Published online Feb 14, 2020. doi: 10.3748/wjg.v26.i6.627
Peer-review started: October 10, 2019
First decision: November 27, 2019
Revised: December 3, 2019
Accepted: December 22, 2019
Article in press: December 22, 2019
Published online: February 14, 2020
Processing time: 127 Days and 10.2 Hours
Colorectal cancer (CRC) is one of the most common malignancies worldwide.
To explore the expression of microRNA miR-19a-3p and Forkhead box F2 (FOXF2) in patients with CRC and the relevant mechanisms.
Sixty-two CRC patients admitted to the hospital were enrolled into the study group, and sixty healthy people from the same period were assigned to the control group. Elbow venous blood was sampled from the patients and healthy individuals, and blood serum was saved for later analysis. MiR-19a-3p mimics, miR-19a-3p inhibitor, miR-negative control, small interfering-FOXF2, and short hairpin-FOXF2 were transfected into HT29 and HCT116 cells. Then quantitative polymerase chain reaction was performed to quantify the expression of miR-19a-3p and FOXF2 in HT29 and HCT116 cells, and western blot (WB) analysis was conducted to evaluate the levels of FOXF2, glycogen synthase kinase 3 beta (GSK-3β), phosphorylated GSK-3β (p-GSK-3β), β-catenin, p-β-catenin, α-catenin, N-cadherin, E-cadherin, and vimentin. The MTT, Transwell, and wound healing assays were applied to analyze cell proliferation, invasion, and migration, respectively, and the dual luciferase reporter assay was used to determine the correlation of miR-19a-3p with FOXF2.
The patients showed high serum levels of miR-19a-3p and low levels of FOXF2, and the area under the curves of miR-19a-3p and FOXF2 were larger than 0.8. MiR-19a-3p and FOXF2 were related to sex, tumor size, age, tumor-node-metastasis staging, lymph node metastasis, and differentiation of CRC patients. Silencing of miR-19a-3p and overexpression of FOXF2 suppressed the epithelial-mesenchymal transition, invasion, migration, and proliferation of cells. WB analysis revealed that silencing of miR-19a-3p and FOXF2 overexpression significantly suppressed the expression of p-GSK-3β, β-catenin, N-cadherin, and vimentin; and increased the levels of GSK-3β, p-β-catenin, α-catenin, and E-cadherin. The dual luciferase reporter assay confirmed that there was a targeted correlation of miR-19a-3p with FOXF2. In addition, a rescue experiment revealed that there were no differences in cell proliferation, invasion, and migration in HT29 and HCT116 cells co-transfected with miR-19a-3p-mimics+sh-FOXF2 and miR-19a-3p-inhibitor+si-FOXF2 compared to the miR-negative control group.
Inhibiting miR-19a-3p expression can upregulate the FOXF2-mediated Wnt/β-catenin signaling pathway, thereby affecting the epithelial-mesenchymal transition, proliferation, invasion, and migration of cells. Thus, miR-19a-3p is likely to be a therapeutic target in CRC.
Core tip: Colorectal cancer (CRC) has a high rate of mortality, and patients with this disease often miss the optimal treatment period due to the lack of clinical symptoms of early CRC, which affects their prognosis. At present, CRC is extremely difficult to prevent and treat. Therefore, this study investigated the changes in biological functions of CRC cells from the perspective of the CRC mechanism, with the goal of evaluating the effects of microRNA miR-19a-3p-mediated regulation of the Forkhead box F2-mediated Wnt/β-catenin signaling pathway on the biological functions of CRC cells.
- Citation: Yu FB, Sheng J, Yu JM, Liu JH, Qin XX, Mou B. MiR-19a-3p regulates the Forkhead box F2-mediated Wnt/β-catenin signaling pathway and affects the biological functions of colorectal cancer cells. World J Gastroenterol 2020; 26(6): 627-644
- URL: https://www.wjgnet.com/1007-9327/full/v26/i6/627.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i6.627
Colorectal cancer (CRC) is a malignant tumor of the digestive system, also known as the gastrointestinal tract, with an incidence second only to gastric cancer. With the development of the social economy, an increasing number of young people suffer from CRC due to unhealthy living and eating habits[1,2]. Early detection, diagnosis, and radical therapy are pivotal to the treatment of CRC, but most patients already have advanced-stage disease at the time of diagnosis due to the insidious clinical characteristics in the early stage; thus, CRC patients suffer a poor prognosis and extremely high mortality[3,4]. At present, CRC is mainly treated with surgical resection, radiotherapy, or chemoradiotherapy in clinical practice[5,6]. In recent years, based on in-depth studies on the mechanism of CRC, it has been found that the regulation of tumor-suppressor or tumor-promoting genes can affect the occurrence and development of tumors, thereby offering novel insights into CRC therapy and prognosis[7]. Studies on tumors have concluded that the invasion and metastasis of cancer cells are strongly linked to activation of the epithelial-mesenchymal transition (EMT) pathway as follows. With the EMT, tumor epithelial cells have enhanced invasion and migration ability, transform into mesenchymal-like cells, and then fall off from the primary foci to form new metastatic foci. The EMT of tumors involves a series of complex molecular events and regulatory pathways[8]. MicroRNA (miRNAs), as endogenous non-coding single-stranded RNAs with cancer-promoting or cancer-inhibiting functions, may participate in the EMT of tumors through a variety of signaling pathways[9]. Studies have shown that miR-19a-3p is an miRNA with abnormal expression in a variety of tumor cells[10]. For example, a study revealed that highly expressed miR-19a-3p could regulate cell adhesion molecule 2 and affect the proliferation and migration mechanism of renal carcinoma cells through the AKT signaling pathway[11]. In addition, a study found that miR-19a-3p reversed the tumor-associated macrophage phenotype by regulating Fos-related antigen 1/IL-6/signal transducer and activator of transcription 3 signaling pathways, thus inhibiting breast cancer metastasis[12]. Although these studies have demonstrated that miR-19a-3p is extensively involved in cancer such as renal cell carcinoma and breast cancer, the specific mechanism underlying its role in CRC is not completely understood[13,14]. Forkhead box F2 (FOXF2) is a homeobox protein and an inducer of the EMT. A previous study showed that FOXF2 can promote the EMT of CRC cells by mediating the Wnt/β-catenin signaling pathway[15]. Wnt/β-catenin is a classical signaling pathway in the pathological process of cancer, which is overexpressed in tumor cells and has a very close relationship with their proliferation and apoptosis[16].
We predicted that FOXF2 is probably the target site of miR-19a-3p using online biological prediction software (http://www.targetscan.org/vert_72/). Based on the abovementioned studies, the goal of this study was to determine if miR-19a-3p influences the biological functions of CRC cells by targeting the FOXF2-mediated Wnt/β-catenin signaling pathway.
Sixty-two CRC patients in the Fourth Affiliated Hospital of Kunming Medical University between March 2017 and July 2018 were enrolled as the study group, and sixty healthy people from the same period were enrolled as the control group. There were 32 males and 30 females (57.8 ± 3.80 years-old) in the study group, and 34 males and 26 females (57.5 ± 3.50 years-old) in the control group. Though comparable, there were no significant differences regarding sex and age between groups. The inclusion criteria of the study were: CRC patients with confirmed disease based on pathological diagnosis[17], and patients with expected survival longer than 3 mo. The exclusion criteria were: Patients who received preoperative chemoradiotherapy and immunotherapy. All patients and their families signed an informed consent form after understanding this study, and the study was conducted with permission from the Ethics Committee of the Fourth Affiliated Hospital of Kunming Medical University.
The human normal colonic epithelial cell line and human CRC cell lines (HT29, SW480, SW620, and HCT116) (Nos. BNCC338003, BNCC337731, BNCC100604, BNCC337664, and BNCC337692; Beijing BeNa Culture Collection, Beijing); TransScript/TransScript IIGreen miRNA Two-Step qRT Polymerase Chain Reaction (PCR) SuperMix (AQ202-01/AQ301-01; Beijing TransGen Biotech, Beijing, China); MTT Kit (SY0502; Beijing Biolab Technology Co., Ltd., Beijing, China); Trizol reagent (10296010; Invitrogen Company, Carlsbad, CA, United States); Dual Luciferase Reporter Gene Determination Kit (KFS303; Beijing Biolab Technology); fetal bovine serum (FBS), Transwell Kit, and phosphate-buffered saline (PBS) (1142802, 10010049, 10437028; Gibco Company, Gaithersburg, MD, United States), radio-immunoprecipitation assay (JN0190; Beijing Biolab Technology); Bicinchoninic Acid Protein Assay Kit (A53225; Thermo Fisher Scientific, Waltham, MA, United States); FOXF2, glycogen synthase kinase 3 beta (GSK-3β), phosphorylated GSK-3β (p-GSK-3β), β-catenin, p-β-catenin, α-catenin, N-cadherin, E-cadherin, vimentin, and β-actin antibodies (Cell Signaling Technology, Danvers, MA, United States); goat anti-rabbit IgG secondary antibody (Wuhan BOSTER Biological Technology Co., Ltd., Wuhan, China); electrochemiluminescence developer (Thermo Fisher Scientific), and PCR instrument (7500; Applied Biosystems, Waltham, MA, United States). The design and synthesis of all primers were carried out by Shanghai Sangon Biotech Co., Ltd. (Shanghai, China).
CRC cell lines were incubated in Dulbecco's Modified Eagle’s Medium containing 10% PBS in an incubator with CO2 at 37 °C for transfection. When cell growth and fusion under transfection reached 50%, 25% pancreatin was added to the cell lines for digestion, and after digestion, the cell lines were cultured in medium again to complete passage. Cells in the logarithmic phase were grouped and then transfected with an inhibition sequence (miR-19a-3p-inhibitor), overexpression sequence (miR-19a-3p-mimics), miR negative control (miR-NC), targeted inhibited FOXF2 RNA (si-FOXF2), targeted overexpressed FOXF2 RNA (sh-FOXF2), and NC RNA with the Lipofectamine™ 2000 kit, in accordance with the instructions of the corresponding kit.
Quantitative PCR assay: The quantitative PCR (qPCR) assay was employed to determine the level of mRNA in serum and cells. Total serum RNA was extracted from the patients as per the operating instructions of Trizol reagent and dissolved in 20 μL diethylpyrocarbonate water, followed by reverse transcription using a reverse transcription kit in 15 μL total reaction volume containing 1 μL M-MLV, 1 μL Oligo (d T), 0.5 μL RNA enzyme inhibitor, 1 μL NTP, and RNAse-free water to adjust the volume. The RNA was incubated at 38 °C for 60 min. cDNA (1 μL) was sampled and synthesized at 85 °C for 5 s. The synthesized cDNA was adopted as a qPCR amplification template. The PCR reaction system was prepared to a 25 μL total reaction volume with 2.5 μL of 10 × PCR buffer, 1 μL dNTPs, 1 μL upstream and downstream primers, 0.25 μL Taq DNA Polymerase, and dd H2O to adjust the volume. The reaction conditions were as follows: 95 °C for 15 min, 95 °C for 15 s, 58 °C for 30 min (a total of 35 cycles), and then 72 °C for 15 min. Three replicate wells were made for each sample for three repeated experiments. U6 was applied as an internal reference for miR-19a-3p, and β-actin as an internal reference for FOXF2. After reaction, the amplification and dissociation curves of the qPCR were confirmed, and the relative quantity of target gene was calculated based on obtained parameters. The relative quantification of target genes was analyzed using 2-ΔCt.
Western blot assay: Cultured cells were collected from each group, and the total protein was extracted using the radioimmunoprecipitation assay lysis method. The bicinchoninic acid method was adopted to determine the level of total protein, and was adjusted to 4 μg/μL. The total protein was transferred to a PVDF membrane through 6%-12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Corresponding protein band was selected according to the target protein, and the membrane was blocked in 5% skim milk for 2 h, followed by the addition of primary antibody (Beyotime Biology Co., Ltd., Jiangsu, China) at a dilution of 1:1000. Then the membrane was washed and stored at 4 °C for one night. The next day, the membrane was incubated with secondary antibody (Beyotime Biology Co., Ltd.) for 1 h with the same procedures, after which developer was added in a dark room. The PVDF membrane was imaged by a Tocan240 motored molecular imaging system (Shanghai Tocan Bio-technology Co., Ltd., Shanghai, China), and the imaging results were analyzed using Image LabTM software.
MTT assay: Cells transfected for 24 h were collected, and seeded into a 96-well plate at 5 × 103 cells/well, followed by the addition of 20 μL of MTT solution (5 μg/mL) at 24, 48, and 72 h after being incubated at 37 °C. Then the cells were cultured at 37 °C for 4 h. A total of 200 μL dimethyl sulfoxide was added to each well, and then the optical density of each group of cells under 570 nm was measured using a spectrophotometer.
Transwell assay: Cells transfected for 24 h were collected and seeded into a 24-well plate at 3 × 104 cells/well. The cells were digested with pancreatin, and transferred to the upper compartment. The upper compartment was added with 200 μL of RPMI 1640 culture solution, and the lower compartment was added with 500 mL of RPMI 1640 containing 10% FBS. The plate was cultured at 37 °C for 48 h, and the substrate and cells not passing through the microporous membrane in the upper chamber were removed. The plate was washed three times with PBS, followed by immobilization using paraformaldehyde for 10 min, and then washed three times with double distilled water. Subsequently, 0.5% crystal violet was employed to stain the dry plate, and cells invasion was evaluated using a microscope.
The cells were seeded into a 6-well plate after being diluted to 3 × 105 cells/mL. When the confluency of the cells was up to 85%, a 200 μL sterile loading gun was used to divide the cells to form a cell-free area in the center of the culture plate. The divided cells were cultured in new culture medium after being washed with PBS. At 0 h (W0) and 24 h (W24) after cell division, the cell migration ability was evaluated according to wounds at three different sites using a microscope.
Stable miR-19a-3p-inhibitor and 3 × 106 HCT116 cells transfected with control plasmid were subcutaneously injected into the left abdomen of female BALB/c nude mice (4 wk old) raised in a sterile environment (5 nude mice in each group). The tumor growth of each mouse was evaluated every 7 d, and at 28 d after injection, the mice were executed by cervical dislocation, and their tumor size and mass in vivo were measured.
The collected data were analyzed statistically with SPSS20.0, and visualized with GraphPad 7. Inter-group comparison was conducted using the independent-samples t-test, and multi-group comparisons were conducted by the one-way analysis of variance, and expressed as F. Post hoc pairwise comparison was carried out using the least significant difference t-test, and comparison of data at multiple time points was carried out using the variance of repeated measures, and expressed as F. Post test was performed using Bonferroni. Receiver operating characteristic (ROC) curves of miR-19a-3p and FOXF2 and the value of diagnosing CRC was drawn. Pearson’s correlation analysis was carried out to investigate the relationship between serum miR-19a-3p and serum FOXF2. P < 0.05 indicated a significant difference.
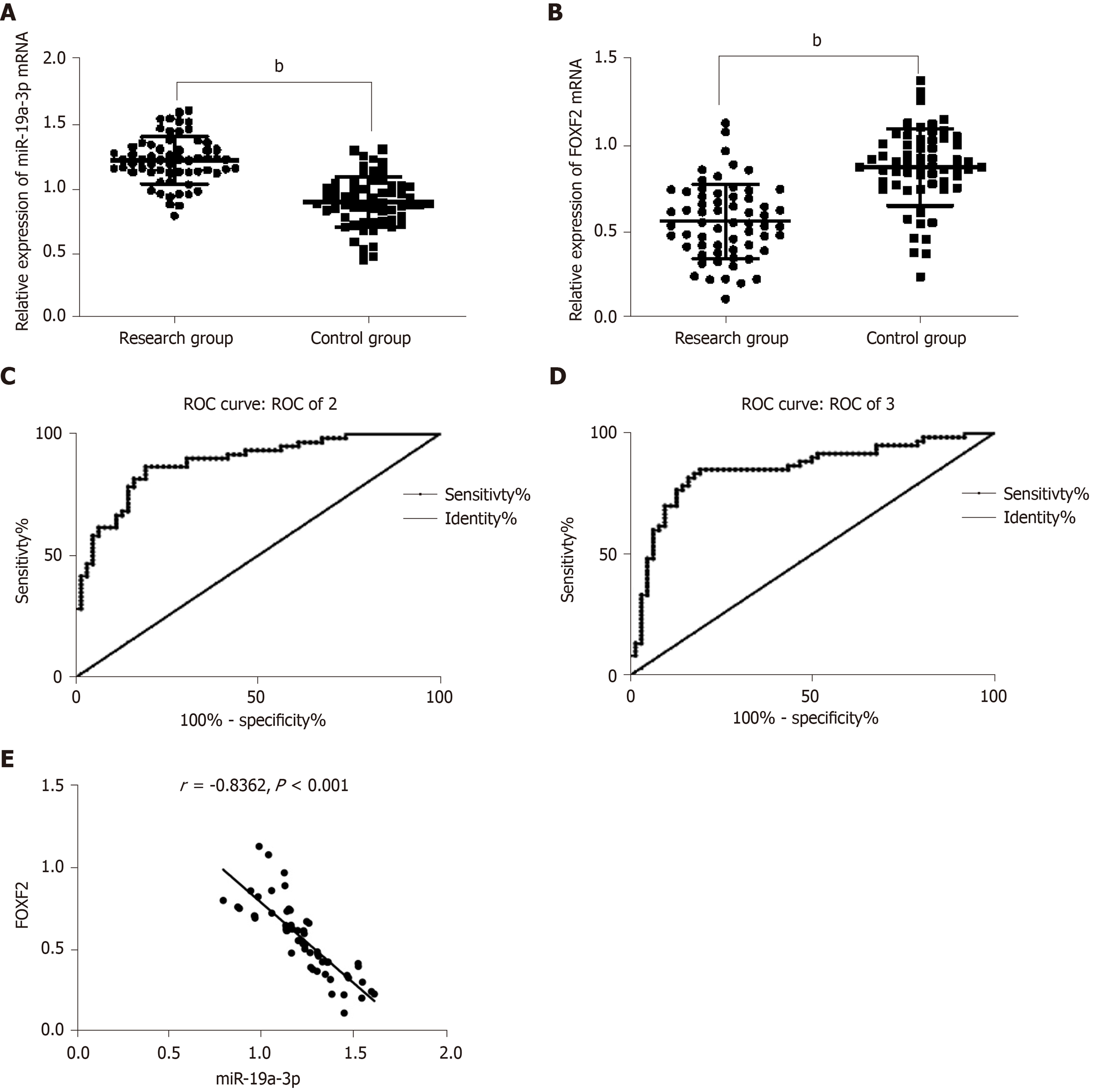
The determination of the levels of serum miR-19a-3p and FOXF2 in the subjects revealed that the study group had a significantly higher level of serum miR-19a-3p, and a significantly lower level of serum FOXF2 than the control group (both P < 0.001). Pearson’s correlation analysis revealed that the level of serum miR-19a-3p in CRC patients was negatively correlated with that of serum FOXF2 (P < 0.001), and the ROC curves showed that the area under the curves (AUC) of miR-19a-3p and FOXF2 were 0.883 and 0.850, respectively. Analysis of the correlation of miR-19a-3p and FOXF2 with pathological data of the patients revealed that the two indexes were strongly linked to age, sex, tumor size, differentiation, tumor-node-metastasis (TNM) staging, and lymph node metastasis (LNM) (all P < 0.05; Table 1 and Figure 1).
| Factors | n | Relative expression of miR-19a-3p | t | P value | Relative expression of FOXF2 | t | P value |
| Sex | |||||||
| Male (n = 32) | 1.08 ± 0.14 | 9.600 | < 0.001 | 0.41 ± 0.13 | 9.686 | < 0.001 | |
| Female (n = 30) | 1.41 ± 0.13 | 0.73 ± 0.13 | |||||
| Age | |||||||
| < 57 years-old (n = 24) | 1.03 ± 0.13 | 9.140 | < 0.001 | 0.36 ± 0.11 | 10.080 | < 0.001 | |
| ≥ 57 years-old (n = 38) | 1.37 ± 0.15 | 0.70 ± 0.14 | |||||
| TNM stage | |||||||
| I, II (n = 35) | 1.1 ± 0.14 | 9.203 | < 0.001 | 0.43 ± 0.13 | 9.910 | < 0.001 | |
| III, IV (n = 27) | 1.42 ± 0.13 | 0.76 ± 0.13 | |||||
| Tumor size | |||||||
| < 3 cm (n = 30) | 1.06 ± 0.13 | 10.290 | < 0.001 | 0.4 ± 0.12 | 10.370 | < 0.001 | |
| ≥ 3 cm (n = 32) | 1.4 ± 0.13 | 0.73 ± 0.13 | |||||
| Lymph node metastasis | |||||||
| Yes (n = 42) | 1.12 ± 0.14 | 10.780 | < 0.001 | 0.46 ± 0.14 | 9.615 | < 0.001 | |
| No (n = 20) | 1.49 ± 0.09 | 0.81 ± 0.12 | |||||
| Differentiation | |||||||
| Low (n = 27) | 1.39 ± 0.14 | 10.290 | < 0.001 | 0.38 ± 0.12 | 10.080 | < 0.001 | |
| Medium and high (n = 35) | 1.05 ± 0.12 | 0.72 ± 0.14 | |||||
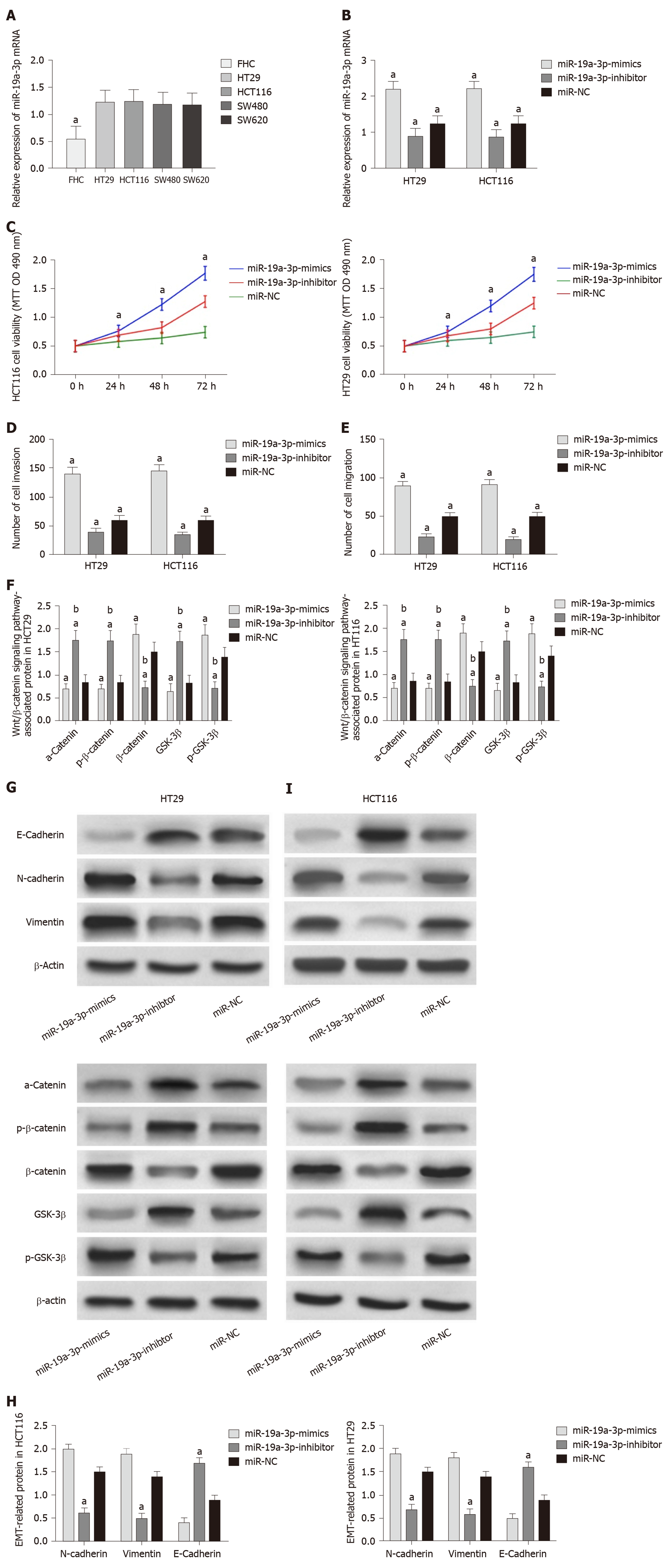
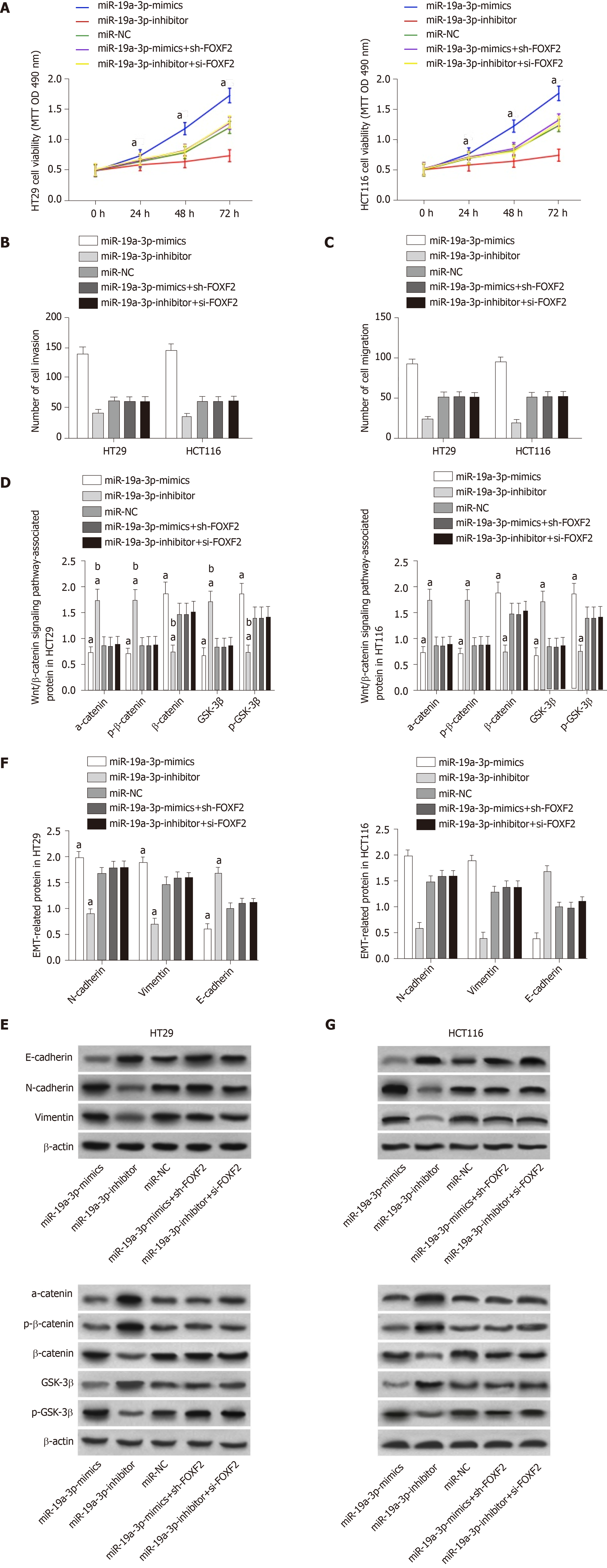
The determination of miR-19a-3p expression in CRC cells revealed that miR-19a-3p expression in the human normal colonic epithelial cell line was significantly higher than that in human CRC cell lines (HT29, SW480, SW620 and HCT116) (P < 0.05). Transfection of miR-19a-3p-mimics, miR-19a-3p-inhibitor, and miR-NC into HCT116 and HT29 cells led to the following results. HCT116 and HT29 cells transfected with miR-19a-3p-mimics had significantly higher miR-19a-3p expression than those transfected with miR-NC, and HCT116 and HT29 cells transfected with miR-19a-3p-inhibitor had significantly lower miR-19a-3p expression than those transfected with miR-NC. Determination of the biological functions of cells in the two groups demonstrated that cells with transfected miR-19a-3p-mimics had significantly more cell proliferation, invasion, and migration abilities than those transfected with miR-NC and also had significantly higher expression of p-GSK-3β, β-catenin, N-cadherin, and vimentin proteins, and significantly lower expression of GSK-3β, p-β-catenin, a-catenin, and E-cadherin. In addition, cells transfected with miR-19a-3p-inhibitor had significantly weaker cell proliferation, invasion, and migration abilities than those transfected with miR-NC, and also had significantly higher expression of p-GSK-3β, β-catenin, N-cadherin, and vimentin and significantly lower expression of GSK-3β, p-β-catenin, α-catenin,, and E-cadherin (Figure 2).
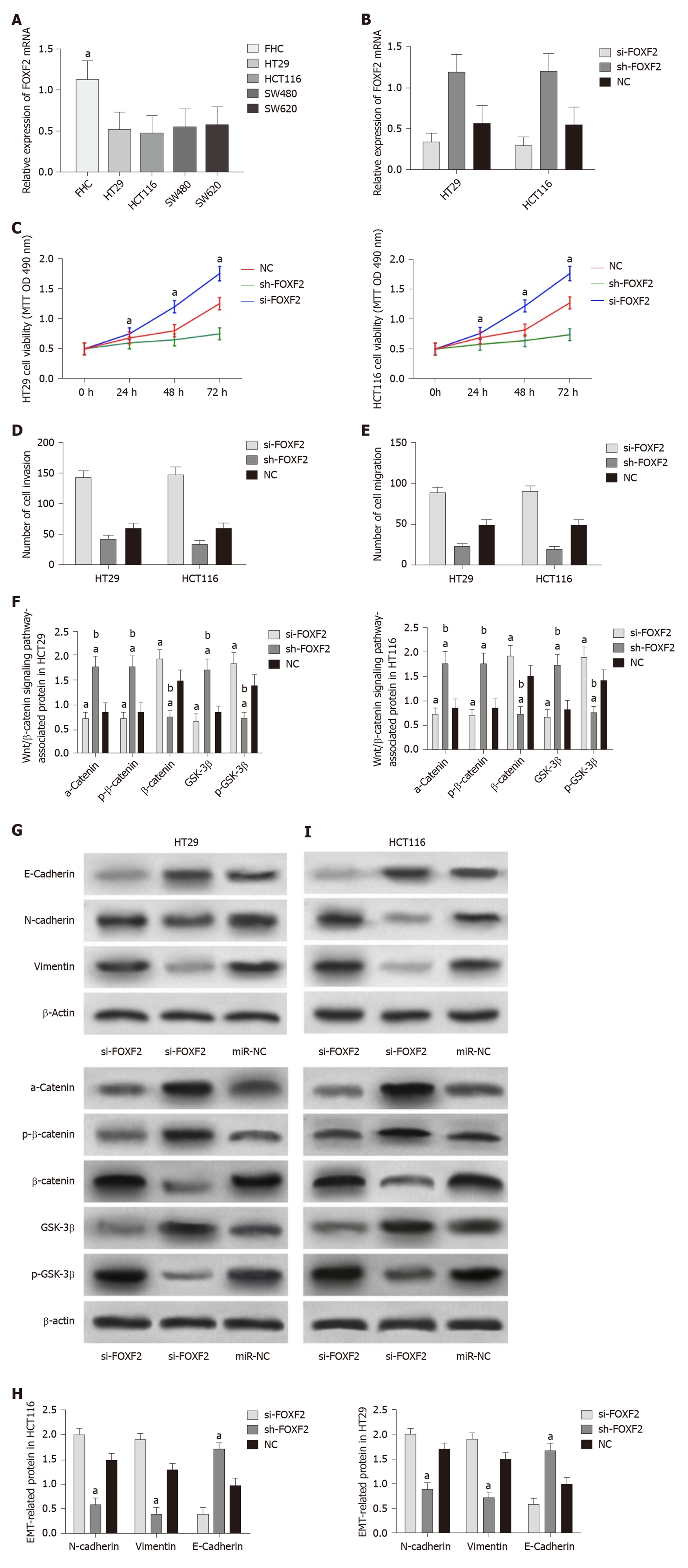
The determination of FOXF2 expression in CRC cells revealed that human CRC cell lines (HT29, SW480, SW620 and HCT116) showed significantly lower expression of FOXF2 than the human normal colonic epithelial cell line (FHC) (P < 0.05). Transfection of si-FOXF2, sh-FOXF2, and miR-NC into HCT116 and HT29 cells transfected with si-FOXF2 showed significantly lower FOXF2 expression than those transfected with NC, while HCT116 and HT29 cells transfected with sh-FOXF2 showed significantly higher FOXF2 expression than those transfected with NC. Determination of the biological functions of cells in the two groups revealed that cells transfected with si-FOXF2 showed significantly stronger cell proliferation, invasion, and migration abilities than those transfected with NC, and also showed significantly higher expression of p-GSK-3β, β-catenin, N-cadherin, and vimentin, and significantly lower expression of GSK-3β, p-β-catenin, α-catenin,, and E-cadherin than those transfected with NC. In addition, cells transfected with sh-FOXF2 showed significantly weaker cell proliferation, invasion, and migration abilities than those transfected with NC, and also showed significantly lower expression of p-GSK-3β, β-catenin, N-cadherin, and vimentin and significantly higher levels of GSK-3β, p-β-catenin, α-catenin, and E-cadherin than those transfected with NC (all P < 0.05; Figure 3).
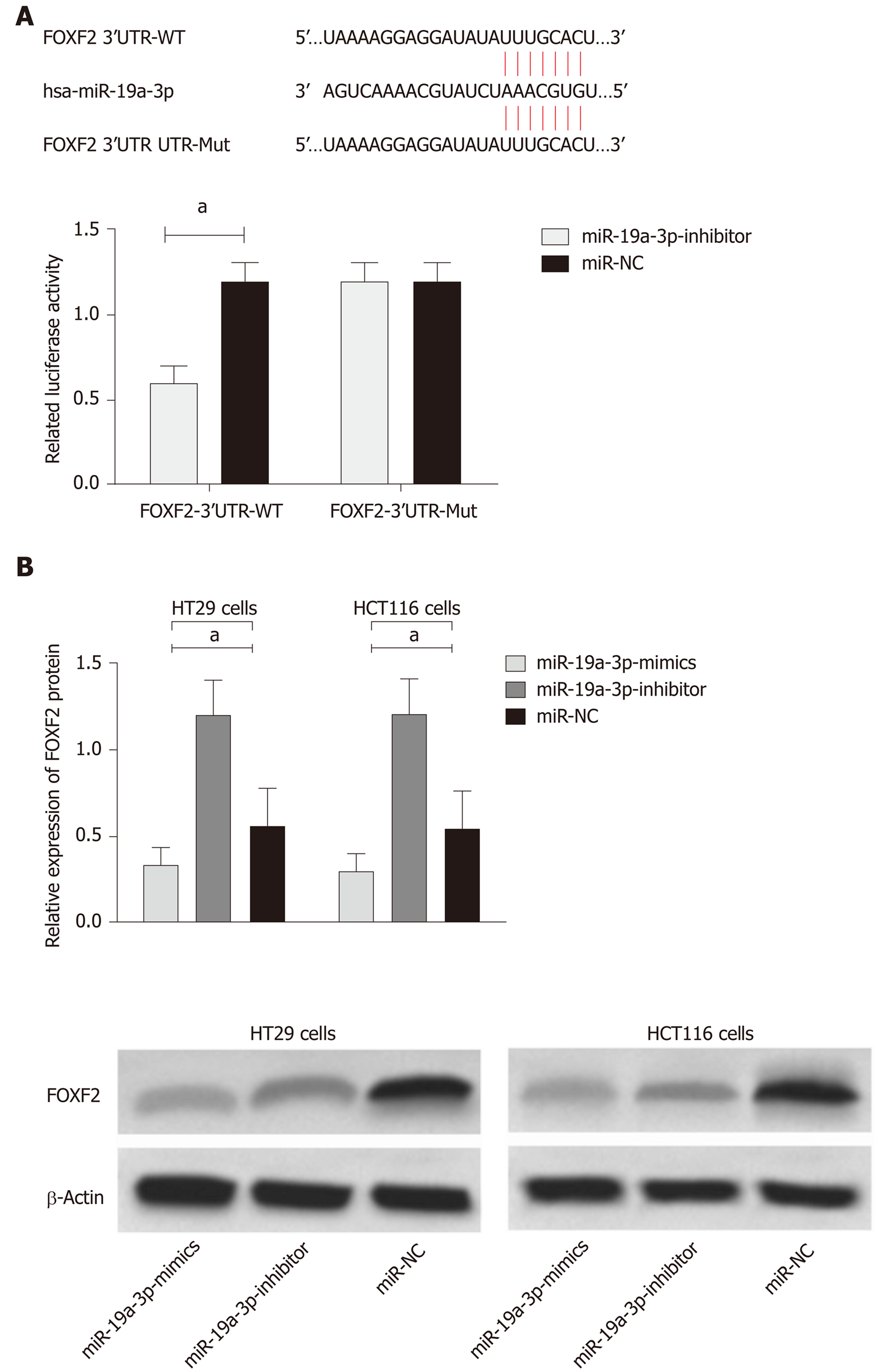
In order to verify the relationship between miR-19a-3p and FOXF2, we predicated the downstream target genes of miR-19a-3p through Targetscan7.2, and found targeted binding sites between FOXF2 and miR-19a-3p. Therefore, we conducted dual luciferase detection, and found that inhibiting miR-19a-3p significantly lowered the luciferase activity of pmirGLO-FOXF2-3’UT Wt luciferase activity (P < 0.001), but had no effect on pmirGLO-FOXF2-3’UTR Mut. WB analysis revealed that HT29 and HCT116 cells transfected with miR-19a-3p-inhibitor had significantly upregulated FOXF2 expression, and those transfected with miR-19a-3p-mimics had significantly downregulated FOXF2 expression (both P < 0.05; Figure 4).
We transfected miR-19a-3p-mimics + sh-FOXF2 and miR-19a-3p-inhibitor + si-FOXF2 into HT29 and HCT116 cells, and detected their biological functions. There were no significant differences between these transfected cells and those transfected with miR-NC regarding cell proliferation, invasion, and migration. Cells transfected with one of those constructs showed significantly stronger proliferation, invasion, and migration abilities than those transfected with miR-19a-3p-mimics, and showed significantly weaker proliferation, invasion, and migration abilities than those transfected with miR-19a-3p-inhibitor (all P < 0.05). The EMT-related protein assay revealed that there were no significant differences between cells transfected with miR-19a-3p-mimics + sh-FOXF2 or miR-19a-3p-inhibitor + si-FOXF2 and those transfected with miR-NC regarding the expression of E-cadherin, N-cadherin, and vimentin, while cells transfected with miR-19a-3p-mimics + sh-FOXF2 or miR-19a-3p-inhibitor + si-FOXF2 showed a significantly lower level of E-cadherin, and significantly higher levels of N-cadherin and vimentin than those transfected with miR-19a-3p-mimics, and showed a significantly higher level of E-cadherin, and significantly lower levels of N-cadherin and vimentin than those transfected with miR-19a-3p-inhibitor (all P < 0.05; Figure 5).
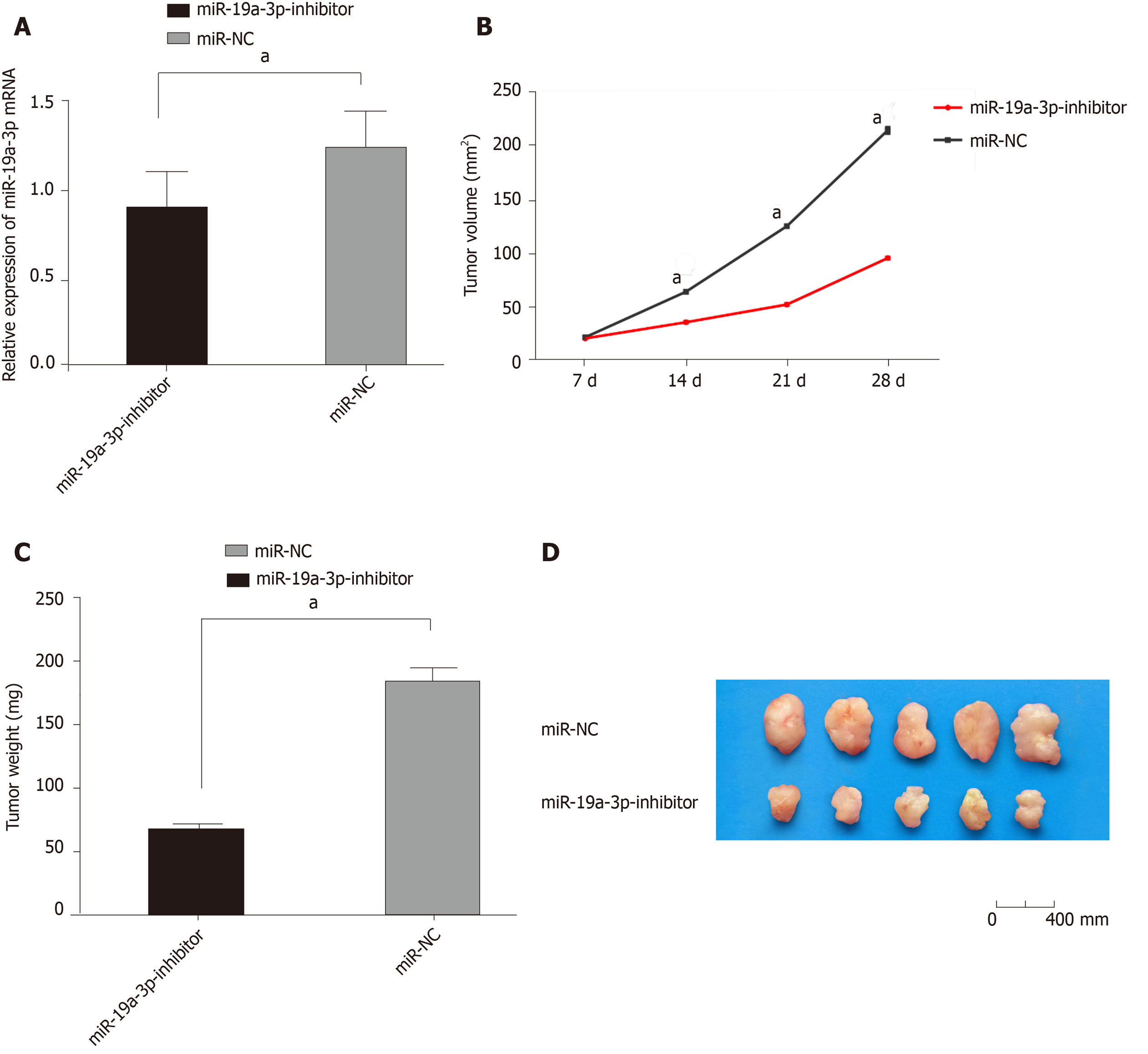
We constructed a tumor formation model by injecting HCT116 cells transfected with miR-19a-3p-inhibitor or miR-NC into the abdomen of nude mice. Nude mice injected with miR-19a-3p-inhibitor had a significantly slower growth rate than those injected with miR-NC, and after the mice were sacrificed, the tumor size and weight of mice with miR-19a-3p-inhibitor were significantly smaller than those with miR-NC (all P < 0.001). In addition, mice infected with miR-19a-3p-inhibitor had significantly lower miR-19a-3p expression than those infected with NC (P < 0.001), which suggested that inhibiting miR-19a-3p expression can suppress tumor growth in vivo (Figure 6).
The biological characteristics of CRC cells affect the development of CRC. For example, the changes in proliferation, invasion, migration, and apoptosis of tumor cells affect and control tumor growth[17]. A large number of studies have concluded that cell proliferation is an important vital sign of biological organisms, and consistent proliferation, invasion and metastasis of tumor cells are the main causes of death for patients with malignant tumor[18]. Recent studies have indicated that miRNAs are strongly linked to various tumors, and play important roles in carcinogenesis and tumor progression, because they can affect the proliferation, invasion, migration, and apoptosis of tumor cells. One study confirmed that miR-19a-3p is abnormally and increasingly expressed in CRC tissues, and the changes in miR-19a-3p expression can affect the EMT of CRC cells[19], but the specific mechanism is not completely understood. This study determined that one biological function of miR-19a-3p in CRC cells is to regulate the FOXF2-mediated Wnt/β-catenin signaling pathway, thereby providing a novel theoretical basis for diagnosing and treating CRC in molecular biology.
In this study, we conducted qPCR to evaluate the expression of serum miR-19a-3p in CRC patients and healthy individuals, and found that serum miR-19a-3p was abnormally increased in CRC patients, in line with a study conducted by Jiang et al[20]. We also carried out relevant analyses on the clinicopathological features of the patients, finding that high miR-19a-3p expression was correlated with age, sex, tumor size, differentiation, TNM stage, and LNM of CRC patients. We drew a ROC curve for miR-19a-3p, and found that its AUC was more than 0.8, suggesting that miR-19a-3p had a high diagnostic value for CRC. Those findings indicated that miR-19a-3p is correlated with the development and progression of CRC. A large number of studies have confirmed that miRNA can affect the biological functions of cells by regulating their target genes. For example, miR-19a-3p is reportedly able to target downstream gene pairs to regulate the biological functions of cells[21,22]. We carried out our analyses based on the TargetScan database, finding that there was a targeted relationship between miR-19a-3p and FOXF2. We also analyzed the relationship between FOXF2 and clinicopathological characteristics of the patients, finding that low expression of FOXF2 was correlated with clinicopathologic feature of CRC patients. Furthermore, we drew a ROC curve for FOXF2, and found that its AUC was more than 0.8. FOXF2, as a transcription factor, regulates the progression and differentiation of tumors, and is involved in the changes of biological functions of various tumor cells[23]. It can affect the cycle and other biological functions of CRC cells by regulating CRC cell genes[24]. In this study, we analyzed FOXF2 expression in CRC cells based on the data from TCGA, and found that CRC cells showed increased miR-19a-3p and decreased FOXF2 expression, which was consistent with the experimental results of this study, suggesting that both may play regulatory roles in CRC. However, it remains unclear whether there is a regulatory relationship between the two and their effects on the EMT of CRC cells.
In order to verify these results, we performed additional cell experiments. We compared human CRC cell lines (HT29, SW480, SW620, and HCT116) with the human normal colonic epithelial cell line (FHC), and found that CRC cells showed higher miR-19a-3p expression and lower FOXF2 expression, consistent with our previous results. After the comparison, we regulated the expression of miR-19a-3p and FOXF2 in HT29 and HCT116 cells, and transfected miR-19a-3p-mimics, miR-19a-3p-inhibitor, sh-FOXF2, and si-FOXF2 into HT29 and HCT116 cells, finding that cells with inhibited miR-19a-3p expression or overexpressed FOXF2 expression showed significantly suppressed cell proliferation and invasion abilities, and also showed decreased expression of N-cadherin and vimentin, and significantly increased E-cadherin expression during EMT. However, HT29 and HCT116 transfected with miR-19a-3p-mimics and FOXF2-inhibitor showed opposite results. The occurrence and development of tumors are closely related to the EMT, and can promote proliferation, invasion, and migration of tumor cells. Therefore, the above results indicate that miR-19a-3p acts as a potential target for CRC, and silencing of miR-19a-3p can suppress the proliferation, invasion, migration, and EMT of CRC cells. Tumorigenicity in nude mice in vitro revealed that miR-19a-3p overexpression was able to strongly promote tumor formation and growth, which further indicated the function of miR-19a-3p changes in CRC. However, it is not clear how miR-19a-3p affects the biological functions of CRC cells and EMT.
One study demonstrated that activating the Wnt/β-catenin signaling pathway exerts great effects on promoting EMT[25], and some studies have shown that, by regulating the Wnt/β-catenin signaling pathway, FOXF2 can inhibit the proliferation, migration, invasion, and EMT of cervical cancer Hela cells[26]. In this study, we observed the Wnt/β-catenin signaling pathway-related proteins after inhibiting or overexpressing miR-19a-3 in CRC cells, finding that cells with underexpressed miR-19a-3p or overexpressed FOXF2 showed significantly downregulated levels of p-GSK-3β, β-catenin, N-cadherin, and vimentin, and significantly upregulated levels of GSK-3β, p-β-catenin, α-catenin, and E-cadherin. However, cells with overexpressed miR-19a-3p or underexpressed FOXF2 presented with the opposite results. It suggests that miR-19a-3p is able to suppress the activation of the Wnt/β-catenin signaling pathway by regulating FOXF2, thereby inhibiting the EMT of cells. Studies on the Wnt signaling pathway have also revealed that activating the Wnt/β-catenin signaling pathway can promote the invasion and proliferation of esophageal squamous cell carcinoma, nasopharyngeal carcinoma, as well as breast cancer cells, and induce their EMT [27-29].
At the end of the study, we performed a rescue experiment, and found no significant differences between HT29 and HCT116 cells transfected with miR-19a-3p-mimics + sh-FOXF2 and miR-19a-3p-inhibitor + si-FOXF2 and those transfected with miR-NC regarding cell proliferation, invasion and migration, but cells transfected with miR-19a-3p-mimics + sh-FOXF2 and miR-19a-3p-inhibitor + si-FOXF2 showed significantly stronger abilities than those transfected with miR-19a-3p-inhibitor regarding cell proliferation, invasion and migration, and significantly weaker cell proliferation, invasion and migration abilities than those transfected with miR-19a-3p-mimics. The results suggested that miR-19a-3p can target FOXF2. Therefore, the dual luciferase reporter assay was conducted to verify the correlation of miR-19a-3 with FOXF2. The results showed that overexpressing miR-19a-3 significantly increased the luciferase activity of FOXF2-3’UT Wt, but had no effect on the activity of FOXF2-3’UTR Mut. Moreover, cells transfected with miR-19a-3p-inhibitor showed significantly upregulated FOXF2 expression, which indicated that there was a targeted regulatory relationship between miR-19a-3p and FOXF2. Therefore, we believe that inhibiting the expression of miR-19a-3p can affect the biological functions of CRC cells by promoting the expression of FOXF2.
This study proved that with high expression in CRC cells, miR-19a-3p can inhibit FOXF2-mediated cell proliferation. However, this study had some limitations. For example, other factors that may affect the growth of tumors in mice were not been further analyzed. The regulatory network of miR-19a-3p is still under investigation, and whether it can affect the development and progression of tumors through other mechanisms requires more research. Therefore, bioinformatics analysis is still ongoing to explore the regulatory network, in order to provide more references for our experiments. To summarize, miR-19a-3p can mediate changes of CRC cells by regulating the FOXF2 expression, and it is of great significance to study molecular targeted treatments for CRC.
Colorectal cancer (CRC) has a high rate of mortality, and patients with this disease often miss the optimal treatment period due to the lack of clinical symptoms of early CRC, which affects their prognosis. At present, CRC is extremely difficult to prevent and treat.
In this study, we studied the effects of regulating the Forkhead box F2 (FOXF2)-mediated Wnt/β-catenin signaling pathway by miR-19a-3p on the biological functions of CRC cells from the perspective of the mechanism of CRC, so as to explore the changes in biological functions of CRC cells.
This study evaluated the expression of miR-19a-3p and FOXF2 in patients with CRC and the relevant mechanisms.
Elbow venous blood was sampled from CRC patients and healthy individuals, and blood serum was saved for later analysis. MiR-19a-3p-mimics, miR-19a-3p-inhibitor, miR-NC, si-FOXF2, and sh-FOXF2 were transfected into HT29 and HCT116 cells. Then quantitative polymerase chain reaction was applied to determine the expression of miR-19a-3p and FOXF2 in HT29 and HCT116 cells, and Western blotting was conducted to determine the expression of FOXF2, GSK-3β, p-GSK-3β, β-catenin, p-β-catenin, α-catenin, N-cadherin, E-Cadherin, and vimentin. The MTT, Transwell, and wound-healing assays were applied to detect cell proliferation, invasion, and apoptosis, respectively, and the dual luciferase reporter assay was used to determine the relationship between miR-19a-3p and FOXF2.
MiR-19a-3p was highly expressed in the serum of the patients, while FOXF2 was lowly expressed in them. MiR-19a-3p and FOXF2 were related to age, sex, tumor size, tumor, node, metastasis staging, lymph node metastasis, and differentiation of patients with CRC. Silencing of miR-19a-3p and over-expression of FOXF2 suppressed epithelial-mesenchymal transition, proliferation, invasion, and migration of cells, and Western blot assay supported that silencing of miR-19a-3p and over-expression of FOXF2 significantly suppressed epithelial-mesenchymal transition. Dual luciferase reporter assay confirmed that there was a targeted relationship between miR-19a-3p and FOXF2. Therefore, inhibiting the expression of miR-19a-3p can affect the biological functions of CRC cells by promoting the expression of FOXF2.
Inhibiting the expression of miR-19a-3p can affect the biological functions of CRC cells by promoting the expression of FOXF2.
It has been confirmed that inhibiting the expression of miR-19a-3p can up-regulate the FOXF2-mediated Wnt/β-catenin signaling pathway, thus affecting the epithelial-mesenchymal transition, proliferation, invasion, and migration of cells, so miR-19a-3p is expected to be a potential therapeutic target for CRC.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Jeong KY, Katuchova J, Maric I S-Editor: Wang J L-Editor: Filipodia E-Editor: Qi LL
| 1. | Vukovic B, Beheshti B, Park P, Lim G, Bayani J, Zielenska M, Squire JA. Correlating breakage-fusion-bridge events with the overall chromosomal instability and in vitro karyotype evolution in prostate cancer. Cytogenet Genome Res. 2007;116:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 2. | Bahreini F, Soltanian AR. Identification of A Gene Set Associated with Colorectal Cancer in Microarray Data Using The Entropy Method. Cell J. 2019;20:569-575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 3. | Moridikia A, Mirzaei H, Sahebkar A, Salimian J. MicroRNAs: Potential candidates for diagnosis and treatment of colorectal cancer. J Cell Physiol. 2018;233:901-913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 150] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 4. | Oberthür R, Seemann H, Gehrig J, Rave-Fränk M, Bremmer F, Halpape R, Conradi LC, Scharf JG, Burfeind P, Kaulfuß S. Simultaneous inhibition of IGF1R and EGFR enhances the efficacy of standard treatment for colorectal cancer by the impairment of DNA repair and the induction of cell death. Cancer Lett. 2017;407:93-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 5. | Rejhová A, Opattová A, Čumová A, Slíva D, Vodička P. Natural compounds and combination therapy in colorectal cancer treatment. Eur J Med Chem. 2018;144:582-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 254] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 6. | Pugh S, Thiébaut R, Bridgewater J, Grisoni ML, Moutasim K, Rousseau F, Thomas GJ, Griffiths G, Liebaert F, Primrose J, Laurent-Puig P. Association between miR-31-3p expression and cetuximab efficacy in patients with KRAS wild-type metastatic colorectal cancer: a post-hoc analysis of the New EPOC trial. Oncotarget. 2017;8:93856-93866. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 7. | Weissfeld JL, Schoen RE, Pinsky PF, Bresalier RS, Doria-Rose VP, Laiyemo AO, Church T, Yokochi LA, Yurgalevitch S, Rathmell J, Andriole GL, Buys S, Crawford ED, Fouad M, Isaacs C, Lamerato L, Reding D, Prorok PC, Berg CD; PLCO Project Team. Flexible sigmoidoscopy in the randomized prostate, lung, colorectal, and ovarian (PLCO) cancer screening trial: added yield from a second screening examination. J Natl Cancer Inst. 2012;104:280-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 8. | Wei SC, Fattet L, Tsai JH, Guo Y, Pai VH, Majeski HE, Chen AC, Sah RL, Taylor SS, Engler AJ, Yang J. Matrix stiffness drives epithelial-mesenchymal transition and tumour metastasis through a TWIST1-G3BP2 mechanotransduction pathway. Nat Cell Biol. 2015;17:678-688. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 527] [Cited by in RCA: 699] [Article Influence: 69.9] [Reference Citation Analysis (0)] |
| 9. | Su HT, Weng CC, Hsiao PJ, Chen LH, Kuo TL, Chen YW, Kuo KK, Cheng KH. Stem cell marker nestin is critical for TGF-β1-mediated tumor progression in pancreatic cancer. Mol Cancer Res. 2013;11:768-779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 67] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 10. | Zou M, Wang F, Gao R, Wu J, Ou Y, Chen X, Wang T, Zhou X, Zhu W, Li P, Qi LW, Jiang T, Wang W, Li C, Chen J, He Q, Chen Y. Autophagy inhibition of hsa-miR-19a-3p/19b-3p by targeting TGF-β R II during TGF-β1-induced fibrogenesis in human cardiac fibroblasts. Sci Rep. 2016;6:24747. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 95] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 11. | Jiang XM, Yu XN, Liu TT, Zhu HR, Shi X, Bilegsaikhan E, Guo HY, Song GQ, Weng SQ, Huang XX, Dong L, Janssen HLA, Shen XZ, Zhu JM. microRNA-19a-3p promotes tumor metastasis and chemoresistance through the PTEN/Akt pathway in hepatocellular carcinoma. Biomed Pharmacother. 2018;105:1147-1154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 82] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 12. | Yang J, Zhang Z, Chen C, Liu Y, Si Q, Chuang TH, Li N, Gomez-Cabrero A, Reisfeld RA, Xiang R, Luo Y. MicroRNA-19a-3p inhibits breast cancer progression and metastasis by inducing macrophage polarization through downregulated expression of Fra-1 proto-oncogene. Oncogene. 2014;33:3014-3023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 147] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 13. | Lee S, Lee H, Bae H, Choi EH, Kim SJ. Epigenetic silencing of miR-19a-3p by cold atmospheric plasma contributes to proliferation inhibition of the MCF-7 breast cancer cell. Sci Rep. 2016;6:30005. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 14. | Pellegrini KL, Gerlach CV, Craciun FL, Ramachandran K, Bijol V, Kissick HT, Vaidya VS. Application of small RNA sequencing to identify microRNAs in acute kidney injury and fibrosis. Toxicol Appl Pharmacol. 2016;312:42-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 15. | Zhang J, Zhang C, Sang L, Huang L, Du J, Zhao X. FOXF2 inhibits proliferation, migration, and invasion of Hela cells by regulating Wnt signaling pathway. Biosci Rep. 2018;38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 16. | Shah R, Jones E, Vidart V, Kuppen PJ, Conti JA, Francis NK. Biomarkers for early detection of colorectal cancer and polyps: systematic review. Cancer Epidemiol Biomarkers Prev. 2014;23:1712-1728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 17. | He F, Song Z, Chen H, Chen Z, Yang P, Li W, Yang Z, Zhang T, Wang F, Wei J, Wei F, Wang Q, Cao J. Long noncoding RNA PVT1-214 promotes proliferation and invasion of colorectal cancer by stabilizing Lin28 and interacting with miR-128. Oncogene. 2019;38:164-179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 87] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 18. | Zhang Y, Wang T, Huang HQ, Li W, Cheng XL, Yang J. Human MALAT-1 long non-coding RNA is overexpressed in cervical cancer metastasis and promotes cell proliferation, invasion and migration. J BUON. 2015;20:1497-1503. [PubMed] |
| 19. | Fu F, Jiang W, Zhou L, Chen Z. Circulating Exosomal miR-17-5p and miR-92a-3p Predict Pathologic Stage and Grade of Colorectal Cancer. Transl Oncol. 2018;11:221-232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 163] [Article Influence: 23.3] [Reference Citation Analysis (1)] |
| 20. | Zhang X, Chen Y, Zhao P, Zang L, Zhang Z, Wang X. MicroRNA-19a functions as an oncogene by regulating PTEN/AKT/pAKT pathway in myeloma. Leuk Lymphoma. 2017;58:932-940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 21. | Qiao F, Gong P, Song Y, Shen X, Su X, Li Y, Wu H, Zhao Z, Fan H. Downregulated PITX1 Modulated by MiR-19a-3p Promotes Cell Malignancy and Predicts a Poor Prognosis of Gastric Cancer by Affecting Transcriptionally Activated PDCD5. Cell Physiol Biochem. 2018;46:2215-2231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 22. | Li X, Yan X, Wang F, Yang Q, Luo X, Kong J, Ju S. Down-regulated lncRNA SLC25A5-AS1 facilitates cell growth and inhibits apoptosis via miR-19a-3p/PTEN/PI3K/AKT signalling pathway in gastric cancer. J Cell Mol Med. 2019;23:2920-2932. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 58] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 23. | Hauptman N, Jevšinek Skok D, Spasovska E, Boštjančič E, Glavač D. Genes CEP55, FOXD3, FOXF2, GNAO1, GRIA4, and KCNA5 as potential diagnostic biomarkers in colorectal cancer. BMC Med Genomics. 2019;12:54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 24. | Hernández R, Sánchez-Jiménez E, Melguizo C, Prados J, Rama AR. Downregulated microRNAs in the colorectal cancer: diagnostic and therapeutic perspectives. BMB Rep. 2018;51:563-571. [PubMed] |
| 25. | Sohn SH, Kim B, Sul HJ, Kim YJ, Kim HS, Kim H, Seo JB, Koh Y, Zang DY. INC280 inhibits Wnt/β-catenin and EMT signaling pathways and its induce apoptosis in diffuse gastric cancer positive for c-MET amplification. BMC Res Notes. 2019;12:125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 26. | Higashimori A, Dong Y, Zhang Y, Kang W, Nakatsu G, Ng SSM, Arakawa T, Sung JJY, Chan FKL, Yu J. Forkhead Box F2 Suppresses Gastric Cancer through a Novel FOXF2-IRF2BPL-β-Catenin Signaling Axis. Cancer Res. 2018;78:1643-1656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 27. | Yang L, Ye Y, Chu J, Jia J, Qu Y, Sun T, Yin H, Ming L, Wan J, He F. Long noncoding RNA FEZF1-AS1 promotes the motility of esophageal squamous cell carcinoma through Wnt/β-catenin pathway. Cancer Manag Res. 2019;11:4425-4435. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 28. | Luo M, Wu C, Guo E, Peng S, Zhang L, Sun W, Liu D, Hu G, Hu G. FOXO3a knockdown promotes radioresistance in nasopharyngeal carcinoma by inducing epithelial-mesenchymal transition and the Wnt/β-catenin signaling pathway. Cancer Lett. 2019;455:26-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 29. | Hseu YC, Lin YC, Rajendran P, Thigarajan V, Mathew DC, Lin KY, Way TD, Liao JW, Yang HL. Antrodia salmonea suppresses invasion and metastasis in triple-negative breast cancer cells by reversing EMT through the NF-κB and Wnt/β-catenin signaling pathway. Food Chem Toxicol. 2019;124:219-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |