Published online Feb 21, 2015. doi: 10.3748/wjg.v21.i7.2011
Peer-review started: September 4, 2014
First decision: October 29, 2014
Revised: November 6, 2014
Accepted: December 14, 2014
Article in press: December 16, 2014
Published online: February 21, 2015
Processing time: 160 Days and 9.9 Hours
AIM: To identify the genes induced and regulated by the MYC protein in generating tumors from liver stem cells.
METHODS: In this study, we have used an immortal porcine liver stem cell line, PICM-19, to study the role of c-MYC in hepatocarcinogenesis. PICM-19 cells were converted into cancer cells (PICM-19-CSCs) by overexpressing human MYC. To identify MYC-driven differential gene expression, transcriptome sequencing was carried out by RNA sequencing, and genes identified by this method were validated using real-time PCR. In vivo tumorigenicity studies were then conducted by injecting PICM-19-CSCs into the flanks of immunodeficient mice.
RESULTS: Our results showed that MYC-overexpressing PICM-19 stem cells formed tumors in immunodeficient mice demonstrating that a single oncogene was sufficient to convert them into cancer cells (PICM-19-CSCs). By using comparative bioinformatics analyses, we have determined that > 1000 genes were differentially expressed between PICM-19 and PICM-19-CSCs. Gene ontology analysis further showed that the MYC-induced, altered gene expression was primarily associated with various cellular processes, such as metabolism, cell adhesion, growth and proliferation, cell cycle, inflammation and tumorigenesis. Interestingly, six genes expressed by PICM-19 cells (CDO1, C22orf39, DKK2, ENPEP, GPX6, SRPX2) were completely silenced after MYC-induction in PICM-19-CSCs, suggesting that the absence of these genes may be critical for inducing tumorigenesis.
CONCLUSION: MYC-driven genes may serve as promising candidates for the development of hepatocellular carcinoma therapeutics that would not have deleterious effects on other cell types in the liver.
Core tip: It is well-established that cancer stem cells not only drive tumor growth in the liver, in general, but also that the proto-oncogene c-MYC plays a critical role in that process. However, little is known about genes induced and regulated by MYC to generate tumors, and, in particular, those involved in liver stem cells. In this study, we examined the role of MYC protein in hepatocarcinogenesis using an immortal porcine liver stem cell line, PICM-19. Interestingly, MYC-overexpression silenced the expression of six genes in PICM-19 cells (CDO1, C22orf39, DKK2, ENPEP, GPX6, SRPX2), suggesting that they may be critical in inducing tumorigenesis.
- Citation: Aravalli RN, Talbot NC, Steer CJ. Gene expression profiling of MYC-driven tumor signatures in porcine liver stem cells by transcriptome sequencing. World J Gastroenterol 2015; 21(7): 2011-2029
- URL: https://www.wjgnet.com/1007-9327/full/v21/i7/2011.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i7.2011
Hepatocellular carcinoma (HCC) is the most common form of liver cancer, and is the third leading cause of cancer-related deaths worldwide[1]. It occurs as a consequence of exposure to a variety of risk factors, and appears primarily in liver disease patients with underlying cirrhosis[2]. While surgical resection and whole liver transplantation are the most effective treatment options currently available for the treatment of HCC, recurrence is quite common in patients who have had a resection, and the 5-year survival rate post-surgery is < 50%[3].
Hepatocarcinogenesis is a multistep process involving a number of cellular and molecular factors. Among these, the proto-oncogene c-MYC (herein, referred to as MYC) has been shown to be a critical regulator of malignant transformation during the early stages of liver cancer[4]. Dysregulation of MYC expression correlates with poor prognosis in human cancers, including HCC[5]. Its overexpression, and subsequent induction of its target genes, causes the malignant conversion of preneoplastic liver lesions[4]. Conversely, silencing of MYC results in the inhibition of migration, invasion and proliferation of human liver cancer cells[6]. Therefore, the study of MYC-expressing liver stem cells could be highly relevant to our understanding of the pathogenesis in human HCC. Due to the involvement of the MYC oncoprotein in angiogenesis and various cellular processes such as cell cycle regulation, differentiation, growth, metabolism, adhesion and motility, MYC, and its downstream target genes have become attractive therapeutic aims of cancer research[7].
It is now apparent that the capacity to sustain tumor growth resides in a small population of stem cells [termed “cancer stem cells” (CSCs)] present in most cancers, including HCC[8-10]. CSCs possess greater colony-forming efficiency, higher proliferation potential, and the necessary phenotype of “stemness” to form tumors in animals[5]. To support this, CD133-expressing progenitor cells were shown to drive the recurrence of liver cancer following chemotherapy, suggesting that CSCs are resistant to treatment[11]. A possible mechanism for the enhanced survival of CSCs in tumors is the activation of the Akt (also known as protein kinase B) and B-cell lymphoma 2 (Bcl-2) signaling pathways[11]. Therefore, a critical step towards developing effective cancer treatments is to understand cellular, molecular and biochemical differences between normal stem cells and CSCs.
To decipher the role of MYC and MYC-targeted genes in HCC, we have developed a liver cell model of MYC oncogene transformation based on the overexpression of MYC in a porcine liver stem cell line, PICM-19[12]. The PICM-19 cell line originated from the spontaneous differentiation of cultured pig epiblast tissue and was, therefore, derived from pig embryonic stem cells[13]. The cell line is unique in its ability to differentiate into either of the two cell types that comprise the parenchyma of the developing liver, i.e., fetal hepatocytes or bile duct epithelium, and grows indefinitely without change in stem cell character. In this study, we have MYC-transformed the PICM-19 cell line into CSCs, and studied their transcriptome profiles by RNA sequencing (RNA-seq). We determined that MYC induced the expression of a large number of genes with diverse cellular processes in PICM-19 cells, most of which were not previously implicated in HCC. Our data identified novel therapeutic targets while underscoring the importance of MYC in tumor generation in primary liver stem cells.
The PICM-19 porcine liver stem cell line was established from a pig embryo’s inner cell mass, i.e., embryonic stem cells, and was cultured as described previously[12,13]. Briefly, cells were grown in 10% DMEM/199 medium and plated as feeder-free cultures using Matrigel (BD Biosciences, San Jose, CA). This growth medium is a 50:50 mixture of DMEM low glucose (Invitrogen, Carlsbad, CA) and Medium 199 (Invitrogen) supplemented with 10% fetal bovine serum, 2-mercaptoethanol and nucleosides. In subsequent steps, cells were fed daily with conditioned medium obtained from γ-irradiated STO feeder cells, a mouse embryonic fibroblast cell line (Cat. #CRL-1503, ATCC, Rockville, MD). PICM-19-CSCs, generated by transfecting the PICM-19 cell line (see below), were also maintained under these growth conditions. All cells were maintained at 37 °C in a 5% CO2 humidified incubator.
The expression vector pUNO1-hMYC was generated by cloning the human c-MYC open reading frame (ORF) into the multiple cloning site of the plasmid pUNO1-mcs (InvivoGen, San Diego, CA) downstream of a strong elongation factor (EF)-1α/human T-lymphotropic virus (HTLV) hybrid promoter active in most cell types. pUNO1-mcs contains the blasticidin resistance gene driven by a CMV promoter and enhancer in tandem with the bacterial EM7 promoter. This allows the amplification of the plasmid, and, after transfection into mammalian cells, the blasticidin selection of stable transfectants. Another plasmid, pUNO1-MYC-IRES-Luc was also constructed by cloning the firefly luciferase ORF downstream of MYC ORF separated by an internal ribosome entry site (IRES) sequence to maintain expression of both MYC and luciferase (luc) genes under the control of the hybrid promoter in this plasmid. Large scale preparations of both vectors were prepared by transforming them into competent cells of Escherichia coli strain DH5α, and by extracting them using the Qiagen Plasmid Maxi kit (Qiagen, Valencia, CA).
PICM-19 cells were successfully transfected with the pUNO1-MYC-IRES-Luc plasmid using the mouse macrophage nucleofection kit (Amaxa Biosystems, Gaithersburg, MD) and the program A-13 on the nucleofector I device (Amaxa). Following nucleofection, cells were plated in 12-well plates and incubated overnight at 37 °C. Growth medium was then replaced with the fresh medium containing 5 μg/mL blasticidin (InvivoGen) to select for positive transfectants. Individual colonies that formed were further grown, and assessed for MYC expression using reverse transcription polymerase chain reaction (RT-PCR) (data not shown). The clone that showed the highest MYC expression was then used in further experiments mentioned below. Next, cells of this clone were plated in 6-well plates, and 24 h post-plating they were resuspended in fresh medium and were treated with the Bright-Glo luciferase assay substrate (Promega, Madison, WI) to measure luciferase activity using the IVIS® Imaging System (Xenogen Corporation, Alameda, CA).
Western blot analysis of cellular proteins extracted from PICM-19 and PICM-19-CSCs was performed using mouse anti-human c-MYC antibody (Cat. #sc-40, Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Ten μg of total protein from these cells was loaded onto a 10% denaturing SDS-PAGE gel. Following separation, proteins were transferred to a nitrocellulose filter and treated with blocking solution containing 5% milk powder. Primary antibody at a concentration of 1:500 was then added to the filter and incubated for 2 h in a rotating chamber at 4 °C. After several washes in buffer, the secondary anti-mouse IgG antibody (Santa Cruz Biotechnology) was added at a concentration of 1:1000 and the filter incubated for 2 h at RT. The blot was washed and probed with ECL solution to visualize protein bands (Thermo Fischer Scientific, Rockford, IL).
Five-week old NOD/SCID mice were purchased from Charles River Breeding Laboratories (Wilmington, MA). Freshly cultured PICM-19 and PICM-19-CSCs were treated with trypsin to detach and harvest them in PBS. After washing twice in PBS, 1 × 106 cells were resuspended in 100 μL PBS, and were injected into the flanks of the immunodeficient mice (n = 3). This study was approved by the University of Minnesota Institutional Animal Care and Use Committee. Animals were observed on a daily basis for signs of declining health and lethargy. Tumor formation was monitored twice weekly by measuring the width and length of tumors. Animals with tumors that grew to a diameter of 1.5 cm, as measured by calipers, were sacrificed. To alleviate pain and stress in animals, carbon dioxide was administered in the fume hood prior to euthanasia. Tumors were then harvested and immediately fixed in 10% formalin for 24 h. These tissue samples were cut in 5 μm-thick sections on a microtome. Sections were deparaffinized with xylene, rehydrated in three five-minute steps with 100% ethanol, 90% ethanol, and 70% ethanol, and stained with hematoxylin and eosin (HE) for 1 h. Slides were rinsed with RNase-free water and mounted under glass coverslips with Clarion mounting medium (Santa Cruz Biotechnology). They were observed using a Nikon DXM1200C microscope.
Total cellular RNAs were isolated from PICM-19 and PICM-19-CSCs using the RNeasy Mini kit (Qiagen). They were quantified using a fluorimetric RiboGreen assay. Total RNA integrity was assessed by capillary electrophoresis using BioAnalyzer 2100 (Agilent Technologies, Santa Clara, CA, United States). For samples to pass the initial quality control (QC) step, it was necessary to quantify > 1 μg and to have a RNA Integrity Number of ≥ 8. They were then converted to Illumina sequencing libraries using the Truseq RNA Sample Preparation Kit (Cat. # RS-122-2001, Illumina, San Diego, CA). For amplification and library preparation, 1 μg of total RNA was purified using oligo-dT coated magnetic beads, fragmented and reverse transcribed into cDNA. The cDNA was fragmented, blunt-ended, and ligated to indexed (barcoded) adaptors and amplified using 15 cycles of PCR. Final library size distribution was validated using capillary electrophoresis and quantified using fluorimetry (PicoGreen) and by Q-PCR. Indexed libraries were normalized, pooled and size selected to 320bp +/- 5% using Caliper’s XT instrument.
Truseq libraries were hybridized to a paired-end flow cell and individual fragments were clonally amplified by bridge amplification on the Illumina cBot. Once clustering was complete, the flow cell was loaded onto the Illumina HiSeq 2000 sequencer and sequenced using Illumina’s SBS chemistry. Upon completion of read 1, a 7 base pair index read was performed. Finally, the library fragments were resynthesized in the reverse direction and sequenced from the opposite end of the read 1 fragment thus producing the template for paired end read 2.
Base call (.bcl) files for each cycle of sequencing were generated by Illumina Real Time Analysis software. The base call files and run folders were then exported to servers maintained at the Minnesota Supercomputing Institute, where primary analysis and de-multiplexing were performed using Illumina’s CASAVA software 1.8.2. De-multiplexed FASTQ files generated by the CASAVA workflow were analyzed by the mapping and aligning software as described below.
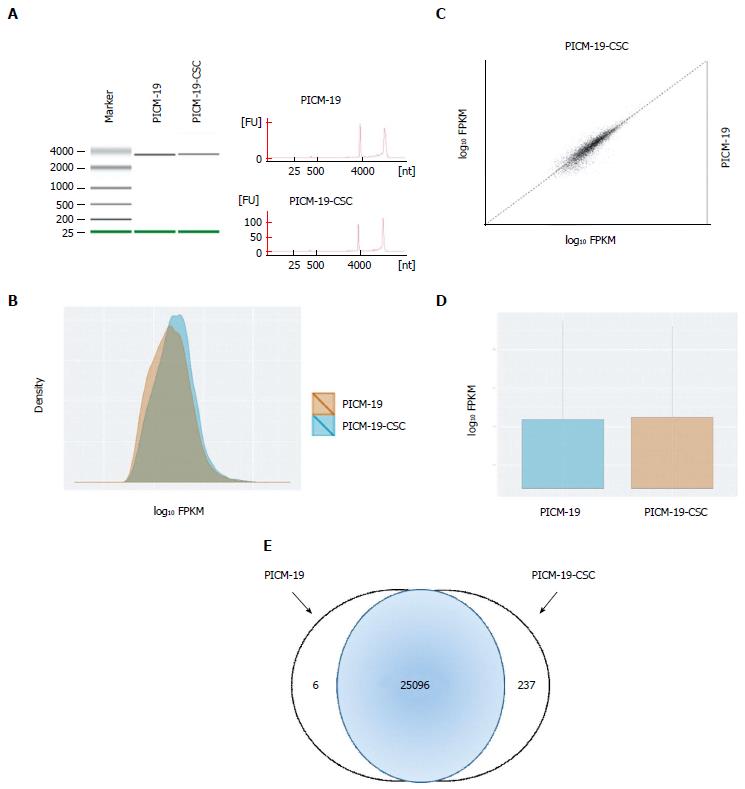
Raw paired-end RNA-seq reads in FASTQ format were assessed for base call quality, cycle uniformity and contamination using FastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/). The QC-filtered paired-end reads were mapped to pig genome (UCSC susScr3 assembly) via Tophat 2[14] using the UCSC pig annotation in GTF format. Resulting Binary Alignment and Mapping (BAM) files were then visualized using the Integrative Genomics Viewer (IGV)[15] to ensure reasonable behavior of expected control invariant genes [e.g., glyceraldehyde 3-phosphate dehydrogenase (GAPDH)], and to confirm expected behavior of positive control genes (e.g., MYC). BAM files were subsequently analyzed via Cuffdiff (default parameters using the iGenomes GTF file as shown previously[16]) to quantify the expression level(s) of each known gene in units of FPKM (Fragment mapped per kilobase of genes per million mapped reads), and tested for differential expression. A matrix of FPKM transcript abundance levels for each gene was extracted and downloaded into CummeRbund[17] to assess sample clustering using Multi-Dimensional Scaling and Principle Component Analysis, which revealed sample outliers. Differential gene sets were filtered to remove those with fold changes < 1.5 (up- or downregulated) and with a BH FDR-corrected P-value > 0.05.
The cDNA from PICM-19 and PICM-19-CSCs was synthesized with 1 μg of total RNA using SuperScript II reverse transcriptase (Invitrogen Life Technologies, Carlsbad, CA) and oligo dT6-12 primers (Sigma-Genosys, The Woodlands, TX). Quantitative real-time PCR was performed using the FullVelocity SYBR Green QPCR master mix (Stratagene, La Jolla, CA) following the manufacturer’s specifications. The 25 μL final reaction volume consisted of pre-made reaction mix (SYBR Green I dye, reaction buffer, Taq DNA polymerase, and dNTPs), 0.3 mmol/L of each primer, and 0.5 ng cDNA in water. Reaction conditions for PCR for the Mx3000P QPCR System (Stratagene) were as follows: polymerase activation at 95 °C for 5 min, 40 denaturation cycles of 95 °C for 10 s, annealing at 60 °C for 10 s and elongation at 72 °C for 10 s. A melting curve was performed to assess primer specificity and product quality using the same conditions. The relative product levels were quantified using the 2(-∆∆CT) method[18]. Data are presented as relative induction of each porcine gene, normalized to β-actin, and are representative of three independent experiments.
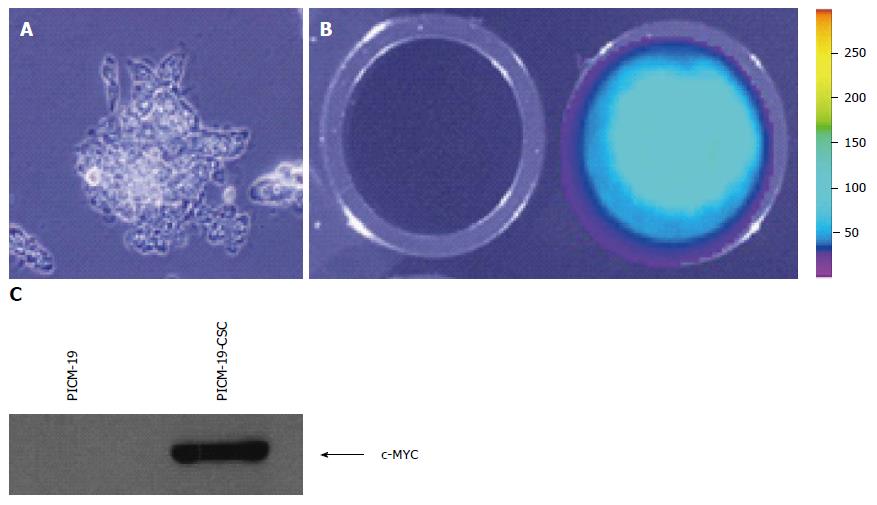
We initially determined that PICM-19 cells can be transfected with an expression vector driven by the EF-1α/HTLV hybrid promoter. pUNO1-MYC-IRES-Luc was constructed by cloning the open reading frame (ORF) of firefly luciferase (luc) gene downstream of the MYC ORF separated by an IRES sequence. This vector also contains the blasticidin resistance gene as a selectable marker. The function of the genetic construct was tested by assaying the expression of luciferase in transfected cells. After transfection, feeder-independent PICM-19 cells were cultured (Figure 1A) and transfectants were selected using blasticidin. Individual clones were then screened for MYC expression, and the clone that had the highest MYC expression as detected by RT-PCR (data not shown) was assayed for luciferase activity in 24-well plates using D-luciferin as the substrate. Transfected PICM-19 cells expressed luciferase demonstrating that they took up and incorporated the plasmid DNA (Figure 1B). This finding also indicated that the stem cells were expressing the MYC ORF located upstream of luc ORF in the expression vector. This was confirmed by Western blotting assay using anti-human MYC antibody (Figure 1C).
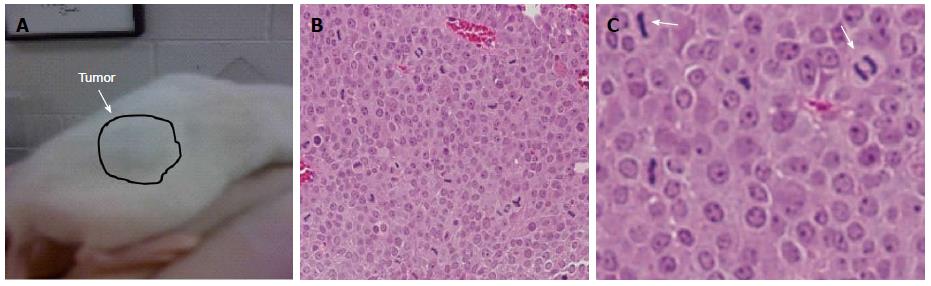
We subcutaneously injected 1 × 106 cells into the back flanks of NOD-SCID mice to establish that MYC-overexpressing PICM-19 cells (PICM-19-CSCs) can generate tumors in mice. PICM-19-CSCs, but not the control PICM-19 stem cells, generated tumors in vivo (Figure 2). When the tumors reached 1.5 cm in diameter, animals were sacrificed and the tumors were sectioned to be analyzed by HE staining. The tumors were composed of a uniform mass of cells that were actively undergoing cell division. This experiment demonstrated the proof-of-principle that PICM-19-CSCs are capable of producing tumors in vivo.
To understand the differences between PICM-19 and PICM-19-CSCs at the molecular level, we examined the RNA-seq profiles of both these cell types, and found sets of genes that were expressed in each cell line (Tables 1 and 2). These genes were normalized to the expression of housekeeping gene GAPDH, and were analyzed. Only a two-fold change or greater was considered significant. In this process, we identified a set of six genes that were expressed only in PICM-19 stem cells (Table 1). The expression of these six genes (CDO1, C22orf39, DKK2, ENPEP, GPX6 and SRPX2) was undetectable in PICM-19-CSCs, as a result of MYC protein overexpression. Their absence in PICM-19-CSCs suggested their potential use as therapeutic targets for liver cancers associated with MYC overexpression. Among these, CDO1 is a tumor suppressor[19], while Dickkopf-related protein 2 (DKK2) is an inhibitor of the Wnt signaling pathway[20]. Both these genes are epigenetically silenced in human cancers. While the overexpression of DKK2 led to an inhibition in renal cancer progression through apoptotic and cell cycle pathways[20], forced expression of cysteine dioxygenase type 1 (CDO1) decreased tumor cell growth of different tumor cell lines and in mice[19]. Lack of DDK2 expression in PICM-19-CSCs suggested that a putative epigenetic control mechanism to silence DKK2 also exists in MYC-induced liver cancer.
| Gene ID | Gene abbreviation | Function | Fold Δ1 |
| C22ORF39 | Human chromosome 22 orf39 homolog | Unknown | 11.33 |
| CDO1 | Cysteine dioxygenase type 1 | Tumor suppressor | 6.28 |
| DKK2 | Dickkopf Wnt signaling pathway inhibitor 2 | Cell signaling | 12.20 |
| ENPEP | Glutamyl aminopeptidase | Proteolysis | 4.82 |
| GPX6 | Glutathione peroxidase 6 | Detoxification | 2.18 |
| SRPX2 | Sushi-repeat containing protein, X-linked 2 | Angiogenesis | 17.22 |
| Gene ID | Gene abbreviation | Fold Δ1 |
| VEGF | Vascular endothelial growth factor | 41718.7 |
| snoZ40 | Small nucleolar RNA Z40 | 16658.2 |
| ssc-mir-195a-1 | MicroRNA mir-195a-1 | 6514.83 |
| ssc-mir-125b-1 | MicroRNA mir-125b-1 | 3627.42 |
| ssc-mir-423 | MicroRNA mir-423 | 3627.42 |
| SNORD53 | Small nucleolar RNA, C/D box53 | 2811.10 |
| ssc-mir-421 | MicroRNA mir-421 | 2811.10 |
| ssc-mir-133a-2 | MicroRNA mir-133a-2 | 2558.12 |
| ssc-mir-365-1 | MicroRNA mir-365-1 | 2558.12 |
| SNORD16 | Small nucleolar RNA, C/D box 16 | 1497.27 |
| ssc-mir-31 | MicroRNA mir-31 | 1144.09 |
| SERP2 | Stress-associated ER protein family member 2 | 1137.16 |
| SNORD94 | Small nucleolar RNA, C/D box 94 | 1126.85 |
| SNORD67 | Small nucleolar RNA, C/D box 67 | 816.01 |
| Y_RNA | Non-coding RNA | 727.41 |
| GRIA | FAD-dependent oxygenase | 370.13 |
| CH242-74M17.6 | GAS locus protein, porcine-specific | 322.43 |
| CH242-74M17.6 | GNAS complex locus, porcine-specific | 322.43 |
| SNORA36 | Small nucleolar RNA, C/D box 36 | 199.10 |
| RBP1 | Retinol-binding protein 1 | 182.72 |
| TIMP-3 | TIMP metallopeptidase inhibitor 3 | 173.55 |
| RNF130 | Ring finger protein 130 | 147.15 |
| MT-ND6 | Mitochondrial NADH dehydrogenase 6 | 141.82 |
| PCDH7 | Protocadherin 7 | 90.60 |
| ND4L | NADH dehydrogenase subunit 4L | 89.11 |
| U12 | Non-coding RNA | 85.94 |
| MT1A | Metallothionein 1A | 84.09 |
| ZFX | Zinc finger protein X-linked | 79.94 |
| CH242-17J7.1 | GNAS complex locus, porcine-specific | 65.88 |
| RNPC1 | RNA binding motif protein 38 | 65.10 |
| GEM1 | Gremlin 1 | 54.41 |
| CADM4 | Cell adhesion molecule 4 | 39.92 |
| CYP1A1 | Cytochrome P450, family 1, subfamily A1 | 39.89 |
| TFAP2A | Transcription factor AP-2 alpha | 39.27 |
| AFP | Alpha-fetoprotein | 33.23 |
| CFD | Complement factor D | 26.62 |
| GNG | Two-component system response regulator GnG/NtrC | 24.20 |
| RNF128 | Ring finger protein 128 | 23.79 |
| MIF | Macrophage migration inhibitory factor | 23.69 |
| IFI27l2 | Interferon, alpha-inducible protein 27-like 2 | 22.72 |
| PDE6H | Phosphodiesterase 6H, cGMP-specific | 22.54 |
| KLF5 | Krüppel-like factor 5 | 22.53 |
| PRKD1 | Protein kinase D1 | 21.27 |
| SPINT2 | Serpin peptidase inhibitor, Kunitz type, 2 | 18.54 |
| MYC | Myelocytomatosis oncogene | 17.64 |
| CD44 | Cell surface glycoprotein 44 | 17.14 |
| GPC3 | Glypican 3 | 16.54 |
| ECHS1 | Enoyl-coA hydratase, short-chain, 1 | 15.42 |
| S100A14 | S100 calcium binding protein A14 | 15.41 |
| LONRF3 | LON peptidase N-terminal domain and ring finger 3 | 15.25 |
| CU463271.1 | Protein encoded by CU463271.1 locus | 14.86 |
| MMP16 | Matrix metalloproteinase 16 | 14.28 |
| S100A16 | S100 calcium binding protein A16 | 14.14 |
| SLC22A17 | Solute carrier family 22 member A17 | 14.07 |
| ID1 | Inhibitor of DNA binding 1 | 13.34 |
| ATP5E | ATP synthase H+ transporting epsilon | 13.14 |
| ISG12(a) | ISG12 protein-like | 13.03 |
| UQCRH | Uniquinol-cytochrome c reductase hinge protein | 12.44 |
| DKK2 | Dickkopf homolog 2 | 12.20 |
| IL17RA | Interleukin 17 receptor A | 12.13 |
| STXBP6 | Syntaxin binding protein 6 | 11.63 |
| F2 | Coagulation factor II | 11.57 |
| COX17 | Cytochrome c oxidase copper chaperone | 10.05 |
| MGMT | O-6-methylguanine-DNA methyltransferase | 9.96 |
| SDF4 | Stromal cell derived factor 4 | 9.50 |
| PDPN | Podoplanin | 9.46 |
| DLK2 | Delta-like 2 homolog | 9.24 |
| WNT1 | Wingless-related MMTV integration site family member 1 | 9.03 |
| CISD1 | CDGSH iron sulfur domain 1 | 9.00 |
| MYD88 | Myeloid differentiation primary response 88 | 8.71 |
| SDC4 | Syndecan 4 | 8.68 |
| EPCAM | Epithelial cell adhesion molecule | 8.59 |
| RPS29 | Ribosomal protein S29 | 8.32 |
| P14ARF | p14 ARF protein (porcine-specific) | 8.29 |
| ELAVL2 | ELAV-like neuron-specific RNA binding protein 2 | 8.23 |
| FZD3 | Frizzled homolog 3 | 8.17 |
| XBP1 | X-box binding protein 1 | 8.09 |
| TRIAP1 | TP53 regulated inhibitor of apoptosis 1 | 8.08 |
| ATP5O | ATP synthase H+ transporting O subunit | 8.02 |
| VKORC1 | Vitamin K epoxide reductase complex, subunit 1 | 7.80 |
| FAM84B | Family with sequence similarity 84, member B | 7.70 |
| ECI2 | Enoyl-coA delta isomerase 2 | 7.69 |
| ARHGDIB | Rho GDP dissociation inhibitor beta | 7.41 |
| C20ORF27 | Human chromosome 22 orf39 homolog | 7.31 |
| RAB6B | RAS oncogene family member | 7.28 |
| DDT | D-dopachrome tautomerase | 7.14 |
| AGMAT | Agmatine ureohydrolase (Agmatinase) | 7.12 |
| CHCHD2 | Coiled-coil-helix-coil-helix domain 2 | 6.92 |
| EPHA7 | Eph receptor A7 | 6.72 |
| OSR2 | Odd-skipped related 2 | 6.71 |
| CFI | Complement factor I | 6.55 |
| NDUFB11 | NADH dehydrogenase 1 alpha subcomplex 11 | 6.37 |
| SLC35F1 | Solute carrier family 35 member F1 | 6.37 |
| NUDT14 | Nucleoside diphosphate linked moiety X-type motif 14 | 6.35 |
| SH3GL3 | SH3 domain GRB2-like 3 | 6.32 |
| NDUFA12 | NADH dehydrogenase 1 alpha subcomplex 12 | 6.30 |
| CDO1 | Cysteine dioxygenase type 1 | 6.28 |
| DYNLT1 | Dynein, light-chain, Tctex-type | 6.27 |
| HSBP1 | Heat shock factor binding protein 1 | 6.20 |
| CCK | Chloecystokinin | 5.98 |
| ANXA1 | Annexin A2 | 5.85 |
| FCGRT | Fc fragment of IgG, receptor, transporter, alpha | 5.84 |
| AK5 | Adenylate kinase 5 | 5.83 |
| RDH11 | Retinol dehydrogenase 11 | 5.58 |
| RPS18 | Ribosomal protein S18 | 5.56 |
| TGOLN2 | Trans-golgi network protein 2 | 5.55 |
| RNF150 | Ring finger protein 150 | 5.54 |
| HIFX | Hypoxia-inducible factor X | 5.47 |
| CFH | Complement factor H | 5.43 |
| NPC2 | Niemann-Pick disease, type C2 | 5.34 |
| DCTPP1 | dCTP pyrophosphatase 1 | 5.25 |
| CLVS1 | Clavesin 1 | 5.24 |
| HSF4 | Heat shock transcription factor 4 | 5.23 |
| SERINC2 | Serine incorporator 2 | 5.08 |
| MSMB | Microseminoprotein, beta | 5.03 |
| CHID1 | Chitinase domain containing 1 | 4.98 |
| LYSMD2 | LysM, putative peptidoglycan-binding, domain 2 | 4.95 |
| SH3BGRL2 | SH3 domain binding glutamic acid-rich protein-like 2 | 4.86 |
| POU3F1 | POU domain, class 3, transcription factor 1 | 4.77 |
| TCN2 | Transcobalamin II | 4.76 |
| SORT1 | Sortilin 1 | 4.75 |
| PTGES | Prostaglandin E synthase | 4.65 |
| CCND2 | Cyclin D2 | 4.63 |
| GXYLT2 | Glucoside xylosyltransferase 2 | 4.50 |
| CRTAC1 | Cartilage acidic protein 1 | 4.33 |
| MFGE8 | Milk fat globule-EGF factor 8 | 4.29 |
| TNC | Tenascin C | 4.28 |
| PCBD1 | Pterin-4 alpha-carbinolamine | 4.27 |
| PDE10A | Phosphodiesterase 10A | 4.22 |
| RAB27B | RAS oncogene family member | 4.18 |
| INCENP | Inner centromere protein antigens 135/155kDa | 4.14 |
| GJB1 | Gap junction protein, beta 1 | 4.09 |
| CEBPG | CCAAT/enhancer binding protein, gamma | 4.04 |
| DUROC-SLA1 | Src-like adaptor 1 (pig-specific) | 4.04 |
| ITIH4 | Inter-alpha-trypsin inhibitor heavy chain family, member 4 | 4.03 |
| PEBP1 | Phosphatidylethanolamine binding protein 1 | 4.02 |
| ABTB2 | Ankyrin repeat and BTB (POZ) domain containing 2 | 4.00 |
| GJB5 | Gap junction protein, beta 5 | 3.95 |
| CREB3 | cAMP responsive element binding protein 3 | 3.86 |
| DPM2 | Dolichyl-phasphate mannosyltranferase polypeptide 2 | 3.85 |
| GEM | GTP binding protein overexpressed in skeletal muscle | 3.85 |
| DOCK8 | Dedicator of cytokinesis 8 | 3.84 |
| TMP-SLA-2 | Transmembrane protein, pig-specific | 3.83 |
| CYB5B | Cytochrome b5 type B | 3.80 |
| TSAN6 | Tsan6, Pig-specific protein | 3.75 |
| RNASEH2C | Ribonuclease H2, subunit C | 3.71 |
| NARS | Asparaginyl-tRNA synthetase | 3.70 |
| LSM6 | U6 snRNP-associated protein | 3.69 |
| MTFP1 | Mitochondrial fission process 1 | 3.66 |
| SLC16A1 | Solute carrier family 16 member A1 | 3.65 |
| RARB | Retinoic acid receptor, beta | 3.64 |
| SDSL | Serine dehydrarase-like | 3.64 |
| MPC2 | Mitochondrial pyruvate carrier 2 | 3.63 |
| PARVB | Parvin, beta | 3.60 |
| RENBP | Renin binding protein | 3.55 |
| POC1A | POC1 centrolar protein A | 3.54 |
| GUSB | Glucuronidase, beta | 3.53 |
| CH242-9E10.1 | GNAS complex locus, porcine-specific | 3.52 |
| REEP3 | Receptor accessory protein 3 | 3.51 |
| CPT1A | Carnitine palmitoyltransferase 1A | 3.40 |
| BHLHB9 | Basis helix-loop-helix domain containing B9 | 3.37 |
| FAAH | Fatty acid amide hydrolase | 3.37 |
| MRPL20 | Mitochondrial ribosomal protein L20 | 3.37 |
| HTR2A | 5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled | 3.36 |
| TRERF1 | Transcriptional regulating factor 1 | 3.35 |
| XYLB | Xylulokinase | 3.32 |
| RBPMS2 | RNA binding protein with multiple splicing 2 | 3.28 |
| FGFR3 | Fibroblast growth factor receptor 3 | 3.27 |
| SLC22A23 | Solute carrier family 22 member A23 | 3.21 |
| NDUFB1 | NADH dehydrogenase 1 alpha subcomplex 1 | 3.18 |
| ZFAND2A | Zinc finger, AN1-type domain 2A | 3.16 |
| EPB41 | Erythrocyte membrane protein band 4.1 | 3.11 |
| IGSF23 | Immunoglobulin superfamily, member 23 | 3.11 |
| FAM122C | Family with sequence similarity 122, member C | 3.10 |
| AMIGO2 | Adhesion molecule with Ig-like domain | 3.03 |
| SHROOM4 | Shroom family member 4 | 3.02 |
| MAT1A | Methionine adenosyltransferase 1A | 3.01 |
| PARN | Poly(A)-specific ribonuclease | 3.00 |
| PFKFB1 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 | 3.00 |
| TCF25 | Transcription factor 25 | 2.98 |
| PRODH2 | Proline dehydrogenase (oxidase) 2 | 2.95 |
| CHCHD10 | Coiled-coil-helix-coil-helix domain 10 | 2.91 |
| CH242-217H9.3 | GNAS complex locus, porcine-specific | 2.85 |
| CASC5 | Cancer susceptibility candidate 5 | 2.84 |
| IGLON5 | IgLON family member 5 | 2.84 |
| CDH17 | Cadherin 17 | 2.82 |
| SLC44A4 | Solute carrier family 44 member A4 | 2.82 |
| LRCH2 | Leucine-rich repeats and calponin homology domain 2 | 2.76 |
| DUROC-NEU1 | Sialidase 1 (pig-specific) | 2.75 |
| CDC45 | Cell division cycle 45 | 2.71 |
| BCKDHB | Branched chain keto acid dehydrogenase E1 | 2.69 |
| HNF4A | Hepatocyte nuclear factor 4, alpha | 2.67 |
| SCG5 | Secretogranin V | 2.66 |
| BOLA1 | BolA family member 1 | 2.65 |
| PYGL | Phosphorylase, glycogen, liver-specific | 2.64 |
| SCARB1 | Scavenger receptor class B, member 1 | 2.64 |
| SERPIND1 | Serpin peptidase inhibitor, clade D, member 1 | 2.63 |
| GAS2L3 | Growth arrest-specific 2 like 3 | 2.61 |
| EFNA2 | Ephrin A2 | 2.60 |
| PDZK1IP1 | PDZK1 interacting protein 1 | 2.59 |
| VIMP | VCP-interacting membrane protein | 2.59 |
| PRR5L | Proline-rich 5-like | 2.57 |
| ACVR2B | Activin A receptor, type IIB | 2.56 |
| ATP9A | ATPase, class II, type 9A | 2.55 |
| PLAUR | Plasminogen activator, urokinase receptor | 2.53 |
| LDLR | Low density lipoprotein receptor | 2.51 |
| FLRT3 | Fibronectin leucine-rich transmembrane protein 3 | 2.50 |
| ADAMTSL3 | ADAMTS-like 3 | 2.49 |
| CINP | Cyclin-dependent kinase 2 interacting protein | 2.48 |
| WBSCR22 | Williams Beuren syndrome chromosome region 22 | 2.48 |
| ALDH9A1 | Aldehyde dehydrogenase 9 family member 1 | 2.46 |
| SSC.82438 | Sus scrofa Amphiregulin | 2.44 |
| NME3 | NME/NME23 nucleoside diphosphate kinase 3 | 2.43 |
| CCS | Copper chaperone for superoxide dismutase | 2.40 |
| PITX2 | Paired-like homeodomain transcription factor 2 | 2.38 |
| MRPL3 | Mitochondrial ribosomal protein L3 | 2.33 |
| BNC1 | Basonuclin 1 | 2.32 |
| RPS16 | Ribosomal protein S16 | 2.32 |
| TM7SF2 | Transmembrane 7 superfamily member 2 | 2.30 |
| CRELD2 | Cystein-rich with EGF-like domains 2 | 2.22 |
| HHEX | Hematopoietically expressed homeobox | 2.20 |
| RPL31 | Ribosomal protein L31 | 2.20 |
| TGFA | Transforming growth factor, alpha | 2.20 |
| IDS | Iduronate 2-sulfatase | 2.18 |
| NRG4 | Neuregulin 4 | 2.17 |
| DGAT1 | Diacylglycerol O-acyltransferase 1 | 2.16 |
| GNAZ | Guanine nucleotide binding protein | 2.14 |
| ARL3 | ADP-ribosylation factor-like 3 | 2.12 |
| FABP5 | Fatty acid binding protein 5 | 2.12 |
| KIF12 | Kinesin family member 12 | 2.12 |
| IKBKG | Inhibitor of kappa B | 2.09 |
| PDZRN3 | PDZ domain containing ring finger 3 | 2.09 |
| PHF19 | PHD finger protein 19 | 2.09 |
| MRPL24 | Mitochondrial ribosomal protein L24 | 2.07 |
| ARAP2 | ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 | 2.04 |
| ITGF3 | ItgF3-like protein | 2.02 |
| POLA2 | Polymerase (DNA directed), alpha 2 | 2.02 |
| ADAMTS7 | ADAM metallopeptidase with thrombo- Spondin type 1 motif 7 | 1.45 |
In contrast, ectopic expression of MYC protein upregulated the expression of 237 genes in PICM-19-CSCs not found in the PICM-19 stem cell profile (Table 3). New genes upregulated by MYC included those involved in cell growth and proliferation, adhesion, migration, protein transport, cell cycle, metabolism, inflammation, and tumorigenesis underscoring the broad reach of MYC protein in regulating diverse cellular processes. However, genes coding RNAs involved in transcriptional and post-transcriptional regulatory mechanisms dominated the top 25 MYC-upregulated genes (Table 2), consistent with the active cell growth and proliferative nature of PICM-19-CSCs.
| Gene ID | Gene abbreviation | Function | log2 ratio |
| CH242-278B18.2 | GNAS Complex, porcine-specific | Unknown | 10.25 |
| NFKB1 | Nuclear factor κB | Inflammation | 8.94 |
| SEMA3F | Semaphorin 3F | Cell signaling | 8.27 |
| NME1-NME2 | NME1-NME2 read-through | Inflammation | 8.22 |
| SERPINA1 | Serpin protease inhibitor, clade A1 | Inflammation | 8.17 |
| VLDLR | Very low density lipoprotein receptor | Metabolism | 8.01 |
| APOE | Apolipoprotein E | Metabolism | 7.57 |
| ITGA3 | Integrin, alpha 3 | Migration, Cell adhesion, Invasion | 7.41 |
| IGF2BP2 | Insulin-like growth factor 2 mRNA binding protein 2 | Gene regulation | 7.33 |
| MT-ATP6 | Mitochondrial ATP synthase 6 | Metabolism | 7.21 |
| HIF1A | Hypoxia-inducible factor 1α | Angiogenesis | 7.12 |
| APOA1 | Apolipoprotein A1 | Metabolism | 7.06 |
| ITGB7 | Integrin, beta 7 | Cell adhesion | 7.03 |
| NKAIN1 | Na+/K+ transporting ATPase | Ion transport | 7.00 |
| FGG | Fibrinogen gamma chain | Cell adhesion | 6.79 |
| CTSB | Cathepsin B | Carcinogenesis | 6.74 |
| NTN1 | Netrin 1 | Inflammation | 6.65 |
| JUP | Junction plakoglobin | Cell adhesion | 6.61 |
| PLG | Plasminogen | Cell proliferation | 6.60 |
| MT-ND2 | Mitochondrial NADH dehydrogenase 2 | Metabolism | 6.37 |
| SERPINA3-2 | Serpin protease inhibitor, clade A3-2 | Inflammation | 6.31 |
| RBP4 | Retinol binding protein 4 | Metabolism | 6.18 |
| FTL | Ferritin, light polypeptide | Iron metabolism | 6.08 |
| DUSP4 | Dual specificity phosphatase 4 | Cell proliferation | 6.02 |
| MT-CO1 | Mitochondrial cytochrome c oxidase I | Inflammation | 6.00 |
The farnesoid X receptor (FXR), a member of the nuclear receptor superfamily, plays an important role in regulating the progression of several cancers including HCC; and miR-421, which was highly upregulated in PICM-19-CSCs, suppresses the expression of FXR to induce tumorigenesis and migration in HCC cells[21]. MiR-421 also regulates deleted in pancreatic cancer 4 (DPC4; also known as Smad4), a critical tumor suppressor shown to be involved in the progression of pancreatic cancer[22].
In addition to these regulatory RNAs, protocadherin-7, tissue inhibitor of metalloproteinase 3 and vascular endothelial growth factor (VEGF) were highly expressed in PICM-19-CSCs compared to PICM-19 parental stem cells. The ratio between the matrix metalloproteinsases (MMPs) and their inhibitors (TIMPs) is an important factor in balancing tissue homeostasis. Recent studies have shown that the TIMP family of proteins not only inhibits several disintegrin-metalloproteinases (ADAMs and ADAMTSs) but also has contrasting functions including anti- and pro-apoptotic[23]. The sequestration of TIMP-3 by binding to the extracellular matrix (ECM) or through endocytosis may help to protect cells from apoptosis while still allowing it to control pericellular proteolysis via its potent inhibition of MMPs, ADAMs and ADAMTSs[23]. In PICM-19-CSCs, only TIMP-3 was upregulated (Table 2), and ADAM23 and ADAMTS2 were suppressed (Table 4).
| Gene ID | Gene abbreviation | Function | log2 ratio |
| ASPN | Asporin | Extracellular matrix | 8.20 |
| IGF1 | Insulin-like growth factor 1 | Cell growth | 7.44 |
| OGN | Osteoglycin | Metabolism | 6.52 |
| GPM6B | Glycoprotein M6B | Membrane trafficking, cell-to-cell communication | 6.23 |
| ISLR | Immunoglobulin superfamily containing leucine-rich repeat | Unknown | 6.19 |
| ANGPT1 | Angiopoietin 1 | Angiogenesis | 5.53 |
| WDR66 | WD repeat domain 66 | EMT modulator | 5.49 |
| RSPO2 | R-spondin 2 homolog | Wnt modulator | 5.46 |
| COL6A2 | Collagen, type VI, alpha 2 | Fibrosis | 5.14 |
| TNR | Tenascin R | Extracellular matrix | 5.03 |
| FAM5C | Family with sequence similarity 5, member C | Unknown | 4.97 |
| FIGF | c-Fos iduced growth factor | Angiogenesis | 4.92 |
| ADAM23 | ADAM metallopeptidase domain 23 | Metabolism | 4.84 |
| MAB21L2 | Mab-21-like 2 | Cell survival | 4.83 |
| PDE1A | Phosphodiesterase 1A | Metabolism, Apoptosis | 4.83 |
| CLMP | CXADR-like membrane protein | Cell adhesion | 4.74 |
| COL28A1 | Collagen, type XXVIII, alpha 1 | Fibrosis | 4.74 |
| GJA1 | Gap junction protein, alpha 1 | Oxidative stress | 4.67 |
| SELENBP1 | Selenium binding protein 1 | Tumor suppressor | 4.58 |
| CPED1 | Cadherin-like and PC-esterase domain containing 1 | Unknown | 4.52 |
| COL5A3 | Collagen, type V, alpha 3 | Fibrosis | 4.51 |
| ADAMTS2 | ADAM metallopeptidase with thrombospondin typ1 motif 2 | Metabolism | 4.50 |
| PDE8B | Phosphodiesterase 8B | Metabolism | 4.47 |
| SGCE | Sarcoglycan, epsilon | Cytoskeleton | 4.42 |
| GDPD2 | Glycerophosphodiester phosphodiesterase domain containing 2 | Cell growth | 4.00 |
It has been well established that VEGF is highly produced in HCC, and the inhibition of VEGF results in the suppression of angiogenesis[24]. Moreover, miR-26a targets VEGF and hepatocyte growth factor (HGF) to cause this effect via the HGF-cMet pathway[24] as well as the phosphatidylinositol 3-kinase (PIK3C2α)/Akt/hypoxia-inducible factor (HIF)-1α pathway in HCC cells[25]. In our study, downregulation of miR-26a and upregulation of HIF-1α in PICM-19-CSCs demonstrates that MYC is also a player in this tumor-inducing molecular mechanism of HCC.
Next, we studied the profiles of PICM-19 and PICM-19-CSCs to identify the genes whose expression was not altered in stem cells due to MYC overexpression. For this, we selected genes that showed more than a two-fold increase when normalized with the GAPDH. We found that the expression of 28 genes was not affected by the MYC protein (Table 5). These genes included a number of microRNAs (miRNAs) and small nucleolar RNAs (SnoRNAs) that regulate post-transcriptional gene expression, and are expressed at very high levels. As MYC has no influence on their expression, it appears that the genes of this 28-gene set may be primarily involved in maintaining the cellular integrity and stemness.
| Gene ID | Gene abbreviation | Function | Fold Δ1 |
| SSC-MIR-425 | MicroRNA mir-425 | Gene regulation | 7925.28 |
| SNORD53 | Small nucleolar RNA, C/D box 53 | Gene regulation | 4969.50 |
| SSC-MIR-378-1 | MicroRNA mir-378-1 | Gene regulation | 3962.64 |
| SSC-MIR-99A | MicroRNA mir-99a | Gene regulation | 3761.27 |
| SSC-MIR-101-2 | MicroRNA mir-101-2 | Gene regulation | 3400.56 |
| SSC-LET-7F-2 | MicroRNA let-7f-2 | Gene regulation | 2814.68 |
| 5s rRNA | 5S ribosomal RNA | Protein synthesis | 2255.63 |
| SSC-MIR-210 | MicroRNA mir-210 | Gene regulation | 1730.53 |
| U6 | Small nucleolar RNA U6 | RNA interference | 1187.43 |
| SSC-MIR-196A-2 | MicroRNA mir-196a-2 | Gene regulation | 1127.81 |
| SCARNA15 | Small Cajal body-specific RNA 15 | Gene regulation | 287.24 |
| SNORA72 | Small nucleolar RNA, H/ACA box 72 | Gene regulation | 255.25 |
| SCARNA11 | Small Cajal body-specific RNA 11 | Gene regulation | 163.50 |
| RAMP2 | Receptor (G protein-coupled) activity modifying protein 2 | Protein transport | 56.66 |
| CCNI2 | Cyclin I family member 2 | Cell cycle | 10.67 |
| P4HTM | Prolyl 4-hydroxylase, transmembrane | Hypoxia | 6.25 |
| SPDYA | Speedy/RINGO cell cycle regulator family member A | Cell cycle | 5.56 |
| PARK2 | Parkin RBR E3 ubiquitin ligase | Unknown | 5.09 |
| GRPR | Gastrin-releasing peptide receptor | Cell proliferation | 4.52 |
| ATG10 | Autophagy related 10 | Autophagy | 3.76 |
| PHYHIP | Phytanoyl-CoA hydroxylase interacting protein | Cell proliferation | 3.41 |
| DPEP1 | Dipeptidase 1 | Metabolism | 2.72 |
| DUROC-C2 | Nuclear receptor corepressor 2 | DNA binding | 2.64 |
| HIST1H2BH | Histone cluster 1, H2bh | Histone modification | 2.58 |
| PHKG1 | Phosphorylase kinase, gamma 1 | Glycogenolysis | 2.52 |
| TMEM79 | Transmembrane protein 79 | Protein secretion | 2.04 |
| APH1B | APH1B gamma secretase subunit | Protein transport | 2.01 |
| IRAK4 | Interleukin-1 receptor-associated kinase 4 | Cell signaling | 2.01 |
In order to identify genes that are involved in MYC-induced oncogenesis, we studied global transciptome profiles of PICM-19-CSCs and compared them with those obtained with PICM-19 stem cells. While these genes are expressed by both PICM-19 and PICM-19-CSCs, a significant number of genes were differentially regulated by MYC. While MYC upregulated 4173 genes in PICM-19-CSCs, it downregulated 4248 genes. Statistical analysis of the data has revealed 616 upregulated and 391 downregulated genes that had a log2 ratio of at least 2. Analysis of gene expression of these two cell types showed that MYC overexpression resulted in changes in gene expression among genes involved in diverse cellular processes. This is not surprising as it is known that MYC regulates about 15% of the genes in mammalian genomes[26].
Among the genes upregulated by MYC are ribosomal genes demonstrating active protein synthesis in CSCs which is associated with cell growth, proliferation, and metabolism. While our data show only 8 of these ribosomal genes, others such as the initiation factors (elF-2a, eIF4A, eIF4E, eIF4G, eIF5A), MrDb, nucleolin, and ribosomal proteins (S11, S16) were also induced > 1.5 fold (data not shown). These genes were previously shown to be regulated by the MYC protein[27].
MYC is also considered an important regulator of the cell cycle. We found that the expression of several cell cycle genes (cdc45, CCND2, CIPN) was enhanced in PICM-19-CSCs. Adhesion molecule with Ig-like domain 2 (AMIGO2), a leucine-rich repeat family member implicated in decreased cell adhesion and migration, and an increase in chromosomal instability and tumorigenicity in human gastric adenocarcinoma[28], was also upregulated in PICM-19-CSCs.
The majority of the top 25 genes upregulated by MYC are those involved in metabolism, adhesion and inflammation (Table 3). Integrin genes ITGA3 and ITGB4, and plakoglobin JUP were upregulated in PICM-19-CSCs. These three genes were recently found to be responsible for metastasis seen in oral squamous cell carcinoma[29]. Their expression in CSCs is significant because integrin adhesion receptors link the ECM to actin cytoskeleton and transmit biochemical signals across the plasma membrane[30]. Binding of integrins to their cognitive receptors trigger signaling cascades to induce diverse cellular processes such as cell survival, apoptosis, proliferation, and motility[29]. JUP expression was also associated with colorectal tumor metastasis[31].
Interestingly, markers of inflammation in the liver, apolipoproteins (APOA1 and APOE) and the SERPINA1 gene encoding alpha1-antitypsin (AAT), and another member of the AAT family, SERPINA3-2 were also induced in PICM-19-CSCs. While APOE was shown to confer protection against liver damage caused by hepatitis C virus (HCV) infection[32], its expression level was found to be significantly higher in HBV associated liver cirrhosis[33]. In a recent HCC study on familial aggregation, APOA1 and APOE were identified by mass spectroscopy of the proteins as being among nine highly overexpressed in individuals with HCC[34]. Previous studies have shown that the protein level of APOE was increased in human HCC[35], and that human hepatoma-derived cell lines express high levels of APOE and very low density lipoprotein (VLDL). Consistent with this, PICM-19-CSCs displayed an upregulated expression of VLDLR (Table 3). AAT is the most abundant liver-derived glycoprotein in the plasma, and a diagnostic and prognostic marker of HCC[36,37]. It is associated with the progression from liver cirrhosis to cancer in HCV-infected individuals[38]. Collectively, upregulation of APOA1, APOE, VLDLR and AAT in PICM-19-CSCs suggested their involvement in MYC-induced liver cancer.
We found that MYC overexpression resulted in downregulation of a number of genes (Table 4) that were previously implicated in tumor reduction and/or inhibition of human cancers. These genes differ from those that were silenced; and the reduced expression could be a result of either direct or indirect MYC targeting (Table 1). Among these, c-Fos induced growth factor, FIGF (also known as vascular endothelial growth factor D), is active in angiogenesis and endothelial cell growth. Its putative role in human cancers is unclear. While it was shown to enhance lymphatic node metastasis in tissue sarcomas[39], its expression was significantly lower in the serum of metastatic patients with differentiated thyroid sarcoma suggesting that FIGF may prevent metastasis. Interestingly, MYC caused the downregulation of FIGF in PICM-19-CSCs (Table 4) indicating that, in liver stem cells, FIGF has an anti-tumor function. Osteoglycin, a protein that decreases gelatinase activity and suppresses lymphatic metastasis[40], was also downregulated in PICM-19-CSCs.
Collagens are structural components of the ECM and are primarily synthesized by hepatic stellate cells in the liver to induce fibrosis[41]. A recent proteomic study with platelet-derived growth factor C transgenic and phosphatase and tensin homolog null mouse models of liver fibrosis and its progression to HCC demonstrated that the protein levels of collagens COL6A2 and COL28A1 were downregulated in fibrosis, and COL5A3 was upregulated in fibrosis; whereas only COL6A2, but not COL5A3 and COL28A1, was increased in tumors generated in both mouse models[42]. We found that COL5A3, COL6A2, COL28A1 were significantly downregulated in PICM-19-CSCs. Thus, in liver cells, MYC may be involved in the regulation of COL5A3 and COL28A1.
Among regulatory RNAs upregulated in PICM-19-CSCs, miR-125b was recently shown as a negative regulator of p53[43]. While the knockdown of miR-125b elevated p53 levels and induced apoptosis in zebra fish and human cells[43], its overexpression promoted human glioma cell proliferation and inhibited apoptosis by targeting the pro-apoptotic Bcl-2 modifying factor[44].
MiR-31 is an oncomir that is upregulated in PICM-19-CSCs. In colorectal cancer cells, miR-31 activated the rat sarcoma (RAS) pathway and inhibited the function of the RAS p21 GTPase activating protein 1 (RASA1) in promoting tumorigenesis and cancer cell growth[45]. It was also shown to promote oncogenesis in the cholangiocarcinoma HCCC-9810 cell line by inhibiting RASA1, thereby downregulating the mitogen-activated protein kinase signaling pathway[46]. Based on these findings, it appears that miR-31 may also be inhibiting the RASA1 in liver cancers. Paradoxically, miR-195, which was shown to suppress tumorigenicity and cause cell cycle arrest by blocking the G1/S transition via the repression of retinoblastoma-E2 transcription factor signaling through cyclin D1, CDK6, and E2F3 in HCC cell lines[47], was upregulated in PICM-19-CSCs.
High levels of miR-365 were found in PICM-19-CSCs. The expression levels of miR-365 vary in different malignancies. It is was overexpressed in both cells and clinical specimens of cutaneous squamous cell carcinoma[48], where it was found to suppress to Mybl2 activity[49]. siRNA silencing of Mybl2 in proliferating Caco-2 cells increased the expression of the cell-cycle regulators cdk2, cyclin D2, and c-MYC; and decreased the expression of cdc25B and cyclin B2[50]. Thus, it is possible that MYC protein suppresses the expression of Mybl2 through the upregulation of miR-365, as seen in the PICM-19-CSCs.
Another key and common feature of CSCs was activation of NF-κB signaling, which is known to increase stress resistance and survival[10]. PICM-19-CSCs had a high level of NF-κB expression when compared to PICM-19 cells. Similarly, the expression of anti-apoptotic genes Bcl-xl and Mcl-1 were elevated in PICM-19-CSCs. Thus, the blocking of anti-apoptotic effects of these proteins may be a viable strategy to eliminate tumor cells. Sabutoclax, a pan-inhibitor of Bcl-2 family of protein that was shown recently to target Mcl-1[51], may be a suitable drug for this purpose.
Selenium-binding protein (SELENBP1) is a tumor suppressor and lower SELENBP1 levels are associated with poor prognosis of liver, lung, colorectal and stomach cancers[52,53]. Decreased SELENBP1 expression has been associated with increased cell motility, proliferation and inhibition of apoptosis, and vascular invasion in HCC[53]. Its expression was suppressed in PICM-19-CSCs suggesting that MYC regulates this gene in the liver to promote cell proliferation and invasion. In a recent proteomic study conducted with serum and tumor tissue specimens of c-MYC transgenic mice, it was shown that APOE, SELENBP1 and retinol binding protein 4 are differentially regulated[54]. Our data support these findings and confirms that MYC differentially regulates the expression of these proteins in liver cells.
Previous studies have also identified MYC as a downstream target of the Wnt signaling, a pathway that is highly active in HCC[55,56]. Dysregulated Wnt signaling occurs in 30% of HCCs. Also, deletion of adenomatous polyposis coli (APC), a negative regulator of Wnt, in the adult mouse liver, leads to an increase in the nuclear β-catenin and MYC[57]. Gene expression array analysis of colorectal cancer cells has shown that MYC is required for the majority of Wnt targeted gene activation after the APC loss[58]. In our study, we found a high level of Wnt expression (Table 2) demonstrating that the Wnt pathway was activated in PICM-19-CSCs.
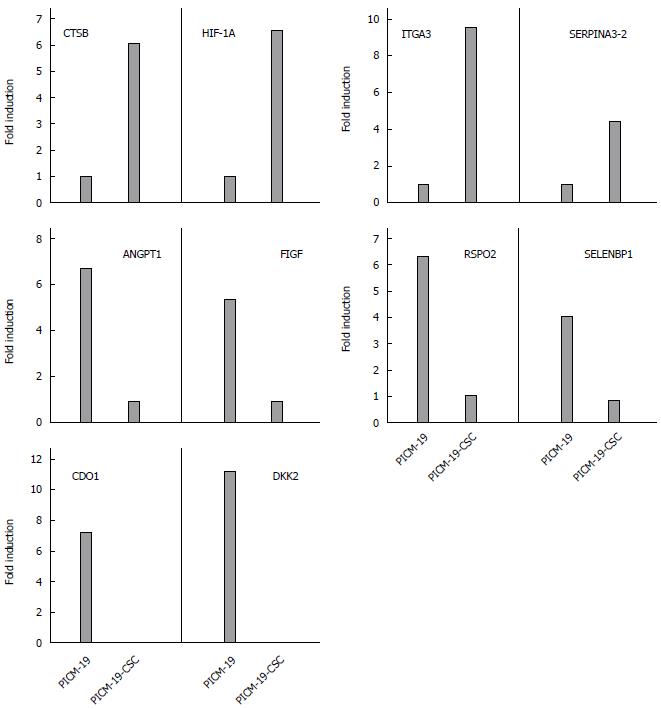
To further validate these RNA-seq data, four genes that were upregulated, four genes downregulated, and two genes that were silenced in PICM-19-CSCs, when compared with PICM-19 cells (Tables 1, 3-4) were selected for quantitative real-time PCR studies on the basis of their roles in diverse cellular processes. PCR was performed using porcine-specific primers for CTSB, HIF-1A α-subunit, SERPINA3-2, ITGA3, SELENBP1, RSPO2, FIGF, ANGPT1, CDO1 and DKK2 (Table 6). While two of them (HIF-1α and SELENBP1) were suggested as biomarkers for HCC, others were identified in studies with multiple human cancers. As shown in Figure 3, induced expression of MYC in PICM-19-CSCs resulted in the alteration of gene expression for all ten genes studies. These findings clearly illustrate differences in the expression of these genes between PICM-19 stem cells and PICM-19-CSCs.
| Gene | Gene ID | GenBank# | Primer sequences | Product size (bp) |
| CTSB | 100370961 | NM_001097458.1 | F: AGCCTGGAACTTCTGGACAA | 190 |
| R: TTTGTAGGACGGGGTGTAGC | ||||
| HIF-1A α-subunit | 396696 | NM_001123124.1 | F: GCCAGAACCTCCTGTAACCA | 204 |
| R: CCTTTTCCTGCTCTGTTTGG | ||||
| SERPINA3-2 | 396686 | NM_213787.1 | F: TGAAAGTTTCCCAGGTGGTC | 200 |
| R: CTAGGGCTTGGTGACTTTGC | ||||
| ITGA3 | 100517053 | XM_005668879.1 | F: CTTCAAACGGAACCAGAGGA | 203 |
| R: AGAAGCTTTGTAGCCGGTGA | ||||
| SELENBP1 | 100152724 | XM_001929643.3 | F: GGTACCAGCCTCGACACAAT | 203 |
| R: GTTGTGCAGGAATCGGATCT | ||||
| RSPO2 | 100154008 | NM_001293141.1 | F: GTCCAGATGGTTTTGCACCT | 202 |
| R: CTCCTGGATTCAGCAATGGT | ||||
| FIGF | 100155670 | XM_001928382.3 | F: TCCCCAACCCTGTCATTTTA | 202 |
| R: TCCTTCTCCATTCCTTGGTG | ||||
| ANGPT1 | 397009 | NM_213959.1 | F: GGGGGAGGTTGGACTGTAAT | 203 |
| R: TGTGAATAGGCTCGGTTTCC | ||||
| CDO1 | 100312964 | NM_001167643.1 | F: TCTGAAGATGCTGCAAGGAA | 208 |
| R: CGTATCGAAGGGTGGACTGT | ||||
| DKK2 | 100519672 | XM_003129269.2 | F: CTGGTACCCGCTGCAATAAT | 198 |
| R: GACAGGGATCTCCTTCATGC | ||||
| β-actin | 45269029 | AY550069.1 | F: GGACCTGACCGACTACCTCA | 180 |
| R: GGCAGCTCGTAGCTCTTCTC |
Collectively, the vast repertoire of genes that are either directly or indirectly induced by MYC protein expression in PICM-19-CSCs underscores the importance of its involvement in the induction of diverse cellular pathways that ultimately may lead to liver cancer.
CSCs have been identified in a number of cancers, including HCC, as a unique population of cells that possess self-renewing and tumor-initiating properties[5,8]. Mounting evidence strongly suggests that CSCs are resistant to anti-cancer therapies[11,59]. Therefore, targeting CSCs may be an effective strategy to eliminate the drug resistant and metastatic tumor cells[60]. Here, we have developed CSCs that are capable of forming tumors in mice by overexpressing human MYC in the liver stem cell line, PICM-19. The involvement of MYC in murine and human HCC malignant transformation has been well documented[2,4,7]. Cellular targets of MYC include genes from almost every biochemical and regulatory pathway, including growth, metabolism, cell cycle progression, differentiation and apoptosis[26,61]. Our results provide a snapshot of differential gene expression induced by the MYC protein in an in vitro model of liver stem cells. PICM-19-CSCs were able to form tumors in immunodeficient mice within three weeks, demonstrating their elevated proliferation and tumor-producing potential in vivo. Analysis of gene expression profiles in PICM-19-CSCs has revealed novel “molecular signatures” that were either up- or downregulated as a consequence of MYC overexpression. A number of genes were both up- and downregulated by MYC overexpression in PICM-19 cells demonstrating their potential involvement in MYC-induced tumorigenesis. We also found that MYC expression silenced six genes completely (Table 1), suggesting that the inhibition of these genes is potentially crucial for tumor induction by MYC. To our knowledge, this is the first report to investigate transcriptome profiles driven by a single oncogene in liver stem cells, and cells of porcine origin, an important animal for modeling human physiology and disease.
The use of MYC to generate CSCs is highly relevant to study HCC because MYC is a key component of hepatocarcinogenesis. High-level expression of MYC was also shown to be essential for the proliferation and survival of other CSCs, such as in glioma[62]. In a recent report, chemoresistant CSCs were isolated from MYC-driven mouse liver tumors[63]. Side-population (SP) cells in these tumors expressed CD44, a marker of hepatic progenitor cells, and displayed an upregulation in the expression of multidrug resistance gene 1 (MDR1), which conferred resistance to doxorubicin, paclitaxel and SN38[63]. We found that PICM-19-CSCs had a significant upregulated expression of MDR1 when compared to PICM-19 cells, suggesting that MDR1 can be an effective therapeutic target for MYC-driven liver cancers. Interestingly, CD44 expression was found only in PICM-19-CSCs but not in PICM-19 cells, demonstrating that MYC regulates CD44.
We discovered several notable similarities in comparing our RNA-seq data with those recently published from CD90+ liver CSCs isolated from HCC patients[64]. Among these, upregulation of glypican 3 (GPC3), apolipoprotein E (APOE), Polo-like kinase 2 and peroxisome proliferator-activated receptor γ, and downregulation of interleukins 8 and 12, suppressor of cytokine signaling 3, and chemokine ligands CXCL12 and CCL2 also occurred in PICM-19-CSCs. Interestingly, neither PICM-19 nor PICM-19-CSCs expressed the progenitor cell markers CD90 and CD133. This observation suggests species-specific differences between cells of human and porcine origin.
MYC also induced the expression of hepatic stem cell marker EpCAM, a target of the Wnt/β-catenin signaling pathway, in PICM-19-CSCs. EpCAM-expressing cells in HCC tumor specimens were previously shown to possess stem-like features and were able to initiate tumors within the liver[65]. PICM-19 cells expressed MYC, Oct4, Sox2, Klf4, and Nanog, which are factors that are commonly used to generate induced pluripotent stem cells. When MYC protein was overexpressed in PICM-19 cells the expression of these proteins was elevated, suggesting that such reprogramming factors are critical for the proliferation of CSCs, and may play a role in tumor-initiation, as shown recently in Ewing sarcoma[66] and breast cancer[67].
By comparing global transcriptome sequencing data between PICM-19 and PICM-19-CSCs, we have confirmed the involvement of a number of MYC-regulated genes involved in development (Sox2, Klf4, Nanog, Oct4, Amd1, Med12), stemness and self-renewal (Wnt/β-catenin), cell cycle (Cyclin D1, GPC3), metabolism (APOA1, APOE), and markers of hepatic stem/progenitor cells (EpCAM, KRT19). In fact, some of these were found to be expressed by side-population (SP) cells isolated from liver cancer cell lines[68]. Their upregulation in CSCs makes them potential candidates as therapeutic targets in liver cancers where MYC is overexpressed. In addition to these genes, PICM-19-CSCs also displayed high levels of a novel protein Sal-like protein 2 (SALL2), a member of the zinc finger transcription factor family. Recently, a family member SALL4 was found to be expressed in liver cancers[69], where it regulates the stemness of EpCAM+ CSCs[70]. While we did not detect SALL4 in PICM-19-CSCs, the induction of SALL2 suggests a certain degree of redundancy in the function of these proteins.
Hypoxia plays an important role in the recurrence of HCC, and HIF-1α is critical part of this process. After insufficient radiofrequency ablation of HCC or sustained treatment with sorafenib, tumor recurrence occurs via signaling pathways involving HIF-1α[71,72], and the expression level of HIF-1α was recently shown to correlate with unfavorable outcomes in patients with HCC[73]. We found that the expression of HIF-1α was significantly upregulated in CSCs. Another protein overexpressed in CSCs is cathepsin B (CTSB). Even though the role of CTSB in HCC is unclear, it has been shown to interact directly with the hepatitis B spliced protein-encoded by the HBV in hepatoma cell lines promoting tumor cell migration and invasion[74]. The alpha-1-antichymotrypsin 2 (SERPINA3-2) is a porcine-specific gene that is also significantly upregulated in CSCs (Figure 4). It shares 71% identity with human SERPINA3, a serine protease inhibitor[75]. While its involvement in HCC is unknown, it has been reported recently that the expression of SEPRINA3 is elevated in cholangiocarcinoma[76].
Epithelial-mesenchymal transition is a crucial event in the tumor progression of HCC. It has been reported recently that the expression of integrin α3 (ITGA3) is a molecular marker of cells undergoing EMT and of cancer cells with aggressive phenotypes[77]. ITGA3 expression is upregulated in breast cancer[77], and colorectal cancer[78]; and ITGA3-knockout mice were found to be considerably less susceptible to skin carcinogenesis[79], suggesting that ITGA3 is a critical gene in multiple cancers. While its involvement has not been demonstrated in HCC, we observed that its expression is enhanced in CSCs. This result indicated that ITGA3 may play a role in MYC-induced liver cancer.
The R-spondin proteins are secreted agonists of the canonical Wnt/β-catenin signaling pathway. Although they are unable to initiate Wnt signaling, they can enhance responses to low-dose Wnt proteins[80]. One of these proteins, R-spondin 2 (RSPO2) possesses tumor-suppressive activity and was recently shown to inhibit tumor growth in colorectal cancer cells by inhibiting Wnt signaling[81]. Induction of Wnt signaling and downregulation of RSPO2 was also observed in human neurofibromas and malignant peripheral nerve sheath tumors[82]. We found that RSPO2 was downregulated in CSCs, suggesting that it might act as a tumor suppressor in liver cells as well.
The expression of SELENBP1 is markedly reduced in multiple tumor types. Downregulation of SELENBP1 correlated with increased DNA damage and oxidative stress in tissue samples of patients with HCC[83]. Recent studies have shown that the reduced expression of SELENBP1 correlated with poor survival in breast cancer patients[84], and with the degree of colorectal cancer differentiation[85], suggesting that its expression could serve as a potential biomarker for these cancers. We found down-regulation of SELENBP1 in CSCs, demonstrating that that MYC-overexpression can result in this effect.
Tumor metastasis is associated with accelerated cell proliferation, dedifferentiation and loss of cell-cell contacts. FIGF and ANGPT1, two genes involved in angiogenesis, were found to be down-regulated in canine mammary carcinoma[86], breast cancer[87], and prostate cancer[88]. In our studies, the expression of both these genes was significantly lowered in CSCs, suggesting that their reduced expression is a factor in MYC-induced tumorigenesis in liver cells.
Aberrant DNA methylation was shown to play a role in carcinogenesis and disease progression in many cancers[89], and DNA methylation analysis of cancer genes might reveal novel prognostic biomarkers[90]. Cysteine dioxygenase 1 (CDO1), a tumor suppressor, is one such gene that is silenced by promoter methylation in multiple human cancers[19]. For instance, in lymph node-positive breast cancer patients, promoter DNA methylation of CDO1 causes its downregulation, making it an ideal biomarker for disease outcome prediction[91]. A recent DNA methylation study on human lung squamous cell carcinoma has also shown that CDO1 is hypermethylated and downregulated in lung cancer[92]. Even though forced expression of CDO1 resulted in markedly decreased tumor cells growth in human cancer cell lines and in mice[19], little is known about its role in HCC. In our studies, we found that CDO1 is silenced in CSCs, indicating that CDO1 may play a role in MYC-induced carcinogenesis.
Among Wnt antagonists, the Dickkopf (DKK) family of proteins act as tumor suppressors and are frequently silenced in various cancers[93,94]. It has been previously shown that DKK2 is epigenetically silenced in renal cancer, where it inhibits tumor progression through apoptotic and cell cycle pathways involving Bcl2 and cyclin D1[20]. In our study, we found that the expression of DKK2 is silenced in CSCs after MYC-overexpression (Figure 4), suggesting that the inhibition of Wnt antagonist is a mechanism of carcinogenesis mediated by the MYC protein.
Approximately, 70000 cancer deaths in the United States each year are associated with changes in the MYC gene or its expression[61]. Despite enormous progress achieved during the past several decades, long-term patient survival remains very low. Advances in stem cell biology provide opportunities to develop novel approaches to reduce morbidity and mortality associated with liver cancer. Our study is an important contribution to understanding the potential of CSC-targeted therapy for HCC. Novel genes identified in our investigation may serve as promising candidates to develop effective therapeutic options for HCC.
We thank Dr. Aaron Becker (BioMedical Genomics Center, University of Minnesota) for RNA library preparation and sequencing, and Drs. John Garbe and Ying Zhang (Minnesota Supercomputing Institute) for help with Galaxy Suite software analysis.
It is now well-established that cancer stem cells (CSCs) not only drive tumor growth in the liver, in general, but also that the proto-oncogene c-Myc plays a critical role in that process. However, little is known about the genes that are induced and regulated by the MYC protein to generate tumors, and, in particular, those involved in liver stem cells.
Hepatocellular carcinoma is a multifactorial disease in which the MYC protein plays a central role. However, a number of different proteins and signaling pathways are activated in liver cancer. Cancer stem cells were isolated from hepatocellular carcinoma (HCC) specimens and by culturing human HCC cell lines. But it is unclear as to which cellular pathways are activated by the MYC protein.
This is the first study to demonstrate the regulation of global gene expression by a single oncoprotein (MYC) in porcine liver stem cells using a novel stem cell line, PICM-19, that is capable of propagating in cell culture for a prolonged time period without losing its phenotypic characteristics. By overexpressing the MYC protein, these stem cells were converted into cancer cells. By using a cutting-edge deep sequencing technique (RNA-seq), the authors have identified six genes (CDO1, C22orf39, DKK2, ENPEP, GPX6, SRPX2) that might be critical in carcinogenesis induced by the MYC protein in PICM-19 cells.
MYC-driven genes identified in this study may serve as promising candidates for the development of HCC therapeutics without the deleterious effects on non-cancerous stem cells and on other cells present in the liver.
CSC - cancer stem cells; RNA-seq - RNA sequencing.
In this study, the authors identify the overall genes expression in a liver stem cell after induction by c-MYC plasmid transfection in generating hepatic tumors. The authors used an immortal porcine liver stem cell line, PICM-19, to identify MYC-driven differential gene expression. Transcriptome sequencing was carried out by RNA sequencing and genes identified by this method were validated using real-time PCR. MYC-driven genes identified in this study may serve as promising candidates for the development of HCC therapeutics that would not have deleterious effects on other cell types in the liver.
P- Reviewer: Cao GW, Leardkamolkarn V, Welling TH, Wu JL S- Editor: Ma YJ L- Editor: A E- Editor: Liu XM
| 1. | Nordenstedt H, White DL, El-Serag HB. The changing pattern of epidemiology in hepatocellular carcinoma. Dig Liver Dis. 2010;42 Suppl 3:S206-S214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 367] [Cited by in RCA: 404] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 2. | Aravalli RN, Cressman EN, Steer CJ. Cellular and molecular mechanisms of hepatocellular carcinoma: an update. Arch Toxicol. 2013;87:227-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 177] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 3. | Blum HE. Hepatocellular carcinoma: therapy and prevention. World J Gastroenterol. 2005;11:7391-7400. [PubMed] |
| 4. | Kaposi-Novak P, Libbrecht L, Woo HG, Lee YH, Sears NC, Coulouarn C, Conner EA, Factor VM, Roskams T, Thorgeirsson SS. Central role of c-Myc during malignant conversion in human hepatocarcinogenesis. Cancer Res. 2009;69:2775-2782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 144] [Cited by in RCA: 149] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 5. | Ma S, Chan KW, Hu L, Lee TK, Wo JY, Ng IO, Zheng BJ, Guan XY. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542-2556. [PubMed] |
| 6. | Zhao Y, Jian W, Gao W, Zheng YX, Wang YK, Zhou ZQ, Zhang H, Wang CJ. RNAi silencing of c-Myc inhibits cell migration, invasion, and proliferation in HepG2 human hepatocellular carcinoma cell line: c-Myc silencing in hepatocellular carcinoma cell. Cancer Cell Int. 2013;13:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 7. | Lin CP, Liu CR, Lee CN, Chan TS, Liu HE. Targeting c-Myc as a novel approach for hepatocellular carcinoma. World J Hepatol. 2010;2:16-20. [PubMed] |
| 8. | Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105-111. [PubMed] |
| 9. | Wang B, Jacob ST. Role of cancer stem cells in hepatocarcinogenesis. Genome Med. 2011;3:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 10. | Alison MR. Liver stem cells: implications for hepatocarcinogenesis. Stem Cell Rev. 2005;1:253-260. [PubMed] |
| 11. | Ma S, Lee TK, Zheng BJ, Chan KW, Guan XY. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene. 2008;27:1749-1758. [PubMed] |
| 12. | Talbot NC, Blomberg Le Ann, Garrett WM, Caperna TJ. Feeder-independent continuous culture of the PICM-19 pig liver stem cell line. In Vitro Cell Dev Biol Anim. 2010;46:746-757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 13. | Talbot NC, Caperna TJ, Garrett WM. Growth and Development Symposium: Development, characterization, and use of a porcine epiblast-derived liver stem cell line: ARS-PICM-19. J Anim Sci. 2013;91:66-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 14. | Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105-1111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10271] [Cited by in RCA: 9375] [Article Influence: 585.9] [Reference Citation Analysis (0)] |
| 15. | Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178-192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5308] [Cited by in RCA: 6306] [Article Influence: 485.1] [Reference Citation Analysis (0)] |
| 16. | Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511-515. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13157] [Cited by in RCA: 11559] [Article Influence: 770.6] [Reference Citation Analysis (0)] |
| 17. | Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562-578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8810] [Cited by in RCA: 9300] [Article Influence: 715.4] [Reference Citation Analysis (0)] |
| 18. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [PubMed] |
| 19. | Brait M, Ling S, Nagpal JK, Chang X, Park HL, Lee J, Okamura J, Yamashita K, Sidransky D, Kim MS. Cysteine dioxygenase 1 is a tumor suppressor gene silenced by promoter methylation in multiple human cancers. PLoS One. 2012;7:e44951. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 106] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 20. | Hirata H, Hinoda Y, Nakajima K, Kawamoto K, Kikuno N, Kawakami K, Yamamura S, Ueno K, Majid S, Saini S. Wnt antagonist gene DKK2 is epigenetically silenced and inhibits renal cancer progression through apoptotic and cell cycle pathways. Clin Cancer Res. 2009;15:5678-5687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 73] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 21. | Zhang Y, Gong W, Dai S, Huang G, Shen X, Gao M, Xu Z, Zeng Y, He F. Downregulation of human farnesoid X receptor by miR-421 promotes proliferation and migration of hepatocellular carcinoma cells. Mol Cancer Res. 2012;10:516-522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 70] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 22. | Hao J, Zhang S, Zhou Y, Liu C, Hu X, Shao C. MicroRNA 421 suppresses DPC4/Smad4 in pancreatic cancer. Biochem Biophys Res Commun. 2011;406:552-557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 88] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 23. | Brew K, Nagase H. The tissue inhibitors of metalloproteinases (TIMPs): an ancient family with structural and functional diversity. Biochim Biophys Acta. 2010;1803:55-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 799] [Cited by in RCA: 956] [Article Influence: 63.7] [Reference Citation Analysis (0)] |
| 24. | Yang X, Zhang XF, Lu X, Jia HL, Liang L, Dong QZ, Ye QH, Qin LX. MicroRNA-26a suppresses angiogenesis in human hepatocellular carcinoma by targeting hepatocyte growth factor-cMet pathway. Hepatology. 2014;59:1874-1885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 170] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 25. | Chai ZT, Kong J, Zhu XD, Zhang YY, Lu L, Zhou JM, Wang LR, Zhang KZ, Zhang QB, Ao JY. MicroRNA-26a inhibits angiogenesis by down-regulating VEGFA through the PIK3C2α/Akt/HIF-1α pathway in hepatocellular carcinoma. PLoS One. 2013;8:e77957. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 98] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 26. | Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22:2755-2766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 681] [Cited by in RCA: 756] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 27. | Guo QM, Malek RL, Kim S, Chiao C, He M, Ruffy M, Sanka K, Lee NH, Dang CV, Liu ET. Identification of c-myc responsive genes using rat cDNA microarray. Cancer Res. 2000;60:5922-5928. [PubMed] |
| 28. | Rabenau KE, O’Toole JM, Bassi R, Kotanides H, Witte L, Ludwig DL, Pereira DS. DEGA/AMIGO-2, a leucine-rich repeat family member, differentially expressed in human gastric adenocarcinoma: effects on ploidy, chromosomal stability, cell adhesion/migration and tumorigenicity. Oncogene. 2004;23:5056-5067. [PubMed] |
| 29. | Nagata M, Noman AA, Suzuki K, Kurita H, Ohnishi M, Ohyama T, Kitamura N, Kobayashi T, Uematsu K, Takahashi K. ITGA3 and ITGB4 expression biomarkers estimate the risks of locoregional and hematogenous dissemination of oral squamous cell carcinoma. BMC Cancer. 2013;13:410. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 30. | Calderwood DA, Shattil SJ, Ginsberg MH. Integrins and actin filaments: reciprocal regulation of cell adhesion and signaling. J Biol Chem. 2000;275:22607-22610. [PubMed] |
| 31. | Luque-García JL, Martínez-Torrecuadrada JL, Epifano C, Cañamero M, Babel I, Casal JI. Differential protein expression on the cell surface of colorectal cancer cells associated to tumor metastasis. Proteomics. 2010;10:940-952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 75] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 32. | Wozniak MA, Itzhaki RF, Faragher EB, James MW, Ryder SD, Irving WL. Apolipoprotein E-epsilon 4 protects against severe liver disease caused by hepatitis C virus. Hepatology. 2002;36:456-463. [PubMed] |
| 33. | Ahn SJ, Kim DK, Kim SS, Bae CB, Cho HJ, Kim HG, Kim YJ, Lee JH, Lee HJ, Lee MY. Association between apolipoprotein E genotype, chronic liver disease, and hepatitis B virus. Clin Mol Hepatol. 2012;18:295-301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 34. | Zhong DN, Ning QY, Wu JZ, Zang N, Wu JL, Hu DF, Luo SY, Huang AC, Li LL, Li GJ. Comparative proteomic profiles indicating genetic factors may involve in hepatocellular carcinoma familial aggregation. Cancer Sci. 2012;103:1833-1838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 35. | Yokoyama Y, Kuramitsu Y, Takashima M, Iizuka N, Terai S, Oka M, Nakamura K, Okita K, Sakaida I. Protein level of apolipoprotein E increased in human hepatocellular carcinoma. Int J Oncol. 2006;28:625-631. [PubMed] |
| 36. | Topic A, Ljujic M, Radojkovic D. Alpha-1-antitrypsin in pathogenesis of hepatocellular carcinoma. Hepat Mon. 2012;12:e7042. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 37. | Pirisi M, Fabris C, Soardo G, Toniutto P, Vitulli D, Bartoli E. Prognostic value of serum alpha-1-antitrypsin in hepatocellular carcinoma. Eur J Cancer. 1996;32A:221-225. [PubMed] |
| 38. | Comunale MA, Rodemich-Betesh L, Hafner J, Wang M, Norton P, Di Bisceglie AM, Block T, Mehta A. Linkage specific fucosylation of alpha-1-antitrypsin in liver cirrhosis and cancer patients: implications for a biomarker of hepatocellular carcinoma. PLoS One. 2010;5:e12419. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 108] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 39. | Yanagawa T, Shinozaki T, Watanabe H, Saito K, Raz A, Takagishi K. Vascular endothelial growth factor-D is a key molecule that enhances lymphatic metastasis of soft tissue sarcomas. Exp Cell Res. 2012;318:800-808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 40. | Cui X, Song B, Hou L, Wei Z, Tang J. High expression of osteoglycin decreases the metastatic capability of mouse hepatocarcinoma Hca-F cells to lymph nodes. Acta Biochim Biophys Sin (Shanghai). 2008;40:349-355. [PubMed] |
| 41. | Kisseleva T, Brenner DA. Hepatic stellate cells and the reversal of fibrosis. J Gastroenterol Hepatol. 2006;21 Suppl 3:S84-S87. [PubMed] |
| 42. | Lai KK, Shang S, Lohia N, Booth GC, Masse DJ, Fausto N, Campbell JS, Beretta L. Extracellular matrix dynamics in hepatocarcinogenesis: a comparative proteomics study of PDGFC transgenic and Pten null mouse models. PLoS Genet. 2011;7:e1002147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 73] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 43. | Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, Lodish HF, Lim B. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862-876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 538] [Cited by in RCA: 527] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 44. | Xia HF, He TZ, Liu CM, Cui Y, Song PP, Jin XH, Ma X. MiR-125b expression affects the proliferation and apoptosis of human glioma cells by targeting Bmf. Cell Physiol Biochem. 2009;23:347-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 131] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 45. | Sun D, Yu F, Ma Y, Zhao R, Chen X, Zhu J, Zhang CY, Chen J, Zhang J. MicroRNA-31 activates the RAS pathway and functions as an oncogenic MicroRNA in human colorectal cancer by repressing RAS p21 GTPase activating protein 1 (RASA1). J Biol Chem. 2013;288:9508-9518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 156] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 46. | Hu C, Huang F, Deng G, Nie W, Huang W, Zeng X. miR-31 promotes oncogenesis in intrahepatic cholangiocarcinoma cells via the direct suppression of RASA1. Exp Ther Med. 2013;6:1265-1270. [PubMed] |
| 47. | Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP, Zhuang SM. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology. 2009;50:113-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 317] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 48. | Zhou M, Liu W, Ma S, Cao H, Peng X, Guo L, Zhou X, Zheng L, Guo L, Wan M. A novel onco-miR-365 induces cutaneous squamous cell carcinoma. Carcinogenesis. 2013;34:1653-1659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 49. | Papetti M, Augenlicht LH. Mybl2, downregulated during colon epithelial cell maturation, is suppressed by miR-365. Am J Physiol Gastrointest Liver Physiol. 2011;301:G508-G518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 50. | Papetti M, Augenlicht LH. MYBL2, a link between proliferation and differentiation in maturing colon epithelial cells. J Cell Physiol. 2011;226:785-791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 51. | Jackson RS, Placzek W, Fernandez A, Ziaee S, Chu CY, Wei J, Stebbins J, Kitada S, Fritz G, Reed JC. Sabutoclax, a Mcl-1 antagonist, inhibits tumorigenesis in transgenic mouse and human xenograft models of prostate cancer. Neoplasia. 2012;14:656-665. [PubMed] |
| 52. | Yang W, Diamond AM. Selenium-binding protein 1 as a tumor suppressor and a prognostic indicator of clinical outcome. Biomark Res. 2013;1:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 53. | Huang C, Ding G, Gu C, Zhou J, Kuang M, Ji Y, He Y, Kondo T, Fan J. Decreased selenium-binding protein 1 enhances glutathione peroxidase 1 activity and downregulates HIF-1α to promote hepatocellular carcinoma invasiveness. Clin Cancer Res. 2012;18:3042-3053. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 54. | Ritorto MS, Borlak J. Combined serum and tissue proteomic study applied to a c-Myc transgenic mouse model of hepatocellular carcinoma identified novel disease regulated proteins suitable for diagnosis and therapeutic intervention strategies. J Proteome Res. 2011;10:3012-3030. [PubMed] |
| 55. | Merle P, Kim M, Herrmann M, Gupte A, Lefrançois L, Califano S, Trépo C, Tanaka S, Vitvitski L, de la Monte S. Oncogenic role of the frizzled-7/beta-catenin pathway in hepatocellular carcinoma. J Hepatol. 2005;43:854-862. [PubMed] |
| 56. | Quinn LM, Secombe J, Hime GR. Myc in stem cell behaviour: insights from Drosophila. Adv Exp Med Biol. 2013;786:269-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 57. | Reed KR, Athineos D, Meniel VS, Wilkins JA, Ridgway RA, Burke ZD, Muncan V, Clarke AR, Sansom OJ. B-catenin deficiency, but not Myc deletion, suppresses the immediate phenotypes of APC loss in the liver. Proc Natl Acad Sci USA. 2008;105:18919-18923. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 55] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 58. | Sansom OJ, Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR, Vass JK, Athineos D, Clevers H, Clarke AR. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676-679. [PubMed] |
| 59. | Perona R, López-Ayllón BD, de Castro Carpeño J, Belda-Iniesta C. A role for cancer stem cells in drug resistance and metastasis in non-small-cell lung cancer. Clin Transl Oncol. 2011;13:289-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 55] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 60. | Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1349] [Cited by in RCA: 1450] [Article Influence: 103.6] [Reference Citation Analysis (0)] |
| 61. | Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1-11. [PubMed] |
| 62. | Wang J, Wang H, Li Z, Wu Q, Lathia JD, McLendon RE, Hjelmeland AB, Rich JN. c-Myc is required for maintenance of glioma cancer stem cells. PLoS One. 2008;3:e3769. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 290] [Cited by in RCA: 336] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 63. | Chow EK, Fan LL, Chen X, Bishop JM. Oncogene-specific formation of chemoresistant murine hepatic cancer stem cells. Hepatology. 2012;56:1331-1341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 80] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 64. | Ho DW, Yang ZF, Yi K, Lam CT, Ng MN, Yu WC, Lau J, Wan T, Wang X, Yan Z. Gene expression profiling of liver cancer stem cells by RNA-sequencing. PLoS One. 2012;7:e37159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 66] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 65. | Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, Jia H, Ye Q, Qin LX, Wauthier E. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012-1024. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 936] [Cited by in RCA: 957] [Article Influence: 59.8] [Reference Citation Analysis (0)] |
| 66. | Riggi N, Suvà ML, De Vito C, Provero P, Stehle JC, Baumer K, Cironi L, Janiszewska M, Petricevic T, Suvà D. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev. 2010;24:916-932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 222] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 67. | Leis O, Eguiara A, Lopez-Arribillaga E, Alberdi MJ, Hernandez-Garcia S, Elorriaga K, Pandiella A, Rezola R, Martin AG. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene. 2012;31:1354-1365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 411] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 68. | Marquardt JU, Raggi C, Andersen JB, Seo D, Avital I, Geller D, Lee YH, Kitade M, Holczbauer A, Gillen MC. Human hepatic cancer stem cells are characterized by common stemness traits and diverse oncogenic pathways. Hepatology. 2011;54:1031-1042. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 69. | Oikawa T, Kamiya A, Zeniya M, Chikada H, Hyuck AD, Yamazaki Y, Wauthier E, Tajiri H, Miller LD, Wang XW. Sal-like protein 4 (SALL4), a stem cell biomarker in liver cancers. Hepatology. 2013;57:1469-1483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 149] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 70. | Zeng SS, Yamashita T, Kondo M, Nio K, Hayashi T, Hara Y, Nomura Y, Yoshida M, Hayashi T, Oishi N. The transcription factor SALL4 regulates stemness of EpCAM-positive hepatocellular carcinoma. J Hepatol. 2014;60:127-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 110] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 71. | Xu M, Zheng YL, Xie XY, Liang JY, Pan FS, Zheng SG, Lü MD. Sorafenib blocks the HIF-1α/VEGFA pathway, inhibits tumor invasion, and induces apoptosis in hepatoma cells. DNA Cell Biol. 2014;33:275-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 72. | Liang Y, Zheng T, Song R, Wang J, Yin D, Wang L, Liu H, Tian L, Fang X, Meng X. Hypoxia-mediated sorafenib resistance can be overcome by EF24 through Von Hippel-Lindau tumor suppressor-dependent HIF-1α inhibition in hepatocellular carcinoma. Hepatology. 2013;57:1847-1857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 233] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 73. | Yang SL, Liu LP, Jiang JX, Xiong ZF, He QJ, Wu C. The correlation of expression levels of HIF-1α and HIF-2α in hepatocellular carcinoma with capsular invasion, portal vein tumor thrombi and patients’ clinical outcome. Jpn J Clin Oncol. 2014;44:159-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 74. | Chen WN, Chen JY, Jiao BY, Lin WS, Wu YL, Liu LL, Lin X. Interaction of the hepatitis B spliced protein with cathepsin B promotes hepatoma cell migration and invasion. J Virol. 2012;86:13533-13541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 75. | Stratil A, Peelman LJ, Mattheeuws M, Van Poucke M, Reiner G, Geldermann H. A novel porcine gene, alpha-1-antichymotrypsin 2 (SERPINA3-2): sequence, genomic organization, polymorphism and mapping. Gene. 2002;292:113-119. [PubMed] |
| 76. | Chapman MH, Tidswell R, Dooley JS, Sandanayake NS, Cerec V, Deheragoda M, Lee AJ, Swanton C, Andreola F, Pereira SP. Whole genome RNA expression profiling of endoscopic biliary brushings provides data suitable for biomarker discovery in cholangiocarcinoma. J Hepatol. 2012;56:877-885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 40] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 77. | Shirakihara T, Kawasaki T, Fukagawa A, Semba K, Sakai R, Miyazono K, Miyazawa K, Saitoh M. Identification of integrin α3 as a molecular marker of cells undergoing epithelial-mesenchymal transition and of cancer cells with aggressive phenotypes. Cancer Sci. 2013;104:1189-1197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 78. | Denadai MV, Viana LS, Affonso RJ, Silva SR, Oliveira ID, Toledo SR, Matos D. Expression of integrin genes and proteins in progression and dissemination of colorectal adenocarcinoma. BMC Clin Pathol. 2013;13:16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 79. | Sachs N, Secades P, van Hulst L, Kreft M, Song JY, Sonnenberg A. Loss of integrin α3 prevents skin tumor formation by promoting epidermal turnover and depletion of slow-cycling cells. Proc Natl Acad Sci USA. 2012;109:21468-21473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 62] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 80. | de Lau WB, Snel B, Clevers HC. The R-spondin protein family. Genome Biol. 2012;13:242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 219] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 81. | Wu C, Qiu S, Lu L, Zou J, Li WF, Wang O, Zhao H, Wang H, Tang J, Chen L. RSPO2-LGR5 signaling has tumour-suppressive activity in colorectal cancer. Nat Commun. 2014;5:3149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 93] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 82. | Watson AL, Rahrmann EP, Moriarity BS, Choi K, Conboy CB, Greeley AD, Halfond AL, Anderson LK, Wahl BR, Keng VW. Canonical Wnt/β-catenin signaling drives human Schwann cell transformation, progression, and tumor maintenance. Cancer Discov. 2013;3:674-689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 82] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 83. | DI Stasio M, Volpe MG, Colonna G, Nazzaro M, Polimeno M, Scala S, Castello G, Costantini S. A possible predictive marker of progression for hepatocellular carcinoma. Oncol Lett. 2011;2:1247-1251. [PubMed] |
| 84. | Zhang S, Li F, Younes M, Liu H, Chen C, Yao Q. Reduced selenium-binding protein 1 in breast cancer correlates with poor survival and resistance to the anti-proliferative effects of selenium. PLoS One. 2013;8:e63702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 85. | Wang N, Chen Y, Yang X, Jiang Y. Selenium-binding protein 1 is associated with the degree of colorectal cancer differentiation and is regulated by histone modification. Oncol Rep. 2014;31:2506-2514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 86. | Klopfleisch R, Lenze D, Hummel M, Gruber AD. The metastatic cascade is reflected in the transcriptome of metastatic canine mammary carcinomas. Vet J. 2011;190:236-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 46] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 87. | Roberti MP, Arriaga JM, Bianchini M, Quintá HR, Bravo AI, Levy EM, Mordoh J, Barrio MM. Protein expression changes during human triple negative breast cancer cell line progression to lymph node metastasis in a xenografted model in nude mice. Cancer Biol Ther. 2012;13:1123-1140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 40] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 88. | Wong L, Gipp J, Carr J, Loftus CJ, Benck M, Lee S, Mehta V, Vezina CM, Bushman W. Prostate angiogenesis in development and inflammation. Prostate. 2014;74:346-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 89. | Ting AH, McGarvey KM, Baylin SB. The cancer epigenome-components and functional correlates. Genes Dev. 2006;20:3215-3231. [PubMed] |
| 90. | Duffy MJ, Napieralski R, Martens JW, Span PN, Spyratos F, Sweep FC, Brunner N, Foekens JA, Schmitt M. Methylated genes as new cancer biomarkers. Eur J Cancer. 2009;45:335-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 75] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 91. | Dietrich D, Krispin M, Dietrich J, Fassbender A, Lewin J, Harbeck N, Schmitt M, Eppenberger-Castori S, Vuaroqueaux V, Spyratos F. CDO1 promoter methylation is a biomarker for outcome prediction of anthracycline treated, estrogen receptor-positive, lymph node-positive breast cancer patients. BMC Cancer. 2010;10:247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 92. | Kwon YJ, Lee SJ, Koh JS, Kim SH, Lee HW, Kang MC, Bae JB, Kim YJ, Park JH. Genome-wide analysis of DNA methylation and the gene expression change in lung cancer. J Thorac Oncol. 2012;7:20-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 93. | Maehata T, Taniguchi H, Yamamoto H, Nosho K, Adachi Y, Miyamoto N, Miyamoto C, Akutsu N, Yamaoka S, Itoh F. Transcriptional silencing of Dickkopf gene family by CpG island hypermethylation in human gastrointestinal cancer. World J Gastroenterol. 2008;14:2702-2714. [PubMed] |
| 94. | Sato H, Suzuki H, Toyota M, Nojima M, Maruyama R, Sasaki S, Takagi H, Sogabe Y, Sasaki Y, Idogawa M. Frequent epigenetic inactivation of DICKKOPF family genes in human gastrointestinal tumors. Carcinogenesis. 2007;28:2459-2466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 171] [Article Influence: 9.5] [Reference Citation Analysis (0)] |