Published online Nov 7, 2019. doi: 10.3748/wjg.v25.i41.6273
Peer-review started: August 19, 2019
First decision: September 10, 2019
Revised: September 23, 2019
Accepted: October 17, 2019
Article in press: October 17, 2019
Published online: November 7, 2019
Processing time: 79 Days and 22.6 Hours
Increasing evidence demonstrates that by acting as microRNA sponges modulating gene expression at the transcriptional or post-transcriptional level, circular RNAs (circRNAs) participate in the pathogenesis of a variety of diseases and are considered ideal biomarkers of human disease.
To examine the expression of circRNA_103516 in inflammatory bowel disease (IBD) and its associations with clinical phenotypes and inflammatory cytokines.
Peripheral blood mononuclear cells (PBMCs) were obtained from patients with IBD, healthy controls (HCs), and patient controls (PCs). Expression of circRNA_103516 and hsa-miR-19b-1-5p was assessed by quantitative reverse transcription-polymerase chain reaction. Crohn's disease activity index (CDAI), Mayo score, C-reactive protein (CRP) level, and erythrocyte sedimentation rate (ESR) were measured. To assess the inflammatory cytokines tumour necrosis factor α (TNF-α), interferon-γ (IFN-γ), and interleukin-10 (IL-10), blood samples were analysed by flow cytometry.
Ninety Crohn’s disease (CD) and 90 ulcerative colitis (UC) patients, 80 HCs, and 35 PCs were included in the study. CircRNA_103516 was upregulated in CD and UC patients compared with HCs and PCs (P < 0.05). The area under the curve of circRNA_103516 for diagnosing CD and UC was 0.790 and 0.687, respectively. In addition, circRNA_103516 levels were increased in active CD and UC compared with remittent groups (P = 0.027, P = 0.045). Furthermore, in CD, circRNA_103516 correlated positively with CDAI (P < 0.001), CRP (P < 0.001), ESR (P < 0.001), TNFα (P < 0.001), and IFN-γ (P < 0.001) and negatively correlated with IL-10 (P = 0.006). In UC patients, circRNA_103516 correlated with Mayo score (P < 0.001), CRP (P < 0.001), ESR (P < 0.001), TNFα (P < 0.001), IFN-γ (P =0.011), and IL-10 (P = 0.002). Additionally, circRNA_103516 correlated positively with stricturing (P = 0.018) and penetrating (P = 0.031) behaviour. Moreover, hsa-miR-19b-1-5p correlated negatively with circRNA_103516 in CD.
CircRNA_103516 levels in PBMCs can be considered an ideal candidate biomarker for diagnosing IBD. Dysregulation of circRNA_103516 may participate in the molecular mechanism of IBD through hsa-miR-19b-1-5p sponging.
Core tip: The study aimed to assess circular RNA (circRNA)_103516 in peripheral blood mononuclear cells of inflammatory bowel disease (IBD) patients and evaluate its potential as a biomarker in Crohn’s disease and ulcerative colitis patients with regard to clinical phenotype and inflammatory cytokines. Our results showed that PBMC circRNA_103516 levels may be considered an ideal candidate biomarker for the diagnosis of IBD and have the ability to predict clinical outcomes in IBD. Dysregulation of circRNA_103516 may participate in the molecular mechanism of IBD through hsa-miR-19b-1-5p sponging.
- Citation: Ye YL, Yin J, Hu T, Zhang LP, Wu LY, Pang Z. Increased circulating circular RNA_103516 is a novel biomarker for inflammatory bowel disease in adult patients. World J Gastroenterol 2019; 25(41): 6273-6288
- URL: https://www.wjgnet.com/1007-9327/full/v25/i41/6273.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i41.6273
Inflammatory bowel disease (IBD), including ulcerative colitis (UC) and Crohn’s disease (CD), comprises chronic inflammatory disorders of the gastrointestinal tract. IBD pathogenesis is believed to involve a complex interplay among immunology, genetic predisposition, and environmental risk factors[1-3]. Due to the characteristic of repeated recurrence, the disease activity of IBD must be assessed and monitored repeatedly. However, as most conventional detection methods are invasive, current detection methods are not suitable for repeated clinical applications. To overcome this limitation, serological biomarkers may constitute an optimal alternative choice for evaluating and screening disease activity in IBD.
Circular RNAs (circRNAs) are covalently closed continuous single-stranded RNA molecules that have advantageous properties because their circular structure enables rolling circle RNA replication and producing multiple genomic copies once a single initiation event occurs[4,5]. However, the characteristics of circRNAs are much less clear than those of microRNAs (miRNAs) and long noncoding RNAs (lncRNAs). Nonetheless, research on their functions has recently emerged, and increasing evidence demonstrates that circRNAs can modulate gene expression at the transcriptional or post-transcriptional level by sponging miRNAs or by interacting with other molecules[6]. Furthermore, circRNAs are evolutionally conserved, and their expression is relatively stable in the cytoplasm; these features indicate that circRNAs may be ideal biomarkers in human disease[7]. Although much remains to be revealed regarding circRNA biology and gene regulatory mechanisms, studies have elucidated their function in a variety of cancers[8], cardiovascular disease[9], autoimmune disease[10], and nervous system disorders[11], among others.
To date, little is known about the relationships between circRNAs and IBD. Our previous research based on microarray analysis identified 155 upregulated circRNAs and 229 downregulated circRNAs in peripheral blood mononuclear cells (PBMCs) from CD patients compared with healthy controls (HCs). Moreover, some aberrantly expressed circRNAs were chosen for evaluation of their potential use in the diagnosis of CD[12].
In this study, we focused on circRNA_103516, which is located at chr3: 171969049-172028671 and spliced from FNDC3B. It was reported that FNDC3B might play a role in the epithelial-to-mesenchymal transition and activates several cancer pathways, including phosphoinositide 3-kinase/Akt, retinoblastoma 1, and transforming growth factor (TGF) β signalling[13]. Our bioinformatics analysis showed that hsa_circRNA_103516 is predicted to harbour hsa-miR-147b, hsa-miR-19b-1-5p, hsa-miR-134-3p, hsa-miR-576-5p, and hsa-miR-493-5p. Among them, miR-19b was found to be decreased in the serum and intestinal tissue of IBD patients[14]. Thus, in the present study, we explored the link between circRNA_103516 and hsa-miR-19b-1-5p in PBMCs from IBD patients and determined its possible correlations with the clinical phenotypes of CD and UC.
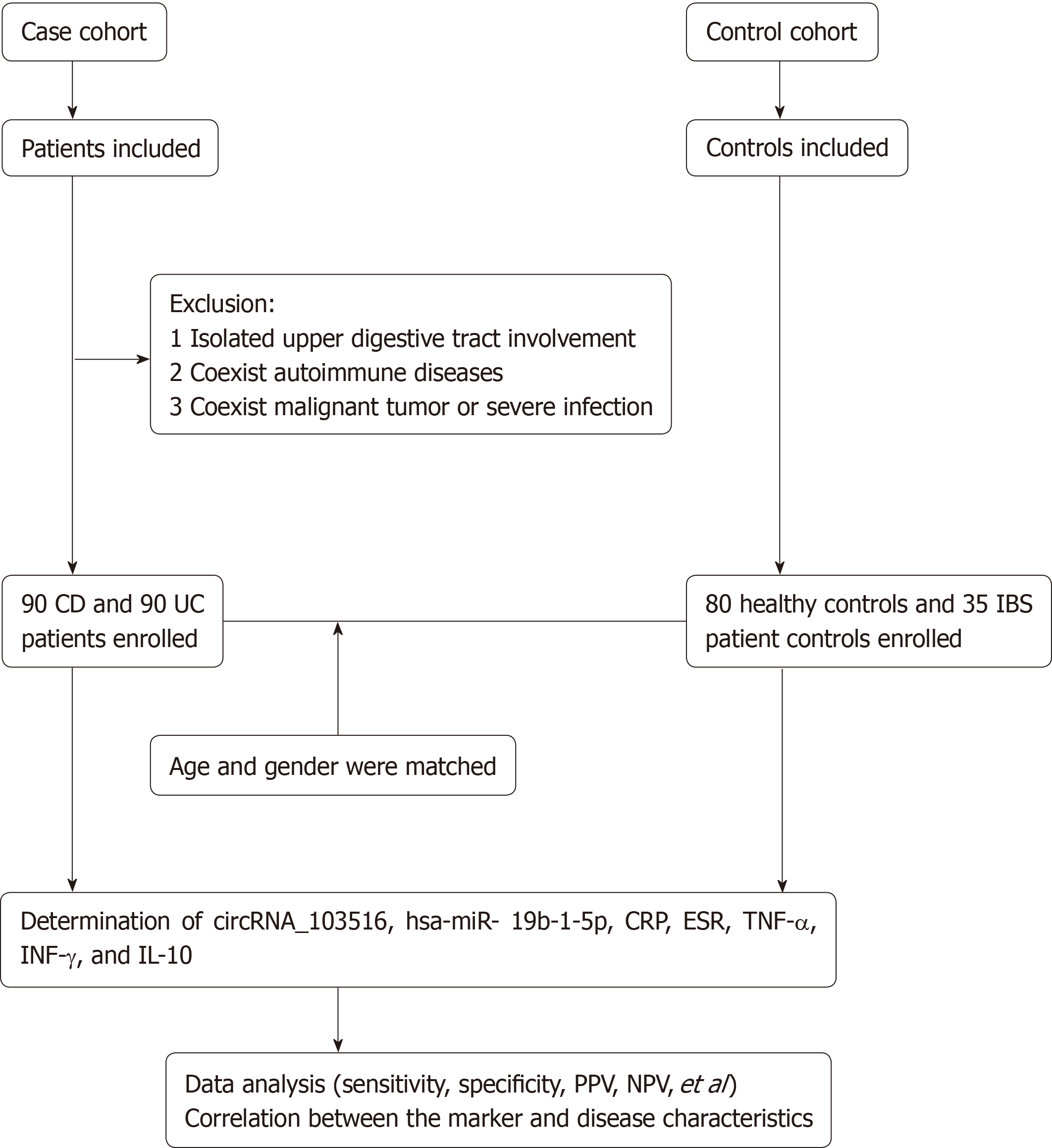
Between January 2018 and December 2018, 180 IBD patients (90 with CD and 90 with UC) were prospectively recruited from the Department of Gastroenterology of the North District of the Affiliated Suzhou Hospital of Nanjing Medical University (Jiangsu, China). Eighty HCs and 35 patient controls (PCs) were also included. The age and sex of all groups were matched. The inclusion criteria were: (1) A diagnosis of IBD established based on clinical manifestations, radiological findings, and endoscopic and histological criteria; (2) Age from 17 to 75 years old; and (3) Demographic and clinical information collected by experienced clinicians reviewing and completing medical questionnaires. Patients with irritable bowel syndrome were used as the PCs. The exclusion criteria were as follows: (1) Isolated upper digestive tract involvement; (2) Other coexisting autoimmune diseases, including systemic lupus erythematosus, rheumatoid arthritis, and asthma; and (3) A history of malignant tumour or severe infection (Figure 1). Disease activity was identified according to the CD activity index (CDAI) for CD and the Mayo score for UC[15,16]. Active IBD was defined as CDAI above 150 points or Mayo clinical score above 2. The sites and behaviour of the disease were defined according to the Montreal classification[17]. For assessing circRNA_103516, hsa-miR-19b-1-5p, C-reactive protein (CRP), erythrocyte sedimentation rate (ESR), TNF-α, interferon-γ (IFN-γ), and interleukin-10 (IL-10), blood samples were collected into ethylenediamine tetraacetic acid tubes for testing within one week before or after endoscopy.
The present study was approved by the Ethics Committees of the Affiliated Suzhou Hospital of Nanjing Medical University (Jiangsu, China). All IBD patients and control subjects signed an informed consent form, in accordance with the relevant guidelines and regulations.
PBMCs were immediately separated after blood sample collection from each donor according to the manufacturer’s protocol (GE Healthcare, Uppsala, Sweden). Two millilitres of blood diluted in 2 mL of saline solution was layered onto 4 mL of Ficoll-Paque PLUS. After centrifugation for 30 min at 400 g at room temperature, the interlayer was collected by washing twice with the same volume of saline solution. The precipitate was gathered by centrifuging for 15 min at 90 g at room temperature. PBMCs were then frozen at -80 °C. TRIzol reagent (Invitrogen, Carlsbad CA, United States) was utilized for total RNA extraction from the PBMCs.
Total RNA was reverse transcribed using a PrimerScript Realtime Reagent Kit (Takara Bio Inc., TaKaRa, Shiga, Japan), and expression of circRNA_103516 was quantitated with TB Green™ Premix Ex Taq™ II (Tli RNaseH Plus; TaKaRa, Shiga, Japan) and a LightCycler 480II real-time PCR system (Roche, Rotkreuz, Switzerland). The 2-∆∆Ct method was employed to analyse the data. β-actin was used as an internal reference. The primer sequences are presented in Table 1. In total, the cycling parameters for PCR were 30 s for 95 °C, followed by 40 cycles of denaturation at 95 °C for 5 s and annealing and extension at 60 °C for 30 s.
| Gene | Primer sequence | Produce size (bp) |
| β-actin | F: 5'‑GTGGCCGAGGACTTTGATTG 3' | 73 |
| R: 5'CCTGTAACAACGCATCTCATATT-3' | ||
| circRNA_103516 | F: 5'-GCACCAATTGACAACGGTTC-3' | 123 |
| R: 5'-CTGGTCTTCTCGGGTGATGT-3' |
According to the manufacturer’s instructions, TNFα, IFN-γ, and IL-10 levels in plasma samples from IBD patients were measured using flow cytometry (FCM) kits (Hangzhou Cellgene Biotechco, LTD, China).
TargetScan (http://www.targetscan.org/) and miRanda (http://www. microrna.org/) were used to predict circRNA/miRNA interactions. Differential expression of circRNA_103516, as identified by qRT-PCR, was annotated in detail with the circRNA/miRNA interactions. Additionally, the sequences of MREs and predicted miRNA targets were examined.
Expression of miR-19b-1-5p (predicted from annotations) in PBMCs from the 90 patients with CD and 90 patients with UC was detected using a Hairpin-itTM qRT-PCR miRNA Kit (GenePharma, LTD, China). U6 was used as an internal reference. The primer sequences used for hsa-miR-19b-1-5p are: 5’-UGUGCAAAUCCA UGCAAAACUG-3’ (forward) and 5’-GCTCACTGCAACCTCCTCCTCC-3’ (reverse). The primer sequences used for U6 are 5’-GCTTCGGCAGCACATA-TACTAAAAT-3’ (forward) and 5’-CGCT-TCACGAATTTGCGTGTCAT-3’ (reverse). The PCR conditions included predenaturation at 95 °C for 3 min, followed by 40 cycles of denaturation at 95 °C for 12 s and annealing and elongation at 62 °C for 40 s. The 2-∆∆Ct method was used to analyse the data.
The results are reported as the mean ± standard deviation or median (25%75%); assays were performed in triplicate. The unpaired t-test or Mann-Whitney U-test was applied to compare continuous variables for two groups. Multiple comparisons were assessed by one-way analysis of variance or the χ2 test. Spearman’s analysis was performed to determine linear correlation in different groups. Receiver operating characteristic curve analysis was employed to evaluate the clinical diagnostic value of candidate circRNAs. Logistic regression was used to identify risk factors. A P value < 0.05 was considered statistically significant. The statistical analyses were performed using the package SPSS 19.0 (SPSS Inc., IBM, United States) for windows and GraphPad Prism 7.04 (GraphPad Software, San Diego, CA, United States).
The demographic and clinical data of the IBD patients and health controls and PCs are listed in Table 2. There were 90 CD patients [48 males (53.3%), mean age 39.94 years], 90 UC patients [38 males (42.2%), mean age 41.69 years], 80 HCs [46 males (57.5%), mean age 37.64 years], and 35 PCs [19 males (54.3%), mean age 42.40 years]. Age and sex were not significantly different among the four groups (P > 0.05). CD and UC activity was classified as remission, mild, moderate, or severe according to CDAI and the Mayo score. Laboratory test results (CRP, ESR, TNF-α, INF-γ, and IL-10) and disease activity are shown in Table 3.
| Characteristic | CD (n = 90) | UC (n = 90) | HCs (n = 80) | PCs (n = 35) |
| Male/female | 48/42 | 38/52 | 46/34 | 19/16 |
| Mean age (yr) | 39.94 ± 11.80 | 41.69 ± 12.40 | 37.64 ± 9.30 | 42.40 ± 10.20 |
| Range (yr) | 17-67 | 20-73 | 20-72 | 18-70 |
| Tobacco smoking (n) | ||||
| Never | 60 | 67 | 50 | 20 |
| Past or current use | 30 | 23 | 30 | 15 |
| Disease duration (yr) | 5.1 ± 3.1 | 5.1 ± 3.4 | ||
| Disease location: CD, n (%) | ||||
| L1 | 37 (41.1) | |||
| L2 | 20 (22.2) | |||
| L3 | 33 (36.7) | |||
| Disease activity: CDAI, n (%) | ||||
| Remission | 16 (17.8) | |||
| Mild | 19 (21.1) | |||
| Moderate | 40 (44.4) | |||
| Severe | 15 (16.7) | |||
| Disease behavior:CD, n (%) | ||||
| B1 | 39 (43.3) | |||
| B2 | 33 (36.7) | |||
| B3 | 18 (20.0) | |||
| Disease location: UC, n (%) | ||||
| E1 | 36 (40.0) | |||
| E2 | 29 (32.2) | |||
| E3 | 25 (27.8) | |||
| Disease severity, n (%) | ||||
| Remission | 14 (15.6) | |||
| Mild | 30 (33.3) | |||
| Moderate | 31 (34.4) | |||
| Severe | 15 (16.7) | |||
| Medications, n (%) | ||||
| 5-ASA | 80 (88.9) | 87 (96.7) | ||
| Corticosteroids | 50 (11.7) | 41 (45.5) | ||
| Immunosuppressants | 28 (31.1) | 14 (15.6) | ||
| Anti-TNF-α | 13 (14.4) | 4 (4.4) | ||
| Surgery | 4 (4.4) | 1 (1.1) | ||
| Parameter | CD (n = 90) | UC (n = 90) | HCs (n = 80) | PCs (n = 35) |
| CRP (mg/L) | 79.95 (18.47-121.22) | 58.10 (16.43-123.67) | 3.49 (3.30-6.09) | 5.73 (3.84-7.94) |
| ESR (mm/H) | 38.50 (17.75-67.00) | 30.00 (15.00-58.00) | 12.02 (4.87-14.25) | 17.32 (3.87-26.85) |
| TNF-α (pg/mL) | 6.83 (3.89-12.26) | 6.70 (3.45-9.45) | -- | |
| INF-γ (pg/mL) | 9.29 (5.77-12.35) | 6.70 (4.4-10.65) | -- | |
| IL-10 (pg/mL) | 10.99 (6.21-19.27) | 11.3 (6.66-25.08) | -- | |
| CDAI score | 270.15 (170.60-376.58) | -- | -- | |
| Mayo score | -- | 5.00 (3.00-10.00) | -- |
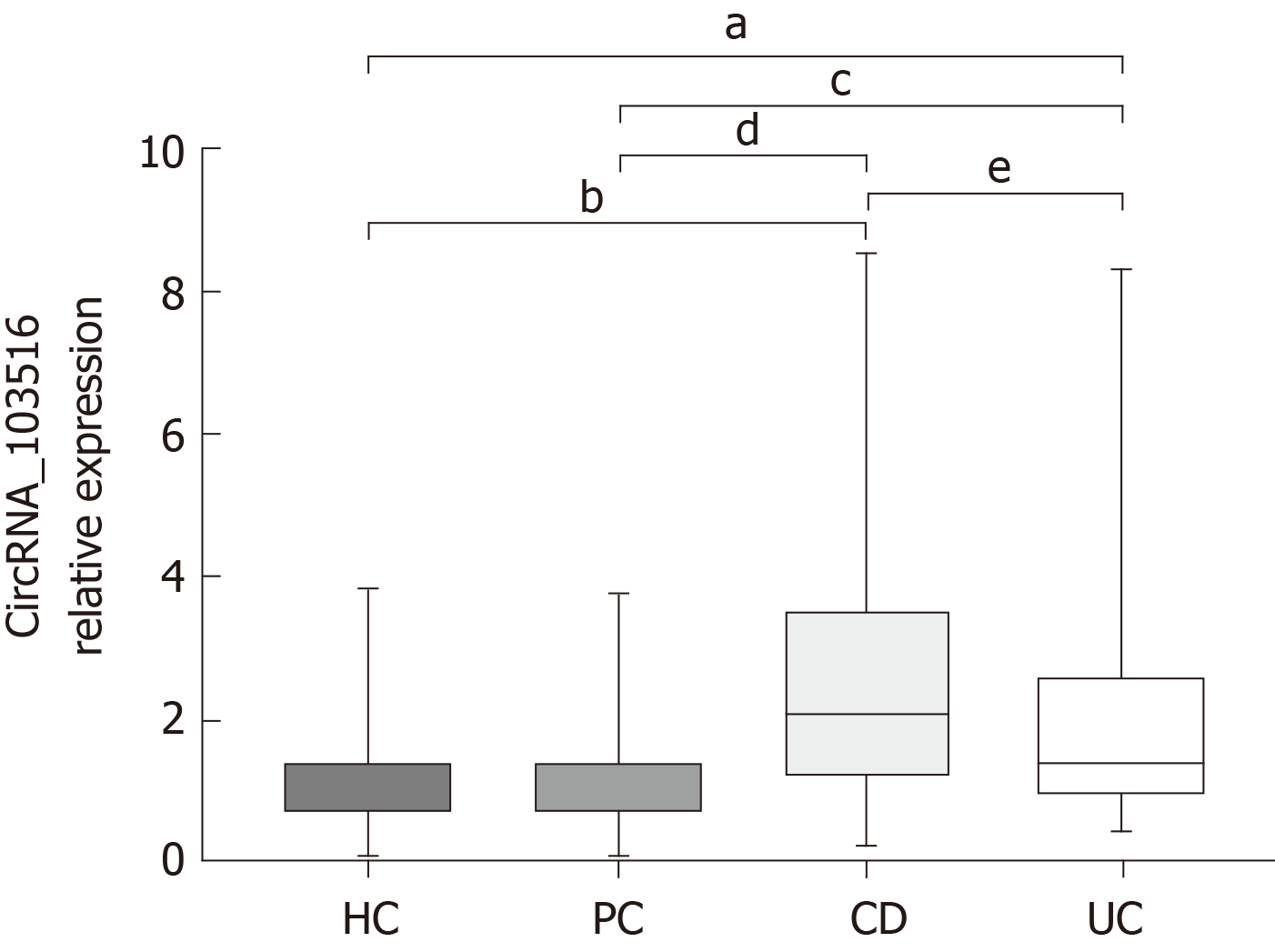
Expression of circRNA_103516 in PBMCs was increased in CD patients [2.085 (1.1953.510)] compared with that in UC patients [1.385 (0.9252.599), P = 0.003], HCs [0.901 (0.7241.376), P < 0.001], and PCs [0.901 (0.7421.375), P < 0.001]. The expression level of circRNA_103516 was higher in PBMCs from the UC group compared with those of the HC (P = 0.004) and PC (P = 0.025) groups. However, there was no significant difference between the HC and PC groups (P = 0.998) (Figure 2).
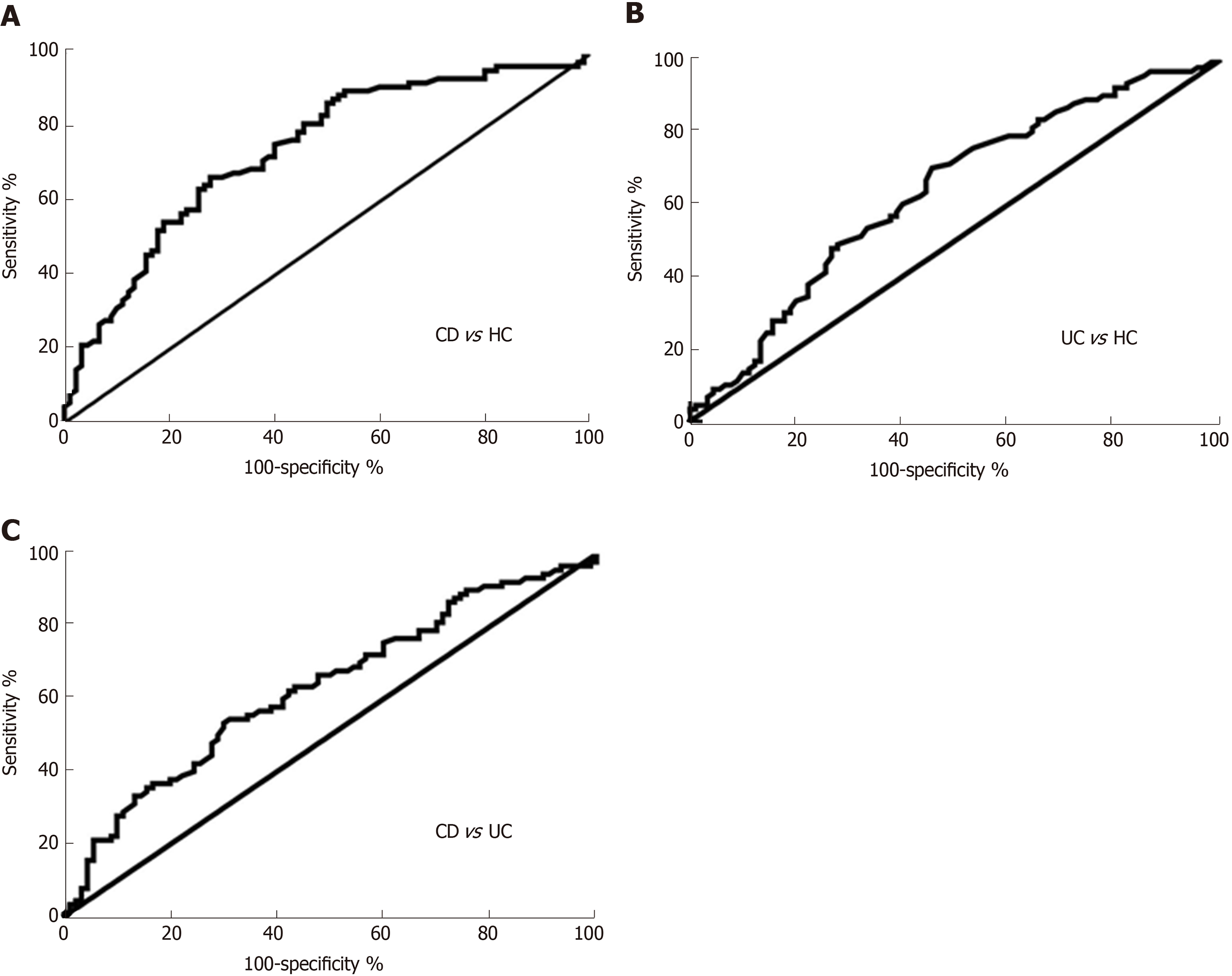
The diagnostic value of circRNA_103516 in differentiating the CD group from the HC group [area under the curve (AUC) = 0.790, 95% confidence interval (CI): 0.722-0.857] and the UC group (AUC = 0.631, 95%CI: 0.550-0.712) was also examined (Figure 3A and C, Table 4). Overall, circRNA_103516 may be considered a predictive factor in differentiating UC from HC (AUC = 0.687, 95%CI: 0.608-0.767) (Figure 3B, Table 4). The level of circRNA_103516 was considered positive at a cutoff value > 1.412 for CD and > 1.151 for UC (Table 4).
| Group | AUC (95%CI) | P value | Cutoff value | Sensitivity (95%CI) | Specificity (95%CI) | PPV | NPV | LR + | LR - | Diagnostic accuracy |
| CD vs HC | 0.790 (0.722-0.857) | < 0.001 | 1.412 | 66.67 (55.95%-76.26%) | 78.75 (68.17%-87.11%) | 77.63% | 67.02% | 3.137 | 0.423 | 71.76% |
| UC vs HC | 0.687 (0.608-0.767) | < 0.001 | 1.151 | 66.67 (55.95%-76.26%) | 62.50 (50.96%-73.08%) | 66.67% | 62.50% | 1.778 | 0.533 | 64.71% |
| CD vs UC | 0.631 (0.550-0.712) | 0.002 | 1.963 | 54.44 (43.6%- 64.98%) | 68.89 (58.26% to 78.23%) | 55.05% | 55.75% | 1.750 | 0.661 | 59.41% |
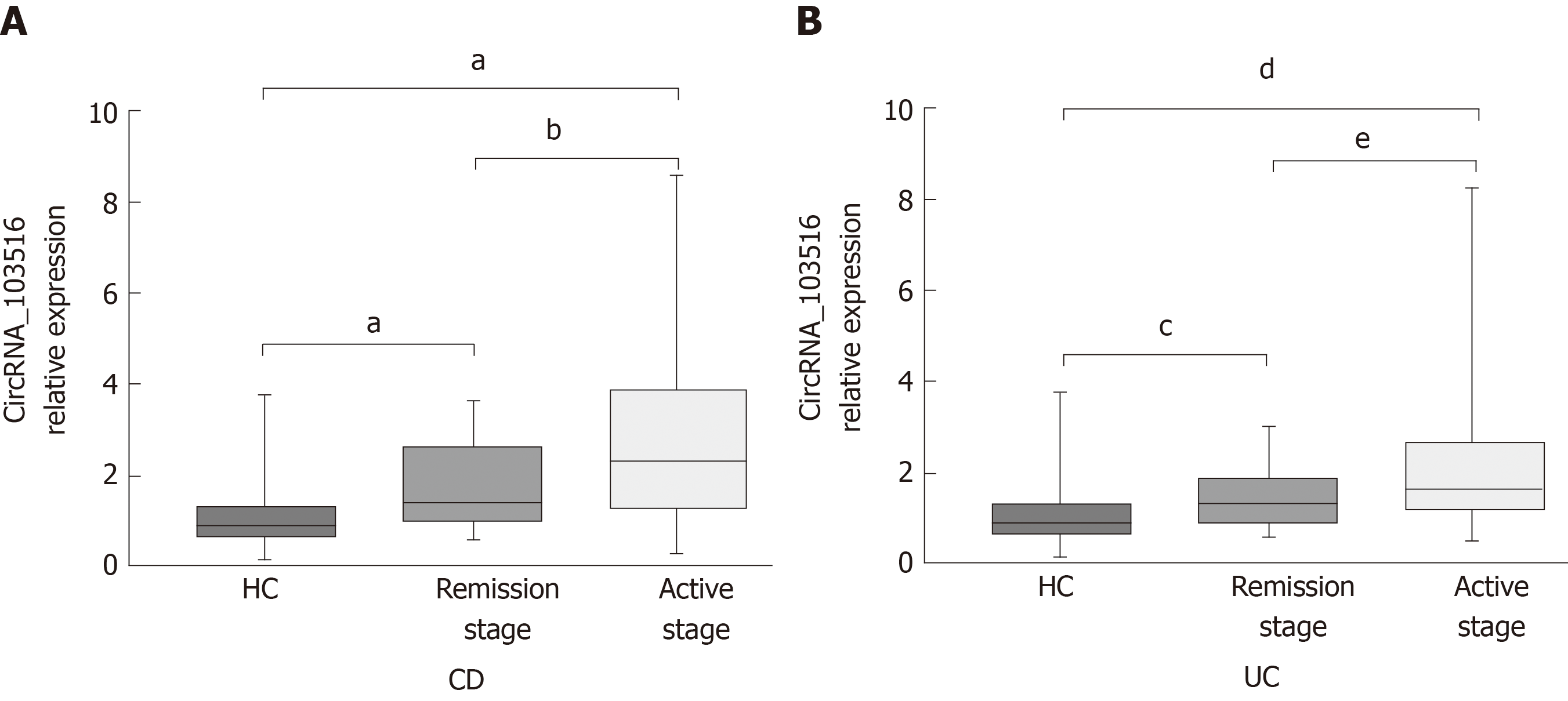
The expression level of circRNA_103516 was markedly increased, with values 2.614-fold and 1.953-fold higher in PBMCs from active CD and active UC, respectively (P < 0.001 and P < 0.001, respectively). However, expression was only 1.639-fold and 1.319-fold higher in CD and UC patients who were in remission, respectively (P < 0.001 and P = 0.024, respectively) (Figure 4A), and circRNA_103516 levels in active CD and UC were higher than those in remittent CD and UC (P = 0.027, P = 0.045) (Figure 4A and B).
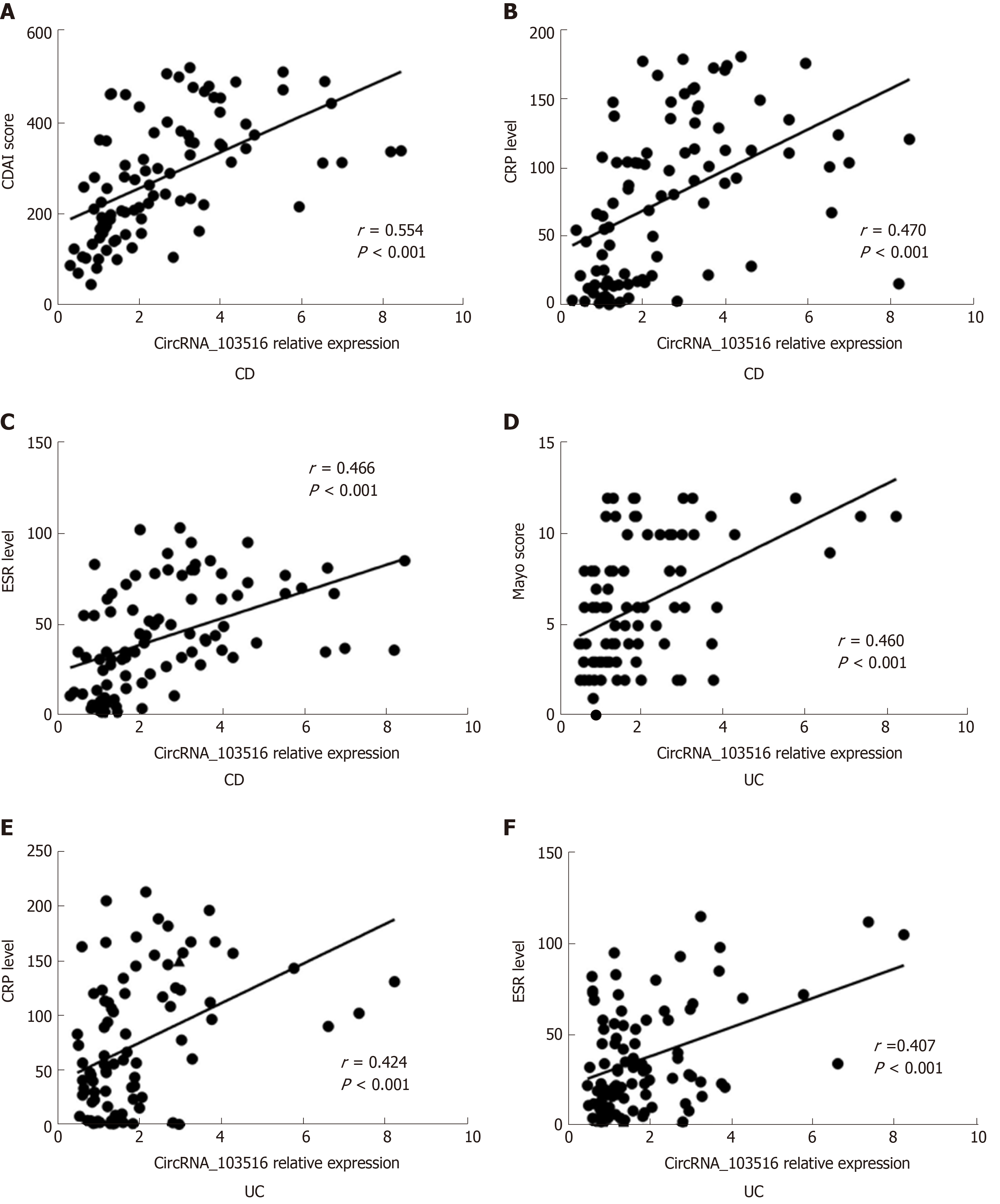
Additionally, circRNA_103516 levels exhibited a positive correlation with disease activity and laboratory tests. In CD patients, circRNA_103516 correlated positively with CDAI (r = 0.554, P < 0.001), CRP (r = 0.470, P < 0.001) and ESR (r = 0.466, P < 0.001) (Figure 5A-C); in UC patients, circRNA_103516 correlated with the Mayo score (r = 0.460, P < 0.001), CRP (r = 0.424, P < 0.001), and ESR (r = 0.407, P < 0.001) (Figure 5D and F).
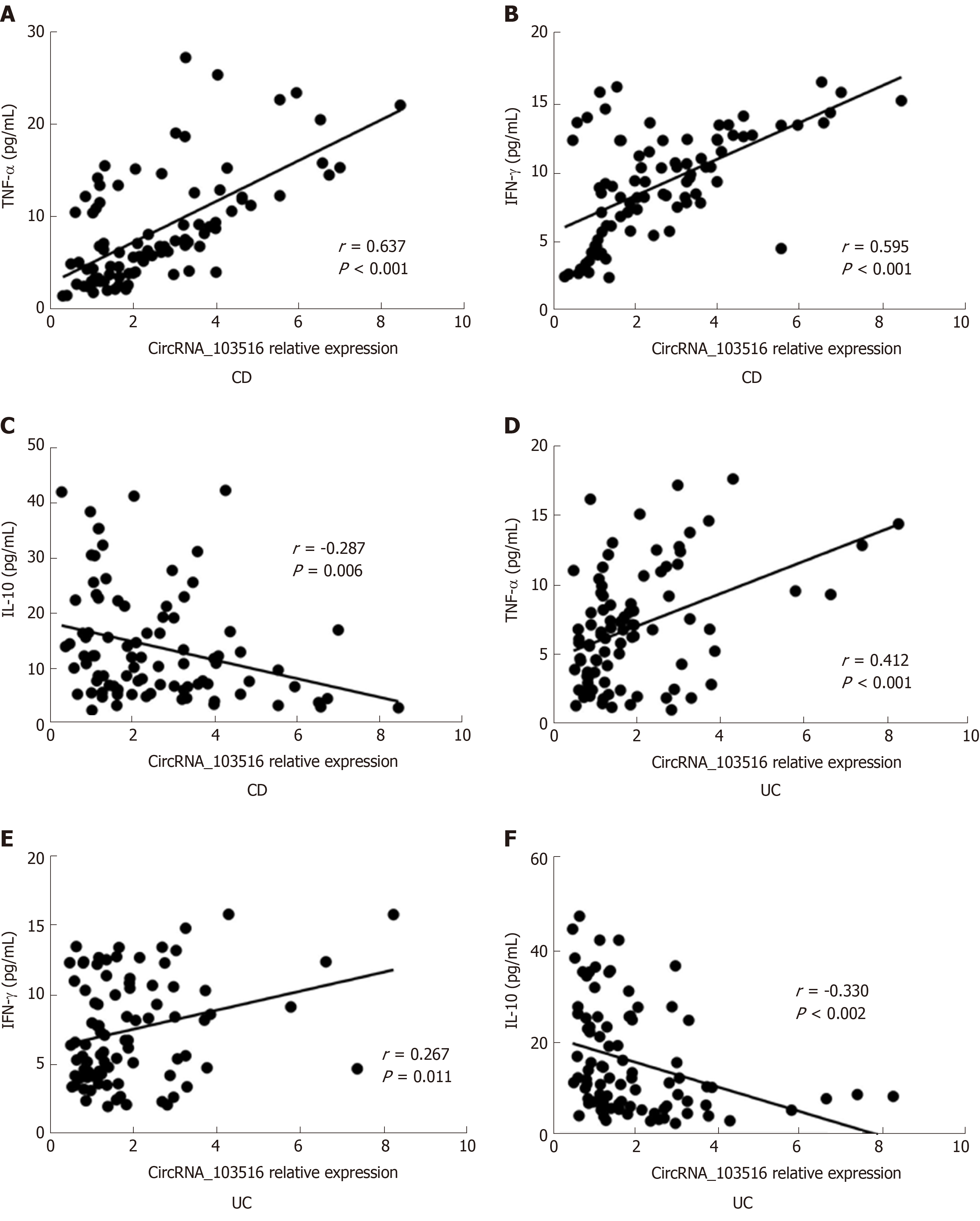
CircRNA_103516 levels showed a positive correlation with TNFα and IFN-γ, though negative correlations were found for circRNA_103516 with IL-10.
In CD patients, circRNA_103516 correlated positively with TNFα (r = 0.637, P < 0.001) and IFN-γ (r = 0.595, P < 0.001) and negatively with IL-10 (r = -0.287, P = 0.006) (Figure 6A-C). Similarly, we observed a correlation of circRNA_103516 with TNFα (r = 0.412, P < 0.001), IFN-γ (r = 0.267, P = 0.011), and IL-10 (r = -0.330, P = 0.002) in UC patients (Figure 6E and F).
The disease phenotypes of CD compared with the corresponding circRNA_103516 status are listed in Table 5. The prevalence of circRNA_103516 positivity was higher in patients with moderate and severe CD than in patients with mild disease (82.5%, 80.0% vs 47.4%, P = 0.008, P = 0.025). After adjustment for sex, age, and smoking, bivariate logistic analysis showed that positive circRNA_103516 was independently associated with an increased risk of disease activity [moderate: odds ratio (OR) = 4.886, 95%CI: 1.383-17.251, P = 0.014; severe: OR = 4.416, 95%CI: 0.084-22.054, P = 0.043]. Additionally, circRNA_103516 positivity was more frequently detected in patients with complicated CD (B2/B3) compared with those with the simple phenotype (B1) (78.8%, 77.8% vs 48.7%, P = 0.011, P = 0.045). After adjusting for clinical factors, logistic analysis also identified that circRNA_103516 correlated independently with an increased risk of stricturing (OR = 3.641, 95%CI: 1.245-10.650, P = 0.018) and penetrating (OR = 4.375, 95%CI: 1.147-16.690, P = 0.031) behaviour. In contrast, circRNA_103516 showed no correlation with disease location or medications.
| Clinical variable | n | CircRNA_103765 + (%) | Crude | Adjusted | ||||
| OR | 95%CI | P value | OR | 95%CI | P value | |||
| Gender | ||||||||
| Male | 48 | 66.7 | ||||||
| Female | 42 | 64.3 | 1.11 | 0.465-2.655 | 0.813 | NS | ||
| Age | ||||||||
| < 40 yr | 47 | 57.4 | ||||||
| ≥ 40 yr | 43 | 74.4 | 2.155 | 0.879-5.281 | 0.093 | NS | ||
| Smoking status | ||||||||
| Never | 60 | 68.3 | ||||||
| Past or current use | 30 | 60.0 | 1.439 | 0.579-3.577 | 0.434 | NS | ||
| Disease location of CD | ||||||||
| L1 | 37 | 64.7 | ||||||
| L2 | 20 | 64.7 | 0.524 | 0.094-2.930 | 0.462 | NS | ||
| L3 | 33 | 63.3 | 0.522 | 0.082-3.364 | 0.496 | |||
| Disease activity of CD | ||||||||
| Mild | 19 | 47.4 | ||||||
| Moderate | 40 | 82.5 | 5.238 | 1.554-17.653 | 0.008 | 4.886 | 1.384-17.251 | 0.014 |
| Severe | 15 | 80.0 | 4.444 | 0.941-21.001 | 0.025 | 4.416 | 0.084-22.054 | 0.043 |
| Disease behavior of CD | ||||||||
| B1 | 39 | 48.7 | ||||||
| B2 | 33 | 78.8 | 3.910 | 1.376-11.110 | 0.011 | 3.641 | 1.245-10.650 | 0.018 |
| B3 | 18 | 77.8 | 3.684 | 1.028-13.202 | 0.045 | 4.375 | 1.147-16.690 | 0.031 |
| Medications | ||||||||
| 5-ASA | 80 | 73.8 | 1.309 | 0.340-5.035 | 0.696 | NS | ||
| Corticosteroids | 50 | 74.0 | 2.329 | 0.959-5.655 | 0.062 | NS | ||
| Immunosuppressants | 28 | 67.9 | 1.161 | 0.450-2.998 | 0.758 | NS | ||
| Anti-TNF-α | 13 | 76.9 | 1.905 | 0.483-7.505 | 0.357 | NS | ||
| Surgery | 4 | 75.0 | 1.607 | 0.160-16.130 | 0.687 | NS | ||
In UC, circRNA_103516 positivity was associated with an increased risk of more severe disease behaviour (OR = 10.803, 95%CI: 1.171-94.687) (Table 6), though statistical power was not high due to the small number of patients with severe UC. No correlation of circRNA_103516 positivity with any other variable in UC was observed.
| Clinical variable | n | CircRNA_103765 + (%) | Crude | Adjusted | ||||
| OR | 95%CI | P value | OR | 95%CI | P value | |||
| Gender | ||||||||
| Male | 38 | 52.6% | ||||||
| Female | 52 | 59.6% | 1.329 | 0.571-3.090 | 0.509 | NS | ||
| Age | ||||||||
| < 40 yr | 40 | 60.0% | ||||||
| ≥ 40 yr | 50 | 54.0% | 0.783 | 0.337-1.817 | 0.568 | NS | ||
| Smoking status | ||||||||
| Never | 67 | 69.7% | ||||||
| Past or current use | 23 | 47.8% | 0.619 | 0.239-1.604 | 0.323 | NS | ||
| Disease location of UC | ||||||||
| E1 | 36 | 44.4% | ||||||
| E2 | 30 | 60.0% | 1.875 | 0.702-5.009 | 0.210 | NS | ||
| E3 | 24 | 70.8% | 3.036 | 1.012-9.107 | 0.048 | |||
| Disease severity of UC | ||||||||
| Mild | 30 | 60.0% | ||||||
| Moderate | 31 | 70.9% | 1.630 | 0.562-4.729 | 0.369 | NS | ||
| Severe | 15 | 93.3% | 9.333 | 1.080-80.627 | 0.042 | 10.803 | 1.171-94.687 | 0.036 |
| Medications: n (%) | ||||||||
| 5-ASA | 87 | 57.5% | 0.786 | 0.143-2.037 | 0.342 | NS | ||
| Corticosteroids | 41 | 70.7% | 1.827 | 0.762-5.234 | 0.084 | NS | ||
| Immunosuppressants | 14 | 71.4% | 1.234 | 0.673-3.238 | 0.705 | NS | ||
| Anti-TNF-α | 4 | 50.0% | 0.385 | 0.051-2.919 | 0.355 | NS | ||
| Surgery | 1 | 0.0% | 0.000 | 0.000- | 1.000 | NS | ||
Using miRNA target prediction software targetScan and miRanda, we investigated the top five predicted miRNA targets for circRNA_103516. The candidate hsa_circRNA_103516 is predicted to harbour sites for hsa-miR-147b, hsa-miR-19b-1-5p, hsa-miR-134-3p, hsa-miR-576-5p, and hsa-miR-493-5p, with different seed region types (i.e., 8mer, 7mer-m8, offset 6mer, and imperfect, respectively). We chose hsa-miR-19b-1-5p for detailed investigation of a correlation with circRNA_103516.
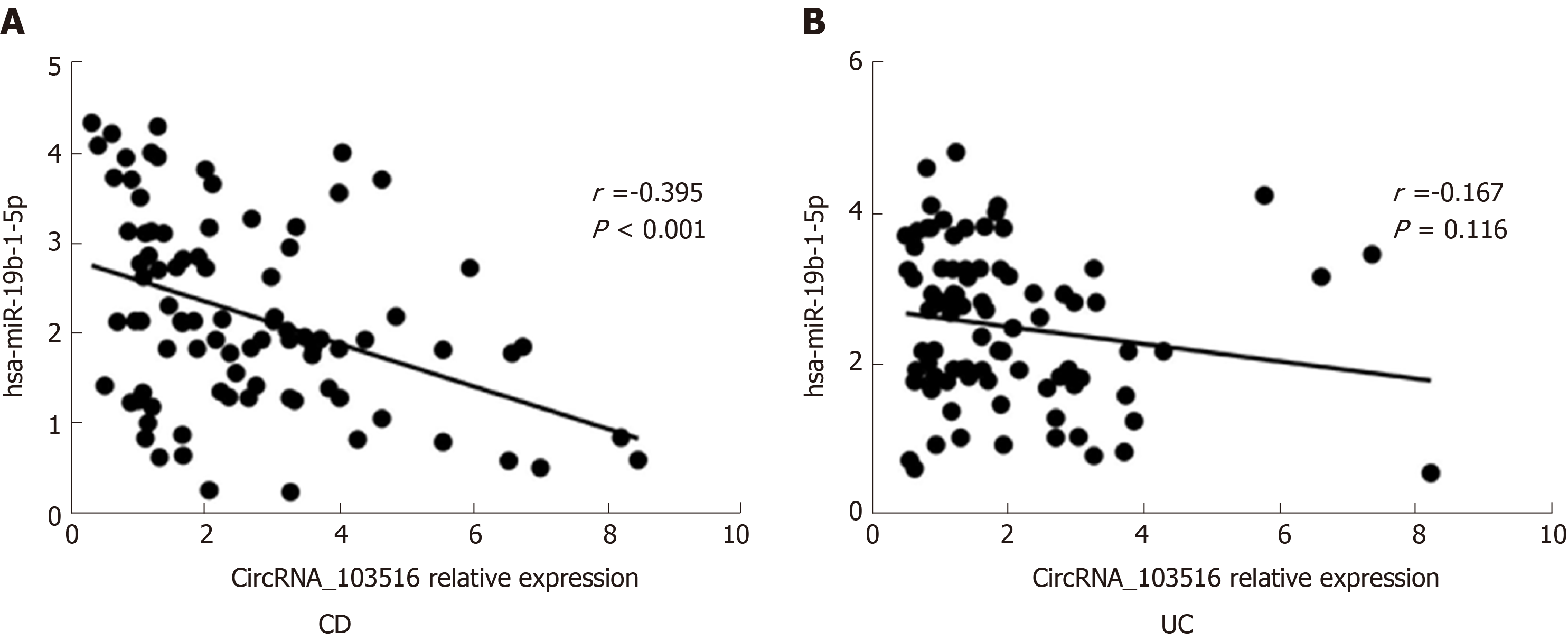
The expression level of circRNA_103516 correlated negatively with hsa-miR-19b-1-5p.
In CD patients, circRNA_103516 showed a negative correlation with hsa-miR-19b-1-5p (r = -0.395, P < 0.001) (Figure 7A). In contrast, there was no significant correlation between circRNA_103516 and hsa-miR-19b-1-5p in patients with UC (r = -0.167, P = 0.116) (Figure 7B).
The diagnosis and therapy of IBD involve enormous challenges that can only be addressed by obtaining a better understanding of the molecular mechanisms of IBD. Currently, owing to the advent of high-density microarrays, numerous noncoding RNAs are being considered as novel and potent regulators of gene expression. Noncoding RNAs comprise miRNAs, lncRNAs, circular RNAs, ribosomal RNAs, and snRNPs as well as a variety of regulatory RNAs[18]. Among them, circRNAs are ideal diagnostic and therapeutic candidates due to their stability and conservation[19]. The most profound studies between circRNAs and diseases are with regard to tumours[20]. For example, circEXOC6B and circN4BP2L2 are considered novel prognostic biomarkers in epithelial ovarian cancer[21]. Circ_0001451 was found to be downregulated in clear cell renal cell carcinoma and a potential biomarker for treatment of the disease[22]. Furthermore, sponging of miR-200a by circular RNA 101368 regulates the migration of hepatocellular carcinoma cells via the HMGB1/RAGE pathway[23].
Hundreds of miRNAs and lncRNAs have been reported to participate in IBD pathogenesis and can be considered as biomarkers for these patients[24,25]. Nevertheless, the roles of circRNA in IBD have only begun to be elucidated.
Our previous research investigated circRNA microarray profiles in peripheral blood from adults with CD, revealing 384 differential circRNAs in CD vs HCs, with 155 increased circRNAs and 229 decreased circRNAs; thus, these molecules have great potential for investigating the pathogenesis of CD. In the present study, we focused on circRNA_103516 and validated it by real-time PCR in a larger IBD cohort. First, circRNA_103516 exhibited strong clinical diagnostic value for CD and UC, suggesting that circRNA_103516 may be considered a novel biomarker for IBD. Second, circRNA_103516 was significantly more upregulated in the active stage than in the remission stage of both CD and UC, with positive correlations with disease activity (CDAI, Mayo, CRP, and ESR). Third, circRNA_103516 correlated positively with proinflammatory cytokines (TNF-α and INF-γ) in CD and UC patients and negatively with anti-inflammatory cytokines (IL-10). Fourth, logistic regression analysis revealed that circRNA_103516 was independently associated with an increased risk of disease activity in CD and UC. These data indicate that circRNA_103516 may have a proinflammatory role.
Given the roles of circRNAs as miRNA sponges and gene regulators[6,26], we investigated the top five predicted miRNA targets for circRNAs and identified that circRNA_103516 may target hsa-miR-147b, hsa-miR-19b-1-5p, hsa-miR-134-3p, hsa-miR-576-5p, and hsa-miR-493-5p, competitively restraining miRNA activity. We chose hsa-miR-19b-1-5p to further evaluate the correlation with circRNA_103516 in detail. The current study showed that circRNA_103516 correlated negatively with hsa-miR-19b-1-5p in CD patients.
Interestingly, previous studies have identified that miR-19b levels are decreased in the serum and intestinal tissue of IBD patients[14,27], which is consistent with our study. Research has also shown that miR-19b may participate in abnormal inflammatory reactions by inhibiting suppressor of cytokine signalling 3 to regulate chemokine production in intestinal epithelial cells[27]. Furthermore, miR-19 and its family members modulate the chronic inflammatory process by activating NF-κB signalling in rheumatoid arthritis[28]. The NF-κB signalling pathway plays a vital role in autoimmune diseases by regulating T cell and macrophage development and function, ultimately participating in the inflammatory process and immune regulation[29]. Moreover, miRNA19b modulates Th17 cell development by targeting thymic stromal lymphopoietin[30] and has a vital function by suppressing the IL-10 level in peripheral dendritic cells[31]. Thus, we suggest that circRNA_103516 acts as a proinflammatory gene by sponging miRNA19b and is involved in the pathogenesis of IBD by mediating inflammation and immune-related signalling pathways.
Previous studies have shown that serum levels of miR-19 correlated negatively with stricturing CD[14]. The groups of Zhao and Zou attempted to explain these results by uncovering a new mechanism in which miR-19b depresses fibrogenesis by targeting TGF-beta receptor 2 in biliary atresia-related fibrosis[32] and myocardial fibrosis[33]. However, our study identified that circRNA_103516 positivity may be an independent risk factor for CD stricture and penetrating behaviour. Indeed, a higher prevalence of circRNA_103516 positivity was found in patients with complicated CD (stricturing and penetrating). These results further suggest that circRNA_103516 may play a significant role in the development and progression of CD by acting as a microRNA19b sponge. Nevertheless, elucidation of the precise function of miRNA-circRNA interactions in CD pathogenesis requires further research.
To the best of our knowledge, this is the first study to identify the relationship between circRNA_103516 expression in PBMCs and disease activity and the risk of IBD and inflammatory cytokines. Nonetheless, certain limitations in our study should not be neglected. First, the number of subjects was not large, which might have affected the results. In addition, a more diverse disease control group is needed for a more applicable conclusion. Second, we identified miRNA-circRNA interactions only by functional analysis and not by experimental verification. Thus, further research is needed.
In conclusion, we verified that increased circRNA_103516 levels are a prospective candidate marker for IBD diagnosis and correlate positively with disease activity and behaviour in CD patients. Moreover, circRNA_103516 may participate in the molecular mechanism of CD by sponging miR-19b. The precise molecular mechanisms underlying circRNA functions in CD require further investigation.
Inflammatory bowel disease (IBD), including ulcerative colitis (UC) and Crohn’s disease (CD), comprises chronic gastrointestinal tract inflammatory disorders. Due to the characteristic of repeated recurrence, the disease activity of IBD must be assessed and monitored repeatedly. Accordingly, non-invasive serological biomarkers may constitute an optimal alternative choice for evaluating and screening disease activity in IBD. Increasing evidence demonstrates that circular RNAs (circRNAs) participate in the pathogenesis of a variety of diseases by modulating gene expression at the transcriptional or post-transcriptional level and are considered ideal biomarkers in human disease.
To date, little is known about the relationships between circRNAs and IBD. Our previous research identified 155 upregulated circRNAs and 229 downregulated circRNAs by microarray analysis of peripheral blood mononuclear cells (PBMCs) from CD patients compared with healthy controls (HCs). Moreover, bioinformatics analysis predicted that hsa_circRNA_103516 might participate in the pathogenesis of IBD by sponging hsa-miR-19b-1-5p.
The aim of this study was to identify expression of circRNA_103516 in IBD and its association with clinical phenotypes and inflammatory cytokines.
We performed an observational study. PBMCs were obtained from patients with IBD, HCs, and patient controls. Expression of circRNA_103516 and hsa-miR-19b-1-5p was assessed by quantitative reverse transcription-polymerase chain reaction. Crohn's disease activity index (CDAI), Mayo score, C-reactive protein (CRP) level, and erythrocyte sedimentation rate (ESR) were measured. Flow cytometry was used to examine blood samples for the measurement of the inflammatory cytokines tumour necrosis factor-α, interferon-γ (IFN-γ), and interleukin-10 (IL-10). Spearman correlation analysis was performed to determine linear correlations for different groups. Receiver operating characteristic curve analysis was used to assess the clinical diagnostic value of candidate circRNAs. Logistic regression analysis was used to identify risk factors.
CircRNA_103516 was upregulated in CD and UC. Furthermore, circRNA_103516 correlated positively with CDAI, Mayo score, CRP, ESR, TNFα, and IFN-γ and negatively with IL-10 in IBD. Additionally, circRNA_103516 correlated positively with stricturing and penetrating behaviour. The predicted hsa-miR-19b-1-5p correlated negatively with circRNA_103516 in CD.
CircRNA_103516 in PBMCs can be considered an ideal candidate for the diagnosis of IBD. Dysregulation of circRNA_103516 may participate in the molecular mechanism of IBD through hsa-miR-19b-1-5p sponging.
More large prospective multicentre clinical studies need to be performed for a more applicable conclusion. In addition, miRNA-circRNA interactions were conducted by functional analysis and not by experimental verification. It is imperative to further explore their respective molecular mechanisms and genetic changes to achieve better diagnostic and treatment strategies in the future.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Berezin AE, Gazouli M S-Editor: Tang JZ L-Editor: Wang TQ E-Editor: Ma YJ
| 1. | Karantanos T, Gazouli M. Inflammatory bowel disease: recent advances on genetics and innate immunity. Ann Gastroenterol. 2011;24:164-172. [PubMed] |
| 2. | Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2894] [Cited by in RCA: 3350] [Article Influence: 186.1] [Reference Citation Analysis (11)] |
| 3. | Sartor RB. Genetics and environmental interactions shape the intestinal microbiome to promote inflammatory bowel disease versus mucosal homeostasis. Gastroenterology. 2010;139:1816-1819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 141] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 4. | Diener TO. Circular RNAs: relics of precellular evolution? Proc Natl Acad Sci U S A. 1989;86:9370-9374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 122] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 5. | Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1186] [Cited by in RCA: 1499] [Article Influence: 136.3] [Reference Citation Analysis (0)] |
| 6. | Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4631] [Cited by in RCA: 6043] [Article Influence: 503.6] [Reference Citation Analysis (0)] |
| 7. | Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1572] [Cited by in RCA: 2072] [Article Influence: 207.2] [Reference Citation Analysis (0)] |
| 8. | Xin Z, Ma Q, Ren S, Wang G, Li F. The understanding of circular RNAs as special triggers in carcinogenesis. Brief Funct Genomics. 2017;16:80-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 9. | Gomes CPC, Salgado-Somoza A, Creemers EE, Dieterich C, Lustrek M, Devaux Y; Cardiolinc™ network. Circular RNAs in the cardiovascular system. Noncoding RNA Res. 2018;3:1-11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 10. | Zheng F, Yu X, Huang J, Dai Y. Circular RNA expression profiles of peripheral blood mononuclear cells in rheumatoid arthritis patients, based on microarray chip technology. Mol Med Rep. 2017;16:8029-8036. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 98] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 11. | Kumar L, Shamsuzzama, Haque R, Baghel T, Nazir A. Circular RNAs: the Emerging Class of Non-coding RNAs and Their Potential Role in Human Neurodegenerative Diseases. Mol Neurobiol. 2017;54:7224-7234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 146] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 12. | Yin J, Hu T, Xu L, Li P, Li M, Ye Y, Pang Z. Circular RNA expression profile in peripheral blood mononuclear cells from Crohn disease patients. Medicine (Baltimore). 2019;98:e16072. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 13. | Cai C, Rajaram M, Zhou X, Liu Q, Marchica J, Li J, Powers RS. Activation of multiple cancer pathways and tumor maintenance function of the 3q amplified oncogene FNDC3B. Cell Cycle. 2012;11:1773-1781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 65] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 14. | Lewis A, Mehta S, Hanna LN, Rogalski LA, Jeffery R, Nijhuis A, Kumagai T, Biancheri P, Bundy JG, Bishop CL, Feakins R, Di Sabatino A, Lee JC, Lindsay JO, Silver A. Low Serum Levels of MicroRNA-19 Are Associated with a Stricturing Crohn's Disease Phenotype. Inflamm Bowel Dis. 2015;21:1926-1934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 15. | Best WR, Becktel JM, Singleton JW, Kern F. Development of a Crohn's disease activity index. National Cooperative Crohn's Disease Study. Gastroenterology. 1976;70:439-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2405] [Cited by in RCA: 2334] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 16. | Paine ER. Colonoscopic evaluation in ulcerative colitis. Gastroenterol Rep (Oxf). 2014;2:161-168. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 17. | Satsangi J, Silverberg MS, Vermeire S, Colombel JF. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut. 2006;55:749-753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1970] [Cited by in RCA: 2346] [Article Influence: 123.5] [Reference Citation Analysis (2)] |
| 18. | Yang S, Sun Z, Zhou Q, Wang W, Wang G, Song J, Li Z, Zhang Z, Chang Y, Xia K, Liu J, Yuan W. MicroRNAs, long noncoding RNAs, and circular RNAs: potential tumor biomarkers and targets for colorectal cancer. Cancer Manag Res. 2018;10:2249-2257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 69] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 19. | Zhang Z, Yang T, Xiao J. Circular RNAs: Promising Biomarkers for Human Diseases. EBioMedicine. 2018;34:267-274. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 535] [Cited by in RCA: 611] [Article Influence: 87.3] [Reference Citation Analysis (0)] |
| 20. | Xu XY, Zhou LL, Yu C, Shen B, Feng JF, Yu SR. Advances of circular RNAs in carcinoma. Biomed Pharmacother. 2018;107:59-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 21. | Ning L, Long B, Zhang W, Yu M, Wang S, Cao D, Yang J, Shen K, Huang Y, Lang J. Circular RNA profiling reveals circEXOC6B and circN4BP2L2 as novel prognostic biomarkers in epithelial ovarian cancer. Int J Oncol. 2018;53:2637-2646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 22. | Wang G, Xue W, Jian W, Liu P, Wang Z, Wang C, Li H, Yu Y, Zhang D, Zhang C. The effect of Hsa_circ_0001451 in clear cell renal cell carcinoma cells and its relationship with clinicopathological features. J Cancer. 2018;9:3269-3277. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 23. | Li S, Gu H, Huang Y, Peng Q, Zhou R, Yi P, Chen R, Huang Z, Hu X, Huang Y, Tang D. Circular RNA 101368/miR-200a axis modulates the migration of hepatocellular carcinoma through HMGB1/RAGE signaling. Cell Cycle. 2018;17:2349-2359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 24. | Cao B, Zhou X, Ma J, Zhou W, Yang W, Fan D, Hong L. Role of MiRNAs in Inflammatory Bowel Disease. Dig Dis Sci. 2017;62:1426-1438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 25. | Zacharopoulou E, Gazouli M, Tzouvala M, Vezakis A, Karamanolis G. The contribution of long non-coding RNAs in Inflammatory Bowel Diseases. Dig Liver Dis. 2017;49:1067-1072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 26. | Salzman J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet. 2016;32:309-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 477] [Cited by in RCA: 648] [Article Influence: 72.0] [Reference Citation Analysis (0)] |
| 27. | Cheng X, Zhang X, Su J, Zhang Y, Zhou W, Zhou J, Wang C, Liang H, Chen X, Shi R, Zen K, Zhang CY, Zhang H. miR-19b downregulates intestinal SOCS3 to reduce intestinal inflammation in Crohn's disease. Sci Rep. 2015;5:10397. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 28. | Gantier MP, Stunden HJ, McCoy CE, Behlke MA, Wang D, Kaparakis-Liaskos M, Sarvestani ST, Yang YH, Xu D, Corr SC, Morand EF, Williams BR. A miR-19 regulon that controls NF-κB signaling. Nucleic Acids Res. 2012;40:8048-8058. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 159] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 29. | Yue Y, Stone S, Lin W. Role of nuclear factor κB in multiple sclerosis and experimental autoimmune encephalomyelitis. Neural Regen Res. 2018;13:1507-1515. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 30. | Wang Z, Chen Y, Xu S, Yang Y, Wei D, Wang W, Huang X. Aberrant decrease of microRNA19b regulates TSLP expression and contributes to Th17 cells development in myasthenia gravis related thymomas. J Neuroimmunol. 2015;288:34-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 31. | Luo XQ, Shao JB, Xie RD, Zeng L, Li XX, Qiu SQ, Geng XR, Yang LT, Li LJ, Liu DB, Liu ZG, Yang PC. Micro RNA-19a interferes with IL-10 expression in peripheral dendritic cells of patients with nasal polyposis. Oncotarget. 2017;8:48915-48921. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 32. | Zhao D, Luo Y, Xia Y, Zhang JJ, Xia Q. MicroRNA-19b Expression in Human Biliary Atresia Specimens and Its Role in BA-Related Fibrosis. Dig Dis Sci. 2017;62:689-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 33. | Zou M, Wang F, Gao R, Wu J, Ou Y, Chen X, Wang T, Zhou X, Zhu W, Li P, Qi LW, Jiang T, Wang W, Li C, Chen J, He Q, Chen Y. Autophagy inhibition of hsa-miR-19a-3p/19b-3p by targeting TGF-β R II during TGF-β1-induced fibrogenesis in human cardiac fibroblasts. Sci Rep. 2016;6:24747. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 95] [Article Influence: 10.6] [Reference Citation Analysis (0)] |