Published online Nov 14, 2005. doi: 10.3748/wjg.v11.i42.6694
Revised: April 23, 2005
Accepted: April 26, 2005
Published online: November 14, 2005
AIM: To determine the DNA binding activity and protein levels of the Ku70/80 heterodimer, the functional mediator of the NHEJ activity, in human colorectal carcinogenesis.
METHODS: The Ku70/80 DNA-binding activity was determined by electrophoretic mobility shift assays in 20 colon adenoma and 15 colorectal cancer samples as well as matched normal colonic tissues. Nuclear and cytoplasmic protein expression was determined by immunohistochemistry and Western blot analysis.
RESULTS: A statistically significant difference was found in both adenomas and carcinomas as compared to matched normal colonic mucosa (P<0.00). However, changes in binding activity were not homogenous with approximately 50% of the tumors showing a clear increase in the binding activity, 30% displaying a modest increase and 15% showing a decrease of the activity. Tumors, with increased DNA-binding activity, also showed a statistically significant increase in Ku70 and Ku86 nuclear expression, as determined by Western blot and immunohistochemical analyses (P<0.001). Cytoplasmic protein expression was found in pathological samples, but not in normal tissues either from tumor patients or from healthy subjects.
CONCLUSION: Overall, our DNA-binding activity and protein level are consistent with a substantial activation of the NHEJ pathway in colorectal tumors. Since the NHEJ is an error prone mechanism, its abnormal activation can result in chromosomal instability and ultimately lead to tumorigenesis.
- Citation: Mazzarelli P, Parrella P, Seripa D, Signori E, Perrone G, Rabitti C, Borzomati D, Gabbrielli A, Matera MG, Gravina C, Caricato M, Poeta ML, Rinaldi M, Valeri S, Coppola R, Fazio VM. DNA end binding activity and Ku70/80 heterodimer expression in human colorectal tumor. World J Gastroenterol 2005; 11(42): 6694-6700
- URL: https://www.wjgnet.com/1007-9327/full/v11/i42/6694.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i42.6694
Colorectal cancer is a significant cause of morbidity and mortality in Western populations[1]. This cancer progresses through a series of defined histopathological stages, going from a small benign tumor (adenomatous polyps) to a malignant cancer (carcinoma)[2]. During tumor progression, a stepwise accumulation of genetic changes is observed, leading to inactivation of tumor suppressor genes (e.g. APC, p53) and activation of oncogenes (e.g. K-ras, β-catenin)[2]. The number of genomic alterations in cancer appears to exceed the level possibly due to the accumulation of mutations in cells with normal mutation rates. A number of intricate networks have evolved in eukaryotic cells to respond to exogenous and endogenous genotoxic stimuli[3]. Genes involved in these pathways play a crucial role in maintaining DNA integrity and a defect in these processes may result in hypersensitivity to DNA damaging agents and genomic instability[4]. Two main forms of genetic instability are associated with tumors. One arises from the inactivation of DNA mismatch repair (MMR) genes[5], leading to instability at the nucleotide sequence level (microsatellite instability, MSI). The other results from a disruption of the pathways intending to protect the cells from chromosomal breakage (double-strand breaks, DSBs), which leads to gross chromosomal rearrangements (chromosomal instability, CIN)[6]. Of the many types of DNA damage, DSBs are the most dangerous, because of the intrinsic difficulty of their repair as compared to other types of DNA damage[6]. In physiological conditions, DNA-DSBs are generated by homologous recombination (HR) during meiosis and occur in other events, such as V(D)J recombination and immunoglobulin class switch[6]. In addition, DSBs can result from both exogenous agents such as ionizing radiation or chemotherapeutic agents and endogenously generated reactive oxygen species[6]. Erroneous rejoining of the broken DNA-DSBs may cause loss or amplification of chromosomal material and even translocations, ultimately leading to tumorigenesis[4].
There are two distinct and complementary mechanisms for DNA-DSB repair: the NHEJ and the HR[4]. Recently, a caretaker role in preventing carcinogenesis has been proposed for the NHEJ pathway[7-9]. At present, five proteins involved in the NHEJ pathway have been identified; namely, the ligase IV and its associated protein XRCC1, and the three components of the DNA-dependent protein kinase (DNA-PK) complex, Ku70, Ku86, and the catalytic subunit PKcs[10]. Mutational analysis has shown that activation of the DNA-PK well correlates with Ku protein heterodimerization and DNA-end binding[11]. Once anchored to the DNA, the Ku70/80 heterodimer translocates along the molecule and facilitates recruitment of the catalytic subunit to the site of the break, to form an activated DNA-PK complex[11]. Numerous studies have investigated the role of MMR pathway in colorectal carcinogenesis, but little is known about the involvement of the DSB repair pathway in the adenoma/carcinoma sequence[11-13]. In the attempt to better understand this role, we analyzed the DNA-binding activity of the Ku70/80 heterodimer and the protein expression of the two Ku subunits in colon adenomas, colorectal cancers and matched normal tissues. Ku70/80 DNA binding activity was increased in approximately 50% of adenoma and carcinoma samples, as compared to matched normal tissues. In tumors with increased DNA-binding activity, Ku70 and Ku86 protein expression correlated with the heterodimer binding activity.
Twenty patients with colon adenoma and 15 patients with colorectal carcinoma were recruited in the study. The patients underwent endoscopic polypectomy or surgical resection between 1999 and 2002, at the Departments of Gastroenterology and General Surgery, Campus Bio-Medico University of Rome, Italy. None of the patients were affected by familial polyposis or HNPCC. Subjects included 20 males (57%) and 15 females (43%), with a mean age of 69.5±12 years (range 35-82 years). None had pre-operative chemotherapy or irradiation. All the patients gave informed consent for the study. Thirty-one lesions (89% including adenomas and carcinomas) were located in the colon and 4 (11%) in the rectum. Adenomas were classified according to the National Polyp Study Cohort and WHO recommendations on the basis of size and grade of dysplasia[14,15]. Clinical staging of colorectal cancer was assessed according to the Dukes’ classification[13].
Following surgical resection, tissue samples were im-mediately frozen at -80 °C. For each case, before protein extraction, one 3-µm hematoxylin-eosin stained slide was analyzed to ensure that tumor samples contained at least 70% cancer cells. Protein extraction was performed as previously described[16-18]. Briefly, frozen samples were mechanically fractionated to obtain a cellular suspension. Nuclear and cytoplasmic fractions were separated by centrifugation at 10 000 g and stored at -80 °C.
Protein content in nuclear and cytoplasmic extracts was determined in triplicate by Bradford assay (Bio-Rad Protein Assay, Bio-Rad Laboratories, Munchen).
Electrophoretic mobility shift assay (EMSA) was performed as described previously[18,19]. Briefly, DNA binding reactions contained 50 000 cpm of the labeled probe, nuclear (2 µg) or cytoplasmic (5 µg) extracts with closed circular plasmid DNA pUC-19 (1 µg) as the unspecific competitor. For each sample, three single shift assays were performed. As controls, 33 normal human tissues from patients without colon tumor were analyzed (mammary gland n = 8, bladder mucosa n = 8, and skin n = 17).
To normalize all the samples, electrophoretic mobility shift assays were performed by incubating the nuclear extracts (3 µg) with 50 000 cpm/sample of 32P-end labeled Sp-1 oligonucleotide (Promega Corporation, Madison, WI, USA) in a binding buffer, with 1 µg of poly (dI-dC) as the unspecific competitor. The correction factor (CF) was calculated as follows: SP1 binding activity in the sample/mean SP1 binding activity. Data were normalized using the following formula: Mean Ku70/80 binding activity/CF[20].
For gel supershift experiments, goat polyclonal anti-Ku70 and anti-Ku86 antibodies (M-19, M-20: Santa Cruz Biotechnologies Inc., CA, USA) were incubated with protein extracts for 30 min at room temperature, before the other components were added to the binding reaction. Complexes were separated on 6% non-denaturing polyacrylamide gels and exposed to X-ray films (Amersham-Pharmacia Biotech, England HP7 9NA). The optical densities (OD) were obtained by scanning densitometry using colon carcinoma cell line CaCo-2 (ATCC) as internal control (OD = 10.7±6.4).
Supershift control experiments confirmed the specificity of the results in each gelshift assay experiment.
Paraffin sections from matched normal colonic mucosa were available for all the colon cancer cases entered in the study, whereas paraffin embedded blocks from matched normal tissues were available only for 10 adenomas. Paraffin sections from five subjects who showed normal colonic mucosa at colonoscopy were also analyzed. Consecutive two micron sections were immunostained for Ku70 and Ku86 following the streptavidin-biotin method, as described previously[20] . In brief, sections were deparaffinized, rehydrated in decreasing alcohol and microwave treated. Endogenous peroxidase activity was quenched by treatment with 0.03% hydrogen peroxide in absolute methanol for 30 min at room temperature. The primary antibodies were goat polyclonal anti-Ku70 and anti-Ku86 antibodies specifically validated for immunohistochemical analysis of paraffin embedded tissues (M-19, M-20: Santa Cruz Biotechnology Inc., CA, USA), in a 1:200 dilution. Biotinylated swine antigoat/mouse/rabbit IgG (Dako A/S, Denmark) was used as secondary antibody. After washing, sections were treated with streptavidin-peroxidase reagent (Dakopatts A\S, Denmark), incubated with diaminobenzidine (DAB) and counterstained with hematoxylin. Slides were examined under a two-head microscope by two pathologists, unaware of the clinical data and molecular results. No discrepant results were identified. Results of the nuclear immunostaining were expressed as percentage of positively stained cells. For cytoplasmic staining the microscopic analysis was not able to discriminate one cell from another. Thus the immunoreactivity was classified into four staining levels (SL): 0 SL (no staining); 1 SL (1-33% of positively stained area); 2 SL (33-66% of the area), and 3 SL (66-99% of the area)[21].
Protein extracts (10 µg) were separated in 10% SDS-PAGE, transferred to a PVDF membrane (Hybond-P, Amersham-Pharmacia Biotech, UK HP7 9NA) using an electroblotting apparatus, and incubated for 1 h at room temperature in 1% BSA, 1% skim milk (Difco Lab., Detroit, MI, USA), and 0.5% Tween 20 (USB, Cleveland, OH, USA). Membranes were stained with Ponceau S dye, to check for equal loading and homogeneous transfer. Immunodetection experiments were performed as described previously[17]. Filters were reprobed with anti β-actin (Sigma-Aldrich, St. Louis, MO 63103, USA) mouse IgG1 monoclonal antibody, to normalize the nuclear protein levels. Filters were washed and developed using an enhanced chemiluminescence system (ECL, Amersham-Pharmacia Biotech, UK HP7 9NA). The optical densities (OD) were obtained by scanning densitometric analysis of the bands normalized for the β-actin levels, as reference protein.
All values provided in the text and figures are means of three independent experiments±standard deviations (SD). Variation rates (VR) were defined by the following formula: [(pathological sample value–normal sample value)/pathological sample value]×100. Mean values were compared using the one- or two-tailed Student’s t-test, for independent samples. P<0.05 was considered statistically significant.
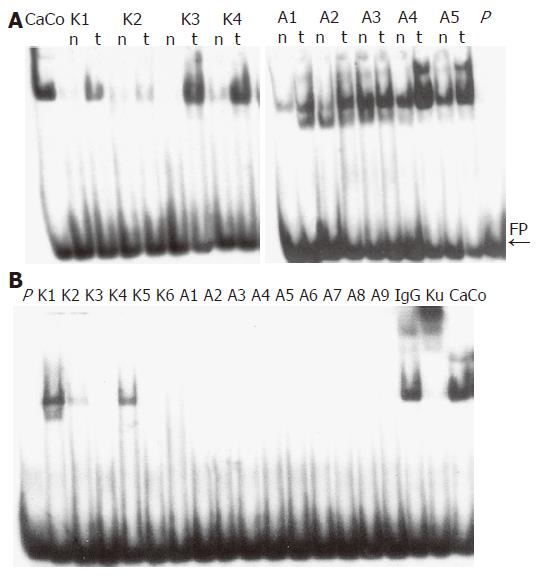
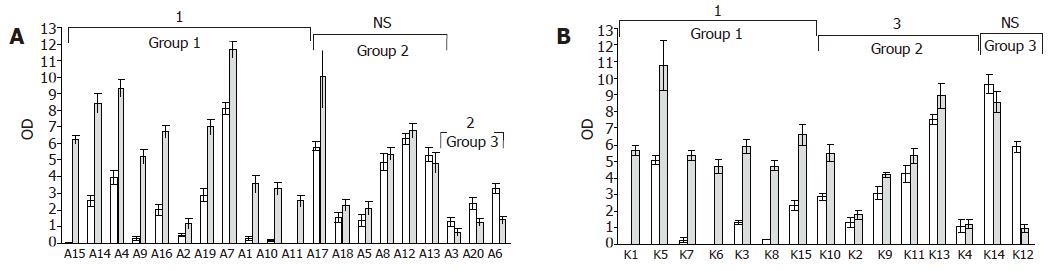
EMSA were performed on nuclear extracts of colon tissues, to compare the DNA binding activity of the Ku70/80 heterodimer in adenoma and carcinoma samples as well as matched normal colonic mucosa samples (Figure 1A). We found that adenoma and carcinoma samples had an overall increase in DNA binding activity as compared to matched normal samples (P<0.01, Table 1). However, these variations were not homogeneous among the tumors that were tested. Since the overall increase in binding activity compared to normal was approximately 50% (Table 1), we set this value as the cut-off for better classifying our pathological samples. Of the 20 adenomas, 11 (55%) showed an increase in DNA binding activity higher than 50% (P = 0.000) (Figure 1A, Group I), 6 (30%) displayed an increase in DNA binding activity lower than 50% (P = NS) (Figure 2A, Group II), and 3 (15%) showed reduced levels of activity (P = 0.04) (Figure 2A, Group III). Of the 15 carcinomas, 7 (47%) displayed an increase in DNA binding activity higher than 50% (Figure 2B, Group I), 6 (40%) showed an increase in DNA binding activity lower than 50% (P<0.02) (Figure 2B, Group II), and 2 (13%) showed reduced levels of activity (P = ND) (Figure 2B, Group III).
| Colon | Ku70/80 DNA binding activity | Ku70 protein expression | ||||
| Tissue | Normal | Tumor | P | Normal | Tumor | P |
| Adenomas | 2.66±2.32 | 5.01±3.15 | <0.01 | 0.88±0.59 | 1.28±0.45 | <0.01 |
| Carcinomas | 2.99±2.84 | 5.35±2.75 | <0.001 | 1.09±0.80 | 1.77±0.83 | <0.01 |
No significant differences were found when the Ku70/80 DNA-binding activity was compared in adenoma and carcinoma samples (Table 1). We also determined, if functional Ku70/80 heterodimer was present in cytoplasmic protein extracts. Cytoplasmic Ku70/80 DNA binding activity was only found in three colorectal cancers, whereas adenomas did not show cytoplasmic activity (Figure 1B). No statistically significant correlation was found when results of nuclear and cytoplasmic Ku70/80 DNA-binding activity were compared with clinical parameters.
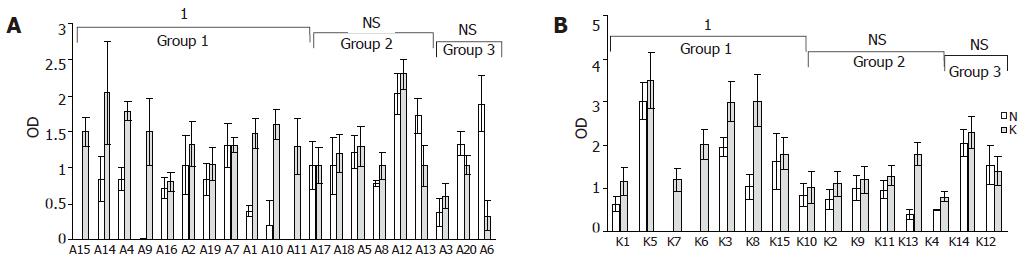
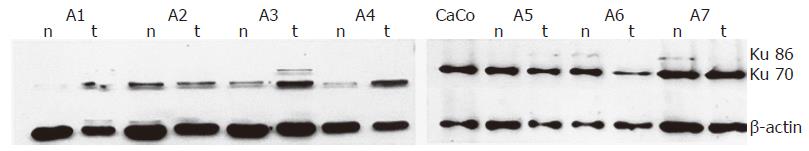
In adenomas and carcinomas with increased DNA-binding activity, the Ku70 nuclear levels were markedly increased as compared to matched normal samples (P<0.000) (Figures 3A and B, Group I), whereas protein level changes were not statistically significant in tumors with minimal increases or reduced binding activity (Figures 3A and B, Groups II and III). Representative results are shown in Figure 4.
In all the samples, Ku86 protein levels were always lower than Ku70 and in most cases it was undetectable (data not shown). Analysis of cytoplasmic protein extracts showed barely detectable Ku70 protein in normal colon tissues and a significant increase in pathologic samples with increased DNA binding activity (0.22±0.3 OD and 0.80±0.2 OD, respectively, P = 0.001) (data not shown). Ku86 protein subunit was undetectable by Western blot analysis on cytoplasmic extracts Table 2.
| Adenomas | Ku70 | Ku86 | ||
| Normal | Tumor | Normal | Tumor | |
| A1 | 61 | 93 | 40 | 81 |
| A2 | 65 | 81 | 33 | 70 |
| A3 | 60 | 83 | 31 | 63 |
| A4 | 61 | 82 | 35 | 71 |
| A5 | 65 | 79 | 32 | 75 |
| A6 | 58 | 68 | 39 | 62 |
| A7 | 64 | 74 | 26 | 62 |
| A16 | 51 | 75 | 25 | 71 |
| A17 | 79 | 92 | 24 | 63 |
| A19 | 61 | 87 | 29 | 62 |
| Carcinomas | ||||
| K1 | 66 | 95 | 27 | 78 |
| K2 | 48 | 76 | 37 | 62 |
| K3 | 66 | 84 | 26 | 71 |
| K4 | 47 | 86 | 40 | 86 |
| K5 | 72 | 85 | 34 | 70 |
| K6 | 76 | 86 | 26 | 61 |
| K7 | 72 | 88 | 28 | 76 |
| K8 | 51 | 92 | 17 | 79 |
| K9 | 62 | 96 | 44 | 65 |
| K10 | 65 | 90 | 51 | 71 |
| K11 | 56 | 94 | 51 | 66 |
| K12 | 52 | 82 | 34 | 73 |
| K13 | 65 | 85 | 25 | 66 |
| K14 | 52 | 92 | 18 | 64 |
| K15 | 65 | 73 | 28 | 65 |
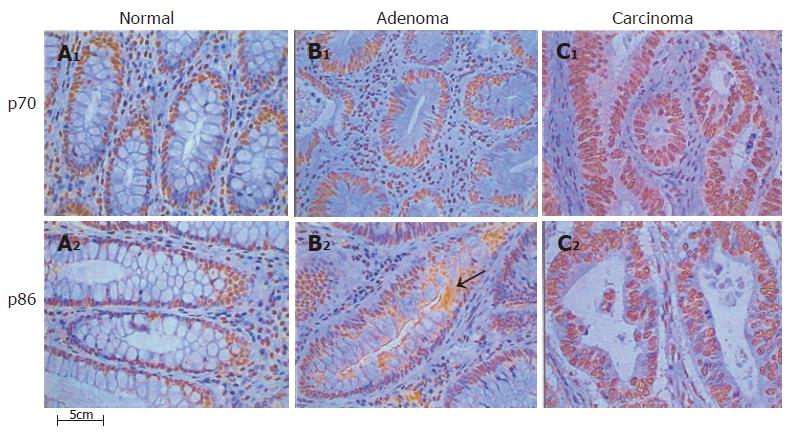
A significant increase in the percentage of stained nuclei was found for Ku70 and Ku86 subunits in all adenoma and carcinoma samples, as compared to matched normal tissues (P<0.000) but no statistically significant differences were found between adenoma and carcinoma samples (Table 3). In all cases, Ku subunits were not detected in the cytoplasm of normal colonic mucosa (n = 25) from patients or healthy donors (n = 5) (SL 0). For Ku70, a low level (SL 1) of cytoplasmic staining was detected in 11 of the 20 adenomas (55%), and in 3 of the 15 carcinomas (20%, P = 0.089). For Ku86, 17 of the 20 adenomas (85%) showed medium levels of cytoplasmic staining (SL 2-3), and 5 of the 15 carcinomas (34%) displayed cytoplasmic staining with SL 1-2 (P = 0.004). Ku86 cytoplasmic staining showed a typical granular pattern, probably due to the presence of protein subunit in cytoplasmic vesicles (Figure 5).
| Colontissue | Ku70 | Ku86 | ||||
| Normal | Tumor | P | Normal | Tumor | P | |
| Adenomas | 63.39±7.72 | 80.22±7.52 | 0.000 | 30.48±5.43 | 67.57±7.64 | 0.000 |
| Carcinomas | 61.42±9.66 | 87.34±6.97 | 0.000 | 32.86±11.29 | 70.36±7.55 | 0.000 |
| Healthy | 74.20±6.38 | - | - | 32.00±5.10 | - | - |
The NHEJ pathway plays a pivotal role in the repair of DSBs. Several studies on cell lines and knock-out mice suggest that a non-functional NHEJ can induce an increased level of genomic instability and lead to cancer progression[22-24]. In a previous study, we found a downregulation of the Ku70/80 DNA binding activity in advanced breast and bladder human cancers, as compared to non-invasive or low stage tumors[17]. This observation was also confirmed in a patient affected by sporadic multiple basal cell carcinoma, where we demonstrated a differential modulation of the NHEJ pathway in non-aggressive and aggressive tumors, showing that the first is an upregulation system, and the latter a strong downregulation system[20].
In the present study, we investigated the involvement of the ku70/80 heterodimer during progression from adenoma to carcinoma. Our results indicated that the NHEJ pathway was activated in approximately half of the cases, with no differences in binding activity between adenomas and carcinomas. In these tumors, the increase in DNA binding activity correlated with protein levels of the Ku subunits as determined by Western blot and immunohistochemical analysis. Moreover the cytoplasmic accumulation of the Ku70 and Ku86 protein subunits only in neoplastic tissues is also indicative of an upregulation of the NHEJ system. The NHEJ pathway repairs DSBs by modifying the two DNA broken ends prior to rejoining, and few nucleotides at each end of the DNA break are lost during this process[23]. Thus, the repair through the NHEJ can potentially lead to chromosomal alterations. Rothkamm et al[25] found that after X-ray exposure, NHEJ-proficient cells form misrejoinings and multiple DSBs more frequently than NHEJ deficient cells. Also, myeloid leukemia cells characterized by gross chromosomal abnormalities show a higher end-joining efficiency but a lower DSB repair fidelity, as compared to lymphocytes from healthy donors[24]. Thus, the upregulation of the NHEJ system in adenoma and colon carcinoma may be responsible for an increase in genomic rearrangements and chromosomal defects, contributing to tumor progression.
Although increase in DNA binding activity was the more frequent abnormality detected in our series of adenomas and carcinomas, we also identified a subset of colon neoplasia (approximately 15%) that showed reduced levels of DNA binding activity. Rigas et al[26] have previously analyzed protein expression of the DNA-PK subunits in adenomas and colorectal cancers by IHC assay. By using a score that correlates the intensity of staining to the percentage of stained cells, a decrease in protein expression levels is demonstrated for all DNA-PK subunits in adenoma and carcinoma samples, as compared to normal tissues[26]. In our study the reduction in DNA binding activity did not correlate completely with Ku subunit protein levels. While three of the adenomas and one of the cancers showed a decrease of Ku70 expression by Western blot analysis, the remaining adenoma and carcinoma samples displayed an increase in protein expression. IHC data also showed an increase in the percentage of nuclear stained cells in the tumor as compared to the normal colonic mucosa. We can speculate that in this subset of tumors the NHEJ proteins are expressed and localized in the nuclei, but they are not functional, due to post-translational regulation mechanisms or mutations in one or more components of the NHEJ pathway.
Overall, our results indicate that the NHEJ pathway is activated in colon adenoma and carcinoma, with only a subset of tumors showing decreased binding activity. These results however do not exclude an involvement of the NHEJ pathway in colon carcinogenesis, rather suggest the presence of colon cancer subsets that may differ in their biological behavior. Since Ku70 and Ku80 protein levels are correlated to tumor radiosensitivity and response to chemotherapy in human colorectal cancer and experimental models[27-30], the clarification of the mechanisms involved in DNA repair may ultimately lead to an improved management of colorectal cancer patients.
The authors thank Miss Simona Virga for skilful help with artwork and manuscript preparation.
Science Editor Wang XL and Guo SY Language Editor Elsevier HK
| 1. | DeVita VT, Hellmann S, Rosemberg SA, eds . Cancer. Principles and Practice of Oncology. Philadelphia, New York: Lippincott-Raven 1997; 1144-97. |
| 2. | Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8087] [Cited by in RCA: 8005] [Article Influence: 228.7] [Reference Citation Analysis (1)] |
| 3. | Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297:547-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 512] [Cited by in RCA: 524] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 4. | Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1683] [Cited by in RCA: 1754] [Article Influence: 73.1] [Reference Citation Analysis (0)] |
| 5. | Loeb LA. A mutator phenotype in cancer. Cancer Res. 2001;61:3230-3239. [PubMed] |
| 6. | van Gent DC, Hoeijmakers JH, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet. 2001;2:196-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 821] [Cited by in RCA: 854] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 7. | Difilippantonio MJ, Zhu J, Chen HT, Meffre E, Nussenzweig MC, Max EE, Ried T, Nussenzweig A. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 411] [Cited by in RCA: 418] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 8. | Ferguson DO, Sekiguchi JM, Chang S, Frank KM, Gao Y, DePinho RA, Alt FW. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc Natl Acad Sci USA. 2000;97:6630-6633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 273] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 9. | Gao Y, Ferguson DO, Xie W, Manis JP, Sekiguchi J, Frank KM, Chaudhuri J, Horner J, DePinho RA, Alt FW. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature. 2000;404:897-900. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 438] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 10. | Jackson SP. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002;23:687-696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 793] [Cited by in RCA: 784] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 11. | Wang J, Dong X, Reeves WH. A model for Ku heterodimer assembly and interaction with DNA. Implications for the function of Ku antigen. J Biol Chem. 1998;273:31068-31074. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 12. | Jin S, Weaver DT. Double-strand break repair by Ku70 requires heterodimerization with Ku80 and DNA binding functions. EMBO J. 1997;16:6874-6885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 128] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 13. | Li GC, He F, Shao X, Urano M, Shen L, Kim D, Borrelli M, Leibel SA, Gutin PH, Ling CC. Adenovirus-mediated heat-activated antisense Ku70 expression radiosensitizes tumor cells in vitro and in vivo. Cancer Res. 2003;63:3268-3274. [PubMed] |
| 14. | O'Brien MJ, Winawer SJ, Zauber AG, Gottlieb LS, Sternberg SS, Diaz B, Dickersin GR, Ewing S, Geller S, Kasimian D. The National Polyp Study. Patient and polyp characteristics associated with high-grade dysplasia in colorectal adenomas. Gastroenterology. 1990;98:371-379. [PubMed] |
| 15. | Jass JR, Sobin LH. Histological typing of intestinal tumours: World Health Organization. 1989. Springer, Berlin. . [DOI] [Full Text] |
| 16. | Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475-1489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8442] [Cited by in RCA: 10166] [Article Influence: 242.0] [Reference Citation Analysis (0)] |
| 17. | Pucci S, Mazzarelli P, Rabitti C, Giai M, Gallucci M, Flammia G, Alcini A, Altomare V, Fazio VM. Tumor specific modulation of KU70/80 DNA binding activity in breast and bladder human tumor biopsies. Oncogene. 2001;20:739-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 77] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 18. | Lahiri DK, Ge Y. Electrophoretic mobility shift assay for the detection of specific DNA-protein complex in nuclear extracts from the cultured cells and frozen autopsy human brain tissue. Brain Res Brain Res Protoc. 2000;5:257-265. [RCA] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 68] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 19. | Frasca D, Barattini P, Goso C, Pucci S, Rizzo G, Bartoloni C, Costanzo M, Errani A, Guidi L, Antico L. Cell proliferation and ku protein expression in ageing humans. Mech Ageing Dev. 1998;100:197-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 20. | Mazzarelli P, Rabitti C, Parrella P, Seripa D, Persichetti P, Marangi GF, Perrone G, Poeta ML, Delfino M, Fazio VM. Differential modulation of Ku70/80 DNA-binding activity in a patient with multiple basal cell carcinomas. J Invest Dermatol. 2003;121:628-633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 21. | Kumagai I, Masuda T, Sato S, Ishikawa K. Immunoreactivity to monoclonal antibody, Hep Par 1, in human hepatocellular carcinomas according to histopathological grade and histological pattern. Hepatol Res. 2001;20:312-319. [RCA] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 22. | Łanuszewska J, Widłak P. The truncation of Ku86 in human lymphocytes. Cancer Lett. 2004;205:197-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 23. | Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712-720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 714] [Cited by in RCA: 713] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 24. | Choi EK, Lee YH, Choi YS, Kwon HM, Choi MS, Ro JY, Park SK, Yu E. Heterogeneous expression of Ku70 in human tissues is associated with morphological and functional alterations of the nucleus. J Pathol. 2002;198:121-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 25. | Rothkamm K, Löbrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci U S A. 2003;100:5057-5062. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1235] [Cited by in RCA: 1216] [Article Influence: 55.3] [Reference Citation Analysis (0)] |
| 26. | Rigas B, Borgo S, Elhosseiny A, Balatsos V, Manika Z, Shinya H, Kurihara N, Go M, Lipkin M. Decreased expression of DNA-dependent protein kinase, a DNA repair protein, during human colon carcinogenesis. Cancer Res. 2001;61:8381-8384. [PubMed] |
| 27. | Komuro Y, Watanabe T, Hosoi Y, Matsumoto Y, Nakagawa K, Tsuno N, Kazama S, Kitayama J, Suzuki N, Nagawa H. The expression pattern of Ku correlates with tumor radiosensitivity and disease free survival in patients with rectal carcinoma. Cancer. 2002;95:1199-1205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 69] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 28. | Komuro Y, Watanabe T, Hosoi Y, Matsumoto Y, Nakagawa K, Saito S, Ishihara S, Kazama S, Tsuno N, Kitayama J. Prediction of tumor radiosensitivity in rectal carcinoma based on p53 and Ku70 expression. J Exp Clin Cancer Res. 2003;22:223-228. [PubMed] |
| 29. | Achanta G, Pelicano H, Feng L, Plunkett W, Huang P. Interaction of p53 and DNA-PK in response to nucleoside analogues: potential role as a sensor complex for DNA damage. Cancer Res. 2001;61:8723-8729. [PubMed] |