Published online Sep 24, 2019. doi: 10.5306/wjco.v10.i9.307
Peer-review started: March 20, 2019
First decision: April 15, 2019
Revised: August 19, 2019
Accepted: September 4, 2019
Article in press: September 5, 2019
Published online: September 24, 2019
Processing time: 203 Days and 9.3 Hours
Nucleic acid isolation from formalin-fixed, paraffin-embedded tissue (FFPET) samples is a daily routine in molecular pathology laboratories, but extraction from FFPET is not always easily achieved. Choosing the right extraction technique is key for further examinations.
To compare the performance of four commercially available kits used for DNA extraction in routine practice.
DNA isolation was performed on 46 randomly selected formalin-fixed, paraffin-embedded (FFPE) colorectal adenocarcinoma (CRC) surgical specimens. Four commercially available extraction kits were used: two for manual DNA extraction (the PureLink Genomic DNA Mini Kit from Invitrogen and the High Pure FFPE DNA Isolation Kit from Roche) and two for automated DNA extraction (the iPrep Genomic DNA Kit from Invitrogen and the MagnaPure LC DNA Isolation Kit from Roche). The DNA concentration and quality (odds ratio) among the four systems were compared. The results were correlated with the clinicopathological aspects of CRC cases: age, gender, localization, macro- and microscopic features, lymph node metastases, and the lymph node ratio.
The highest DNA concentration was obtained using the manual kits: 157.24 ± 62.99 ng/µL for the PureLink Genomic DNA Mini Kit and 86.64 ng/µL ± 43.84 for the High Pure FFPE DNA Isolation Kit (P < 0.0001). Lower concentrations were obtained with automated systems: 20.39 ± 21.19 ng/µL for the MagnaPure LC DNA Isolation Kit and 8.722 ± 6.408 ng/µL for the iPrep Genomic DNA Kit, with differences between the systems used (P < 0.0001). The comparison between age, gender, tumor localization, pT or pN stage and the lymph node ratio indicated no statistically significant difference in DNA concentration using any of the nucleic acid isolation kits. DNA concentration was influenced by the macroscopic features and grade of differentiation. A higher DNA concentration was obtained for well-differentiated polypoid colorectal adenocarcinomas (CRCs), compared with undifferentiated ulcero-infiltrative carcinomas, irrespective of the kit used.
For research or diagnosis that needs high DNA concentrations, manual methods of DNA isolation should be used. A higher amount of DNA can be obtained from polypoid-type differentiated CRCs. Automated systems confer comfort and a lower amount of DNA that is, however, sufficient for classic polymerase chain reaction (PCR) and real-time quantitative PCR molecular examinations. All four commercially available kits can be successfully used in daily practice.
Core tip: In this paper we proved the advantages and disadvantages of manual vs automated methods of DNA extraction. The original aspect refers to the correlation between DNA concentration and colorectal adenocarcinoma (CRC) features. We conclude that manual methods are more suitable for obtaining high DNA concentrations, especially from differentiated polypoid-type CRCs. In CRC samples, a higher DNA concentration is associated with a lower OD value.
- Citation: Kovacs Z, Jung I, Csernak E, Szentirmay Z, Banias L, Rigmanyi G, Gurzu S. DNA extraction from paraffin embedded colorectal carcinoma samples: A comparison study of manual vs automated methods, using four commercially kits. World J Clin Oncol 2019; 10(9): 307-317
- URL: https://www.wjgnet.com/2218-4333/full/v10/i9/307.htm
- DOI: https://dx.doi.org/10.5306/wjco.v10.i9.307
Friedrich Miescher[1] performed the first isolation of DNA in 1868/1869. In 1988, Miller et al[2] described the simple salting-out procedure of DNA extraction from human cells compared to the classic phenol-chloroform method. They found that the salting-out procedure was as good as the classic method using chloroform. In 1991, Lahiri et al[3] demonstrated that the salting-out procedure is even better for RFLP (restriction fragment length polymorphism). Regarding the type of preserved tissue, although fresh tissues are preferred, Goelz et al[4] demonstrated in 1985 that DNA can also be isolated from formalin-fixed, paraffin-embedded tissues (FFPETs).
Irrespective of the source of commercially available kits, manual nucleic acid isolation methods are based on the same principle: cells must be disrupted and digested with Proteinase K and proteins and other contaminants need to be washed out in order to achieve pure DNA. Extraction from FFPETs needs an additional deparaffinization step with 100% xylene in order to get rid of the paraffin[5].
Automated magnetic bead methods are time-saving procedures. Starting from sample lysis to DNA elution, everything is done by a machine. However, isolation from formalin-fixed, paraffin-embedded tissue requires the same additional step as manual methods, namely deparaffinization[6].
The aim of this paper was to compare the advantages and disadvantages of four DNA extraction kits used in daily practice for DNA isolation from formalin-fixed, paraffin-embedded (FFPE) colorectal adenocarcinoma (CRC) surgical specimens: two manual and two automated magnetic bead kits. An analysis of the correlation between the results and the clinicopathological features of colorectal adenocarcinomas (CRCs) was also conducted.
Forty-six consecutive cases of CRC were randomly selected for DNA isolation, with the approval of the Ethical Committee of Clinical County Emergency Hospital and the University of Medicine, Pharmacy, Science and Technology of Tirgu-Mures, Romania. No preoperative chemo- or radiotherapy was administered in any of the examined cases. The used paraffin blocks from CRC surgical specimens were archived at the Department of Pathology of Clinical County Emergency Hospital of Tirgu-Mures, Romania, during the period 2010-2015.
DNA was extracted from FFPE-CRCs. Hematoxilin and eosin stains were first performed to mark the most appropriate area. The selection of the tumor area was based on the presence of tumor cells in over 80% of the marked tissue, without necroses, hemorrhages, inflammatory or highly fibrotic stroma. After macrodissection of the tumor, 3 x 5 µm sections were created and inserted into 1.5 mL Eppendorf SafeLock tubes (Eppendorf, Hamburg, Germany).
For manual DNA isolation, two commercially available kits were used: the PureLink Genomic DNA Mini Kit from Invitrogen Carlsbad, CA 92008, United States and the High Pure FFPE DNA Isolation Kit from Roche GmbH, Mannheim, Germany. For each of the 46 included cases, two manual isolations per case were performed according to the manufacturers’ protocols (Table 1).
| PureLink Genomic DNA mini kit (Invitrogen–ma-nual) | Time | iPrep genomic DNA kit (Invitrogen–aut-omated) | Time | High Pure FFPE DNA isolation kit (Rochemanual) | Time | MagnaPure LC DNA Isolation kit (Roche–automated) | Time |
| 3 sections of 5 µm | 5-10 min | 3 sections of 5 µm | 5-10 min | 3 sections of 5 µm | 5-10 min | 3 sections of 5 µm | 5-10 min |
| Deparaffinizatio-n using Xylol 3 times | 3 x 10 min | Deparaffinization using Xylol 3 times | 3 x 10 min | Deparaffinization using Xylol 3 times | 3 x 10 min | Deparaffinization using Xylol 3 times | 3 x 10 min |
| Lysis and digestion (20 µL of ProtK) | From 30 min to overnight | Sample lysis | Lysis and digestion (70 µL ProtK) | 30-90 min | Sample lysis and binding | ||
| Binding DNA to silica membranes (columns) | 15 min | Binding DNA to magnetic beads | 30 min | Binding DNA to silica membranes (columns) | 15 min | ProtK | 30 min |
| Washing out contaminants | 15 min | Magnetic separation of beads | Washing out contaminants | 15 min | Binding DNA to beads | ||
| Eluting | 3 min | Separation of liquid solutions from beads Washing beads eluting | Eluting | 3 min | Washing out contaminants Washing beads eluting | ||
| ~ 1.96 Euro/sample | ~ 7.8 Euro/sample | ~ 3.66 Euro/sample | ~ 6.81 Euro/sample |
Invitrogen kits were tested at the Pathology Department of Mures County Hospital, while Roche kits were tested at the Molecular Pathology Laboratory of the National Institute of Oncology, Budapest. The same team performed all isolations.
The manual methods (for both kits) were performed on an anion-exchange resin. It is a macroporous silica-based resin with a high density of diethylaminoethyl (DEAE) groups. The purification is based on the interaction between the negatively charged phosphates of the nucleic acid backbone and the positively charged DEAE groups on the surface of the resin[7].
For automated DNA isolation, two commercially available automated magnetic bead kits were used: the iPrep Genomic DNA kit from Invitrogen and the MagnaPure LC DNA Isolation kit from Roche. Similar to the manual methods, for each of the 46 included cases, two automated isolations per case were performed according to the manufacturers’ protocols (Table 1).
Both of the automated purification techniques use magnetic bead isolation principles. Positively charged magnetic beads can form an ionic bond with the negatively charged DNA backbone at low pH values. At high pH values, the magnetic beads lose their charge and DNA binding ability. In deparaffinized tissues, after a standard automatic tissue lysis step which takes 15 min, the genomic DNA is isolated in a 15-minute procedure that involves binding the genomic DNA to the magnetic beads in a low pH buffer, immobilizing the beads with a magnet, washing and finally, elution in a higher pH buffer (Table 1).
DNA parameters (concentration and quality) were determined using a Nanodrop machine (ThermoScientific, United States). Readings were taken at wavelengths of 260 nm and 280 nm. The optical density (OD) ratio (A260/A280) was automatically calculated.
As a standard parameter for purity, an OD ratio value of 1.8-2.0 was used. A ratio less than 1.8 indicated protein contamination, while a ratio above 2.0 indicated contamination by chloroform, phenol or other organic compounds.
Statistical analysis of the data took into account the DNA parameters, which were compared for all four systems used. They were then correlated with tumor localization, macroscopic and microscopic features, the depth of infiltration, the lymph node ratio, and the tumor stage, which were determined according to the latest classification rules[8]. Chi-squared and Fisher’s exact tests were used for statistical analysis using GraphPad Prism 8.0.1 software. A P value lower than 0.05 with a 95% confidence interval (CI) was considered statistically significant. GraphPad Prism 8.0.1 software, using Chi Square test and Fisher’s exact test, was used for statistically assessment. A P value lower than 0.05, at 95%CI, was considered statistically significant.
In all 46 cases, after the deparaffinization step with xylene, the time required for manual DNA isolation for both manual kits according to the manufacturers’ protocols (Table 1) ranged from 60 min to over 12 h when overnight lysis was necessary. This time was respected for the isolation of a few probes (< 5). In cases of incomplete lysis, re-centrifugation was conducted and Proteinase K was added. These supplementary steps prolonged the isolation time irrespective of the kit used.
The major difference between the two manual isolation kits in terms of the indicated protocol (Table 1) is the quantity of Proteinase K that should be added to the tissue lysis buffer. While Invitrogen suggests using 20 µL of Proteinase K, Roche indicates 70 µL.
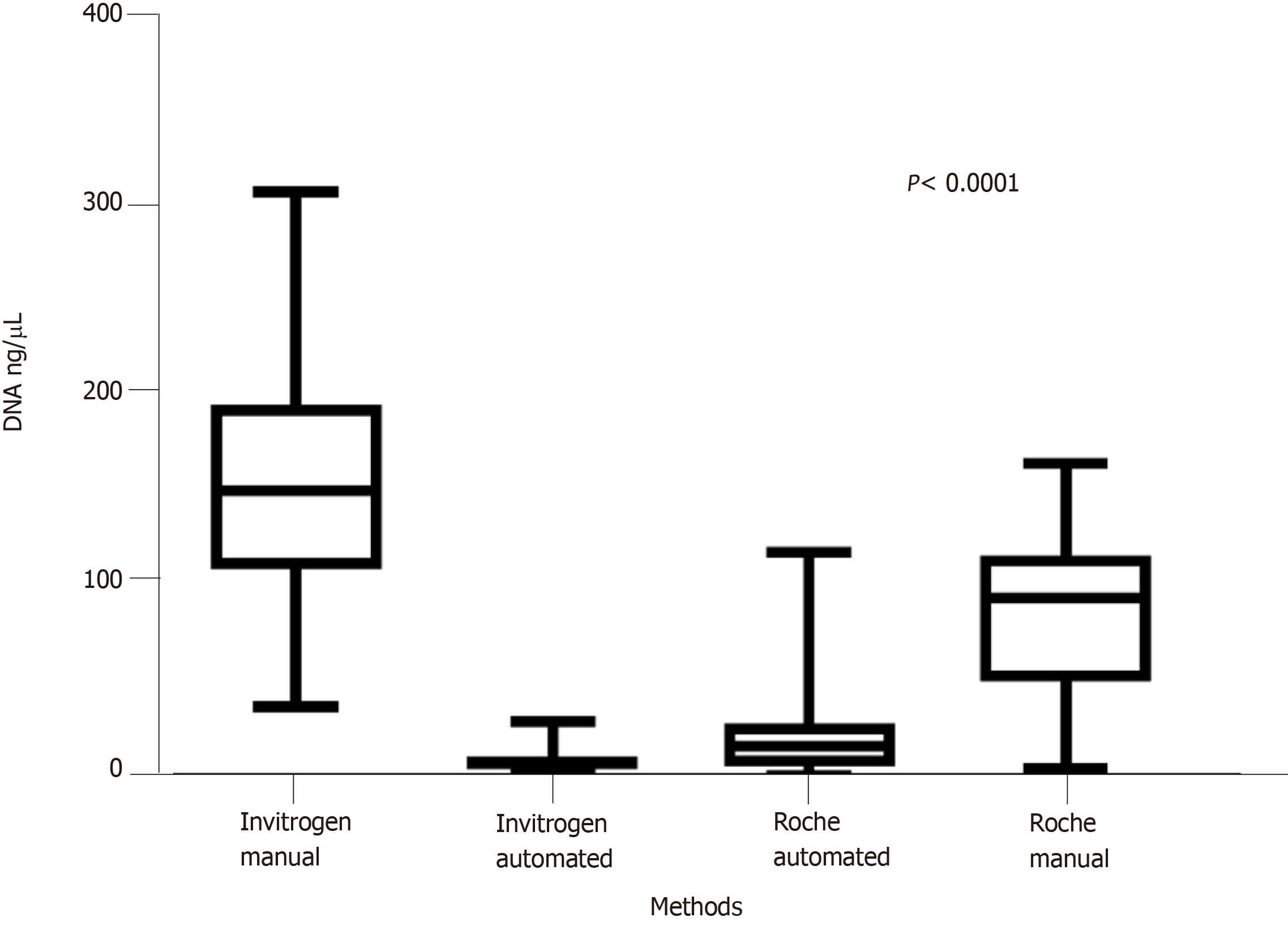
The average DNA concentration isolated with the Invitrogen manual kit was 157.24 ± 62.99 ng/µL (37.6-316 ng/µL), while with the Roche kit a lower median value was obtained (P < 0.0001) at 86.64 ± 43.84 ng/µL (4.2-168.9 ng/µL). In three of the 46 cases, a higher DNA concentration was obtained with the Roche kit, compared with the Invitrogen manual kit (Table 2).
| PureLink Genomic DNA mini kit (Invitrogen–manual) | iPrep genomic DNA kit (Invitrogen–automated) | MagnaPure LC DNA Isolation kit (Roche–automated) | High Pure FFPE DNA isolation kit (Roche–manual) | |
| Mean | 157.24 | 8.72 | 20.39 | 86.64 |
| Standard deviation | 62.99 | 6.40 | 21.19 | 43.84 |
| Minimum | 37.60 | 0.70 | 0.30 | 4.20 |
| Maximum | 316 | 29.80 | 121 | 168.90 |
DNA purity was adequate at 1.8-2.0, without any protein or organic compound contamination, irrespective of the method used. Only four out of the 46 cases had an OD ratio lower than 1.8. A significant difference in the OD value was found between the two manual methods (P = 0.019).
In three of the 46 probes (6.46%), the OD ratio was lower than 1.8 using the Roche manual kit, while using the Invitrogen kit, 16 of the 46 DNA samples (37.78%) had a low OD value. A higher OD value (> 2.00) was found in 29 of the 46 cases using the Roche system and in no cases using the Invitrogen system (Table 3).
| PureLink Genomic DNA mini kit (Invitrogen–manual) | iPrep genomic DNA kit (Invitrogen–automated) | MagnaPure LC DNA Isolation kit (Roche–automated) | High Pure FFPE DNA isolation kit (Roche–manual) | |
| Mean | 1.80 | 1.78 | 1.84 | 2.19 |
| Standard deviation | 0.04 | 0.14 | 0.52 | 0.37 |
| Minimum | 1.67 | 1.42 | 0.99 | 1.60 |
| Maximum | 3.10 | 2.25 | 4.36 | 3.10 |
For both automated methods, the protocol indicated by Invitrogen and Roche, using the magnetic beads principle, was similar (Table 1). For one run, the total time was 30 min for 12 probes with the iPrep Genomic DNA kit from Invitrogen and 30 min for 11 probes with the MagnaPure LC DNA Isolation Kit from Roche. For each run, one template control was used to check the probes for contamination.
Compared with the manual kits, the DNA concentration obtained was significantly lower (P < 0.0001) irrespective of the automatic system used (Figure 1).
A significantly lower (P < 0.0001) DNA concentration (8.72 ± 6.4 ng/µL, 0.70-29.80) was obtained with the automatic iPrep Genomic DNA Kit from Invitrogen, compared with the automatic MagnaPure LC DNA Isolation Kit from Roche (20.39 ± 21.19 ng/µL, 0.30-121) (Tables 2 and 3).
Regarding DNA purity, no significant difference in the OD value was found between the two automated methods (P = 0.56).
In 19 of the 46 probes (41.30%), the OD ratio was lower than 1.8 using the Roche automated system, while using the Invitrogen automated system, 21 of the 46 DNA samples (45.65%) had low OD values. Higher OD values (> 2) were found in 12 of the 46 cases using the Roche system and in four of the 46 cases using the Invitrogen system (Table 3).
Compared to manual isolation methods, the OD values obtained with automated systems were similar for Invitrogen kits (P = 0.32), whereas automated DNA extraction was associated with lower OD values (P < 0.0001).
The comparison between age, gender, tumor localization, the depth of infiltration (pT), lymph node status (pN stage), and the lymph node ratio found no statistically significant difference in DNA concentration using any of the nucleic acid isolation kits (Tables 4-7).
| Characteristics | Number | DNA concentration PureLink Genomic DNA mini kit (manual) | P value | ||
| ≤ 150 ng/µL | > 150 ng/µL | ||||
| Age | ≤ 60 | 20 | 12 | 8 | 0.373 |
| > 60 | 26 | 11 | 15 | ||
| Gender | Male | 26 | 15 | 11 | 0.373 |
| Female | 20 | 8 | 12 | ||
| Macroscopic aspect | Polypoid | 10 | 4 | 15 | 0.00103 |
| Ulcero-infiltrative | 36 | 19 | 8 | ||
| Microscopic aspect | G1 | 13 | 3 | 10 | 0.043 |
| G2 + G3 | 33 | 20 | 13 | ||
| Localization | Proximal colon | 8 | 5 | 3 | 0.892 |
| Distal colon | 21 | 11 | 10 | ||
| Rectum | 17 | 7 | 10 | ||
| Depth of infiltration (pT stage) | T2-T3 T4 | 40 6 | 20 3 | 20 3 | > 0.992 |
| Lymph node metastasis (pN stage) | Present (pN1-3) | 17 | 13 | 4 | 0.063 |
| Absent (pN0) | 29 | 10 | 19 | ||
| Lymph node ratio | < 0.15 | 36 | 17 | 19 | 0.723 |
| ≥ 0.15 | 10 | 6 | 4 | ||
| Characteristics | Number | iPrep genomic DNA kit (magnetic beads) | P value | ||
| ≤ 10 ng/µL | > 10 ng/µL | ||||
| Age | ≤ 60 | 20 | 12 | 8 | 0.093 |
| > 60 | 26 | 22 | 4 | ||
| Gender | Male | 26 | 19 | 7 | 0.993 |
| Female | 20 | 15 | 5 | ||
| Macroscopic aspect | Polypoid | 10 | 3 | 7 | 0.00043 |
| Ulcero-infiltrative | 36 | 31 | 5 | ||
| Microscopic aspect | G1 | 13 | 4 | 9 | < 0.00013 |
| G2 + G3 | 33 | 30 | 3 | ||
| Localization | Proximal colon | 8 | 6 | 2 | 0.522 |
| Distal colon | 21 | 17 | 4 | ||
| Rectum | 17 | 11 | 6 | ||
| Depth of infiltration (pT stage) | T2-T3 T4 | 40 6 | 28 6 | 12 0 | 0.112 |
| Lymph node metastasis (pN stage) | Present (pN1-3) | 17 | 14 | 3 | 0.313 |
| Absent (pN0) | 29 | 20 | 9 | ||
| Lymph node ratio | < 0.15 | 36 | 25 | 11 | 0.253 |
| ≥ 0.15 | 10 | 9 | 1 | ||
| Characteristics | Number | High Pure FFPE DNA isolation kit (manual) | P value | ||
| < 100 ng/µL | ≥ 100 ng/µL | ||||
| Age | ≤ 60 | 20 | 12 | 8 | 0.993 |
| > 60 | 26 | 15 | 11 | ||
| Gender | Male | 26 | 15 | 11 | 0.993 |
| Female | 20 | 12 | 8 | ||
| Macroscopic aspect | Polypoid | 10 | 8 | 17 | 0.123 |
| Ulcero-infiltrative | 36 | 19 | 2 | ||
| Microscopic aspect | G1 | 13 | 3 | 10 | 0.0023 |
| G2 + G3 | 33 | 24 | 9 | ||
| Localization | Proximal colon | 8 | 5 | 3 | 0.962 |
| Distal colon | 21 | 12 | 9 | ||
| Rectum | 17 | 10 | 7 | ||
| Depth of infiltration (pT stage) | T2-T3 T4 | 40 6 | 24 3 | 16 3 | 0.642 |
| Lymph node metastasis (pN stage) | Present (pN1-3) | 17 | 8 | 9 | 0.353 |
| Absent (pN0) | 29 | 19 | 10 | ||
| Lymph node ratio | < 0.15 | 36 | 23 | 13 | 0.273 |
| ≥ 0.15 | 10 | 4 | 6 | ||
| Characteristics | P value | MagnaPure LC DNA Isolation kit (magnetic beads) | P value | ||
| < 20 ng/µL | ≥ 20 ng/µL | ||||
| Age | ≤ 60 | 20 | 12 | 8 | 0.383 |
| > 60 | 26 | 12 | 14 | ||
| Gender | Male | 26 | 12 | 14 | 0.383 |
| Female | 20 | 12 | 8 | ||
| Macroscopic aspect | Polypoid | 10 | 8 | 2 | 0.043 |
| Ulcero-infiltrative | 36 | 16 | 20 | ||
| Microscopic aspect | G1 | 13 | 4 | 9 | 0.023 |
| G2 + G3 | 33 | 23 | 10 | ||
| Localization | Proximal colon | 8 | 3 | 5 | 0.602 |
| Distal colon | 21 | 11 | 10 | ||
| Rectum | 17 | 10 | 7 | ||
| Depth of infiltration (pT stage) | T2-T3 | 40 | 20 | 20 | 0.442 |
| T4 | 6 | 4 | 2 | ||
| Lymph node metastasis (pN stage) | Present (pN1-3) | 17 | 5 | 12 | 0.033 |
| Absent (pN0) | 29 | 19 | 10 | ||
| Lymph node ratio | < 0.15 | 36 | 18 | 18 | 0.723 |
| ≥ 0.15 | 10 | 6 | 4 | ||
DNA concentration was influenced by the macroscopic aspects and grade of differentiation. A higher concentration of DNA was obtained for polypoid in comparison to ulcero-infiltrative carcinomas, with both Roche systems (Tables 6 and 7) and using the automated system from Invitrogen (Table 5). The manual kit from Invitrogen allowed good concentrations to be extracted, but in half of the cases (23 of 46 cases) a value below 150 ng/µL was obtained (Table 4). For this reason, the P value was considered to be at the limit of statistical significance.
Regarding the microscopic aspect of CRC, the concentration of nucleic acids was higher in well-differentiated (G1) carcinomas, compared with G2 + G3 cases (Tables 4-7).
In FFPETs, after deparaffinization, the first step in DNA isolation is cell disruption/lysis[3]. After DNA exposure, membrane lipid removal is conducted by adding detergents, proteins and even proteases (an optional step, but almost always included). Precipitation of the DNA is then performed with alcohol (usually ice-cold ethanol or isopropanol). At the end of the procedure, solubilizing the DNA must be conducted in an alkaline buffer or in ultra-pure water.
During DNA isolation, a chelating agent can be added in order to bind divalent cations and stop DNase activities. Cellular or histone proteins bound to DNA can be removed by adding a protease or by precipitating proteins with sodium/ammonium acetate, or extracting them with a phenol-chloroform mixture prior to the DNA precipitation.
The most commonly used protease in DNA extraction is Proteinase K (protease K or endopeptidase K), which is a broad-spectrum serine protease. It digests and removes proteins as a nucleic acid decontamination step. Proteinase K also inactivates nucleases that might induce DNA or RNA degradation during DNA purification. In this study, it was observed that protein contamination was the same when manual protocols were used, highlighting the fact that it is not affected by the amount of Proteinase K (20 µL vs 70 µL). On the other hand, the manual probes showed a higher median DNA concentration (157 ng/µL vs 87 ng/µL). Irrespective of the manufacturer, the automated DNA extraction was associated with a higher protein contamination rate (OD < 1.8). In these cases, it related to a shorter Proteinase K exposure time, which cannot be modified in-house. Better tissue lysis might induce a lower protein contamination rate.
One original aspect that could be useful in daily practice concerns the correlation obtained in this study between DNA concentration and the clinicopathological parameters of CRCs. Patient age and gender did not influence the DNA concentration, as well as most of the tumor parameters (localization, macroscopic features, pT and pN stage, and lymph node ratio).
We successfully proved that the highest concentration of DNA can be obtained from FFPE well-differentiated CRCs with a polypoid aspect, irrespective of their localization. As ulcero-infiltrative tumors are usually associated with a higher grade of macroscopic lysis, this parameter can influence DNA parameters.
Tumor dedifferentiation might be associated with a high cell division rate[9,10], which could lead to a lower rate of successful DNA lysis.
There are several commercial kits available that include manual and automated isolation procedures. However, although time-consuming, nucleic acid isolation can be done by in-house preparation of all the buffers and solutions necessary for extraction[9,10]. The method used should take into account the quantity of DNA needed [e.g., for adductomics studies or polymerase chain reaction (PCR)] but also the human component, as manual systems need to be managed by well-prepared technicians or biologists.
Fully automated methods can be used successfully for PCR reactions. Although the DNA concentration obtained is lower than by manual methods, it is sufficient for PCR. The costs are higher than for the manual methods.
All of the probes from this study were successfully amplified for real-time PCR reactions. The literature data show that both DNA and RNA can be isolated by automated methods from FFPETs[11-13]. The authors applied a fully automated xylene-free isolation with iron oxide beads coated with a nanolayer of silica[11-13].
An important step in performing DNA isolation from FFPETs is the pre-isolation protocol. Deparaffinization can be performed in tubes (such as in this study) or using slide-digestion (overnight or 72 h) based on in-house protocols. Both methods can be successfully adopted. DNA concentration obtained after 72 h on slide-deparaffinization can be over 500 ng/µL[11-13].
In 2015, Kocjan et al[14] compared 69 commercially available DNA extraction kits from 43 companies. They showed that deparaffinization and supplementary lysis can induce a lower DNA concentration[14]. In this study, we have shown that a lower amount of Proteinase K with longer tissue exposure (which is possible for manual kits) leads to a higher concentration of DNA. Although manual extraction confers a higher yield and DNA concentration, automated isolation will replace it in short time, when the costs decrease significantly[15].
The unresolved issue refers to the imbalance between concentration and quality. We have obtained a reverse correlation between concentration and OD value, which could help researchers in their decisions regarding the most appropriate methods (manual vs automatic) and provide explanations for the understandable problems encountered daily in the laboratory. Similar to our findings, it was previously demonstrated that DNA integrity is higher with manual purification, for both tissues and whole blood[16].
The present study has some limitations. Firstly, the number of included cases is small and originates from a single department, with the same techniques used for tissue preparation. Secondly, only CRC samples were used. The above-mentioned aspects should be investigated in larger cohorts with sample size calculations.
In conclusion, the results of this single-center study highlight the importance of the quality of nucleic acid isolation techniques. Manual methods proved to be more controllable and permit in-house adaptation of the protocol, while the obtained DNA concentrations and purity were higher. On the other hand, automated methods are a time-saving option for PCR and real-time quantitative PCR reactions. For CRC samples, it is expected that a higher DNA concentration would be obtained from differentiated polypoid carcinomas.
Nucleic acid isolation from formalin-fixed, paraffin-embedded tissue (FFPET) samples is a daily routine in molecular pathology laboratories, but extraction from FFPET is not always easily achieved. Choosing the right extraction technique is key for further examinations. Several commercial kits are available on the molecular biology market, including both manual isolation procedures and automated extraction. When choosing the right method for isolation, consideration must be given to the aspects of time, precision, downstream applications and price. Choosing the right technique is key for success in molecular biology, because nucleic acid isolation is always the first step in molecular biology and molecular pathology.
The aim of this paper was to compare the advantages and disadvantages of four DNA extraction kits used in daily practice for DNA isolation from formalin-fixed, paraffin-embedded (FFPE) colorectal adenocarcinoma (CRC) surgical specimens: two manual and two automated magnetic bead kits. A correlation of the results with the clinicopathological features of CRCs was also performed.
By comparing the advantages and disadvantages of nucleic acid isolation techniques used in daily routines, precise decisions can be made regarding the most suitable DNA extraction approach for molecular applications.
DNA was extracted from FFPE-CRCs. The selection of tumor area was based on the presence of tumor cells in over 80% of the marked tissue, without necroses, hemorrhages, inflammatory, or highly fibrotic stroma. For manual DNA isolation, two commercially available kits were used: The PureLink Genomic DNA Mini Kit from Invitrogen Carlsbad, CA 92008, United States and the High Pure FFPE DNA Isolation Kit from Roche GmbH, Mannheim, Germany. For automated DNA isolation, two commercially available automated magnetic bead kits were used: The iPrep Genomic DNA Kit from Invitrogen and the MagnaPure LC DNA Isolation Kit from Roche. DNA parameters (concentration and quality) were determined using a Nanodrop machine (ThermoScientific, United States). Readings were taken at wavelengths of 260 nm and 280 nm. The optical density (OD) ratio (A260/A280) was automatically calculated, before being correlated with tumor localization, macroscopic and microscopic features, the depth of infiltration, the lymph node ratio, and tumor stage, which were determined according to the latest classification rules.
DNA concentration was influenced by the macroscopic features and grade of differentiation. A higher DNA concentration was obtained for polypoid compared with ulcero-infiltrative carcinomas, with both Roche systems and using the automated system from Invitrogen. The manual kit from Invitrogen allowed good concentrations to be extracted, but in half of the cases (23 of 46 cases) a value below 150 ng/µL was obtained. For this reason, the P value was considered to be at the limit of statistical significance.
Manual methods of DNA extraction are more controllable and allow the in-house adaptation of the protocol. The obtained DNA concentrations and purity are higher. Automated methods are a time-saving option for polymerase chain reaction (PCR) and real-time quantitative PCR reactions. For CRC samples, a higher DNA concentration is expected to be obtained from differentiated polypoid carcinomas.
DNA integrity is higher when manual purification is performed, for both tissues and whole blood. The unresolved issue refers to the imbalance between concentration and quality. The above-mentioned aspects should be investigated in larger cohorts with sample size calculations.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country of origin: Romania
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Vynios D, Isik A, Kopljar M S-Editor: Tang JZ L-Editor: A E-Editor: Liu MY
| 1. | Dahm R. Discovering DNA: Friedrich Miescher and the early years of nucleic acid research. Hum Genet. 2008;122:565-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 91] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 2. | Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13387] [Cited by in RCA: 14454] [Article Influence: 390.6] [Reference Citation Analysis (0)] |
| 3. | Lahiri DK, Nurnberger JI. A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res. 1991;19:5444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1594] [Cited by in RCA: 1713] [Article Influence: 50.4] [Reference Citation Analysis (0)] |
| 4. | Goelz SE, Hamilton SR, Vogelstein B. Purification of DNA from formaldehyde fixed and paraffin embedded human tissue. Biochem Biophys Res Commun. 1985;130:118-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 560] [Cited by in RCA: 553] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 5. | Ludyga N, Grünwald B, Azimzadeh O, Englert S, Höfler H, Tapio S, Aubele M. Nucleic acids from long-term preserved FFPE tissues are suitable for downstream analyses. Virchows Arch. 2012;460:131-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 141] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 6. | Barresi V, Cinnirella G, Valenti G, Spampinato G, Musso N, Castorina S, Condorelli DF. Gene expression profiles in genome instability-based classes of colorectal cancer. BMC Cancer. 2018;18:1265. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 7. | Smith C, Otto P, Bitner R, Shiels G. A silica membrane-based method for the isolation of genomic DNA from tissues and cultured cells. CSH Protoc. 2006;2006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 8. | Fulop ZZ, Gurzu S, Bara T, Dragus E, Bara T, Voidazan S, Banias L, Jung I. Lymph node ratio, an independent prognostic factor for patients with stage II-III rectal carcinoma. Pathol Res Pract. 2019;215:152384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 9. | Jordan P. Colorectal Cancer Subtypes - The Current Portrait. Adv Exp Med Biol. 2018;1110:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 10. | Tan SC, Yiap BC. DNA, RNA, and protein extraction: the past and the present. J Biomed Biotechnol. 2009;2009:574398. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 413] [Cited by in RCA: 346] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 11. | Hennig G, Gehrmann M, Stropp U, Brauch H, Fritz P, Eichelbaum M, Schwab M, Schroth W. Automated extraction of DNA and RNA from a single formalin-fixed paraffin-embedded tissue section for analysis of both single-nucleotide polymorphisms and mRNA expression. Clin Chem. 2010;56:1845-1853. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 64] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 12. | Bohmann K, Hennig G, Rogel U, Poremba C, Mueller BM, Fritz P, Stoerkel S, Schaefer KL. RNA extraction from archival formalin-fixed paraffin-embedded tissue: a comparison of manual, semiautomated, and fully automated purification methods. Clin Chem. 2009;55:1719-1727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 97] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 13. | Sengüven B, Baris E, Oygur T, Berktas M. Comparison of methods for the extraction of DNA from formalin-fixed, paraffin-embedded archival tissues. Int J Med Sci. 2014;11:494-499. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 14. | Kocjan BJ, Hošnjak L, Poljak M. Commercially available kits for manual and automatic extraction of nucleic acids from formalin-fixed, paraffin-embedded (FFPE) tissues. Acta Dermatovenerol Alp Pannonica Adriat. 2015;24:47-53. [RCA] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 15. | Kalmár A, Péterfia B, Wichmann B, Patai ÁV, Barták BK, Nagy ZB, Furi I, Tulassay Z, Molnár B. Comparison of Automated and Manual DNA Isolation Methods for DNA Methylation Analysis of Biopsy, Fresh Frozen, and Formalin-Fixed, Paraffin-Embedded Colorectal Cancer Samples. J Lab Autom. 2015;20:642-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 16. | Riemann K, Adamzik M, Frauenrath S, Egensperger R, Schmid KW, Brockmeyer NH, Siffert W. Comparison of manual and automated nucleic acid extraction from whole-blood samples. J Clin Lab Anal. 2007;21:244-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 53] [Article Influence: 2.9] [Reference Citation Analysis (0)] |