Copyright
©The Author(s) 2025.
World J Gastrointest Oncol. Aug 15, 2025; 17(8): 105321
Published online Aug 15, 2025. doi: 10.4251/wjgo.v17.i8.105321
Published online Aug 15, 2025. doi: 10.4251/wjgo.v17.i8.105321
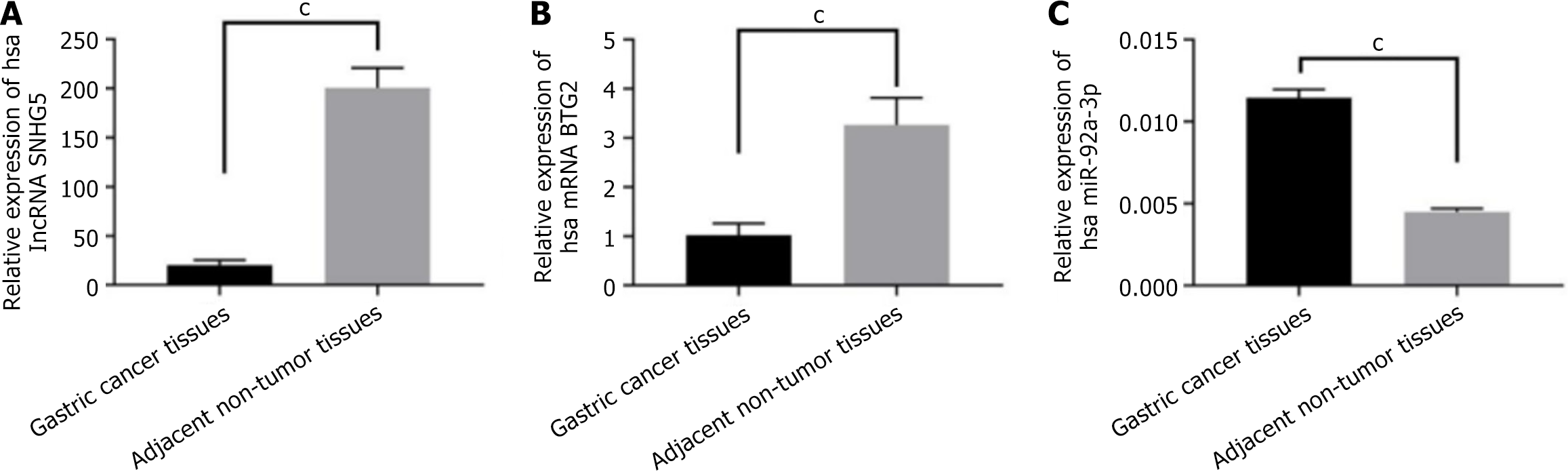
Figure 1 Expression of long non-coding RNA small nucleolar RNA host gene 5, miR-92a-3p, and B-cell translocation gene 2 in gastric cancer.
Quantitative reverse transcription PCR was performed to detect the mRNA level of long non-coding RNA small nucleolar RNA host gene 5, BTG2, and miR-92a-3p in gastric cancer tissues and adjacent non-tumor tissues. A: Long non-coding RNA small nucleolar RNA host gene 5; B: B-cell translocation gene 2; C: MiR-92a-3p. cP < 0.001. lncRNA: Long non-coding RNA; SNHG5: Small nucleolar RNA host gene 5; BTG2: B-cell translocation gene 2; hsa: Human; miR: MicroRNA.
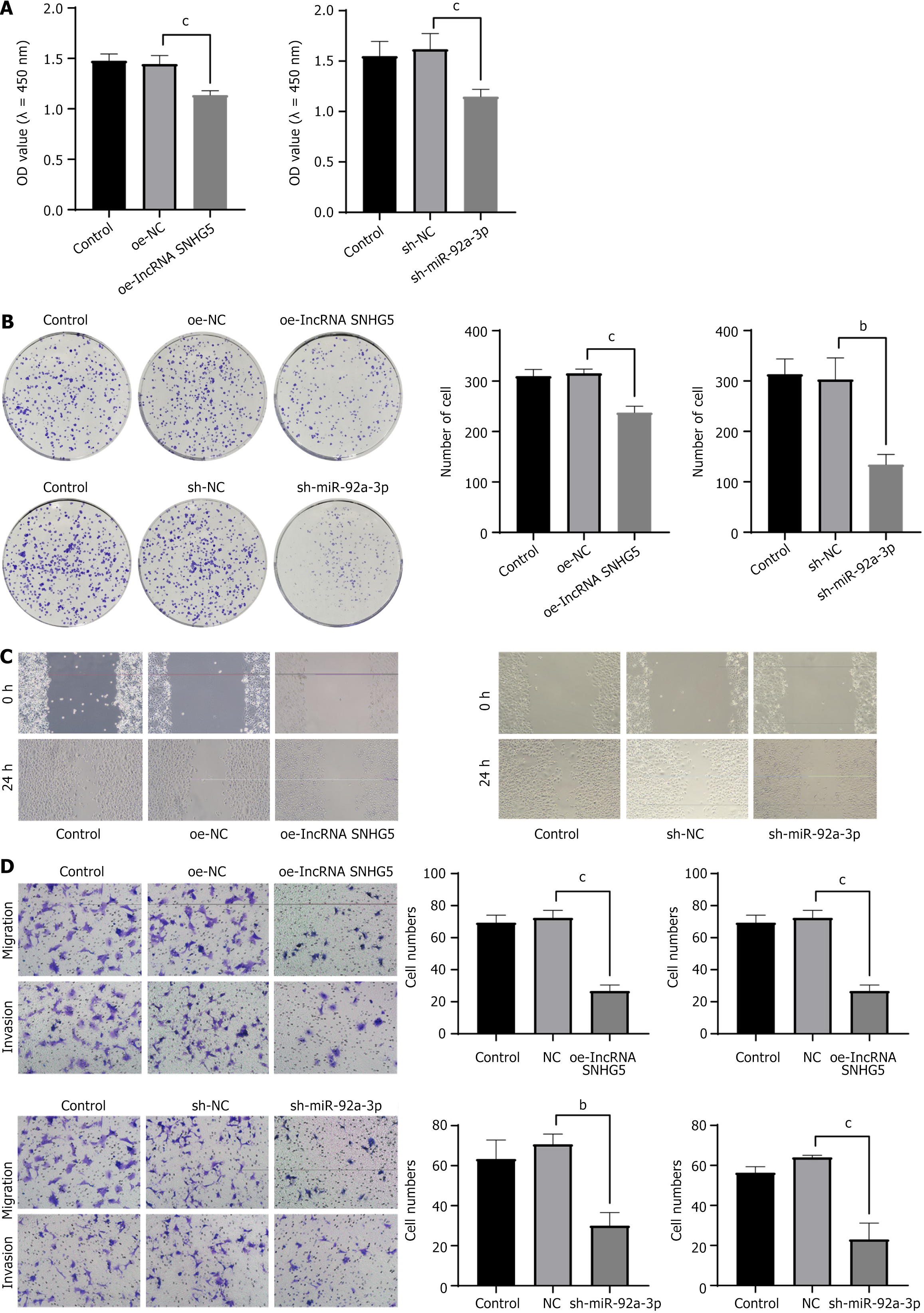
Figure 2 Long non-coding RNA small nucleolar RNA host gene 5 overexpression and miR-92a-3p knockdown inhibited the proliferation and migration of gastric cancer cells.
A: CCK-8 assay was used to detect the cell activity after long non-coding RNA (lncRNA) small nucleolar RNA host gene 5 (SNHG5) overexpression and miR-92a-3p knockdown; B: The colony formation assay was performed to detect the clonogenic ability of gastric cancer (GC) cells after lncRNA SNHG5 overexpression and miR-92a-3p knockdown; C: Scratch assay was used to detect the wound healing ability of GC cells after lncRNA SNHG5 overexpression and miR-92a-3p knockdown; D: The Transwell assay detected the migration and invasion ability of GC cells after lncRNA SNHG5 overexpression and miR-92a-3p knockdown (× 200 magnification). Scale bar: 100 μm. bP < 0.01; cP < 0.001; lncRNA: Long non-coding RNA; SNHG5: Small nucleolar RNA host gene 5; NC: Negative control; OE-lncRNA SNHG5: Overexpression of long non-coding RNA small nucleolar RNA host gene 5; sh: Short hairpin; miR: MicroRNA.
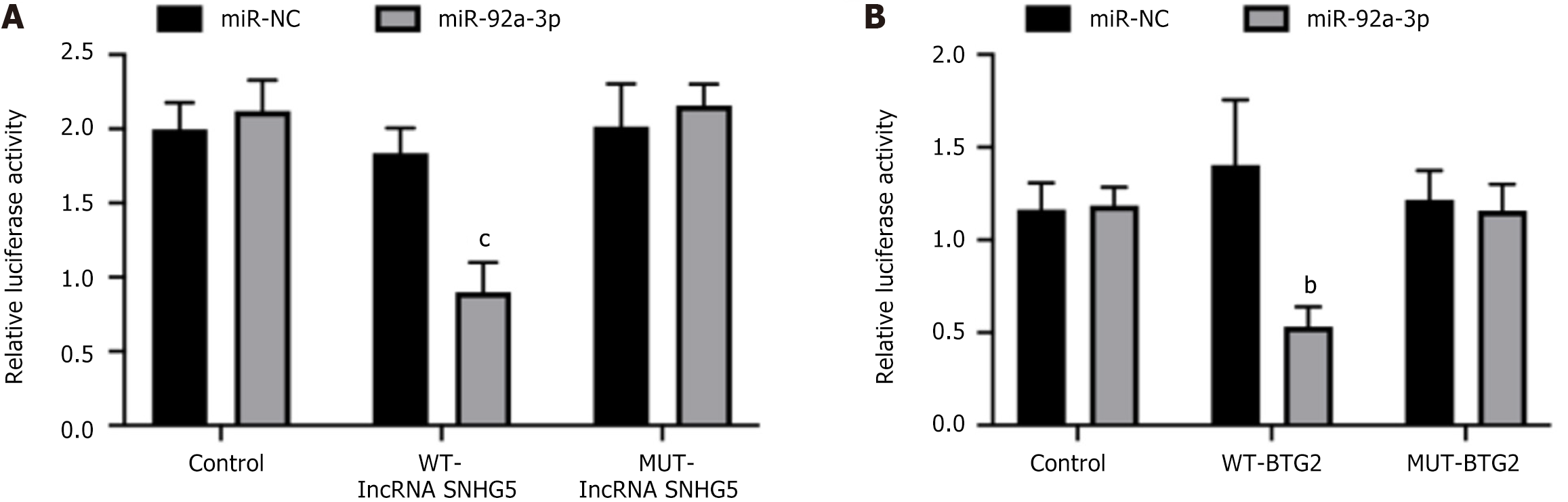
Figure 3 Luciferase report showed the relationship between small nucleolar RNA host gene 5/miR-92a-3p and miR-92a-3p/B-cell translocation gene 2.
A: Small nucleolar RNA host gene 5/miR-92a-3p; B: MiR-92a-3p/B-cell translocation gene 2. bP < 0.01; cP < 0.001; NC: Negative control; WT-lncRNA SNHG5: Wild-type long non-coding RNA small nucleolar RNA host gene 5 vector; MUT-lncRNA SNHG5: Mutant long non-coding RNA small nucleolar RNA host gene 5 vector; WT-BTG2: Wild-type B-cell translocation gene 2 vector; MUT-BTG2: Mutant B-cell translocation gene 2 vector; miR: MicroRNA.
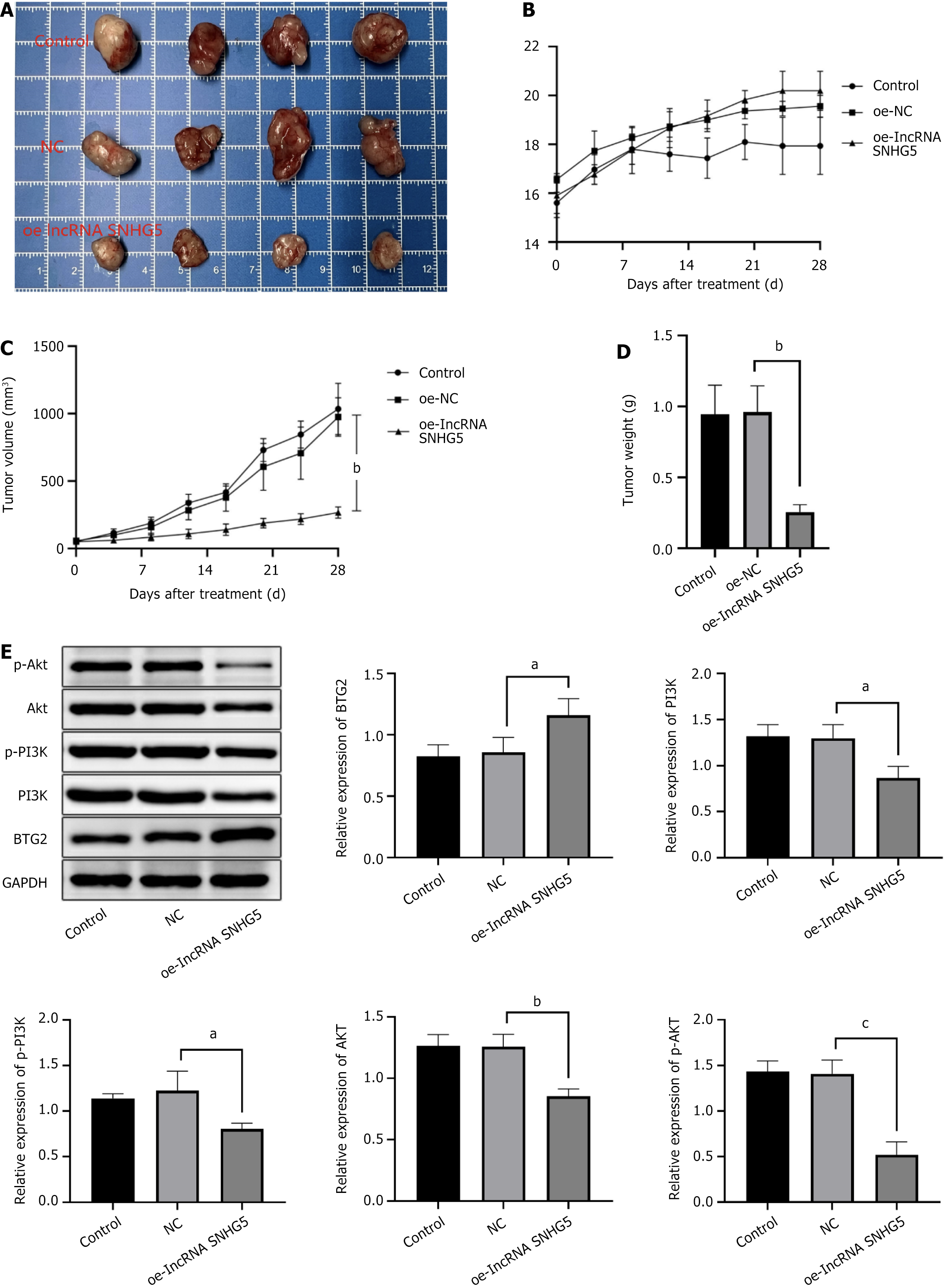
Figure 4 Long non-coding RNA small nucleolar RNA host gene 5 overexpression improved the severity of gastric cancer in vivo.
A: Tumor photographs; B: Body weight change (g); C: Tumor volume (mm3); D: Tumor weight (g); E: Western blot analysis was performed to determine the expression of B-cell translocation gene 2, PI3K, p-PI3K, AKT, and p-AKT. With the ratio of B-cell translocation gene 2/GAPDH, PI3K/GAPDH, p-PI3K/GAPDH, AKT/GAPDH, and p-AKT/GAPDH, the control was defined as 1. aP < 0.05; bP < 0.01; cP < 0.001; NC: Negative control; OE-lncRNA SNHG5: Overexpression of long non-coding RNA small nucleolar RNA host gene 5; BTG2: B-cell translocation gene 2.
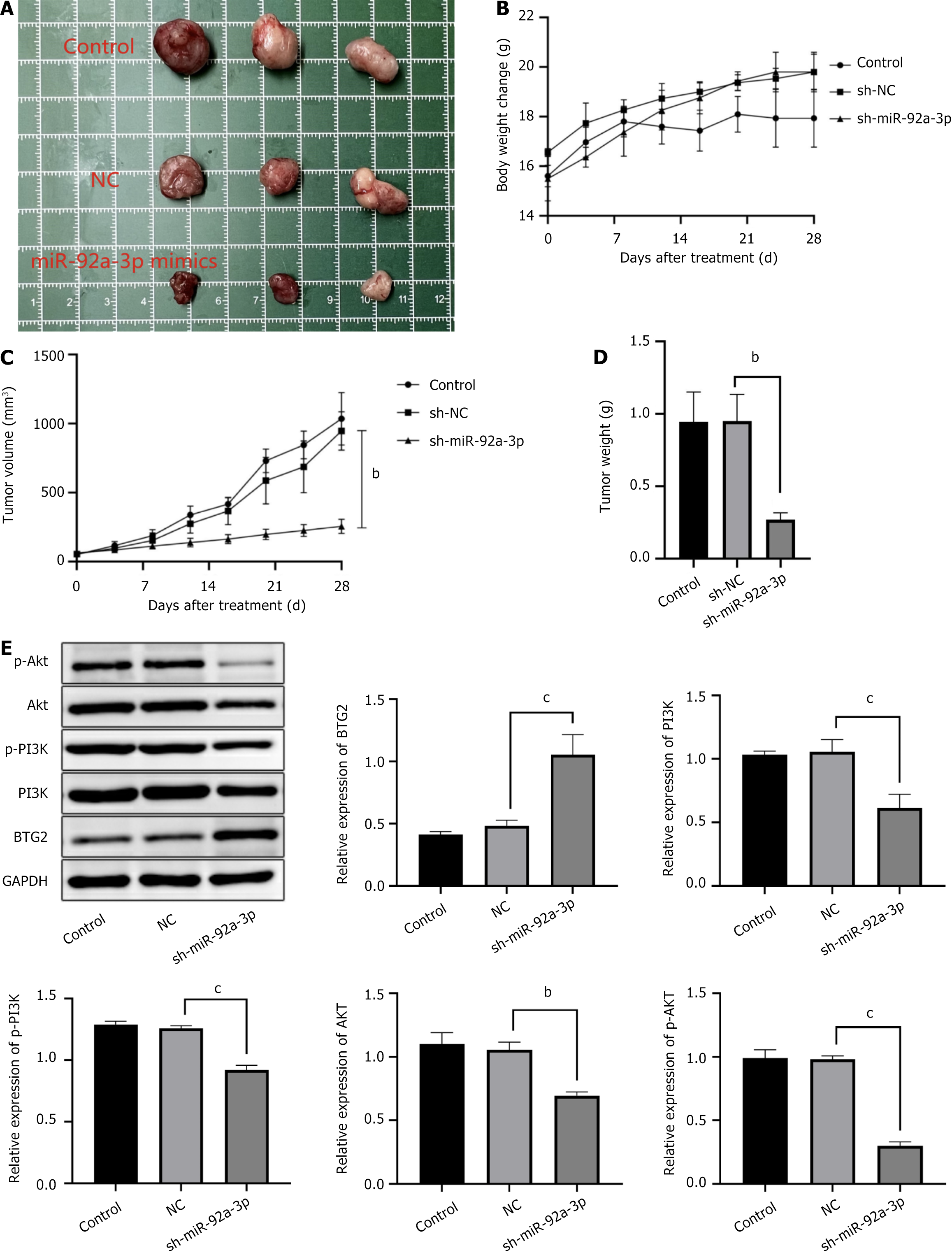
Figure 5 miR-92a-3p knockdown improved the severity of gastric cancer in vivo.
A: Tumor photographs; B: Body weight change (g); C: Tumor volume (mm3); D: Tumor weight (g); E: Western blot analysis was performed to determine the expression of B-cell translocation gene 2, PI3K, p-PI3K, AKT, and p-AKT. With the ratio of B-cell translocation gene 2/GAPDH, PI3K/GAPDH, p-PI3K/GAPDH, AKT/GAPDH, and p-AKT/GAPDH, the control was defined as 1. bP < 0.01; cP < 0.001; NC: Negative control; BTG2: B-cell translocation gene 2; sh: Short hairpin; miR: MicroRNA.
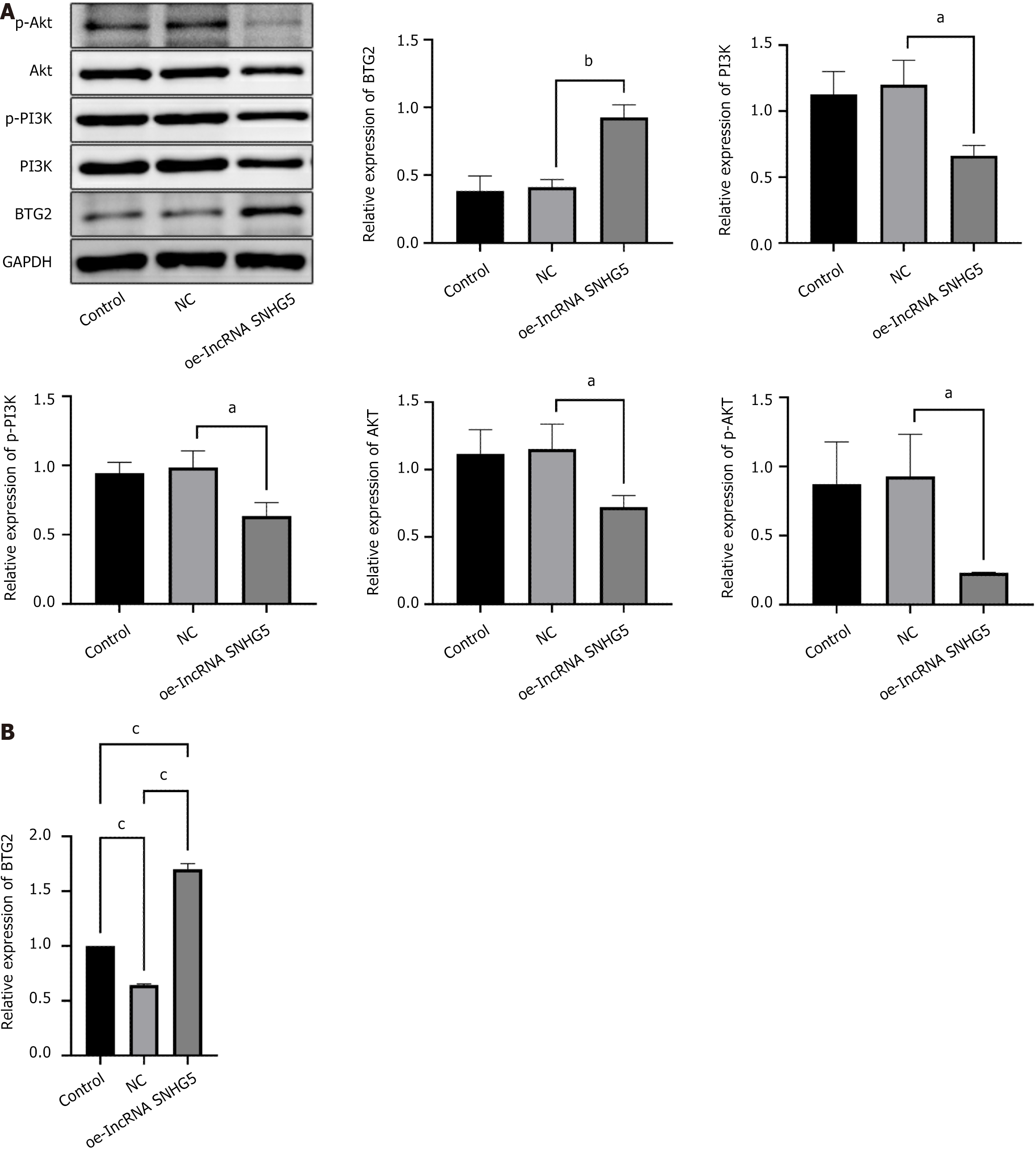
Figure 6 Long non-coding RNA small nucleolar RNA host gene 5 overexpression was related to miR-92a-3p, B-cell translocation gene 2, and the PI3K/AKT signaling pathway.
A: Western blot analysis was performed to determine the expression of B-cell translocation gene 2 (BTG2), PI3K, p-PI3K, AKT, and p-AKT. With the ratio of BTG2/GAPDH, PI3K/GAPDH, p-PI3K/GAPDH, AKT/GAPDH, and p-AKT/GAPDH, the control was defined as 1; B: Quantitative reverse transcription PCR was applied to detect BTG2. aP < 0.05; bP < 0.01; cP < 0.001; NC: Negative control; BTG2: B-cell translocation gene 2; OE-lncRNA SNHG5: Overexpression of long non-coding RNA small nucleolar RNA host gene 5.
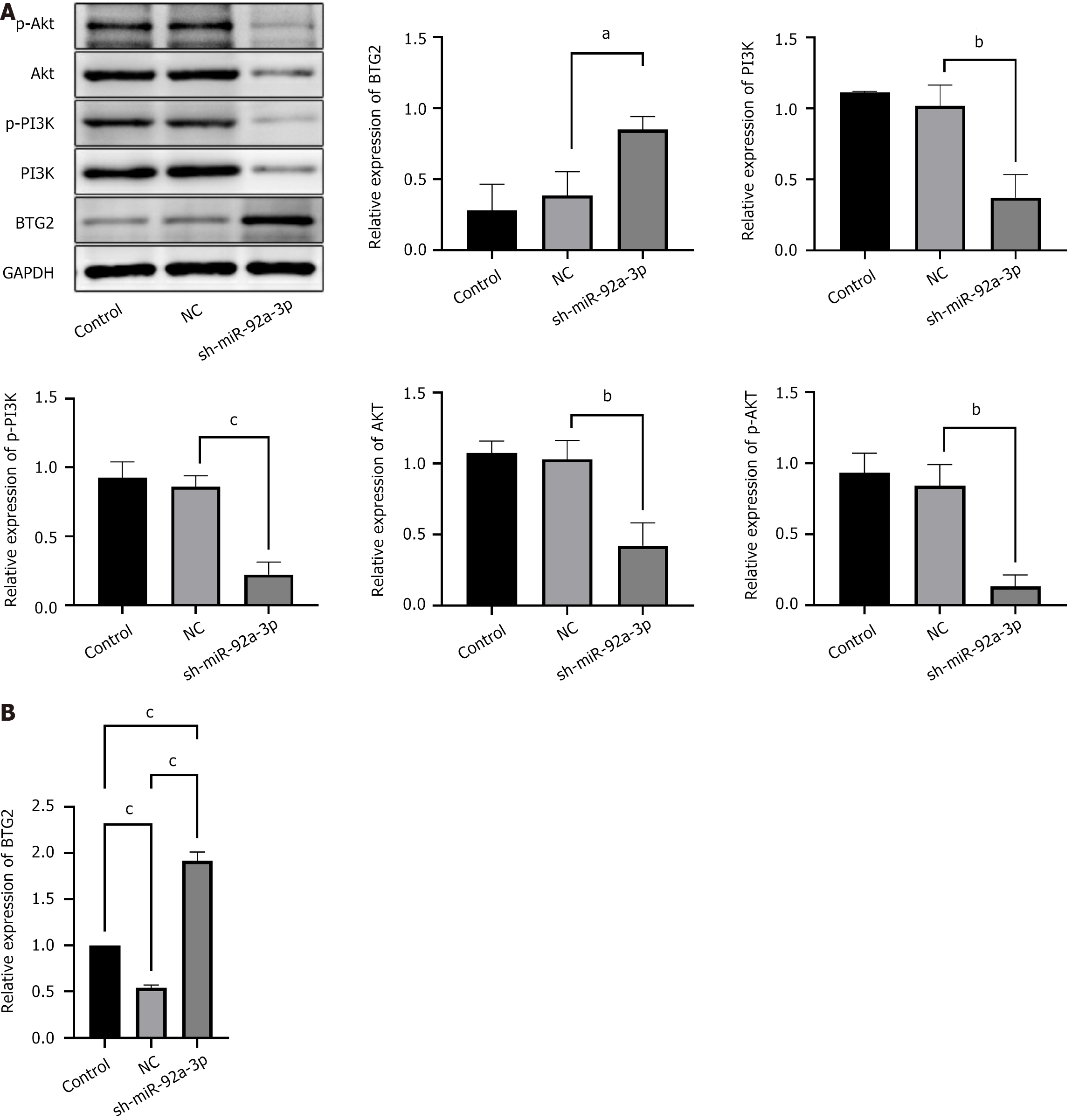
Figure 7 Effects of miR-92a-3p knockdown on B-cell translocation gene 2 and the PI3K/AKT signaling pathway.
A: Western blot analysis was performed to determine the expression of B-cell translocation gene 2 (BTG2), PI3K, p-PI3K, AKT, and p-AKT. With the ratio of BTG2/GAPDH, PI3K/GAPDH, p-PI3K/GAPDH, AKT/GAPDH, and p-AKT/GAPDH, the control was defined as 1; B: Quantitative reverse transcription PCR was applied to detect BTG2. aP < 0.05; bP < 0.01; cP < 0.001; NC: Negative control; BTG2: B-cell translocation gene 2; sh: Short hairpin; miR: MicroRNA.
- Citation: Mao QQ, Zhang ML, Zhong L, Xu XD, Wang XH, Pan DY, Zhou FS, Huang JX, Zhao XG, Chen JJ, Jiang XY, Sun X, Ding WQ. LncRNA SNHG5 modulates cell proliferation and migration through the miR-92a-3p/BTG2 axis in gastric cancer by the PI3K/AKT pathway. World J Gastrointest Oncol 2025; 17(8): 105321
- URL: https://www.wjgnet.com/1948-5204/full/v17/i8/105321.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i8.105321