Published online Nov 16, 2021. doi: 10.12998/wjcc.v9.i32.9699
Peer-review started: April 18, 2021
First decision: June 3, 2021
Revised: June 5, 2021
Accepted: September 8, 2021
Article in press: September 8, 2021
Published online: November 16, 2021
Processing time: 205 Days and 19.9 Hours
Hepatocellular carcinoma (HCC) remains one of the most frequent types of liver cancer and is characterized by a high recurrence rate. Recent studies have proposed that long non-coding RNAs (lncRNAs) are potential biomarkers in several recurrent tumor types. It is now well understood that invasion, migration, and metastasis are important factors for tumor recurrence. Moreover, some of the known risk factors for HCC may affect the expression levels of several types of lncRNAs and thus affect the recurrence of liver cancer through lncRNA regula
Core Tip: Hepatocellular carcinoma (HCC) is one of the most recurring malignant tumors in the world. Intrahepatic metastasis and multicenter occurrence are two ways of recurrence of HCC. Currently, a growing number of studies have shown that long non-coding RNAs (lncRNAs), regulators of human gene expression, are abnormally expressed and influence the development of HCC. So, we need to further understand the significance of lncRNAs in the clinical diagnosis and treatment of recurrent HCC and explore the regulatory mechanisms of lncRNAs in the occurrence and treatment of recurrent HCC.
- Citation: Fang Y, Yang Y, Li N, Zhang XL, Huang HF. Emerging role of long noncoding RNAs in recurrent hepatocellular carcinoma. World J Clin Cases 2021; 9(32): 9699-9710
- URL: https://www.wjgnet.com/2307-8960/full/v9/i32/9699.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i32.9699
In most countries, the trend of hepatocellular carcinoma (HCC) mortality has increased in recent decades[1]. It is also the third leading cause of cancer-related deaths worldwide. For all stages combined, the 5-year relative survival rate is lowest for cancers of the liver (18%)[2]. Owing to insidious symptoms and early metastases, most HCC patients are diagnosed at an advanced stage, resulting in limited or ineffec
The recurrence of intrahepatic HCC is caused by two different ways: (1) Intrahepatic metastasis (IM) descending from the primary cancer; and (2) Independent carcinogenesis leading to multicentric occurrence (MO)[5-7]. It is worth noting that these two mechanisms are not mutually exclusive, and both factors can lead to the recurrence of intrahepatic HCC. So far, there has been no definite standard to accurately distinguish the origin of multifocal HCC from IM or MO. Hence, histopathological features are still the most convenient strategy. Treatment options for recurrent intrahepatic HCC include repeat liver resection and ablative therapy[8] (Figure 1). Generally, after radical resection, the overall survival (OS) and recurrence-free survival (RFS) rates of MO-HCC patients are better than those of IM-HCC patients[9]. IM is more metastatic and has greater migratory ability than MO. HCC with IM recurs earlier and has a poorer prognosis than HCC with MO[10]. In recent decades, OS and RFS after hepatectomy have remained unsatisfactory due to the high rates of IM and MO[11]. Histopathological analysis is still the most convenient strategy, and it is objective and accurate. Pathology remains a cornerstone in the clinical treatment of patients with HCC, as it allows a definitive diagnosis and provides prognostic information. However, most current studies focus on primary liver cancer, but there are few studies on recurrent HCC. Therefore, precise diagnostic/prognostic biomarkers are urgently needed to improve the clinical outcomes of recurrent intrahepatic HCC.
Long non-coding RNAs (lncRNAs) are defined as transcripts of more than 200 nucleotides that are not translated into proteins and act as important regulators in gene expression networks[12]. In recent years, with the development of next-generation sequencing, lncRNAs have become the focus of research[13]. A few studies suggest that annotated lncRNA transcripts in the whole human genome are involved in the biological processes of recurrent HCC.
In this review, we summarize the current understanding of the molecular mecha
We often divide noncoding RNAs into two categories: (1) Noncoding RNAs shorter than 200 nucleotides, including PIWI-interacting RNAs (piRNAs), small interfering RNAs (siRNAs), tRNA-derived small RNAs (tsRNAs), and microRNAs (miRNAs); and (2) RNAs longer than 200 nucleotides (long ncRNAs; lncRNAs), including large intergenic ncRNAs (lincRNAs) and very long ncRNAs (vlncRNAs)[14]. LncRNA characteristics cover unique regulatory mechanisms, alternative forms of biogenesis, cis-regulatory activities, and functional structured RNA domains[15]. Therefore, lncRNAs are emerging as important regulators of tissue physiology and disease processes, including cancer[16]. Increasing evidence has shown that lncRNAs play important roles in transcriptional regulation, cell growth, and tumorigenesis through a variety of mechanisms[17]. Some studies have shown that certain lncRNAs are potential targets and biomarkers for the diagnosis and prognosis of malignant tumors. For instance, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) was included in the first batch of lncRNAs to be employed in the lncRNA-targeted therapy of lung cancer[18]. Shi et al[19] discovered that AC069513.4 and four other lncRNAs could be used as independent prognostic biomarkers to predict the survival of patients with clear cell renal cell carcinoma. The lncRNA PCAL7 is overexpressed in prostate cancer (PCa) and promotes PCa by strengthening androgen receptor signaling[20]. An increasing number of studies have shown that lncRNAs play vital roles in the pathogenesis and therapeutic response of IM and MO[21,22].
We believe that elucidating the molecular mechanisms related to liver cancer invasion and metastasis can help prevent and treat liver cancer recurrence. The molecular mechanisms by which lncRNAs play an important role in the regulation of approximately all steps of cancer progression include epigenetic regulation, miRNA regulation, cell growth, and epithelial-mesenchymal transition (EMT)[23] (Figure 2).
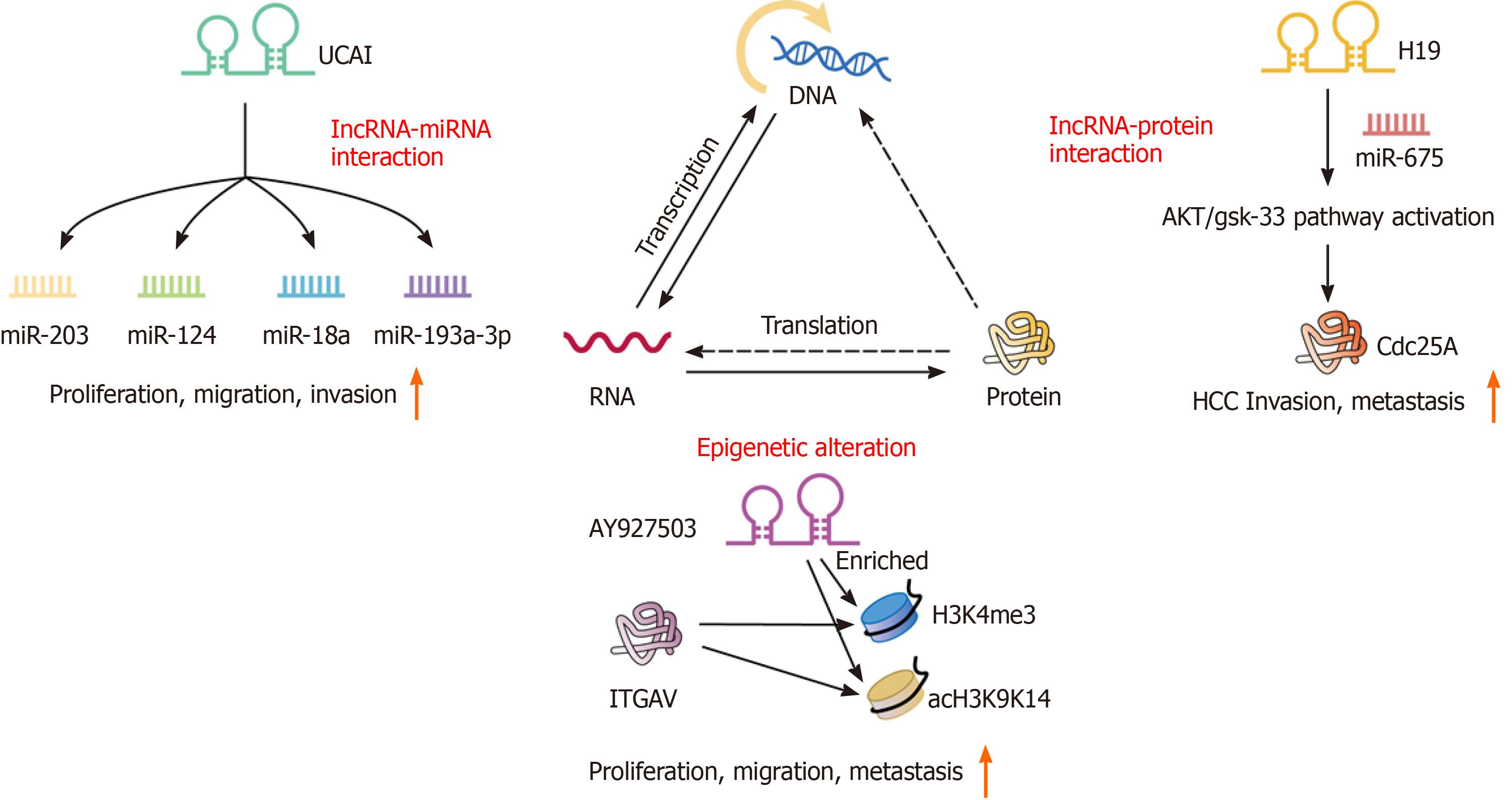
Epigenetics involves the modification of DNA molecules that regulate gene activity uninfluenced by the DNA sequence, and mitosis is stable. To date, in recurrent HCC, the most recognized epigenetic mechanisms are chromatin modification and DNA methylation. It is widely acknowledged that epigenetic regulation plays a key role in invasion and metastasis in diverse types of cancer, including recurrent HCC. In colorectal neoplasia, the lncRNA CRNDE directly binds to EZH2, SUZ12, and SUV39H1, and mediated their inhibition of tumor suppressor genes, including CELF2 and LATS2[24]. Kang et al[25] showed that a high level of AY927503 could promote HCC metastasis and is related to the poor prognosis of HCC patients. The promotion of metastasis by AY927503 is related to the activation of ITGAV transcription by recruiting chromatin modification mechanisms to the ITGAV promoter and reducing H1FX binding[25].
Furthermore, through bioinformatics analyses using the TCGA and GEO databases, it has been found that the mutual regulatory network between lncRNAs and miRNAs is involved in the progression of cancer[26]. The recurrence and metastasis of tumors are closely related to complex regulatory networks among protein-coding genes, lncRNAs, and miRNAs. Recently, many studies have reported that miRNAs and lncRNAs are involved in miRNA regulation[27]. Xu et al[28] and Chen et al[29] demonstrated the molecular mechanisms by which lncRNAs and miRNAs act in the process of recurrence in low-grade glioma and ovarian cancer. Similarly, the complex regulatory network between lncRNAs and miRNAs will help clarify the molecular mechanism of lncRNAs in recurrent liver cancer. H19 is a 2.3-kb lncRNA that is composed of five exons and four small introns and is located on ch11p15.5 as an imprinting gene with maternal expression[30]. H19 has been characterized to work either as a tumor suppressor or an oncogene in vitro and in vivo[31]. Lv et al[32] revealed that the inhibition of H19 and miR-675 promoted the invasion and metastasis of HCC by activating the AKT/GSK-3β/Cdc25A signaling pathway. Interestingly, Sui et al[33] found that through contact with EZH2 and miR-200b/a/429, GIHCG recruits EZH2 and DNMT1 to the promoter of miR-200b/a/429 and increases H3K27me3 and DNA methylation levels in the promoter of miR-200b/a/429. Functional experiments showed that GHICG promotes the proliferation, migration, and invasion of HCC cells in vitro and promotes the growth and metastasis of xenografts in vivo[33]. This result is fully in line with the characteristics of IM and OM. Moreover, the discovery of this pathway showed a new mechanistic link between lncRNAs, epigenetic modulations, and miRNAs.
Research has revealed that lncRNAs play an irreplaceable role in the development of recurrent HCC through EMT. EMT is a crucial cell remodeling process during embryonic development and organogenesis. During EMT, epithelial cells lose their polarized structure and gain migration and invasion capabilities[34]. A large amount of evidence has revealed the activation of EMT in cancer metastasis, which contributes to metastasis to the surrounding tissue and distant organs. Huang et al[35] explored whether the cancer susceptibility candidate 2 (CASC2)/miR-367/FBXW7 axis suppresses the migration, invasion, and EMT progression of HCC cells. Among the players in this pathway, the lncRNA CASC2 was determined to inhibit the migration and invasion abilities of HCC cells in vitro and in vivo.
The list of lncRNAs is still under development, and their molecular mechanisms are continuously being elucidated. Therefore, in recurrent HCC, lncRNAs act not only through a certain mechanism but through multiple molecular mechanisms. Next, we discuss the role of lncRNA expression in recurrent HCC.
Early HCC-related research focused mainly on protein-coding genes because of their central position in the regulation of biological processes. However, increasing evidence indicates that lncRNAs play an important role in diverse physiological and pathological processes. These lncRNAs are differentially expressed in different tissues and cancers, thereby affecting cancer invasion and metastasis. Aberrant expression of lncRNAs is associated with epigenetic reprogramming during tumor development and progression, mainly due to their ability to interact with DNA, RNA, or proteins to regulate gene expression[36]. The following section of this review discusses characteristics of the candidate lncRNAs in recurrent HCC according to their expression (upregulated or downregulated) (Table 1).
| Gene | Gene ID | Location | Expression | miRNA | Processes | Clinical association | Ref. |
| PTTG3P | 26255 | 8q13.1 | ↑ | miR-383 | Proliferation, migration, invasion, metastasis, cell apoptosis, cell cycle progression, tumorigenesis, EMT | Tumor size, TNM stage, poor prognosis, metastasis | [40,41] |
| PDIA3P1 | 171423 | 1q21.1 | ↑ | miR-125a/b, miR-124 | Proliferation, migration, invasion | Tumor size, metastasis, TNM stage, poor RFS and OS | [43,67] |
| MALAT1 | 378938 | 11q13.1 | ↑ | miR-146a, miR-22, miR-3064-5p, miR-125a-3p, miR-140, miR-124-3p, miR-124, miR-30a-5p, miR-195 | Proliferation, apoptosis, autophagy, proliferation, migration, invasion, angiogenesis, immunosuppression, glucose metabolism | Poor RFS and OS, metastasis | [68-79] |
| UCA1 | 652995 | 19p13.12 | ↑ | miRNA-193a-3p, miR-18a, miR-124, miR-203, miR-216B | Proliferation, migration, invasion, apoptosis, EMT | TNM stage, intrahepatic metastasis, postoperative recurrence, postoperative survival, shorter OS, tumor size, vascular invasion | [49,50,80-84] |
| MEG3 | 55384 | 14q32.2 | ↓ | miR-9-5p, miR-10a-5p, miR-493-5p, miR-483-3p, miR-26a, miR-29a | Cell apoptosis, growth inhibition, proliferation, apoptosis, cell cycle progression, migration, invasion, EMT | Poor RFS and OS, metastasis, | [55-58,61] |
| GAS5 | 60674 | 1q25.1 | ↓ | miR-21, miR-1323, miR-182, miR-135B | Proliferation, invasion, apoptosis, metastasis | Drug resistance, metastasis, shorter RFS, poor prognosis, TNM stage, differentiation, glucose levels, portal vein tumor thrombosis, tumor size, lymph node metastasis | [63,65,66,85-88] |
| CASC2 | 255082 | 10q26.11 | ↓ | miR-183, miR-362-5p, miR-24-3p, miR-367 | Proliferation, migration, invasion, colony formation, cell cycle, apoptosis, metastasis, EMT | Tumor size, metastasis | [89-94] |
| H19 | 283120 | 11p15.5 | ↑ | miR675, miR-193B, miR-15b, miR-675, miR-326 | Proliferation, motility, migration, invasion, apoptosis, EMT | Differentiation, drug resistance, metastasis, growth, shorter survival time, lymph node metastasis, distant metastasis | [32,95-103] |
PTTG3P: Previous studies have suggested that pituitary tumor-transforming 3, pseudogene (PTTG3P) serves as an oncogene in human cancers. The PTTG3P gene is mapped to ch8q13.1 in humans. PTTG3P is upregulated in several types of cancer. In addition, PTTG3P regulates migration and invasion in multiple types of tumors, such as gastric cancer, colorectal cancer, and breast cancer[37-39]. Similarly, several studies have shown that PTTG3P is upregulated in HCC tissues and cells. To date, PTTG3P has been shown to affect PI3K/AKT signaling by upregulating PTTG1 and the PTTG3P-miR-383-CCND1/PARP2 axis in HCC[40,41]. Bai et al[42] showed that high PTTG3P expression was an independent indicator associated with a short OS and RFS regardless of the pathological stage or tumor grade, suggesting the potential usage as a prognostic biomarker for recurrence.
PDIA3P1: Protein disulfide isomerase family A member 3 pseudogene 1 [PDIA3P1 (gene ID: 171423)] is located on ch1q21.1 with a 2099-bp segment and is located primarily in the cytoplasm. PDIA3P1 has been reported to be upregulated in HCC and is highly expressed under hypoxic conditions. PDIA3P1 regulates the p53 pathway to promote cell proliferation, migration, and invasion and suppresses apoptosis in HCC. Moreover, in patients with HCC, high PDIA3P1 expression is significantly related to tumor size, metastasis, TNM stage, and a poorer survival outcome than patients with low PDIA3P1 expression. Furthermore, Xie et al[43] found the hMTR4-PDIA3P1-miR-125/124-TRAF6 axis and studied its function in NF-κB signal transduction activated by DNA damage. The study on this axis implied that the upregulation of PDIA3P1 may confer chemoresistance. Targeting PDIA3P1 represents a promising strategy to inactivate NF-κB signaling and enhance cancer cell chemosensitivity. The same study also verified one of the main working models of a cytoplasmic lncRNA: As a combination of a ceRNA and miRNA, it upregulates the expression of miRNA targets. In addition, PDIA3P1 may be useful as a new biomarker for multidrug resistance and progression of recurrent HCC.
MALAT1: MALAT1, also known as NEAT2, is a key lncRNA gene that is located on ch11q13.1 and encodes a 6-kb protein. A meta-analysis of a transcriptome dataset showed that MALAT1 is upregulated in several cancers, including lung cancer, prostate cancer, and breast cancer[44]. Additionally, high MALAT1 expression is associated with invasion and metastasis in lung, breast, and liver cancers, suggesting the pivotal role of MALAT1 in MO and IM[45-47]. In both HCC cell lines and clinical tissue samples, MALAT1 is upregulated and is associated with invasion, metastasis, migration, cell proliferation, apoptosis, and a short OS and RFS. In particular, Lai et al[48] demonstrated that higher MALAT1 expression was associated with a shortened RFS and suggested that MALAT1 could serve as an independent prognostic factor for predicting HCC recurrence. These findings suggest that MALAT1 may selectively affect the spread of cancer cells or residual cancer cells after surgery to cause liver cancer recurrence, showing important clinical significance.
UCA1: Urothelial carcinoma associated 1 (UCA1), a novel vital oncogenic lncRNA, is located on ch19p13.12 with a TATA box at its 5' end and a poly A tail at its 3' end. UCA1 was first discussed in bladder cancer and is highly expressed in multiple cancers. In HCC, Wang et al[49] found that UCA1 was significantly upregulated in tumor tissues and associated with TNM stage, metastasis, and a poor survival. In addition, high UCA1 expression in HCC was positively associated with tumor size, vascular invasion, and American Joint Committee on Cancer stage (P < 0.05)[50]. Furthermore, gain-of-function and loss-of-function analyses showed that UCA1 knockdown inhibited HCC cell proliferation, migration, and invasion in vitro and xenograft tumor growth in vivo. At the molecular level, miR-203, miR-124, miR-18a, and miR-193a-3p may affect the proliferation, migration, and invasion of cancer cells in hepatocellular carcinoma by altering UCA1. Consequently, these data indicate that UCA1 could serve as an oncogene in tumorigenesis and act as a novel serum biomarker for the diagnosis and prognosis of HCC recurrence.
MEG3: Maternally expressed 3 (MEG3) is a maternally imprinted gene localized on ch14q32.2 that has been reported to be downregulated in multiple cancer tissues compared with nontumoral tissues of the same origin. MEG3 overexpression can reinforce cell apoptosis and decrease proliferation, migration, and invasion in breast cancer[51], colorectal carcinoma[52], and oral cancer stem cells[53]. In liver cancer, the loss of methylation at the MEG3 locus is linearly related to the overall loss of DNA methylation[54]. Several studies suggested that the methylation-dependent tissue-specific regulation of MEG3 by miR-29a and miR-10a-5p may contribute to HCC growth and that miR-9-5p, miR-493-5p, miR-483-3p, and miR-664 inhibit HCC growth[55-59]. After further evaluation of its biological function, it was determined that the stable overexpression of MEG3 can inhibit migration and invasion by regulating EMT[60,61]. Moreover, Kaplan-Meier analysis demonstrated that patients with low MEG3 expression had a worse OS and RFS than those with high expression[62]. This information provides valuable explanations for literature on the function of MEG3 and provides recommendations for new therapeutic targets.
GAS5: Growth arrest specific 5 (GAS5) is a novel tumor suppressor lncRNA located on ch1q25. Although GAS5 has a short open reading frame, it does not encode a protein and acts as a snoRNA host gene. Notably, GAS5 expression levels are downregulated in a number of human malignancies, and such aberrant expression is negatively associated with disease stage and prognosis. In HCC, low GAS5 expression is significantly associated with differentiation and TNM stage[63]. In addition, Kaplan-Meier survival curves revealed that low GAS5 expression was associated with a poor OS and RFS in HCC patients[64]. Through functional experiments, Chen et al[65] found that GAS5 could significantly inhibit the migration and invasion of HCC cells in vitro and suppress tumor metastasis in vivo. At the molecular level, GAS5 can suppress the migration, invasion, and metastasis of HCC via miR-21, miR-135b, miR-182, and miR-382-3p. Interestingly, GAS5-mediated miR-1323 promotes cell proliferation and invasion and inhibits apoptosis by targeting TP53INP1 in HCC[66]. Therefore, GAS5 may play an important role in the recurrence of liver cancer, and its expression is an independent prognostic factor for patients with HCC.
HCC is one of the most common malignant cancers in the world, but the underlying mechanism of the pathogenesis of recurrent HCC is still not clearly understood. However, research on lncRNA-related recurrence in liver cancer is still lacking. Therefore, this review focuses on the molecular mechanisms and expression of lncRNAs, classifies them according to their biological processes, and further subdivides them by their most common modes of molecular interactions in recurrent HCC. Currently, a growing number of studies have shown that lncRNAs, regulators of human gene expression, are abnormally expressed and influence the development of cancers. The lncRNA/miRNA/mRNA axis participates in diverse biological functions, including cancer migration, invasion, and metastasis. Analysis of lncRNAs in recurrent HCC can be interesting and will lead to the identification of novel diagnostic and prognostic markers because it is noninvasive and easily accessible. For recurrent HCC, the identification of early and prognostic biomarkers can help reveal the patient’s disease classification, formulate a personalized clinical treatment plan, improve efficacy and prognosis, and extend survival. At present, only a few lncRNAs that have been researched in recurrent HCC may serve as prognostic markers. However, much more research is required to apply lncRNA in clinical practice along with the development of some standards for identifying lncRNA biomarkers in recurrent HCC. In summary, at present and in the future, we still need to further understand the significance of lncRNAs in the clinical diagnosis and treatment of recurrent HCC and explore the regulatory mechanisms of lncRNAs in the occurrence and treatment of recurrent HCC.
Provenance and peer review: Unsolicited article; Externally peer reviewed
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Perse M S-Editor: Wu YXJ L-Editor: Wang TQ P-Editor: Li JH
| 1. | Bertuccio P, Turati F, Carioli G, Rodriguez T, La Vecchia C, Malvezzi M, Negri E. Global trends and predictions in hepatocellular carcinoma mortality. J Hepatol. 2017;67:302-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 380] [Cited by in RCA: 477] [Article Influence: 59.6] [Reference Citation Analysis (0)] |
| 2. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12667] [Cited by in RCA: 15289] [Article Influence: 3057.8] [Reference Citation Analysis (4)] |
| 3. | Zheng J, Kuk D, Gönen M, Balachandran VP, Kingham TP, Allen PJ, D'Angelica MI, Jarnagin WR, DeMatteo RP. Actual 10-Year Survivors After Resection of Hepatocellular Carcinoma. Ann Surg Oncol. 2017;24:1358-1366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 86] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 4. | Erridge S, Pucher PH, Markar SR, Malietzis G, Athanasiou T, Darzi A, Sodergren MH, Jiao LR. Meta-analysis of determinants of survival following treatment of recurrent hepatocellular carcinoma. Br J Surg. 2017;104:1433-1442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 5. | Calderaro J, Ziol M, Paradis V, Zucman-Rossi J. Molecular and histological correlations in liver cancer. J Hepatol. 2019;71:616-630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 367] [Article Influence: 61.2] [Reference Citation Analysis (0)] |
| 6. | Xie DY, Fan HK, Ren ZG, Fan J, Gao Q. Identifying Clonal Origin of Multifocal Hepatocellular Carcinoma and Its Clinical Implications. Clin Transl Gastroenterol. 2019;10:e00006. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 7. | Morimoto O, Nagano H, Sakon M, Fujiwara Y, Yamada T, Nakagawa H, Miyamoto A, Kondo M, Arai I, Yamamoto T, Ota H, Dono K, Umeshita K, Nakamori S, Sasaki Y, Ishikawa O, Imaoka S, Monden M. Diagnosis of intrahepatic metastasis and multicentric carcinogenesis by microsatellite loss of heterozygosity in patients with multiple and recurrent hepatocellular carcinomas. J Hepatol. 2003;39:215-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 90] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 8. | Tabrizian P, Jibara G, Shrager B, Schwartz M, Roayaie S. Recurrence of hepatocellular cancer after resection: patterns, treatments, and prognosis. Ann Surg. 2015;261:947-955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 345] [Cited by in RCA: 654] [Article Influence: 65.4] [Reference Citation Analysis (0)] |
| 9. | Yang SL, Luo YY, Chen M, Zhou YP, Lu FR, Deng DF, Wu YR. A systematic review and meta-analysis comparing the prognosis of multicentric occurrence and vs. intrahepatic metastasis in patients with recurrent hepatocellular carcinoma after hepatectomy. HPB (Oxford). 2017;19:835-842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 10. | Wang B, Xia CY, Lau WY, Lu XY, Dong H, Yu WL, Jin GZ, Cong WM, Wu MC. Determination of clonal origin of recurrent hepatocellular carcinoma for personalized therapy and outcomes evaluation: a new strategy for hepatic surgery. J Am Coll Surg. 2013;217:1054-1062. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 11. | Kang KJ, Ahn KS. Anatomical resection of hepatocellular carcinoma: A critical review of the procedure and its benefits on survival. World J Gastroenterol. 2017;23:1139-1146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 44] [Cited by in RCA: 38] [Article Influence: 4.8] [Reference Citation Analysis (1)] |
| 12. | Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol. 2019;21:542-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 693] [Cited by in RCA: 1034] [Article Influence: 172.3] [Reference Citation Analysis (0)] |
| 13. | Saleembhasha A, Mishra S. Novel molecules lncRNAs, tRFs and circRNAs deciphered from next-generation sequencing/RNA sequencing: computational databases and tools. Brief Funct Genomics. 2018;17:15-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 14. | Osielska MA, Jagodziński PP. Long non-coding RNA as potential biomarkers in non-small-cell lung cancer: What do we know so far? Biomed Pharmacother. 2018;101:322-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 15. | Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2868] [Cited by in RCA: 2716] [Article Influence: 301.8] [Reference Citation Analysis (0)] |
| 16. | Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1788] [Cited by in RCA: 2133] [Article Influence: 213.3] [Reference Citation Analysis (0)] |
| 17. | Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2002] [Cited by in RCA: 2313] [Article Influence: 192.8] [Reference Citation Analysis (0)] |
| 18. | Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, Zörnig M, MacLeod AR, Spector DL, Diederichs S. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180-1189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1087] [Cited by in RCA: 1293] [Article Influence: 99.5] [Reference Citation Analysis (0)] |
| 19. | Shi D, Qu Q, Chang Q, Wang Y, Gui Y, Dong D. A five-long non-coding RNA signature to improve prognosis prediction of clear cell renal cell carcinoma. Oncotarget. 2017;8:58699-58708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 20. | Li Z, Teng J, Jia Z, Zhang G, Ai X. The long non-coding RNA PCAL7 promotes prostate cancer by strengthening androgen receptor signaling. J Clin Lab Anal. 2021;35:e23645. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 21. | Cui C, Lu Z, Yang L, Gao Y, Liu W, Gu L, Yang C, Wilson J, Zhang Z, Xing B, Deng D, Sun ZS. Genome-wide identification of differential methylation between primary and recurrent hepatocellular carcinomas. Mol Carcinog. 2016;55:1163-1174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 22. | Yang Y, Chen L, Gu J, Zhang H, Yuan J, Lian Q, Lv G, Wang S, Wu Y, Yang YT, Wang D, Liu Y, Tang J, Luo G, Li Y, Hu L, Sun X, Guo M, Xi Q, Xi J, Wang H, Zhang MQ, Lu ZJ. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat Commun. 2017;8:14421. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 253] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 23. | Hauptman N, Glavač D. Long non-coding RNA in cancer. Int J Mol Sci. 2013;14:4655-4669. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 244] [Cited by in RCA: 290] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 24. | Xie SC, Zhang JQ, Jiang XL, Hua YY, Xie SW, Qin YA, Yang YJ. LncRNA CRNDE facilitates epigenetic suppression of CELF2 and LATS2 to promote proliferation, migration and chemoresistance in hepatocellular carcinoma. Cell Death Dis. 2020;11:676. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 25. | Kang CL, Qi B, Cai QQ, Fu LS, Yang Y, Tang C, Zhu P, Chen QW, Pan J, Chen MH, Wu XZ. LncRNA AY promotes hepatocellular carcinoma metastasis by stimulating ITGAV transcription. Theranostics. 2019;9:4421-4436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 26. | Song J, Ye A, Jiang E, Yin X, Chen Z, Bai G, Zhou Y, Liu J. Reconstruction and analysis of the aberrant lncRNA-miRNA-mRNA network based on competitive endogenous RNA in CESC. J Cell Biochem. 2018;119:6665-6673. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 79] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 27. | Luo W, Li X, Song Z, Zhu X, Zhao S. Long non-coding RNA AGAP2-AS1 exerts oncogenic properties in glioblastoma by epigenetically silencing TFPI2 through EZH2 and LSD1. Aging (Albany NY). 2019;11:3811-3823. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 28. | Xu H, Mao HL, Zhao XR, Li Y, Liu PS. MiR-29c-3p, a target miRNA of LINC01296, accelerates tumor malignancy: therapeutic potential of a LINC01296/miR-29c-3p axis in ovarian cancer. J Ovarian Res. 2020;13:31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 29. | Chen PY, Li XD, Ma WN, Li H, Li MM, Yang XY, Li SY. Comprehensive Transcriptomic Analysis and Experimental Validation Identify lncRNA HOXA-AS2/miR-184/COL6A2 as the Critical ceRNA Regulation Involved in Low-Grade Glioma Recurrence. Onco Targets Ther. 2020;13:4999-5016. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 30. | Gabory A, Ripoche MA, Yoshimizu T, Dandolo L. The H19 gene: regulation and function of a non-coding RNA. Cytogenet Genome Res. 2006;113:188-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 187] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 31. | Tietze L, Kessler SM. The Good, the Bad, the Question-H19 in Hepatocellular Carcinoma. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 32. | Lv J, Ma L, Chen XL, Huang XH, Wang Q. Downregulation of LncRNAH19 and MiR-675 promotes migration and invasion of human hepatocellular carcinoma cells through AKT/GSK-3β/Cdc25A signaling pathway. J Huazhong Univ Sci Technolog Med Sci. 2014;34:363-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 65] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 33. | Sui CJ, Zhou YM, Shen WF, Dai BH, Lu JJ, Zhang MF, Yang JM. Long noncoding RNA GIHCG promotes hepatocellular carcinoma progression through epigenetically regulating miR-200b/a/429. J Mol Med (Berl). 2016;94:1281-1296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 85] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 34. | Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871-890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6805] [Cited by in RCA: 7736] [Article Influence: 483.5] [Reference Citation Analysis (0)] |
| 35. | Wang Y, Liu Z, Yao B, Li Q, Wang L, Wang C, Dou C, Xu M, Liu Q, Tu K. Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol Cancer. 2017;16:123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 201] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 36. | Ghafouri-Fard S, Shoorei H, Taheri M. The Role of Long Non-coding RNAs in Cancer Metabolism: A Concise Review. Front Oncol. 2020;10:555825. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 37. | Liu N, Dou L, Zhang X. LncRNA PTTG3P Sponge Absorbs microRNA-155-5P to Promote Metastasis of Colorectal Cancer. Onco Targets Ther. 2020;13:5283-5291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 38. | Lou W, Ding B, Fan W. High Expression of Pseudogene PTTG3P Indicates a Poor Prognosis in Human Breast Cancer. Mol Ther Oncolytics. 2019;14:15-26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 39. | Weng W, Ni S, Wang Y, Xu M, Zhang Q, Yang Y, Wu Y, Xu Q, Qi P, Tan C, Huang D, Wei P, Huang Z, Ma Y, Zhang W, Sheng W, Du X. PTTG3P promotes gastric tumour cell proliferation and invasion and is an indicator of poor prognosis. J Cell Mol Med. 2017;21:3360-3371. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 40. | Huang JL, Cao SW, Ou QS, Yang B, Zheng SH, Tang J, Chen J, Hu YW, Zheng L, Wang Q. The long non-coding RNA PTTG3P promotes cell growth and metastasis via up-regulating PTTG1 and activating PI3K/AKT signaling in hepatocellular carcinoma. Mol Cancer. 2018;17:93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 166] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 41. | Zhou Q, Zhang W, Wang Z, Liu S. Long non-coding RNA PTTG3P functions as an oncogene by sponging miR-383 and up-regulating CCND1 and PARP2 in hepatocellular carcinoma. BMC Cancer. 2019;19:731. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 42. | Bai H, Luo X, Liao D, Xiong W, Zeng M, Zheng B. Long Noncoding RNA PTTG3P Expression Is an Unfavorable Prognostic Marker for Patients With Hepatocellular Carcinoma. Technol Cancer Res Treat. 2019;18:1533033819887981. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 43. | Xie C, Zhang LZ, Chen ZL, Zhong WJ, Fang JH, Zhu Y, Xiao MH, Guo ZW, Zhao N, He X, Zhuang SM. A hMTR4-PDIA3P1-miR-125/124-TRAF6 Regulatory Axis and Its Function in NF kappa B Signaling and Chemoresistance. Hepatology. 2020;71:1660-1677. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 102] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 44. | Li J, Cui Z, Li H, Lv X, Gao M, Yang Z, Bi Y, Zhang Z, Wang S, Zhou B, Yin Z. Clinicopathological and prognostic significance of long noncoding RNA MALAT1 in human cancers: a review and meta-analysis. Cancer Cell Int. 2018;18:109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 45. | Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, Kim Y, MacLeod AR, Krainer AR, Norton L, Brogi E, Egeblad M, Spector DL. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34-51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 485] [Cited by in RCA: 467] [Article Influence: 51.9] [Reference Citation Analysis (0)] |
| 46. | Malakar P, Shilo A, Mogilevsky A, Stein I, Pikarsky E, Nevo Y, Benyamini H, Elgavish S, Zong X, Prasanth KV, Karni R. Long Noncoding RNA MALAT1 Promotes Hepatocellular Carcinoma Development by SRSF1 Upregulation and mTOR Activation. Cancer Res. 2017;77:1155-1167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 256] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 47. | Jadaliha M, Zong X, Malakar P, Ray T, Singh DK, Freier SM, Jensen T, Prasanth SG, Karni R, Ray PS, Prasanth KV. Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget. 2016;7:40418-40436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 132] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 48. | Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810-1816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 476] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 49. | Wang F, Ying HQ, He BS, Pan YQ, Deng QW, Sun HL, Chen J, Liu X, Wang SK. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6:7899-7917. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 271] [Cited by in RCA: 314] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 50. | Xiao JN, Yan TH, Yu RM, Gao Y, Zeng WL, Lu SW, Que HX, Liu ZP, Jiang JH. Long non-coding RNA UCA1 regulates the expression of Snail2 by miR-203 to promote hepatocellular carcinoma progression. J Cancer Res Clin Oncol. 2017;143:981-990. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 78] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 51. | Zhu M, Wang F, Mi H, Li L, Wang J, Han M, Gu Y. Long noncoding RNA MEG3 suppresses cell proliferation, migration and invasion, induces apoptosis and paclitaxel-resistance via miR-4513/PBLD axis in breast cancer cells. Cell Cycle. 2020;19:3277-3288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 52. | Wang G, Ye Q, Ning S, Yang Z, Chen Y, Zhang L, Huang Y, Xie F, Cheng X, Chi J, Lei Y, Guo R, Han J. LncRNA MEG3 promotes endoplasmic reticulum stress and suppresses proliferation and invasion of colorectal carcinoma cells through the MEG3/miR-103a-3p/PDHB ceRNA pathway. Neoplasma. 2021;68:362-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 53. | Chen PY, Hsieh PL, Peng CY, Liao YW, Yu CH, Yu CC. LncRNA MEG3 inhibits self-renewal and invasion abilities of oral cancer stem cells by sponging miR-421. J Formos Med Assoc. 2021;120:1137-1142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 54. | Anwar SL, Krech T, Hasemeier B, Schipper E, Schweitzer N, Vogel A, Kreipe H, Lehmann U. Loss of imprinting and allelic switching at the DLK1-MEG3 locus in human hepatocellular carcinoma. PLoS One. 2012;7:e49462. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 111] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 55. | Liu Z, Chen JY, Zhong Y, Xie L, Li JS. lncRNA MEG3 inhibits the growth of hepatocellular carcinoma cells by sponging miR-9-5p to upregulate SOX11. Braz J Med Biol Res. 2019;52:e8631. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 56. | Zhang Y, Liu J, Lv Y, Zhang C, Guo S. LncRNA meg3 suppresses hepatocellular carcinoma in vitro and vivo studies. Am J Transl Res. 2019;11:4089-4099. [PubMed] |
| 57. | Gailhouste L, Liew LC, Yasukawa K, Hatada I, Tanaka Y, Kato T, Nakagama H, Ochiya T. MEG3-derived miR-493-5p overcomes the oncogenic feature of IGF2-miR-483 loss of imprinting in hepatic cancer cells. Cell Death Dis. 2019;10:553. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 58. | He JH, Han ZP, Liu JM, Zhou JB, Zou MX, Lv YB, Li YG, Cao MR. Overexpression of Long Non-Coding RNA MEG3 Inhibits Proliferation of Hepatocellular Carcinoma Huh7 Cells via Negative Modulation of miRNA-664. J Cell Biochem. 2017;118:3713-3721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 76] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 59. | Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S, Negrini M, Miotto E, Croce CM, Patel T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750-4756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 487] [Cited by in RCA: 545] [Article Influence: 38.9] [Reference Citation Analysis (0)] |
| 60. | Zhang Z, Wang S, Liu W. EMT-related long non-coding RNA in hepatocellular carcinoma: A study with TCGA database. Biochem Biophys Res Commun. 2018;503:1530-1536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 61. | Fan Z, He J, Fu T, Zhang W, Yang G, Qu X, Liu R, Lv L, Wang J. Arsenic trioxide inhibits EMT in hepatocellular carcinoma by promoting lncRNA MEG3 via PKM2. Biochem Biophys Res Commun. 2019;513:834-840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 62. | Zhuo H, Tang J, Lin Z, Jiang R, Zhang X, Ji J, Wang P, Sun B. The aberrant expression of MEG3 regulated by UHRF1 predicts the prognosis of hepatocellular carcinoma. Mol Carcinog. 2016;55:209-219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 114] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 63. | Wang Y, Jing W, Ma W, Liang C, Chai H, Tu J. Down-regulation of long non-coding RNA GAS5-AS1 and its prognostic and diagnostic significance in hepatocellular carcinoma. Cancer Biomark. 2018;22:227-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 64. | Tu ZQ, Li RJ, Mei JZ, Li XH. Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7:4303-4309. [PubMed] |
| 65. | Chen F, Li Y, Li M, Wang L. Long noncoding RNA GAS5 inhibits metastasis by targeting miR-182/ANGPTL1 in hepatocellular carcinoma. Am J Cancer Res. 2019;9:108-121. [PubMed] |
| 66. | Zhang F, Yang C, Xing Z, Liu P, Zhang B, Ma X, Huang L, Zhuang L. LncRNA GAS5-mediated miR-1323 promotes tumor progression by targeting TP53INP1 in hepatocellular carcinoma. Onco Targets Ther. 2019;12:4013-4023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 67. | Kong Y, Zhang L, Huang Y, He T, Zhao X, Zhou X, Zhou D, Yan Y, Zhou J, Xie H, Zhou L, Zheng S, Wang W. Pseudogene PDIA3P1 promotes cell proliferation, migration and invasion, and suppresses apoptosis in hepatocellular carcinoma by regulating the p53 pathway. Cancer Lett. 2017;407:76-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 68. | Liu D, Zhu Y, Pang J, Weng X, Feng X, Guo Y. Knockdown of long non-coding RNA MALAT1 inhibits growth and motility of human hepatoma cells via modulation of miR-195. J Cell Biochem. 2018;119:1368-1380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 69. | Pan Y, Tong S, Cui R, Fan J, Liu C, Lin Y, Tang J, Xie H, Lin P, Zheng T, Yu X. Long Non-Coding MALAT1 Functions as a Competing Endogenous RNA to Regulate Vimentin Expression by Sponging miR-30a-5p in Hepatocellular Carcinoma. Cell Physiol Biochem. 2018;50:108-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 70. | He B, Peng F, Li W, Jiang Y. Interaction of lncRNA-MALAT1 and miR-124 regulates HBx-induced cancer stem cell properties in HepG2 through PI3K/Akt signaling. J Cell Biochem. 2019;120:2908-2918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 71. | Malakar P, Stein I, Saragovi A, Winkler R, Stern-Ginossar N, Berger M, Pikarsky E, Karni R. Long Noncoding RNA MALAT1 Regulates Cancer Glucose Metabolism by Enhancing mTOR-Mediated Translation of TCF7L2. Cancer Res. 2019;79:2480-2493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 128] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 72. | Zhao ZB, Chen F, Bai XF. Long Noncoding RNA MALAT1 Regulates Hepatocellular Carcinoma Growth Under Hypoxia via Sponging MicroRNA-200a. Yonsei Med J. 2019;60:727-734. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 73. | Cui RJ, Fan JL, Lin YC, Pan YJ, Liu C, Wan JH, Wang W, Jiang ZY, Zheng XL, Tang JB, Yu XG. miR-124-3p availability is antagonized by LncRNA-MALAT1 for Slug-induced tumor metastasis in hepatocellular carcinoma. Cancer Med. 2019;8:6358-6369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 74. | Yuan LT, Chang JH, Lee HL, Yang YC, Su SC, Lin CL, Yang SF, Chien MH. Genetic Variants of lncRNA MALAT1 Exert Diverse Impacts on the Risk and Clinicopathologic Characteristics of Patients with Hepatocellular Carcinoma. J Clin Med. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 75. | Hou ZH, Xu XW, Fu XY, Zhou LD, Liu SP, Tan DM. Long non-coding RNA MALAT1 promotes angiogenesis and immunosuppressive properties of HCC cells by sponging miR-140. Am J Physiol Cell Physiol. 2020;318:C649-C663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 82] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 76. | Liu S, Qiu J, He G, Liang Y, Wang L, Liu C, Pan H. LncRNA MALAT1 acts as a miR-125a-3p sponge to regulate FOXM1 expression and promote hepatocellular carcinoma progression. J Cancer. 2019;10:6649-6659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 77. | Zhang P, Ha M, Li L, Huang X, Liu C. MicroRNA-3064-5p sponged by MALAT1 suppresses angiogenesis in human hepatocellular carcinoma by targeting the FOXA1/CD24/Src pathway. FASEB J. 2020;34:66-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 78. | Chen S, Wang G, Tao K, Cai K, Wu K, Ye L, Bai J, Yin Y, Wang J, Shuai X, Gao J, Pu J, Li H. Long noncoding RNA metastasis-associated lung adenocarcinoma transcript 1 cooperates with enhancer of zeste homolog 2 to promote hepatocellular carcinoma development by modulating the microRNA-22/Snail family transcriptional repressor 1 axis. Cancer Sci. 2020;111:1582-1595. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 79. | Peng N, He J, Li J, Huang H, Huang W, Liao Y, Zhu S. Long noncoding RNA MALAT1 inhibits the apoptosis and autophagy of hepatocellular carcinoma cell by targeting the microRNA-146a/PI3K/Akt/mTOR axis. Cancer Cell Int. 2020;20:165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 61] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 80. | Qin LT, Tang RX, Lin P, Li Q, Yang H, Luo DZ, Chen G, He Y, Li P. Biological function of UCA1 in hepatocellular carcinoma and its clinical significance: Investigation with in vitro and meta-analysis. Pathol Res Pract. 2018;214:1260-1272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 81. | Zhao B, Lu Y, Cao X, Zhu W, Kong L, Ji H, Zhang F, Lin X, Guan Q, Ou K, Zhang X, Chen Q. MiRNA-124 inhibits the proliferation, migration and invasion of cancer cell in hepatocellular carcinoma by downregulating lncRNA-UCA1. Onco Targets Ther. 2019;12:4509-4516. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 82. | Zhou Y, Li Y, Wang N, Li X, Zheng J, Ge L. UPF1 inhibits the hepatocellular carcinoma progression by targeting long non-coding RNA UCA1. Sci Rep. 2019;9:6652. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 83. | Zhang Z, Li JZ, Wei ZW, Li F, Li HM, Xiao Y, Qin YQ. Correlation between expression levels of lncRNA UCA1 and miR-18a with prognosis of hepatocellular cancer. Eur Rev Med Pharmacol Sci. 2020;24:3586-3591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 84. | Wang HZ, Liu L, Xu Y, Zhang GY, Wang YY. LncRNA UCA1 Affects the Cell Proliferation, Migration, Invasion and Apoptosis of Hepatic Carcinoma Cells by Targeting MicroRNA-193a-3p. Cancer Manag Res. 2020;12:10897-10907. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 85. | Zhu Q, Yang H, Cheng P, Han Q. Bioinformatic analysis of the prognostic value of the lncRNAs encoding snoRNAs in hepatocellular carcinoma. Biofactors. 2019;45:244-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 86. | Yang L, Jiang J. GAS5 Regulates RECK Expression and Inhibits Invasion Potential of HCC Cells by Sponging miR-135b. Biomed Res Int. 2019;2019:2973289. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 87. | Wang C, Ke S, Li M, Lin C, Liu X, Pan Q. Downregulation of LncRNA GAS5 promotes liver cancer proliferation and drug resistance by decreasing PTEN expression. Mol Genet Genomics. 2020;295:251-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 88. | Wang X, Li FY, Zhao W, Gao ZK, Shen B, Xu H, Cui YF. Long non-coding RNA GAS5 overexpression inhibits M2-like polarization of tumour-associated macrophages in SMCC-7721 cells by promoting PTEN expression. Int J Exp Pathol. 2020;101:215-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 89. | Fan JC, Zeng F, Le YG, Xin L. LncRNA CASC2 inhibited the viability and induced the apoptosis of hepatocellular carcinoma cells through regulating miR-24-3p. J Cell Biochem. 2018;119:6391-6397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 90. | Zhao L, Zhang Y. Long noncoding RNA CASC2 regulates hepatocellular carcinoma cell oncogenesis through miR-362-5p/Nf-κB axis. J Cell Physiol. 2018;233:6661-6670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 91. | Gao X, Du H, Zhang R, Li C, Wang H, Xuan Q, Liu D. Overexpression of cancer susceptibility candidate 2 inhibited progression of hepatocellular carcinoma cells. J Cell Physiol. 2019;234:9008-9018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 92. | Refai NS, Louka ML, Halim HY, Montasser I. Long non-coding RNAs (CASC2 and TUG1) in hepatocellular carcinoma: Clinical significance. J Gene Med. 2019;21:e3112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 93. | Sun J, Liu L, Zou H, Yu W. The Long Non-Coding RNA CASC2 Suppresses Cell Viability, Migration, and Invasion in Hepatocellular Carcinoma Cells by Directly Downregulating miR-183. Yonsei Med J. 2019;60:905-913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 94. | Li QY, Yang K, Liu FG, Sun XG, Chen L, Xiu H, Liu XS. Long noncoding RNA CASC2c inhibited cell proliferation in hepatocellular carcinoma by inactivated ERK1/2 and Wnt/β-catenin signaling pathway. Clin Transl Oncol. 2020;22:302-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 95. | Wei LQ, Li L, Lu C, Liu J, Chen Y, Wu H. Involvement of H19/miR-326 axis in hepatocellular carcinoma development through modulating TWIST1. J Cell Physiol. 2019;234:5153-5162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 96. | Ge L, Wang Q, Hu S, Yang X. Rs217727 polymorphism in H19 promotes cell apoptosis by regulating the expressions of H19 and the activation of its downstream signaling pathway. J Cell Physiol. 2019;234:7279-7291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 97. | Zhou Y, Fan RG, Qin CL, Jia J, Wu XD, Zha WZ. LncRNA-H19 activates CDC42/PAK1 pathway to promote cell proliferation, migration and invasion by targeting miR-15b in hepatocellular carcinoma. Genomics. 2019;111:1862-1872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 98. | Li L, Han T, Liu K, Lei CG, Wang ZC, Shi GJ. LncRNA H19 promotes the development of hepatitis B related hepatocellular carcinoma through regulating microRNA-22 via EMT pathway. Eur Rev Med Pharmacol Sci. 2019;23:5392-5401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 99. | Ye Y, Guo J, Xiao P, Ning J, Zhang R, Liu P, Yu W, Xu L, Zhao Y, Yu J. Macrophages-induced long noncoding RNA H19 up-regulation triggers and activates the miR-193b/MAPK1 axis and promotes cell aggressiveness in hepatocellular carcinoma. Cancer Lett. 2020;469:310-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 89] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 100. | Yi T, Wang T, Shi Y, Peng X, Tang S, Zhong L, Chen Y, Li Y, He K, Wang M, Zhao H, Li Q. Long noncoding RNA 91H overexpression contributes to the growth and metastasis of HCC by epigenetically positively regulating IGF2 expression. Liver Int. 2020;40:456-467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 101. | Xu Y, Liu Y, Li Z, Li H, Li X, Yan L, Mao J, Shen J, Chen W, Xue F. Long noncoding RNA H19 is involved in sorafenib resistance in hepatocellular carcinoma by upregulating miR675. Oncol Rep. 2020;44:165-173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 102. | Wang D, Xing N, Yang T, Liu J, Zhao H, He J, Ai Y, Yang J. Exosomal lncRNA H19 promotes the progression of hepatocellular carcinoma treated with Propofol via miR-520a-3p/LIMK1 axis. Cancer Med. 2020;9:7218-7230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 103. | Sun Z, Xue S, Zhang M, Xu H, Hu X, Chen S, Liu Y, Guo M, Cui H. Aberrant NSUN2-mediated m5C modification of H19 lncRNA is associated with poor differentiation of hepatocellular carcinoma. Oncogene. 2020;39:6906-6919. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 192] [Article Influence: 38.4] [Reference Citation Analysis (0)] |