Published online Nov 15, 2022. doi: 10.4251/wjgo.v14.i11.2138
Peer-review started: July 9, 2022
First decision: July 31, 2022
Revised: August 30, 2022
Accepted: October 31, 2022
Article in press: October 31, 2022
Published online: November 15, 2022
Processing time: 128 Days and 16.4 Hours
Several genes, important for development, are reduced or silenced in adulthood, and their abnormal expression has been related to the occurrence and deve
To explore the expression pattern of six SIX proteins in CRCs and their re
The expression level of SIXs in normal tissues of different organs and related cancerous tissues was analyzed in the Human Protein Atlas. Kaplan-Meier Plotter and GEPIA2 were used to analyze the prognostic values of SIXs. To analyze the potential signaling pathways with SIX family involvement, LinkedOmics was used to perform Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses of SIX4-related genes. Subsequently, immunohistochemical experiments were performed on CRC tissues and adjacent normal tissues, and we examined the SIX4 expression level in 87 pairs of patients with tissue microarray. The relationship between SIX4 and clinicopathological parameters in CRC patients was tested using the χ2 test and Fisher’s exact probability to verify the results of the database analysis.
The RNA levels of SIX1-4 and SIX6 were relatively low in normal human tissues, while SIX5 was highly expressed at both the RNA and protein levels. However, the protein level of SIX4 was found to be elevated in various malignancies. In CRC tissues, SIX1, SIX2 and SIX4 were elevated in cancer tissues compared with adjacent normal tissue. Among all SIXs, a high level of SIX4 was found to be associated with poor overall and disease-free survival in patients with CRC. For different clinicopathological parameters, increased SIX4 expression was positively correlated with advanced CRC. The top 50 SIX4-related genes were involved with oxidative phosphorylation and the respiratory chain signaling pathways.
The current results provided a comprehensive analysis of the expression and prognostic values of SIX family members in CRC. Among different SIXs, SIX4 plays an oncogenic role in CRC to promote the development of malignancy. In CRC, SIX4 mRNA and protein expression is higher than that in normal tissues and associated with shorter CRC patient survival, suggesting that SIX4 may be a potential therapeutic target for treatment of CRC patients.
Core Tip: This study systematically analyzed the expression pattern and prognostic value of sine oculis homeobox homolog (SIX) family members in colorectal cancer (CRC). It was found that the expression of SIX4 in CRC tissues positively correlated with the development of CRC and negatively correlated with overall survival. The top 50 SIX4-related genes were involved with oxidative phosphorylation and the respiratory chain signaling pathways. SIX4 may be a novel and potential therapeutic target for CRC.
- Citation: Fang ZX, Li CL, Wu Z, Hou YY, Wu HT, Liu J. Comprehensive analysis of the potential role and prognostic value of sine oculis homeobox homolog family in colorectal cancer. World J Gastrointest Oncol 2022; 14(11): 2138-2156
- URL: https://www.wjgnet.com/1948-5204/full/v14/i11/2138.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i11.2138
Although the study of early human development is limited due to the small number of samples, it is well known how the genome activates or silences transcriptional programs to govern organ formation[1]. Gerrard et al[2] focused on histone modifications during human organogenesis and found that key developmental gene sets are actively repressed outside of the appropriate organ. Interestingly, the abnormal expression of organogenesis-related genes at inappropriate developmental stages has been reported in different diseases, especially malignant tumors[3].
Human sine oculis homeobox homolog (SIX) proteins comprise a group of six family members that act as key regulators of organogenesis in the kidney, limbs, eye, brain and craniofacial structure[4-7]. Defects in these genes result in hypoplastic disorders, such as autosomal dominant deafness type 23, branchiootic syndrome type 3, holoprosencephaly type 2, branchiootorenal syndrome type 2 and isolated microphthalmia with cataract type 2[8-12].
Colorectal cancer (CRC) is ranked third in incidence among malignant tumors worldwide and is the second leading cause of cancer-related mortality[13]. In China, the annual average increase of new CRC cases has been estimated to be 4.2%[14]. Although sex and regional differences are reported to be the prognostic factors for CRC patients[15], the etiology of CRC oncogenesis and development is still complex and unclear. The high mortality of patients with CRC and the limitations of the traditional tumor-node-metastasis staging emphasize the need to explore key genes closely associated with CRC development and prognosis[16].
Unsurprisingly, abnormal SIX levels have been reported to participate in the regulation of human cancers. Several studies report that mutations or aberrant expression of the SIX family play an important role in colorectal tumorigenesis through multiple processes, such as transformation, proliferation, angiogenesis, migration and metastasis[17-19]. Song et al[20] showed that SIX1 was highly expressed in CRC patients who have short overall survival (OS) and enhances proliferation and migration of CRC cells through activation of Wnt/β-catenin signaling. Human SIX2 levels are higher in non-metastatic CRC, and targeting this gene can modulate CRC metastasis and immunity, thereby improving the survival of CRC patients[18]. In glioblastoma, where SIX3 is transcriptionally silenced by DNA hypermethylation, SIX3 functions as a tumor suppressor[21], and SIX4 promotes development and progression in CRC through the PI3K-AKT signaling pathways[17]. SIX5 is an important paralog of SIX4 and promotes lung adenocarcinoma progression through transcriptional activation of LINC01468 and its downstream pathways[22], and SIX5 cooperates with hypoxia-induced EYA3 and P3000 to mediate tumorigenesis and cancer progression[19]. However, SIX6 is poorly studied in CRC than in other tumor types. Hence, the current study conducted a comprehensive evaluation of the potential roles of SIX family members and provided novel therapeutic targets for further investigation.
The Human Protein Atlas (HPA) (https://www.proteinatlas.org/), a rich resource database with more than 5000 types of human protein expression data and high-definition images of human normal and cancerous cells[23], was used to obtain and analyze the mRNA and protein levels of SIX family members in normal tissues and different types of cancers. RNA expression levels were evaluated using consensus normalized expression (NX) combined with three transcriptome datasets (HPA, GTEx and FANTOM5), as described before[24]. RNA expression was divided into four levels: Not detected (NX < 1), low expression (1 ≤ NX < 15), medium expression (15 ≤ NX < 30) and high expression (NX ≥ 30)[24]. Similarly, protein expression was also categorized into four groups: Negative (-), low expression (+), medium expression (++) and high expression (+++). The Cancer Genome Atlas (TCGA) datasets were evaluated through the Tumor Immune Estimation Resource 2.0 online resource (http://timer.cistrome.org/) for pan-cancer analysis[25] and the UCSC Xena database (https://genome-cancer.ucsc.edu/)[26] for colon and rectal adenocarcinomas and related normal tissues.
Kaplan-Meier Plotter (http://kmplot.com/analysis/), a dataset containing gene expression and survival of cancer patients[27], was assessed for the prognostic value regarding OS of SIXs in patients with CRC. For disease-free survival (DFS) analysis, GEPIA2 (http://gepia2.cancerpku.cn/) information, containing 9736 tumors and 8587 normal samples[28], was used to detect the correlation between SIX levels and DFS in CRC patients.
LinkedOmics (http://www.linkedomics.org), a multi-omics dataset that includes data from 32 TCGA tumor types and 10 clinical proteomic tumor analysis cohorts[29], was used to predict potential SIX4-related genes and explore their related signaling pathways through the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analyses. The LinkFinder module in the LinkedOmics dataset was applied to analyze the correlation between the expression level of SIX4 and clinicopathological parameters of CRC patients.
A tissue microarray with 87 matched primary colon cancer tissues and their corresponding adjacent normal colon tissue samples and 6 extra samples of cancer cases were purchased from the Shanghai OUTDO Biotech Company (Shanghai, China). This study was approved by the Ethics Committee of Shantou University Medical college (SUMC-2022-045).
Immunohistochemical staining of SIX4 was performed as described previously[30]. Deparaffinization in xylene, hydration in graded alcohols and epitope retrieval by microwaving in EDTA (Fuzhou Maixin Biotechnology Development Co. LTD, Fuzhou, China) was conducted sequentially on the tissue microarray slide. After blocking endogenous peroxidase with 3% H2O2, the slide was incubated with anti-SIX4 antibody (dilution: 1:400, bs-17503R, Bioss, China,) at 4 °C overnight. Stained tissues were mounted by nuclear counterstaining with hematoxylin for visualization.
The sections were visualized and evaluated independently under a bright-field microscope (PerkinElmer Vectra, PerkinElmer, United States) by two investigators with no prior knowledge of the patient information. For evaluating SIX4 expression, the staining intensities with colorless, light yellow, brown yellow and dark brown were labeled as 0, 1, 2 and 3, respectively, while the percentage of positive cells corresponding to 0%, 1%-25%, 26%-50%, 51%-75% and 76%-100% were recorded as 0, 1, 2, 3 and 4, respectively. The final staining score for SIX4 expression was calculated as the sum of staining intensity and percentage of positive cells and divided into low (0-4) and high (5-7) expression groups.
SPSS 25.0 statistical software was used to do the statistical analyses. The relationship between expression levels of SIX4 in 87 paired CRC and paracancerous tissues was performed using the χ2 test. A total of 93 case numbers from the tissue microarrays were recruited to analyze the relationship between SIX4 and clinicopathological parameters of CRC patients using the χ2 or Fisher’s exact probability test. To investigate the prognostic value of SIX4 in CRC patients, the Kaplan-Meier survival curve and log-rank test were used. The difference was considered statistically significant at P < 0.05.
As shown in Table 1, the results from the HPA database revealed that in normal tissues, SIX1-4 and SIX6 were expressed at relatively low levels, except for SIX1 in the testis, prostate and uterine cervix. SIX5 RNA and protein were highly expressed in the thyroid gland, stomach, colon/rectum, kidney, urinary bladder, prostate, endometrium and breast tissues. The protein levels of SIX2-4 have yet to be examined.
| Organs | SIX1 | SIX2 | SIX3 | SIX4 | SIX5 | SIX6 | ||||||
| RNA | Protein | RNA | Protein | RNA | Protein | RNA | Protein | RNA | Protein | RNA | Protein | |
| Cerebral cortex | + | - | + | NE | ++ | NE | + | NE | + | + | - | - |
| Thyroid gland | + | - | + | NE | - | NE | + | NE | ++ | ++ | - | - |
| Lung | + | - | + | NE | - | NE | + | NE | + | - | - | - |
| Esophagus | + | - | + | NE | - | NE | + | NE | + | + | - | - |
| Stomach | + | - | + | NE | - | NE | + | NE | ++ | ++ | - | - |
| Colon/rectum | - | - | + | NE | - | NE | + | NE | +++ | ++ | - | - |
| Liver | - | - | - | NE | - | NE | + | NE | ++ | ++ | - | - |
| Pancreas | + | + | + | NE | + | NE | + | NE | +++ | + | - | - |
| Kidney | - | - | + | NE | + | NE | + | NE | ++ | ++ | - | - |
| Urinary bladder | + | + | + | NE | - | NE | + | NE | ++ | ++ | - | - |
| Testis | + | ++ | + | NE | + | NE | + | NE | + | ++ | + | - |
| Prostate | + | ++ | + | NE | - | NE | + | NE | +++ | ++ | - | - |
| Ovary | + | - | + | NE | - | NE | + | NE | +++ | - | - | - |
| Endometrium | - | - | + | NE | - | NE | + | NE | +++ | ++ | - | - |
| Uterine cervix | + | +++ | + | NE | + | NE | + | NE | +++ | - | - | - |
| Breast | + | - | + | NE | + | NE | + | NE | ++ | ++ | - | - |
| Skin | - | - | + | NE | - | NE | + | NE | + | - | - | - |
| Lymph node | + | - | + | NE | + | NE | - | NE | + | - | - | - |
| Bone marrow | - | - | - | NE | - | NE | + | NE | + | - | - | - |
To explore the different expression patterns of SIXs in normal and malignant tissues, the expression levels of SIXs in different types of malignant tumors was collected from the HPA (Table 2). Interestingly, the levels of SIX1-3 and SIX6 were still relatively low in different types of cancers, except for SIX1 in prostate, cervix uterine and breast cancers. It was found that the RNA levels of SIX5 were low, but the protein levels of SIX5 were quite high in cancers of the thyroid gland, prostate, ovary and breast and moderately in cancers of the colon/rectum, liver and endometrium. Although there is still no data on the protein level of SIX2 and SIX3, the protein level of SIX4 has been evaluated in a series of studies and found to be high in cancers of the thyroid gland, breast and skin, moderately in cancers of the colon/rectum, liver, prostate and uterine cervix and low in cancers of the lung, pancreas, kidney, urinary bladder, testis, ovary, endometrium and lymph node.
| Organs | SIX1 | SIX2 | SIX3 | SIX4 | SIX5 | SIX6 | ||||||
| RNA | Protein | RNA | Protein | RNA | Protein | RNA | Protein | RNA | Protein | RNA | Protein | |
| Cerebral cortex | - | - | - | NE | - | NE | - | - | - | - | - | - |
| Thyroid gland | + | - | + | NE | + | NE | + | +++ | + | +++ | + | - |
| Lung | + | + | + | NE | + | NE | + | + | + | + | + | - |
| Esophagus | - | - | - | NE | - | NE | - | - | - | - | - | |
| Stomach | - | - | + | NE | + | NE | + | - | + | + | + | - |
| Colon/rectum | + | - | + | NE | + | NE | + | ++ | + | ++ | + | - |
| Liver | + | - | + | NE | + | NE | + | ++ | + | ++ | + | - |
| Pancreas | + | + | + | NE | + | NE | + | + | + | ++ | + | - |
| Kidney | + | - | + | NE | + | NE | + | + | + | + | + | - |
| Urinary bladder | + | - | + | NE | + | NE | + | + | + | + | + | - |
| Testis | + | - | + | NE | + | NE | + | + | + | + | + | - |
| Prostate | + | ++ | + | NE | + | NE | + | ++ | + | +++ | + | - |
| Ovary | + | - | + | NE | + | NE | + | + | + | +++ | + | + |
| Endometrium | + | + | + | NE | + | NE | + | + | + | ++ | + | - |
| Cervix uterine | + | ++ | + | NE | + | NE | + | ++ | + | + | + | - |
| Breast | + | ++ | + | NE | + | NE | + | +++ | + | +++ | + | - |
| Skin | - | - | - | NE | - | NE | - | +++ | - | - | - | - |
| Lymph node | - | - | - | NE | - | NE | - | + | - | - | - | - |
| Bone marrow | - | - | - | NE | - | NE | - | - | - | - | - | |
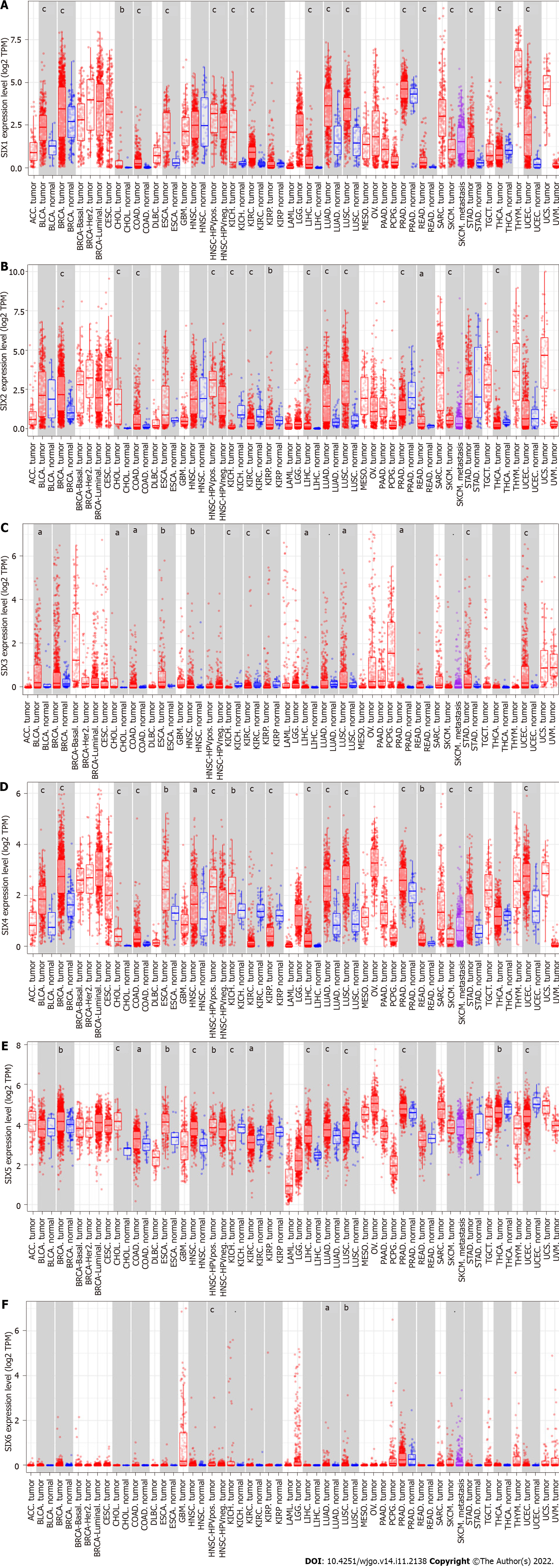
To confirm the above findings, the Tumor Immune Estimation Resource 2.0 with TCGA datasets was used to evaluate the expression of SIXs in different cancerous tissues and corresponding normal tissues (Figure 1). SIX1 and SIX4 were highly expressed in almost all types of cancer tissues compared with their corresponding normal tissues (P < 0.001), except for SIX1 in head and neck squamous cell carcinoma, kidney renal papillary cell carcinoma and thyroid carcinoma and SIX4 in kidney renal clear cell carcinoma, kidney renal papillary cell carcinoma and thyroid carcinoma. In contrast, almost no difference was found in the expression of SIX6 in cancers, except for lung adenocarcinoma and lung squamous cell carcinoma (P < 0.05). The expressions of SIX2, SIX3 and SIX5 were inconsistent in the different types of cancer tissues compared with their corresponding normal tissues.
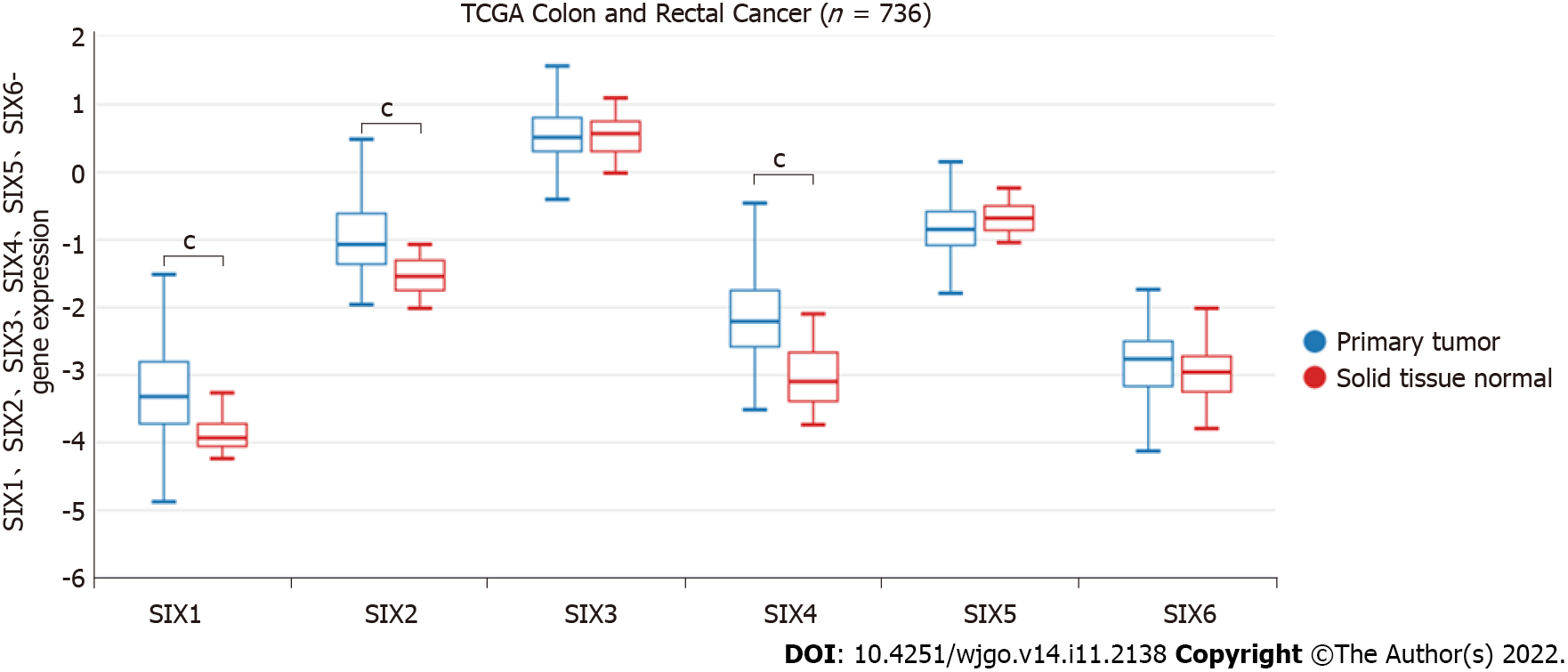
The expression patterns of SIXs in CRC were verified with the UCSC Xena database (Figure 2). In TCGA colon and rectal cancer cohort (n = 736), the expression of SIX1, SIX2 and SIX4 in CRC tissues was higher than that in normal tissues (P < 0.001), while no significant difference was found for the levels of SIX3 and SIX6.
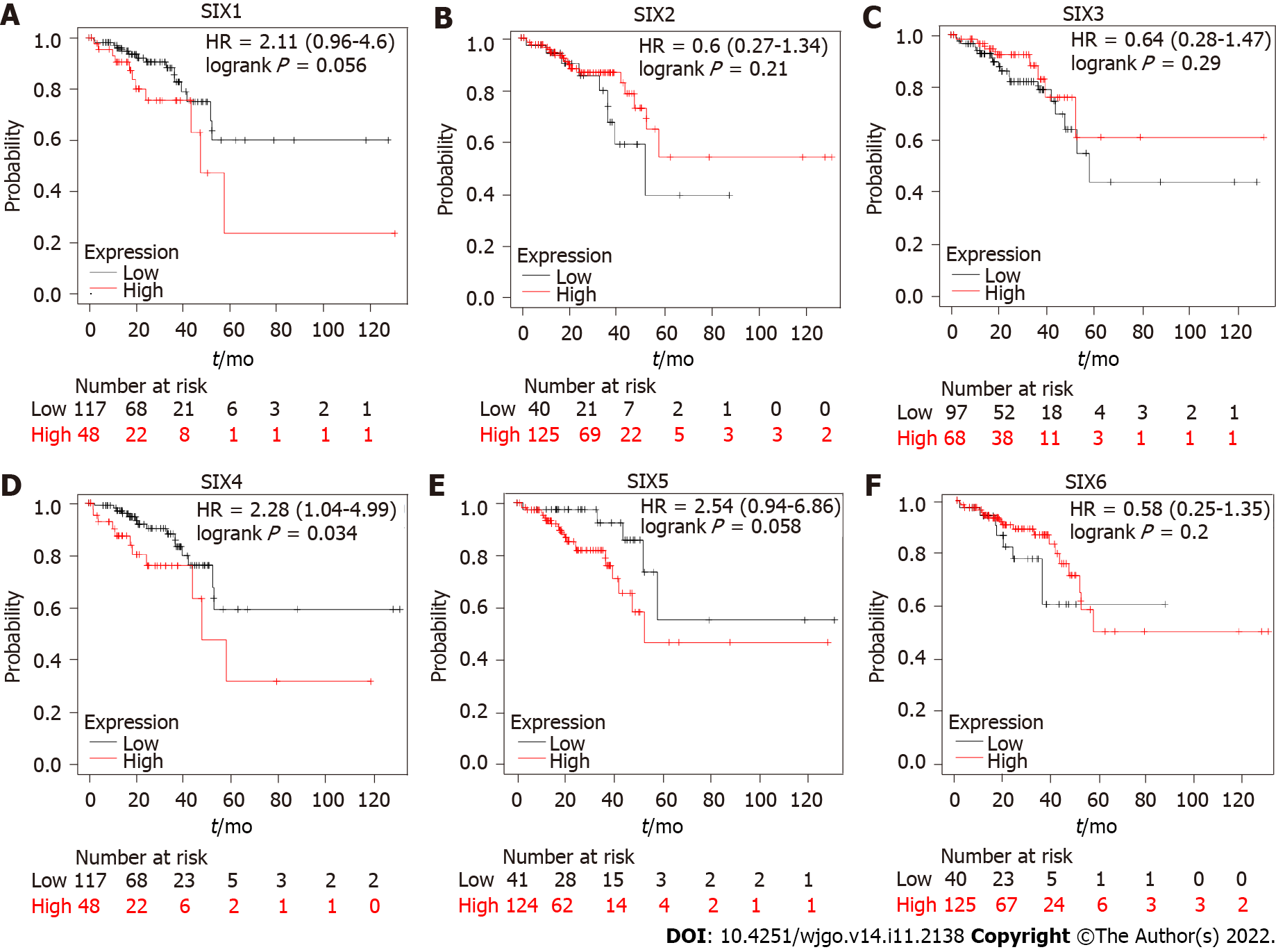
To estimate the prognostic function of SIXs in CRC patients, OS and DFS were analyzed. CRC patients with high SIX4 levels had poor OS, with hazard ratio = 2.28 (1.04-4.99), predicting a potential oncogenic role for SIX4 in CRC development (Figure 3). Although no statistical significance was found for the other SIXs, CRC patients with high SIX1 or SIX5 tended to display poor OS, and the hazard ratio was predicted to be 2.11 (0.96-4.6) and 2.54 (0.94-6.86), respectively.
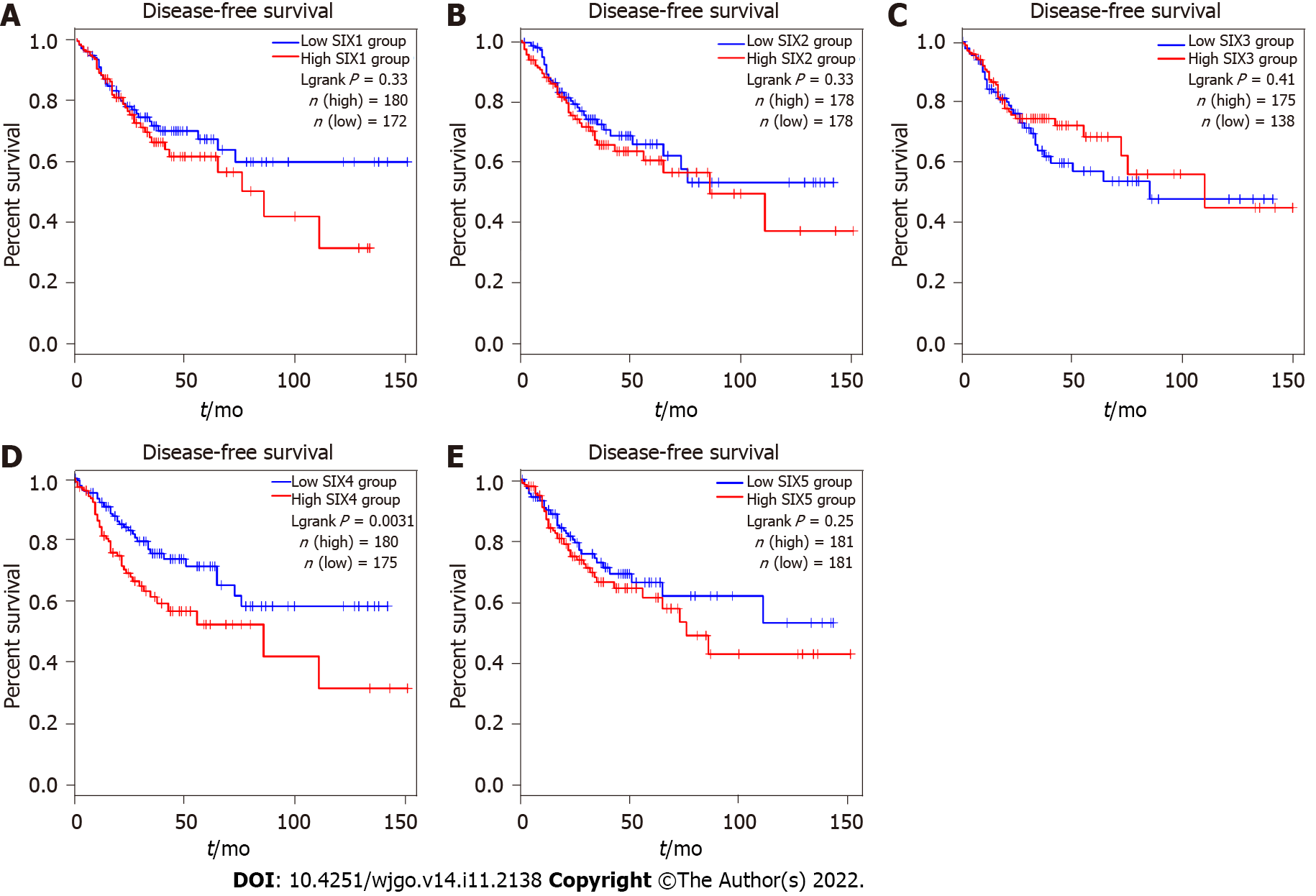
As shown in Figure 4, only the SIX4 expression level was found to be associated with progression of CRC. High levels of SIX4 in CRC patients predicted short DFS, indicating that the expression of SIX4 might promote the recurrence or relapse of CRC. Interestingly, CRC patients with high SIX1 or SIX5 also tended to have short DFS but were without statistical significance. As of now, no results have been collected for SIX6.
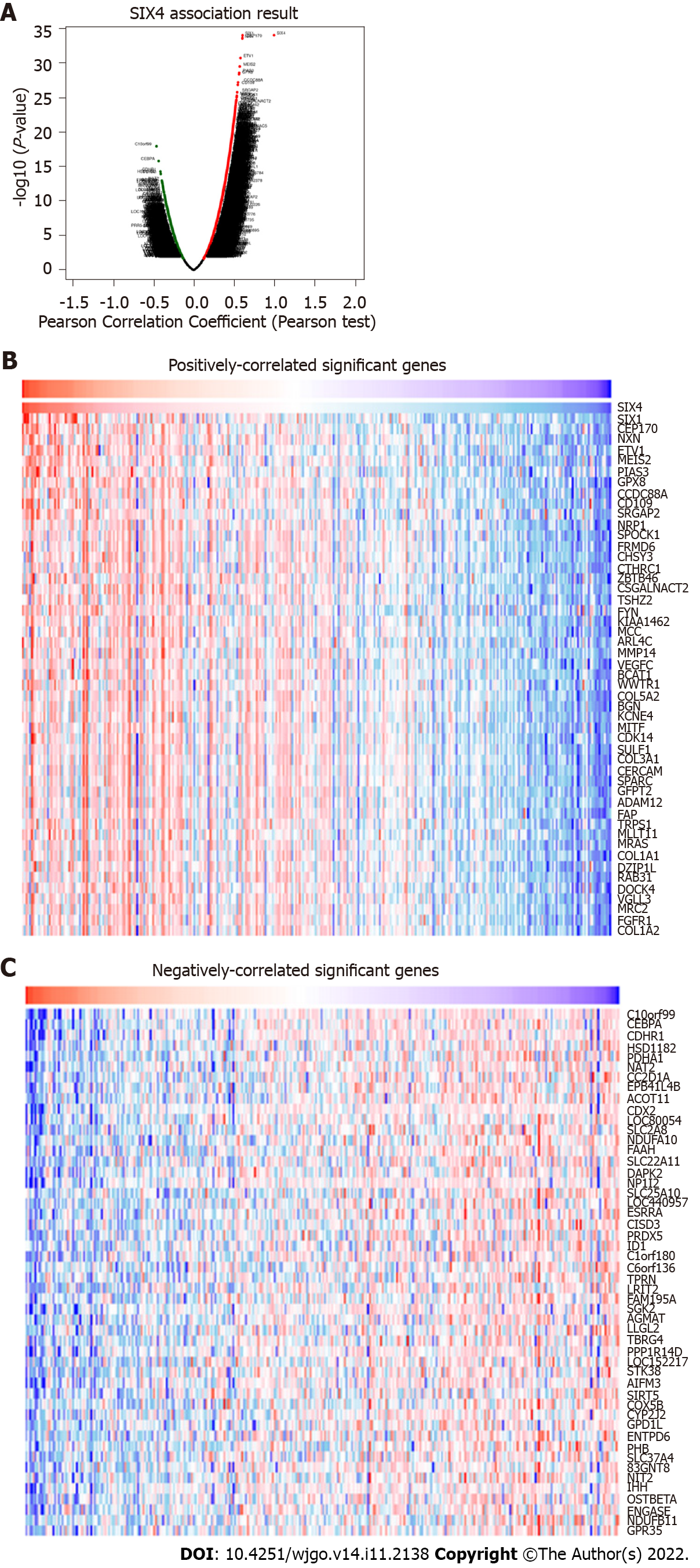
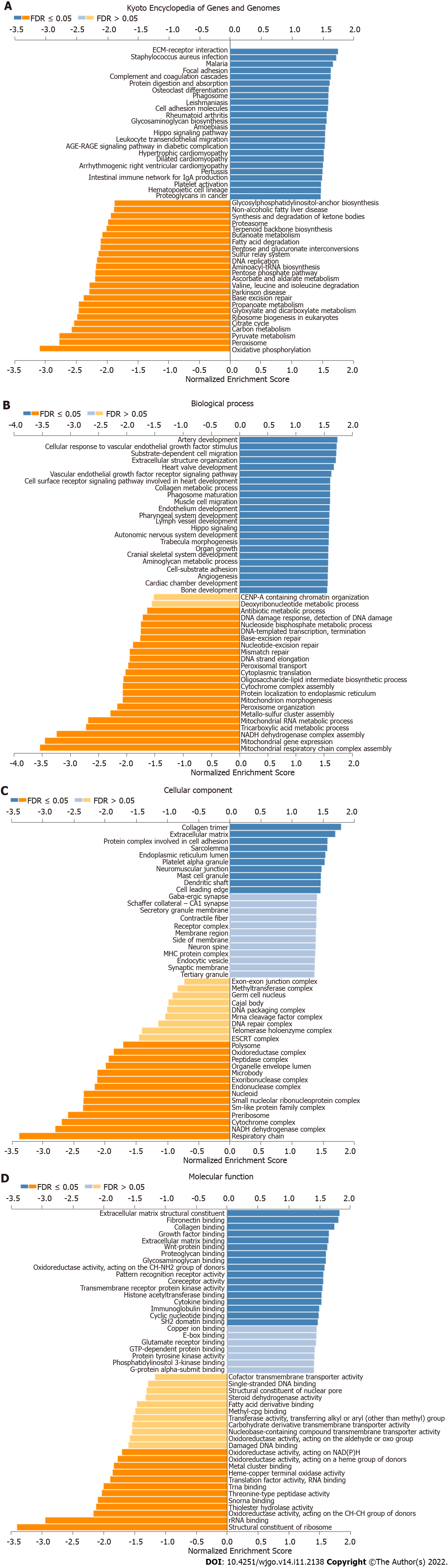
The above results indicated that SIX4 could have a potential oncogenic role in CRC. Therefore, further investigation was conducted to delineate potential roles. Potential SIX4-related genes were collected through LinkedOmics (Figure 5A), and the top 50 positively- and negatively-correlated genes were recruited for KEGG and GO analysis to identify related signaling pathways (Figures 5B and C).
In Figure 6, the top 50 positively- and negatively-correlated genes were analyzed. KEGG analysis revealed that SIX4-related genes were mainly involved in oxidative phosphorylation, peroxisome, pyruvate metabolism and carbon metabolism pathways (Figure 6A). For biological process annotation, SIX4-related genes were associated with mitochondrial respiratory chain complex assembly, mitochondrial gene expression, NADH dehydrogenase complex assembly and tricarboxylic acid metabolic process (Figure 6B). In cellular component analysis, the respiratory chain was the significant category for SIX4-related genes (Figure 6C), whereas for molecular function, structural constituent of ribosome and rRNA binding were found to be associated with SIX4-related genes (Figure 6D).
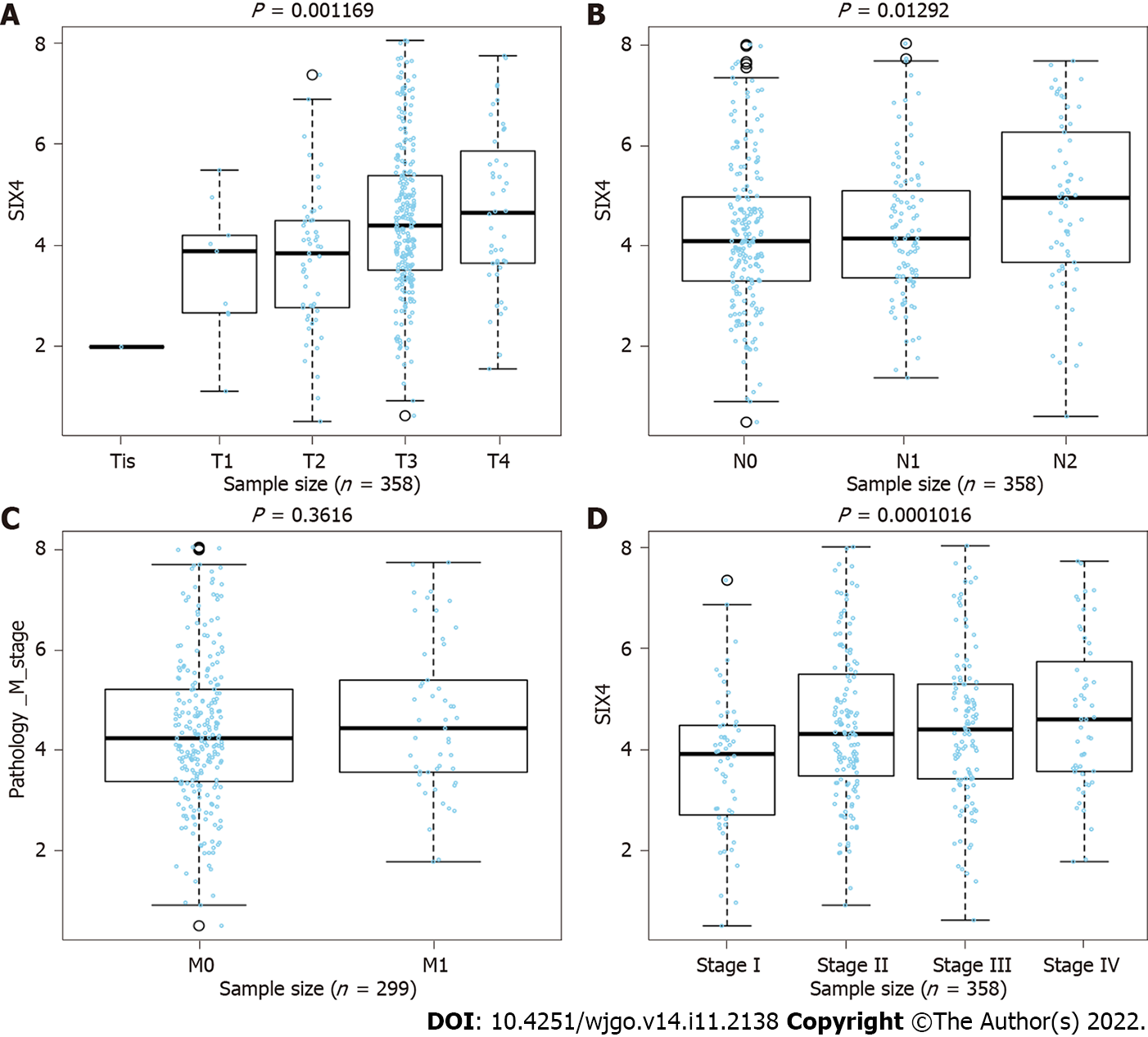
Using the LinkFinder module in the LinkedOmics dataset, expression of SIX4 was found to be positively related to tumor size and lymph node metastasis in CRC, meaning that the progression of CRC may be driven by high expression of SIX4 (Figures 7A and B). However, no association was found between SIX4 expression and CRC patient metastasis status (Figure 7C). Importantly, considering the progression of CRC, the expression of SIX4 was positively correlated with the stage of CRC patients, consistent with the results for tumor size and lymph node status (Figure 7D).
The tissue microarray included 93 colon cancer patients, 87 of whom had corresponding adjacent tissue (Table 3). There were 44 male patients and 49 female patients. The average age of the patients was 67.7 ± 9.8 years-old. Among the pathological stages, stage II patients accounted for the largest proportion (82.8%). Most patients did not have lymph node metastasis. During long-term follow-up, 45 of the patients died of CRC.
| Characteristic | |
| Patients, n (M/F) | 93 (44/49) |
| Type of tumor (colon tumor/others) | 93 (93/0) |
| age in yr, mean ± SD (range) | 67.7 ± 9.8 (43-90) |
| Histologic type, n (carcinoma/adjacent) | 93 (93/87) |
| Pathological stage, n (I/II/III) | 93 (1/77/15) |
| Primary tumor, n (T1/T2/T3/T4)1 | 92 (1/4/70/15) |
| Lymph node status, n (N0/N1/N2) | 93 (58/26/9) |
| Survival state, n (life/death) | 93 (48/45) |
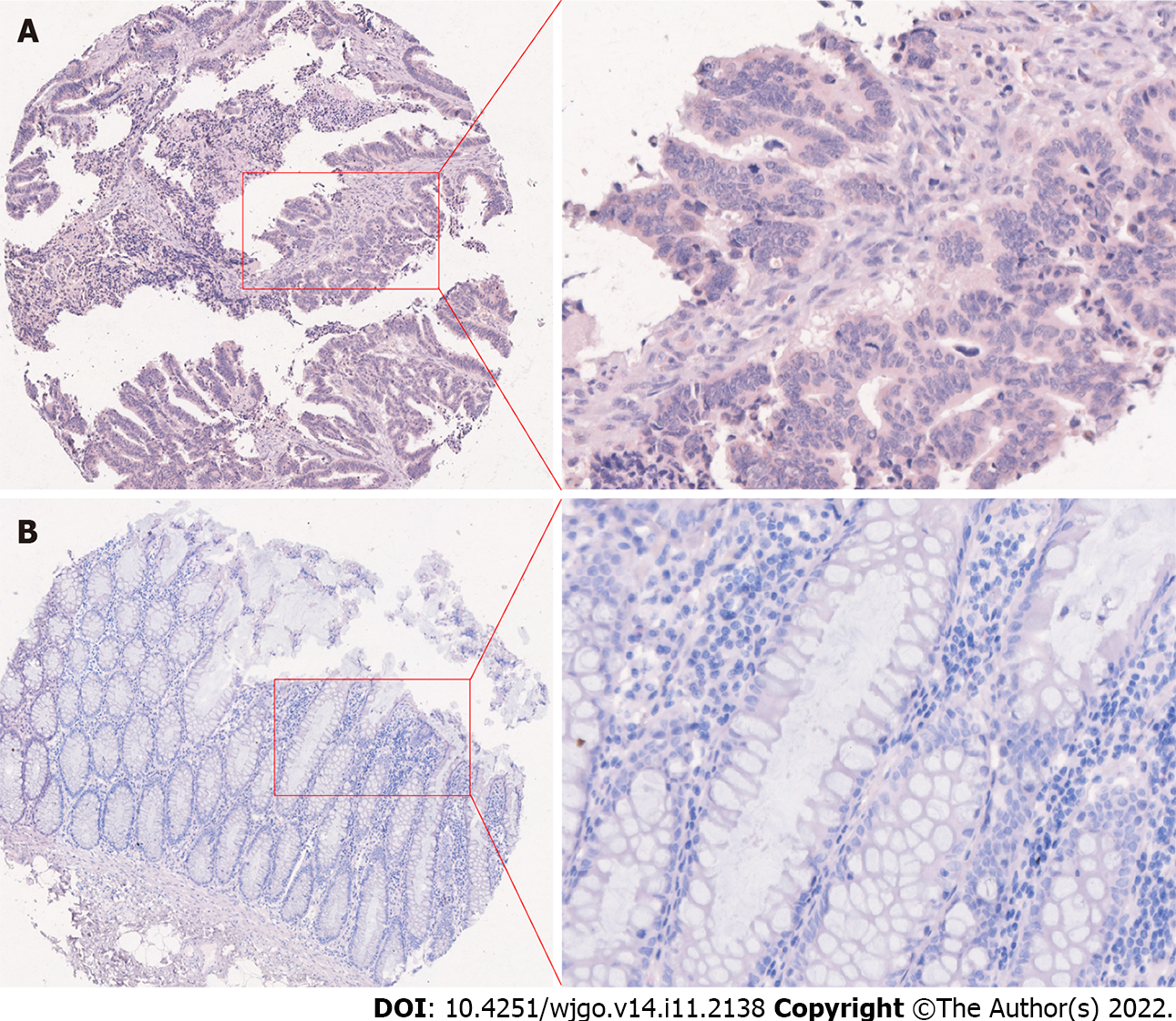
As an important transcription factor involved in development, SIX4 was predicted to be subcellularly located mainly in the nucleus but also in the cytosol. Immunohistochemical staining confirmed the subcellular location of SIX4 (Figure 8). The protein level of SIX4 was consistently high in CRC tissues compared with adjacent normal tissues (P < 0.0001) (Table 4).
| Tissue type | n | Low (%) | High (%) | χ2 | P value |
| Adjacent tissues | 87 | 60 (69.0) | 27 (31.0) | 22.10 | < 0.0001 |
| CRC tissues | 87 | 29 (33.3) | 58 (66.7) |
Further analysis of the expression pattern of SIX4 was conducted in 93 CRC patients (Table 5). However, no statistical significance was found between SIX4 levels and clinicopathological parameters, such as the onset age, sex, primary tumor characteristics and lymph node status (P > 0.05). Although no difference was found, the percentage of patients with high SIX4 levels tended to be increased at advanced pathological stage, while high expression of SIX4 tended to promote lymph node metastasis, which is similar to the results from analysis of SIX4 mRNA levels.
| Clinicopathological parameter | n | SIX4 expression | χ2 | P value | |
| Low | High | ||||
| Age | |||||
| ≤ 60 yr | 22 | 10 (45.5%) | 12 (54.5%) | 1.558 | 0.2120 |
| > 60 yr | 71 | 22 (31.0%) | 49 (69.0%) | ||
| Sex | |||||
| Male | 44 | 17 (38.6%) | 27 (61.4%) | 0.6614 | 0.4161 |
| Female | 49 | 15 (30.6%) | 34 (69.4%) | ||
| Maximum tumor diameter1 | |||||
| < 5 cm | 37 | 13 (35.1%) | 24 (64.9%) | 0.0034 | 0.9563 |
| ≥ 5 cm | 55 | 19 (34.5%) | 36 (65.5%) | ||
| Pathological stage | |||||
| I-II | 78 | 29 (37.2%) | 49 (62.8%) | - | 0.2470 |
| III | 15 | 3 (20.0%) | 12 (80.0%) | ||
| Primary tumor | |||||
| T1-T2 | 5 | 2 (40.0%) | 3 (60.0%) | 0.0696 | 0.9658 |
| T3 | 70 | 24 (34.3%) | 46 (65.7%) | ||
| T4 | 17 | 6 (35.3%) | 11 (64.7%) | ||
| N | |||||
| N0 | 58 | 24 (41.4%) | 34 (58.6%) | 3.318 | 0.0685 |
| N1-N2 | 35 | 8 (22.9%) | 27 (77.1%) | ||
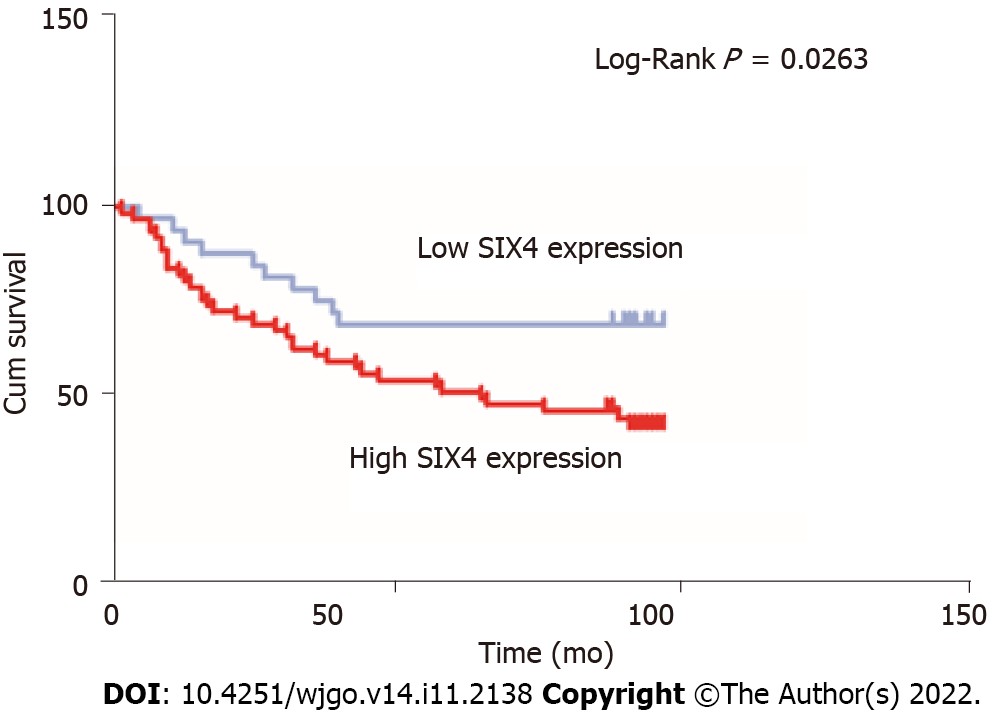
Survival analysis, based on the expression level of SIX4, was also conducted in 93 CRC patients (Figure 9). Unsurprisingly, high levels of SIX4 predicted poor OS for patients with CRC, suggesting that the SIX4 level is negatively correlated with the survival of CRC patients (P = 0.0263).
The SIX family members were originally identified as members of the homeobox family, SIX subfamily and are required for organogenesis during human development. Previous reports have demonstrated that members of the SIX family not only regulate progenitor cell proliferation and differentiation but also contribute to tumorigenesis[31]. In this study, bioinformatics was used to comprehensively analyze the expression and clinical significance of the SIX family in CRC.
Members of the SIX family are homologs of the Drosophila sine oculis, optix and Dsix4 genes, which play important roles in organ formation and mesoderm derivatives in Drosophila[32]. Interestingly, the genes encoding SIXs are located at three loci:14q23.1 for SIX1, SIX4 and SIX6, 2q21 for SIX2 and SIX3 and 19q13.32 for SIX5, although they can be divided into three subgroups (SIX1 and SIX2, SIX3 and SIX6 and SIX4 and SIX5) based on their SIX-type homeodomains and SIX domains[33]. All SIX proteins are predicted to have DNA-binding transcription factor and chromatin-binding activity[34].
SIX1 has been reported to be a strong factor for many diseases, with important roles in tumorigenesis[35]. Jiang et al[36] showed that SIX1 was highly expressed in gastric cancer patients who have short OS and is the target by which the ginsenocide Rh4 suppresses metastasis of gastric cancer through inhibition of the SIX1-stimulated transforming growth factor-β/Smad2/Smad3 signaling pathway. The transforming growth factor-β/Smad2/3 signaling pathway also has been found to be the downstream target of SIX2, which is strongly expressed in hepatocellular carcinoma and negatively related to the prognosis of hepatocellular carcinoma patients[37].
In glioblastoma, where SIX3 is transcriptionally silenced by DNA hypermethylation, SIX3 functions as a tumor suppressor. Elevated expression of SIX4 has been found in human hepatocellular carcinoma and to be positively correlated with loss of tumor encapsulation, microvascular invasion, higher tumor-node-metastasis stage and poor prognosis[38]. SIX5 is an important paralog of SIX4 and promotes lung adenocarcinoma progression through transcriptional activation of LINC01468 and its downstream pathways[22]. In T cell acute lymphoblastic leukemia, SIX6 has been shown to belong to a relevant regulatory transcription factor in T cell acute lymphoblastic leukemia to regulate gene networks[39]. However, research about the function of the SIX family members is still limited, especially for the novel family members.
The present study systematically analyzes the expression pattern of SIXs in normal and cancerous tissues. It is interesting that the expression level of SIXs is relatively low in a majority of organs, indicating they are relatively silent or suppressed in adults. In contrast, the expression of SIX1, SIX2 and SIX4 was found to be higher in CRC than in corresponding normal tissues, based on analysis of the UCSC Xena database, while the expression levels of SIX3 and SIX6 were not significantly different. Further survival analysis, using Kaplan-Meier Plotter, showed that the OS and DFS of CRC patients with high SIX4 expression were significantly worse, and there was no significant difference in other members. Previous studies simply demonstrated that SIX4 promoted the development of CRC cells by activating the PI3K-AKT pathway at the cellular level. To better understand the functional mechanism of SIX4, we used the LinkedOmics database to show that SIX4 expression was positively correlated with clinical stage, T stage and N stage in CRC, and the top 50 SIX4-related genes were involved in oxidative phosphorylation and respiratory chain signaling pathways, suggesting that SIX4 is involved in promoting the growth and metastasis of CRC.
CRC threatens the health and life quality of patients and needs more specific therapeutic targets and novel treatment strategies[40]. Using different databases, the expression of SIX4 was found to be increased in CRC tissues compared with adjacent normal ones. As a transcription factor, abnormal increases in SIX4 may enhance the expression of downstream oncogenes, such as AKT, YAP1 and c-MET[38,41]. SIX4 promotes activation of the STAT3 signaling pathway in breast cancer and plays an important role in epithelial-mesenchymal transition. Furthermore, SIX4 has been reported to play a carcinogenic role in CRC by activating the Akt signaling pathway[17]. KEGG and GO annotation predicted SIX4 and its correlated genes to be involved in oxidative phosphorylation, respiratory chain and metabolism, which are also related to classical signaling pathways, including AKT and YAP1. Therefore, the Akt signaling pathway is extremely likely to be the mechanism of SIX4-enhanced development of CRC.
The expression of SIX4 was also evaluated in clinical trials to provide solid evidence for the use of SIX4 in clinical testing. Significantly, lymph node metastasis was found to be positively associated with SIX4 level in CRC patients. The development of CRC may be at least partially due to high SIX4 expression and its transcription of downstream genes, subsequently promoting the migratory ability of tumor cells through the lymph node. As a solid tumor, the recurrence and relapse of CRC could severely affect patient survival[42]. The present study demonstrated that CRC patients with low SIX4 expression have longer survival, suggesting a novel therapy to inhibit or suppress SIX4 activity would be useful in prolonging CRC patient survival. This study provides the first comprehensive analysis of the potential role and prognostic value of the SIX family in CRC. However, the current study is limited to analyzing the expression and clinicopathological parameters of SIXs in CRC, and the sample size is relatively limited. Therefore, further in-depth analysis will be conducted in future studies. Although SIX4 may be involved in oxidative phosphorylation, respiratory chain activity and metabolism, further studies on these can be investigated.
Among the SIX family members, SIX4 was found to be a tumor-promoting factor in the intestinal tract and could be a prognostic indicator and treatment target for patients with CRC. Further investigation of novel therapeutic strategies targeting SIX4 in CRC patients could provide important potential for treatment of CRC.
Human sine oculis homeobox homolog (SIX) protein belongs to the homeobox family, which plays an important role in different developmental organs, and the abnormal expression of most development-related genes is closely related to the occurrence and development of malignant tumors. However, no studies have comprehensively analyzed SIXs in colorectal cancer (CRC).
SIXs has been found to be closely related to the occurrence and development of cancer. However, there are few studies on SIXs in CRC. SIXs may become a new potential prognostic indicator of CRC.
To comprehensively analyze the expression and the prognosis of the SIX family in CRC tissues and to explore the potential role of the SIX family as a new prognostic indicator of CRC.
In this study, the RNA and protein expression levels of the SIX family in CRC were analyzed by various online databases and immunohistochemically. Then the relationship between the SIX family and the prognosis of CRC was further analyzed. In order to better understand the mechanism of SIX4, the positive correlation between SIX4 expression and tumor-node-metastasis stage of CRC was analyzed. Then, the relationship between SIX4 mRNA levels and clinicopathological parameters in CRC patients was analyzed.
The expression levels of SIXs in most organs were relatively low in the Human Protein Atlas. UCSC Xena database analysis showed that the expression levels of SIX1, SIX2 and SIX4 in CRC were higher than those in corresponding normal tissue. Further survival analysis with Kaplan-Meier Plotter showed that the relation between poor overall survival and disease-free survival of CRC patients and high SIX4 expression were significant. Using the LinkedOmics database, the expression of SIX4 was positively correlated with the clinical stage, T stage and N stage of CRC, and the top 50 SIX4-related genes were involved in oxidative phosphorylation and respiratory chain signaling pathway, suggesting that SIX4 was involved in promoting the growth and metastasis of CRC.
As a member of the SIX family, SIX4 played a role in promoting tumor development in the intestine, which may serve as a potential prognostic indicator and therapeutic target for CRC patients. Therefore, targeting SIX4 may serve as a new therapeutic strategy for CRC patients, providing important potential for the treatment of CRC.
This is the first comprehensive analysis of the potential role and prognostic value of the SIX family in CRC. However, the current research is limited, and future studies need to further explore the oxidative phosphorylation, respiratory chain activity and metabolism of SIX4 in CRC.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Brkanović S, Croatia; Janmohammadi P, Iran; Jeong KY, South Korea S-Editor: Wang JJ L-Editor: Filipodia P-Editor: Wang JJ
| 1. | Töhönen V, Katayama S, Vesterlund L, Jouhilahti EM, Sheikhi M, Madissoon E, Filippini-Cattaneo G, Jaconi M, Johnsson A, Bürglin TR, Linnarsson S, Hovatta O, Kere J. Novel PRD-like homeodomain transcription factors and retrotransposon elements in early human development. Nat Commun. 2015;6:8207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 88] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 2. | Gerrard DT, Berry AA, Jennings RE, Birket MJ, Zarrineh P, Garstang MG, Withey SL, Short P, Jiménez-Gancedo S, Firbas PN, Donaldson I, Sharrocks AD, Hanley KP, Hurles ME, Gomez-Skarmeta JL, Bobola N, Hanley NA. Dynamic changes in the epigenomic landscape regulate human organogenesis and link to developmental disorders. Nat Commun. 2020;11:3920. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 3. | Hong S, Shen X, Luo C, Sun F. Comparative analysis of the testes from wild-type and Alkbh5-knockout mice using single-cell RNA sequencing. G3 (Bethesda). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 4. | Xu PX, Zheng W, Huang L, Maire P, Laclef C, Silvius D. Six1 is required for the early organogenesis of mammalian kidney. Development. 2003;130:3085-3094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 275] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 5. | Xu J, Li J, Ramakrishnan A, Yan H, Shen L, Xu PX. Six1 and Six2 of the Sine Oculis Homeobox Subfamily are Not Functionally Interchangeable in Mouse Nephron Formation. Front Cell Dev Biol. 2022;10:815249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 6. | Ghanbari H, Seo HC, Fjose A, Brändli AW. Molecular cloning and embryonic expression of Xenopus Six homeobox genes. Mech Dev. 2001;101:271-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 107] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 7. | Chen R, Hou Y, Connell M, Zhu S. Homeodomain protein Six4 prevents the generation of supernumerary Drosophila type II neuroblasts and premature differentiation of intermediate neural progenitors. PLoS Genet. 2021;17:e1009371. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 8. | Mosrati MA, Hammami B, Rebeh IB, Ayadi L, Dhouib L, Ben Mahfoudh K, Hakim B, Charfeddine I, Mnif J, Ghorbel A, Masmoudi S. A novel dominant mutation in SIX1, affecting a highly conserved residue, result in only auditory defects in humans. Eur J Med Genet. 2011;54:e484-e488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 9. | Ruf RG, Xu PX, Silvius D, Otto EA, Beekmann F, Muerb UT, Kumar S, Neuhaus TJ, Kemper MJ, Raymond RM Jr, Brophy PD, Berkman J, Gattas M, Hyland V, Ruf EM, Schwartz C, Chang EH, Smith RJ, Stratakis CA, Weil D, Petit C, Hildebrandt F. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc Natl Acad Sci U S A. 2004;101:8090-8095. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 297] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 10. | Geng X, Speirs C, Lagutin O, Inbal A, Liu W, Solnica-Krezel L, Jeong Y, Epstein DJ, Oliver G. Haploinsufficiency of Six3 fails to activate Sonic hedgehog expression in the ventral forebrain and causes holoprosencephaly. Dev Cell. 2008;15:236-247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 137] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 11. | Wang SH, Wu CC, Lu YC, Lin YH, Su YN, Hwu WL, Yu IS, Hsu CJ. Mutation screening of the EYA1, SIX1, and SIX5 genes in an East Asian cohort with branchio-oto-renal syndrome. Laryngoscope. 2012;122:1130-1136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 12. | Panagiotou ES, Fernandez-Fuentes N, Farraj LA, McKibbin M, Elçioglu NH, Jafri H, Cerman E, Parry DA, Logan CV, Johnson CA, Inglehearn CF, Toomes C, Ali M. Novel SIX6 mutations cause recessively inherited congenital cataract, microcornea, and corneal opacification with or without coloboma and microphthalmia. Mol Vis. 2022;28:57-69. [PubMed] |
| 13. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8287] [Cited by in RCA: 11936] [Article Influence: 2984.0] [Reference Citation Analysis (4)] |
| 14. | Xing XL, Yao ZY, Zhang T, Zhu N, Liu YW, Peng J. MicroRNA-Related Prognosis Biomarkers from High-Throughput Sequencing Data of Colorectal Cancer. Biomed Res Int. 2020;2020:7905380. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 15. | Zahnd WE, Josey MJ, Schootman M, Eberth JM. Spatial accessibility to colonoscopy and its role in predicting late-stage colorectal cancer. Health Serv Res. 2021;56:73-83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 16. | Gong B, Kao Y, Zhang C, Sun F, Gong Z, Chen J. Identification of Hub Genes Related to Carcinogenesis and Prognosis in Colorectal Cancer Based on Integrated Bioinformatics. Mediators Inflamm. 2020;2020:5934821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 17. | Li G, Hu F, Luo X, Hu J, Feng Y. SIX4 promotes metastasis via activation of the PI3K-AKT pathway in colorectal cancer. PeerJ. 2017;5:e3394. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 18. | Yang Y, Feng M, Bai L, Liao W, Zhou K, Zhang M, Wu Q, Wen F, Lei W, Zhang P, Zhang N, Huang J, Li Q. Comprehensive analysis of EMT-related genes and lncRNAs in the prognosis, immunity, and drug treatment of colorectal cancer. J Transl Med. 2021;19:391. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 19. | Yang C, Liu H. Both a hypoxia-inducible EYA3 and a histone acetyltransferase p300 function as coactivators of SIX5 to mediate tumorigenesis and cancer progression. Ann Transl Med. 2022;10:752. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 20. | Song W, Ma J, Lei B, Yuan X, Cheng B, Yang H, Wang M, Feng Z, Wang L. Sine oculis homeobox 1 promotes proliferation and migration of human colorectal cancer cells through activation of Wnt/β-catenin signaling. Cancer Sci. 2019;110:608-616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 21. | Yu Z, Feng J, Wang W, Deng Z, Zhang Y, Xiao L, Wang Z, Liu C, Liu Q, Chen S, Wu M. The EGFR-ZNF263 signaling axis silences SIX3 in glioblastoma epigenetically. Oncogene. 2020;39:3163-3178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 22. | Yuan Y, Zhou D, Chen F, Yang Z, Gu W, Zhang K. SIX5-activated LINC01468 promotes lung adenocarcinoma progression by recruiting SERBP1 to regulate SERPINE1 mRNA stability and recruiting USP5 to facilitate PAI1 protein deubiquitylation. Cell Death Dis. 2022;13:312. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 23. | Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7696] [Cited by in RCA: 10536] [Article Influence: 1053.6] [Reference Citation Analysis (0)] |
| 24. | Wu HT, Chen WJ, Xu Y, Shen JX, Chen WT, Liu J. The Tumor Suppressive Roles and Prognostic Values of STEAP Family Members in Breast Cancer. Biomed Res Int. 2020;2020:9578484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 25. | Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77:e108-e110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2728] [Cited by in RCA: 4092] [Article Influence: 511.5] [Reference Citation Analysis (0)] |
| 26. | Goldman M, Craft B, Swatloski T, Cline M, Morozova O, Diekhans M, Haussler D, Zhu J. The UCSC Cancer Genomics Browser: update 2015. Nucleic Acids Res. 2015;43:D812-D817. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 209] [Cited by in RCA: 229] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 27. | Lánczky A, Győrffy B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J Med Internet Res. 2021;23:e27633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 1045] [Article Influence: 261.3] [Reference Citation Analysis (0)] |
| 28. | Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556-W560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1991] [Cited by in RCA: 3076] [Article Influence: 512.7] [Reference Citation Analysis (0)] |
| 29. | Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956-D963. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 808] [Cited by in RCA: 1537] [Article Influence: 256.2] [Reference Citation Analysis (0)] |
| 30. | Liu J, Wei XL, Huang WH, Chen CF, Bai JW, Zhang GJ. Cytoplasmic Skp2 expression is associated with p-Akt1 and predicts poor prognosis in human breast carcinomas. PLoS One. 2012;7:e52675. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 31. | Ronca R, Benkheil M, Mitola S, Struyf S, Liekens S. Tumor angiogenesis revisited: Regulators and clinical implications. Med Res Rev. 2017;37:1231-1274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 137] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 32. | Kumar JP. The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cell Mol Life Sci. 2009;66:565-583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 208] [Cited by in RCA: 202] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 33. | Laclef C, Hamard G, Demignon J, Souil E, Houbron C, Maire P. Altered myogenesis in Six1-deficient mice. Development. 2003;130:2239-2252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 203] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 34. | Rafiq A, Aashaq S, Jan I, Beigh MA. SIX1 transcription factor: A review of cellular functions and regulatory dynamics. Int J Biol Macromol. 2021;193:1151-1164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 35. | Wu W, Ren Z, Li P, Yu D, Chen J, Huang R, Liu H. Six1: a critical transcription factor in tumorigenesis. Int J Cancer. 2015;136:1245-1253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 36. | Jiang H, Ma P, Duan Z, Liu Y, Shen S, Mi Y, Fan D. Ginsenoside Rh4 Suppresses Metastasis of Gastric Cancer via SIX1-Dependent TGF-β/Smad2/3 Signaling Pathway. Nutrients. 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 26] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 37. | Wan ZH, Ma YH, Jiang TY, Lin YK, Shi YY, Tan YX, Dong LW, Wang HY. Six2 is negatively correlated with prognosis and facilitates epithelial-mesenchymal transition via TGF-β/Smad signal pathway in hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2019;18:525-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 38. | He Q, Lin Z, Wang Z, Huang W, Tian D, Liu M, Xia L. SIX4 promotes hepatocellular carcinoma metastasis through upregulating YAP1 and c-MET. Oncogene. 2020;39:7279-7295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 39. | Laukkanen S, Oksa L, Nikkilä A, Lahnalampi M, Parikka M, Seki M, Takita J, Degerman S, de Bock CE, Heinäniemi M, Lohi O. SIX6 is a TAL1-regulated transcription factor in T-ALL and associated with inferior outcome. Leuk Lymphoma. 2020;61:3089-3100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 40. | Biller LH, Schrag D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA. 2021;325:669-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 1438] [Article Influence: 359.5] [Reference Citation Analysis (0)] |
| 41. | Sun X, Hu F, Hou Z, Chen Q, Lan J, Luo X, Wang G, Hu J, Cao Z. SIX4 activates Akt and promotes tumor angiogenesis. Exp Cell Res. 2019;383:111495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 42. | Dueland S, Smedman TM, Røsok B, Grut H, Syversveen T, Jørgensen LH, Line PD. Treatment of relapse and survival outcomes after liver transplantation in patients with colorectal liver metastases. Transpl Int. 2021;34:2205-2213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |