Published online May 15, 2024. doi: 10.4251/wjgo.v16.i5.2006
- This article has been corrected.
- See: World J Gastrointest Oncol. Feb 15, 2025; 17(2): 99728
Peer-review started: September 13, 2023
First decision: December 14, 2023
Revised: January 10, 2024
Accepted: March 18, 2024
Article in press: March 18, 2024
Published online: May 15, 2024
Processing time: 239 Days and 12.1 Hours
N6-methyladenosine (m6A) modification represents the predominant alteration found in eukaryotic messenger RNA and plays a crucial role in the progression of various tumors. However, despite its significance, the comprehensive investigation of METTL5, a key m6A methyltransferase, in colorectal cancer (CRC) remains limited.
To investigate the role of METTL5 in CRC.
We assessed METTL5 expression levels in clinical samples obtained from CRC patients as well as in CRC cell lines. To elucidate the downstream targets of METTL5, we performed RNA-sequencing analysis coupled with correlation analysis, leading us to identify Toll-like receptor 8 (TLR8) as a potential downstream target. In vitro functional assessments of METTL5 and TLR8 were conducted using CCK-8 assays, scratch assays, as well as assays measuring cell migration and invasion.
Our findings reveal a pronounced upregulation of METTL5 expression in both CRC cells and tissues, which correlated significantly with an unfavorable prognosis. In vitro experiments unequivocally demonstrated the oncogenic role of METTL5, as evidenced by its promotion of CRC cell proliferation, invasion, and migration. Notably, we identified TLR8 as a downstream target of METTL5, and subsequent down-regulation of TLR8 led to a significant inhibition of CRC cell proliferation, invasion, and tumor growth.
The heightened expression of METTL5 in CRC is strongly associated with clinicopathological features and a poor prognosis, thereby underscoring its potential utility as a critical marker for facilitating early diagnosis and prognostication in CRC.
Core Tip: N6-methyladenosine is one of the most common post-transcriptional RNA modifications in mammals and one of the major methylation modifications in mRNA and non-coding RNAs, which can affect RNA splicing, translation, stability, and epigenetic effects of certain non-coding RNAs. This is the first fundamental study to examine the role of METTL5 in colorectal cancer (CRC), which is important for understanding the impact of the METTL family on the development of CRC.
- Citation: Kong LS, Tao R, Li YF, Wang WB, Zhao X. METTL5 promotes cell proliferation, invasion, and migration by up-regulating Toll-like receptor 8 expression in colorectal cancer. World J Gastrointest Oncol 2024; 16(5): 2006-2017
- URL: https://www.wjgnet.com/1948-5204/full/v16/i5/2006.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i5.2006
Colorectal cancer (CRC) continues to be a serious health problem worldwide, ranking third in both incidence and mortality, with over 1.8 million new CRC cases and 881000 deaths estimated in 2018[1]. Despite numerous early screening and prevention efforts for CRC, the anticipated burden is set to rise. Apart from surgical resection, CRC treatment options encompass chemotherapy, immunotherapy, and targeted therapy. While personalized treatment can extend the overall survival (OS) of CRC patients, the 5-year survival rate remains notably low due to the elusive clinical manifestations of early-stage CRC and the prevalence of advanced cases[2-5]. Moreover, the precise mechanisms underlying tumorigenesis and distant metastasis remain elusive. Consequently, there is an urgent need for further research to delve into the molecular underpinnings of CRC.
In recent years, there has been increasing evidence that the accumulation of various genetic and epigenetic changes activates a variety of carcinogenic signals that are critical to the pathogenesis of CRC. N6-methyladenosine (m6A) modification, the most prevalent internal modification in eukaryotes, influences RNA splicing, transcription, translation, localization, metabolism, and stability, and it plays a key role in many biological processes, such as nervous system development, circadian rhythms, DNA damage response, heat shock response, cell signal transduction, and tumorigenesis. Among them, the role of m6A in tumors has become the focus of current research. As a reversible RNA modification, RNA m6A methylation is regulated by a series of epigenetic regulatory enzymes called “writer”, “reader”, and “eraser”. The expression levels of these m6A regulatory proteins have been found to be dysregulated in many kinds of tumors, including respiratory tumors[6], digestive system tumors[7], reproductive system tumors[8], hematological tumors[9], nervous system tumors[10], etc. Despite much progress, however, the role of some family members in tumor pathogenesis remains unclear.
METTL5, functioning as a methyltransferase, demonstrates targeted activity on the UAACA motif within 18S A1832, altering the conformation between ribosomal RNA and messenger RNA (mRNA), and thereby governing ribosomal translation via m6A modifications in this particular region[11]. Prior research indicated that the absence of METTL5 in mouse embryonic stem cells results in diminished cell translation rates, leading to the natural loss of pluripotency and impaired differentiation potential. Similarly, METTL5 deficiency in Caenorhabditis elegans adversely affects translation. Furthermore, biallelic variants of METTL5 can cause autosomal recessive conditions, potentially resulting in mental retardation and microcephaly[12]. Unlike other METTL family members, the relationship between METTL5 and disease, especially tumors, has not been revealed, and its regulatory role in tumors remains to be further explored, which will help in the development of targeted cancer treatments in the future.
A total of 164 patients who underwent surgical treatment for CRC at the First Affiliated Hospital of Anhui Medical University from January 2018 to December 2018 were included in this study. All patients had complete clinical and pathological data, and their survival was observed through telephone follow-up every 6 months. Finally, the correlation of clinicopathological parameters and prognosis of 150 CRC patients was analyzed. OS was defined as the time from surgery to death. The last follow-up occurred in June 2023. Postoperative specimens (both cancerous and para-cancerous tissues) from all patients were obtained and preserved in liquid nitrogen, and tumor-node-metastasis (TNM) staging of CRC was determined in postoperative pathological diagnosis using the Union International Against Cancer/American Joint Committee on Cancer (8th edition, 2018)[13].
The tissue sections were dewaxed with xylene, hydrated with ethanol, and antigen-recovered. Endogenous peroxidase was quenched with hydrogen peroxide for 30 min, and tissue collagen was blocked with 10% normal goat serum for 30 min. This was followed by incubation with rabbit anti-METTL5 polyclonal antibody at room temperature (25-28 °C) for 60 min. Then, the tissue sections were incubated with secondary antibody and streptavidin-biotin-peroxidase for 15 min, respectively. Color was developed with DAB reagent for 60 s, and then the slides were re-dyed with hematoxylin. Immunohistochemical (IHC) results were evaluated by two experienced pathologists.
RNA was extracted with Trizol method, and reverse transcribed according to the instructions of the reverse transcriptional premix. Polymerase chain reaction (PCR) amplification was performed using primers for METTL5 provided by Kingsley Corporation. The reaction system and cycle parameters were carried out according to SYBR specifications. The data results were analyzed by a relatively quantitative method.
Briefly, migration and invasion assays were performed using NEST cell culture inserts (24-well plates, 8 μm pore size). For the migration test, 1 × 104 cells (per well) in 200 μL serum-free medium were loaded into the base of the insert and then the lower chamber was filled with anhydrous medium. For invasion assay, Matrigel was packaged into the insert and then loaded with 8 × 104 cells (per well) in serum-free medium. A medium containing 10% foetal bovine serum was added to the lower chamber. After incubation for 12 h, the cells on the lower side of the membrane were fixed and stained with 0.5% crystal violet solution. Five random fields of view were counted in each well under the microscope.
Cells at a density of 5 × 105 per well were inoculated into a 6-well culture plate. After incubation for 24 h, a sterile 200 μL pipette tip was used to make a scratch on the middle slide. The cells were allowed to grow for another 24 h and photographs were taken to estimate the closing of the gap. The gap distance was quantitatively evaluated using ImageJ.
For cell proliferation assays, 2 × 103 cells per well were seeded into a 96-well plate. Following cell adhesion, 10 μL of CCK-8 reagent was added to each well on days 1, 2, 3, 4, and 5, and absorbance was measured by spectrophotometry at a wavelength of 450 nm, 2 h post-addition. In the colony formation assay, 500 cells per well were plated in a 6-well dish. After a duration of 2 wk, the cells were fixed using 4% paraformaldehyde, stained with crystal violet, and enumerated under a microscope.
The protein was extracted and resolved via 10% sodium-dodecyl sulfate gel electrophoresis. Subsequently, the protein was transferred onto a polyvinylidene fluoride membrane with a pore size of 0.45 μm. To mitigate nonspecific binding, the membrane was treated with 5% bovine serum albumin for 1 h. Following this, the membrane was initially incubated with the primary antibody overnight at 4 °C, followed by incubation with the secondary antibody. Finally, an ultra-sensitive ECL assay kit was used to demonstrate the immune response, and a two-color infrared fluorescence imaging system was used to image the spots.
Total RNA was isolated from cells using RNAiso plus (#9109, Takara), and its purity and integrity were assessed with the ND-1000 NanoDrop and Agilent 2200 TapeStation, respectively. rRNA was removed with the Epicentre Ribo-Zero rRNA removal kit (Illumina), followed by cDNA synthesis, adapter ligation, and low-cycle enrichment using Illumina’s NEBNext® Ultra™ RNA library preparation kit. Sequencing (2 × 150 bp) was performed on the HiSeq 3000 platform. Clean reads were obtained after filtering, and differential expression analysis was conducted using DEseq with the Benjamini-Hochberg multiple test correction method (fold change > 2, adjusted P value < 0.05). Kyoto Encyclopedia of Genes and Genomes enrichment analysis was performed with a significance threshold of P < 0.05.
SPSS 22.0 and GraphPad Prism 9 were used to process the data. Adobe Illustrator software was used to further manipulate images. All statistical criteria were P < 0.05. Quantitative real-time PCR (RT-qPCR) results were analyzed by paired sample t test, and IHC results were stratified data and compared by rank-transformation nonparametric test. Kaplan-Meier survival analysis was used to analyze the prognostic data of CRC patients.
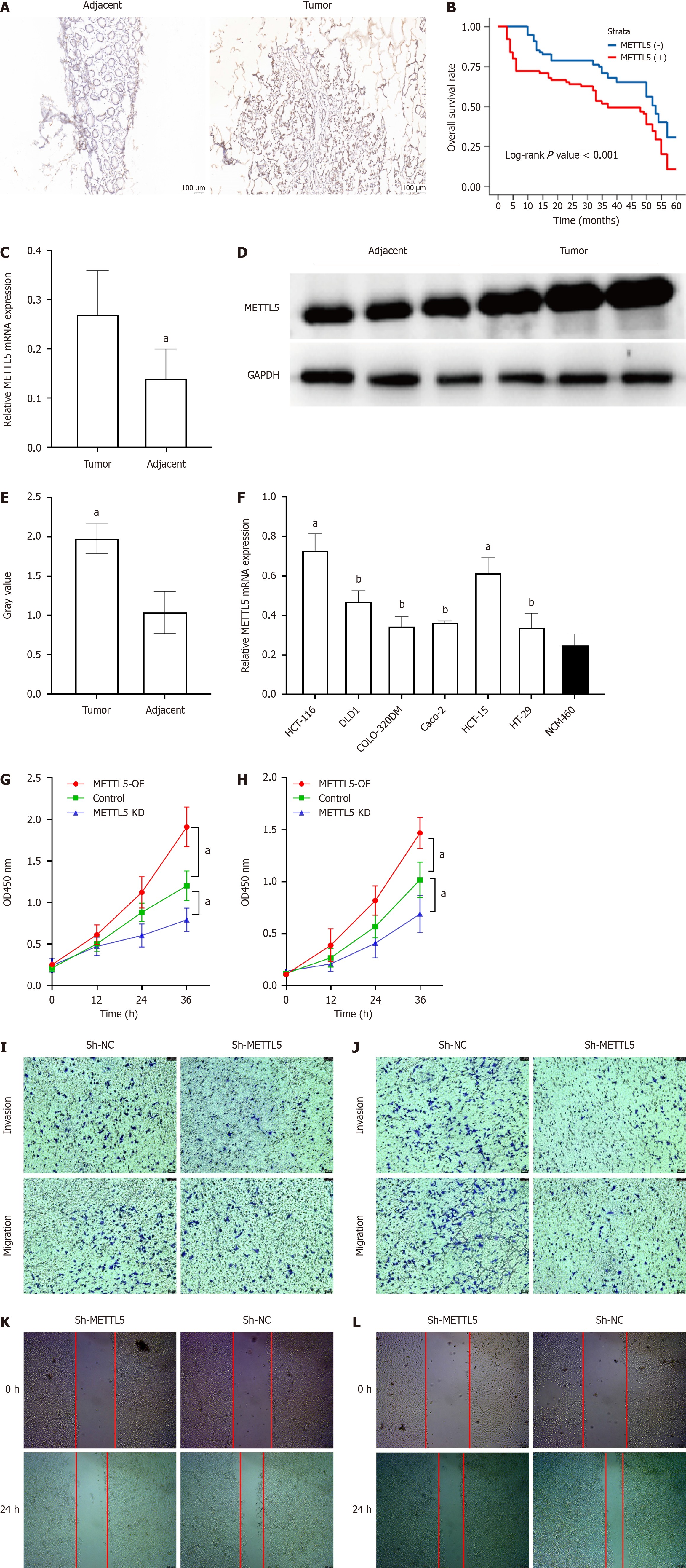
To investigate the expression and clinical significance of METTL5 in CRC, IHC was performed on postoperative paraffin sections of tissues from all patients included in the study. The results showed that METTL5 was highly expressed in 98 cases (65.33%) of CRC tissues (Figure 1A). The χ2 test showed that the expression of METTL5 in CRC and paracancer tissues was significantly different. CRC patients were divided into low expression group and high expression group according to the median immunohistochemical score (75 vs 75). After analyzing the clinical, pathological, and prognostic data of patients, we observed that the expression of METTL5 was correlated with the degree of tumor differentiation and TNM stage in CRC patients (Table 1). METTL5 expression had no significant correlation with gender, age, tumor size, depth of infiltration, positive lymph nodes, vascular and nerve invasion. Univariate and multivariate analyses showed that the expression of METTL5 protein was an independent prognostic factor in CRC patients (Table 2). Patients exhibiting high METTL5 expression had a significantly lower OS compared to those with low expression (median survival and 5-year survival, Figure 1B). The expression of METTL5 mRNA in CRC tissues and adjacent normal tissues was detected by RT-qPCR in 50 random samples of CRC patients. The results showed that the expression of METTL5 mRNA in CRC tissues was higher than that in corresponding adjacent tissues (P < 0.01, Figure 1C). The samples of three patients were randomly selected and western blot method was used to determine the protein level, and the same result was obtained (Figure 1D and E). These results all confirm that METTL5 is highly expressed in CRC tissues, and overexpression of METTL5 is associated with a poor prognosis in CRC patients.
| Variable | METTL5 expression | P value | ||
| Low (n = 75) | High (n = 75) | |||
| Gender | Female | 31 | 40 | 0.141 |
| Male | 44 | 35 | ||
| Age (yr) | > 55 | 48 | 51 | 0.605 |
| ≤ 55 | 27 | 24 | ||
| Tumor size (cm) | > 3.5 | 55 | 56 | 0.852 |
| ≤ 3.5 | 20 | 19 | ||
| Vascular invasion | Positive | 35 | 37 | 0.744 |
| Negative | 40 | 38 | ||
| Nerve invasion | Positive | 31 | 36 | 0.412 |
| Negative | 44 | 39 | ||
| Differentiation degree | High/median | 25 | 45 | 0.001 |
| Low | 50 | 30 | ||
| Depth of infiltration | T1/T2 | 50 | 49 | 0.863 |
| T3/T4 | 25 | 26 | ||
| Positive lymph nodes | ≥ 3 | 10 | 16 | 0.196 |
| < 3 | 65 | 59 | ||
| TNM stage | I/II | 57 | 26 | < 0.001 |
| III/IV | 18 | 49 | ||
| Variable | Univariate analysis | Multivariate analysis | ||||
| HR | 95%CI | P value | HR | 95%CI | P value | |
| Age (yr) | ||||||
| > 55 vs ≤ 55 | 1.217 | 0.827-1.789 | 0.319 | |||
| Gender | ||||||
| Female vs male | 0.845 | 0.589-1.211 | 0.36 | |||
| Tumor site | ||||||
| Colon vs rectum | 0.698 | 0.463-1.053 | 0.087 | |||
| Tumor size | ||||||
| > 3.5 cm vs ≤ 3.5 cm | 1.09 | 0.717-1.657 | 0.687 | |||
| Differentiation degree | ||||||
| Poor vs high/median | 1.483 | 1.034-2.128 | 0.032a | 1.485 | 1.029-2.143 | 0.035a |
| Vascular invasion | ||||||
| Positive vs negative | 1.011 | 0.705-1.45 | 0.951 | |||
| Nerve invasion | ||||||
| Positive vs negative | 1.139 | 0.793-1.635 | 0.482 | |||
| TNM stage | ||||||
| I/II vs III/IV | 1.659 | 1.157-2.38 | 0.006a | 1.498 | 1.024-2.193 | 0.037a |
| METTL5 expression | ||||||
| Decreased vs non-decreased | 1.831 | 1.272-2.635 | 0.001a | 1.582 | 1.078-2.323 | 0.019a |
To further confirm the role of METTL5 in CRC progression, we first evaluated the expression of METTL5 mRNA in different CRC cell lines using RT-PCR, and we found that METTL5 expression was generally increased in CRC cell lines relative to normal colorectal mucosal epithelial cells (Figure 1F). After that, lentivirus transfection was used to establish stable cell lines with METTL5 knockdown. CCK-8 results showed that down-regulated METTL5 could significantly inhibit the proliferation ability of CRC cells (Figure 1G and H).
The transwell assay was employed to examine the impact of METTL5 on the invasion and migration capabilities of CRC cells. The findings indicated that in HCT-15 and HCT-116 cell lines, compared with the control group, the expression of METTL5 was knocked down to significantly reduce the invasion and migration ability of CRC cells (Figure 1I and J). The scratch test obtained the same results (Figure 1K and L).
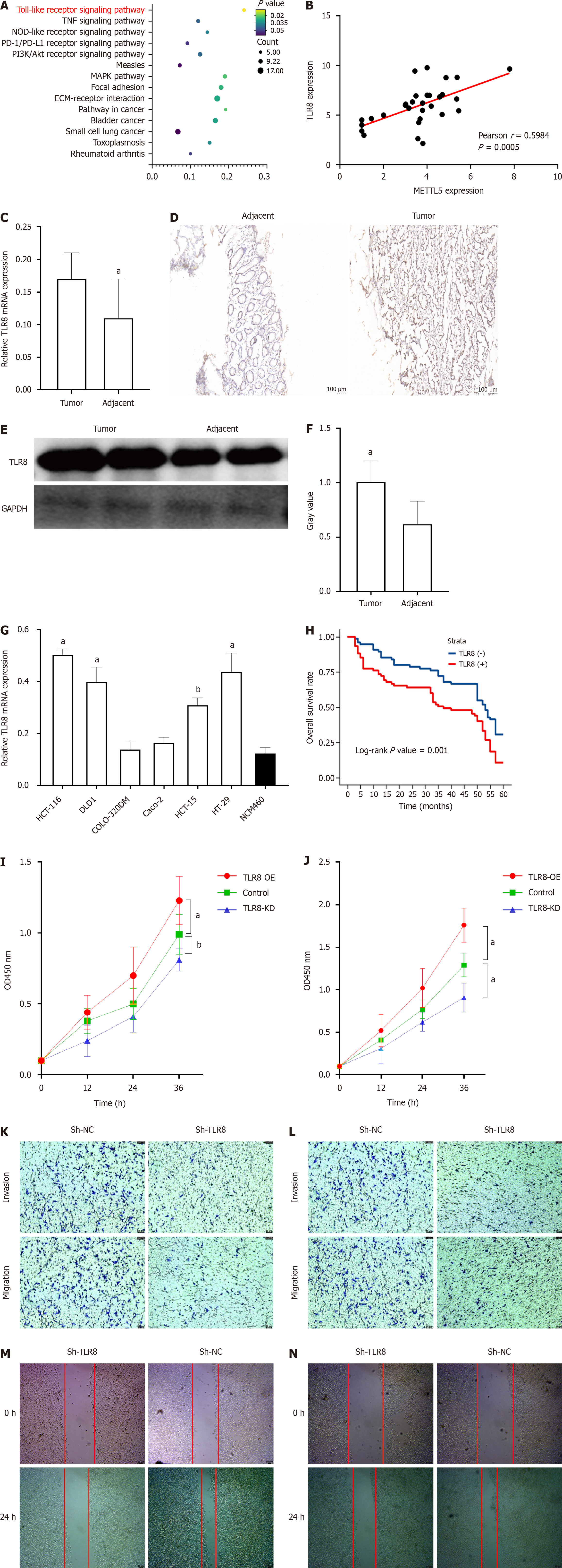
To identify additional molecular targets of METTL5, transcriptome sequencing (mRNA-seq) was conducted on HCT-116 cells following METTL5 depletion. Differentially expressed genes were then selected and ranked based on their P-values. These genes are mainly concentrated in the Toll-like receptor (TLR) pathway (Figure 2A). Among the 203 upregulated genes identified in HCT-116 cell line with METTL5 knockout, a molecule related to TLR was selected as the main effective downstream target, the correlation analysis was completed by PCR assay in 30 randomly selected CRC tissues, and finally TLR8 was selected as the candidate molecule (Figure 2B). Together, all these findings suggest that TLR8 is a downstream target of METTL5.
To investigate the expression and clinical significance of TLR8 in CRC, immunohistochemical staining was performed on postoperative paraffin sections of all patients included in the study. The results showed that TLR8 was highly expressed in 87 cases (58%) of CRC (Figure 2D). The χ2 test showed that TLR8 expression was significantly different between CRC and para-cancer tissues. The results of PCR and western blot analysis confirmed that further (Figure 2C, E, and F). Patients with high expression levels of TLR8 exhibited significantly lower OS rates compared to those with low TLR8 expression (median survival and 5-year survival, Figure 2H). These results all confirm that METTL5 is highly expressed in CRC tissues, and TLR8 overexpression is associated with a poor prognosis in CRC patients.
In CRC cell lines, we first evaluated TLR8 mRNA expression using PCR and showed that TLR8 expression was generally increased in CRC cell lines relative to normal colorectal mucosal epithelial cells (Figure 2G). After that, lentivirus transfection was used to establish a stable TLR8 knockdown cell line. CCK-8 results showed that down-regulation of TLR8 could significantly inhibit the proliferation of CRC cells (Figure 2I and J). The transwell assay was employed to assess the impact of TLR8 on the invasion and migration abilities of CRC cells. The results demonstrated that in HCT-116 and HT-29 cell lines, the knockdown of TLR8 expression significantly reduced the invasion and migration of CRC cells compared with the control group (Figure 2K and L). The scratch test obtained the same results (Figure 2M and N).
Along with the progress of targeted therapy, the prognosis of CRC patients has improved to some extent, but advanced CRC remains a great challenge. There is an urgent need for new diagnosis and therapeutic targets and further understanding of the pathogenesis of CRC. There is increasing evidence that dysregulation of m6A modifications plays an important role in the pathogenesis of many different types of diseases, especially cancer. M6A methyltransferases are important molecules in the m6A modification process, which has important physiological and biological functions. METTL5 is a member of the METTL family, and unlike other family members such as METTL3, few previous studies have described its relationship with tumors, and the role of METTL5 in CRC remains puzzling.
Currently, the expression patterns of METTL5 in CRC and its prognostic implications remain obscure. In this study, IHC staining was conducted on 100 pairs of paraffin sections comprising CRC tissues and their corresponding para-cancerous tissues. Subsequent PCR and western blot analyses were employed to validate METTL5 mRNA and protein levels in CRC tissues. Our findings revealed a significant upregulation of METTL5 expression in CRC tissues compared to para-cancerous tissues. Moreover, in CRC patients, elevated METTL5 expression correlated negatively with OS and exhibited close associations with factors such as TNM stage and differentiation degree, thereby establishing it as an independent prognostic indicator for CRC. Furthermore, quantitative analyses of METTL5 mRNA and protein levels in CRC cell lines yielded results consistent with those obtained from tissue experiments.
In cancer studies, METTL5 has been reported to promote the activation of p70-S6K translation initiation, and the loss of METTL5 significantly reduces the abundance of the polymer. METTL5 is expressed elevated in breast cancer samples and cell lines, promoting translation initiation and cell growth[14]. METTL5 has been identified as an oncogene in pancreatic cancer, and its tumorigenic role may be linked to the upregulation of c-Myc translation[15]. Studies by Xia et al[16] have demonstrated that METTL5 drives glucose metabolic reprogramming, thereby facilitating proliferation and metastasis in hepatocellular cancer (HCC). Mechanistically, increased METTL5 expression stabilizes c-Myc. Additionally, Xu et al[17] observed that downregulation of METTL5 led to decreased expression of programmed death ligand 1 and suppressed malignant cell behaviors in HCC by inhibiting the Myc pathway. Jiang et al[18] used bioinformatics methods to comprehensively analyze RNA-seq data and somatic mutation data of The Cancer Genome Atlas Colon Adenocarcinoma and The Cancer Genome Atlas Rectal Adenocarcinoma, and found that in CRC, except METTL14, YTHDF2, and YTHDF3, the expression of m6a related genes (including METTL5) in CRC was significantly different from that in normal control group. This is highly consistent with our findings. Wang and Peng[19] searched several public databases and concluded through analysis that the occurrence and development of liver cancer are closely related to the expression of METTL5, and the overexpression of METTL5 leads to poor survival outcomes in HCC patients by regulating the tumor immune microenvironment. Similar results have also been found in renal, gastric, and lung cancers[20-22].
TLRs were initially identified as a class of proteins expressed on the membrane of innate immune cells such as macrophages and dendritic cells. It has been proved that TLRs are an important part of the innate immune system, which play a connecting role between innate and adaptive immunity, and can regulate pathogen infection and inflammatory response. TLR-mediated signaling pathways include MyD88-dependent pathways and TRIF-dependent pathways, both of which can induce the expression of relevant genes. Mammalian TLRs recognize a variety of microbial products: TLR2 is specific to peptidoglycans and lipoproteins, and it can cooperate with TLR1 and TLR6 to recognize different ligands; TLR3 is specific to double-stranded RNA. TLR4 is specific to lipopolysaccharide and lipoteichoic acid. In addition to MYD88-dependent signaling, TLR4 also signals through an additional adaptor protein TIRAP (an adaptor with TIR domain). TLR5 was specific to bacterial flagellin. TLR9 is specific to CpG-DNA.
TLR8 acts as an important sentinel in response to infection, which makes them irreplaceable in the activation of mammalian innate immune cells. It was also found that this receptor is involved in the regulation of host adaptive immunity. Over the past decade, the emergence of novel small molecule drugs targeting TLR8 has greatly boosted cancer immunotherapy research by harnessing their immune activation potential[23-26]. TLR stimulation leads to the activation of nuclear factor-kappaB, a key regulator that drives cancer inflammation and mitogen-activated protein kinase, and genetic analysis shows that TLR7-TLR10 and COX-2 expression are significantly upregated in CRC tumor tissues. Analysis of tumor cells isolated from the primary tumor showed that TLR7 and TLR8 were co-expressed with CD133 and provided evidence of colon cancer initiation cell subsets. TLR8 expression was found to be an independent prognostic factor in multivariate analysis[27]. Notably, multiple studies have demonstrated the great potential of TLR7/8 agonists as a novel immune adjuvant in oxaliplatin-resistant CRC chemotherapy as well as radiation therapy, inducing a strong local and profound systemic immune response to tumor antigens released by conventional therapy[28,29]. The study of Lu et al[29] confirmed that the expression levels of TLR1, TLR2, TLR4, TLR8, interleukin (IL)-6, and IL-8 in CRC tissues were higher than those in normal colon mucosa, which was consistent with our findings. The authors further found that their upregulation may be related to inflammatory factors such as IL-6.
In summary, this study explored a previously overlooked area, namely, the role of METTL5 in CRC. Finally, it was confirmed that METTL5 plays a role as an oncogenic protein in CRC progression through its interaction with TLR8, providing new evidence and ideas for further understanding the role of m6A’s epigenetic characteristics in malignant tumors.
The high expression of METTL5 in CRC is associated with clinicopathological features and a poor prognosis, and can be used as an important marker for the early diagnosis and prognosis of CRC.
As a key N6-methyladenosine (m6A) methyltransferase, METTL5 has not been thoroughly studied in colorectal cancer (CRC).
For the first time, METTL5, a popular m6A component, was studied in CRC, which confirmed the potential of METTL5 as a possible new therapeutic target.
To look for a new target that can accurately diagnose and treat CRC.
Cell biological behavior experiments were used to demonstrate the effect of METTL5 knockout on tumor cell behavior, and RNA-sequencing was used to search for downstream molecules of METTL5.
In CRC cells and tissues, METTL5 expression was significantly upregulated, which was significantly associated with a poor prognosis. Toll-like receptor 8 may be a downstream target of METTL5.
METTL5 can be used as an important marker for the early diagnosis and prognosis of CRC.
Larger population sample validation and more accurate m6A quantification techniques need to be applied.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Ricci AD, Italy S-Editor: Wang JJ L-Editor: Wang TQ P-Editor: Zheng XM
| 1. | GBD 2017 Colorectal Cancer Collaborators. The global, regional, and national burden of colorectal cancer and its attributable risk factors in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2019;4:913-933. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 283] [Cited by in RCA: 279] [Article Influence: 46.5] [Reference Citation Analysis (0)] |
| 2. | Santoni M, Rizzo A, Mollica V, Matrana MR, Rosellini M, Faloppi L, Marchetti A, Battelli N, Massari F. The impact of gender on The efficacy of immune checkpoint inhibitors in cancer patients: The MOUSEION-01 study. Crit Rev Oncol Hematol. 2022;170:103596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 118] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 3. | Brandi G, Ricci AD, Rizzo A, Zanfi C, Tavolari S, Palloni A, De Lorenzo S, Ravaioli M, Cescon M. Is post-transplant chemotherapy feasible in liver transplantation for colorectal cancer liver metastases? Cancer Commun (Lond). 2020;40:461-464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 4. | Mollica V, Rizzo A, Marchetti A, Tateo V, Tassinari E, Rosellini M, Massafra R, Santoni M, Massari F. The impact of ECOG performance status on efficacy of immunotherapy and immune-based combinations in cancer patients: the MOUSEION-06 study. Clin Exp Med. 2023;23:5039-5049. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 108] [Article Influence: 54.0] [Reference Citation Analysis (0)] |
| 5. | Rizzo A, Nannini M, Novelli M, Dalia Ricci A, Scioscio VD, Pantaleo MA. Dose reduction and discontinuation of standard-dose regorafenib associated with adverse drug events in cancer patients: a systematic review and meta-analysis. Ther Adv Med Oncol. 2020;12:1758835920936932. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 6. | Teng PC, Liang Y, Yarmishyn AA, Hsiao YJ, Lin TY, Lin TW, Teng YC, Yang YP, Wang ML, Chien CS, Luo YH, Chen YM, Hsu PK, Chiou SH, Chien Y. RNA Modifications and Epigenetics in Modulation of Lung Cancer and Pulmonary Diseases. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 99] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 7. | Cai X, Liang C, Zhang M, Xu Y, Weng Y, Li X, Yu W. N6-methyladenosine modification and metabolic reprogramming of digestive system malignancies. Cancer Lett. 2022;544:215815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 8. | Chen J, Fang Y, Xu Y, Sun H. Role of m6A modification in female infertility and reproductive system diseases. Int J Biol Sci. 2022;18:3592-3604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 60] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 9. | Wang WJ, Xu TT, Bao J. N6-methyladenosine in hematological malignancies: a concise review. Curr Opin Hematol. 2023;30:4-13. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 10. | Zhang N, Ding C, Zuo Y, Peng Y, Zuo L. N6-methyladenosine and Neurological Diseases. Mol Neurobiol. 2022;59:1925-1937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 98] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 11. | Turkalj EM, Vissers C. The emerging importance of METTL5-mediated ribosomal RNA methylation. Exp Mol Med. 2022;54:1617-1625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 20] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 12. | Sendinc E, Shi Y. RNA m6A methylation across the transcriptome. Mol Cell. 2023;83:428-441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 209] [Reference Citation Analysis (0)] |
| 13. | Chen K, Collins G, Wang H, Toh JWT. Pathological Features and Prognostication in Colorectal Cancer. Curr Oncol. 2021;28:5356-5383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 122] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 14. | Rong B, Zhang Q, Wan J, Xing S, Dai R, Li Y, Cai J, Xie J, Song Y, Chen J, Zhang L, Yan G, Zhang W, Gao H, Han JJ, Qu Q, Ma H, Tian Y, Lan F. Ribosome 18S m(6)A Methyltransferase METTL5 Promotes Translation Initiation and Breast Cancer Cell Growth. Cell Rep. 2020;33:108544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 98] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 15. | Huang H, Li H, Pan R, Wang S, Khan AA, Zhao Y, Zhu H, Liu X. Ribosome 18S m(6)A methyltransferase METTL5 promotes pancreatic cancer progression by modulating cMyc translation. Int J Oncol. 2022;60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 25] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 16. | Xia P, Zhang H, Lu H, Xu K, Jiang X, Jiang Y, Gongye X, Chen Z, Liu J, Chen X, Ma W, Zhang Z, Yuan Y. METTL5 stabilizes c-Myc by facilitating USP5 translation to reprogram glucose metabolism and promote hepatocellular carcinoma progression. Cancer Commun (Lond). 2023;43:338-364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 81] [Reference Citation Analysis (0)] |
| 17. | Xu W, Liu S, Zhang G, Liu J, Cao G. Knockdown of METTL5 inhibits the Myc pathway to downregulate PD-L1 expression and inhibits immune escape of hepatocellular carcinoma cells. J Chemother. 2023;35:455-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 18. | Jiang T, Xing L, Zhao L, Ye Z, Yu D, Lin S. Comprehensive analysis of m6A related gene mutation characteristics and prognosis in colorectal cancer. BMC Med Genomics. 2023;16:105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 19. | Wang L, Peng JL. METTL5 serves as a diagnostic and prognostic biomarker in hepatocellular carcinoma by influencing the immune microenvironment. Sci Rep. 2023;13:10755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 20. | Wei Zhang, Chen Y, Zeng Z, Peng Y, Li L, Hu N, Gao X, Cai W, Yin L, Xu Y, Zhang X, Tang D, Dai Y. The novel m6A writer METTL5 as prognostic biomarker probably associating with the regulation of immune microenvironment in kidney cancer. Heliyon. 2022;8:e12078. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 21. | Wang Z, Liu J, Yang Y, Xing C, Jing J, Yuan Y. Expression and prognostic potential of ribosome 18S RNA m(6)A methyltransferase METTL5 in gastric cancer. Cancer Cell Int. 2021;21:569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 22. | Zhang Z, Zhang C, Yang Z, Zhang G, Wu P, Luo Y, Zeng Q, Wang L, Xue Q, Zhang Y, Sun N, He J. m(6)A regulators as predictive biomarkers for chemotherapy benefit and potential therapeutic targets for overcoming chemotherapy resistance in small-cell lung cancer. J Hematol Oncol. 2021;14:190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 23. | McGowan DC. Latest Advances in Small Molecule TLR 7/8 Agonist Drug Research. Curr Top Med Chem. 2019;19:2228-2238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 24. | Clark RA, Huang SJ, Murphy GF, Mollet IG, Hijnen D, Muthukuru M, Schanbacher CF, Edwards V, Miller DM, Kim JE, Lambert J, Kupper TS. Human squamous cell carcinomas evade the immune response by down-regulation of vascular E-selectin and recruitment of regulatory T cells. J Exp Med. 2008;205:2221-2234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 195] [Cited by in RCA: 173] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 25. | Ye J, Ma C, Hsueh EC, Dou J, Mo W, Liu S, Han B, Huang Y, Zhang Y, Varvares MA, Hoft DF, Peng G. TLR8 signaling enhances tumor immunity by preventing tumor-induced T-cell senescence. EMBO Mol Med. 2014;6:1294-1311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 130] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 26. | Michaelis KA, Norgard MA, Zhu X, Levasseur PR, Sivagnanam S, Liudahl SM, Burfeind KG, Olson B, Pelz KR, Angeles Ramos DM, Maurer HC, Olive KP, Coussens LM, Morgan TK, Marks DL. The TLR7/8 agonist R848 remodels tumor and host responses to promote survival in pancreatic cancer. Nat Commun. 2019;10:4682. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 141] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 27. | Grimm M, Kim M, Rosenwald A, Heemann U, Germer CT, Waaga-Gasser AM, Gasser M. Toll-like receptor (TLR) 7 and TLR8 expression on CD133+ cells in colorectal cancer points to a specific role for inflammation-induced TLRs in tumourigenesis and tumour progression. Eur J Cancer. 2010;46:2849-2857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 61] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 28. | Liu Z, Xie Y, Xiong Y, Liu S, Qiu C, Zhu Z, Mao H, Yu M, Wang X. TLR 7/8 agonist reverses oxaliplatin resistance in colorectal cancer via directing the myeloid-derived suppressor cells to tumoricidal M1-macrophages. Cancer Lett. 2020;469:173-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 122] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 29. | Lu CC, Kuo HC, Wang FS, Jou MH, Lee KC, Chuang JH. Upregulation of TLRs and IL-6 as a marker in human colorectal cancer. Int J Mol Sci. 2014;16:159-177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |