- Citation: Yang LX, Luo M, Li SY. Tanshinone IIA improves Alzheimer’s disease via RNA nuclear-enriched abundant transcript 1/microRNA-291a-3p/member RAS oncogene family Rab22a axis. World J Psychiatry 2024; 14(4): 563-581
- URL: https://www.wjgnet.com/2220-3206/full/v14/i4/563.htm
- DOI: https://dx.doi.org/10.5498/wjp.v14.i4.563
Alzheimer’s disease (AD), the most common cause of dementia worldwide, is a progressive neurological condition that affects millions of individuals and presents a serious public health issue[1,2]. The amyloid-beta 1-42 (Aβ1-42) peptide is a primary component of the amyloid plaques found in the brains of individuals with AD. It is believed to play a critical role in the neuropathology of AD by initiating a cascade of events that leads to neuronal dysfunction and death[3]. Memory loss and cognitive decline are caused by the buildup of Aβ plaques, neurofibrillary tangles, and synaptic and neuronal loss in individuals with AD[4-6]. Despite extensive research on AD, current treatment options only focus on symptomatic relief rather than disease remission, and the development of new therapeutic agents and targets is required to improve the disease prognosis.
Growing evidence points to neuroinflammation and oxidative stress being key factors in the etiology of AD[7,8]. Nitric oxide (NO) gas is produced in greater amounts when there is an excessive buildup of reactive oxygen species (ROS) in the body. An increased production of NO causes oxidative stress, which damages neurons and exacerbates AD[9]. This process lowers the levels of the antioxidants superoxide dismutase (SOD) and glutathione (GSH), making them less effective at scavenging ROS[10,11]. In addition, the release of neuroinflammatory factors, such as tumor necrosis factor-alpha (TNF-α), interleukin (IL)-1β, and IL-6, exacerbates neuronal damage and contributes to the progression of AD[12,13]. Long non-coding RNA (lncRNA) furthermore acts as a molecular sponge for microRNA (miR, miRNA) adsorption to regulate miRNA expression[14], and such changes in miRNA expression critically affect the transcription and translation of downstream targets[15]. For example, a study by Zhao et al[16] showed that the lncRNA nuclear-enriched abundant transcript 1 (NEAT1) promotes the development of AD through the miR-124/beta-site amyloid precursor protein-cleaving enzyme axis. In addition, miR-291a-3p, which is associated with inflammation, oxidative stress, and apoptosis, may be downregulated in neural injury[17,18]; however, its role and mechanism of function in AD remain unclear.
RAB22A, member RAS oncogene family (Rab22a) is another important tumor regulator[19,20], but its mechanism of action in AD is unclear. Its role in promoting neuroinflammation and oxidative stress[21] warrants the inclusion of Rab22a in the present study to explore its role in AD, especially since it affects Aβ accumulation[22]. Additionally, the nuclear factor kappa-B (NF-κB) pathway is triggered by the neuroinflammatory response in AD, which can aggravate the disease progression. The role of the B-cell lymphoma 2 (Bcl-2) protein in this process is significantly inhibited in contrast to that of pro-apoptotic proteins; the greater the ratio of Bcl-2/Bcl-2-associated X protein (Bax), the greater the anti-apoptotic ability. However, the role of Rab22a on NF-κB and the regulation of downstream pro- and anti-apoptotic proteins require elucidation.
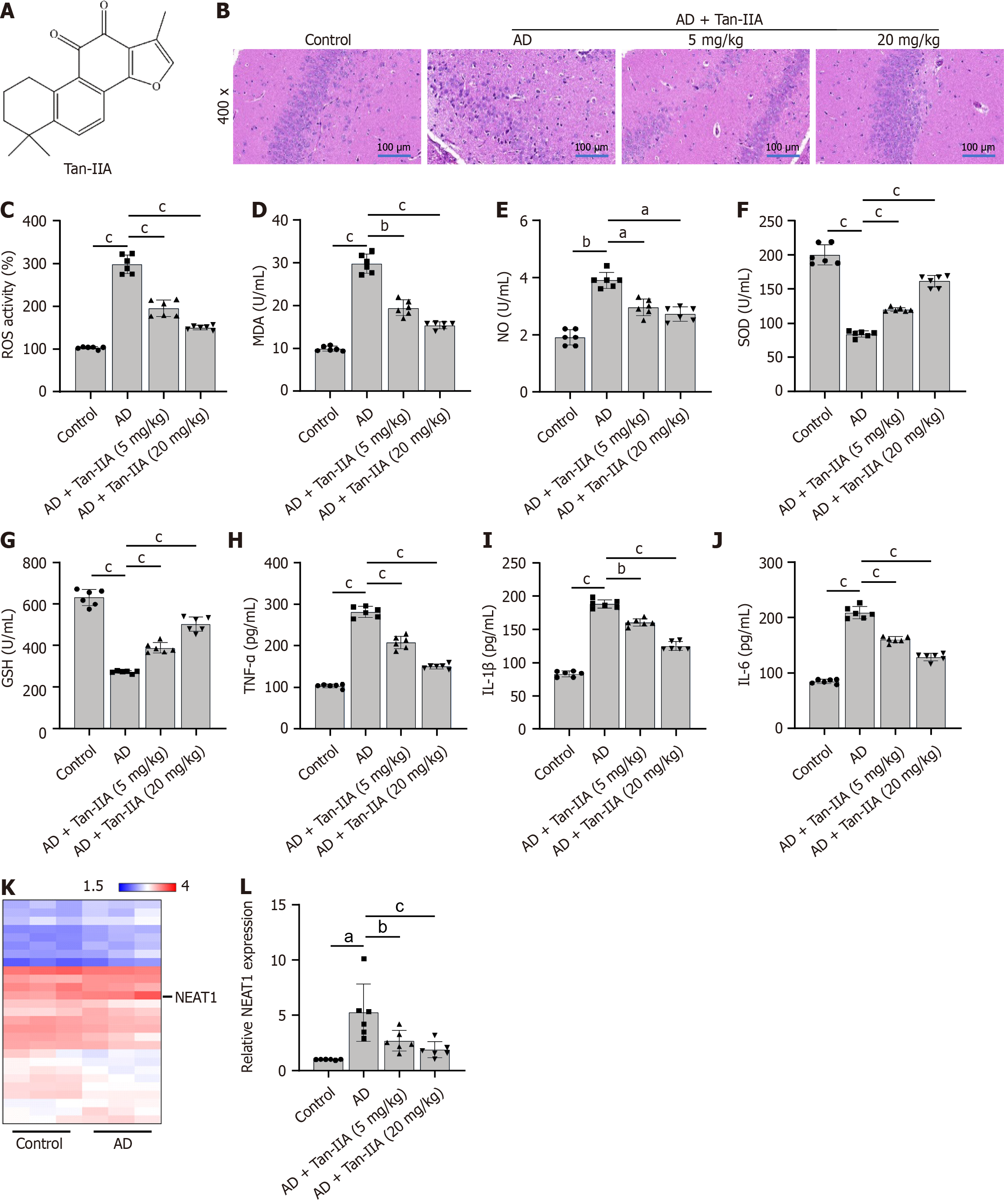
Salvia miltiorrhiza, also known as danshen or red sage, produces a bioactive molecule called tanshinone IIA (Tan-IIA; chemical structure depicted in Figure 1A) with reportedly anti-inflammatory, antioxidant, and anti-apoptotic activities[23,24]. Previous studies demonstrated that Tan-IIA reduces oxidative stress and neuroinflammation, two factors known to worsen AD[25,26]. Although Tan-IIA has been shown to improve AD, the mechanism underlying this improvement is not well understood.
In this study, we investigate processes involving the NEAT1/miR-291a-3p/Rab22a/NF-κB signaling pathway that may underlie the neuroprotective benefits of Tan-IIA, using both in vivo and in vitro models of AD. Our results help to clarify the intricate regulatory network that controls oxidative stress and neuroinflammation connected to AD and may serve as a foundation for the creation of new treatment options for the disease.
The Guangdong Medical Experimental Animal Center (Guangzhou, China) provided all mice; AD was induced in some mice via a bilateral injection of Aβ1-42 oligomers into the CA1 region of the hippocampus, as previously described[27]. All animal treatments were approved by the Guangxi Medical University’s Animal Care and Use Committee [approval no. 2021(KY-E-292); Nanning, China] and were carried out in accordance with the Guide for the Care and Use of Laboratory Animals.
The mice were maintained under a 12-h light/dark cycle and provided with unlimited access to food and water. Mice were randomly placed in one of four treatment groups (n = 6 each): A control group, comprising healthy mice; an AD group, in which AD was induced without further treatment; and two Tan-IIA groups, with AD mice receiving either 5 or 20 mg/kg of Tan-IIA (HY-N0135; MedChemExpress LLC, NJ, United States). Tan-IIA was dissolved in dimethyl sulfoxide (DMSO; Solarbio, Beijing, China) and administered intraperitoneally once daily for 4 wk in mice of the relevant treatment groups. Subsequently, the mice were anesthetized and sacrificed, and their brain tissues were removed and placed in Hank’s balanced salt solution (Gibco, Thermo Fisher Scientific, MA, United States) with 1% penicillin-streptomycin (Gibco) to isolate the neural stem cells (NSCs). Tissues from the hippocampus and subventricular zone were carefully removed, chopped, and subjected to enzymatic digestion with trypsin-EDTA (Gibco) for 15 min at 37 °C. Fetal bovine serum (Gibco) was used to terminate digestion, after which the cell solution was filtered through a 40 m mesh cell strainer (BD Biosciences, CA, United States) to remove debris. The cells were centrifuged at 300 rpm for 5 min, and the resulting cell pellet was resuspended and cultured in NSC culture medium (Procell, Wuhan, China) at 37 °C and in 5% CO2. The medium was replaced every 2 d, and the NSCs were passaged until 80%-90% confluence was reached before being used for subsequent experiments.
The obtained NSCs were incubated with bovine serum albumin for 30 min and incubated further (overnight, at 4 °C and in the dark) with microtubule-associated protein 2 (MAP2; 1:500, ab254264; Abcam, Cambridge, United Kingdom), β III tubulin (3 μg/mL, ab18207; Abcam), and nuclear factor erythroid 2-related factor 2 (Nrf2; 1:100, ab62352; Abcam) antibodies. Thereafter, the cells were incubated at 37 °C for 1 h with either goat anti-rabbit Alexa Fluor® 488 (1:250, ab150077; Abcam) or 647-conjugated secondary antibodies. Finally, the NSCs were mounted on microscope slides using a gold antifade medium containing DAPI stain (ProLong™, Thermo Fisher Scientific). MAP2 and βIII tubulin were used to confirm the isolation of NSCs, and Nrf2 was used to analyze their subcellular localization.
NSCs at 1, 2, 5, and 10 μM were treated for 24 h with Aβ1-42 (Sigma-Aldrich, St Louis, MO, United States) dissolved in DMSO; DMSO alone was used as treatment for a control group. Before Aβ1-42 treatment, the NSCs were pretreated with Tan-IIA at doses of 1, 5, 10, 20, and 40 μM for 1 h. The NEAT1 overexpression plasmid (ov-NEAT1), Rab22a small interfering RNAs (siRNAs) (si-Rab22a-1, si-Rab22a-2, and si-Rab22a-3), and miR-291a-3p mimic/inhibitor were purchased from Sangon Biotech Co., Ltd. (Shanghai, China) and transfected into the NSCs (following their Aβ1-42 treatment) using Lipofectamine 3000 Reagent (Invitrogen, Waltham, MA, United States); all the relevant sequences are listed in Table 1.
| Sequences of si-Rab22a 5’-3’ | |
| si-Rab22a-1 | GGAAATGATCACAAGTAGAGG |
| si-Rab22a-2 | GGAAATGGTAATAAAGACATA |
| si-Rab22a-3 | CGATCTTACTGATGTCAGAGA |
| si-NC | TTCTCCGAACGTGTCACGTTT |
| Sequences of the miR-291a-3p mimic and inhibitor 5’-3’ | |
| Mimic NC | ATTGATTTGTTCCGAAGGCCCT |
| miR-291a-3p mimic | AAAGTGCTTCCACTTTGTGTGC |
| Inhibitor NC | TCGCTCTATCCTGATCGAATGAA |
| miR-291a-3p inhibitor | GCACACAAAGTGGAAGCACTTT |
Mouse brains were sectioned into 5 μm thick slices, which were immersed in paraffin and preserved in 4% paraformaldehyde. The sections were deparaffinized, rehydrated, and submitted to hematoxylin and eosin (HE) staining. Histological images were captured at a 400 × magnification using an Olympus light microscope (IX73, Olympus, Shinjuku, Tokyo, Japan).
Enzyme-linked immunosorbent assay (ELISA) kits (Solarbio) were used in accordance with the manufacturer’s instructions to determine the levels of malondialdehyde (MDA), NO, SOD, GSH, TNF-α, IL-1, and IL-6 in brain tissue homogenates and NSC supernatants. Absorbance was determined at 450 nm using a microplate reader (Thermo Fisher Scientific).
The GSE150696 dataset was analyzed using the DataSet analysis tool (GEO2R, Gene Expression Omnibus 2). The differential expression of lncRNA between the prefrontal cortex of patients with AD and that of elderly individuals without neurological or psychiatric diseases was normalized for log2 |fold change| > 1.5 and P < 0.05.
Cells were collected, rinsed with phosphate buffered saline, and then resuspended in binding buffer in order to analyze apoptosis. Following the manufacturer’s instructions, they were stained with propidium iodide (PI) and Annexin V-fluorescein isothiocyanate (FITC) using an Apoptosis Detection Kit (BD Biosciences, California, United States). Cells were subjected to a 15-min at dark incubation period with Annexin V-FITC and PI at 21 °C, followed by analysis using a BD Biosciences flow cytometry.
First, 10 M of 2’,7’-dichlorofluorescin (DCF) diacetate (Sigma-Aldrich) was added to the brain tissue homogenates and NSCs lysates, which were then allowed to incubate in the dark for 30 min at 37 °C. The quick oxidation of DCF in the presence of ROS produces extreme fluorescence, of which the intensity was measured with a fluorescence microplate reader (Thermo Fisher Scientific) at excitation and emission wavelengths of 485 and 530 nm, respectively.
Following treatment of the NSCs with Aβ1-42 and Tan-IIA, their cell viability was measured using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Briefly, the treated NSCs were seeded into 96-well plates at a density of 3 × 104 cells per well. Each well was supplemented with 20 μL of MTT solution (5 mg/mL), and the plates underwent a 4 h incubation period at 37 °C. After carefully removing the media, DMSO (150 μL) was used to dissolve the formazan crystals that the living cells had generated. Cell viability was then determined (and compared to that of the untreated control group) using a microplate reader to detect absorbance at 570 nm.
Total RNA was extracted using TRIzol reagent (Invitrogen). A PrimeScript RT Reagent Kit (Takara Bio, Shiga, Japan) was used to construct complementary DNA from 1 μg of total RNA. Polymerase chain reaction (PCR) was conducted using a QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific) with SYBR Green PCR Master Mix (Applied Biosystems, CA, United States). Thermal cycling conditions were as follows: 40 cycles of denaturation at 95 °C for 15 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s. A preliminary denaturation was then performed at 95 °C for 10 min. The expression levels of NEAT1, Rab22a, and miR-291a-3p were assessed using the 2-ΔΔCt method, with GAPDH or U6 as internal controls for standardization. Primer sequences for all RNAs are shown in Table 2.
| Primers | Forward primer 5’-3’ | Reverse primer 5’-3’ |
| NEAT1 | TGGAGATTGAAGGCGCAAGT | AAGCACGGAACCTAGGCAAA |
| miR-291a-3p | ACACTCCAGCTGGGAAAGTGCTTCCACTTT | CTCAACTGGTGTCGTGGA |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
| Rab22a | ATCAATCCAACCATAGGGGCAT | TTGGTGCCAATGCACGAAATC |
| GAPDH | GTGGCAAAGTGGAGATTGTTGCC | GATGATGACCCGTTTGGCTCC |
Nuclear and cytoplasmic fractions were separated according to the manufacturer’s instructions using an NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Fisher Scientific). The cell pellet was briefly resuspended in CER I buffer obtained from the kit, followed by the addition of CER II buffer. The mixture was vortexed and then rested on ice for an additional 5 min. The cytoplasmic fraction (supernatant) and nuclear pellet were separated from the homogenate via centrifugation at 16000 rpm for 5 min at 4° C. The purified nuclear and cytoplasmic fractions were then subjected to reverse transcription quantitative PCR (RT-qPCR).
Biotin-labeled NEAT1 RNA (bio-NEAT1) and mutant NEAT1 RNA (bio-mut) were synthesized by Sangon Biotech. The NSCs lysates were incubated overnight with bio-NEAT1, bio-mut, or bio-NC at 4 °C. Streptavidin-coated magnetic beads were added to and incubated with the reaction mixture for 1 h at 4° C to allow the formation of RNA-protein complexes. The beads were then washed, and bound RNA was eluted from them for RT-qPCR analysis to assess enrichment.
The miR-291a-3p binding sites from the 3’ untranslated regions of NEAT1 and Rab22a were cloned into a psiCHECK-2 dual-luciferase reporter vector (Promega, WI, United States). Using Lipofectamine 3000 reagent (Invitrogen), NSCs were then co-transfected with the reporter plasmids with miR-291a-3p mimic or mimic NC. Luciferase activity was assessed after 48 h using the Dual-Luciferase Reporter Assay System (Promega), in accordance with the manufacturer’s instructions. Activity of the Renilla luciferase gene was used to normalize firefly luciferase activity.
Total protein was extracted from the mouse brain tissues and NSCs using RIPA lysis buffer (Abcam) along with protease and phosphatase inhibitors (Thermo Fisher Scientific). The protein content was determined using a BCA Protein Assay Kit (Thermo Fisher Scientific). Similar quantities of proteins were separated via sodium-dodecyl sulfate gel electrophoresis (Millipore, MA, United States) and then transferred to polyvinylidene fluoride membranes. The membranes were incubated overnight and at 4 °C with primary antibodies (all from Abcam, Cambridge, United Kingdom) against Rab22a (1:1000, ab137093), p65 (1:1000, ab32536), phospho (p)-p65 (1:1000, ab76302), Bax (1:1000, ab32503), Bcl-2 (1:2000, ab182858), and GAPDH (1:2000, ab181602). Next, the membranes were treated with HRP-conjugated secondary antibodies (1:3000, Abcam) for 1 h at 22 °C. Electrochemiluminescence Western Blotting Substrate (NIH, Bethesda, MD, United States) was used to visualize the protein bands, which were quantified using ImageJ software.
Data are presented as the mean and standard deviation. GraphPad Prism 8.0 (GraphPad Software, CA, United States) was used for all statistical analyses. One-way analysis of variance was used to evaluate group differences, followed by Tukey’s post-hoc test. Statistical significance was set at P < 0.05.
In the healthy control group, neurons were neatly arranged in brain tissues, displaying intact cell structures and clearly visible cell membranes and nuclei. In contrast, the neurons in AD mouse brain tissues showed extreme disarray and irregularities in size and shape, with a drastic reduction in cell number and blurred cell structures. These problems were significantly improved in the treatment group that received 20 mg/kg of Tan-IIA, which proved to be more effective than the 5 mg/kg treatment (Figure 1B). Furthermore, the ELISA showed that Tan-IIA effectively alleviated the elevated oxidative stress (via reduced ROS, MDA, and NO and increased SOD and GSH levels) and neuroinflammation (via reduced TNF-α, IL-1β, and IL-6 levels) in the AD mouse brain tissue (Figure 1C-J), confirming that 20 mg/kg of Tan-IIA was more effective than 5 mg/kg. A total of 32 associated lncRNAs were identified in the GSE150696 dataset (Table 3), with NEAT1 displaying the highest fold-change to confirm it as a potential therapeutic target (Figure 1K). This lncRNA was both highly expressed in mouse brain tissues and reduced by Tan-IIA treatment (Figure 1L).
| Gene symbol | Fold change | P value |
| NEAT1 | 2.19 | 0.0170 |
| LINC01377 | 1.98 | 0.0248 |
| EYA transcriptional coactivator and phosphatase 3- intronic transcript 1 | 1.95 | 0.0027 |
| LINC01354 | 1.68 | 0.0119 |
| LINC00537 | 1.64 | 0.0498 |
| LINC01441 | 1.63 | 0.0082 |
| LINC01094 | 1.63 | 0.0328 |
| Cancer susceptibility 16 | 1.6 | 0.0376 |
| LINC01101 | 1.55 | 0.0342 |
| LINC00327 | 1.54 | 0.0028 |
| LINC00323 | 1.54 | 0.0459 |
| LINC00644 | 1.53 | 0.0119 |
| LINC00358 | 1.53 | 0.0411 |
| LINC00347 | 1.51 | 0.0409 |
| LINC00641 | -1.53 | 0.0332 |
| LINC00294 | -1.55 | 0.0471 |
| Maternally expressed 8 | -1.56 | 0.0073 |
| Prader-Willi region non-protein coding RNA 1 | -1.59 | 0.0005 |
| LINC00672 | -1.66 | 0.0010 |
| LINC00969 | -1.72 | 0.0302 |
| Prostate androgen-regulated transcript 1 | -1.8 | 0.0049 |
| LINC00622 | -1.8 | 0.0175 |
| LINC00507 | -1.81 | 0.0000625 |
| LINC00403 | -1.84 | 0.0043 |
| LINC01128 | -1.87 | 0.0179 |
| LINC00889 | -1.92 | 0.0113 |
| LINC00473 | -2.43 | 0.0323 |
| LINC00622 | -2.5 | 0.0037 |
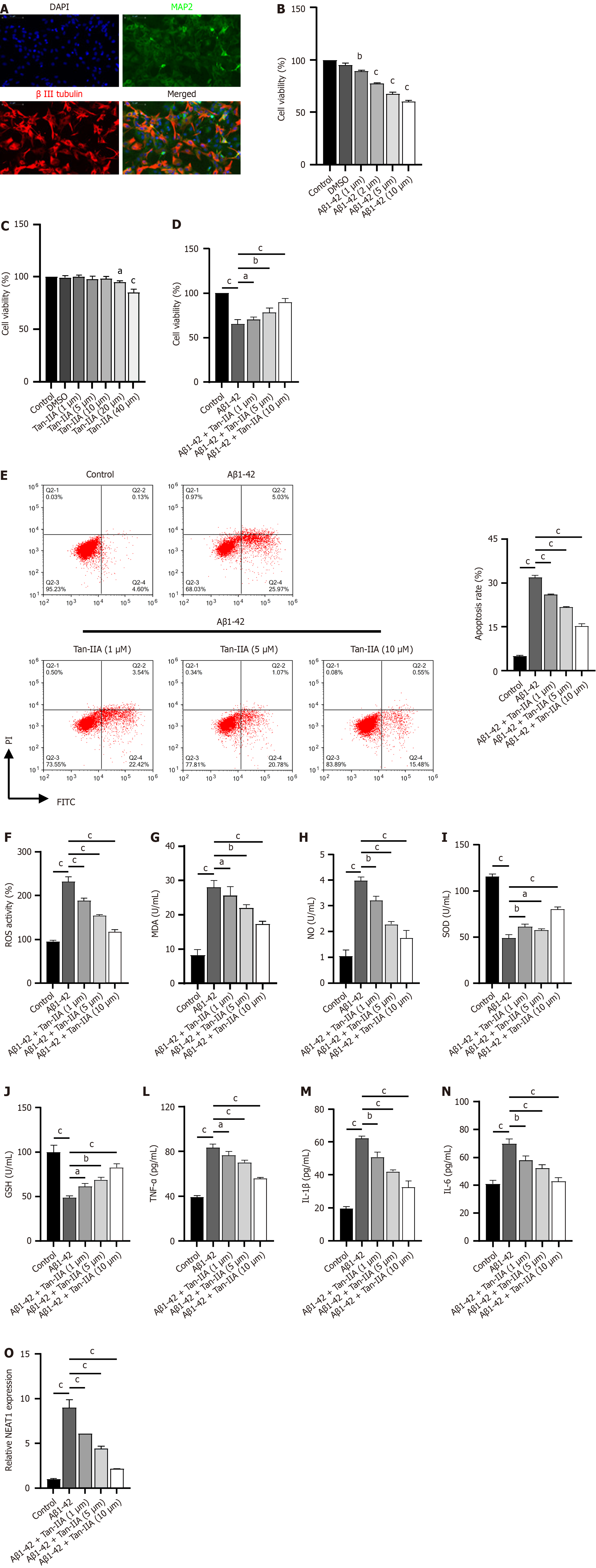
To investigate the protective mechanism of Tan-IIA, we isolated mouse NSCs and used Aβ1-42 induction to establish an in vitro cellular model of AD. Immunofluorescence results showed substantial positivity for MAP2 and β-III tubulin expression in isolated and cultured NSCs (Figure 2A), indicating successful isolation. Aβ1-42 dose-dependently inhibited NSC viability (Figure 2B), with a subsequent dose of 10 μM Aβ1-42 used for validation. After Tan-IIA pretreatment, doses of 20 and 40 μM reduced NSC viability (Figure 2C); therefore, safe doses of 1, 5, and 10 μM were chosen for subsequent experiments to exclude factors intrinsic to Tan-IIA. The validation results confirmed that Tan-IIA dose-dependently ameliorated Aβ1-42-induced reductions in cell viability (Figure 2D), apoptosis (Figure 2E), oxidative stress (increased ROS, MDA, and NO; decreased SOD and GSH) and increases in neuroinflammatory markers (TNF-α, IL-1β, and IL-6) (Figure 2F-N). In the process, the Aβ1-42-induced expression of NEAT1 lncRNA was suppressed (Figure 2O). For subsequent mechanistic studies, a safe dose of 10 μM Tan-IIA was selected, as it ameliorated Aβ1-42-induced effects most efficiently.
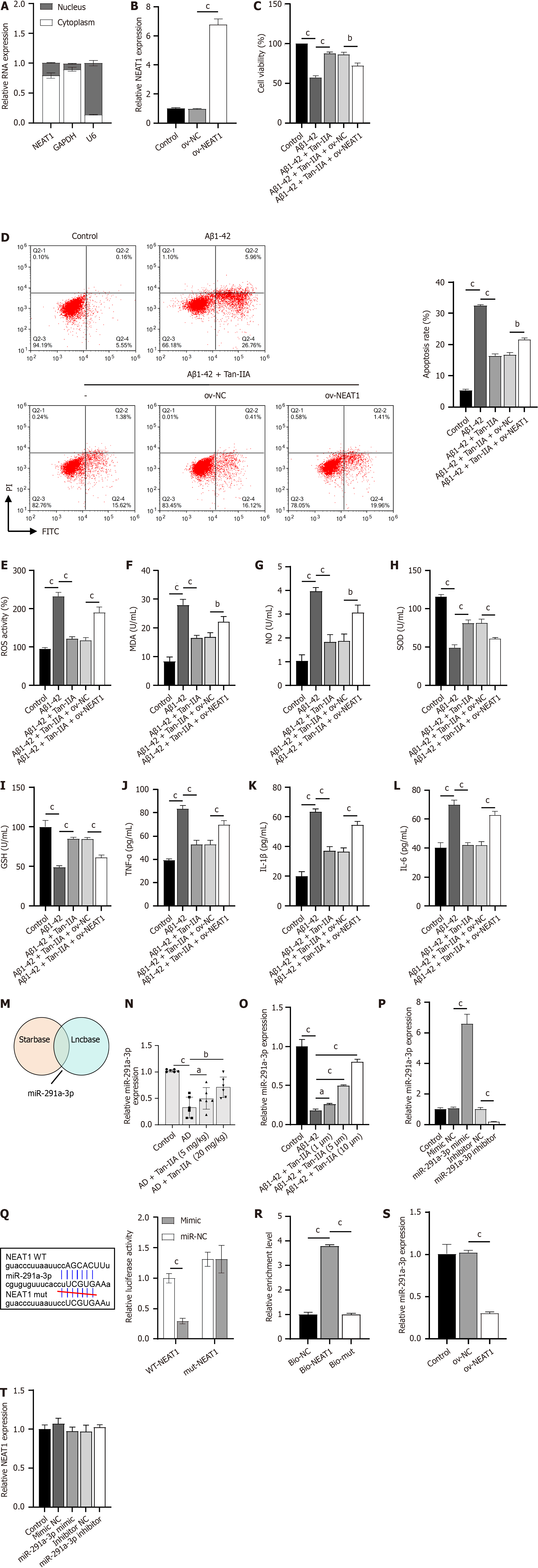
Nucleocytoplasmic separation experiments revealed that the lncRNA NEAT1 was abundantly expressed in the cytoplasm, a result similar to that observed in the positive control GAPDH and opposite to that of U6 (Figure 3A). In verifying the efficacy of the constructed ov-NEAT1 (Figure 3B), it was observed that it partially counteracted the protective effect of Tan-IIA preconditioning on NSCs, partially reversing the Tan-IIA improvement of Aβ1-42-inhibited cell viability (Figure 3C), promoted apoptosis (Figure 3D), oxidative stress (increased ROS, MDA, and NO levels; decreased SOD and GSH levels) and neuroinflammation (increased TNF-α, IL-1β, and IL-6) (Figure 3E-L). A joint analysis using Starbase and Lncbase identified miR-291a-3p as a potential target of NEAT1 (Figure 3M), which was confirmed to be one of the key miRNAs in ameliorating AD[28]. Indeed, miR-291a-3p was downregulated in both the in vivo and in vitro models of AD and upregulated upon Tan-IIA treatment (Figure 3N and O), further supporting this hypothesis. After confirming the efficacy of the synthesized miR-291a-3p mimic and inhibitor (Figure 3P), a dual-luciferase assay was performed. The fluorescence activity in the WT-NEAT1 and miR-291a-3p mimic co-transfected group was significantly lower than that in the WT-NEAT1 and mimic NC co-transfected group, whereas there was no significant difference in fluorescence activity between the mut-NEAT1 and miR-291a-3p mimic or mimic NC co-transfected groups (Figure 3Q). RNA pull-down results showed significant enrichment of miR-291a-3p in bio-NEAT1, with no discernible differences in its expression between the bio-mut and bio-NC groups (Figure 3R). Overexpression of NEAT1 significantly suppressed miR-291a-3p expression (Figure 3S). However, changes in miR-291a-3p expression had no apparent effect on NEAT1 expression (Figure 3T). These results confirm that NEAT1 directly targets and binds to miR-291a-3p.
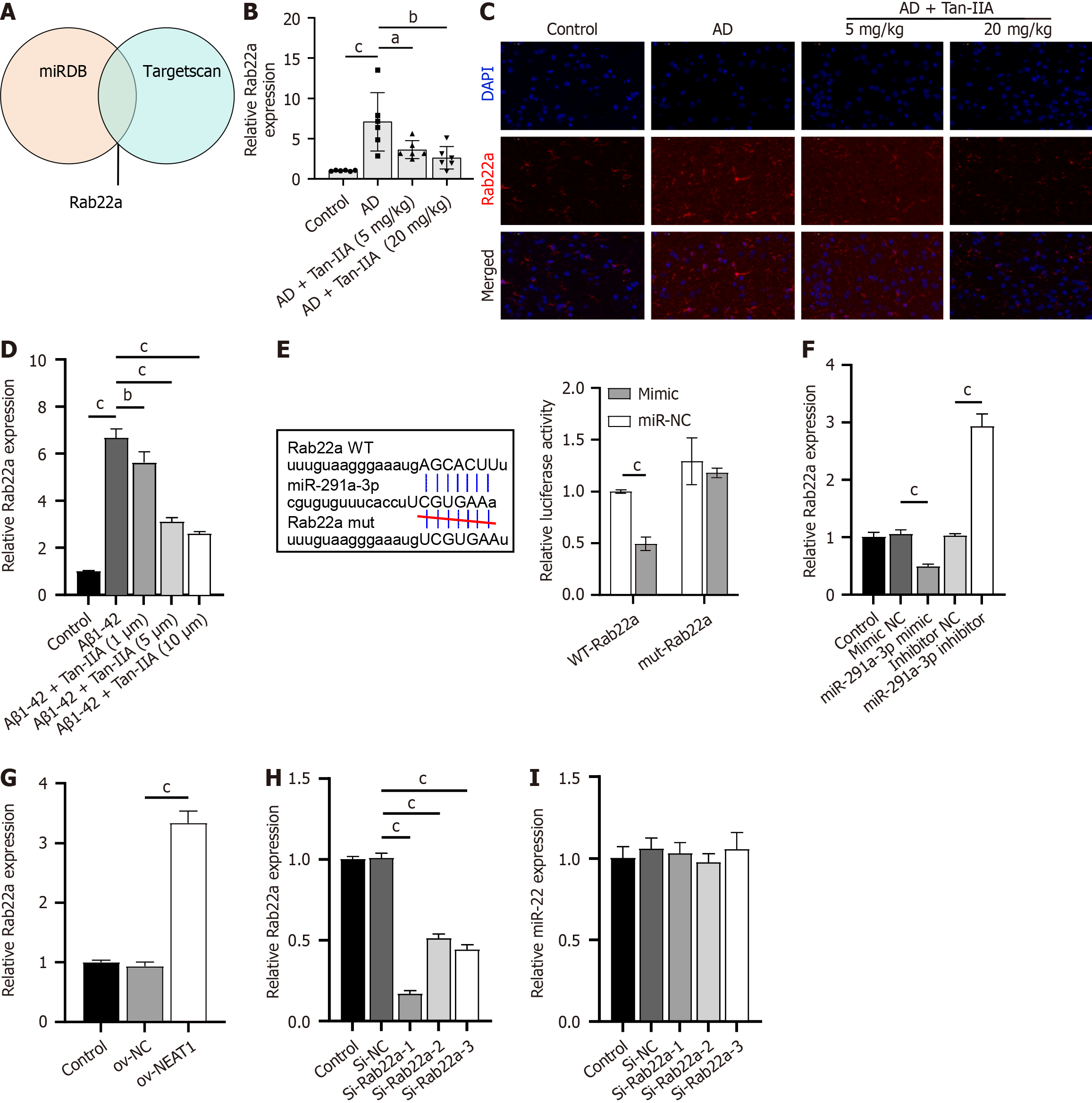
Joint analysis using miRDB and TargetScan databases identified Rab22a, an inhibitor of the AKT pathway, as a potential target of miR-291a-3p (Figure 4A). Rab22a expression increased in both the in vivo and in vitro models and decreased upon Tan-IIA treatment (Figure 4B-D). Dual-luciferase assay results showed that luminescence activity was significantly lower in the WT-NEAT1 and miR-291a-3p mimic co-transfection group than in the WT-Rab22a and NC mimic co-transfection groups, and no significant effects on luminescence activity were found between the mut-Rab22a and miR-291a-3p mimic or NC mimic co-transfection groups (Figure 4E). In addition, Rab22a expression was negatively regulated by miR-291a-3p (Figure 4F) and increased upon NEAT1 overexpression (Figure 4G). Therefore, Rab22a may be a potential target of miR-291a-3p. Among the constructed siRNAs, si-Rab22a-1 was selected for further study because of its high Rab22a silencing efficiency in NSCs (Figure 4H) and because the expression of si-Rab22a did not significantly affect miR-291a-3p (Figure 4I).
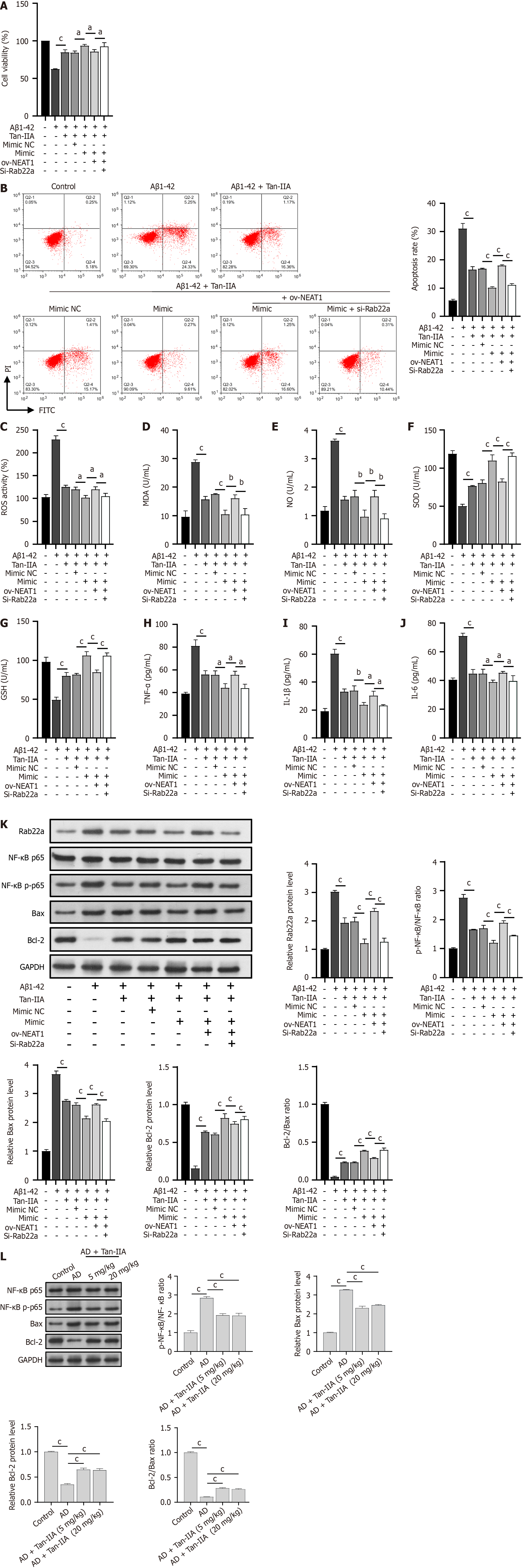
To investigate the protective mechanism of Tan-IIA against Aβ1-42-induced damage, the miR-291a-3p mimic, ov-NEAT1, and si-Rab22a were co-transfected into NSCs. As expected, Tan-IIA ameliorated the Aβ1-42-induced reduction in cell viability, promoted apoptosis, oxidative stress (increased ROS, MDA, and NO levels; decreased SOD and GSH levels), and neuroinflammation (increased TNF-α, IL-1β, and IL-6) (Figure 5A-J). The miR-291a-3p mimic enhanced the effects of Tan-IIA, whereas ov-NEAT1 counteracted or partially counteracted the effects of the miR-291a-3p mimic. However, these effects were reversed by si-Rab22a treatment (Figure 5A-J). Notably, the total p65 protein level was unchanged, and p65 phosphorylation and Bax protein levels were opposite to those of RAB22A and Bcl-2. Tan-IIA reduced Aβ1-42-induced p65 phosphorylation and Bax activation, and the miR-291a-3p mimic further enhanced these effects. In addition, the counteracting effects of ov-NEAT1 on that of miR-291a-3p were inhibited by si-RAB22A (Figure 5K), and the Bcl-2/Bax ratio was consistent with that of Bcl-2. Similar to the in vitro results, Tan-IIA reduced p65 phosphorylation and Bax activation and increased Bcl-2 levels in AD mice in vivo. The enhancing effects of the miR-291a-3p mimic on Tan-IIA were counteracted by NEAT1, but inhibited by si-RAB22A (Figure 5L), and the ratio of Bcl-2/Bax was consistent with that of Bcl-2.
Studies have shown that the dysregulation of lncRNAs - such as lung adenocarcinoma transcripts 1 and NEAT1, which are associated with metastasis - can be critical in the pathogenesis of many degenerative diseases[29-31], including AD. In this study, we investigated the neuroprotective effects of Tan-IIA using both in vivo and in vitro (Aβ1-42-induced NSCs) models of AD to elucidate the underlying mechanisms involving the lncRNA NEAT1/miR-291a-3p/Rab22a signaling axis.
Consistent with previous reports, our results showed that Tan-IIA treatment significantly ameliorated AD-induced histopathological changes, reduced oxidative stress, and attenuated neuroinflammation in the brain tissue of AD mice[26,32]. In particular, the dose-dependent improvements observed in Aβ1-42-induced NSCs in vitro supported the potential of Tan-IIA as a promising AD therapeutic agent. During this process, the expression of NEAT1 - a promising key therapeutic target lncRNA obtained from the GSE150696 dataset analysis - was suppressed in a Tan-IIA dose-dependent manner, suggesting its possible involvement in the protective mechanism of Tan-IIA. Indeed, NEAT1 has been shown to be one of the major lncRNAs that exacerbate AD[33]. This hypothesis was confirmed in our observation that an overexpression of NEAT1 counteracted the ameliorative effect of Tan-IIA on Aβ1-42 induction. This is the first time that the ameliorative effect of Tan-IIA in alleviating AD symptoms has been linked to NEAT1, suggesting the possibility of a new regulatory axis for subsequent lncRNAs, which is important for the development of new therapeutic strategies against AD.
We further revealed a direct interaction between NEAT1 and miR-291a-3p, an important player in nerve injury[34]. Our results show that NEAT1 acts as a sponge for miR-291a-3p, thereby regulating miR-291a-3p availability and function, which is the first confirmation that miR-291a-3p contributes to the regulation of AD mitigation. We also confirmed that Rab22a, a regulator of activated NF-κB signaling, is a direct target of miR-291a-3p. This finding is notable because it is the first time that Tan-IIA regulation of Rab22a via the NEAT1/miR-291a-3p axis has been shown to reduce the activation of NF-κB signaling, a key pathway involved in oxidative stress, neuroinflammation and cell survival in AD. This was confirmed by changes in the levels of factors related to oxidative stress and neuroinflammation. Therefore, we identified the NEAT1/miR-291a-3p/Rab22a axis as an important signaling axis for the Tan-IIA-mediated amelioration of oxidative stress and neuroinflammation levels in both in vivo and in vitro models of AD.
The activation of NF-κB signaling is known to lead to altered activation of the downstream pro- and anti-apoptotic proteins, Bax and Bcl-2[35,36]. In the present study, we observed that Tan-IIA inhibited NF-κB signaling, leading to reduced levels of the pro-apoptotic Bax and activation of the anti-apoptotic Bcl-2 proteins in both our in vivo and in vitro models of AD. The ratio of Bcl-2/Bax was consistent with that of Bcl-2, and these results corresponded to its amelioration of Aβ1-42- induced apoptosis in NSCs, which increases our understanding of the role of Tan-IIA in the mechanism of AD. This study has several limitations. First, it presents no clinical data to confirm the roles of Tan-IIA and NEAT1 in patients with AD. Second, many lncRNAs have not yet been analyzed and verified through GEO data mining. Third, this study was the first to propose miR-291a-3p and Rab22a as novel therapeutic targets for AD, which requires further validation. Finally, our focusing on the NEAT1/miR-291a-3p/Rab22a axis may have overlooked alternative molecular pathways that contribute to the neuroprotective effects of Tan-IIA. The directions and goals of future research should aim to bridge these research gaps.
This study revealed the potential therapeutic role of Tan-IIA in AD by demonstrating its ability to attenuate oxidative stress and neuroinflammation in a mouse model and in Aβ1-42-induced murine NSCs. By elucidating the involvement of the NEAT1/miR-291a-3p/Rab22a signaling axis in the neuroprotective effects of Tan-IIA, this research not only deepens our understanding of the molecular mechanisms underlying AD but also highlights a promising target for the development of new therapeutic strategies.
Alzheimer’s disease (AD) is a prevalent neurodegenerative disorder characterized by cognitive decline and neuronal loss. Oxidative stress and neuroinflammation play pivotal roles in the pathogenesis of this disease. Tanshinone IIA (Tan-IIA), which is derived from Salvia miltiorrhiza, shows potential neuroprotective effects. Understanding the molecular mechanisms underlying these effects is crucial for the development of novel therapeutic strategies.
The motivation for this study was to elucidate the mechanisms by which Tan-IIA exerts neuroprotective effects in AD, focusing on the potential modulation of the long non-coding RNA (lncRNA) nuclear enriched abundant transcript 1 (NEAT1), microRNA (miR)-291a-3p, and RAB22A, member of the RAS oncogene family (Rab22a) signaling pathways. This has important implications for the development of new AD therapies.
The objective of this study was to investigate the neuroprotective effects of Tan-IIA in AD models and elucidate the underlying molecular mechanisms. Specifically, we aimed to determine how Tan-IIA affects oxidative stress, neuroinflammation, and neuronal viability through the NEAT1/miR-291a-3p/Rab22a signaling axis.
The study employed both in vivo and in vitro models of AD using mice and neural stem cells, respectively. Methods included histopathological examinations, enzyme-linked immunosorbent assays, western blotting, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assays, reverse transcription quantitative polymerase chain reaction assays, and various molecular biology techniques to elucidate the role of the NEAT1/miR-291 a-3p/Rab22a pathway in mediating the effects of Tan-IIA.
Tan-IIA ameliorated AD-related pathological changes, reduced oxidative stress, and attenuated neuroinflammation in the mouse models. It modulated the expression of NEAT1, miR-291a-3p, and Rab22a, indicating the involvement of this signaling axis in its neuroprotective effects. This is the first study to link the amelioration of AD symptoms by Tan-IIA with the downregulation of NEAT1.
Tan-IIA has potential therapeutic roles in AD by attenuating oxidative stress and neuroinflammation, primarily through the NEAT1/miR-291a-3p/Rab22a signaling axis. This highlights the intricate molecular interplay involved in AD and identifies lncRNAs and miRNAs as potential therapeutic targets.
Future research should focus on validating the identified therapeutic targets, namely miR-291a-3p and Rab22a, in clinical AD models. It is also crucial to explore other potential molecular pathways affected by Tan-IIA to fully understand its neuroprotective mechanisms. Clinical trials are essential to determine the efficacy and safety of Tan-IIA-based therapies in patients with AD. Expanding our understanding of the role of NEAT1 in AD could open new avenues for RNA-based therapeutic strategies.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Psychiatry
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Jeong KY, South Korea S-Editor: Wang JJ L-Editor: A P-Editor: Zhao S
| 1. | Gibbs DM. Alzheimer's dementia or Alzheimer's disease - What's the difference and why should we care? Ageing Res Rev. 2022;82:101779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 2. | Graff-Radford J, Yong KXX, Apostolova LG, Bouwman FH, Carrillo M, Dickerson BC, Rabinovici GD, Schott JM, Jones DT, Murray ME. New insights into atypical Alzheimer's disease in the era of biomarkers. Lancet Neurol. 2021;20:222-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 357] [Article Influence: 89.3] [Reference Citation Analysis (0)] |
| 3. | Xiao L, Yang X, Sharma VK, Abebe D, Loh YP. Hippocampal delivery of neurotrophic factor-α1/carboxypeptidase E gene prevents neurodegeneration, amyloidosis, memory loss in Alzheimer's Disease male mice. Mol Psychiatry. 2023;28:3332-3342. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 16] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 4. | Tiwari S, Atluri V, Kaushik A, Yndart A, Nair M. Alzheimer's disease: pathogenesis, diagnostics, and therapeutics. Int J Nanomedicine. 2019;14:5541-5554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 782] [Cited by in RCA: 761] [Article Influence: 126.8] [Reference Citation Analysis (0)] |
| 5. | DeTure MA, Dickson DW. The neuropathological diagnosis of Alzheimer's disease. Mol Neurodegener. 2019;14:32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1846] [Cited by in RCA: 1889] [Article Influence: 314.8] [Reference Citation Analysis (0)] |
| 6. | Rajendran L, Paolicelli RC. Microglia-Mediated Synapse Loss in Alzheimer's Disease. J Neurosci. 2018;38:2911-2919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 239] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 7. | Bai R, Guo J, Ye XY, Xie Y, Xie T. Oxidative stress: The core pathogenesis and mechanism of Alzheimer's disease. Ageing Res Rev. 2022;77:101619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 367] [Article Influence: 122.3] [Reference Citation Analysis (0)] |
| 8. | Song T, Song X, Zhu C, Patrick R, Skurla M, Santangelo I, Green M, Harper D, Ren B, Forester BP, Öngür D, Du F. Mitochondrial dysfunction, oxidative stress, neuroinflammation, and metabolic alterations in the progression of Alzheimer's disease: A meta-analysis of in vivo magnetic resonance spectroscopy studies. Ageing Res Rev. 2021;72:101503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 182] [Article Influence: 45.5] [Reference Citation Analysis (0)] |
| 9. | Ionescu-Tucker A, Cotman CW. Emerging roles of oxidative stress in brain aging and Alzheimer's disease. Neurobiol Aging. 2021;107:86-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 377] [Article Influence: 94.3] [Reference Citation Analysis (0)] |
| 10. | Guan L, Mao Z, Yang S, Wu G, Chen Y, Yin L, Qi Y, Han L, Xu L. Dioscin alleviates Alzheimer's disease through regulating RAGE/NOX4 mediated oxidative stress and inflammation. Biomed Pharmacother. 2022;152:113248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 37] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 11. | Abd El-Fatah IM, Abdelrazek HMA, Ibrahim SM, Abdallah DM, El-Abhar HS. Dimethyl fumarate abridged tauo-/amyloidopathy in a D-Galactose/ovariectomy-induced Alzheimer's-like disease: Modulation of AMPK/SIRT-1, AKT/CREB/BDNF, AKT/GSK-3β, adiponectin/Adipo1R, and NF-κB/IL-1β/ROS trajectories. Neurochem Int. 2021;148:105082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 12. | Ganguly U, Kaur U, Chakrabarti SS, Sharma P, Agrawal BK, Saso L, Chakrabarti S. Oxidative Stress, Neuroinflammation, and NADPH Oxidase: Implications in the Pathogenesis and Treatment of Alzheimer's Disease. Oxid Med Cell Longev. 2021;2021:7086512. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 75] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 13. | Yang H, Wang G, Liu J, Lin M, Chen J, Fang Y, Li Y, Cai W, Zhan D. LncRNA JPX regulates proliferation and apoptosis of nucleus pulposus cells by targeting the miR-18a-5p/HIF-1α/Hippo-YAP pathway. Biochem Biophys Res Commun. 2021;566:16-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 14. | Zhang L, Fang Y, Cheng X, Lian YJ, Xu HL. Silencing of Long Noncoding RNA SOX21-AS1 Relieves Neuronal Oxidative Stress Injury in Mice with Alzheimer's Disease by Upregulating FZD3/5 via the Wnt Signaling Pathway. Mol Neurobiol. 2019;56:3522-3537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 71] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 15. | Wang Q, Ge X, Zhang J, Chen L. Effect of lncRNA WT1-AS regulating WT1 on oxidative stress injury and apoptosis of neurons in Alzheimer's disease via inhibition of the miR-375/SIX4 axis. Aging (Albany NY). 2020;12:23974-23995. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 16. | Zhao MY, Wang GQ, Wang NN, Yu QY, Liu RL, Shi WQ. The long-non-coding RNA NEAT1 is a novel target for Alzheimer's disease progression via miR-124/BACE1 axis. Neurol Res. 2019;41:489-497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 91] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 17. | Chen F, Han J, Wang D. Identification of key microRNAs and the underlying molecular mechanism in spinal cord ischemia-reperfusion injury in rats. PeerJ. 2021;9:e11454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 18. | Park S, Lee M, Chun CH, Jin EJ. The lncRNA, Nespas, Is Associated with Osteoarthritis Progression and Serves as a Potential New Prognostic Biomarker. Cartilage. 2019;10:148-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 19. | Gao Y, Zheng X, Chang B, Lin Y, Huang X, Wang W, Ding S, Zhan W, Wang S, Xiao B, Huo L, Yu Y, Chen Y, Gong R, Wu Y, Zhang R, Zhong L, Wang X, Chen Q, Gao S, Jiang Z, Wei D, Kang T. Intercellular transfer of activated STING triggered by RAB22A-mediated non-canonical autophagy promotes antitumor immunity. Cell Res. 2022;32:1086-1104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 63] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 20. | Kong W, Li H, Xie L, Cui G, Gu W, Zhang H, Ma W, Zhou Y. LncRNA MCF2L-AS1 aggravates the malignant development of colorectal cancer via targeting miR-105-5p/RAB22A axis. BMC Cancer. 2021;21:1069. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 21. | Yin X, Chen H, Sun G, Xu Y, Wang L. Circ-C16orf62 Regulates Oxidized low-density Lipoprotein-induced Apoptosis, Inflammation, Oxidative Stress and Cholesterol Accumulation of Macrophages via Mediating RAB22A Expression by Targeting miR-377. Appl Biochem Biotechnol. 2023;195:6586-6606. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 22. | Xie JC, Ma XY, Liu XH, Yu J, Zhao YC, Tan Y, Liu XY, Zhao YX. Hypoxia increases amyloid-β level in exosomes by enhancing the interaction between CD147 and Hook1. Am J Transl Res. 2018;10:150-163. [PubMed] |
| 23. | Bi Z, Wang Y, Zhang W. A comprehensive review of tanshinone IIA and its derivatives in fibrosis treatment. Biomed Pharmacother. 2021;137:111404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 24. | Ansari MA, Khan FB, Safdari HA, Almatroudi A, Alzohairy MA, Safdari M, Amirizadeh M, Rehman S, Equbal MJ, Hoque M. Prospective therapeutic potential of Tanshinone IIA: An updated overview. Pharmacol Res. 2021;164:105364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 106] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 25. | Ba Z, Shi S, Huang N, Li Y, Huang J, You C, Yang X, Liu D, Yu C, He Y, Luo Y. Mesenchymal stem cells after the proprocessing of tanshinone IIA attenuate cognitive deficits and oxidative stress injury in an amyloid β-peptide (25-35)-induced rodent model of Alzheimer's disease. Neuroreport. 2022;33:61-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 26. | Ding B, Lin C, Liu Q, He Y, Ruganzu JB, Jin H, Peng X, Ji S, Ma Y, Yang W. Tanshinone IIA attenuates neuroinflammation via inhibiting RAGE/NF-κB signaling pathway in vivo and in vitro. J Neuroinflammation. 2020;17:302. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 125] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 27. | Tao X, Zhang R, Wang L, Li X, Gong W. Luteolin and Exercise Combination Therapy Ameliorates Amyloid-β1-42 Oligomers-Induced Cognitive Impairment in AD Mice by Mediating Neuroinflammation and Autophagy. J Alzheimers Dis. 2023;92:195-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 28. | Ji Q, Wang X, Cai J, Du X, Sun H, Zhang N. MiR-22-3p Regulates Amyloid β Deposit in Mice Model of Alzheimer's Disease by Targeting Mitogen-activated Protein Kinase 14. Curr Neurovasc Res. 2019;16:473-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 29. | Chanda K, Jana NR, Mukhopadhyay D. Long non-coding RNA MALAT1 protects against Aβ(1-42) induced toxicity by regulating the expression of receptor tyrosine kinase EPHA2 via quenching miR-200a/26a/26b in Alzheimer's disease. Life Sci. 2022;302:120652. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 30. | Varesi A, Carrara A, Pires VG, Floris V, Pierella E, Savioli G, Prasad S, Esposito C, Ricevuti G, Chirumbolo S, Pascale A. Blood-Based Biomarkers for Alzheimer's Disease Diagnosis and Progression: An Overview. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 67] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 31. | Dong LX, Zhang YY, Bao HL, Liu Y, Zhang GW, An FM. LncRNA NEAT1 promotes Alzheimer's disease by down regulating micro-27a-3p. Am J Transl Res. 2021;13:8885-8896. [PubMed] |
| 32. | Peng X, Chen L, Wang Z, He Y, Ruganzu JB, Guo H, Zhang X, Ji S, Zheng L, Yang W. Tanshinone IIA regulates glycogen synthase kinase-3β-related signaling pathway and ameliorates memory impairment in APP/PS1 transgenic mice. Eur J Pharmacol. 2022;918:174772. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 33. | Li K, Wang Z. lncRNA NEAT1: Key player in neurodegenerative diseases. Ageing Res Rev. 2023;86:101878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 70] [Reference Citation Analysis (0)] |
| 34. | Nan A, Zhou X, Chen L, Liu M, Zhang N, Zhang L, Luo Y, Liu Z, Dai L, Jiang Y. A transcribed ultraconserved noncoding RNA, Uc.173, is a key molecule for the inhibition of lead-induced neuronal apoptosis. Oncotarget. 2016;7:112-124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 35. | Hu Q, Zhang W, Wu Z, Tian X, Xiang J, Li L, Li Z, Peng X, Wei S, Ma X, Zhao Y. Baicalin and the liver-gut system: Pharmacological bases explaining its therapeutic effects. Pharmacol Res. 2021;165:105444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 138] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 36. | Mohany M, Ahmed MM, Al-Rejaie SS. The Role of NF-κB and Bax/Bcl-2/Caspase-3 Signaling Pathways in the Protective Effects of Sacubitril/Valsartan (Entresto) against HFD/STZ-Induced Diabetic Kidney Disease. Biomedicines. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 20] [Article Influence: 6.7] [Reference Citation Analysis (0)] |