Published online Nov 15, 2021. doi: 10.4251/wjgo.v13.i11.1709
- This article has been corrected.
- See: World J Gastrointest Oncol. Oct 15, 2022; 14(10): 2085-2087
Peer-review started: April 20, 2021
First decision: June 23, 2021
Revised: June 24, 2021
Accepted: August 23, 2021
Article in press: August 23, 2021
Published online: November 15, 2021
Processing time: 212 Days and 5.9 Hours
Pancreatic cancer (PC) is one of the most lethal malignancies worldwide. It is known that the proliferation of PC cells is a critical process in the disease. Previous studies have failed to identify the key genes associated with PC cell proliferation, using bioinformatic analysis, genome-wide association studies, and candidate gene testing.
To investigate the function of the chromobox 8 (CBX8)/receptor substrate 1 (IRS1)/AKT axis in PC.
A genome-wide CRISPR-Cas9 screening was performed to select genes that could facilitate PC cell proliferation. Quantitative reverse transcription-polymerase chain reaction was used to detect the expression of CBX8 in PC tissues and cells. The regulatory roles of CBX8 in cell proliferation, migration, and invasion were verified by in vivo and in vitro functional assays.
CBX8 was upregulated in PC tissues and shown to drive PC cell proliferation. Higher expression of CBX8 was correlated with worse outcomes of PC patients from two independent cohorts comprising a total of 116 cases. CBX8 was also proved to serve as a promising therapeutic target for a PC xenograft model. We demonstrated that hypoxia-inducible factor (HIF)-1a induced CBX8 transcription by binding to the promoter of CBX8. CBX8 efficiently activated the PI3K/AKT signaling by upregulating insulin IRS1.
CBX8 is a key gene regulated by HIF-1α, and activates the IRS1/AKT pathway, which suggests that targeting CBX8 may be a promising therapeutic strategy for PC.
Core Tip: The authors demonstrated that hypoxia-inducible factor-1a induced chromobox (CBX)8 transcription by binding to the promoter of CBX8. CBX8 efficiently activated the PI3K/AKT signaling by upregulating insulin receptor substrate 1. The newly identified signaling axis may support the development of new therapeutic strategies for pancreatic cancer.
- Citation: Teng BW, Zhang KD, Yang YH, Guo ZY, Chen WW, Qiu ZJ. Genome-wide CRISPR-Cas9 screening identifies that hypoxia-inducible factor-1a-induced CBX8 transcription promotes pancreatic cancer progression via IRS1/AKT axis. World J Gastrointest Oncol 2021; 13(11): 1709-1724
- URL: https://www.wjgnet.com/1948-5204/full/v13/i11/1709.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v13.i11.1709
Pancreatic cancer (PC) is one of the most lethal cancers worldwide. It has become the second most fatal cancer in the United States. The 5-year survival rate of pancreatic ductal adenocarcinoma (PDAC) is < 10% due to late diagnosis and resistance to systemic therapies[1]. Many studies have shown that the prognosis of patients with PC remains poor after complete surgical resection. At the same time, due to the difficulty of diagnosis, many patients with PC have no chance of surgery after diagnosis. Therefore, it is important to study the occurrence and development of PC itself and the corresponding targeted therapy.
Hypoxia is one of the important characteristics of PC[2]. Activation of hypoxia-inducible factors (HIFs; particularly HIF-1a and HIF-2a) is an important mechanism for tumor cells to adapt to a hypoxic microenvironment. Our previous studies indicated that HIFs not only regulate the growth and metastasis of PC cells, but also mediate the immunosuppression[3] and angiogenesis[4] in PC.
Chromobox 8 (CBX8) (also known as human polyclonal 3), together with CBX2, CBX4, CBX6, and CBX7, are members of the CBX protein family. CBX8 plays an oncogenic role in different types of cancer. Zhang et al[5] reported that CBX8 upre
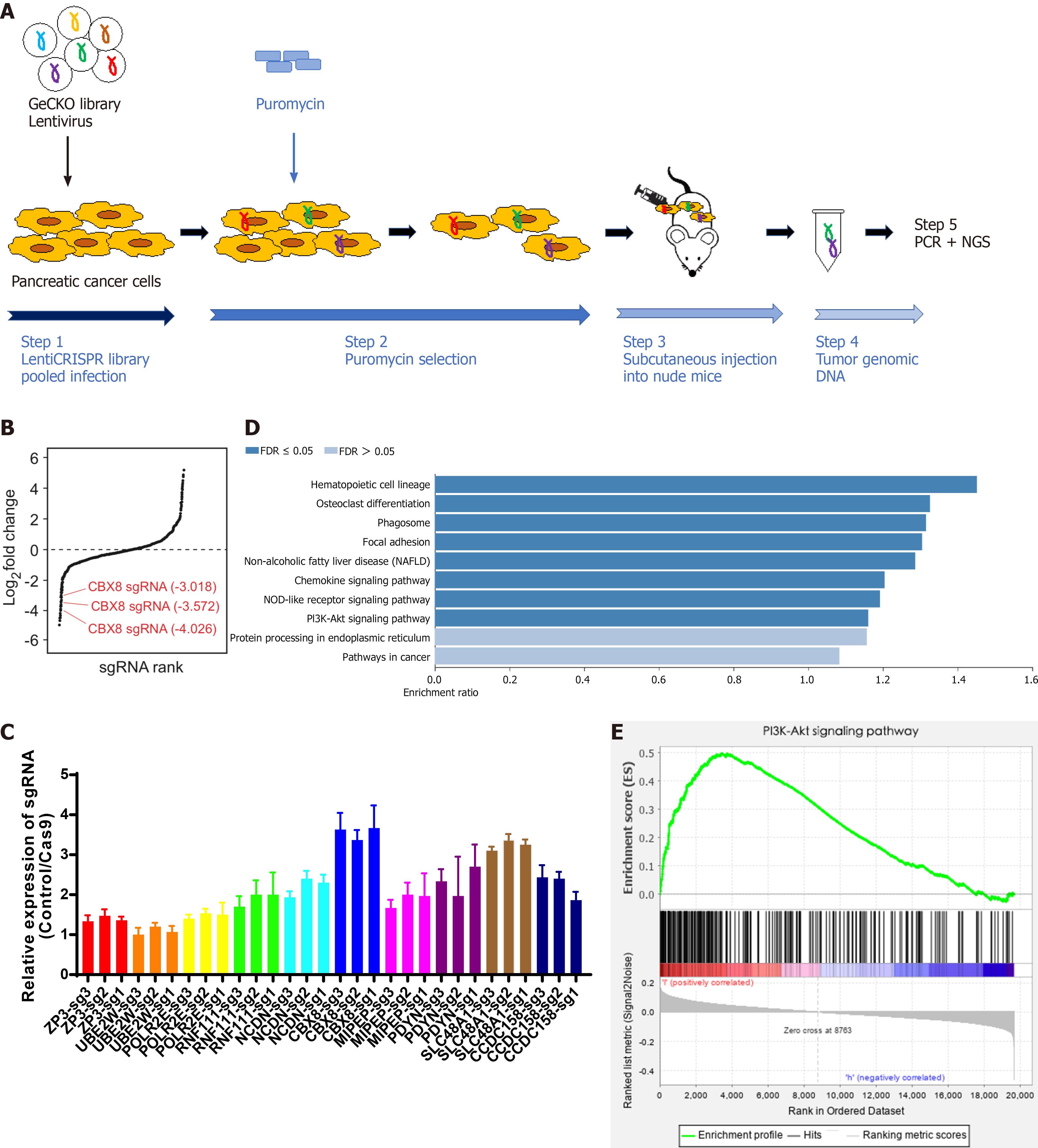
Genome-scale CRISPR-Cas9 knockout (GeCKO) library is a powerful tool for the assessment of gene function[8] and the screening for genes involved in cancer cell proliferation and metastasis. In this study, Bxpc-3 and PANC1 cells were transduced with library lentiviruses. The cells were then injected subcutaneously into nude mice and removed 21 d later to sequence the single guide RNA (sgRNA) in the tumor tissue. Among all candidate genes, CBX8 was selected for further analysis because of its upregulation in cells. Furthermore, higher CBX8 expression was correlated with worse clinical outcomes of PDAC patients from two independent cohorts. We also showed that CBX8 was a key gene that was regulated by HIF-1α, and could activate the IRS1/AKT pathway. The above findings suggest that targeting CBX8 may be a promising therapeutic strategy for PC.
PC cell lines (PANC1 and Bxpc-3) and 293T cells were purchased from American Type Culture Collection (Manassas, VA, United States) and maintained in RPMI-1640 medium containing 10% fetal bovine serum and 1% penicillin/streptomycin in a 95% air/5% CO2 environment at 37 °C. Cells at passages 3–15 were used in this study. For hypoxic culture, cells were cultured under 1% O2.
The Human CRISPR Knockout Pooled Library was obtained from Addgene (http://www.addgene.org/crispr/Libraries/geckov2/, United States). GeCKO library plasmids, pVSVg (AddGene) and psPAX2 (AddGene), were added to 100 μL Opti-MEM at a ratio of 1:0.5:1.5. After 15-min incubation with Lipofectamine 3000 (Invitrogen, Carlsbad, CA, United States), the mixture was added to 293T cells. Forty-eight hours later, cell supernatants containing lentiviruses were collected. PANC1 and BXPC-3 cells were transduced at a low multiplicity of infection (0.3) to ensure that most cells received only one viral construct. The cells were selected with puromycin (1 μg/mL) for 14 d. Only cells transduced with a LentiCRISPR construct could survive. After transfection with GeCKO library, cells were transplanted subcutaneously into the right flank of 4-wk-old male nude mice. Twenty-one days later, the mice were killed and primary tumors were removed for sgRNA sequencing.
Following infection, cells were treated with gemcitabine for 48 h. Cell viability was assessed by CCK-8 assay (Beyotime, China) and the IC50 values were calculated using GraphPad Prism 7 (GraphPad, La Jolla, CA, United States). All measurements were performed in triplicate. The abundance of sgRNA was determined by deep sequencing.
Cells were treated with DMSO or chemotherapeutic agents for 24 h and then seeded in six-well plates (200/well). The cultures were maintained in a 5% CO2 incubator at 37 °C for 2 wk. The colonies were fixed with 4% paraformaldehyde, followed by staining with 0.5% crystal violet. The number of colonies in each group was counted under a microscope. Independent experiments were performed in triplicate.
The promoter regions of CBX8 and IRS1 were amplified by polymerase chain reaction (PCR) and cloned into RB reporter plasmids (Ribo, China). CBX8 knockdown plasmids were cotransfected into 293T cells. Twenty-four hours later, cells were harvested and Renilla and firefly luciferase activities were detected using a dual-luciferase reporter assay kit. The Renilla luciferase activity was used for normalization.
A chromatin immunoprecipitation (ChIP) assay was carried out using an EZ-ChIP Chromatin Immunoprecipitation Kit (Millipore, Bedford, MA, United States). Formaldehyde (1%) was used to crosslink proteins and DNA for 10 min. Cell lysates were sonicated to obtain DNA fragments, which were subjected to IP with primary antibodies or negative control IgG. Purified DNA was analyzed by quantitative reverse transcription PCR (qRT-PCR) with SYBR Green Master Mix (Promega, Beijing, China). The relative enrichment values were calculated through normalization of the results to the input values and are expressed relative to the values obtained with normal IgG.
All animal experimental procedures were approved by the Ethics Committee for Animal Research of Shanghai Jiaotong University School of Medicine (Shanghai, China). Four-week-old male nude mice were subcutaneously transplanted in the right flank with CBX8-silenced or control PC cells (PANC1, Bxpc-3, 2 × 106 cells per mouse) in 100 μL PBS mixed 1:1 with BD Matrigel Basement Membrane Matrix (Corning, Corning, NY, United States) as previously reported. Tumor size was measured every 3 d with a digital caliper. When tumor volume reached approximately 100 mm3 (day 6 postinoculation), mice were randomly assigned to each group. After 21 d, the tumor weight was detected and mice were killed.
This study was approved by the Human Research Ethics Committee of Shanghai General Hospital (Shanghai, China) and performed following the United States Common Rule. Each patient provided written informed consent. A total of 116 archived PDAC specimens were collected. Patients were followed over time.
All data are shown as the mean ± SD. Statistical analyses were performed using Graphpad Prism 7. Student’s t test or analysis of variance was used to compare continuous variables.
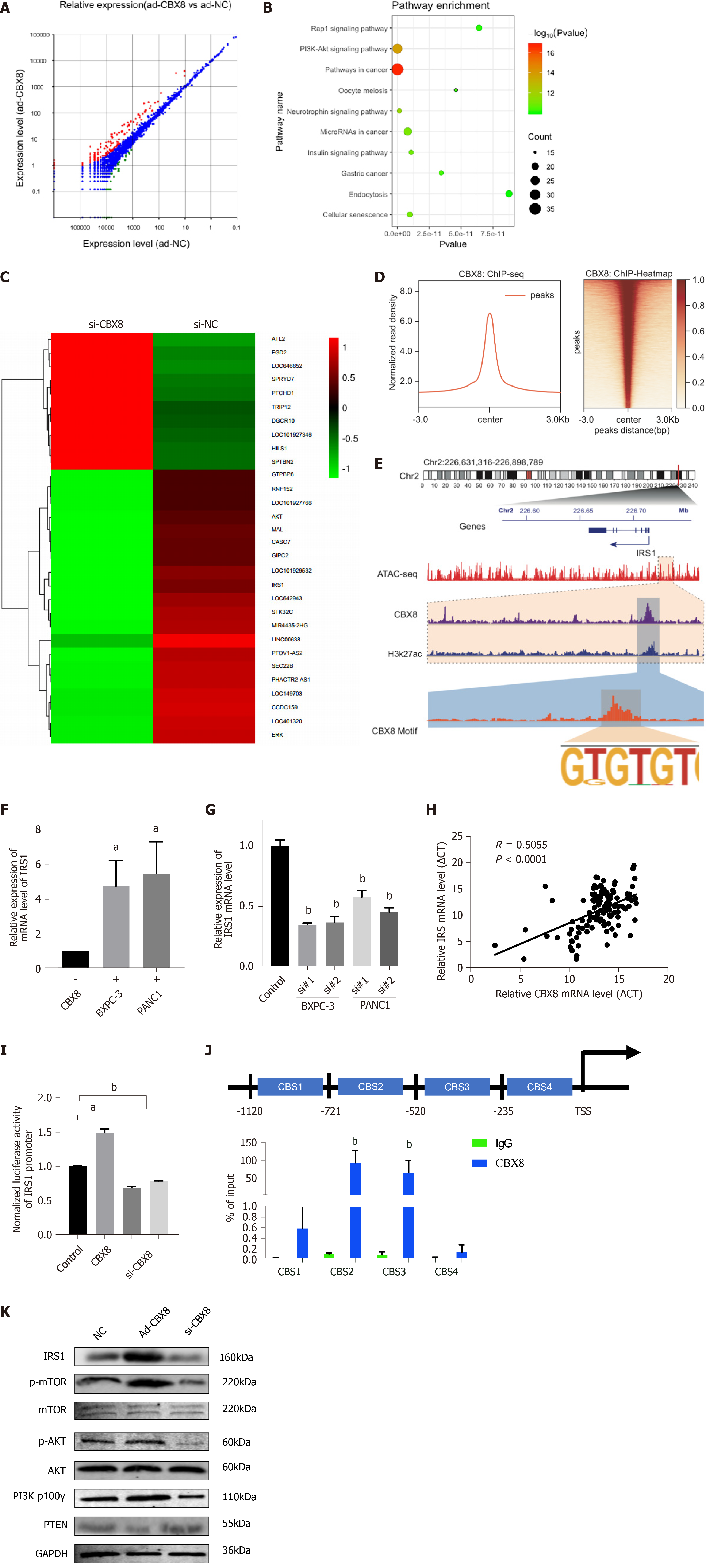
To identify the genes responsible for growth of PC cells, we transduced PANC1 and Bxpc-3 cells with GeCKO library lentiviruses. After 14 d of puromycin selection, cells were transplanted subcutaneously into the right flank of 4-wk-old male nude mice. Twenty-one days later, the mice were killed and primary tumors were removed for sgRNA sequencing (Figure 1A). About 1.6 × 106 sgRNA sequences were obtained from each tumor sample. Genomic DNA was extracted from tumor tissues for PCR and next-generation sequencing (NGS) analysis. The expression of genes in each sample was measured. The genes were ranked according to the numbers of sgRNA and NGS reads. Compared to the control group, 872 cell growth related candidate genes were identified in PANC1 cells, while 819 were found in Bxpc-3 cells. There were 244 genes identified in both groups with differentially enriched sgRNA. We performed qPCR to confirm the candidate top 10 differently expressed sgRNAs from NGS analysis (Figure 1B), which indicated that CBX8 had the lowest expression level of sgRNA (Figure 1C). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and gene set enrichment analysis (GSEA) showed that these universal differentially expressed sgRNAs were enriched in the PI3K/AKT pathway (Figure 1D and E).
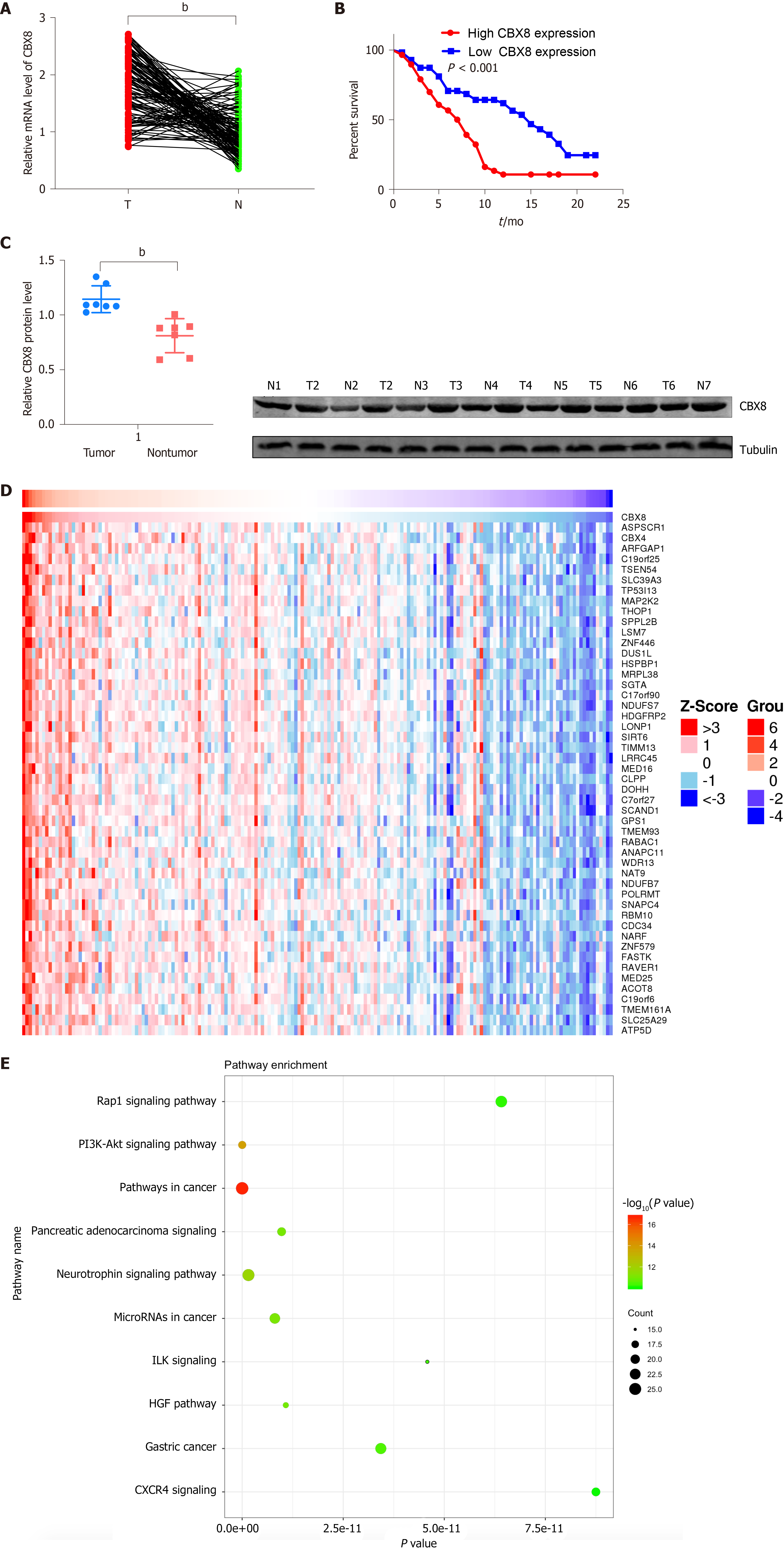
To determine whether CBX8 is involved in PC progression, we examined the mRNA level of CBX8 in 116 paired PC and adjacent nontumor tissue samples and found that CBX8 expression was significantly increased in PC tissues compared with corresponding noncancer tissues (Figure 2A). Kaplan–Meier survival analysis indicated that patients with higher CBX8 expression had a shorter overall survival than those with low CBX8 expression (Figure 2B). The protein content of CBX8 in these tissue samples was obtained by Western blot analysis (Figure 2C). Compared to normal neighboring tissues, the CBX8 protein content in PC tissues was increased.
We analyzed the expression of CBX8 in The Cancer Genome Atlas (TCGA) database to confirm its potential regulatory pathways and cellular functions. KEGG pathway analysis found that genes positively correlated to CBX8 (Figure 2D) were enriched in the PI3K/AKT and pancreatic adenocarcinoma pathway (Figure 2E).
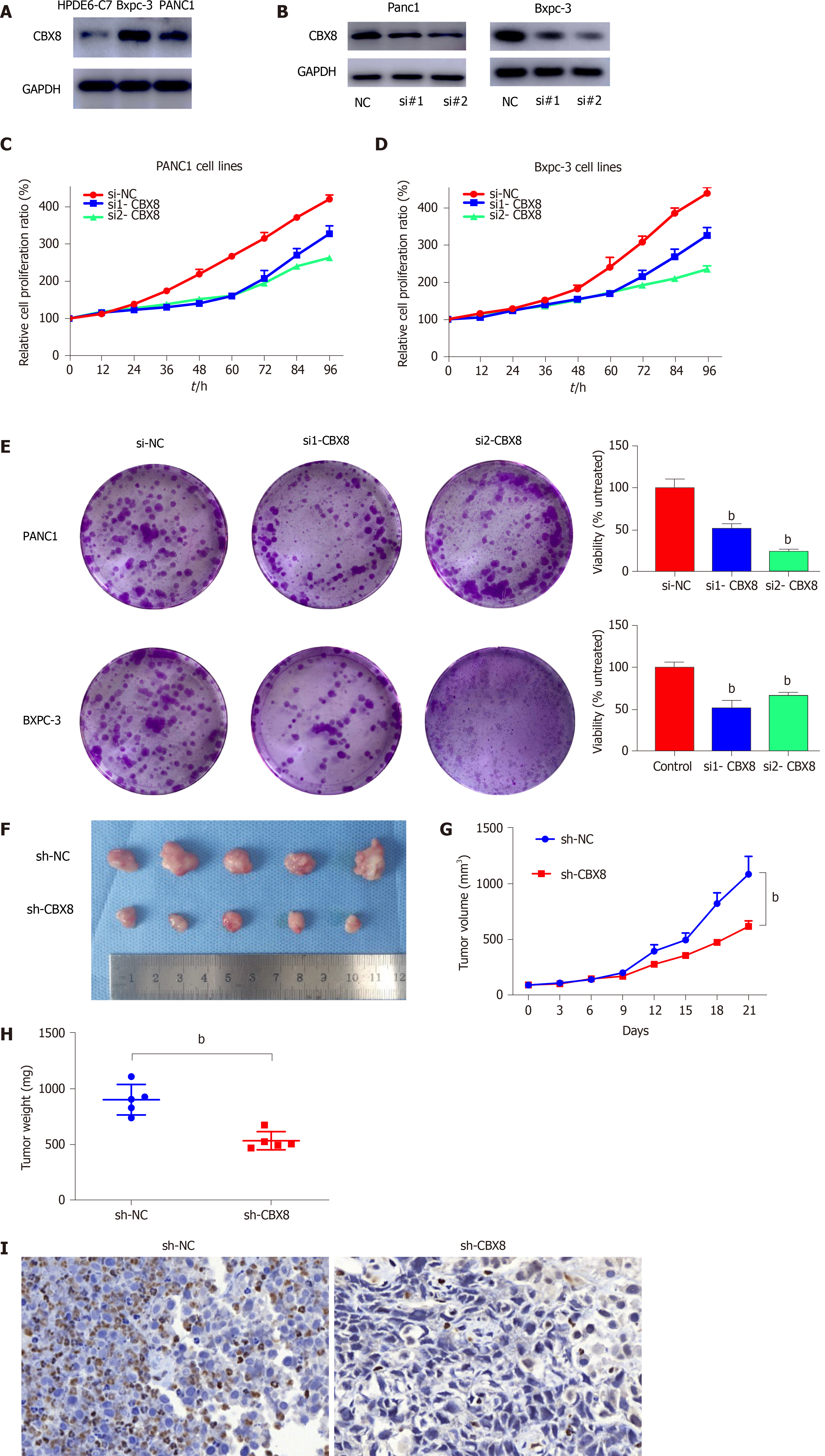
We measured the protein level of CBX8 in PC cell lines and found that CBX8 was upregulated in PANC1 and BXPC-3 cells compared with normal pancreatic epithelial cells (Figure 3A). CCK-8 assay was used to assess the effect of CBX8 on PC cell proliferation, which indicated that CBX8 knockdown (Figure 3B) reduced the proliferation of PC cells (Figure 3C and D). Colony formation assay revealed that CBX8 silencing impeded the proliferation of PC cells (Figure 3E).
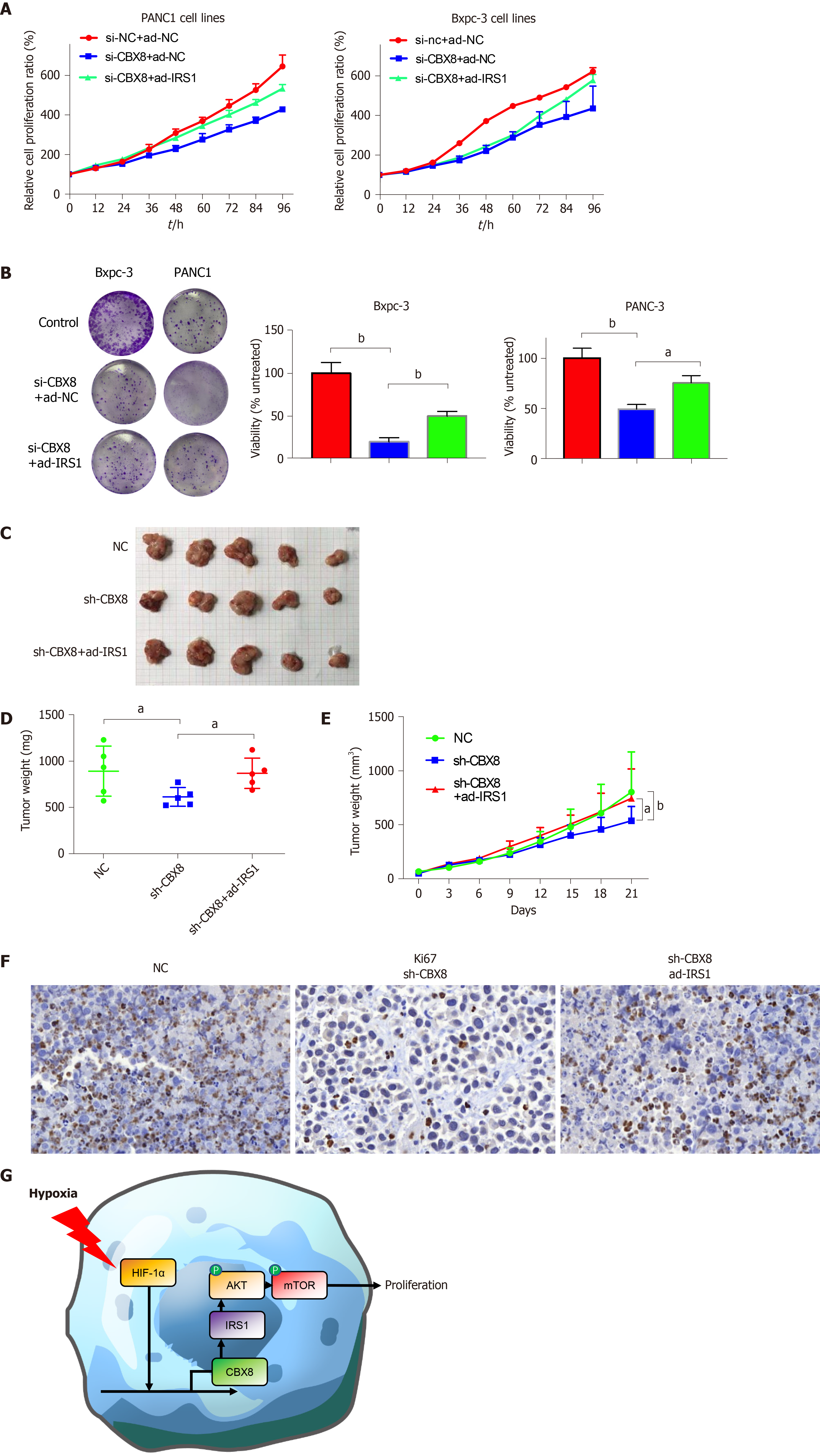
To further explore the role of CBX8 in PC tumorigenesis, BXPC-3 cells were transfected with CBX8 knockdown lentiviral vectors (sh-CBX8) or empty vectors (sh-NC). A PC mouse model was established by subcutaneous injection of two groups of cells into the right flank of nude mice. The tumor size of the sh-CBX8 group was significantly smaller than that in the control group (Figure 3F). The kinetics of tumor growth of each group are shown in Figure 3H. A significant difference was found in the tumor weight between the two groups (Figure 3I).
For further investigation, immunohistochemistry (IHC) for Ki67 was performed and indicated that CBX8 knockdown could restrain the proliferative ability of PC cells (Figure 3J).
Hypoxia is one of the important characteristics of PC. Among these complex mechanisms, HIF-1α is an important molecule for cells to adapt to hypoxia. We sought to investigate the regulatory mechanism of HIF and CBX8.
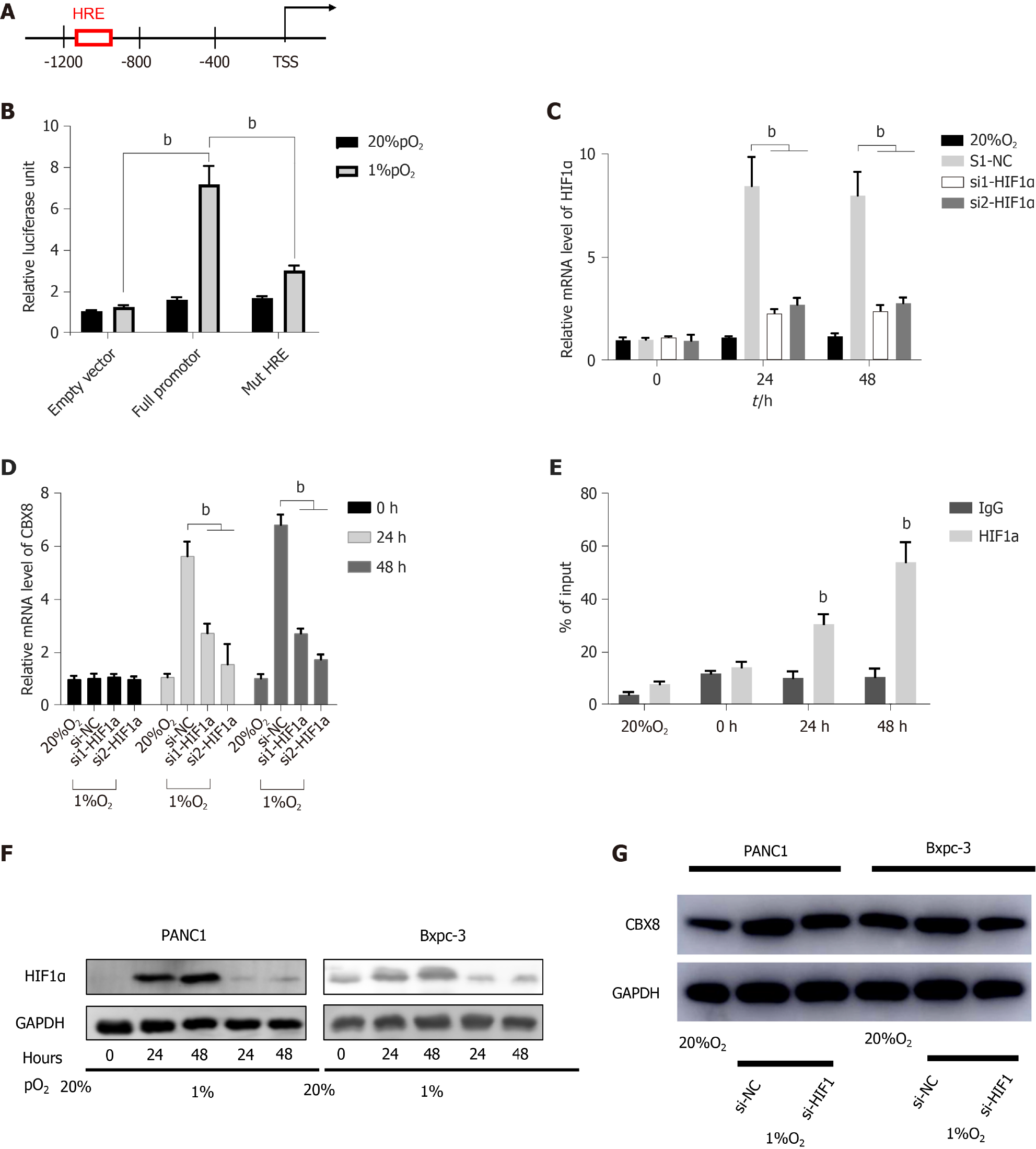
By using active chromatin markers in the University of California Santa Cruz Genome Browser (http://genome.ucsc.edu/), we identified the proximal CBX8 promoter and found the potential hypoxia-responsive elements (HREs) between -1100 and -875 bp before the transcriptional start site (TSS) (Figure 4A). Although HRE sequences are widely distributed in all genes, < 1% show hypoxia-dependent binding of HIFs; therefore, we evaluated the function of these HREs. We transfected the wild-type and mutant CBX8 promoter into 293T cells and exposed them to normoxia or hypoxia. In mutant CBX8 promoter, we deleted the -875 bp site. This demonstrated that the HRE of CBX8 was responsive to hypoxia (Figure 4B). We knocked down HIF-1α (Figure 4C) and detected CBX8 mRNA level under normoxia or hypoxia, which indirectly proved the transcriptional regulation of CBX8 by HIF-1α (Figure 4D). Consistently, ChIP and qPCR demonstrated that HIF-1α bound to CBX8 promoter under hypoxia (Figure 4E). These data revealed that CBX8 was a transcriptional target of HIF-1a under hypoxia. Knockdown of HIF-1α with two different siRNAs prevented accumulation of HIF-1α protein under hypoxia (Figure 4F), and the protein expression of CBX8 under hypoxia was not significantly different from that under normoxia (Figure 4G).
To explore the specific mechanism of the regulation of CBX8 on PC cell proliferation, we analyzed the transcriptome of wild-type and CBX8 knockdown PC cells. The analysis revealed 312 differently expressed genes (DEGs) between the two groups and the heatmap showed the top 30 genes (Figure 5A and C). KEGG analysis showed that the DEGs were significantly enriched in the PI3K/AKT, Rap1, and neurotrophin signaling pathways (Figure 5B and C). To investigate the target genes regulated by CBX8, ChIP-seq was performed and we found 62 135 peaks compared to input signals. The pie chart indicates the CBX8-binding distribution (Figure 5D). Overlapping genes between CBX8 knockdown and the ChIP-seq data were investigated and there were 35 downregulated genes among the CBX8 target genes. IRS1, which regulates PI3K/AKT pathway activation, was in the set of target genes. We performed ATAC-seq and integrated the multiple tracks in the IGV diagram, where CBX8 and H3K27ac were found to co-occupy the IRS1 enhancer region (Figure 5E). Upregulation of CBX8 increased the expression of IRS1 (Figure 5F), whereas CBX8 silencing led to a significant downregulation of IRS1 in PC cells (Figure 5G).
We measured CBX8 and IRS1 expression levels in PC tissues, which showed that the endogenous IRS1 level was positively correlated with CBX8 in PC tissues (Figure 5H). We conducted a luciferase reporter assay in 293T cells. The luciferase activity of IRS1 promoter was enhanced by overexpression of CBX8 but reduced by CBX8 silencing (Figure 5I). Combined ChIP and qPCR analysis revealed that CBX8 bound to regions 2–3 in the IRS1 promoter (Figure 5J), which were included in the -721/235 fragment. The expression of IRS1 in BXPC-3 cells was downregulated after CBX8 knockdown and upregulated after CBX8 overexpression. The phosphorylation levels of AKT and mammalian target of rapamycin (mTOR) in CBX8-overexpressing BXPC-3 cells were increased, while CBX8 knockdown suppressed the phosphorylation of AKT and mTOR (Figure 5K).
After identifying IRS1 as the possible target of CBX8, we explored whether CBX8 promoted the proliferation of PC cells through IRS1. CCK-8 and colony formation assays were performed to assess the effects of CBX8 and IRS1 on PC cell proliferation, which indicated that CBX8 knockdown reduced the proliferation of PC cells, while IRS1 overexpression inhibited the facilitating effect of CBX8 on PC cell proliferation (Figure 6A and B). IRS1 overexpression and CBX8 knockdown plasmids were cotransfected into PC cells. IRS1 overexpression partly rescued the anti-tumorigenicity of CBX8 knockdown in PC cells (Figure 6C-E). IHC for Ki67 indicated that CBX8 knockdown decreased the proliferation of PC cells, but this was reversed partly by IRS1 overexpression (Figure 6F). In conclusion, our results showed that the CBX8/IRS1 axis regulated PC cell proliferation and HIF-1α promoted the expression of CBX8 in PC cells under hypoxia (Figure 6G).
In recent years, CRISPR screening has made great progress in cancer research. Many studies have used CRISPR systems to screen key genes that mediate tumor drug resistance or immune escape[8-10]. In our study, we used the CRISPR screening system to screen genes related to tumor growth. We screened many growth-related genes, among which CBX8 was associated with a poor clinical prognosis of PC, and CBX8 deletion can slow down the proliferation of PC cells. Our data showed that knockdown of CBX8 decreased PC cell proliferation, as demonstrated by weakened colony formation in vitro and in mouse xenografts. The role of CBX8 in PC has not been studied before, and it may become a new therapeutic target.
CBX8 is associated with the stemness and chemosensitivity of many types of cancer. It has been reported that CBX8 upregulates LGR5 expression in a noncanonical manner by interacting with KMT2b and Pol II, leading to increased cancer stemness and decreased chemosensitivity in CC[5]. CBX8 also promotes the proliferation of HCC cells through YBX1-mediated cell cycle progression, and high CBX8 expression is related to a poor prognosis of HCC patients[11]. A recent study showed that CBX8 recruited KMT2b to the LGR5 promoter and modulated H3K27me3 in the promoter of bone morphogenetic protein (BMP)4, resulting in increased BMP4 transcription and subsequent activation of Smads and mitogen-activated protein kinases. In our study, the level of CBX8 in PC tissues was higher than that in adjacent normal tissues. The CRISPR/Cas9 screening also indicated that CBX8 knockout might decrease the proliferation of PC cells.
Hypoxia is a significant feature of PC. Due to the rapid growth of tumor, the blood vessels in PC often show immature microvascular lumina. This results in hypoxic features in PC tissues. In our previous studies, we found that hypoxia mediated the invasion, metastasis, and angiogenesis of PC by promoting the secretion of exosomes[4,12]. In the present study, we found that hypoxia-upregulated HIF-1α bound to CBX8 promoter region and regulated CBX8 expression at the transcriptional level.
The IRS family is composed of four proteins (IRS1–IRS4), which were initially considered as typical cytolytic adaptor proteins. They are involved in IR and insulin-like growth factor I receptor signal transduction[13,14]. Post-translational modification of IRS1 can activate the mTORC1 signaling pathway through chronic elevation of multiple serine phosphorylation sites[15].
In our study, CBX8 modulated H3K27me3 in the promoter of IRS1, resulting in increased IRS1 transcription and subsequent activation of PI3K and AKT. At the same time, in vitro colony formation assay and mouse xenograft model confirmed that CBX8 promoted the growth of PC cells through IRS1.
Through CRISPR screening, we identified a group of genes related to the growth of PC. Combined analysis of clinical samples of patients with PC demonstrated that CBX8 was higher in PC tumor tissue and high expression of CBX8 predicted a poor clinical outcome. HIF-1α regulated expression of CBX8 transcriptionally under hypoxia and CBX8 induced PC cell proliferation by targeting IRS1, which activated the PI3K/AKT pathway. These studies revealed the mechanism of the promotion effect of CBX8 on the development of PC, and provided potential therapeutic targets.
Our results suggest that CBX8 could function as an oncogenic factor in PC progression. High CBX8 expression is correlated with poor clinical outcomes of PDAC patients from two independent cohorts. We also showed that CBX8 is a key gene that is regulated by HIF-1α, and can activate the IRS1/AKT pathway. The above findings suggest that targeting CBX8 may be a promising therapeutic strategy for PC.
Pancreatic cancer (PC) is one of the most lethal cancers worldwide. It has become the second most fatal cancer in the United States. Chromobox (CBX)8 promotes tumor growth and metastasis in other cancers. However, whether CBX8 is involved in the proliferation of PC cells remains unknown.
Many studies have shown that the prognosis of patients with PC remains poor after complete surgical resection. Therefore, it is important to study the occurrence and development of PC and the corresponding targeted therapy. We hope to provide a novel therapeutic target for patients with PC.
The present study aimed to investigate the function of the CBX8/IRS1/AKT axis in PC.
Genome-wide CRISPR-Cas9 screening was performed to select genes that could facilitate PC cell proliferation. A total of 244 candidate genes were identified as being responsible for proliferation of PC cells using deep single guide RNA sequencing. Quantitative reverse transcription-polymerase chain reaction was used to detect the expression of CBX8 in PC tissues and cells. The regulatory roles of CBX8 in cell proliferation, migration, and invasion were verified by CCK-8 and Transwell assays.
CBX8 was upregulated in pancreatic tumor tissues and shown to drive PC cell proliferation. Higher expression of CBX8 was correlated with worse outcomes of PC patients from two independent cohorts with a total of 116 cases. CBX8 also served as a promising therapeutic target for a PC xenograft model. We demonstrated that HIF-1a induced CBX8 transcription by binding to the promoter of CBX8. CBX8 efficiently activated the PI3K/AKT signaling pathway by upregulating insulin receptor substrate (IRS)1.
CBX8 promotes PC cell progression by activating the IRS1/AKT pathway.
CBX8 could promote PC progression, which might provide a potential treatment strategy for this malignancy.
We are grateful to the members of Peng Cao’s laboratory for the critical inputs and suggestions, and Guangzhou Genedenovo Biotechnology Co., Ltd. for assisting in sequencing analysis.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Specialty type: Biochemistry and molecular biology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Casella C S-Editor: Ma YJ L-Editor: Wang TQ P-Editor: Li JH
| 1. | Balachandran VP, Beatty GL, Dougan SK. Broadening the Impact of Immunotherapy to Pancreatic Cancer: Challenges and Opportunities. Gastroenterology. 2019;156:2056-2072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 324] [Article Influence: 54.0] [Reference Citation Analysis (0)] |
| 2. | Choudhry H, Harris AL. Advances in Hypoxia-Inducible Factor Biology. Cell Metab. 2018;27:281-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 536] [Cited by in RCA: 596] [Article Influence: 85.1] [Reference Citation Analysis (0)] |
| 3. | Wang X, Luo G, Zhang K, Cao J, Huang C, Jiang T, Liu B, Su L, Qiu Z. Hypoxic Tumor-Derived Exosomal miR-301a Mediates M2 Macrophage Polarization via PTEN/PI3Kγ to Promote Pancreatic Cancer Metastasis. Cancer Res. 2018;78:4586-4598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 420] [Cited by in RCA: 532] [Article Influence: 76.0] [Reference Citation Analysis (0)] |
| 4. | Guo Z, Wang X, Yang Y, Chen W, Zhang K, Teng B, Huang C, Zhao Q, Qiu Z. Hypoxic Tumor-Derived Exosomal Long Noncoding RNA UCA1 Promotes Angiogenesis via miR-96-5p/AMOTL2 in Pancreatic Cancer. Mol Ther Nucleic Acids. 2020;22:179-195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 136] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 5. | Zhang Y, Kang M, Zhang B, Meng F, Song J, Kaneko H, Shimamoto F, Tang B. m6A modification-mediated CBX8 induction regulates stemness and chemosensitivity of colon cancer via upregulation of LGR5. Mol Cancer. 2019;18:185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 103] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 6. | Zhang CZ, Chen SL, Wang CH, He YF, Yang X, Xie D, Yun JP. CBX8 Exhibits Oncogenic Activity via AKT/β-Catenin Activation in Hepatocellular Carcinoma. Cancer Res. 2018;78:51-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 62] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 7. | Chung CY, Sun Z, Mullokandov G, Bosch A, Qadeer ZA, Cihan E, Rapp Z, Parsons R, Aguirre-Ghiso JA, Farias EF, Brown BD, Gaspar-Maia A, Bernstein E. Cbx8 Acts Non-canonically with Wdr5 to Promote Mammary Tumorigenesis. Cell Rep. 2016;16:472-486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 80] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 8. | Huang K, Liu X, Li Y, Wang Q, Zhou J, Wang Y, Dong F, Yang C, Sun Z, Fang C, Liu C, Tan Y, Wu X, Jiang T, Kang C. Genome-Wide CRISPR-Cas9 Screening Identifies NF-κB/E2F6 Responsible for EGFRvIII-Associated Temozolomide Resistance in Glioblastoma. Adv Sci (Weinh). 2019;6:1900782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 9. | Li Z, Zhou X, Wei M, Gao X, Zhao L, Shi R, Sun W, Duan Y, Yang G, Yuan L. In Vitro and in Vivo RNA Inhibition by CD9-HuR Functionalized Exosomes Encapsulated with miRNA or CRISPR/dCas9. Nano Lett. 2019;19:19-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 209] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 10. | Dufva O, Koski J, Maliniemi P, Ianevski A, Klievink J, Leitner J, Pölönen P, Hohtari H, Saeed K, Hannunen T, Ellonen P, Steinberger P, Kankainen M, Aittokallio T, Keränen MAI, Korhonen M, Mustjoki S. Integrated drug profiling and CRISPR screening identify essential pathways for CAR T-cell cytotoxicity. Blood. 2020;135:597-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 154] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 11. | Xiao L, Zhou Z, Li W, Peng J, Sun Q, Zhu H, Song Y, Hou JL, Sun J, Cao HC, Zhongyi D, Wu D, Liu L. Chromobox homolog 8 (CBX8) Interacts with Y-Box binding protein 1 (YBX1) to promote cellular proliferation in hepatocellular carcinoma cells. Aging (Albany NY). 2019;11:7123-7149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 12. | Keshavarzi F, Golsheh S. IRS1- rs10498210 G/A and CCR5-59029 A/G polymorphisms in patients with type 2 diabetes in Kurdistan. Mol Genet Genomic Med. 2019;7:e631. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 13. | Esposito DL, Aru F, Lattanzio R, Morgano A, Abbondanza M, Malekzadeh R, Bishehsari F, Valanzano R, Russo A, Piantelli M, Moschetta A, Lotti LV, Mariani-Costantini R. The insulin receptor substrate 1 (IRS1) in intestinal epithelial differentiation and in colorectal cancer. PLoS One. 2012;7:e36190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 14. | Ardestani A, Maedler K. mTORC1 and IRS1: Another Deadly Kiss. Trends Endocrinol Metab. 2018;29:737-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |