Published online Mar 28, 2015. doi: 10.3748/wjg.v21.i12.3499
Peer-review started: July 22, 2014
First decision: August 27, 2014
Revised: September 29, 2014
Accepted: November 7, 2014
Article in press: November 11, 2014
Published online: March 28, 2015
AIM: To characterize the influence of location, species and treatment upon RNA degradation in tissue samples from the gastrointestinal tract.
METHODS: The intestinal samples were stored in different medium for different times under varying conditions: different species (human and rat), varying temperature (storage on crushed ice or room temperature), time point of dissection of the submucous-mucous layer from the smooth muscle (before or after storage), different rinsing methods (rinsing with Medium, PBS, RNALater or without rinsing at all) and different regions of the gut (proximal and distal small intestine, caecum, colon and rectum). The total RNA from different parts of the gut (rat: proximal and distal small intestine, caecum, colon and rectum, human: colon and rectum) and individual gut layers (muscle and submucosal/mucosal) was extracted. The quality of the RNA was assessed by micro capillary electrophoresis. The RNA quality was expressed by the RNA integrity number which is calculated from the relative height and area of the 18 S and 28 S RNA peaks. From rat distal small intestine qPCR was performed for neuronal and glial markers.
RESULTS: RNA obtained from smooth muscle tissue is much longer stable than those from submucosal/mucosal tissue. At RT muscle RNA degrades after one day, on ice it is stable at least three days. Cleaning and separation of gut layers before storage and use of RNALater, maintains the stability of muscle RNA at RT for much longer periods. Different parts of the gut show varying degradation periods. RNA obtained from the submucosal/mucosal layer always showed a much worse amplification rate than RNA from muscle tissue. In general RNA harvested from rat tissue, either smooth muscle layer or submucosal/mucosal layer is much longer stable than RNA from human gut tissue, and RNA obtained from smooth muscle tissue shows an increased stability compared to RNA from submucosal/mucosal tissue. At RT muscle RNA degrades after one day, while the stability on ice lasts at least three days. Cleaning and separation of gut layers before storage and use of RNALater, maintains the stability of muscle RNA at RT for much longer periods. Different parts of the gut show varying degradation periods. The RNA from muscle and submucosal/mucosal tissue of the proximal small intestine degrades much faster than the RNA of distal small intestine, caecum or colon with rectum. RNA obtained from the submucosal/mucosal layer always showed a much more reduced amplification rate than RNA from muscle tissue [β-Tubulin III for muscle quantification cycle (Cp): 22.07 ± 0.25, for β-Tubulin III submucosal/mucosal Cp: 27.42 ± 0.19].
CONCLUSION: Degradation of intestinal mRNA depends on preparation and storage conditions of the tissue. Cooling, rinsing and separating of intestinal tissue reduce the degradation of mRNA.
Core tip: The quality of RNA is crucial for an appropriate RNA analysis. Especially when working with human material, precious samples will often be used for different purposes and can therefore not be frozen immediately. Gut tissue is especially fragile and RNA degrades rapidly if not treated adequately. Under these aspects RNA degradation of different gut sections and gut wall layers regarding their treatment was investigated in this study. Storage, rinsing and preparation conditions are essential for RNA stability. Sufficient and permanent cooling as well as the removal of bacterial contamination leads to a reduced degradation in muscle tissue of the gut.
- Citation: Heumüller-Klug S, Sticht C, Kaiser K, Wink E, Hagl C, Wessel L, Schäfer KH. Degradation of intestinal mRNA: A matter of treatment. World J Gastroenterol 2015; 21(12): 3499-3508
- URL: https://www.wjgnet.com/1007-9327/full/v21/i12/3499.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i12.3499
RNA isolation followed by PCR analysis has become a major tool in modern research[1-4]. Working with RNA requires the control of RNA integrity[5]. This is a critical first step in obtaining meaningful gene expression data. Working with low quality RNA might have a significant impact on the experimental results of downstream applications, which are often labour-intensive, time-consuming, and expensive. Using intact RNA is a key factor for the successful application of modern molecular biological methods, such as qRT-PCR or microarray analysis[6,7]. Samples are usually immediately frozen to avoid prolonged storage under suboptimal conditions. The analysis and evaluation of human samples depends on a stable supply and often only small amounts of tissue can be provided. In many cases these rare samples are used for various experiments[8], including the isolation of living cells[9], thus making it necessary to use alternative storage and transportation conditions of vital tissue. Especially in cases, where the samples are received at different locations, or even from abroad, transportation time might exceed 24 h. In these cases, the samples cannot simply be frozen; they have to be transported as vital tissue on crushed ice, which is not an ideal condition for RNA conservation. The integrity of the sample RNA depends strongly on the individual tissue[10]. When working with the sensible gut tissue, several major drawbacks came into the focus of attention. The amount of digestive enzymes and a broad range of microorganisms[11,12] are rendering the gut a problematic tissue. Moreover, it contains different compartments or layers (muscle, submucosal and mucosal layer)[13] with individual properties. In the actual study the integrity of the RNA was investigated concerning transportation or storage time and condition as well as the influence of the gut content upon the individual layer.
Intestinal tissue from children (age ranged from two month to two years) was obtained from two surgical facilities. A total of ten gut tissue samples of the colon which were sectioned in one centimetre segments were included in the study. These sections were analysed as a whole or separated into muscle and submucosal/mucosal layer. All samples were collected after written informed consent of the parents of the patients, according to the Declaration of Helsinki and with the approval of the local ethics committee.
Sprague Dawley rats of one to two weeks of age were used. Rats were killed by decapitation and the small intestine, caecum and colon were removed. The small intestine was sectioned into equal segments. Animal protocols were approved by the internal Veterinary Inspection Office in Mannheim.
For all experiments performed in this study human or rat tissue samples (each one cm long) were used. The individual experiments differ concerning species, location, gut layer, storage conditions (time and temperature), time of separation of the layers and different rinsing methods.
In the first experiment the differences in RNA degradation between human and rat gut tissues have been investigated. The tissues were temporarily stored in a Hepes buffered minimal essential medium (MEM-Hepes) with a stable glutamine dipeptide (Glutamax) on ice. After different storage times the tissue was separated in muscle and submucosal/mucosal layer: 6 h, 24 h and 72 h. The 0 h samples were frozen immediately in liquid nitrogen and stored until further processing for RNA extraction, as described below.
The second experiment was performed to demonstrate the influence of the temperature. The samples from rat small intestine were flushed first in PBS, then separated into muscle and submucosal/mucosal layer and then stored for different times (0 h, 0.5 h, 1 h, 1.5 h, 3 h, 6 h, 8 h, 24 h, 48 h and 72 h) in MEM-Hepes either on ice or at room temperature (RT) before storage until further processing for RNA extraction, as described below.
A third experiment was designed to investigate more precisely the influence of separation of the gut different layers prior to processing upon RNA degradation. The samples from rat small intestine were rinsed in MEM-Hepes with antibiotics (Gentamycin and Metronidazol) before being temporarily stored (0 h, 0.5 h, 1 h, 1.5 h, 3 h, 6 h, 8 h, 24 h, 48 h and 72 h) in MEM-Hepes (Glutamax) on ice or at RT. After storage samples were separated in muscle and submucosa/mucosa. A second group of tissue was investigated where muscle and submucosal/mucosal layer had been separated before storage (same times as above) until further processing for RNA extraction, as described below.
A forth experiment should evaluate the influence of different rinsing solutions. The samples from rat small intestine were rinsed after dissection in PBS or MEM-Hepes with antibiotics (Gentamycin and Metronidazol), in RNALater or not rinsed at all. Then they were temporarily stored in the individually used rinsing solutions on ice or at RT. The tissue was separated in muscle and submucosal/mucosal layer at different times: 0 h, 8 h, 24 h and 48 h before storage until further processing for RNA extraction, as described below.
A fifth experiment was performed to analyze whether the gut region alters the effect of RNA degradation. The whole rat gut was separated into proximal and distal small intestine, caecum, colon and rectum. Then the gut tissues were temporarily stored in a Hepes buffered MEM with a stable glutamine dipeptide (Glutamax) at RT. The tissue was separated into muscle and submucosal/mucosal layer at different times (0 h, 8 h, 24 h and 48 h) before stored until further processing for RNA extraction, as described below.
Total cellular RNA was extracted from one centimetre tissue segments using an “Isolate RNA Mini Kit” (Bioline, Luckenwalde, Germany) following the manufacturer’s protocol. The contaminating genomic DNA was digested with DNase I (Invitrogen, Karlsruhe, Germany). RNA concentration was measured spectrophotometrically using an infinite M200 micro plate reader (Tecan, Mainz-Kastel, Germany).
RNA quality assessment was tested by micro capillary electrophoresis using an Agilent Bioanalyzer 2100 (Agilent Technologies, Waldbronn, Germany) with the RNA 6000 Nano Kit (Agilent Technologies, Waldbronn, Germany) following the manufacturer’s protocol[14]. The RNA quality was determined by the RNA integrity number (RIN), which is calculated from the relative height and area of the 18 S and 28 S RNA peaks and follows a numbering system from 1 to 10, with 1 being the most degraded profile and 10 being the best preserved[15,16]. For the degradation test we used RNA with a concentration of 50 ng/μL.
BioScript TM was used to generate cDNAs (Bioline) from 100 ng RNA according to the manufacturer’s protocol. For real time PCR the SensiMixSYBR Low Rox Kit (Bioline) was used on a MX3005 (Stratagene). 100 ng cDNA was used for each sample. The individual tests were performed in triplicates and repeated 3 times. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as housekeeping gene. The PCR conditions were as follows: initial denaturation 10 min, 95 °C, 40 cycles of denaturation; 30 s, 95 °C; annealing, 30 s, 55 °C; elongation 30 s, 72 °C. The primer sequences (F: forward; R: reverse) were as follows: r-S100 (F: 5’-TTGCCCTCATTGATGTCTTCCA-3’, R: 5’-TCTGCCTTGATTCTTACAGGTGAC-3’) from Lisachev et al[17], r-Tubulin-β III (F: 5’-AGACCTACTGCATCGACAATGAAG-3’, R: 5’-GCTCATGGTAGCAGACACAAGG-3’) from Schwindt et al[18], PGP9.5 (F: 5’-CCCTGAAGACAGAGCCAAGTG-3’, R: 5’-GAGTCATGGGCTGCCTGAA-3’), and GAPDH (F: 5’-GTATGACTCTACCCACGGCAAGT-3’, R: 5’-TTCCCGTTGATGACCAGCTT-3’) from Du et al[19].
For time course measurement on degradation effect of muscle and submucosal/mucosal human and rat tissue in student’s t-test was applied, after testing for normal distribution. The results were considered significant with a P < 0.05.
The response RINs were fit using standard least-square regression using JMP version 10 to explore the impact of temperature, tissue, time, species and tissue separation of the gut tissue in muscle and submucosal/mucosal layer before or after incubation. The contribution of the different formulation variables was compared using analysis of variance (ANOVA) at P < 0.05 significance level.
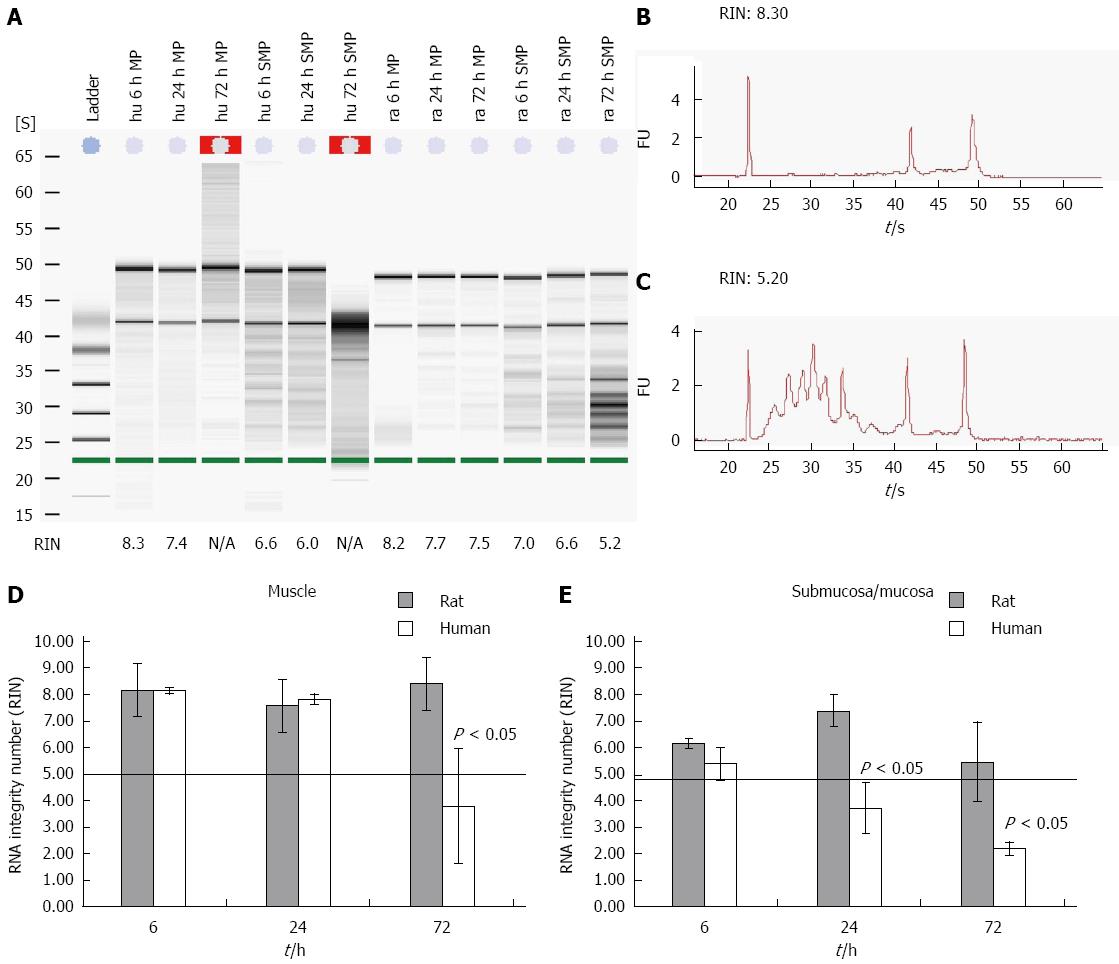
Degradation of RNA was assessed using an Agilent 2100 bioanalyzer. This approach was applied to a large collection of electrophoretic RNA measurements. The resulting algorithm is a user-independent, automated and reliable procedure for standardization of RNA quality control that allows the calculation of an RNA integrity number (RIN)[16]. A minimum level of RNA integrity is necessary to obtain reliable qPCR data. RNA with a RIN below five is highly degraded and should not be used within further qPCR experiments[10]. The needs for microarray experiments are even higher. Here you need a RIN higher than 7. Moreover, a RIN that might be adequate for a 3’ amplification might not work for a 5’ amplification. To test the RNA degradation progress of different tissues gained from human and rodent gut on ice, the samples were separated into muscle and submucosal/mucosal layers (Figure 1A-C). The RNA of the human muscle was intact for more than 24 h. After 6 h the RNA had a RIN of 8.2 ± 0.11 and a RIN of 7.8 ± 0.2 after 24 h. After 72 h the RNA of the muscle was degraded and the RIN dropped to 3.8 ± 2.2 (Figure 1D). The RNA of the submucosal/mucosal layer degraded much faster than the RNA of the muscle layer. The RIN of the submucosal/mucosal layer showed a still acceptable value of 5.4 ± 0.6 after six h and was fully degraded after 24 h with a RIN of 3.7 ± 1.0 and a RIN of 2.2 ± 0.2 after 72 h (Figure 1E).
Comparing rat and human tissue reveals significant differences. While the RIN of rat smooth muscle tissue still reaches 8.4 ± 0.4 (Figure 1D) after 72 h of storage, the human tissue yields not more than a RIN of 3.8 ± 2.2 (Figure 1D). Rat derived submucosal/mucosal layer RNA degraded even faster, already after 24 h. At this point of time the RNA delivered a RIN value of 7.4 ± 0.6. After 48 and 72 h the RIN was still acceptable with values around 5.5 ± 2 (Figure 1E). Human derived submucosal/mucosal tissue degraded already after 6 h with a RIN of 5.4 ± 0.6 and 3.7 ± 1 after 24 h. At this time point the RNA of human submucosal/mucosal layer is completely degraded compared to RNA of rat submucosal/mucosal layer (Figure 1E).
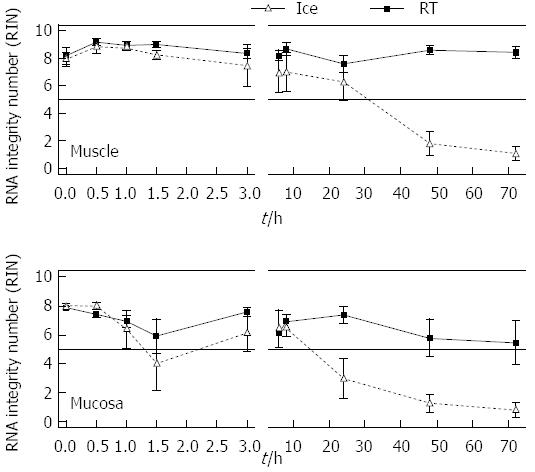
In order to investigate when exactly the RNA of the gut degrades and which role temperature and time are playing, a more detailed analysis was performed with rat derived intestinal tissue. The amount of tissue was not limited. Furthermore individual short periods of time could be explored. Rat tissue samples were collected and stored temporarily in MEM-Hepes on ice or at RT until RNA was extracted and measured. Full thickness samples as well as individual muscle or submucosal/mucosal layers were analysed after separation (Figure 2). Muscle RNA kept its stability for more than 72 h when stored on ice. Here the RIN values varied between 9.2 at “zero” time and 7.6 after 72 h. The RNA quality was still sufficient for further experiments. After 24 h at RT the RNA was 6.2 ± 1.3, after 48 h it dropped to 1.8 ± 0.9 and after 72 h the RIN reached a value of 1.1 ± 0.5 (Figure 2). The RNA of the submucosal/mucosal layer remained intact on ice over 72 h with a RIN of 5.5 ± 1.5 while at room temperature it already degraded after 8 h. Here the RIN dropped to 6.5 ± 0.6 and 3 ± 1.4 after 24 h. Degradation kept on until it reached a value of 1.0 ± 0.5 at 72 h (1.3 ± 0.6, 48 h, Figure 2).
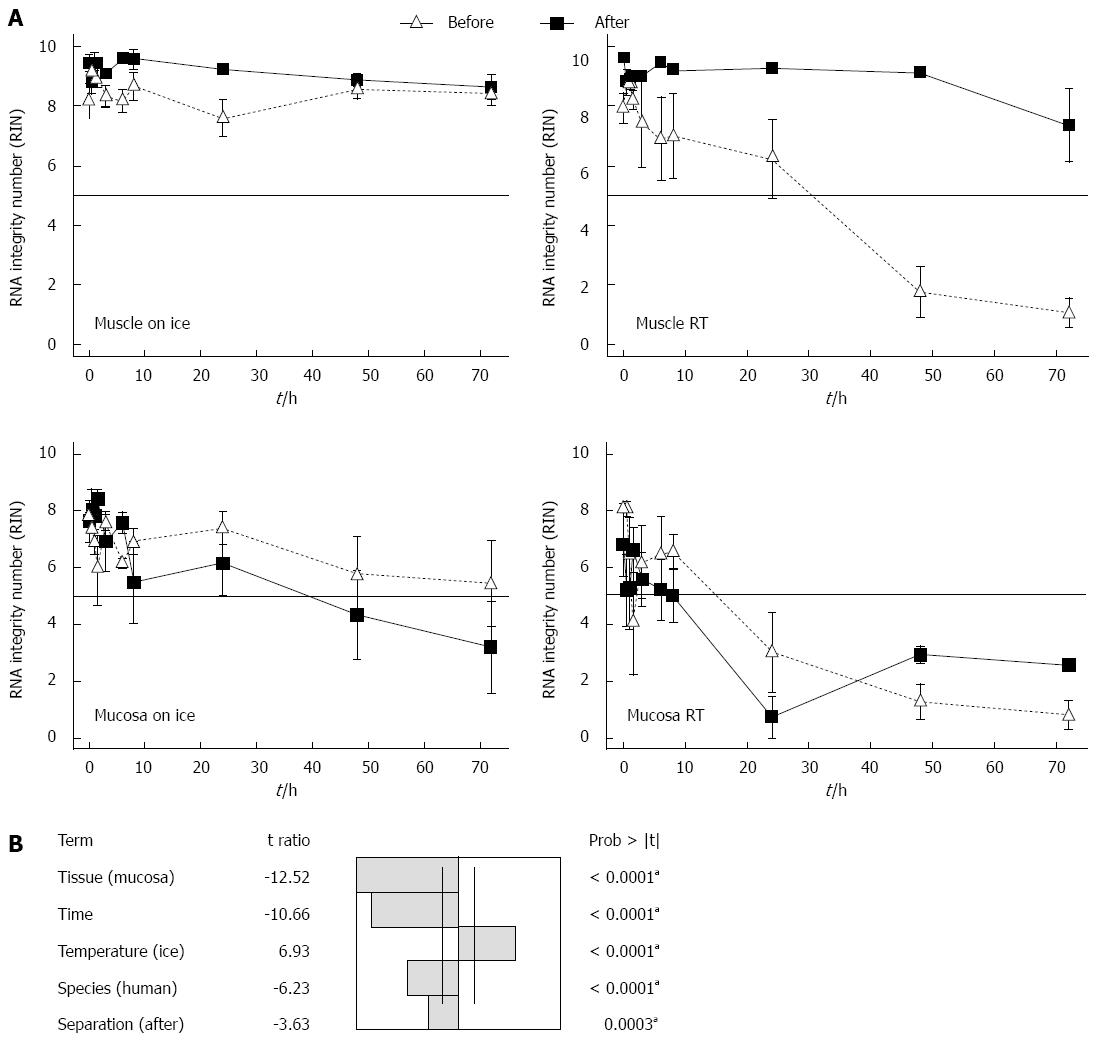
To study whether a special tissue treatment will protect the RNA from degradation, the gut was rinsed with sterile PBS before separating the layers into muscle und submucosal/mucosal tissue. The tissues were either separated immediately before or after storage. Storage took place in MEM-Hepes, either on ice or at RT. After different times the RNA of the samples was isolated and the degradation evaluated. After 72 h the RNA of the muscle layer showed similar RIN values on ice (RIN 7.3 ± 1.2) as at RT (RIN 8.6 ± 0.4) (Figure 3A). So it seems that rinsing and separating the tissue in muscle and submucosal/mucosal layers, prevents the RNA of the muscle in case the cold chain is interrupted. Submucosal/mucosal RNA degraded after 24 h on ice. The RNA after 24 h had a RIN of 6.2 ± 1.1 and of 4.3 ± 1.6 after 48 h, respectively 3.2 ± 1.6 after 72 h (Figure 3A). At RT the RNA of the submucosal/mucosal layer degraded at 8-h incubation in medium and yielded a RIN of 4.9 ± 1.3, while there was a complete degradation to be seen after 24 h with a RIN of 1 ± 0.2 (Figure 3A). Separating muscle and submucosal/mucosal layer makes no difference to the quality of the RNA from the submucosal/mucosal layer in contrast to the muscle.
A multifactorial analysis of variances of the RIN in response to several factors: temperature, tissue, time, species and tissue separation of gut in muscle and submucosal/mucosal layer before or after incubation was undertaken. To identify the impact factors to the RIN, an analysis of variance (ANOVA) at P = 0.05 significant level was used.
The factors temperature, tissue, time, species and tissue separation of the gut tissue in muscle and submucosal/mucosal layer before or after incubation had all a significant influence on the RIN of the RNA (Figure 3B). The factors submucosal/mucosal layer, RT, time, human and separation of gut tissue after incubation time, influence the RIN negatively (Figure 3B). The most important factor for the RIN with the t ratio of 12.52 is the tissue, followed by the time with a t ratio of 10.66, temperature with a t ratio of 6.93, species with a t ratio of 6.23 and finally the separation of the tissue in muscle and submucosal/mucosal layer before or after incubation with a t ratio of 3.63.
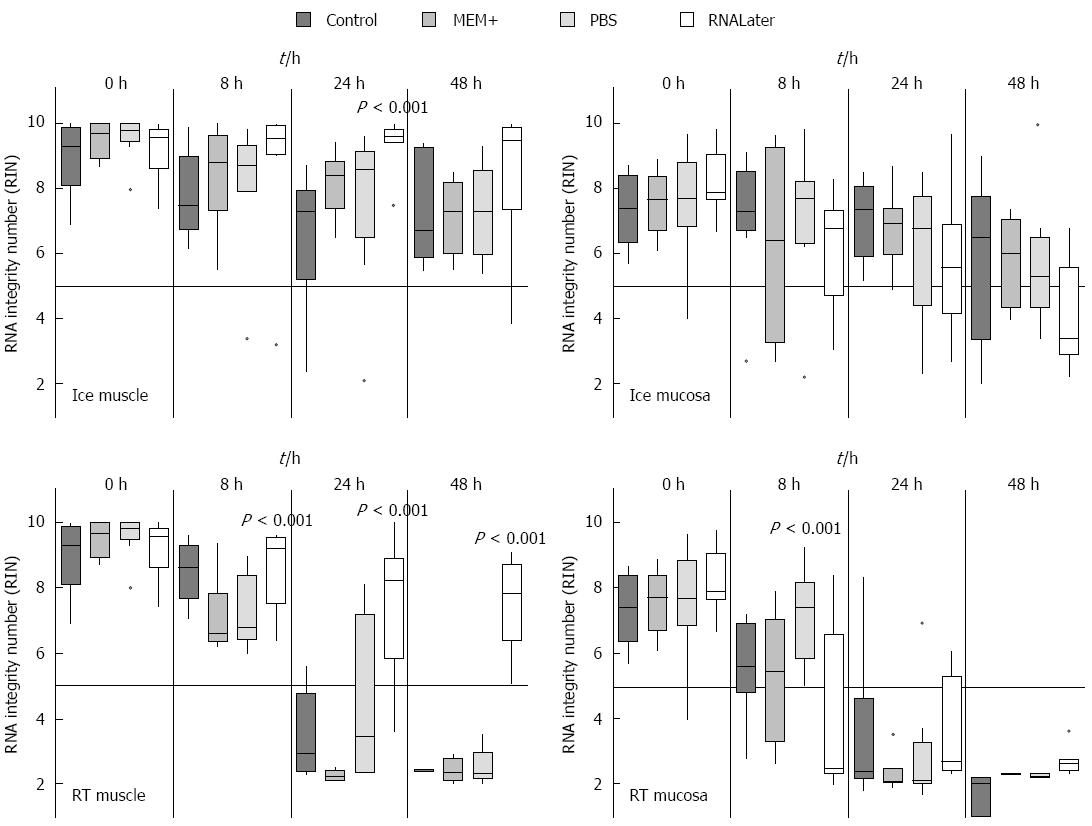
As the previous experiments revealed that rinsing the specimen protects the muscle from RNA degradation, the rinsing media might also be critical. We therefore tried various media for the initial rinsing step. Three different rinsing solutions (MEM+, PBS or RNALater) were used and the individual tissues were compared with tissues which were not rinsed at all and just stored in MEM plus. The tissues were stored in the same media in which they have been rinsed for various incubation times (0 h, 8 h, 24 h and 48 h) on ice or at RT. Regarding the tissues stored in control medium, MEM+ or PBS no visible difference after incubation were observed (Figure 4). However, the tissue that had been washed and stored in RNALater did shrink and became very sticky. This tissue was therefore very hard to dissect in muscle and submucosal/mucosal layer. Due to this heavy sticking the tissue could not be pipetted appropriately. Moreover the tissue was very fragile and easily destroyed. In this experiment it could clearly be demonstrated that the RNA from muscle and submucosal/mucosal layer was best protected by cooling (Figure 4). As soon as the samples were stored on ice, only one significant difference in the muscle was seen after 24 h. Here the tissue stored in RNALater had a significant (P = 0.001) higher RIN. All other washing and storing methods displayed no significant different effects, even after 48 h. However, when the samples were stored at RT, we noticed significant differences for the muscle up to 24 h (Figure 4). The samples which were washed and stored in RNALater had a much better RIN value (eight) than the control samples. Also the samples stored in MEM+ or PBS had a RIN below five. This effect was even more pronounced after 48 h, where the RNALater samples still had a RIN value greater seven, while all others provided only completely degraded RNA (Figure 4).
In the submucosal/mucosal layer samples a similar protective effect of cooling could be demonstrated as seen in the muscle tissue, while there was no difference between the individual media used. At RT RNALater did not have any protective effect on submucosal/mucosal tissue. Interestingly, the best protection effect after 8 h could be seen with PBS (aP = 0.001). After 24 h and 48 h, the RNA was completely degraded in submucosa/mucosal tissue in all rinsing solutions (Figure 4).
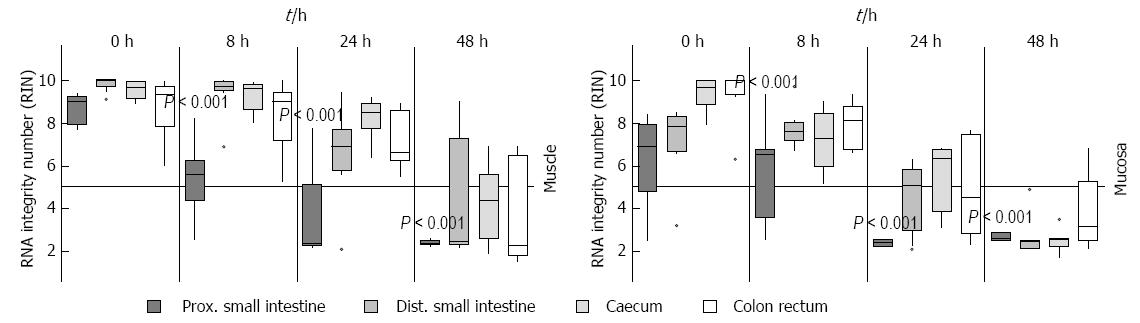
To verify subsequently whether the individual sections of the intestine have an influence on the stability of RNA and whether bacteria or digestive enzymes, which are different in each section, have influence upon the RNA degradation, they were examined separately on RNA degradation. To test this, the intestine was separated into proximal and distal small intestine, caecum and colon with the rectum. These pieces were stored at RT about 0 h, 8 h, 24 h and 48 h in MEM and separated into muscle and submucosal/mucosal layer afterwards. It could be clearly demonstrated that the RNA from the proximal small intestine degraded much faster (P = 0.001, 8 h), as those from other locations (Figure 5). This RNA had already a RIN value about 5 after 8 h in both submucosal/mucosal and muscle layer while the other sections still had a value above seven or ten in both muscle and submucosal/mucosal layer respectively. The muscle RNA after 24 h was degraded only in the proximal small intestine. All other parts had a RIN higher than 5. After 48 h there was no difference between either region or tissue type, all RNA was degraded (Figure 5).
In order to analyse whether the expression of genes from the enteric nervous system (ENS) depends on RNA degradation, the RNA of muscle and of submucosal/mucosal tissue with different RIN’s (two to nine) was analysed with qPCR. Neuronal and glial genes (β-Tubulin III, PGP9.5 and S100), as well as GAPDH as an independent gene for degradation[6] were investigated. When the RIN of the muscle RNA decreases, the quantification cycle (Cp) of the qPCR increases for the neuronal and glial genes (Table 1). So it seems that the Cp from muscle layer depends on the RIN.
| Tissue | Gene | Cp (RIN = 9) | Cp (RIN = 2) |
| Muscle | β-Tubulin III | 22.07 ± 0.25 | 24.41 ± 0.22 |
| Submucosal/mucosal | β-Tubulin III | 27.42 ± 0.19 | 26.00 ± 0.93 |
| Muscle | PGP 9.5 | 20.28 ± 0.09 | 23.67 ± 0.58 |
| Submucosal/mucosal | PGP 9.5 | 25.43 ± 0.54 | 25.28 ± 0.58 |
| Muscle | S100b | 26.65 ± 0.47 | 29.07 ± 0.02 |
| Submucosal/mucosal | S100b | 28.53 ± 0.63 | 29.75 ± 1.30 |
| Muscle | GAPDH | 18.85 ± 0.05 | 18.68 ± 0.08 |
| Submucosal/mucosal | GAPDH | 18.93 ± 0.04 | 19.53 ± 0.20 |
The Cp of the submucosal/mucosal layer with a RIN of nine equals the RIN of five from muscle tissue. The Cp from the submucosal/mucosal layer seems not to be dependent on the RIN of the RNA.
While quality control of RNA is routinely being performed prior to microarray-based gene expression profiling, it is often missing with regard to PCR-based quantification methods. Indeed, even on degraded RNA samples, a nice amplification curve can be obtained[20]. Nevertheless, excellent RNA quality is essential to obtain reliable results. The inclusion of samples with degraded RNA may influence the statistical analysis and hence the interpretation of gene expression levels in relation to biological and/or clinical data. Results should reflect real biological differences and not differences due to poor RNA integrity[21].
The gut is a highly delicate tissue that is filled with digestive enzymes and a microbiome of varying quality and quantity. This leads to a rapid post mortem or even post dissection degradation of the whole tissue. Obviously, human RNA degrades faster than the one derived from rat. RNA from post mortem brain is much more stable than RNA from post mortem intestinal tissue[22]. We demonstrated similar differences between rat and human tissue from surgical gut samples. The storage and dissection before RNA processing changes their individual quality. This was most pronounced in the smooth muscle from both rat and human.
Generally, RNA from the muscle layer remains much longer stable than RNA from the submucosal/mucosal layer. In the submucosal/mucosal layer usually more immune cells are to be found, and due to the neighbourhood to the lumen and the loose arrangement of the tissue the bacterial translocation takes place earlier and to a greater extent.
We have shown in the intestinal samples that measures such as rinsing the tissue and separating the bowel in its submucosal/mucosal and muscle compartments, reduces especially the RNA degradation in the muscle. If the cold chain is not interrupted the RNA of the smooth muscle remains stable for a minimum of 72 h. In contrast, rinsing and separating the tissue wall does not protect the RNA from submucosal/mucosal layers. Here, only cooling reduces degradation. These effects might even be more pronounced when the intestinal tissue is inflamed[3]. We could also demonstrate that different rinsing methods have no impact upon RNA integrity, if the tissue is cooled. But the rinsing method has an influence if the tissue is stored at RT. RNALater prevents RNA degradation in the muscle at all-time points investigated, while also PBS hade a preventing influence upon the submucosal/mucosal layer after 8 h at RT. This might be due to an inhibition of degradating enzymes which needs calcium or magnesium for being active[23,24].
Not only that RNA from the intestine degrades faster than RNA from blood cells, or cell lines, there are also local differences. The RNA of the proximal part of the small intestine (duodenum) degrades much faster than the ones from the distal parts of the small intestine (ileum and jejunum) or the large bowel (caecum and colon with rectum). This demonstrates that digestive enzymes might also play an important role for RNA degradation. Other groups showed that the RNA from ileum and colon degraded slower than the one from jejunum[10]. This is consistent with our data concerning the duodenum. In rodents, proximal small intestine samples might still harbour parts of the pancreas, which is (unless in human) - not arranged in a compact organ. Bits of pancreatic tissue are embedded in the mesenterial wall along the duodenum, and might easily be overseen during dissection. Pancreatic tissue will yield a surplus of degradating enzymes. Moreover, degradation of the RNA does also depend on the amount of connective tissue or fat, or as in our case, in increased enzyme activity[10]. It can lead to a faster degradation of the RNA[22] or caused by inadequate sample processing and storage[20]. Using RNase inhibitors can reduce the problem of RNA degradation, but is not consistent among individuals and RNA degradation even occurred when frozen samples were thawed immediately before nucleic acid extraction[25].
Adequate sample storage in medium as well as cooling reduces RNA degradation at least in muscle and submucosal/mucosal layer.
Also it is known that in some mammalian tissue or blood or even the food in the gut contains inhibiting factors which interferes with the PCR assays[26-30]. So there might be some unknown factors in the submucosal/mucosal layer that can produce a false negative result for the qPCR. In general, rinsing, cooling and separation make the difference concerning RNA quality. While RNALater increases RNA quality, its use makes only sense provided a sole RNA extraction is performed. As soon as the tissue has to be processed further, i.e., by separating the individual gut wall layers, RNALater interferes negatively with the dissection process. In order to avoid misinterpretations, especially with a limited amount of crucial material, optimal pre-treatment and processing is an indispensable prerequisite to obtain reliable and standardized RNA analysis data.
We thank Mrs. Claudia Finzelberg-Bozoklu for the excellent technical support.
The RNA quality of each sample has a great influence on a variety of experimental measurement methods. It is therefore important to test the quality before the individual experiment starts, especially in tissues from the gastrointestinal tract, where both secreted enzymes or fecal and microbial content of the gut can interfere with the quality of RNA.
RNA from the gastrointestinal tract is particularly interesting in various diseases, be it inflammatory or developmental. A standardized protocol for optimal RNA treatment allows the comparison of results from different laboratories in a more reliable way.
A detailed protocol of sample preparation for optimal RNA harvest is provided. Different species have been compared and variations and influences of both species and gut segments investigated.
The provided data can be used for a more standardized investigation of RNA changes in health and disease, based on optimized techniques.
RNA degradation means that the information sequence of the RNA is destroyed and can therefore not be used for a reliable identifications of specific genes, involved in diseases or developmental changes.
This is a study in which the authors analysed the effect of different treatment and storage conditions of intestinal tissue upon the RNA quality derived from these tissues. The results are interesting and show that cooling and separation of tissue prevents the RNA of degradation.
P- Reviewer: Goetze TO, Grizzi F S- Editor: Yu J L- Editor: A E- Editor: Ma S
| 1. | Hagl CI, Heumüller S, Klotz M, Subotic U, Wessel L, Schäfer KH. Smooth muscle proteins from Hirschsprung’s disease facilitates stem cell differentiation. Pediatr Surg Int. 2012;28:135-142. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 2. | Moran GW, O’Neill C, Padfield P, McLaughlin JT. Dipeptidyl peptidase-4 expression is reduced in Crohn’s disease. Regul Pept. 2012;177:40-45. [PubMed] [Cited in This Article: ] |
| 3. | Sánchez-Muñoz F, Fonseca-Camarillo G, Villeda-Ramírez MA, Miranda-Pérez E, Mendivil EJ, Barreto-Zúñiga R, Uribe M, Bojalil R, Domínguez-López A, Yamamoto-Furusho JK. Transcript levels of Toll-Like Receptors 5, 8 and 9 correlate with inflammatory activity in Ulcerative Colitis. BMC Gastroenterol. 2011;11:138. [PubMed] [Cited in This Article: ] |
| 4. | Kontos CK, Papadopoulos IN, Scorilas A. Quantitative expression analysis and prognostic significance of the novel apoptosis-related gene BCL2L12 in colon cancer. Biol Chem. 2008;389:1467-1475. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 5. | Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611-622. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9915] [Cited by in F6Publishing: 10181] [Article Influence: 678.7] [Reference Citation Analysis (0)] |
| 6. | Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech. 2004;15:155-166. [PubMed] [Cited in This Article: ] |
| 7. | Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27:126-139. [PubMed] [Cited in This Article: ] |
| 8. | Holland NT, Smith MT, Eskenazi B, Bastaki M. Biological sample collection and processing for molecular epidemiological studies. Mutat Res. 2003;543:217-234. [PubMed] [Cited in This Article: ] |
| 9. | Rauch U, Hänsgen A, Hagl C, Holland-Cunz S, Schäfer KH. Isolation and cultivation of neuronal precursor cells from the developing human enteric nervous system as a tool for cell therapy in dysganglionosis. Int J Colorectal Dis. 2006;21:554-559. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 82] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 10. | Fleige S, Walf V, Huch S, Prgomet C, Sehm J, Pfaffl MW. Comparison of relative mRNA quantification models and the impact of RNA integrity in quantitative real-time RT-PCR. Biotechnol Lett. 2006;28:1601-1613. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 421] [Cited by in F6Publishing: 430] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 11. | Raybould HE. Gut microbiota, epithelial function and derangements in obesity. J Physiol. 2012;590:441-446. [PubMed] [Cited in This Article: ] |
| 12. | Larsson E, Tremaroli V, Lee YS, Koren O, Nookaew I, Fricker A, Nielsen J, Ley RE, Bäckhed F. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut. 2012;61:1124-1131. [PubMed] [Cited in This Article: ] |
| 13. | Kierszenbaum AL, Tres LL. Histology and Cell Biology: An introduction to pathology. St. Louis: Mosby 2012; . [Cited in This Article: ] |
| 14. | Hawtin P, Hardern I, Wittig R, Mollenhauer J, Poustka A, Salowsky R, Wulff T, Rizzo C, Wilson B. Utility of lab-on-a-chip technology for high-throughput nucleic acid and protein analysis. Electrophoresis. 2005;26:3674-3681. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 15. | Imbeaud S, Graudens E, Boulanger V, Barlet X, Zaborski P, Eveno E, Mueller O, Schroeder A, Auffray C. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res. 2005;33:e56. [PubMed] [Cited in This Article: ] |
| 16. | Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. [PubMed] [Cited in This Article: ] |
| 17. | Lisachev PD, Shtark MB, Sokolova OO, Pustylnyak VO, Salakhutdinova MY, Epstein OI. A Comparison of the Dynamics of S100B, S100A1, and S100A6 mRNA Expression in Hippocampal CA1 Area of Rats during Long-Term Potentiation and after Low-Frequency Stimulation. Cardiovasc Psychiatry Neurol. 2010;2010:pii 720958. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 18. | Schwindt TT, Trujillo CA, Negraes PD, Lameu C, Ulrich H. Directed differentiation of neural progenitors into neurons is accompanied by altered expression of P2X purinergic receptors. J Mol Neurosci. 2011;44:141-146. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 19. | Du F, Wang L, Qian W, Liu S. Loss of enteric neurons accompanied by decreased expression of GDNF and PI3K/Akt pathway in diabetic rats. Neurogastroenterol Motil. 2009;21:1229-e114. [PubMed] [Cited in This Article: ] |
| 20. | Pérez-Novo CA, Claeys C, Speleman F, Van Cauwenberge P, Bachert C, Vandesompele J. Impact of RNA quality on reference gene expression stability. Biotechniques. 2005;39:52, 54, 56. [PubMed] [Cited in This Article: ] |
| 21. | Strand C, Enell J, Hedenfalk I, Fernö M. RNA quality in frozen breast cancer samples and the influence on gene expression analysis--a comparison of three evaluation methods using microcapillary electrophoresis traces. BMC Mol Biol. 2007;8:38. [PubMed] [Cited in This Article: ] |
| 22. | Lee J, Hever A, Willhite D, Zlotnik A, Hevezi P. Effects of RNA degradation on gene expression analysis of human postmortem tissues. FASEB J. 2005;19:1356-1358. [PubMed] [Cited in This Article: ] |
| 23. | Lu Y, Yeung N, Sieracki N, Marshall NM. Design of functional metalloproteins. Nature. 2009;460:855-862. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 566] [Cited by in F6Publishing: 568] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 24. | Niki I, Yokokura H, Sudo T, Kato M, Hidaka H. Ca2+ signaling and intracellular Ca2+ binding proteins. J Biochem. 1996;120:685-698. [PubMed] [Cited in This Article: ] |
| 25. | Cardona S, Eck A, Cassellas M, Gallart M, Alastrue C, Dore J, Azpiroz F, Roca J, Guarner F, Manichanh C. Storage conditions of intestinal microbiota matter in metagenomic analysis. BMC Microbiol. 2012;12:158. [PubMed] [Cited in This Article: ] |
| 26. | Akane A, Matsubara K, Nakamura H, Takahashi S, Kimura K. Identification of the heme compound copurified with deoxyribonucleic acid (DNA) from bloodstains, a major inhibitor of polymerase chain reaction (PCR) amplification. J Forensic Sci. 1994;39:362-372. [PubMed] [Cited in This Article: ] |
| 27. | Al-Soud WA, Jönsson LJ, Râdström P. Identification and characterization of immunoglobulin G in blood as a major inhibitor of diagnostic PCR. J Clin Microbiol. 2000;38:345-350. [PubMed] [Cited in This Article: ] |
| 28. | Al-Soud WA, Rådström P. Purification and characterization of PCR-inhibitory components in blood cells. J Clin Microbiol. 2001;39:485-493. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 694] [Cited by in F6Publishing: 623] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 29. | Bickley J, Short JK, McDowell DG, Parkes HC. Polymerase chain reaction (PCR) detection of Listeria monocytogenes in diluted milk and reversal of PCR inhibition caused by calcium ions. Lett Appl Microbiol. 1996;22:153-158. [PubMed] [Cited in This Article: ] |
| 30. | Rossen L, Nørskov P, Holmstrøm K, Rasmussen OF. Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solutions. Int J Food Microbiol. 1992;17:37-45. [PubMed] [Cited in This Article: ] |