Published online Dec 15, 2022. doi: 10.4251/wjgo.v14.i12.2313
Peer-review started: July 14, 2022
First decision: September 26, 2022
Revised: October 17, 2022
Accepted: November 22, 2022
Article in press: November 22, 2022
Published online: December 15, 2022
Processing time: 151 Days and 4.4 Hours
Invasion and migration are the irreversible stages of colorectal cancer (CRC). The key is to find a sensitive, reliable molecular marker that can predict the migration of CRC at an early stage. N-myc downstream regulated gene 1 (NDRG1) is a multifunctional gene that has been tentatively reported to have a strong relationship with tumor invasion and migration, however the current molecular role of NDRG1 in CRC remains unknown.
To explore the role of NDRG1 in the development of CRC.
NDRG1 stably over-expressed Caco2 cell line was established by lentiviral infection and NDRG1 knock-out Caco2 cell line was established by CRISPR/Cas9. Furthermore, the mRNA and protein levels of NDRG1 in Caco2 cells after NDRG1 over-expression and knockout were detected by real-time polymerase chain reaction and western blot. The cell proliferation rate was measured by the cell counting kit-8 method; cell cycle and apoptosis were detected by flow cytometry; invasion and migration ability were detected by the 24-transwell method.
NDRG1 over-expression inhibited Caco2 proliferation and the cell cycle could be arrested at the G1/S phase when NDRG1 was over-expressed, while the number of cells in the G2 phase was significantly increased when NDRG1 was knocked out. This suggests that NDRG1 inhibited the proliferation of Caco2 cells by arresting the cell cycle in the G1/S phase. Our data also demo
NDRG1 inhibits tumor progression in Caco2 cells which may represent a potential novel therapeutic strategy for the treatment of CRC.
Core Tip: This study investigated the molecular functions of N-myc downstream regulated gene 1 (NDRG1) in the process of colorectal cancer (CRC) migration through stable over-expression or knockout of NDRG1 in the Caco2 CRC cell line. Our results showed that NDRG1 over-expression arrested the cell cycle at the G1/S phase, while its knock-out significantly increased the number of G2 phase cells. Altogether, our results highlight the fact that NDRG1 inhibits tumor progression in Caco2 cells which may provide a novel diagnostic or therapeutic tool in inhibiting the migration of CRC.
- Citation: He YX, Shen H, Ji YZ, Hua HR, Zhu Y, Zeng XF, Wang F, Wang KX. N-myc downstream regulated gene 1 inhibition of tumor progression in Caco2 cells. World J Gastrointest Oncol 2022; 14(12): 2313-2328
- URL: https://www.wjgnet.com/1948-5204/full/v14/i12/2313.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i12.2313
Malignant neoplasms are currently the leading cause of death in the world and the biggest obstacle to longer lives. Among new cancer cases in the world in 2018[1], colorectal cancer (CRC) accounted for 10.2% of new cancer cases and 9.2% of cancer deaths. CRC has become the third most common cancer and the incidence of CRC is increasing year by year.
Invasion and metastasis are two malignant features of CRC, involving the spreading of tumor cells from the primary tumor, penetrating into blood and lymphatic vessels and extravasation to the metastatic site. These are the main issues affecting the efficacy of treatment and the prognosis of CRC, and are the main cause of death in patients with CRC. Recent studies on the molecular biological mechanism of CRC indicate that the invasion and migration of CRC are associated with genes as well as microRNA[2]. It is the result of the combined action of multiple transfer-related genes and transfer-inhibitory genes. Many new biological indicators of prognosis and potential therapeutic targets related to CRC are being studied, however, early and sensitive biological indicators that can predict the migration of CRC at an early stage have not yet been discovered.
N-myc downstream regulated gene 1 (NDRG1) is a multifunctional gene. Recent studies have shown that NDRG1 is related to tumor invasion and migration[3], apoptosis[4], tumor cell proliferation[5], drug response and drug resistance of tumor cells[6]. Its expression in tumors is tissue-specific; it acts as a metastasis suppressor gene in prostate cancer[7] and ovarian cancer[8], but in lung cancer and esophageal squamous cell carcinoma, NDRG1 promotes tumor development[9]. The current role of NDRG1 in CRC is controversial. Many researchers have found that NDRG1 inhibits tumor invasion and migration in CRC[10-12], however, Wang et al[13] and Shah et al[14] found that NDRG1 promoted the development of CRC. Koshiji et al[15] found that the expression of NDRG1 differed with race and pathological stage of CRC patients.
To further investigate the role of NDRG1 in the development of CRC, we over-expressed and knocked out NDRG1 in the Caco2 CRC cell line, and then assessed cell proliferation, apoptosis, invasion and migration in vitro. We conducted these experiments to analyze the role of the NDRG1 gene in CRC, thus providing a theoretical basis for the early diagnosis and prognosis of CRC metastasis and a potential new molecular target for treatment of CRC.
A Caco2 CRC cell line was purchased from the Kunming Cell Bank of the Kunming Institute of Zoology, Chinese Academy of Sciences. The Caco2 cell line was derived from a 72-year-old Caucasian male with colorectal adenocarcinoma. The 293T cells were provided by the Kunming Institute of Zoology, Chinese Academy of Sciences. The media required for culturing Caco2 and 293T cells was a Dulbecco’s Minimum Essential Medium media (DMEM; Gibco, Thermo Fisher Scientific, Waltham, MA, United States) containing 10% fetal bovine serum (FBS; Gibco), 100 U/mL penicillin and 100 mg/mL streptomycin. All cell lines were incubated at 37 °C in a humidified atmosphere with 5% CO2.
The lentiviral plasmid used for NDRG1 over-expression was GV358-NDRG1 (Genechem, Shanghai, China). The plasmid vector used for NDRG1 knockout was pL-CRISPR.EFS.GFP, which was kindly provided by the Kunming Institute of Zoology, Chinese Academy of Sciences. We designed three single-guide RNAs (sgRNAs) which were both located in exon 3 of the NDRG1 gene: sgRNA1, sgRNA2 and sgRNA3 (Table 1), inserting these three sgRNAs into the pL-CRISPR.EFS.GFP plasmid. All constructed plasmids were identified by sequencing. The empty GV358 and pL-CRISPR.EFS.GFP plasmids were used as a control. The plasmid (GV358-NDRG1, pL-CRISPR.EFS.GFP-sgRNA1, pL-CRISPR.EFS.GFP-sgRNA2, pL-CRISPR.EFS.GFP-sgRNA3, pL-control, GV358-control) and its corresponding packaging plasmid (Pspax2, PMD2.G) were co-transfected into 293T cells for virus packaging. Virus supernatants were then collected, concentrated and purified and finally, cells were infected with the virus. The cells successfully infected with the virus were sorted by flow cytometry. Then, the monoclonal NDRG1 knockout cells were sorted and isolated using flow cytometry. The monoclonal cells were expanded and cultured to extract DNA. The third exon of NDRG1 was amplified by polymerase chain reaction (PCR), the PCR product was sequenced and identified and the successfully identified cells were subjected to T-A cloning. The T-A cloning product was also sent for sequencing. All the primers used are shown in Table 1.
| Gene | Forward primer | Reverse primer |
| sgRNA1 | 5’-ATCCTCACCTACCATGACAT-3’ | 5’-ATGTCATGGTAGGTGAGGAT-3’ |
| sgRNA2 | 5’-ACGCTGTGTGGGACTCCCAA-3’ | 5’-TTGGGAGTCCCACACAGCGT-3’ |
| sgRNA3 | 5’-GTTCATGCCGATGTCATGG-3’ | 5’-ACCATGACATCGGCATGAAC-3’ |
| NDRG1 | 5’-TTTGGTGCATTTAACAGCGCAGTCT-3’ | 5’-CAGGAAGTCCCAGGCAAAAAGAAAC-3’ |
The total cellular RNA was extracted (miRNeasy Kit; Qiagen, Hilden, Germany) and converted into cDNA (RevertAid First Strand cDNA Synthesis Kit; Thermo Fisher Scientific) according to the kit instructions. The ABI qPCR instrument was used for amplification of the reactants. The reaction procedure was: 95 °C for 10 min, 95 °C for 15 s, 60 °C for 1 min, 95 °C for 15 s for 40 cycles and 60 °C for 1 min. The results were normalized using GAPDH, and relative gene expression levels were calculated by the ΔΔCt method. Three replicate wells were repeated for each sample. All the primers used are shown in Table 2.
| Gene | Forward primer | Reverse primer |
| GAPDH | 5’-GCACCGTCAAGGCTGAGAA-3’ | 5’-TGGTGAAGACGCCAGTGGA-3’ |
| NDRG1 | 5’-ATGTCTCGGGAGATGCAGGATGTAG-3’ | 5’-CTAGCAGGAGACCTCCATGGACTTG-3’ |
High-efficiency RIPA lysate was used to extract total cellular protein and protein concentrations were quantified using the BCA assay. An aliquot of 80 μg protein was separated by electrophoresis on a 10% SDS-polyacrylamide gel. Proteins were electrotransferred from the gel to a PVDF membrane, and then blocked with 5% non-fat milk solution for 2 h. Membranes were incubated with an anti-human NDRG1 monoclonal antibody (1:500, Cat. No. 9485S; CST, Danvers, MA, United States), β-tubulin (1:5000, Cat. No. 6046; Abcam, Cambridge, United Kingdom) was incubated for 1 h, and then incubated at 4 °C overnight. The next day, the corresponding secondary antibody (NDRG1 1:500, β-tubulin 1:10000, Cat. No. AS014; ABclonal, Woburn, MA, United States) was incubated for 1 h at room temperature. The membrane was washed and underwent detection using an enhanced enterochromaffin-like detection system.
GV358-control, GV358-NDRG1, pL-control, and pL-NDRG1-knockout cells were seeded in 96-well plates at 2 × 103 cells per well for the cell counting kit-8 (CCK-8) cell proliferation assay (CCK-8 kit; Beyotime, Beijing, China), then cultured for 1, 2, 3, and 4 d, respectively. According to the manufacturer’s instructions, the cells were incubated with the CCK-8 reagent at 37 °C for 1 h, with the absorbance of each sample scanned on a microplate reader equipped to read absorbance values at 450 nm.
For cell cycle synchronization, 5 × 105 cells were plated in 6-well plates. After the cells were completely adherent, the cells were cultured in the serum-free medium for 24 h and then cultured in a serum medium for 24 h. Cells were collected and fixed with pre-cooled 75% ethanol at 4 °C for 14-24 h. After fixation, propidium iodide and RNAase were added to the stained cells for 30 min in the dark, then detected by flow cytometry.
Cells were trypsinized without EDTA then washed twice with cell staining buffer and resuspended with Annexin V Binding Buffer to adjust the cell concentration to (0.25-1.0) × 107/mL. 100 μL of the cell suspension was transferred into a new 1.5 mL centrifuge tube and 5 μL of APC Annexin V and 10 μL of propidium iodide solution were added, then incubated at room temperature for 15 min in the dark and tested by flow cytometry.
Invasion experiments were done using the Chamber Matrigel Invasion 24-well DO (Cat. No. 354480; Biocoat Inc., Horsham, PA, United States), and migration experiments were done using a Transwell Chamber (Cat. No. 3422; Corning Inc., Corning, NY, United States). All experimental steps were carried out according to the given protocols. Cells were seeded at a concentration of 1 × 105 in the upper chamber. The lower chamber was filled with a DMEM medium containing 20% FBS. After 48 h, chambers were fixed in 4% paraformaldehyde, stained with 0.1% crystal violet, and the number of cells that passed through the Filter were observed. Cells stained with crystal violet were eluted using 33% acetic acid, and the absorbance of the eluate was measured by a microplate reader at 570 nm.
The data were statistically analyzed using GraphPad Prism 7.0 (La Jolla, CA, United States). Student’s t-test was used for statistical analysis. Data are presented as a mean ± SD. P < 0.05 was considered statistically significant.
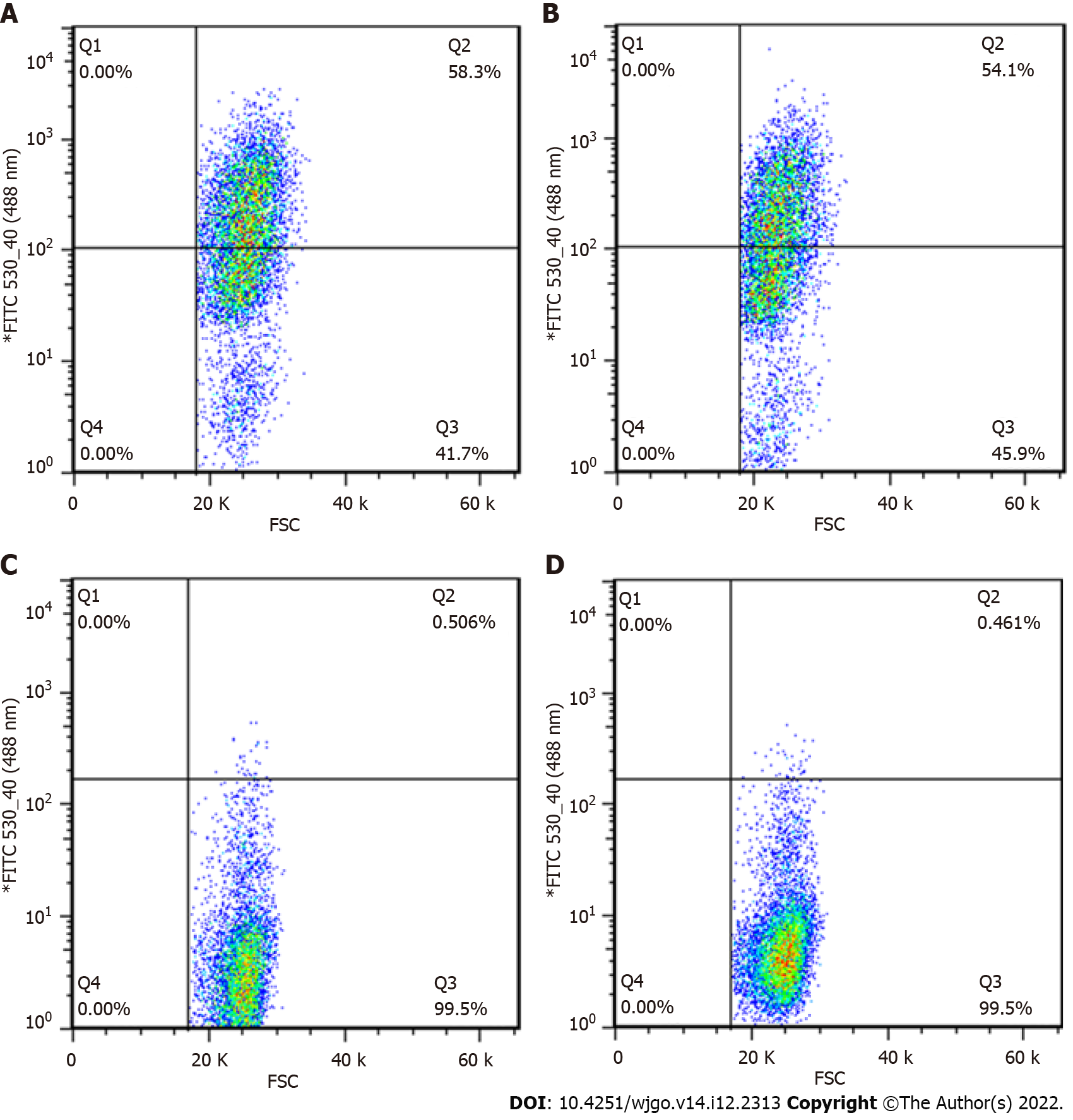
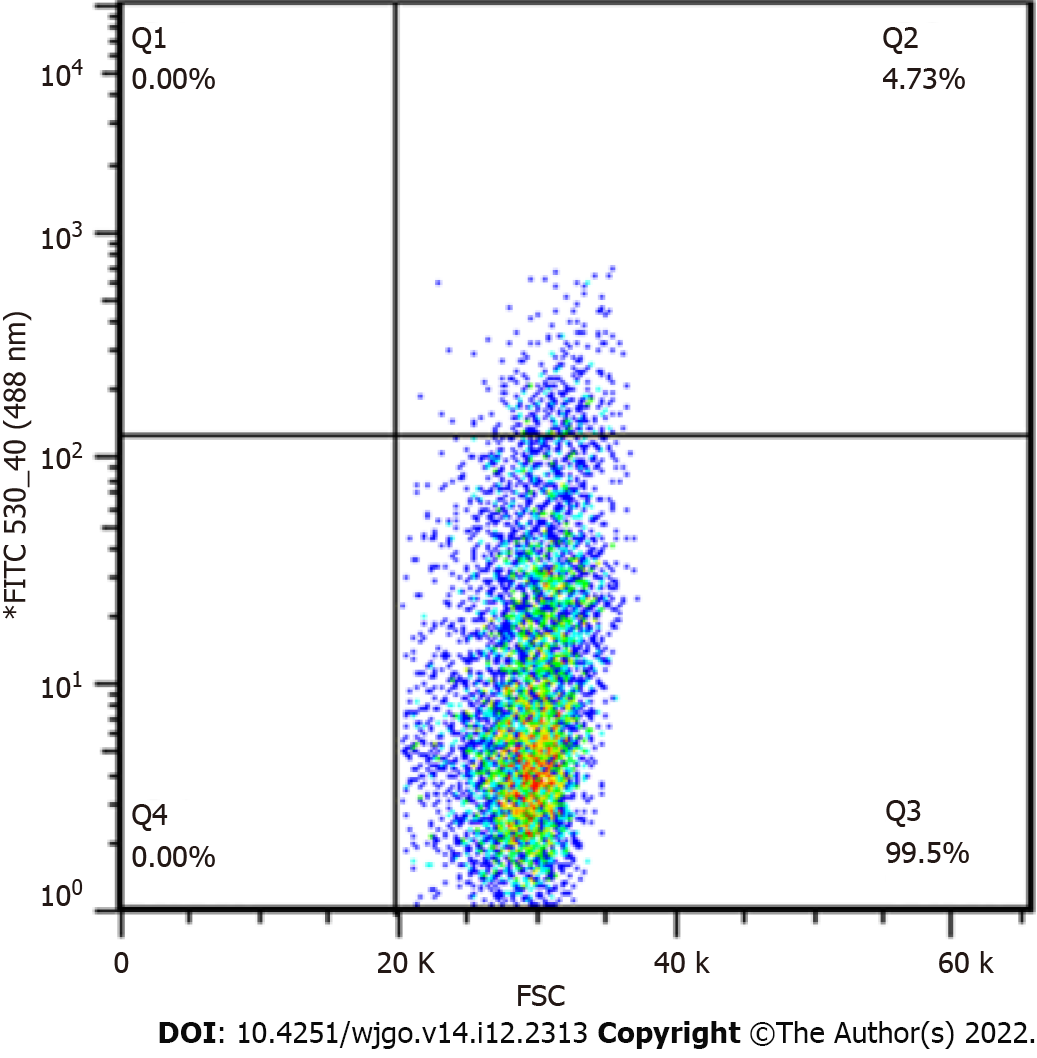
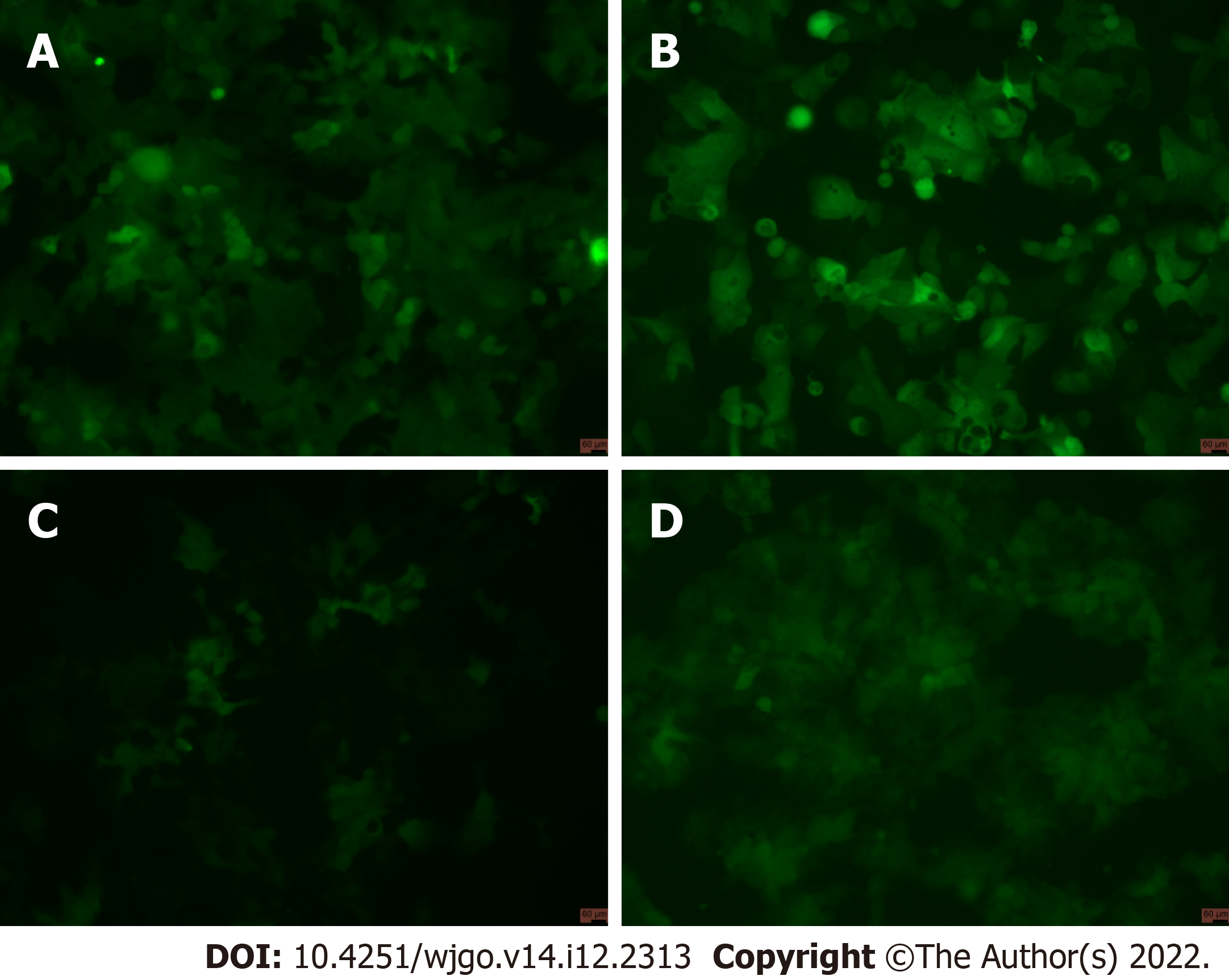
After viral infection of cells, positive cells (GFP-positive) were sorted by flow cytometry. The results showed that positive rates of GV358-control cells, GV358-NDRG1 cells, pL-control cells, and pL-NDRG1-knockout cells were 58.3%, 54.1%, 0.506%, and 0.461%, respectively (Figures 1A-D). The cells in the selected NDRG1 over-expression group (GV358-control cells, GV358-NDRG1 cells) were expanded and cultured. For knockout cells (pL-NDRG1-knockout cells), we used flow cytometry to sort for monoclonal cells with an efficiency of 4.73% (Figure 2).
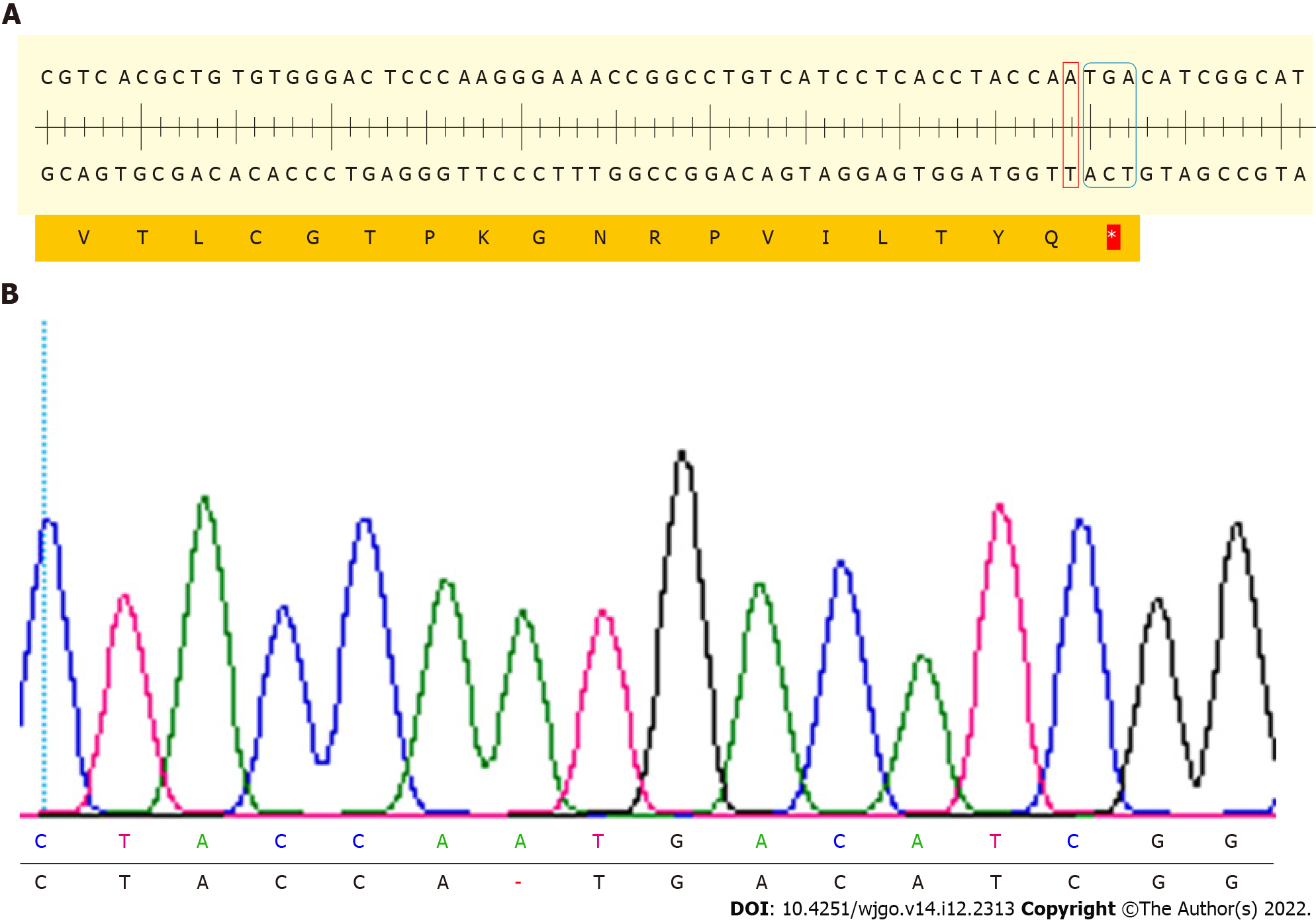
After monoclonal cells were expanded and cultured, cell DNA was extracted, the third exon of NDRG1 was amplified and products were sent for sequencing. Sequencing results showed that a DNA strand nick was generated on the third exon of NDRG1, then an A base was inserted. An analysis of the NDRG1 sequence in which the A base was inserted was performed using SnapGene software, and it was found that a TGA stop codon was formed after the inserted A base, thereby terminating the translation of NDRG1. The amplified exon of NDRG1 was identified by T-A cloning and sent for sequencing again. The sequencing results were consistent with previous results (Figure 3).
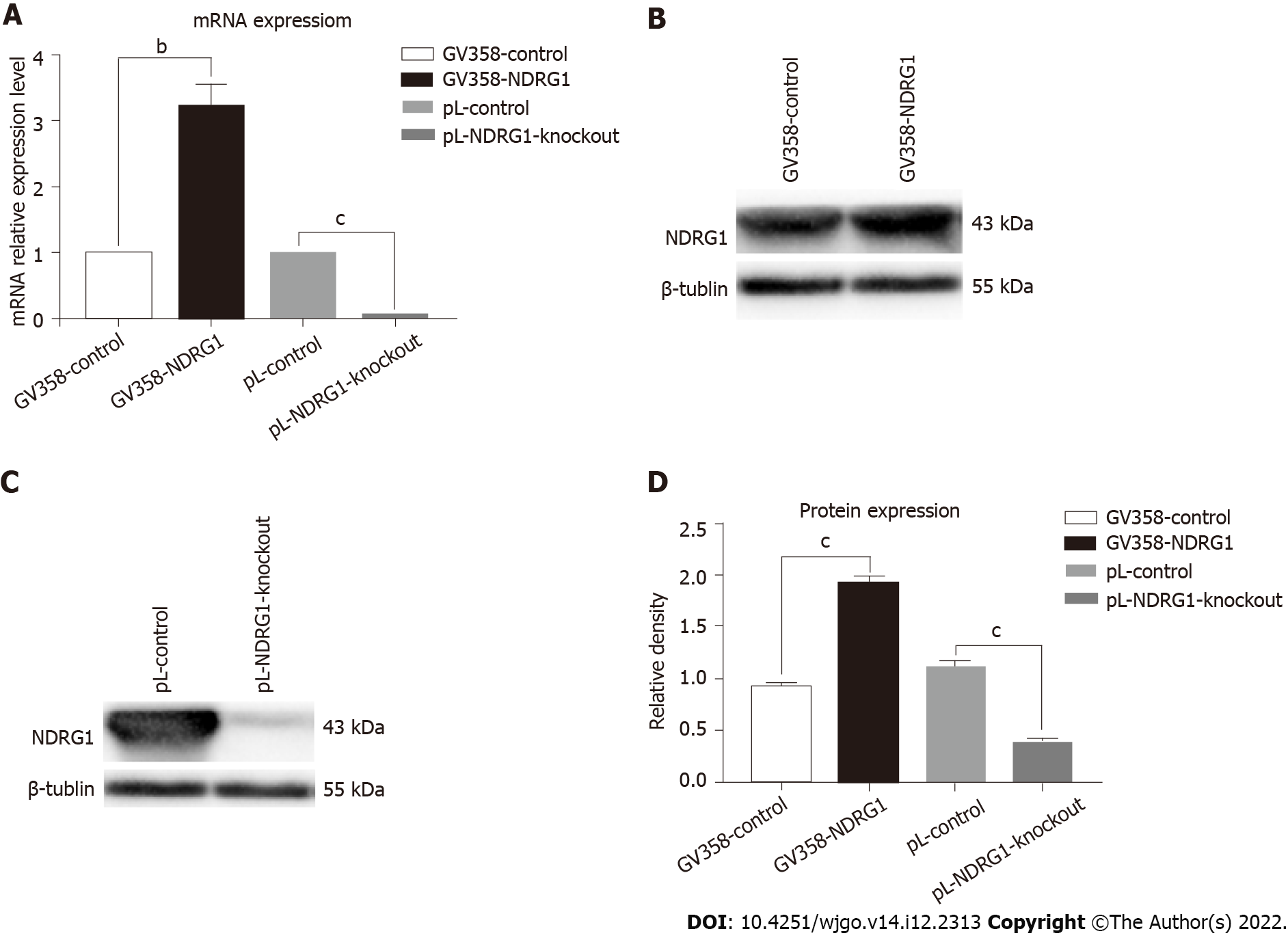
After sorting GFP-positive cells by flow cytometry and sequencing identification, we detected the changes in NDRG1 mRNA and protein expression levels by quantitative PCR (qPCR) and western blotting. The relative expression level of NDRG1 mRNA of GV358-NDRG1 cells increased about three-fold compared with the control group (P = 0.0006). In the pL-NDRG1-knockout cells, NDRG1 mRNA levels were clearly diminished relative to the controls (P < 0.0001) (Figure 4A). Western blotting revealed that the protein levels in the over-expressed cells approximately doubled (P < 0.0001). After NDRG1 gene knockout, the level reduced significantly (P < 0.0001) (Figures 4B-D). All cells were expanded (Figure 5).
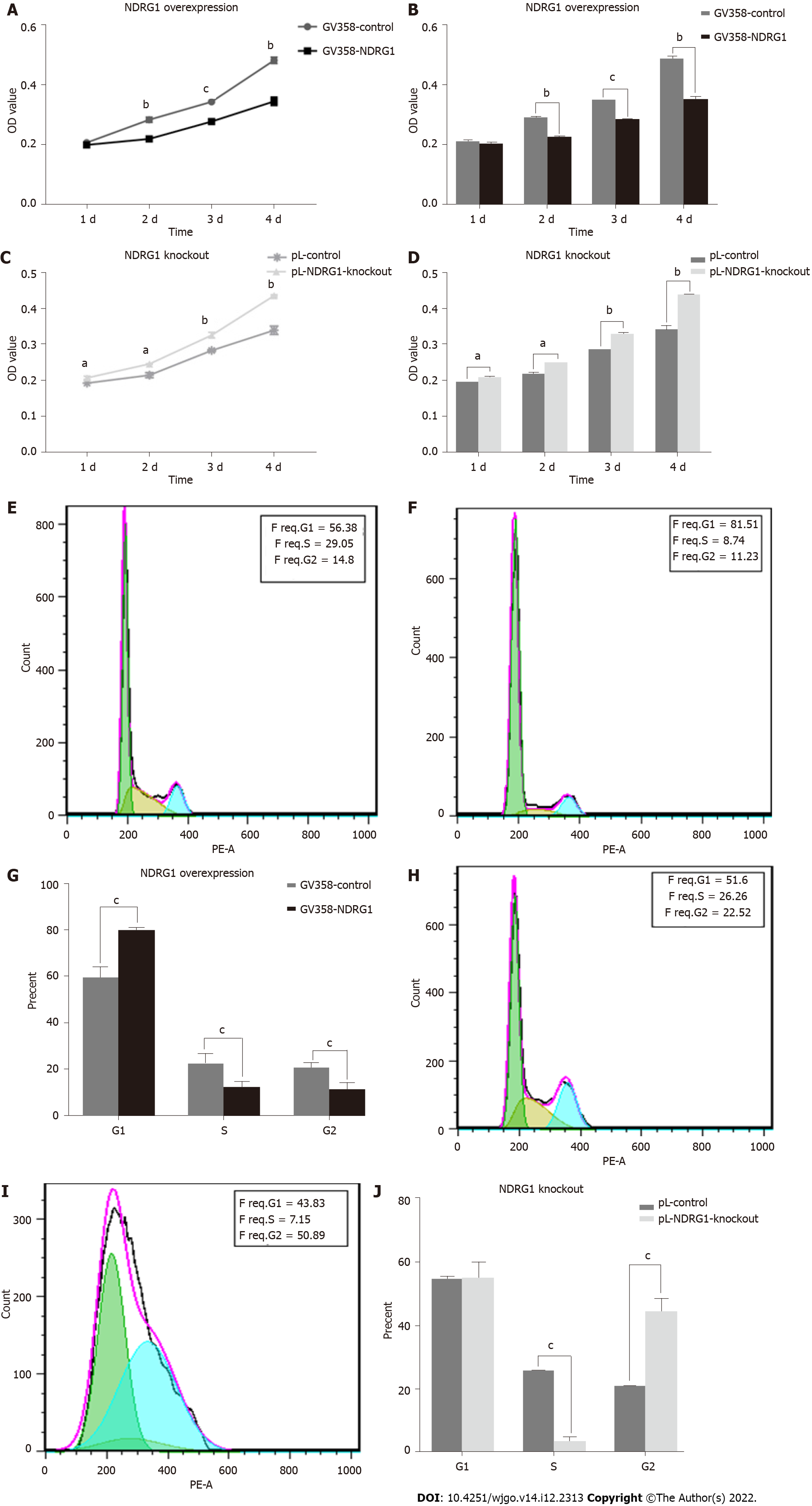
In different tumor cells, NDRG1 can either promote or inhibit the proliferation of tumor cells. To confirm the role of NDRG1 in Caco2 cells, we detected cell proliferation using the CCK-8 assay and flow cytometry. As shown in Figure 6, the relative proliferative activity of GV358-NDRG1 cells was decreased at 48 h, 72 h and 96 h compared with the GV358-control cells (P < 0.001) (Figures 6A and 6B), while pL-NDRG1-knockout cells were higher than pL-control cells at 24 h, 48 h, 72 h and 96 h (P < 0.01) (Figures 6C and 6D), which indicates that over-expression of NDRG1 could inhibit the proliferation of Caco2 cells. Flow cytometry may illuminate the underlying mechanism. The proportion of G1 phase cells was significantly increased (P < 0.0001) (Figures 6E-G) after over-expression of NDRG1, while G2 phase cells were increased after NDRG1 knockout (P < 0.0001) (Figures 6H-J). The above results show that NDRG1 inhibits the proliferation of Caco2 cells by arresting the cell cycle in the G1/S phase.
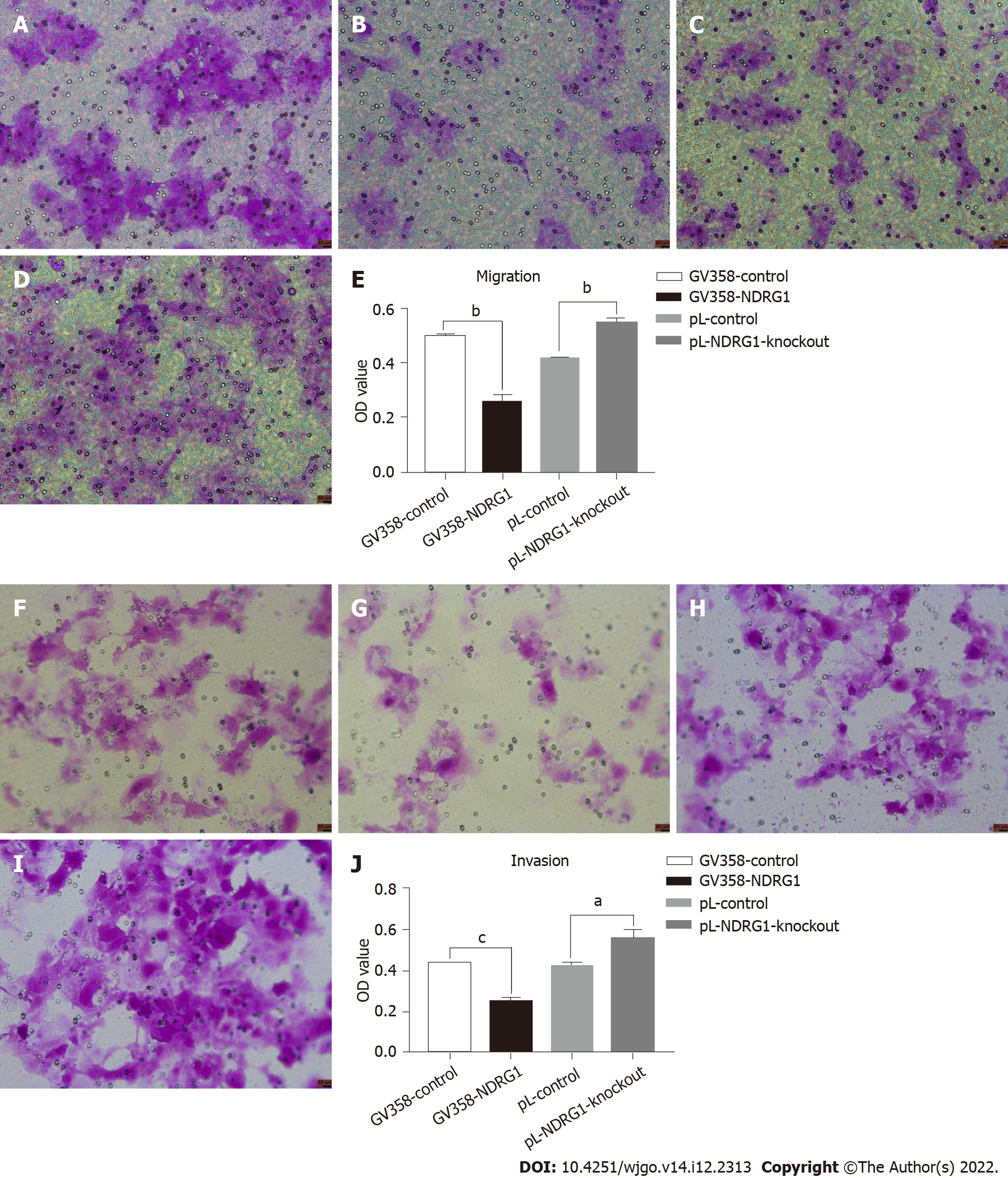
NDRG1 was also involved in tumor invasion and metastasis. Metastatic ability is also called exercise ability. Invasion and metastasis are two complementary processes. Invasive ability can be regarded as the basis of tumor cell metastasis. Thus, we also detected changes in the invasion and migration ability of cells using a 24- well transwell chamber. The number of cells passing through the chamber decreased after over-expression of NDRG1 (P = 0.0001) but increased after NDRG1 knockout (P = 0.0001). These results indicate that NDRG1 inhibited migration and invasion of Caco2 cells (Figure 7).
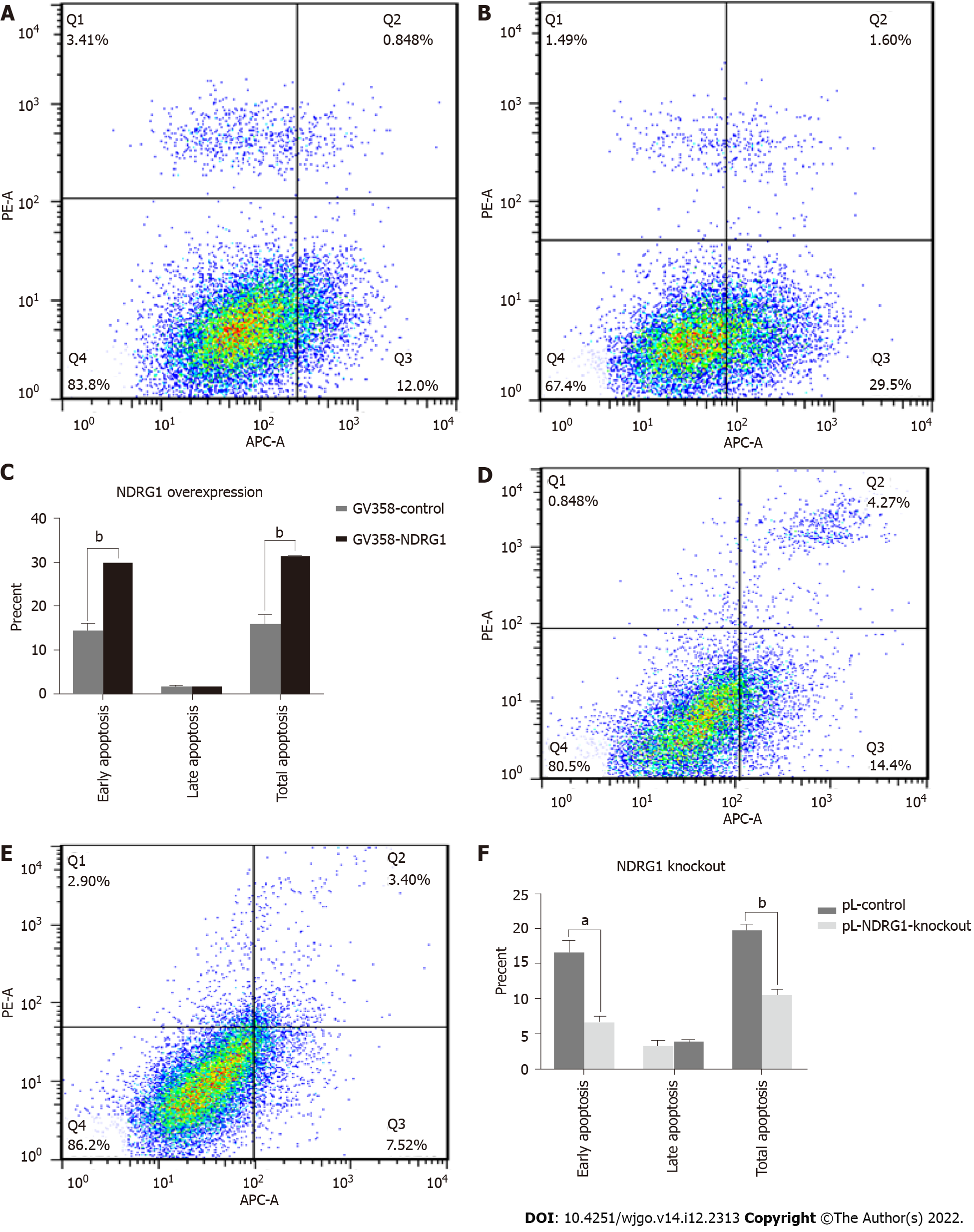
In order to further investigate the effect of NDRG1 on Caco2 cells, we used flow cytometry to detect cell apoptosis. The early apoptosis of the cells was increased after the over-expression of NDRG1 (P = 0.0002) (Figures 8A-C), and the rate of pL-NDRG1-knockout cells was decreased compared with pL-control cells (P = 0.0013) (Figures 8D-F), indicating that NDRG1 promoted early apoptosis of Caco2 cells.
According to the 2018 Global Cancer Statistics Report[1], CRC is the third most common cancer after lung cancer and breast cancer, and represents a serious threat to life and health worldwide. In addition, the incidence and mortality of CRC is on the rise, with the mortality rate reaching 95% for people over 45-years-old. CRC is a common malignant tumor which seriously threatens the life and health of patients. Unfortunately, the cause of this malignant disease is still unclear. Recent studies have shown that an excessive intake of red meat and processed meat increases the risk of CRC[16-18]. A large alcohol intake is also a risk factor[17-18] and a high-fat diet may also be related to CRC[19].
Invasion and metastasis are two malignant features of CRC. Tumor invasion and metastasis are multi-step, multi-stage, complex and orderly processes of interaction between tumor and host. They are the synergistic results of a variety of factors that promote the transfer mechanism of tumor cells in response to changes in the host environment. In addition, in recent years the molecular biology mechanisms of tumor invasion and metastasis research has shown that these processes are related to metastasis promoter gene activation and metastasis suppressor gene inactivation, as mutations of a variety of oncogenes and tumor suppressor genes can induce or enhance the metastatic potential of cancer cells. Therefore, tumor invasion and metastasis are the result of the combined action of multiple genes that promote and inhibit metastasis[20]. Clinically, the vast majority of CRC patients die, not from the primary tumor, but from multiple organ damage caused by tumor invasion and metastasis. Thus, the search for CRC metastasis suppressor genes is particularly important in current research. Further research on metastasis-associated genes in CRC cells will guide the early diagnosis of CRC, predicting early metastasis of cancer which is important for reducing the mortality of patients with CRC.
NDRG 1 is a member of the NDRG family. Human NDRG genes include NDRG1, NDRG2, NDRG3 and NDRG4, which are located on different chromosomes. The NDRG family belongs to an α/β hydrolase superfamily, but there is no hydrolase catalytic site. NDRG1 is located on human chromosome 8q24.3, is about 60 kb in length, and contains 16 exons and 15 introns. The full length of the mRNA is about 3 kb, and is composed of 394 amino acids and is highly conserved. The protein encoded by the gene is 43 kDa, and exists mainly in the cytoplasm, and to a lesser extent in the nucleus. NDRG1 is a multifunctional protein involved in cell growth[21], apoptosis[4], cell cycle regulation[22], tumor cell proliferation[5,23] and tumor invasion and metastasis[3]. It can be induced by a variety of drugs such as induced differentiation agents[24,25], and not only regulates homeostatic and genomic stability[26], but is also involved in the regulation of epidermal growth factor[27]. In addition, NDRG1 is related to the regulation of multiple signaling pathways, including the noncanonical nuclear factor-kappaB pathway[12,25], the PI3K/AKT/mTOR pathway[28,29], the RAS/RAF/MEK/ERK pathway[28,29], the transforming growth factor-beta pathway[30], and the Wnt/β-catenin pathways[3,9], suggesting it plays an important role in the development of tumors.
Recent studies have shown that NDRG1 may promote or inhibit the development of cancers, but its exact function in the process remains undefined. Some scholars believe that NDRG1 promotes the progression of liver cancer[31-33], lung cancer[34-36], bladder cancer[37], and gastric cancer[38,39]. However, other researchers have found that NDRG1 inhibits the development of prostate cancer[7,40], nasopharyngeal carcinoma[41], oropharyngeal squamous cell carcinoma[42], and ovarian cancer[8]. The role of NDRG1 in CRC is not conclusively understood.
Vaes et al[43] found that NDRG1 mRNA levels were significantly reduced in CRC tissues compared to normal colon tissue. Wang et al[10] showed that silencing NDRG1 expression in CRC cells increased cell growth, invasion and migration. Mi et al[11] also found that NDRG1 inhibited epithelial-mesenchymal transition, invasion and migration of CRC cells by promoting ubiquitination of caveolin-1 (Cav1). These studies indicate that NDRG1 is a positive regulator of CRC. Interestingly, Koshiji et al[15] found that the expression of NDRG1 was different in CRC patients of different races and at different pathological stages, which resulted in different clinical outcomes. Some research also supports the premise that NDRG1 is a transfer-promoting gene[13,44]. It has also been observed that NDRG1 locates at the centrosome of CRC cells and participates in the cell cycle as a microtubule-associated protein and mitotic site. In p53 deficient tumor cell lines, NDRG1 inhibits polyploid development and increases the number of cells by inducing cell cycle arrest. Therefore, NDRG1 can protect cells from uncontrolled proliferation, suggesting that NDRG1 possesses different effects in different cancer cells, and may be related to specific cell types and pleiotropic functions of genes.
To further investigate the role of NDRG1 in the development of CRC, lentivirus infection was used to establish stable NDRG1 over-expression in the Caco2 CRC cell line, and CRISPR/Cas9 was used to establish stable NDRG1 knock down in the Caco2 cells. Relative mRNA expression and protein expression of the cloned cells were detected by flow cytometry, qPCR and western blot. The results showed that NDRG1 mRNA levels increased by 3.218 times on average after over-expression, and western blot results showed that protein expression increased by about 2 times on average after NDRG1 over-expression. The relative mRNA expression level after NDRG1 was knockout and was only about 0.07, and the protein expression of NDRG1 after knockout was about 0.3. Following verification of over-expression and knock down, changes in cell proliferative ability, cell cycle, apoptosis, invasion and migratory ability were measured.
The results showed that the rate of cell proliferation was slowed down after NDRG1 over-expression, while the rate of cell proliferation was increased after NDRG1 gene knockout, both of which were statistically significant (P < 0.01). Cell cycle assay results showed that after NDRG1 over-expression, the proportion of cells in G1 phase increased significantly (P < 0.0001), while the proportion of cells in S and G2 phase decreased. After NDRG1 was knocked out, the proportion of cells in S phase decreased significantly, however, there was an increased number of cells in G2 phase indicating that the proportion of cells in the proliferation phase increased, and cell growth remained active. Combining the results of the two experiments, we concluded that NDRG1 over-expression arrested the cell cycle in G1/S phase and sequentially inhibited the cell growth rate of Caco2 cells. In contrast, NDRG1 knockout could enhance the mitosis of Caco2 cells, thus promoting cell growth. Then, flow cytometry was used to analyze the effect of NDRG1 on apoptosis of Caco2 cells. After NDRG1 over-expression, both the rates of early apoptosis and total apoptosis of cells increased (P < 0.05), indicating that NDRG1 over-expression promoted the apoptosis of Caco2 cells. In contrast, after NDRG1 was knocked out, both the rates of early apoptosis and total apoptosis of Caco2 cells decreased (P < 0.05), indicating that NDRG1 knockout can inhibit the apoptosis of Caco2 cells. Invasion and migration are two complementary processes, and migratory ability (also known as motor ability) can be regarded as the basis for invasion of tumor cells. In this experiment, we used a 24-well transwell chamber to detect the invasion and migration of cells at 48 h. The results showed that NDRG1 over-expression inhibited cell invasion and migration, while cell invasion and migration were promoted after knockout. In conclusion, NDRG1 can inhibit the proliferation of Caco2 cells by arresting the cell cycle in G1/S phase, promoting early apoptosis of cells, and inhibiting the invasive and migratory ability of cells. Thus, NDRG1 is a tumor metastasis suppressor gene in Caco2 cells.
NDRG1 has been shown to be a key player in the spread of cancer and the proliferation of cancer cells. However, the effects of NDRG1 on tumor invasion and metastasis, along with the mechanisms behind it are poorly understood. Aikemu et al[45] provided in silico evidence that NDRG1 plays a crucial role in actin reorganization in CRC. They found that NDRG1 loss disrupts the binding between RhoGDIα and CDC42, triggers the activation of CDC42 and the downstream PAK1/Cofilin cascade, thereby promoting the formation of filopodia and invasion of CRC. The knockdown of NDRG1 led to enhanced mobility of CRC cells in vivo and correlates with active CDC42 expression. In addition, Aikemu et al[45] found an elevated level of active CDC42 in patients with advanced T stage cancer that was negatively correlated with NDRG1 expression. In sum, these results uncover a mechanism utilized by NDRG1 to regulate CDC42 activity in coordinating cytoskeleton reorganization, which is crucial in cancer invasion. Claudin-2 (CLDN2), a well-defined component of cellular tight junction, has also been suggested to be associated with CRC progression. Wei et al[46] demonstrated that CLDN2 is up-regulated in CRC samples and associated with poor survival. Additionally, CLDN2 depletion significantly promoted NDRG1 transcription, leading to termination of CRC growth and metastasis in vitro and in vivo. NDRG1 is a key regulator that interacts with many classic tumor signaling pathways, including some molecules downstream of the epidermal growth factor receptor (EGFR). Yang et al[47] demonstrated that NDRG1 inhibited the expression of EGFR, blocked EGFR phosphorylation, and reduced the distribution of EGFR distribution in the cell membrane, cytoplasm and nucleus. NDRG1 suppression of EGFR subsequently suppressed pathways downstream of EGFR, including the RAS/RAF/ERK and PI3k/AKT/mTOR pathways. NDRG1 was also able to attenuate endocytosis and degradation of EGFR induced by Cav1. NDRG1 could be a promising biomarker to predict optimum responses to cyclophosphamide (commonly known as CTX) and a key target to enhance CTX activity in the treatment of metastatic CRC.
To further clarify the mechanism by which NDRG1 inhibits tumorigenesis, we are proceeding to observe the relevant signal pathway and the abilities of NDRG1 in nude mice. This preliminary basic research is useful in providing new potential targets for molecular therapy in CRC.
In conclusion, we conducted analyses on the effect of NDRG1 on the Caco2 cell line. Through the analyses of the NDRG1 on the biological behaviors of Caco2 cell line, we found that NDRG1 would reduce the proliferation, apoptosis, migration and invasion abilities of Caco2 cell line. NDRG1 may have potential application value as a molecular biological index to predict the early invasion and metastasis of CRC.
In American colorectal cancer patients, the expression of N-myc downstream regulated gene 1 (NDRG1) in primary colorectal cancer was always less than that in adjacent normal tissues. Our study on clinical samples showed the opposite. Therefore, the effect of NDRG1 in cancer may be related to the ethnic backgrounds of colorectal cancer. The future research direction is to find out whether the role of NDRG1 in the development of colorectal tumors is related to ethnic differences through in vivo and in vitro experiments.
This study identifies the in vitro role of NDRG1 in Caucasian large intestine tumors as inhibition of tumor cell invasion and migration.
This study only showed the in vitro effect of NDRG1 in Caucasian large intestine tumors, and the question remains to be solved about the effect of NDRG1 in large intestine tumors of other races.
RNA extraction and quantitative polymerase chain reaction, western blotting, cell counting kit-8 assay, assessment of cell cycle by flow cytometric analysis, assessment of apoptosis by flow cytometric analysis, 24-transwell for invasion and migration were used in the experiments. GraphPad Prism 7.0. student’s t-test was used for statistical analysis.
The primary objective was to investigate the role of NDRG1 in the development and progression of colorectal tumors. The objective has been to investigate the in vitro role of NDRG1 in the development of Caucasian human bowel tumors. The significance of achieving these goals for future research in this area is to provide clinicians with a theoretical basis for selecting therapies for colorectal cancer.
The research topic is NDRG1 and the key problem to be solved is the role of NDRG1 in the occurrence and development of colorectal tumors. The results can provide a certain basis for the selection of treatment options for colorectal tumors.
In 2004, our research group published an article in the World Journal of Gastroenterology on our research data with a large clinical sample (150 cases): “Correlation of N-myc downstream-regulated gene 1 over-expression with progressive growth of colorectal neoplasm.”, which carried significance on the association of NDRG1 with colorectal cancer progression.
We thank the Department of Comparative Genomics Group of the Institute of Zoology, the Chinese Academy of Sciences for providing experimental sites and technical support in this study.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ekine-Afolabi B, United Kingdom; Mohamed SY, Egypt S-Editor: Wang JJ L-Editor: Filipodia P-Editor: Wang JJ
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55806] [Article Influence: 7972.3] [Reference Citation Analysis (132)] |
| 2. | Ye H, Pang L, Wu Q, Zhu Y, Guo C, Deng Y, Zheng X. A critical role of mir-199a in the cell biological behaviors of colorectal cancer. Diagn Pathol. 2015;10:65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 3. | Jin R, Liu W, Menezes S, Yue F, Zheng M, Kovacevic Z, Richardson DR. The metastasis suppressor NDRG1 modulates the phosphorylation and nuclear translocation of β-catenin through mechanisms involving FRAT1 and PAK4. J Cell Sci. 2014;127:3116-3130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 78] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 4. | Chen B, Zaveri PG, Longtine MS, Nelson DM. N-myc downstream-regulated gene 1 (NDRG1) mediates pomegranate juice protection from apoptosis in hypoxic BeWo cells but not in primary human trophoblasts. Placenta. 2015;36:847-853. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 5. | Chang X, Xu X, Ma J, Xue X, Li Z, Deng P, Zhang S, Zhi Y, Chen J, Dai D. NDRG1 expression is related to the progression and prognosis of gastric cancer patients through modulating proliferation, invasion and cell cycle of gastric cancer cells. Mol Biol Rep. 2014;41:6215-6223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 6. | Verma N, Müller AK, Kothari C, Panayotopoulou E, Kedan A, Selitrennik M, Mills GB, Nguyen LK, Shin S, Karn T, Holtrich U, Lev S. Targeting of PYK2 Synergizes with EGFR Antagonists in Basal-like TNBC and Circumvents HER3-Associated Resistance via the NEDD4-NDRG1 Axis. Cancer Res. 2017;77:86-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 59] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 7. | Lee JE, Kim JH. Valproic acid inhibits the invasion of PC3 prostate cancer cells by upregulating the metastasis suppressor protein NDRG1. Genet Mol Biol. 2015;38:527-533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 8. | Wang B, Li J, Ye Z, Li Z, Wu X. N-myc downstream regulated gene 1 acts as a tumor suppressor in ovarian cancer. Oncol Rep. 2014;31:2279-2285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 9. | Ai R, Sun Y, Guo Z, Wei W, Zhou L, Liu F, Hendricks DT, Xu Y, Zhao X. NDRG1 overexpression promotes the progression of esophageal squamous cell carcinoma through modulating Wnt signaling pathway. Cancer Biol Ther. 2016;17:943-954. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 10. | Wangpu X, Yang X, Zhao J, Lu J, Guan S, Kovacevic Z, Liu W, Mi L, Jin R, Sun J, Yue F, Ma J, Lu A, Richardson DR, Wang L, Zheng M. The metastasis suppressor, NDRG1, inhibits "stemness" of colorectal cancer via down-regulation of nuclear β-catenin and CD44. Oncotarget. 2015;6:33893-33911. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 11. | Mi L, Zhu F, Yang X, Lu J, Zheng Y, Zhao Q, Wen X, Lu A, Wang M, Zheng M, Ji J, Sun J. The metastatic suppressor NDRG1 inhibits EMT, migration and invasion through interaction and promotion of caveolin-1 ubiquitylation in human colorectal cancer cells. Oncogene. 2017;36:4323-4335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 73] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 12. | Ma J, Gao Q, Zeng S, Shen H. Knockdown of NDRG1 promote epithelial-mesenchymal transition of colorectal cancer via NF-κB signaling. J Surg Oncol. 2016;114:520-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 13. | Wang Z, Wang F, Wang WQ, Gao Q, Wei WL, Yang Y, Wang GY. Correlation of N-myc downstream-regulated gene 1 overexpression with progressive growth of colorectal neoplasm. World J Gastroenterol. 2004;10:550-554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 15] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 14. | Shah MA, Kemeny N, Hummer A, Drobnjak M, Motwani M, Cordon-Cardo C, Gonen M, Schwartz GK. Drg1 expression in 131 colorectal liver metastases: correlation with clinical variables and patient outcomes. Clin Cancer Res. 2005;11:3296-3302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 54] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 15. | Koshiji M, Kumamoto K, Morimura K, Utsumi Y, Aizawa M, Hoshino M, Ohki S, Takenoshita S, Costa M, Commes T, Piquemal D, Harris CC, Tchou-Wong KM. Correlation of N-myc downstream-regulated gene 1 expression with clinical outcomes of colorectal cancer patients of different race/ethnicity. World J Gastroenterol. 2007;13:2803-2810. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 27] [Cited by in RCA: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 16. | Magalhães B, Peleteiro B, Lunet N. Dietary patterns and colorectal cancer: systematic review and meta-analysis. Eur J Cancer Prev. 2012;21:15-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 168] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 17. | Garcia-Larsen V, Morton V, Norat T, Moreira A, Potts JF, Reeves T, Bakolis I. Dietary patterns derived from principal component analysis (PCA) and risk of colorectal cancer: a systematic review and meta-analysis. Eur J Clin Nutr. 2019;73:366-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 65] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 18. | Godos J, Bella F, Torrisi A, Sciacca S, Galvano F, Grosso G. Dietary patterns and risk of colorectal adenoma: a systematic review and meta-analysis of observational studies. J Hum Nutr Diet. 2016;29:757-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 19. | Fu T, Coulter S, Yoshihara E, Oh TG, Fang S, Cayabyab F, Zhu Q, Zhang T, Leblanc M, Liu S, He M, Waizenegger W, Gasser E, Schnabl B, Atkins AR, Yu RT, Knight R, Liddle C, Downes M, Evans RM. FXR Regulates Intestinal Cancer Stem Cell Proliferation. Cell. 2019;176:1098-1112.e18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 323] [Article Influence: 53.8] [Reference Citation Analysis (0)] |
| 20. | Chui MH. Insights into cancer metastasis from a clinicopathologic perspective: Epithelial-Mesenchymal Transition is not a necessary step. Int J Cancer. 2013;132:1487-1495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 92] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 21. | Larkin J, Chen B, Shi XH, Mishima T, Kokame K, Barak Y, Sadovsky Y. NDRG1 deficiency attenuates fetal growth and the intrauterine response to hypoxic injury. Endocrinology. 2014;155:1099-1106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 22. | Matsugaki T, Zenmyo M, Hiraoka K, Fukushima N, Shoda T, Komiya S, Ono M, Kuwano M, Nagata K. N-myc downstream-regulated gene 1/Cap43 expression promotes cell differentiation of human osteosarcoma cells. Oncol Rep. 2010;24:721-725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 23. | Lu WJ, Chua MS, So SK. Suppressing N-Myc downstream regulated gene 1 reactivates senescence signaling and inhibits tumor growth in hepatocellular carcinoma. Carcinogenesis. 2014;35:915-922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 24. | Kalinowski DS, Stefani C, Toyokuni S, Ganz T, Anderson GJ, Subramaniam NV, Trinder D, Olynyk JK, Chua A, Jansson PJ, Sahni S, Lane DJ, Merlot AM, Kovacevic Z, Huang ML, Lee CS, Richardson DR. Redox cycling metals: Pedaling their roles in metabolism and their use in the development of novel therapeutics. Biochim Biophys Acta. 2016;1863:727-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 108] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 25. | Xi R, Pun IH, Menezes SV, Fouani L, Kalinowski DS, Huang ML, Zhang X, Richardson DR, Kovacevic Z. Novel Thiosemicarbazones Inhibit Lysine-Rich Carcinoembryonic Antigen-Related Cell Adhesion Molecule 1 (CEACAM1) Coisolated (LYRIC) and the LYRIC-Induced Epithelial-Mesenchymal Transition via Upregulation of N-Myc Downstream-Regulated Gene 1 (NDRG1). Mol Pharmacol. 2017;91:499-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 26. | Croessmann S, Wong HY, Zabransky DJ, Chu D, Mendonca J, Sharma A, Mohseni M, Rosen DM, Scharpf RB, Cidado J, Cochran RL, Parsons HA, Dalton WB, Erlanger B, Button B, Cravero K, Kyker-Snowman K, Beaver JA, Kachhap S, Hurley PJ, Lauring J, Park BH. NDRG1 links p53 with proliferation-mediated centrosome homeostasis and genome stability. Proc Natl Acad Sci U S A. 2015;112:11583-11588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 27. | Menezes SV, Kovacevic Z, Richardson DR. The metastasis suppressor NDRG1 down-regulates the epidermal growth factor receptor via a lysosomal mechanism by up-regulating mitogen-inducible gene 6. J Biol Chem. 2019;294:4045-4064. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 28. | Kovacevic Z, Chikhani S, Lui GY, Sivagurunathan S, Richardson DR. The iron-regulated metastasis suppressor NDRG1 targets NEDD4L, PTEN, and SMAD4 and inhibits the PI3K and Ras signaling pathways. Antioxid Redox Signal. 2013;18:874-887. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 145] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 29. | McCubrey JA, Steelman LS, Abrams SL, Bertrand FE, Ludwig DE, Bäsecke J, Libra M, Stivala F, Milella M, Tafuri A, Lunghi P, Bonati A, Martelli AM. Targeting survival cascades induced by activation of Ras/Raf/MEK/ERK, PI3K/PTEN/Akt/mTOR and Jak/STAT pathways for effective leukemia therapy. Leukemia. 2008;22:708-722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 189] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 30. | Hu ZY, Xie WB, Yang F, Xiao LW, Wang XY, Chen SY, Li ZG. NDRG1 attenuates epithelial-mesenchymal transition of nasopharyngeal cancer cells via blocking Smad2 signaling. Biochim Biophys Acta. 2015;1852:1876-1886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 31. | Luo Q, Wang CQ, Yang LY, Gao XM, Sun HT, Zhang Y, Zhang KL, Zhu Y, Zheng Y, Sheng YY, Lu L, Jia HL, Yu WQ, Liu J, Dong QZ, Qin LX. FOXQ1/NDRG1 axis exacerbates hepatocellular carcinoma initiation via enhancing crosstalk between fibroblasts and tumor cells. Cancer Lett. 2018;417:21-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 64] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 32. | Zhao K, Zhao Y, Zhu JY, Dong H, Cong WM, Yu Y, Wang H, Zhu ZZ, Xu Q. A Panel of Genes Identified as Targets for 8q24.13-24.3 Gain Contributing to Unfavorable Overall Survival in Patients with Hepatocellular Carcinoma. Curr Med Sci. 2018;38:590-596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 33. | Liu Y, Wang D, Li Y, Yan S, Dang H, Yue H, Ling J, Chen F, Zhao Y, Gou L, Tang P, Huang A, Tang H. Long noncoding RNA CCAT2 promotes hepatocellular carcinoma proliferation and metastasis through up-regulation of NDRG1. Exp Cell Res. 2019;379:19-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 34. | Zhang JZ, Liu ZL, Zhang YX, Lin HJ, Zhang ZJ. Lipoxin A4 Ameliorates Lipopolysaccharide-Induced A549 Cell Injury through Upregulation of N-myc Downstream-Regulated Gene-1. Chin Med J (Engl). 2018;131:1342-1348. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 35. | Dai T, Dai Y, Murata Y, Husni RE, Nakano N, Sakashita S, Noguchi M. The prognostic significance of N-myc downregulated gene 1 in lung adenocarcinoma. Pathol Int. 2018;68:224-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 36. | Chiang CT, Demetriou AN, Ung N, Choudhury N, Ghaffarian K, Ruderman DL, Mumenthaler SM. mTORC2 contributes to the metabolic reprogramming in EGFR tyrosine-kinase inhibitor resistant cells in non-small cell lung cancer. Cancer Lett. 2018;434:152-159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 37. | Li A, Zhu X, Wang C, Yang S, Qiao Y, Qiao R, Zhang J. Upregulation of NDRG1 predicts poor outcome and facilitates disease progression by influencing the EMT process in bladder cancer. Sci Rep. 2019;9:5166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 38. | Kawahara A, Akiba J, Hattori S, Yamaguchi T, Abe H, Taira T, Ureshino H, Murakami Y, Watari K, Koufuji K, Shirouzu K, Kuwano M, Ono M, Kage M. Nuclear expression of N-myc downstream regulated gene 1/Ca(2+)-associated protein 43 is closely correlated with tumor angiogenesis and poor survival in patients with gastric cancer. Exp Ther Med. 2011;2:471-479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 39. | Ureshino H, Murakami Y, Watari K, Izumi H, Kawahara A, Kage M, Arao T, Nishio K, Yanagihara K, Kinoshita H, Kuwano M, Ono M. N-myc downstream regulated gene 1 (NDRG1) promotes metastasis of human scirrhous gastric cancer cells through epithelial mesenchymal transition. PLoS One. 2012;7:e41312. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 40. | Sharma A, Mendonca J, Ying J, Kim HS, Verdone JE, Zarif JC, Carducci M, Hammers H, Pienta KJ, Kachhap S. The prostate metastasis suppressor gene NDRG1 differentially regulates cell motility and invasion. Mol Oncol. 2017;11:655-669. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 41. | Chiang KC, Yang SW, Chang KP, Feng TH, Chang KS, Tsui KH, Shin YS, Chen CC, Chao M, Juang HH. Caffeic Acid Phenethyl Ester Induces N-myc Downstream Regulated Gene 1 to Inhibit Cell Proliferation and Invasion of Human Nasopharyngeal Cancer Cells. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 42. | Lee JC, Chiang KC, Feng TH, Chen YJ, Chuang ST, Tsui KH, Chung LC, Juang HH. The Iron Chelator, Dp44mT, Effectively Inhibits Human Oral Squamous Cell Carcinoma Cell Growth in Vitro and in Vivo. Int J Mol Sci. 2016;17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 42] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 43. | Vaes N, Schonkeren SL, Brosens E, Koch A, McCann CJ, Thapar N, Hofstra RMW, van Engeland M, Melotte V. A combined literature and in silico analysis enlightens the role of the NDRG family in the gut. Biochim Biophys Acta Gen Subj. 2018;1862:2140-2151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 44. | Song Y, Lv L, Du J, Yue L, Cao L. Correlation of N-myc downstream-regulated gene 1 subcellular localization and lymph node metastases of colorectal neoplasms. Biochem Biophys Res Commun. 2013;439:241-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 45. | Aikemu B, Shao Y, Yang G, Ma J, Zhang S, Yang X, Hong H, Yesseyeva G, Huang L, Jia H, Wang C, Zang L, Sun J, Zheng M. NDRG1 regulates Filopodia-induced Colorectal Cancer invasiveness via modulating CDC42 activity. Int J Biol Sci. 2021;17:1716-1730. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 46. | Wei M, Zhang Y, Yang X, Ma P, Li Y, Wu Y, Chen X, Deng X, Yang T, Mao X, Qiu L, Meng W, Zhang B, Wang Z, Han J. Claudin-2 promotes colorectal cancer growth and metastasis by suppressing NDRG1 transcription. Clin Transl Med. 2021;11:e667. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 47. | Yang G, Huang L, Jia H, Aikemu B, Zhang S, Shao Y, Hong H, Yesseyeva G, Wang C, Li S, Sun J, Zheng M, Ma J. NDRG1 enhances the sensitivity of cetuximab by modulating EGFR trafficking in colorectal cancer. Oncogene. 2021;40:5993-6006. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |