Published online Dec 19, 2020. doi: 10.5498/wjp.v10.i12.286
Peer-review started: February 20, 2020
First decision: September 21, 2020
Revised: October 6, 2020
Accepted: November 4, 2020
Article in press: November 4, 2020
Published online: December 19, 2020
Processing time: 298 Days and 10.9 Hours
Post-traumatic stress disorder (PTSD) is a serious stress-related disorder.
To identify the key genes and pathways to uncover the potential mechanisms of PTSD using bioinformatics methods.
Gene expression profiles were obtained from the Gene Expression Omnibus database. The differentially expressed genes (DEGs) were identified by using GEO2R. Gene functional annotation and pathway enrichment were then conducted. The gene-pathway network was constructed with Cytoscape software. Quantitative real-time polymerase chain reaction (qRT-PCR) analysis was applied for validation, and text mining by Coremine Medical was used to confirm the connections among genes and pathways.
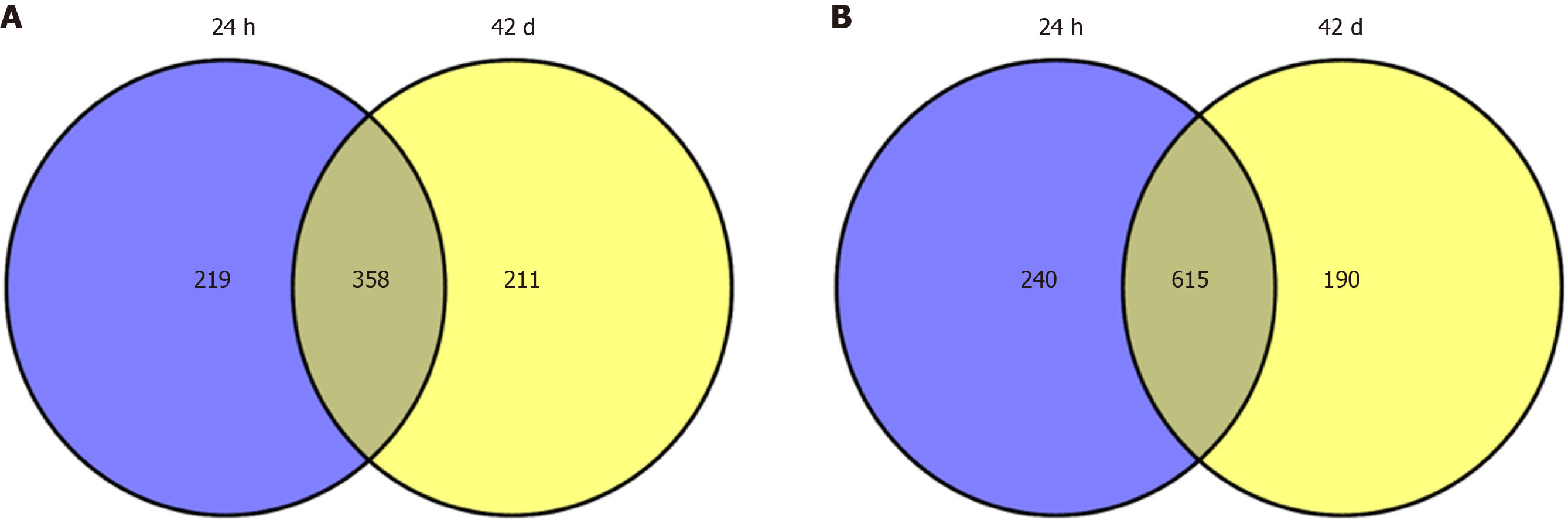
We identified 973 DEGs including 358 upregulated genes and 615 downregulated genes in PTSD. A group of centrality hub genes and significantly enriched pathways (MAPK, Ras, and ErbB signaling pathways) were identified by using gene functional assignment and enrichment analyses. Six genes (KRAS, EGFR, NFKB1, FGF12, PRKCA, and RAF1) were selected to validate using qRT-PCR. The results of text mining further confirmed the correlation among hub genes and the enriched pathways. It indicated that these altered genes displayed functional roles in PTSD via these pathways, which might serve as key signatures in the pathogenesis of PTSD.
The current study identified a panel of candidate genes and important pathways, which might help us deepen our understanding of the underlying mechanism of PTSD at the molecular level. However, further studies are warranted to discover the critical regulatory mechanism of these genes via relevant pathways in PTSD.
Core Tip: Post-traumatic stress disorder (PTSD) is an affective disorder after exposure to trauma or stress directly or indirectly. The pathogenesis of PTSD is not entirely understood. The purpose of this study was to uncover the critical signatures and key pathways to elucidate the underlying mechanisms of PTSD at the molecular level. To address this issue, the closely related genes and the most enriched pathways were identified by using bioinformatics analysis, which was then validated by using basic study in an exploratory approach. Our results showed that a series of significantly expressed genes and relevant pathways might serve as potential biomarkers involved in the pathogenesis of PTSD.
- Citation: Bian YY, Yang LL, Zhang B, Li W, Li ZJ, Li WL, Zeng L. Identification of key genes involved in post-traumatic stress disorder: Evidence from bioinformatics analysis. World J Psychiatr 2020; 10(12): 286-298
- URL: https://www.wjgnet.com/2220-3206/full/v10/i12/286.htm
- DOI: https://dx.doi.org/10.5498/wjp.v10.i12.286
Post-traumatic stress disorder (PTSD), a serious stress-related disorder, is characterized as hypervigilance or hyperarousal, which is caused by traumatic exposure or stressful event directly or indirectly[1]. It presents with a wide range of significant symptoms, e.g., disturbing when reminding the flashbacks, avoidance following re-experiencing the traumatic events, and negative changes in mood and cognitive[2]. It is diagnosed when these symptoms last for at least 1 mo after traumatic or stressor-related exposure[3].
It is estimated that the prevalence of PTSD ranges from 3.1% to 61.6% among 6648 participants in six countries by the International Consortium to Predict PTSD project[4]. In a recent meta-analysis of 96 studies representing 34 countries, a 15.3% prevalence rate of PTSD was found[5]. The lifetime rate of PTSD in the United States population is about 6.8%[6]. A recent cross-sectional study performed in France showed that 35.3% of adults were diagnosed with PTSD[7]. Additionally, about 89.7% of United States adults have exposed at least one traumatic event[8]. Even though PTSD afflicts millions of people throughout the world, it remains largely undiagnosed, particularly in developing countries like China. It is not only comorbid with a wide range of psychological disorders (i.e., depression, anxiety, and substance abuse), which may further cause suicidal ideation or aggressive/addictive behaviors, but also associated with multiple negative psychosocial outcomes such as marital difficulties and unemployment. PTSD has brought a significant burden on individuals, families, and countries. It is reported that the United States spent more than 45 billion dollars annually on the treatment of mental disorders like PTSD[9]. At present, the understanding of the underlying mechanisms of PTSD is not entirely clear, but it is likely related to the hypothalamic-pituitary-adrenal axis, neural circuits, autonomic nervous system, as well as the immune system. Taken together, these facts highlight an urgent need to uncover the molecular mechanisms of PTSD.
To date, with the advances of genomic and transcriptomic study, increasing genes and pathways have been screened out and reported to be related to the occurrence and development of PTSD. For instance, previous studies[10,11] showed that FK506 binding protein 51 was associated with PTSD by deregulating the hypothalamic-pituitary-adrenal axis, indicating a key modulator of PTSD risk. In another instance, a gene-based study[12] found that beta 2 adrenergic receptor gene was related to childhood trauma exposure in predicting PTSD symptoms in adults. Additionally, a variety of signaling pathways were reported to be involved in the developmental progression of PTSD, i.e., NF-κB pathway[13], BDNF-TrkB pathway[14], and JAK/STAT pathway[15], etc. Hence, it is necessary to identify candidate genes and relevant pathways involved in the onset and development of PTSD.
In this study, we screened out a group of candidate signatures and key pathways associated with PTSD from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database by using bioinformatics analysis. Furthermore, the results were verified by using the experimental methods. Our results may provide a significant step forward in understanding the pathogenesis of PTSD at the molecular level.
The gene expression profile of GSE68077 submitted by Muhie et al[16] was obtained for our analysis. In this study, the microarray data of hemibrain samples were collected for analysis. GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r) is an online tool based on the GEO query and Limma R packages. It was conducted to screen differentially expressed genes (DEGs) from different groups of different GEO series[17]. The significant DEGs were obtained by meeting both |logFC| > 1 and P < 0.05. The Venny online tool (available online: http://bioinfogp.cnb.csic.es/tools/venny) was used to identify overlapping DEGs across two groups. The common genes were preserved for further bioinformatics analysis.
Gene ontology (GO) enrichment analysis was used for gene annotation and function description. It consisted of three regulated functions, that is, biological processes (BP), cellular component (CC), and molecular function (MF). The GO terms were identified using the Database for Annotation, Visualization, and Integrated Discovery (DAVID, available online: http://david.abcc.ncifcrf.gov/)[18] to identify the biological significance of genes when P < 0.05 was considered statistically significant. The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was conducted using DAVID[18] to identify pathway enrichment when P < 0.05 was set as the threshold value.
The identified genes and significantly enriched pathways mapped with Cytoscape (http://www.cytoscape.org) software[19] was used to visualize the integrated regulatory networks. Text mining conducted by Coremine Medical (http://www.coremine.com/medical/)[20] was used to visualize the connections among genes and pathways.
Six male SJL albino mice (weighing 30 ± 2 g) provided by the Beijing Vital River Laboratory Animal Technology Co., Ltd (Beijing, China) were trained as aggressor mice according to the study[21]. The mice were housed in home cages that were made of plexiglass material with plenty of water and food. Twelve male C57BL/6N mice (weighing 18 ± 2 g) were purchased from Changzhou Cavens Laboratory Animal Co., Ltd (Changzhou, China). They were randomly assigned to a control or an aggressed-exposed (Agg-E) group. All mice were housed under a standard condition on a 12-h light/darkness cycle in a temperature-controlled room (23 ± 2 ℃). As described previously[22], we used a modified “cage-within-cage resident-intruder” protocol[23] to prepare the social stressed models. Briefly, Agg-E mice were housed in a wire mesh cage (17 cm × 14 cm × 8 cm), which were put inside the aggressor’s larger home cage (50 cm × 30 cm × 20 cm) for 6 h per day. The control mice were treated without aggressor exposure in the same condition. During the exposure time, the experimental mice were given no water and food, while the aggressor mice had free access to them. The direct or indirect exposure lasted for 10 successive days. After 6 wk of recovery, all experimental mice were sacrificed to harvest hemibrain tissues.
All mice underwent three depression-like behavior tests, that is, forced swim test (FST), tail suspension test (TST), and open-field test (OFT). These tests were described previously[23,24]. In brief, the mice were put in a transparent, glass cylindrical tank (2500 mL total volume) filled with 20 cm water (25 ± 1℃) and swam for 6 min. The time when they remained floating passively, stopped struggle, or swimming was regarded as the immobility time. The last 4 min of the immobility time was recorded with a video camera in the FST test. For TST, the mice were suspended by the tail and the last 4 min of the immobility time were measured. While the OFT test was conducted to assess locomotor activity. The mice were allowed to move freely in a specific device for 5 min. The movements of each mouse were measured with a video camera.
Total RNA of hemibrain tissues was isolated according to the instructions of the kit. Polymerase chain reaction was conducted using the SYBR Green Master Mix (ThermoFisher, United States). The gene expression was calculated by 2−ΔΔCT method and normalized against the reference gene, GAPDH. The primers are presented in Supplementary Table 1.
The experimental results are all shown as the mean ± SD. Intergroup differences were analyzed by the t-test by using the SPSS 19.0 software. A P value less than 0.05 was regarded as statistically significant.
First, we retrieved DEGs at two-time points (24 h post 10 d and 42 d post 10 d) by using GEO2R, separately. A total of 1432 DEGs (577 upregulated DEGs and 855 downregulated DEGs) at the time point of 24 h post 10 d, and 1374 DEGs (569 upregulated DEGs and 805 downregulated DEGs) at the time point of 42 d post 10 d were obtained. Based on Venny online tool, a total of 358 overlapping upregulated DEGs and 615 overlapping downregulated DEGs of the two-time points were identified in the social-stress group compared with the control group. The Venny diagram of upregulated and downregulated DEGs was shown in Figure 1.
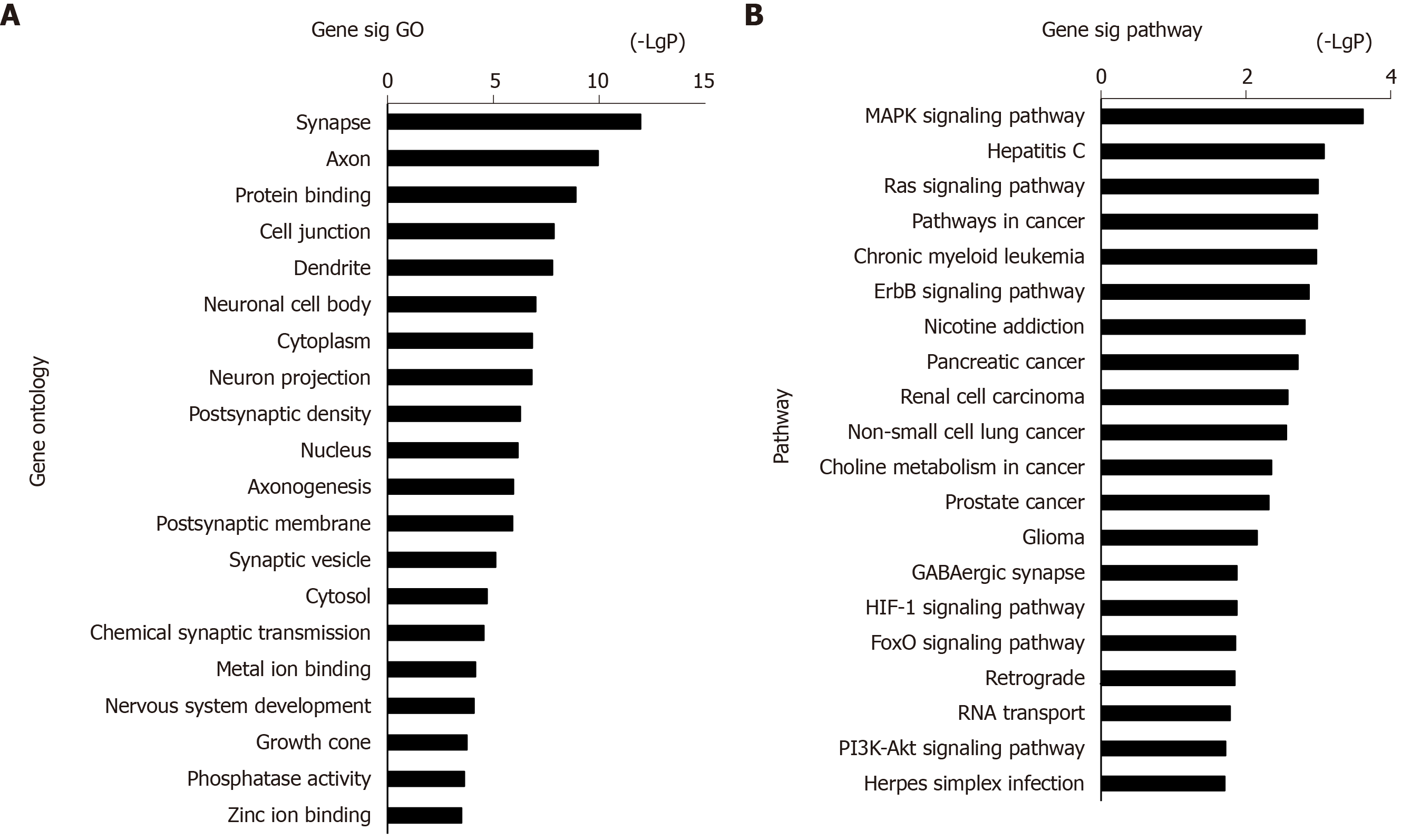
The 973 DEGs (358 upregulated DEGs and 615 downregulated DEGs) were assessed using the DAVID database for functional assignment and pathway enrichment with a P value < 0.05. We identified the most enriched GOs (top 20) ranked by P value, including protein binding (GO: 0005515), cell junction (GO: 0030054), and neuron projection (GO: 0043005) (Figure 2A). Functional enrichment analysis based on GO terms showed that these altered genes significantly participated in PTSD-related pathways (top 20, Figure 2B), i.e., MAPK, Ras, and ErbB signaling pathways.
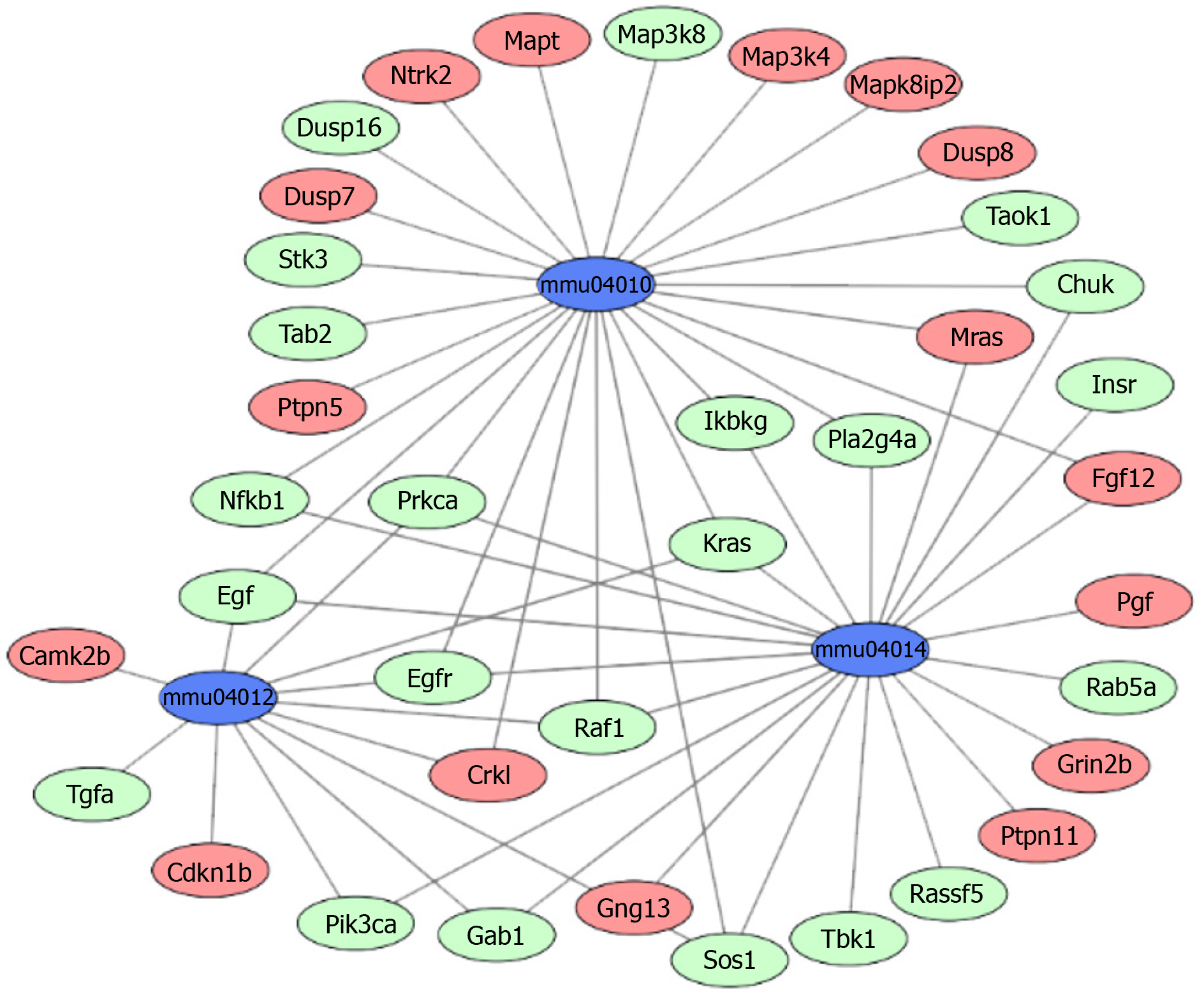
The above 358 upregulated DEGs and 615 downregulated DEGs were analyzed using DAVID, respectively. Based on P value, the upregulated genes in BP terms were markedly enriched in nervous system development, cell differentiation, and positive regulation of neuron projection development. The upregulated genes in CC terms were mainly associated with cell junction, and the upregulated genes in MF terms were markedly enriched in cell adhesion molecule binding and protein domain specific binding. Meanwhile, the downregulated genes in BP terms were markedly enriched in protein transport, cell cycle, and vesicle-mediated transport. The downregulated genes in MF terms were markedly enriched in protein binding, nucleic acid binding, and nucleocytoplasmic transporter activity (Table 1). Furthermore, KEGG enrichment analysis results showed that the upregulated genes were enriched in neuroactive ligand-receptor interaction, calcium signaling pathway, and MAPK signaling pathway, while the downregulated genes were enriched in the ErbB signaling pathway and Toll-like receptor signaling pathway (Table 2). Combined with the results in Figure 2, MAPK, Ras, and ErbB signaling pathways were the main PTSD-related pathways. These markedly enriched pathways with their DEGs are presented in Table 3. Finally, these genetic pathways with their genes were uploaded to Cytoscape to construct the co-expression pattern of the most significantly enriched pathways of DEGs involved in PTSD, as shown in Figure 3.
| Expression | Category | Term | Function | Count | P value |
| Up-regulated | BP | GO: 0007399 | Nervous system development | 25 | 1.21E–08 |
| BP | GO: 0007409 | Axonogenesis | 14 | 1.82E–08 | |
| BP | GO: 0007268 | Chemical synaptic transmission | 16 | 1.33E–07 | |
| BP | GO: 0030154 | Cell differentiation | 32 | 3.75E–06 | |
| BP | GO: 0010976 | Positive regulation of neuron projection development | 12 | 1.51E–05 | |
| CC | GO: 0045202 | Synapse | 43 | 1.25E–18 | |
| CC | GO: 0043025 | Neuronal cell body | 39 | 1.18E–14 | |
| CC | GO: 0030425 | Dendrite | 37 | 2.35E–14 | |
| CC | GO: 0030054 | Cell junction | 44 | 7.70E–14 | |
| CC | GO: 0030424 | Axon | 31 | 3.10E–13 | |
| MF | GO: 0005515 | Protein binding | 99 | 3.15E–05 | |
| MF | GO: 0050839 | Cell adhesion molecule binding | 8 | 1.71E–04 | |
| MF | GO: 0019904 | Protein domain specific binding | 15 | 3.48E–04 | |
| MF | GO: 0005216 | Ion channel activity | 11 | 5.51E–04 | |
| MF | GO: 0008144 | Drug binding | 8 | 0.002625 | |
| Down-regulated | BP | GO: 0015031 | Protein transport | 38 | 7.01E–06 |
| BP | GO: 0007049 | Cell cycle | 38 | 1.59E–05 | |
| BP | GO: 0016192 | Vesicle-mediated transport | 17 | 4.14E–04 | |
| BP | GO: 0007040 | Lysosome organization | 7 | 7.61E–04 | |
| BP | GO: 0006281 | DNA repair | 21 | 8.39E–04 | |
| CC | GO: 0005737 | Cytoplasm | 243 | 1.19E–08 | |
| CC | GO: 0005634 | Nucleus | 219 | 2.55E–07 | |
| CC | GO: 0005829 | Cytosol | 77 | 5.42E–05 | |
| CC | GO: 0005768 | Endosome | 30 | 4.60E–04 | |
| CC | GO: 0005794 | Golgi apparatus | 53 | 4.91E–04 | |
| MF | GO: 0046872 | Metal ion binding | 135 | 4.24E–06 | |
| MF | GO: 0005515 | Protein binding | 156 | 1.17E–05 | |
| MF | GO: 0003676 | Nucleic acid binding | 61 | 1.93E–05 | |
| MF | GO: 0005487 | Nucleocytoplasmic transporter activity | 5 | 0.001069 | |
| MF | GO: 0016874 | Ligase activity | 22 | 0.001335 |
| Expression | Term | Definition | Count | P value |
| Upregulated | mmu04080 | Neuroactive ligand-receptor interaction | 16 | 2.02E–05 |
| mmu05014 | ALS | 6 | 1.08E–04 | |
| mmu04530 | Tight junction | 8 | 1.23E–03 | |
| mmu00430 | Taurine and hypotaurine metabolism | 2 | 1.30E–03 | |
| mmu04020 | Calcium signaling pathway | 9 | 2.08E–03 | |
| mmu00650 | Butanoate metabolism | 3 | 3.77E–03 | |
| mmu04512 | ECM-receptor interaction | 5 | 6.91E–03 | |
| mmu05322 | Systemic lupus erythematosus | 7 | 8.98E–03 | |
| mmu04720 | Long-term potentiation | 4 | 1.25E–02 | |
| mmu04010 | MAPK signaling pathway | 10 | 1.31E–02 | |
| Downregulated | mmu05223 | Non-small cell lung cancer | 9 | 1.48E–05 |
| mmu05212 | Pancreatic cancer | 10 | 2.35E–05 | |
| mmu05160 | Hepatitis C | 14 | 4.31E–05 | |
| mmu05211 | Renal cell carcinoma | 8 | 6.64E–04 | |
| mmu05221 | Acute myeloid leukemia | 7 | 6.55E–04 | |
| mmu04012 | ErbB signaling pathway | 9 | 6.19E–04 | |
| mmu04620 | Toll-like receptor signaling pathway | 10 | 5.05E–04 | |
| mmu03013 | RNA transport | 14 | 3.89E–04 | |
| mmu05214 | Glioma | 8 | 3.55E–04 | |
| mmu05215 | Prostate cancer | 10 | 1.91E–04 |
| Term | Pathway | P value | Genes | |
| Upregulated | Downregulated | |||
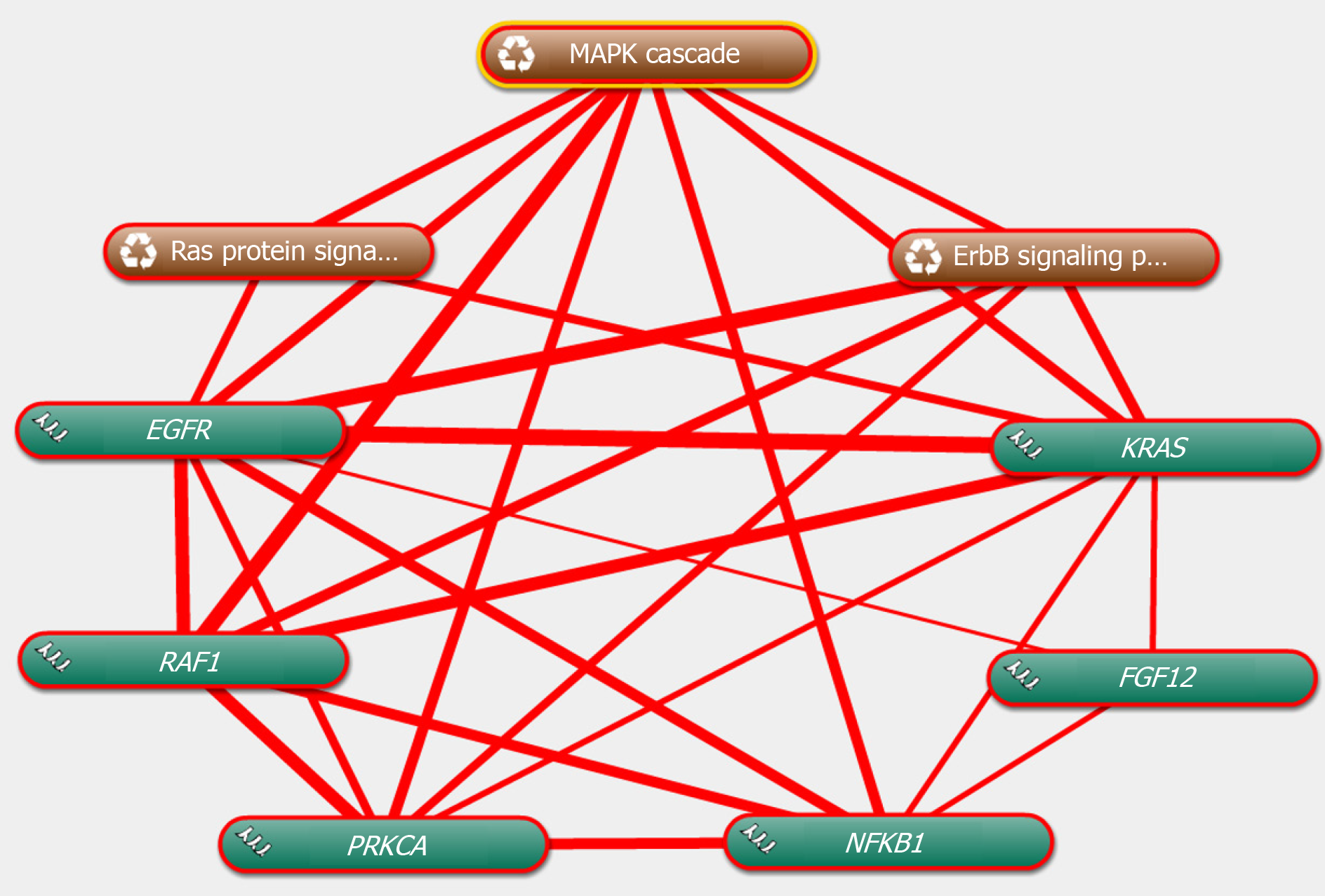
| mmu04010 | MAPK signaling pathway | 1.51E–08 | Crkl, Fgf12, Mras, Mapt, Ntrk2, Dusp8, Ptpn5, Map3k4, Mapk8ip2, Dusp7 | Chuk, Egf, Egfr, Ikbkg, Kras, Nfkb1, Prkca, Pla2g4a, Sos1, Map3k8, Stk3, Tab2, Dusp16, Raf1, Taok1 |
| mmu04014 | Ras signaling pathway | 1.84E–07 | Fgf12, Grin2b, Mras, Pgf, Ptpn11, Gng13 | Chuk, Egf, Egfr, Gab1, Ikbkg, Insr, Kras, Nfkb1, Pik3ca, Prkca, Pla2g4a, Sos1, Rassf5, Tbk1, Raf1, Rab5a |
| mmu04012 | ErbB signaling pathway | 3.85E–06 | Camk2b, Cdkn1b, Crkl | Egf, Egfr, Gab1, Kras, Pik3ca, Prkca, Sos1, Tgfa, Raf1 |
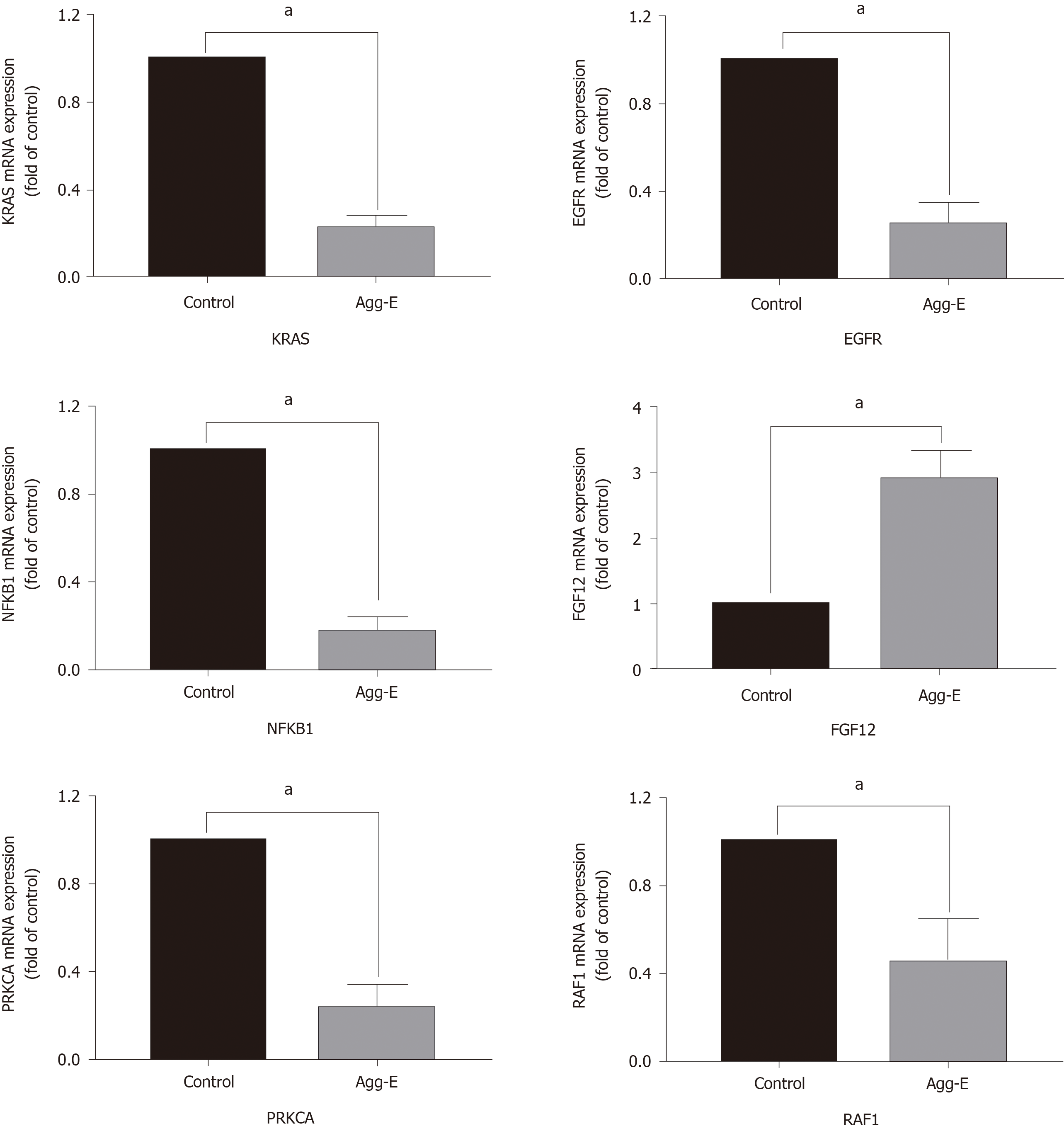
The genes in the central roles connected to the three enriched pathways were considered as hub genes. In our study, we tried to uncover the closely enriched genes related to the most significant pathways in an exploratory approach. Six hub genes were selected based on the literature due to their neurobiology assumption to be related to PTSD. The hub genes were listed as follows, that is, nuclear factor kappa B subunit 1 (NFKB1), KRAS proto-oncogene, GTPase (KRAS), epidermal growth factor receptor (EGFR), fibroblast growth factor 12 (FGF12), protein kinase C alpha (PRKCA), and Raf-1 proto-oncogene, serine/threonine kinase (RAF1). qRT-PCR was used for validation. The above genes in hemibrain tissues were differently expressed between the two groups (Figure 4), which was in line with the results of the microarray datasets. It indicated that these genes played crucial roles in the occurrence and development of PTSD. To further visualize the linear connections between the hub genes and the associated pathways, text mining was performed with Coremine Medical online tool. The above six gene symbols and three pathways were used as search terms, and the co-expressed network is shown in Figure 5. We found that KRAS and EGFR were correlated with the Ras pathway, while four genes (KRAS, EGFR, PRKCA, and RAF1) were related to the ErbB pathway. The genes except FGF12 were associated with the MAPK pathway. More importantly, EGFR and KRAS were connected with three pathways, which further demonstrated that these genes have important biological functions in the signaling pathway.
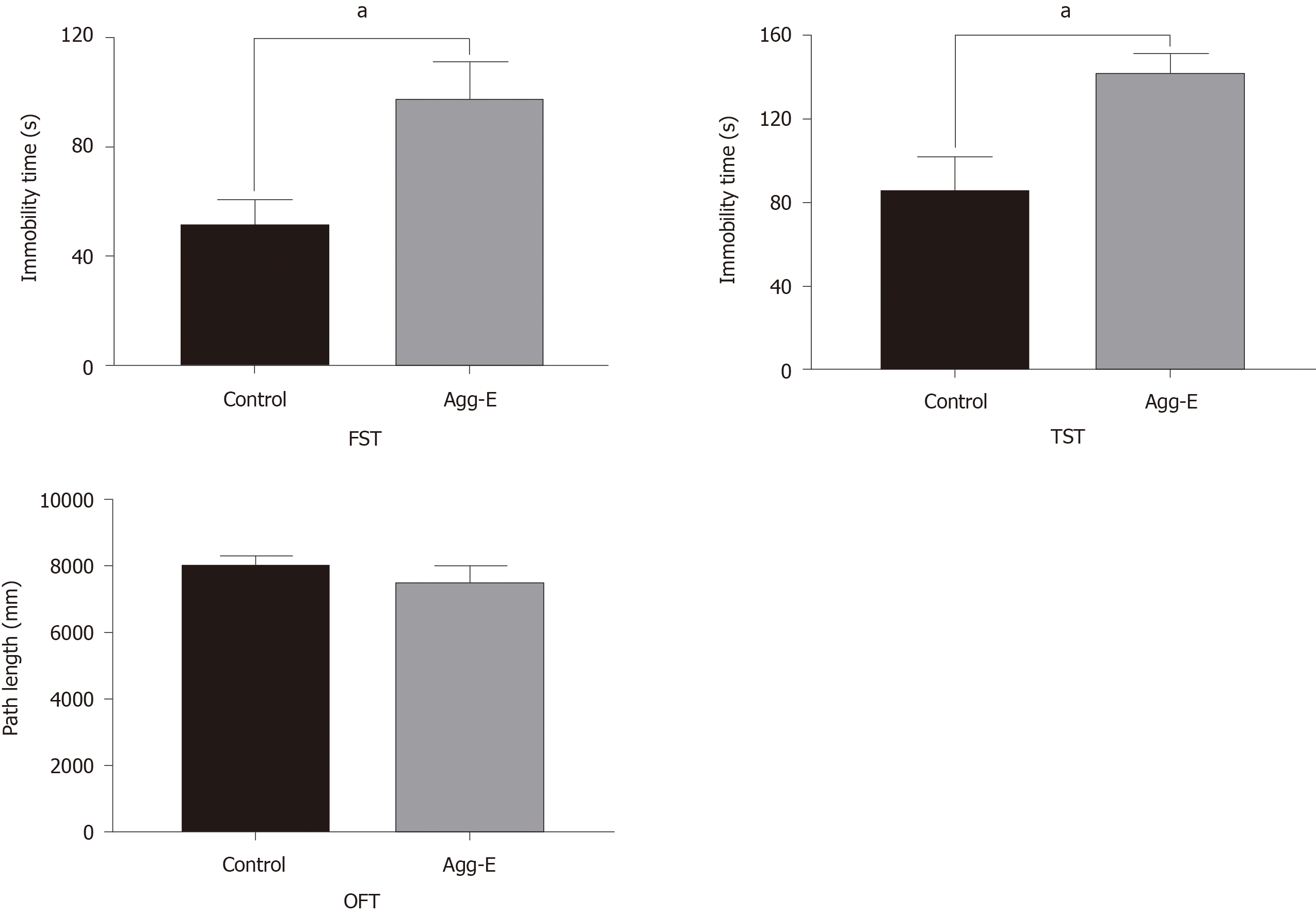
The results of FST, TST, and OFT are presented in Figure 6. We found that the immobility time in the Agg-E group was significantly increased than that of the control group. However, the path length of OFT showed no significant difference between the Agg-E group and control group.
PTSD as a trauma-related or stressor-related disorder is the consequence of direct or indirect stress. It can lead to subsequent response patterns such as avoidance, negative emotion, and hyperarousal[2]. PTSD is diagnosed when the above symptoms last for at least 1 mo after the onset of the event[25]. The genesis and progression of PTSD are complicated and the pathogenesis is not entirely understood. Thus, it is needed to uncover the key targets to elucidate the underlying mechanisms associated with PTSD.
In this study, the gene expression microarrays of GSE68077 were downloaded from the GEO database. A total of 973 DEGs including 358 upregulated genes and 615 downregulated genes were identified. The functional enrichment analysis of DEGs based on GO terms demonstrated that the most functional and molecular work might focus on MAPK, Ras, and ErbB signaling pathways. Based on functional enrichment, we found that the upregulated genes displayed functionalities, such as cell differentiation and positive regulation of neuron projection development, while the downregulated genes had other functionalities, such as protein transport, cell cycle, and DNA repair. The upregulated genes were markedly concentrated in neuroactive ligand-receptor interaction, calcium, and MAPK signaling pathways, and the downregulated genes were markedly enriched in the ErbB and Toll-like receptor signaling pathways. Taken together, we speculated that MAPK, Ras, and ErbB pathways played regulatory roles in the occurrence and progression of PTSD. Consistent with our hypothesis, the MAPK pathway as a kind of stress response signaling cascade, is involved in synaptic plasticity and is connected with depressive-like symptoms or severity of the depression[26]. In line with our findings, a previous study[27] based on bioinformatics analysis showed that the MARK signaling pathway was one of the mainly enriched signaling pathways involved in major depressive disorder. Another study suggested that the ErbB signaling pathway might be associated with cognitive deficiencies after early social isolation[28], and Ras pathway acted as a critical role in the pathogenesis of depression[29].
Currently, we identified the closely related genes involved in the most significant pathways above, as shown in Table 3. Among these genes, we found that a panel of hub genes was closely related to the significantly expressed pathways. In an exploratory approach, several hub genes were selected based on the literature due to their neurobiology assumption to be related to PTSD. To validate our hypothesis, we further performed qRT-PCR to verify our findings. The results were consistent with the microarray analysis. Besides, several genes were reported in previous studies. For example, NFKB1, a transcription regulator, was found to be overexpressed in depressive disorders patients[30]. PRKCA has been implicated in PTSD pathogenesis, and the single nucleotide polymorphism of PRKCA was considered to be significantly associated with traumatic experiences and PTSD symptoms, which can further increase the rate of PTSD[31]. EGFR plays a vital role in cellular growth, differentiation, and apoptosis. Several studies indicated that EGFR genotype is closely correlated to depression or major depressive disorder[32-34]. Its main downstream pathway is MAPK that plays a dominant role in signal transduction in many biological activities such as cell proliferation and inflammatory responses. It has been known that the MAPK pathway is activated by neuropeptide oxytocin in the paraventricular nucleus of the hypothalamus by transactivation of EGFR to regulate anxiety[35]. Interestingly, the connections among hub genes and relevant pathways were supported by the results of the co-occurrence analysis of test mining. We found that the above genes were correlated with the enriched signaling pathways. It indicated that these altered hub genes played functional roles in PTSD via these pathways. Based on the evidence above, we argued that these genes may serve as important contributors to PTSD. Besides, the depression-like behavioral data (FST and TST) indicated a marked increase in immobility time in the Agg-E group. It showed that stress exposure could induce depressive-behavior that may co-occur with PTSD.
In conclusion, the present study identified a series of significantly expressed genes and relevant pathways, which may serve as potential biomarkers involved in the pathogenesis of PTSD. The results were confirmed using experimental verification. We further surmised that the identified genes may be key contributors associated with PTSD. However, future studies on the precise mechanism are warranted.
Post-traumatic stress disorder (PTSD) is a common trauma-related or stressor-related disorder characterized by a wide range of significant symptoms, e.g., avoidance, negative emotion, and hyperarousal. A variety of targeted genes and pathways may be associated with the occurrence and progression of PTSD.
The underlying mechanisms of PTSD are complicated and not entirely clear. We sought to uncover the key targets for the potential mechanisms associated with PTSD at the molecular level.
The main objective was to identify critical genes and key pathways associated with PTSD.
Our study was conducted based on the NCBI Gene Expression Omnibus database by using bioinformatics analysis. The differentially expressed genes were identified by using GEO2R. Gene functional annotation and pathway enrichment were performed using the Database for Annotation, Visualization, and Integrated Discovery. The gene-pathway network was mapped with Cytoscape software. Quantitative real-time polymerase chain reaction (qRT-PCR) analysis was applied for validation, and text mining by Coremine Medical was used to confirm the connections among genes and pathways.
A total of 358 upregulated genes and 615 downregulated genes were obtained. These altered genes were significantly enriched in PTSD-related pathways, such as MAPK, Ras, and ErbB signaling pathways. The co-expression pattern of these enriched pathways with their genes was constructed. And a group of hub genes was obtained from the gene-pathway network. qRT-PCR indicated that the hub genes were differentially expressed. Besides, text mining showed that these genes were linearly connected with the associated pathways, which suggested that these genes might play crucial roles in the signal pathway conduction in the onset and development of PTSD.
The results of this study have contributed to the identification of a panel of candidate genes and important pathways, which may serve as potential biomarkers involved in the pathogenesis of PTSD.
Our findings may provide a significant step forward in understanding the pathogenesis of PTSD at the molecular level.
Manuscript source: Unsolicited manuscript
Specialty type: Psychiatry
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ahboucha S, Vyshka G S-Editor: Chen XF L-Editor: Wang TQ P-Editor: Li JH
| 1. | Watkins LE, Sprang KR, Rothbaum BO. Treating PTSD: A Review of Evidence-Based Psychotherapy Interventions. Front Behav Neurosci. 2018;12:258. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 286] [Cited by in RCA: 291] [Article Influence: 41.6] [Reference Citation Analysis (0)] |
| 2. | Dennis PA, Weinberg JB, Calhoun PS, Watkins LL, Sherwood A, Dennis MF, Beckham JC. An investigation of vago-regulatory and health-behavior accounts for increased inflammation in posttraumatic stress disorder. J Psychosom Res. 2016;83:33-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 3. | Paredes Molina CS, Berry S, Nielsen A, Winfield R. PTSD in civilian populations after hospitalization following traumatic injury: A comprehensive review. Am J Surg. 2018;216:745-753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 4. | Qi W, Ratanatharathorn A, Gevonden M, Bryant R, Delahanty D, Matsuoka Y, Olff M, deRoon-Cassini T, Schnyder U, Seedat S, Laska E, Kessler RC, Koenen K, Shalev A on behalf of the ICPP. Application of data pooling to longitudinal studies of early post-traumatic stress disorder (PTSD): the International Consortium to Predict PTSD (ICPP) project. Eur J Psychotraumatol. 2018;9:1476442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 5. | Charlson F, van Ommeren M, Flaxman A, Cornett J, Whiteford H, Saxena S. New WHO prevalence estimates of mental disorders in conflict settings: a systematic review and meta-analysis. Lancet. 2019;394:240-248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 576] [Cited by in RCA: 741] [Article Influence: 123.5] [Reference Citation Analysis (0)] |
| 6. | Kessler RC, Chiu WT, Demler O, Merikangas KR, Walters EE. Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62:617-627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7240] [Cited by in RCA: 7251] [Article Influence: 362.6] [Reference Citation Analysis (0)] |
| 7. | Husky MM, Mazure CM, Kovess-Masfety V. Gender differences in psychiatric and medical comorbidity with post-traumatic stress disorder. Compr Psychiatry. 2018;84:75-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 8. | Kilpatrick DG, Resnick HS, Milanak ME, Miller MW, Keyes KM, Friedman MJ. National estimates of exposure to traumatic events and PTSD prevalence using DSM-IV and DSM-5 criteria. J Trauma Stress. 2013;26:537-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1038] [Cited by in RCA: 1208] [Article Influence: 100.7] [Reference Citation Analysis (0)] |
| 9. | Kheirbek MA, Klemenhagen KC, Sahay A, Hen R. Neurogenesis and generalization: a new approach to stratify and treat anxiety disorders. Nat Neurosci. 2012;15:1613-1620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 357] [Cited by in RCA: 395] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 10. | Young DA, Inslicht SS, Metzler TJ, Neylan TC, Ross JA. The effects of early trauma and the FKBP5 gene on PTSD and the HPA axis in a clinical sample of Gulf War veterans. Psychiatry Res. 2018;270:961-966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 11. | Hawn SE, Sheerin CM, Lind MJ, Hicks TA, Marraccini ME, Bountress K, Bacanu SA, Nugent NR, Amstadter AB. GxE effects of FKBP5 and traumatic life events on PTSD: A meta-analysis. J Affect Disord. 2019;243:455-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 12. | Hauser MA, Garrett ME, Liu Y, Dennis MF, Kimbrel NA, Veterans Affairs Mid-Atlantic Mental Illness Research Education And Clinical Center Workgroup, Beckham JC, Ashley-Koch AE. Further evidence for a role of the ADRB2 gene in risk for posttraumatic stress disorder. J Psychiatr Res. 2017;84:59-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 13. | Guardado P, Olivera A, Rusch HL, Roy M, Martin C, Lejbman N, Lee H, Gill JM. Altered gene expression of the innate immune, neuroendocrine, and nuclear factor-kappa B (NF-κB) systems is associated with posttraumatic stress disorder in military personnel. J Anxiety Disord. 2016;38:9-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 44] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 14. | Green CR, Corsi-Travali S, Neumeister A. The Role of BDNF-TrkB Signaling in the Pathogenesis of PTSD. J Depress Anxiety. 2013;2013:006. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 15. | Feng D, Guo B, Liu G, Wang B, Wang W, Gao G, Qin H, Wu S. FGF2 alleviates PTSD symptoms in rats by restoring GLAST function in astrocytes via the JAK/STAT pathway. Eur Neuropsychopharmacol. 2015;25:1287-1299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 16. | Muhie S, Gautam A, Meyerhoff J, Chakraborty N, Hammamieh R, Jett M. Brain transcriptome profiles in mouse model simulating features of post-traumatic stress disorder. Mol Brain. 2015;8:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 17. | Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991-D995. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4527] [Cited by in RCA: 6784] [Article Influence: 521.8] [Reference Citation Analysis (0)] |
| 18. | Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2023] [Cited by in RCA: 1801] [Article Influence: 100.1] [Reference Citation Analysis (0)] |
| 19. | Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498-2504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24663] [Cited by in RCA: 33566] [Article Influence: 1598.4] [Reference Citation Analysis (0)] |
| 20. | de Leeuw N, Dijkhuizen T, Hehir-Kwa JY, Carter NP, Feuk L, Firth HV, Kuhn RM, Ledbetter DH, Martin CL, van Ravenswaaij-Arts CM, Scherer SW, Shams S, Van Vooren S, Sijmons R, Swertz M, Hastings R. Diagnostic interpretation of array data using public databases and internet sources. Hum Mutat. 2012;33:930-940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 80] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 21. | Hammamieh R, Chakraborty N, De Lima TC, Meyerhoff J, Gautam A, Muhie S, D'Arpa P, Lumley L, Carroll E, Jett M. Murine model of repeated exposures to conspecific trained aggressors simulates features of post-traumatic stress disorder. Behav Brain Res. 2012;235:55-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 22. | Bian Y, Yang L, Zhao M, Li Z, Xu Y, Zhou G, Li W, Zeng L. Identification of Key Genes and Pathways in Post-traumatic Stress Disorder Using Microarray Analysis. Front Psychol. 2019;10:302. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 23. | Steru L, Chermat R, Thierry B, Simon P. The tail suspension test: a new method for screening antidepressants in mice. Psychopharmacology. 85:367-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2352] [Cited by in RCA: 2585] [Article Influence: 64.6] [Reference Citation Analysis (0)] |
| 24. | Porsolt RD, Le Pichon M, Jalfre M. Depression: a new animal model sensitive to antidepressant treatments. Nature. 1977;266:730-732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3141] [Cited by in RCA: 3288] [Article Influence: 68.5] [Reference Citation Analysis (0)] |
| 25. | Lisieski MJ, Eagle AL, Conti AC, Liberzon I, Perrine SA. Single-Prolonged Stress: A Review of Two Decades of Progress in a Rodent Model of Post-traumatic Stress Disorder. Front Psychiatry. 2018;9:196. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 149] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 26. | Duric V, Banasr M, Licznerski P, Schmidt HD, Stockmeier CA, Simen AA, Newton SS, Duman RS. A negative regulator of MAP kinase causes depressive behavior. Nat Med. 2010;16:1328-1332. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 347] [Cited by in RCA: 330] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 27. | Ji HF, Zhuang QS, Shen L. Genetic overlap between type 2 diabetes and major depressive disorder identified by bioinformatics analysis. Oncotarget. 2016;7:17410-17414. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 28. | Makinodan M, Rosen KM, Ito S, Corfas G. A critical period for social experience-dependent oligodendrocyte maturation and myelination. Science. 2012;337:1357-1360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 573] [Cited by in RCA: 685] [Article Influence: 52.7] [Reference Citation Analysis (0)] |
| 29. | Li H, Linjuan-Li, Wang Y. G-CSF improves CUMS-induced depressive behaviors through downregulating Ras/ERK/MAPK signaling pathway. Biochem Biophys Res Commun. 2016;479:827-832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 30. | Iacob E, Light KC, Tadler SC, Weeks HR, White AT, Hughen RW, Vanhaitsma TA, Bushnell L, Light AR. Dysregulation of leukocyte gene expression in women with medication-refractory depression versus healthy non-depressed controls. BMC Psychiatry. 2013;13:273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 31. | de Quervain DJ, Kolassa IT, Ackermann S, Aerni A, Boesiger P, Demougin P, Elbert T, Ertl V, Gschwind L, Hadziselimovic N, Hanser E, Heck A, Hieber P, Huynh KD, Klarhöfer M, Luechinger R, Rasch B, Scheffler K, Spalek K, Stippich C, Vogler C, Vukojevic V, Stetak A, Papassotiropoulos A. PKCα is genetically linked to memory capacity in healthy subjects and to risk for posttraumatic stress disorder in genocide survivors. Proc Natl Acad Sci USA. 2012;109:8746-8751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 32. | Pirl WF, Traeger L, Greer JA, Bemis H, Gallagher E, Lennes I, Sequist L, Heist R, Temel JS. Tumor epidermal growth factor receptor genotype and depression in stage IV non-small cell lung cancer. Oncologist. 2011;16:1299-1306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 33. | Pirl WF, Traeger L, Greer JA, Jackson V, Lennes IT, Gallagher E, Sequist L, Temel JS. Depression, survival, and epidermal growth factor receptor genotypes in patients with metastatic non-small cell lung cancer. Palliat Support Care. 2013;11:223-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 34. | Jacobs JM, Traeger L, Eusebio J, Simon NM, Sequist LV, Greer JA, Temel JS, Pirl WF. Depression, inflammation, and epidermal growth factor receptor (EGFR) status in metastatic non-small cell lung cancer: A pilot study. J Psychosom Res. 2017;99:28-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 35. | Blume A, Bosch OJ, Miklos S, Torner L, Wales L, Waldherr M, Neumann ID. Oxytocin reduces anxiety via ERK1/2 activation: local effect within the rat hypothalamic paraventricular nucleus. Eur J Neurosci. 2008;27:1947-1956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 204] [Article Influence: 12.0] [Reference Citation Analysis (0)] |