Published online Dec 7, 2020. doi: 10.3748/wjg.v26.i45.7118
Peer-review started: August 1, 2020
First decision: September 30, 2020
Revised: October 10, 2020
Accepted: November 12, 2020
Article in press: November 12, 2020
Published online: December 7, 2020
Processing time: 125 Days and 0.1 Hours
Helicobacter pylori (H. pylori) colonizes the human stomach and is a major cause of peptic ulcer disease and gastric cancer. However, although the prevalence of H. pylori is high in Africa, the incidence of gastric cancer is low, and this phenomenon is called to be African enigma. The CagA protein produced by H. pylori is the most studied virulence factor. The carcinogenic potential of CagA is associated with the Glu-Pro-Ile-Tyr-Ala (EPIYA) patterns and CagA-multimerization (CM) motifs.
To better understand the EPIYA patterns and CM motifs of the cagA gene.
Gastric mucosal biopsy specimens were obtained from 258 patients with dyspepsia living in the Dominican Republic, from which 120 H. pylori strains were cultured. After the bacterial DNA extraction, the EPIYA pattern and CM motif genotypes were determined using a polymerase chain reaction-based sequencing. The population structure of the Dominican Republic strains was analyzed using multilocus sequence typing (MLST). Peptic ulcer disease and gastric cancer were identified via endoscopy, and gastric cancer was confirmed by histopathology. Histological scores of the gastric mucosa were evaluated using the updated Sydney system.
All CagA-positive strains carried the Western-type CagA according to the identified EPIYA patterns. Twenty-seven kinds of CM motifs were observed. Although the typical Western CM motif (FPLKRHDKVDDLSKVG) was observed most frequently, the typical East Asian CM motif (FPLRRSAAVNDLSKVG) was not observed. However, “FPLRRSAKVEDLSKVG”, similar to the typical East Asian CM motif, was found in 21 strains. Since this type was significantly more frequent in strains classified as hpAfrica1 using MLST analysis (P = 0.034), we termed it Africa1-CM (Af1-CM). A few hpEurope strains carried the Af1-CM motif, but they had a significantly higher ancestral Africa1 component than that of those without the Af1-CM motif (P = 0.030). In 30 cagA-positive strains, the "GKDKGPE" motif was observed immediately upstream of the EPIYA motif in the EPIYA-A segment, and there was a significant association between strains with the hpAfrica1 population and those containing the “GKDKGPE” motif (P = 0.018). In contrast, there was no significant association between the CM motif patterns and histological scores and clinical outcomes.
We found the unique African CM motif in Western-type CagA and termed it Africa1-CM. The less toxicity of this motif could be one reason to explain the African enigma.
Core Tip: We found the unique African (hpAfrica1-type) CagA-multimerization (CM) motif in Western-type CagA in the Dominican Republic and termed it Africa1-CM (Af1-CM). In addition, a few hpEurope strains carrying the Af1-CM motif had a significantly higher ancestral Africa1 component than those without the Af1-CM motif. This result suggests that the ancestors of these hpEurope strains having the Af1-CM motif underwent genetic recombination with the hpAfrica1 strains in the past. In contrast, there was no significant association between the CM motif patterns and histological scores and clinical outcomes.
- Citation: Ono T, Cruz M, Nagashima H, Subsomwong P, Akada J, Matsumoto T, Uchida T, Suzuki R, Hosking C, Jiménez Abreu JA, Yamaoka Y. Discovery of unique African Helicobacter pylori CagA-multimerization motif in the Dominican Republic. World J Gastroenterol 2020; 26(45): 7118-7130
- URL: https://www.wjgnet.com/1007-9327/full/v26/i45/7118.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i45.7118
Helicobacter pylori (H. pylori) plays an essential role in the development of various gastroduodenal diseases including gastric cancer[1]. However, only a small proportion of people infected with H. pylori develop these diseases. Interestingly, the incidence of gastric cancer in Africa is much lower than that in other countries, despite the high prevalence of H. pylori infection in this area[1]. This contradictory phenomenon is known as the “African enigma”. Geographic differences in the incidence of gastric cancer can be explained, at least in part, by the presence of different types of H. pylori virulence factor, especially CagA, VacA and OipA[1].
The H. pylori CagA protein is one of the well-known pathogenic virulence factors involved in the pathogenesis of gastric cancer[1]. The cagA gene encodes a 120-145 kDa CagA and is located at one end of the cag pathogenicity island (PAI) that encodes the type IV secretion system (TFSS)[2]. CagA can directly translocate into host gastric epithelial cells via the TFSS and undergoes tyrosine phosphorylation by Src family kinases and then binds to various molecules including the Src homology 2-containing protein tyrosine phosphatase-2 (SHP-2)[3-5]. Gastric mucosal epithelial cells form a monolayer of polarized cells through tight junctions. When CagA binds to the kinase domain of partitioning-defective 1b (PAR1b)/microtubule affinity-regulating kinase 2 (MARK2), it destroys the tight junctions in a tyrosine phosphorylation-independent manner and causes loss of epithelial cell polarity[6]. CagA multimerizes in cells independently of tyrosine phosphorylation, and a CagA-multimerization (CM) sequence consisting of 16 amino acids has been identified as its responsible region[7]. There is also a report that the CM sequence is called conserved repeat responsible for phosphorylation-independent activity[8]. This CM sequence is essential for binding between CagA and PAR1. PAR1 forms a homomultimer in the cell. Thus, CagA forms a multimer indirectly through binding to the PAR1 multimer and binds to SHP-2 after being tyrosine-phosphorylated[7].
The tyrosine phosphorylation site of CagA is characterized by the presence of a unique Glu-Pro-Ile-Tyr-Ala (EPIYA) motif, which is present in multiple numbers in the C-terminal region of CagA[1]. The EPIYA-repeat region is composed of various combinations of four distinct EPIYA segments, EPIYA-A, -B, -C and -D, based on the sequences surrounding each of the EPIYA motifs[9]. H. pylori cagA-positive strains isolated in Western countries possess the Western CagA, which contains EPIYA-A, EPIYA-B, and EPIYA-C segments, whereas that in East Asian countries possess the East Asian CagA, which contains EPIYA-A, EPIYA-B and EPIYA-D segments. We previously showed that the East Asian CagA was more virulent than the Western CagA[10]. The EPIYA-D motif of the East Asian CagA exhibits a stronger SHP-2 binding affinity than the EPIYA-C motif of the Western CagA[9]. In addition, the number of EPIYA-C sites is directly correlated to the level of tyrosine phosphorylation and SHP-2 binding activity among the Western CagA species.
Eleven of the 16 amino acids in the CM motif are well conserved between the Western and East Asian cagA species[7]. The peptide sequence of the typical Western CM motif (W-CM), which is observed in the Western cagA, is “FPLKRHDKVD DLSKVG” and that of the typical East Asian CM motif (E-CM), which is observed in the East Asian cagA, is “FPLRRSAAVNDLSKVG”[11,12]. In the Western cagA species, each EPIYA-C segment contains a single CM motif, and there is also another copy of the CM motif immediately distal to the last repeat of the EPIYA-C segment. In the East Asian cagA species, there is a CM motif that is located immediately distal to each repeat of the EPIYA-D segment. Thus, the Western cagA carries at least two W-CM sequences, whose number increases in parallel with the number of EPIYA-C segments. The ability of the Western CagA to bind to PAR1 is proportional to the number of W-CM sequences[11]. Meanwhile, a single E-CM sequence has almost twice the PAR1 binding ability of the single W-CM sequence.
We have previously reported that all 64 H. pylori strains isolated in 2011 in the Dominican Republic carried the Western cagA specific sequences[13]. However, this report did not mention the CM motif. In the subsequent study in the Dominican Republic, we have already performed a multilocus sequence typing (MLST) analysis of seven housekeeping genes for a total of 119 H. pylori strains, which were collected in 2011 and 2016, followed by population structure analysis using STRUCTURE software[14]. Therefore, H. pylori strains of the Dominican Republic were divided into two populations: 68 strains with hpAfrica1 [46, 5, and 17 with hspWAfrica, hspSAfrica, and a hybrid (hspWAfrica/hpEurope), respectively], and 51 strains with hpEurope (47 and 4 with hspEuropeS and hspEuropeN, respectively). The ethnics in the Dominican Republic consists of 16% of European, 11% of African, and 73% of mixed race[15]. Therefore, it is important to investigate the genetic diversity of the cagA gene in the Dominican Republic, where human demography consists of various ancestries. In this study, we evaluated the correspondence between the CM motif and phylogeo-graphical classification using MLST population structure in H. pylori in the Dominican Republic. We also examined the relationship between these types and gastric mucosal damages.
We recruited outpatients with mild dyspeptic symptoms living in the Dominican Republic. Gastric mucosal biopsy specimens were taken from 258 dyspeptic patients (158 in 2011 and 100 in 2016; 86 males and 172 females; age range, 17-91 years; mean age, 46.2 ± 15.8 years) who underwent endoscopy examination at the Digestive Disease Center (Dr Luis E. Aybar Health and Hygiene City, Santo Domingo, Dominican Republic). Patients with a history of partial gastric resection or previous treatment for H. pylori infection were excluded. During each endoscopy session, 4 gastric biopsy specimens were obtained (three from the antrum and one from the corpus). The three specimens from the antrum were used for H. pylori culture, rapid urease test and histological examination. The specimen from the corpus was used for histological examination.
Gastric biopsy specimens for histological examination were embedded in paraffin after being fixed in 40 g/L formaldehyde. Hematoxylin and eosin and May-Giemsa were selected as stain techniques. The updated Sydney system was used to analyze histological severity[16]. The degree of the bacterial load was classified into four grades: 0, “normal”; 1, “mild”; 2, “moderate”; and 3, “marked”. Peptic ulcer and gastric cancer were identified via endoscopy, and gastric cancer was confirmed by histopathology. Gastritis was diagnosed in the absence of peptic ulcer or gastric malignancy. For H. pylori culture, antral biopsy specimens were homogenized and inoculated onto antibiotic selection plates, and then subcultured on Mueller Hinton II Agar medium (Becton Dickinson, Sparks; MD, United States) supplemented with 7% horse blood without antibiotics. The plates were incubated up to 10 d at 37 °C under microaerophilic conditions (100 mL/L O2, 50 mL/L CO2, and 850 mL/L N2). H. pylori isolates were identified based on colony morphology, Gram staining results, and oxidase, catalase, and urease reactions. Isolated strains were stored at −80 °C in Brucella broth (Becton Dickinson, Sparks; MD, United States) containing 10% dimethyl sulfoxide and 10% horse serum. Bacterial DNA was extracted using a commercially available kit (QIAGEN Inc.; Valencia, CA, United States).
Eventually, 64 strains were cultured from 158 patients in 2011, and 56 strains were cultured from 100 patients in 2016, thus a total of 120 H. pylori strains were obtained. The ethnicity of the 120 patients, based on self-assessment at the time of medical examination, was 114 multiracial (32 males, 82 females) and 6 Africans (3 males, 3 females).
A detailed description of H. pylori population structure methods has already been published in our previous study[14].
The cagA gene was determined using polymerase chain reaction PCR-based sequencing as described previously[10,17]. The absence of cagA was confirmed by the presence of cagA empty site, as previously described[18]. The cagA genotype (East Asian-type and Western-type) was confirmed by sequencing the PCR products as described previously[19]. The EPIYA-A region and CM motif from the strains were compared using the program WebLogo (version 3.7.4) (http://weblogo.threeplusone.com/).
Statistical significance was tested using Fisher’s exact test and Wilcoxon rank-sum test performed in the R package (version 3.4.0). A P value of < 0.05 was accepted as statistically significant.
We performed endoscopy for 258 dyspeptic patients. The prevalence of H. pylori infection determined via histological examination was 67.8% (175/258). Clinical diagnosis of them were 219 cases with chronic gastritis, 38 cases with peptic ulcer and 1 case with gastric cancer. From 258 dyspeptic patients, a total of 120 strains were isolated (120/258, 46.5%). Of these, 93 strains were isolated from subjects with chronic gastritis, 26 from peptic ulcer subjects, and 1 from a gastric cancer subject (Supplementary Table 1).
As we have already reported[14], the Dominican Republic strains were assigned to either hpAfrica1 (57.1%, 68/119) or hpEurope (42.9%, 51/119) (SupplementaryTable 1). From the result of the STRUCTURE run of linkage model at K = 4, the four major ancestral components reported in the previous studies were observed: Ancestral Africa1, ancestral Europe 1, ancestral Europe 2, and ancestral East Asia.
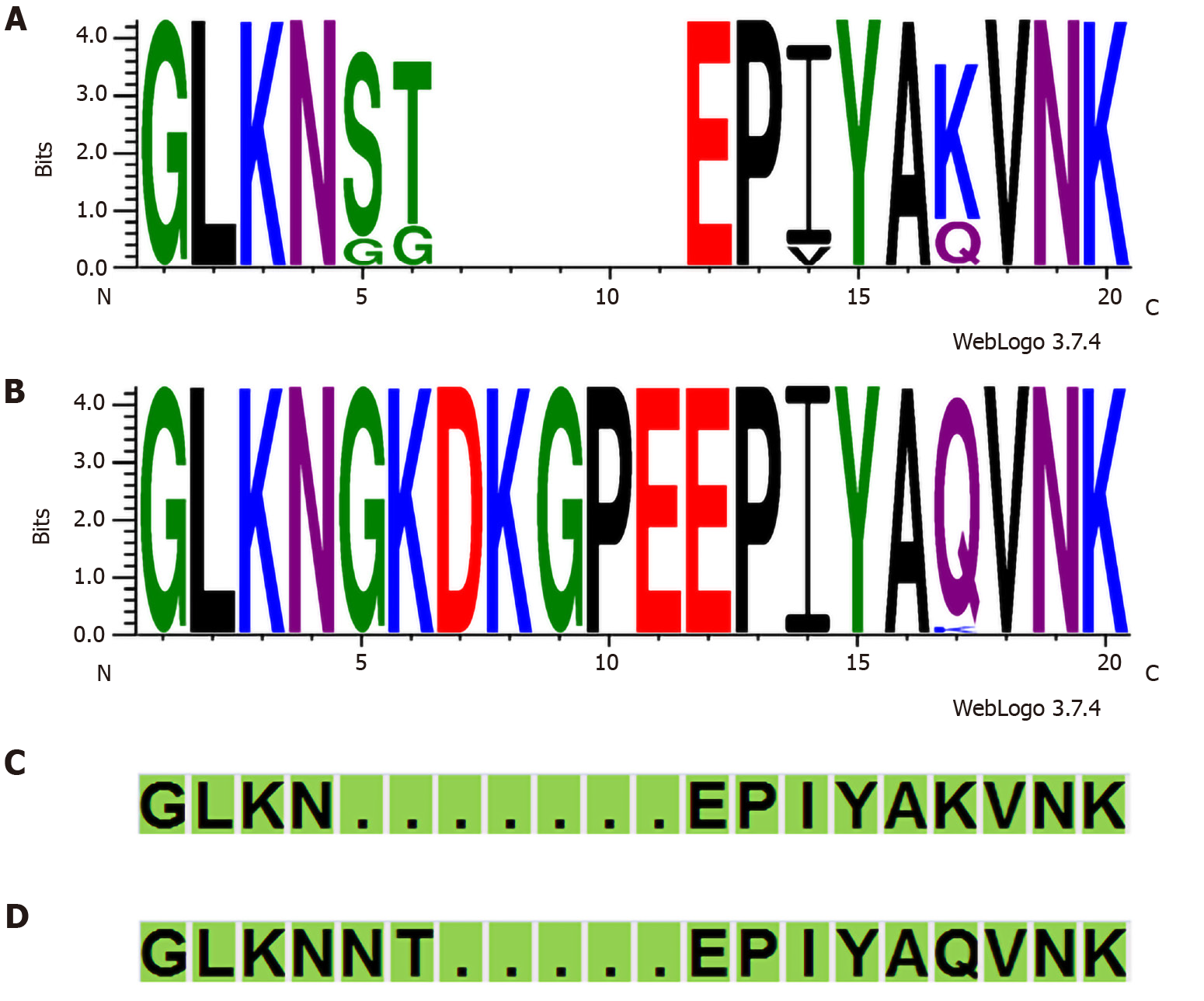
In total, 84 of 120 (70.0%) strains possessed the cagA gene. The cagA gene was not detected in one strain; however, this strain contained partial cag PAI genes, as confirmed by cag PAI empty site. Thus, we considered this strain as “cagA undetermined” (Supplementary Table 1). The prevalence of cagA was compared between the clinical outcomes of patients (Table 1). No significant difference was found between gastritis and peptic ulcer patient groups. All 84 cagA-positive strains were classified as Western CagA: ABC (n = 72), ABCC (n = 7), AC (n = 2), AABC (n = 1), AB (n = 1), and ABBC (n = 1). Interestingly, in 30 cagA-positive strains, the "GKDKGPE" motif was observed immediately upstream of the EPIYA motif in the EPIYA-A segment, which was not seen in the typical Western-type and East Asian-type CagA (Figure 1, Supplementary Table 1).
| Total | Gastritis | Peptic ulcer | Gastric cancer | |
| Number of strains | 120 | 93 | 26 | 1 |
| cagA positive | 84 (70.0%) | 66 (71.0%) | 18 (69.2%) | 0 |
| cagA negative | 35 (29.2%) | 26 (28.0%) | 8 (30.8%) | 1 (100%) |
| cagA undetermined | 1 (0.008%) | 1 (1.1%) | 0 | 0 |
In total, 172 CM motifs were found in 84 cagA-positive strains (82 for 1st CM motif, 83 for 2nd CM motif, and 7 for 3rd CM motif). These CM motifs were classified into 27 types. Twelve types were observed in the 1st CM motif, 19 types in the 2nd CM motif, and 3 types in the 3rd CM motif (Table 2). The CM motif type that had the highest number of appearances was the typical W-CM motif (FPLKRHDKVDDLSKVG) observed in strains circulating in Western countries[20], with 45 found in the 1st CM motif, 31 found in the 2nd CM motif, and 4 found in the 3rd CM motif. Interestingly, no typical E-CM motif (FPLRRSAAVNDLSKVG) was observed in strains circulating in East Asian countries. Furthermore, the Amerindian CM motif (Am-CM: xxLKRxAKVDDLxKxG and YTLKMHAGDDNLRSKVG) of Amerindian CagA was not observed[21,22]. However, "FPLRRSAKVEDLSKVG", which was found in 21 strains similar to the typical E-CM motif, was found in the 2nd and 3rd CM motifs. Amino acid sequence comparison with the GenBank BLAST data (http://www.ncbi.nlm.nih.gov/BLAST/) indicated that the "FPLRRSAKVEDLSKVG" sequence mainly matched with the sequences of strains in the African (Senegal, South Africa, and Gambia) and American (United States, Colombia, Mexico, and Cuba) continents (Supplementary Table 1). In previous studies[12,20,23], this motif was classified as a subtype of the typical E-CM motif. Interestingly, there were no East Asian and Amerind strains in the list of matches from the GenBank BLAST search. As reported in our previous study[14], many of the strains used in this study were African type (hpAfrica1) (Supplementary Table 1). Therefore, we hypothesized that "FPLRRSAKVEDLSKVG" is the typical Africa1 CM (Af1-CM) motif specific to a strain derived from Africa (hpAfrica1 strain). A previous study[11] noted that W-CM and E-CM varied at positions 4, 6, 7, 8 and 10 (FPLxRxxxVxDLSKVG). In the type classification of CM motif in this study, we focused on these five sequences, and when four or more positions of the sequences of a strain matched the typical W-CM, we designated a classification criterion that this strain belongs to W-CM. Similarly, when four or more positions of the sequences of a strain match the typical E-CM or the typical Af1-CM, we designated a classification criterion that this strain belongs to E-CM or Af1-CM, respectively. Other types of sequences not classified by these criteria were assigned as different CM (D-CM).
| First motif | Second motif | Third motif | ||||||
| Peptide sequences | Type | No. | Peptide sequences | Type | No. | Peptide sequences | Type | No. |
| FPLKRHDKVDDLSKVG | W-CM | 45 | FPLKRHDKVDDLSKVG | W-CM | 31 | FPLKRHDKVDDLSKVG | W-CM | 4 |
| FPLKRHDKVEDLSKVG | W-CM | 17 | FPLRRSAKVEDLSKVG | Af1-CM | 21 | FPLRRSAKVEDLSKVG | Af1-CM | 2 |
| FPLKRHAKVDDLSKVG | W-CM | 4 | FPLKKHAKVEDLSKVG | D-CM | 7 | FLLKRHDKVDDLSKVG | W-CM | 1 |
| FPLKKHAKVEDLSKVG | D-CM | 3 | FPLRRSAKVEDLSKAG | Af1-CM | 4 | |||
| FPLKKHDKVDDLSKVG | W-CM | 3 | FPLKKHDKVEDLSKVG | W-CM | 2 | |||
| FPLKKHDKVEDLSKVG | W-CM | 3 | FPLKRSAKVEDLSKVG | Af1-CM | 2 | |||
| FPLKKHAKVDDLSKVG | W-CM | 2 | FPLKRSAKVDDLSKVG | D-CM | 2 | |||
| FLLKRHDKVDNLSKVG | W-CM | 1 | FPLRRSAKVDDLSKVG | Af1-CM | 2 | |||
| FPLRRSDKVDDLSKVG | D-CM | 1 | FPLKRYDKVDNLSKVG | W-CM | 2 | |||
| FPLKKHAKVEDLSEVG | D-CM | 1 | FPLKRHDKVEDLSKVG | W-CM | 1 | |||
| FPLKRHDKIDDLSKVG | W-CM | 1 | FPLKKHDKVDDLSKVG | W-CM | 1 | |||
| FPLKKHAKVEDLSKAG | D-CM | 1 | FPLRRSTKVEDLSKAG | Af1-CM | 1 | |||
| Nothing (Deletion) | 2 | FPLRRSAAVDDLSKVG | E-CM | 1 | ||||
| FPLRRGAKVEDLSKVG | Af1-CM | 1 | ||||||
| FPLRKSAKVEDLSKVG | Af1-CM | 1 | ||||||
| FPLRRSDKVDNLSKVG | D-CM | 1 | ||||||
| FPSKKHAKVEDLSKVG | D-CM | 1 | ||||||
| FPFRRSDKVEDLSKVG | Af1-CM | 1 | ||||||
| FPLRRSDKVEDLSKVG | Af1-CM | 1 | ||||||
| Total | 84 | Total | 83 | Total | 7 | |||
As shown in Table 2, in the 1st CM motif, 76 strains were W-CM and 6 were D-CM. In the 2nd CM motif, 37 strains were W-CM, 34 were Af1-CM, 1 was E-CM and 11 were D-CM. In the 3rd CM motif, 5 strains were W-CM and 2 were Af1-CM. Table 3 shows the combinations of the 1st, 2nd and 3rd CM motifs. The three frequent types were W-CM:W-CM (n = 30), followed by W-CM:Af1-CM (n = 27), and W-CM:D-CM (n = 10).
| CM motif pattern | Occurrence | Frequency (%) |
| W-W | 30 | 35.7 |
| W-Af1 | 27 | 32.1 |
| W-D | 10 | 11.9 |
| D-Af1 | 5 | 6.0 |
| W-W-W | 5 | 6.0 |
| W-Af1-Af1 | 2 | 2.4 |
| -W | 2 | 2.4 |
| D-D | 1 | 1.2 |
| W-E | 1 | 1.2 |
| W- | 1 | 1.2 |
| Total | 84 |
To count the appearance frequencies of 27 kinds of CM motifs observed in the Dominican Republic in each country of the world, a GenBank BLAST search was performed. The results are shown in Supplementary Table 2. The three W-CM peptide sequences of FPLKRHDKVDDLSKVG, FPLKRHDKVEDLSKVG and FPLKKHDKVDDLSKVG had more than 100 matches in the database, therefore, it was difficult to create a detailed list. However, these three peptide sequences were contained in strains around the world, with no regional bias.
Of the 68 hpAfrica1 strains, 61 were cagA positive (92.8%), 6 were cagA negative (9.8%), and one could not determine the presence of cagA (Supplementary Table 1). In contrast, of the 51 hpEurope strains, only 23 were cagA positive (45.1%) and 28 were cagA negative (54.9%). The difference in cagA prevalence between the hpAfrica1 and the hpEurope strains was significant (P < 0.001, Fisher’s exact test). Next, to verify whether the Af1-CM motif was related to the hpAfrica1 strain, it was compared with the phylogeographical classification using MLST population structure. Among 34 strains containing Af1-CM in the 2nd CM motif, 29 were hpAfrica1 (85.3%) and 5 were hpEurope (14.7%). In contrast, of the 37 strains containing the W-CM motif in the 2nd CM motif, 23 were hpAfrica1 (62.2%) and 14 were hpEurope (37.8%). The difference in CM motif type between the hpAfrica1 and the hpEurope strains was significant (P = 0.034, Fisher’s exact test). Next, among 23 cagA-positive strains of hpEurope, the ratio of the ancestral Africa1 component, which was calculated using the STRUCTURE run of linkage model, was compared between 5 strains having Af1-CM motif and 18 strains without Af1-CM motif. From the result, the 5 strains having the Af1-CM motif had a significantly higher ancestral Africa1 component than the 18 strains without the Af1-CM motif (P = 0.030, Wilcoxon rank-sum test; Figure 2).
Harrison et al[24] hypothesized that the "KDKGPE" motif in the EPIYA-A segment of some Western CagA strains could be a unique African motif. Beltrán-anaya et al[25] have also noted the appearance of "GKDKGPE" in some Mexican strains. Amino acid sequence comparison with the GenBank BLAST data (http://www.ncbi.nlm.nih.gov/BLAST//BLAST/) indicated that the "GKDKGPEEPIY AQVNK" sequence observed in many of the strains in this study was mainly matched with the sequences of strains in the African (Senegal, Gambia, and Sudan) and American (United States, Colombia, Mexico, and Cuba) continents. To verify whether the “GKDKGPE” motif was related to the hpAfrica1 strain, it was compared with the phylogeographical classification using MLST population structure. Among 30 strains containing the “GKDKGPE” motif, 28 were hpAfrica1 and 2 were hpEurope. In contrast, after excluding the two AC-type strains, among the 52 strains without the “GKDKGPE” motif, 33 were hpAfrica1 and 19 were hpEurope. The presence of the “GKDKGPE” motif was significant between the hpAfrica1 and the hpEurope strains (P = 0.003, Fisher’s exact test). In addition, another characteristic was observed in the four amino acid sequences immediately upstream of the EPIYA motif in the EPIYA-A segment among the 30 strains containing the “GKDKGPE” motif; 29 were QVNK and 1 was KVNK (Supplementary Table 1). In contrast, among 52 strains in which the “GKDKGPE” motif was not conserved, 12 strains were QVNK, and 40 strains were KVNK. The presence of the “GKDKGPE” motif was significant between the QVNK and KVNK motifs (P < 0.001, Fisher’s exact test).
Among 37 strains containing the W-CM motif in the 2nd CM motif, the "GKDKGPE" motif was conserved in 3 strains and not in 34 strains (Supplementary Table 1). In contrast, among 34 strains containing the Af1-CM motif in the 2nd CM motif, the "GKDKGPE" motif was conserved in 22 strains and not in 12 strains. The presence of the “GKDKGPE” motif was significant between the two CM motif types in the 2nd CM motif (P < 0.001, Fisher’s exact test).
Histological scores according to the CM motif patterns are shown in Supplementary Table 3. No significant difference was observed between the two groups in all histological scores. Association of CM motif patterns with clinical outcomes is shown in Supplementary Table 4. No correlation was found between clinical results and CM motif patterns.
Sequencing data for the cagA genes of H. pylori is available under DDBJ accession numbers AB860373-AB860407, AB860409-AB860411, and LC546765-LC546810.
All 84 cagA-positive strains in this study were classified as Western CagA. The hpAfrica1 strains had a significantly higher ratio of having the cagA gene than the hpEurope strains (P < 0.001, Fisher’s exact test). This ratio was similar to that of the previous study[26]. Twenty-seven kinds of CM motifs were observed in 84 cagA-positive strains isolated in the Dominican Republic, which is higher in number than those of reports in many countries around the world[21,27-30]. In contrast, this rich variety of CM motifs in the Dominican Republic was similar to that of reports in Colombia and New York[12,23]. The CM motif type with the highest number of occurrences was the typical W-CM motif, but no typical E-CM motif or Am-CM was observed. Interestingly, a large number of CM motifs (FPLRRSAKVEDLSKVG) having a sequence similar to that of the typical E-CM motif were observed in the 2nd CM motif (also in some 3rd CM motifs), and we termed it Africa1-CM (Af1-CM). The proportion of Af1-CM in the 2nd CM motif in the hpAfrica1 strains was significantly higher than that in the hpEurope strains (P = 0.034, Fisher’s exact test). This significant difference supported our hypothesis that Af1-CM is a unique CM motif in an African-derived strain (hpAfrica1). This Af1-CM does not appear in the 1st CM motif but seems to appear in the 2nd or 3rd CM motif. Interestingly, no strain with the same sequence as the five kinds of Af1-CM subtypes was found in the African continent using the GenBank BLAST search (Supplementary Table 2). Thus, although similar in sequence to the original Af1-CM motif, these strains may not be of African origin and hence further investigation is required. No typical E-CM (FPLRRSAAVNDLSKVG) was observed in the 84 cagA-positive strains isolated in the Dominican Republic, but a subtype of E-CM (FPLRRSAAVDDLSKVG), one amino acid difference from E-CM, was found. However, the fourth Arginine and the sixth Serine were also included in the sequence of the typical Af1-CM, and the tenth was Aspartic acid instead of Asparagine specific to E-CM. Therefore, if different classification criteria are set, this motif may be classified into the Af1-CM subtype. Seven kinds of CM motifs were classified into D-CM. Since these motifs contain amino acids characteristic of W-CM and Af1-CM respectively, their origin may be a recombinant strain, or a completely different ancestral strain that has not yet been surveyed. The same reason may be assigned to why many types of subtypes of W-CM, E-CM, and Af1-CM appeared.
Olbermann et al[26] compared the CM motifs of 38 representative cagA-positive strains of the world. Among them, strain CC42C (FPLRRSAKVEDLSKVG, hpAfrica1, South Africa) and strain D3a (FPLKRSAKVEDLSKVG, hpAfrica1, Senegal) had the same motif as Af1-CM proposed by our group. The fact that the phylogeographical classification types of two strains are hpAfrica1 supports our Af1-CM hypothesis. Strain LSU2003-1 (FPLRKSAKVDDLSKVG, hpAfrica1, United States) is classified as D-CM according to the classification criteria defined in our study, but this strain also contains some amino acids characteristic of Af1-CM.
Sicinschi et al[12] reported that of the 42 Western CagA (ABC) strains collected in Colombia, 13 had the E-CM motif, of which 12 were collected from the low-risk gastric cancer region (residents of Pacific coast). According to the classification criteria defined in our study, it follows that 13 strains classified as E-CM in their paper are classified as Af1-CM. Thus, these observations in Colombia support our hypothesis. Hatakeyama[20,31] considered in a review paper that the 1st CM and 2nd CM of two strains of Western CagA (ABC) collected in Mexico were both E-CM, which is a rare case. However, this E-CM motif has the same sequence as the typical Af1-CM motif proposed by our group. It makes sense when we consider that these strains are descendants of African strains from the slave trade era. Ogorodnik et al[23] reported that 27 out of 42 Western CagA strains collected at a New York Hospital had an E-CM motif. In addition, when compared with the clinical diagnosis, the strain containing E-CM motif was less toxic than that containing W-CM motif. The main E-CM motifs of these strains are FPLRRSAKVEDLSKVG, FPLRRSAKVDDLSKVG, and FPLRKSAKVELSKVG, which are the same motifs as Af1-CM proposed by our group. The majority of H. pylori infected people treated at this New York hospital are immigrants from the Caribbean, such as Jamaica, the Dominican Republic, and Haiti. Thus, it also makes sense when we consider that these strains are descendants of African strains from the slave trade era.
Ahire et al[32] created both E-CM motif-transfected and W-CM motif-transfected strains and then compared the toxicities of these strains by testing with gastric epithelial cells. The E-CM motif used in that cloning study had the same sequence as the typical Af1-CM motif proposed by our group. This study also reported that strains with the E-CM motif (Af1-CM motif in the classification criteria in our paper) induced less hummingbird elongation than strains with the W-CM motif. Nesić et al[33] showed that the third Leucine, fifth Arginine, ninth Valine, and twelfth Leucine of the 14 amino acid CM motifs were four critical amino acid residues involved in the MARK2 kinase binding domain of CagA, via crystal structure analysis using X-ray diffraction. In the Af1-CM motif that we found in our study, these four amino acids were always conserved, similar to the E-CM and W-CM motifs, and thus it does not seem to affect this interaction.
Harrison et al[24] hypothesized that the "KDKGPE" motif in the EPIYA-A segment of some Western CagA strains could be a unique African motif. Amino acid sequence comparison with the GenBank BLAST data indicated that the "GKDKGPEEPIY
In our study, a few hpEurope strains had the Af1-CM and “GKDKGPE” motifs. H. pylori frequently undergoes recombination among unrelated strains[34,35]. Among the hpEurope strains, having the Af1-CM motif had a significantly higher ancestral Africa1 component than not having it (P = 0.030, Wilcoxon rank-sum test). This result suggests that the ancestors of the hpEurope strains that had the Af1-CM motif underwent genetic recombination with the hpAfrica1 strains in the past.
In this study, no association was found between the CM motif patterns and gastric disorders. According to previous studies[12,23], H. pylori strains with Af1-CM are less toxic compared to strains with the W-CM motif. Thus, the less toxicity of Af1-CM motif could be one reason to explain the African enigma. Twenty-seven kinds of CM motifs were observed in 84 strains of the Western-type CagA in the Dominican Republic. In the future, it is necessary to evaluate the relationship between these various CM motifs and their toxicity by conducting tests such as hummingbird phenotype in vitro.
To our knowledge, this report is the first study to elucidate the diverse characteristics of the CM motif types of the Western-type CagA strains in the Dominican Republic. However, there are several limitations in this study. Firstly, the number of strains involved was small. For a better understanding, further studies should use a large number of samples, balanced for each diagnosis. Secondly, although our samples were taken at a national reference hospital in Santo Domingo city, the capital of the Dominican Republic, our results cannot be generalized across the entire region of the Dominican Republic since the physical and cultural landscape varies by the geographic region in the Dominican Republic.
In conclusion, we found the unique African (hpAfrica1-type) CM motif in the Western-type CagA in the Dominican Republic and termed it Africa1-CM. The less toxicity of this motif could be one reason to explain the African enigma. In addition, a few hpEurope strains carrying the Af1-CM motif had a significantly higher ancestral Africa1 component than those without the Af1-CM motif. This result suggests that the ancestors of these hpEurope strains, having the Af1-CM motif, underwent genetic recombination with the hpAfrica1 strains in the past.
Helicobacter pylori (H. pylori) plays an essential role in the development of peptic ulcer disease and gastric cancer. The CagA protein produced by H. pylori is the most studied virulence factor.
Gastric cancer is one of the most common cancers.
The present study aimed to evaluate the correspondence between the CagA-multimerization (CM) motif and phylogeographical classification using multilocus sequence typing (MLST) population structure in H. pylori in the Dominican Republic, which could be helpful to elucidate the African enigma.
The Glu-Pro-Ile-Tyr-Ala (EPIYA) pattern and CM motif genotypes were determined using a polymerase chain reaction-based sequencing. The population structure was analyzed using MLST. Peptic ulcer disease and gastric cancer were identified via endoscopy, and gastric cancer was confirmed by histopathology.
Many CM motifs, which are the amino acid sequences of "FPLRRSAKVEDLSKVG", were found. This type was significantly more frequent in strains classified as hpAfrica1 using MLST analysis (P = 0.034).
We found the unique African CM motif in the Western-type CagA in the Dominican Republic and termed it Africa1-CM.
In the future, it is necessary to evaluate the relationship between these various CM motifs and their toxicity by conducting tests such as hummingbird phenotype in vitro.
We thank members of our laboratories for the discussions and comments.
Manuscript source: Invited manuscript
Corresponding Author's Membership in Professional Societies: Japanese Society of Internal Medicine, 23593; Japanese Society of Gastroenterological Endoscopy, 20030690; Japanese Society of Gastroenterology, 028098; Japanese Gastroenterology Association, 13201100.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Japan
Peer-review report’s scientific quality classification
Grade A (Excellent): A, A
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Coskun A, Isik A, Perse M, Slomiany BL S-Editor: Chen XF L-Editor: A P-Editor: Ma YJ
| 1. | Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol. 2010;7:629-641. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 500] [Cited by in RCA: 458] [Article Influence: 30.5] [Reference Citation Analysis (1)] |
| 2. | Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993;90:5791-5795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 883] [Cited by in RCA: 933] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 3. | Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648-14653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1375] [Cited by in RCA: 1392] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 4. | Higashi H, Tsutsumi R, Muto S, Sugiyama T, Azuma T, Asaka M, Hatakeyama M. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science. 2002;295:683-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 755] [Cited by in RCA: 782] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 5. | Backert S, Tegtmeyer N, Fischer W. Composition, structure and function of the Helicobacter pylori cag pathogenicity island encoded type IV secretion system. Future Microbiol. 2015;10:955-965. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 155] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 6. | Saadat I, Higashi H, Obuse C, Umeda M, Murata-Kamiya N, Saito Y, Lu H, Ohnishi N, Azuma T, Suzuki A, Ohno S, Hatakeyama M. Helicobacter pylori CagA targets PAR1/MARK kinase to disrupt epithelial cell polarity. Nature. 2007;447:330-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 351] [Cited by in RCA: 395] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 7. | Ren S, Higashi H, Lu H, Azuma T, Hatakeyama M. Structural basis and functional consequence of Helicobacter pylori CagA multimerization in cells. J Biol Chem. 2006;281:32344-32352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 103] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 8. | Suzuki M, Mimuro H, Kiga K, Fukumatsu M, Ishijima N, Morikawa H, Nagai S, Koyasu S, Gilman RH, Kersulyte D, Berg DE, Sasakawa C. Helicobacter pylori CagA phosphorylation-independent function in epithelial proliferation and inflammation. Cell Host Microbe. 2009;5:23-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 266] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 9. | Higashi H, Tsutsumi R, Fujita A, Yamazaki S, Asaka M, Azuma T, Hatakeyama M. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc Natl Acad Sci USA. 2002;99:14428-14433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 434] [Cited by in RCA: 456] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 10. | Matsunari O, Shiota S, Suzuki R, Watada M, Kinjo N, Murakami K, Fujioka T, Kinjo F, Yamaoka Y. Association between Helicobacter pylori virulence factors and gastroduodenal diseases in Okinawa, Japan. J Clin Microbiol. 2012;50:876-883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 11. | Lu HS, Saito Y, Umeda M, Murata-Kamiya N, Zhang HM, Higashi H, Hatakeyama M. Structural and functional diversity in the PAR1b/MARK2-binding region of Helicobacter pylori CagA. Cancer Sci. 2008;99:2004-2011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 37] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 12. | Sicinschi LA, Correa P, Peek RM, Camargo MC, Piazuelo MB, Romero-Gallo J, Hobbs SS, Krishna U, Delgado A, Mera R, Bravo LE, Schneider BG. CagA C-terminal variations in Helicobacter pylori strains from Colombian patients with gastric precancerous lesions. Clin Microbiol Infect. 2010;16:369-378. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 76] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 13. | Shiota S, Cruz M, Abreu JAJ, Mitsui T, Terao H, Disla M, Iwatani S, Nagashima H, Matsuda M, Uchida T, Tronilo L, Rodríguez E, Yamaoka Y. Virulence genes of Helicobacter pylori in the Dominican Republic. J Med Microbiol. 2014;63:1189-1196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 14. | Ono T, Cruz M, Jiménez Abreu JA, Nagashima H, Subsomwong P, Hosking C, Shiota S, Suzuki R, Yamaoka Y. Comparative study between Helicobacter pylori and host human genetics in the Dominican Republic. BMC Evol Biol. 2019;19:197. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 15. | Central Intelligence Agency. The World Factbook. 2020. Available from: https://www.cia.gov/Library/publications/the-world-factbook/geos/dr.html. |
| 16. | Rugge M, Meggio A, Pennelli G, Piscioli F, Giacomelli L, De Pretis G, Graham DY. Gastritis staging in clinical practice: the OLGA staging system. Gut. 2007;56:631-636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 348] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 17. | Yamaoka Y, Osato MS, Sepulveda AR, Gutierrez O, Figura N, Kim JG, Kodama T, Kashima K, Graham DY. Molecular epidemiology of Helicobacter pylori: separation of H. pylori from East Asian and non-Asian countries. Epidemiol Infect. 2000;124:91-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 90] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 18. | Mukhopadhyay AK, Kersulyte D, Jeong JY, Datta S, Ito Y, Chowdhury A, Chowdhury S, Santra A, Bhattacharya SK, Azuma T, Nair GB, Berg DE. Distinctiveness of genotypes of Helicobacter pylori in Calcutta, India. J Bacteriol. 2000;182:3219-3227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 181] [Article Influence: 7.2] [Reference Citation Analysis (1)] |
| 19. | Xia Y, Yamaoka Y, Zhu Q, Matha I, Gao X. A comprehensive sequence and disease correlation analyses for the C-terminal region of CagA protein of Helicobacter pylori. PLoS One. 2009;4:e7736. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 77] [Article Influence: 4.8] [Reference Citation Analysis (1)] |
| 20. | Hatakeyama M. Anthropological and clinical implications for the structural diversity of the Helicobacter pylori CagA oncoprotein. Cancer Sci. 2011;102:36-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 21. | Suzuki M, Kiga K, Kersulyte D, Cok J, Hooper CC, Mimuro H, Sanada T, Suzuki S, Oyama M, Kozuka-Hata H, Kamiya S, Zou QM, Gilman RH, Berg DE, Sasakawa C. Attenuated CagA oncoprotein in Helicobacter pylori from Amerindians in Peruvian Amazon. J Biol Chem. 2011;286:29964-29972. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 22. | Kersulyte D, Kalia A, Gilman RH, Mendez M, Herrera P, Cabrera L, Velapatiño B, Balqui J, Paredes Puente de la Vega F, Rodriguez Ulloa CA, Cok J, Hooper CC, Dailide G, Tamma S, Berg DE. Helicobacter pylori from Peruvian amerindians: traces of human migrations in strains from remote Amazon, and genome sequence of an Amerind strain. PLoS One. 2010;5:e15076. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 66] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 23. | Ogorodnik E, Raffaniello RD. Analysis of the 3'-variable region of the cagA gene from Helicobacter pylori strains infecting patients at New York City hospitals. Microb Pathog. 2013;56:29-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 24. | Harrison U, Fowora MA, Seriki AT, Loell E, Mueller S, Ugo-Ijeh M, Onyekwere CA, Lesi OA, Otegbayo JA, Akere A, Ndububa DA, Adekanle O, Anomneze E, Abdulkareem FB, Adeleye IA, Crispin A, Rieder G, Fischer W, Smith SI, Haas R. Helicobacter pylori strains from a Nigerian cohort show divergent antibiotic resistance rates and a uniform pathogenicity profile. PLoS One. 2017;12:e0176454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 25. | Beltrán-Anaya FO, Poblete TM, Román-Román A, Reyes S, de Sampedro J, Peralta-Zaragoza O, Rodríguez MÁ, del Moral-Hernández O, Illades-Aguiar B, Fernández-Tilapa G. The EPIYA-ABCC motif pattern in CagA of Helicobacter pylori is associated with peptic ulcer and gastric cancer in Mexican population. BMC Gastroenterol. 2014;14:223. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 26. | Olbermann P, Josenhans C, Moodley Y, Uhr M, Stamer C, Vauterin M, Suerbaum S, Achtman M, Linz B. A global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island. PLoS Genet. 2010;6:e1001069. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 155] [Cited by in RCA: 176] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 27. | El Khadir M, Alaoui Boukhris S, Benajah DA, Ibrahimi SA, Chbani L, Bouguenouch L, El Rhazi K, El Abkari M, Nejjari C, Mahmoud M, Bennani B. Helicobacter pylori CagA EPIYA-C motifs and gastric diseases in Moroccan patients. Infect Genet Evol. 2018;66:120-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 28. | Myint T, Miftahussurur M, Vilaichone RK, Ni N, Aye TT, Subsomwong P, Uchida T, Mahachai V, Yamaoka Y. Characterizing Helicobacter pylori cagA in Myanmar. Gut Liver. 2018;12:51-57. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 29. | Subsomwong P, Miftahussurur M, Uchida T, Vilaichone RK, Ratanachu-Ek T, Mahachai V, Yamaoka Y. Prevalence, risk factors, and virulence genes of Helicobacter pylori among dyspeptic patients in two different gastric cancer risk regions of Thailand. PLoS One. 2017;12:e0187113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 30. | Aftab H, Miftahussurur M, Subsomwong P, Ahmed F, Khan AKA, Matsumoto T, Suzuki R, Yamaoka Y. Two populations of less-virulent Helicobacter pylori genotypes in Bangladesh. PLoS One. 2017;12:e0182947. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 31. | Reyes-Leon A, Atherton JC, Argent RH, Puente JL, Torres J. Heterogeneity in the activity of Mexican Helicobacter pylori strains in gastric epithelial cells and its association with diversity in the cagA gene. Infect Immun. 2007;75:3445-3454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 54] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 32. | Ahire D, Alston T, Raffaniello R. Variations in the multimerization region of the Helicobacter pylori cytotoxin CagA affect virulence. Oncol Lett. 2017;13:1444-1450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 33. | Nesić D, Miller MC, Quinkert ZT, Stein M, Chait BT, Stebbins CE. Helicobacter pylori CagA inhibits PAR1-MARK family kinases by mimicking host substrates. Nat Struct Mol Biol. 2010;17:130-132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 121] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 34. | Achtman M, Azuma T, Berg DE, Ito Y, Morelli G, Pan ZJ, Suerbaum S, Thompson SA, van der Ende A, van Doorn LJ. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol. 1999;32:459-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 270] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 35. | Wirth T, Meyer A, Achtman M. Deciphering host migrations and origins by means of their microbes. Mol Ecol. 2005;14:3289-3306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 70] [Article Influence: 3.7] [Reference Citation Analysis (0)] |