Published online Apr 28, 2005. doi: 10.3748/wjg.v11.i16.2508
Revised: October 25, 2003
Accepted: April 13, 2004
Published online: April 28, 2005
AIM: To study the global gene expression of chemotactic genes in macrophage line U937 treated with human monocyte chemoattractant protein-1 (MCP-1) through the use of ExpreeChipTMHO2 cDNA array.
METHODS: Total RNA was extracted from MCP-1 treated macrophage line U937 and normal U937 cells, reversely transcribed to cDNA, and then screened in parallel with HO2 human cDNA array chip. The scanned result was additionally validated using RT-PCR.
RESULTS: The result of cDNA array showed that one chemotactic-related gene was up-regulated more than two-fold (RANTES) and seven chemotactic-related genes were down-regulated more than two-fold (CCR1, CCR5, ccl16, GROβ, GROγ, IL-8 and granulocyte chemotactic protein 2) in MCP-1 treated U937 cells at mRNA level. RT-PCR analysis of four of these differentially expressed genes gave results consistent with cDNA array findings.
CONCLUSION: MCP-1 could influence some chemokine and receptor expressions in macrophages in vitro. MCP-1 mainly down-regulates the expression of chemotactic genes influencing neutrophilic granulocyte expression (GROβ, GROγ, IL-8 and granulocyte chemotactic protein 2), and the mRNA level of CCR5, which plays a critical role in many disorders and illnesses.
- Citation: Bian GX, Miao H, Qiu L, Cao DM, Guo BY. Profiling of differentially expressed chemotactic-related genes in MCP-1 treated macrophage cell line using human cDNA arrays. World J Gastroenterol 2005; 11(16): 2508-2512
- URL: https://www.wjgnet.com/1007-9327/full/v11/i16/2508.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i16.2508
Chemokines are small secreted proteins that function as potent activators and chemoattractants for leukocyte subpopulations and some nonhemopoietic cells. Most chemokines elicit their effects through interactions with seven-transmembrane-domain, G-protein-coupled receptors. The size of this family has grown considerably and now includes dozens of members[1]. According to the position of conserved cysteine residues in their primary sequence, the chemokine superfamily is divided into four subfamilies (C-X-C, C-C, C, and C-X3-C) which attract specific subsets of leukocytes[2]. Chemokine expression secondary to stimulation with proinflammatory cytokines has been reported in many types of diseases[3].
Monocyte chemoattractant protein-1 (MCP-1) was first purified from conditioned medium of baboon aortic smooth muscle cells in culture on the basis of its ability to attract monocytes, but not neutrophils, in vitro. It is a potent chemoattractant for monocytes in vitro, with an ED50 similar to that of IL-8 for neutrophils (500 pmol/L). MCP-1 induces the expression of integrins required for chemotaxis, and has also been reported to attract NK cells as well as T lymphocytes[4-6].
Recently, the function of chemokines has extended far beyond leukocyte physiology. For example, MCP-1 was found to play a pathogenic role in many diseases. Its expression could be detected in human atheromatous plaques and in aortic walls of primates fed with high-cholesterol diets, consistent with a model of atherogenesis in which MCP-1 in the vessel walls attracted monocytes that eventually became foam cells[7]. Similarly, the presence of inflammatory cells in the joints of patients with rheumatoid arthritis has been explained by IL-8 and MCP-1 in synovial fluids[8]. This expression was also documented in glomerulonephritis, asthma, inflammatory bowel disease, and allogeneic transplant rejection[6,9].
The ligand-binding repertoires of different chemokine receptors significantly overlap, so do the sets of receptors expressed by different leukocytes and other target cells, this further adds to the versatility of the chemokine system. There are high complexities among the conditions of chemokine expressions and binding to receptors. For example, among the known CC chemokines, MCP-1, MCP-2, MCP-3, MCP-4, MCP-5, macrophage inflammatory protein (MIP)-1α, MIP-1β, I309, and HCC-1, all have monocyte chemoattractant activities in vitro. Furthermore, monocytes express at least three cloned CC chemokine receptors, namely CCR1, CCR2, and CCR5, and even though MCP-1 binds to only CCR2 with a high affinity, CCR2 also binds to MCP-3 and MCP-5[4,6]. The cDNA array technology has been demonstrated as a very useful tool for identifying differentially expressed genes. In order to study the regulation of MCP-1 on other chemokines and their receptors, we studied the potential regulation function of MCP-1 on the expression of chemotactic-related genes in macrophages.
huMCP-1 was purchased from Dingguo Biotech Corp.[10]. Human macrophage line U937 was reserved by our study group. FCS, chloroform, isopropanol, DEPC, TRIzol were purchased from Huashun Corp.
Macrophage line U937 was incubated in 10 mL RPMI1640 medium containing 10% FCS. When cell count reached 0.5-1×106/mL, cells were centrifuged and the supernates were discarded. The cells were resuspended with the same volume of RPMI1640 medium containing huMCP-1 (10 mg/mL) and incubated overnight.
Culture cells were washed with cooled PBS (pH 7.2) twice, then lysed by TRIzol, extracted with chloroform, precipitated with isopropanol, and washed with 80% ethanol. The deposits were dehydrated by vacuum, then solubilized by Nes buffer. mRNA was purified by an Oligotex mRNA mini kit, then the control mRNA of cells was labeled with cy5-dUTP and the mRNA of stimulated cells was labeled with cy3-dUTP, deposited by ethanol and solubilized in 20 μL hybridization buffer (5×SSC+0.2% SDS). ExpreeChipTMHO2 was made by MERGEN Corp. The chip and probe were degenerated for 5 min at 95 °C, then hybridized for 15-17 h at 60 °C, washed with 2×SSC+0.2% SDS and 0.1%×SSC +0.2% SDS, 0.1%×SSC, and dried at room temperature. The chip was scanned by ScanArray3000, then the result was analyzed by ImaGene3.0. The criteria of gene expression changes were cy3/cy5≥2, or cy3/cy5≤0.5.
cDNA was generated using 1 µg of total RNA from the two U937 cell lines (normal and MCP-1 treated) as templates in a 20-µL reaction mixture, and reverse transcription was carried out at 42 °C for 1 h followed by at 95 °C for 10 min using the preamplification system (GIBCOL). cDNA (2 µL) was amplified in a 25 µL PCR reaction mixture containing 2×PCR buffer (10 mmol/L Tris-HCl, pH 8.3, 50 mmol/L KCl), 1.9 or 2.4 mmol/L of MgCl2, 0.5 µmol/L of primers, 0.18 mmol/L of deoxynucleotide triphosphate, and 1 unit Taq DNA polymerase (TaKaRa). The conditions of hot-start PCR reaction were as follows: at 95 °C for 10 min followed by 25-35 cycles of denaturation at 94 °C for 1 min, annealing at 60 °C for 1 min (for primers of β-actin, GROβ, GROγ and IL-8) or at 50 °C for 1 min (for primers of regulated upon activation, normal T cell expressed and secreted, RANTES), and extension at 72 °C for 1 min. The final step of extension was at 72 °C for 10 min. PCR reagents were purchased from Takara. All of the primers were synthesized by Genecore Corp., Shanghai. The cycle number was optimized for each gene-specific primer pair to ensure the amplification in a linear range, and the results were semiquantitative. PCR products (5 µL) were visualized by electrophoresis on a 2% agarose gel stained with ethidium bromide.
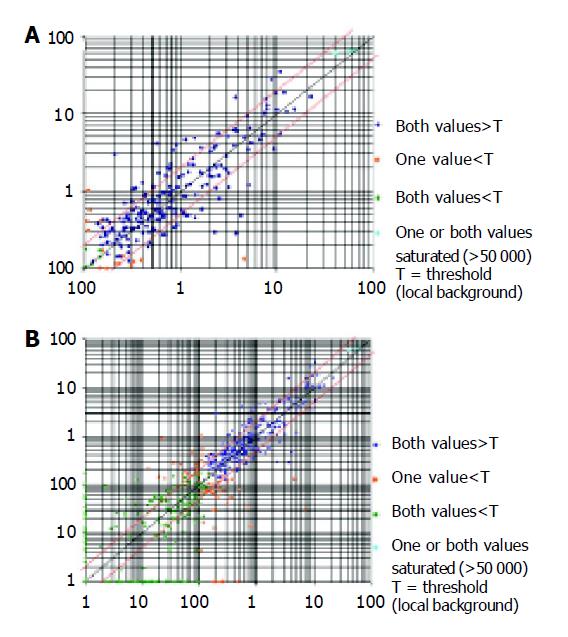
ExpreeChipTMHO2 (invoice: 0102-003) used in this study was made by MERGEN Corp. The chip contained 10 ng of each gene-specific cDNA from 1 152 known genes and 9 housekeeping genes. Several plasmid and bacteriophage DNAs and blank spots were also included as negative and blank controls to confirm hybridization specificity. A complete list of the genes with array positions and GenBank accession numbers of the chip used here could be accessed at the website. Genes were considered to be up-regulated when the intensity ratio between expressions in the MCP-1 treated U937 cell lines compared with normal cell lines was two-fold. Genes were labeled as down-regulated when the ratio between normal and MCP-1 treated cell lines was two-fold. The analysis of scatter diagrams is seen in Figure 1, using EC cells as system controls.
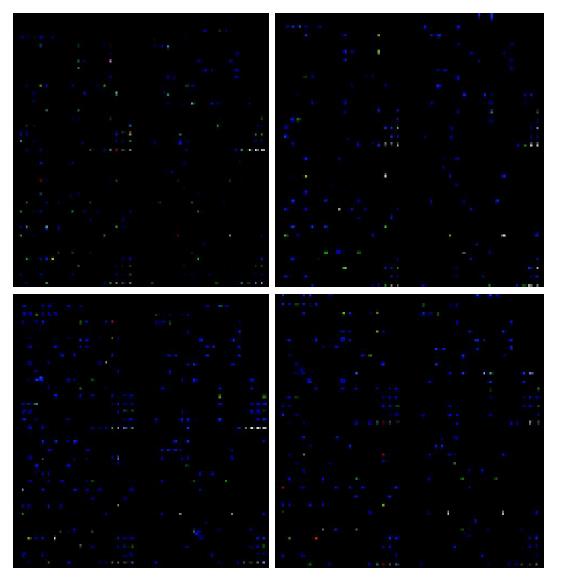
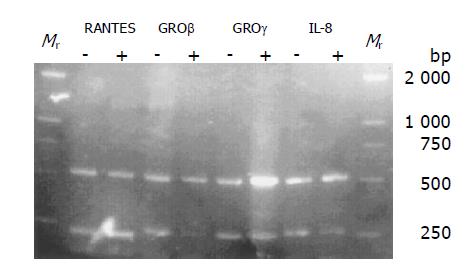
The membranes carrying 1152 cDNA probes of defined human genes, and their accession numbers, names, and the scanned data were given below. Among the 1152 genes, 110 were up-regulated, 91 were down-regulated (Figure 2). We searched for chemokine genes and chemokine receptor genes to study the gene expression changes in chemokine superfamily (Tables 1 and 2). Gene names shown in bold designate that genes with mRNA expression were also analyzed by RT-PCR (Figure 3).
| Sample/Control | Control/Sample | Unigene symbol | Gene description |
| 0.1 | 1.0 | CCR1 | Chemokine (C-C motif) receptor 1 |
| 1.0 | 19.2 | CCR2 | Chemokine (C-C motif) receptor 2 |
| 0.7 | 1.3 | CCR3 | Chemokine (C-C motif) receptor 3 |
| 0.6 | 1.6 | CCR3 | Chemokine (C-C motif) receptor 3 |
| 1.0 | 1.0 | CCR4 | Chemokine (C-C motif) receptor 4 |
| 0.1 | 7.3 | CCR5 | Chemokine (C-C motif) receptor 5 |
| 1.0 | 1.0 | CCR6 | Chemokine (C-C motif) receptor 6 |
| 1.0 | 1.0 | CCR7 | Chemokine (C-C motif) receptor 7 |
| 1.0 | 1.0 | CCR8 | Chemokine (C-C motif) receptor 8 |
| 1.0 | 1.0 | CCRL2 | Chemokine (C-C motif) receptor-like 2 |
| 1.4 | 0.7 | CXCR4 | Chemokine (C-X-C motif), receptor 4 (fusin) |
| Sample/Control | Control/Sample | Unigene symbol | Gene description |
| 1.6 | 0.6 | SCYA3 | Small inducible cytokine A3 (homologous to mouse Mip-1a) |
| 2.6 | 0.4 | SCYA5 | Small inducible cytokine A5 (RANTES) |
| 1.0 | 1.0 | SCYA17 | Small inducible cytokine subfamily A (Cys-Cys), member 17 |
| 1.0 | 1.0 | SCYA11 | Small inducible cytokine subfamily A (Cys-Cys), member 11 (eotaxin) |
| 1.0 | 1.0 | SCYA13 | Small inducible cytokine subfamily A (Cys-Cys), member 13 |
| 1.0 | 1.0 | SCYA14 | Small inducible cytokine subfamily A (Cys-Cys), member 14 |
| 0.4 | 2.8 | SCYA16 | Small inducible cytokine subfamily A (Cys-Cys), member 16 |
| 1.0 | 1.0 | SCYA18 | Small inducible cytokine subfamily A (Cys-Cys), member 18,pulmonary and activation-regulated |
| 1.0 | 1.0 | SCYA19 | Small inducible cytokine subfamily A (Cys-Cys), member 19 |
| 1.0 | 1.0 | SCYA20 | Small inducible cytokine subfamily A (Cys-Cys), member 20 |
| 1.0 | 1.0 | SCYA21 | Small inducible cytokine subfamily A (Cys-Cys), member 21 |
| 1.0 | 1.0 | SCYA22 | Small inducible cytokine subfamily A (Cys-Cys), member 22 |
| 1.0 | 1.0 | SCYA23 | Small inducible cytokine subfamily A (Cys-Cys), member 23 |
| 1.0 | 1.0 | SCYA25 | Small inducible cytokine subfamily A (Cys-Cys), member 25 |
| 0.0 | 469.8 | GRO2 | GROb (melanoma growth stimulating activity, beta) |
| 0.0 | 47.6 | GRO3 | GROg (melanoma growth stimulating activity, gamma) |
| 0.0 | 34.5 | IL8 | Interleukin 8 |
| 1.0 | 1.0 | SCYB11 | Small inducible cytokine subfamily B (Cys-X-Cys), member 11 |
| 1.0 | 1.0 | SCYB5 | Small inducible cytokine subfamily B (Cys-X-Cys), member 5 (epithelial-derived neutrophil-activating peptide 78) |
| 0.0 | 261.6 | SCYB6 | Small inducible cytokine subfamily B (Cys-X-Cys), member 6 (granulocyte chemotactic protein 2) |
| 1.0 | 1.0 | SCYC2 | Small inducible cytokine subfamily C, member 2 |
| 1.0 | 1.0 | SCYD1 | Small inducible cytokine subfamily D (Cys-X3-Cys), member 1 (fractalkine, neurotactin) |
The semiquantitative RT-PCR results showed that RANTES genes were up-regulated, whereas GROβ, GROγ and IL-8 were down-regulated in MCP-1 treated cell lines (Figure 3) .These results were similar to those detected by Human ExpreeChipTMHO2 chip (Tables 1 and 2).
MCP-1 is a CC chemokine that attracts monocytes, memory T lymphocytes, and natural killer cells. The interaction of MCP-1 with its receptor is essential for monocyte activation and induction of chemotaxis during an inflammatory response. Because of its target cell specificity, MCP-1 has been postulated to play a pathogenic role in a variety of diseases characterized by mononuclear cell infiltration, including atherosclerosis, rheumatoid arthritis, and multiple sclerosis[6,11]. MCP-1 may exert these effects by influencing the expression of other chemokines, which is hard to be demonstrated for the complexity of the chemokine network. Genechip is a high-throughput method to evaluate hundreds of genes at one time, so it is the best method to investigate this complex process and the relationship between MCP-1 and other chemokines and receptors.
Although U937 cell line may differ in some aspects from human blood macrophages, it expresses functional chemokines and cytokines as human blood macrophages. The major advantage of using U937 cells is the homogeneity of the cell line, allowing comparison of findings between different experiments. For this reason, we used U937 cells in the present study to examine macrophage responses to MCP-1, even though they were not absolutely identical to human peripheral macrophages.
In the present study, we specifically examined the global gene expression of chemokines and their receptors, and demonstrated that MCP-1 could strongly down-regulate the expression level of the CXC subfamily chemokines: IL-8, GROβ, GROγ and granulocyte chemotactic protein 2. MCP-1 also could up-regulate the expression level of RANTES (CC subfamily), down-regulate the expression level of CCL16 (CC subfamily). It had no effect on the expression of XCL2 (C subfamily) and fractalkine (the only member of CX3C subfamily). In chemokine receptors, it could down-regulate the expression level of CCR2 and CCR5.
IL-8, GROβ, GROγ and granulocyte chemoattractant protein-2 are all members of the CXC chemokine family. The GRO proteins are about 90% identical in amino acid sequence. IL-8 and granulocyte chemoattractant protein-2 are about 40-50% identical to each other and to any of the GRO proteins. The CXC subfamily can be further subdivided into ELR+ and ELR groups, based on the presence or absence of the sequence motif glutamic acid-leucine-arginine (ELR) N-terminal to the first cysteine. They are all ELR+ CXC chemokines. All ELR+ CXC chemokines are powerful activators of neutrophils and induce chemotaxis, shape change, a rise in intracellular free calcium levels, exocytosis, and respiratory burst in vitro and neutrophil accumulation in vivo, whereas the ELR CXC chemokines are not neutrophil chemoattractants[12-14]. Our findings suggest that MCP-1, which activated and chemoattracted monocytes and macrophages, could depress the infiltration of neutrophils in inflammation by rendering macrophages to express less neutrophil chemokines. Many disorders begin as neutrophils infiltrate at the inflammatory location, and further develop as monocytes or macrophages infiltrate, MCP-1 then may be one of the regulating factors of such changes.
RANTES was isolated in a T- vs B-lymphocyte differential screen, and found to be inducible by mitogens or antigens in a variety of T-cell lines and circulating lymphocytes. In vitro, RANTES was nearly as a potent chemoattractant for monocytes as MCP-1, but was much less effective in stimulating exocytosis. In endothelial cell-free assays, RANTES attracted CD4+, CD45R0+ T lymphocytes, but in transendothelial systems it attracted CD8+ cells as well as CD4+, and was the most potent CC chemokine for CD8+ chemoattraction. The first hint about a connection between chemokines and HIV-1 came from the finding that RANTES could prevent infection by macrophage-tropic, nonsyncytium-inducing strains of HIV-1[15].
In vitro ligand binding experiments suggested that the sole cloned receptor of MCP-1 was CCR2. CCR2 responded to MCP-1, MCP-3 and MCP-5, but maximum responses were only obtained to MCP-1. CCR5 could interact with RANTES, MIP-1, or MCP-2 under physiological conditions[16,17]. CCR5 could also act as a co-receptor in HIV-1-mediated infection of CD4-positive lymphocytes and microglia. In addition, the ligands for CCR5 could inhibit infection with certain strains of HIV-1, and decreased susceptibility to HIV-1 infection has been linked with mutations in CCR5 gene[18,19]. CCR5 was also involved in a diverse array of inflammatory diseases[11,20,21]. Our findings suggested that MCP-1 might influence the process of these diseases, although the mechanism is not clear. Detailed data need to be further explored.
In summary, MCP-1 can influence the expression of some chemokines and receptors in macrophages in vitro. MCP-1 can also down-regulate the mRNA level of CCR5, which plays a critical role in many disorders and illnesses. MCP-1 can also greatly change other cytokines of the immune system, such as IL-18, TNF, IFN. Our findings disclose some relationship among MCP-1 and other chemokine-related members, shedding new light on the mechanism of the function of MCP-1 and the pathogenesis of related diseases.
| 1. | Rossi D, Zlotnik A. The biology of chemokines and their receptors. Annu Rev Immunol. 2000;18:217-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1803] [Cited by in RCA: 1792] [Article Influence: 71.7] [Reference Citation Analysis (0)] |
| 2. | Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392:565-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2088] [Cited by in RCA: 2043] [Article Influence: 75.7] [Reference Citation Analysis (0)] |
| 3. | Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2814] [Cited by in RCA: 2818] [Article Influence: 112.7] [Reference Citation Analysis (0)] |
| 4. | Zhang L, Khayat A, Cheng H, Graves DT. The pattern of monocyte recruitment in tumors is modulated by MCP-1 expression and influences the rate of tumor growth. Lab Invest. 1997;76:579-590. [PubMed] |
| 5. | Valente AJ, Rozek MM, Sprague EA, Schwartz CJ. Mechanisms in intimal monocyte-macrophage recruitment. A special role for monocyte chemotactic protein-1. Circulation. 1992;86:III20-III25. [PubMed] |
| 6. | Martinet N, Beck G, Bernard V, Plenat F, Vaillant P, Schooneman F, Vignaud JM, Martinet Y. Mechanism for the recruitment of macrophages to cancer site. In vivo concentration gradient of monocyte chemotactic activity. Cancer. 1992;70:854-860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 7. | Lu B, Rutledge BJ, Gu L, Fiorillo J, Lukacs NW, Kunkel SL, North R, Gerard C, Rollins BJ. Abnormalities in monocyte recruitment and cytokine expression in monocyte chemoattractant protein 1-deficient mice. J Exp Med. 1998;187:601-608. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 806] [Cited by in RCA: 813] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 8. | Corrigall VM, Arastu M, Khan S, Shah C, Fife M, Smeets T, Tak PP, Panayi GS. Functional IL-2 receptor beta (CD122) and gamma (CD132) chains are expressed by fibroblast-like synoviocytes: activation by IL-2 stimulates monocyte chemoattractant protein-1 production. J Immunol. 2001;166:4141-4147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 9. | Koch AE, Kunkel SL, Harlow LA, Johnson B, Evanoff HL, Haines GK, Burdick MD, Pope RM, Strieter RM. Enhanced production of monocyte chemoattractant protein-1 in rheumatoid arthritis. J Clin Invest. 1992;90:772-779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 443] [Cited by in RCA: 486] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 10. | Xie L, Guo J, Qian X, Chen W. Multiple types of chemokines expressed in mouse thymic stromal cell lines. Zhongguo YiXue KeXueYuan XueBao. 2000;22:498-501. [PubMed] |
| 11. | Gu L, Tseng S, Horner RM, Tam C, Loda M, Rollins BJ. Control of TH2 polarization by the chemokine monocyte chemoattractant protein-1. Nature. 2000;404:407-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 633] [Cited by in RCA: 645] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 12. | King AG, Horowitz D, Dillon SB, Levin R, Farese AM, MacVittie TJ, Pelus LM. Rapid mobilization of murine hematopoietic stem cells with enhanced engraftment properties and evaluation of hematopoietic progenitor cell mobilization in rhesus monkeys by a single injection of SB-251353, a specific truncated form of the human CXC chemokine GRObeta. Blood. 2001;97:1534-1542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 96] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 13. | Wang D, Richmond A. Nuclear factor-kappa B activation by the CXC chemokine melanoma growth-stimulatory activity/growth-regulated protein involves the MEKK1/p38 mitogen-activated protein kinase pathway. J Biol Chem. 2001;276:3650-3659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 80] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 14. | Miura M, Fu X, Zhang QW, Remick DG, Fairchild RL. Neutralization of Gro alpha and macrophage inflammatory protein-2 attenuates renal ischemia/reperfusion injury. Am J Pathol. 2001;159:2137-2145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 164] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 15. | Fischer FR, Luo Y, Luo M, Santambrogio L, Dorf ME. RANTES-induced chemokine cascade in dendritic cells. J Immunol. 2001;167:1637-1643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 62] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 16. | Oppermann M, Mack M, Proudfoot AE, Olbrich H. Differential effects of CC chemokines on CC chemokine receptor 5 (CCR5) phosphorylation and identification of phosphorylation sites on the CCR5 carboxyl terminus. J Biol Chem. 1999;274:8875-8885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 148] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 17. | Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, Goedert JJ, Buchbinder SP, Vittinghoff E, Gomperts E. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856-1862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1804] [Cited by in RCA: 1735] [Article Influence: 59.8] [Reference Citation Analysis (0)] |
| 18. | Raport CJ, Gosling J, Schweickart VL, Gray PW, Charo IF. Molecular cloning and functional characterization of a novel human CC chemokine receptor (CCR5) for RANTES, MIP-1beta, and MIP-1alpha. J Biol Chem. 1996;271:17161-17166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 353] [Cited by in RCA: 355] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 19. | Samson M, Labbe O, Mollereau C, Vassart G, Parmentier M. Molecular cloning and functional expression of a new human CC-chemokine receptor gene. Biochemistry. 1996;35:3362-3367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 526] [Cited by in RCA: 521] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 20. | Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955-1958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2146] [Cited by in RCA: 2109] [Article Influence: 72.7] [Reference Citation Analysis (0)] |
| 21. | Vielhauer V, Anders HJ, Mack M, Cihak J, Strutz F, Stangassinger M, Luckow B, Gröne HJ, Schlöndorff D. Obstructive nephropathy in the mouse: progressive fibrosis correlates with tubulointerstitial chemokine expression and accumulation of CC chemokine receptor 2- and 5-positive leukocytes. J Am Soc Nephrol. 2001;12:1173-1187. [PubMed] |