Published online Jan 6, 2021. doi: 10.12998/wjcc.v9.i1.133
Peer-review started: August 6, 2020
First decision: November 3, 2020
Revised: November 7, 2020
Accepted: November 14, 2020
Article in press: November 14, 2020
Published online: January 6, 2021
Processing time: 147 Days and 21.7 Hours
Helicobacter pylori (H. pylori) infection is closely associated with the etiology of a variety of gastric diseases. The effective eradication of H. pylori infection has been shown to reduce the incidence of gastric carcinoma. However, the rate of H. pylori eradication has significantly declined due to its increasing resistance to antibiotics, especially to clarithromycin. Therefore, the detection of clarithromycin resistance is necessary prior to the treatment of H. pylori. Although many studies have been conducted on the use of polymerase chain reaction (PCR)-based tests to detect clarithromycin resistance in stool samples, no accurate data on the feasibility of these tests are available. Here, we performed a meta-analysis to assess the feasibility of these noninvasive tests.
To evaluate the reliability of PCR-based tests for detecting H. pylori clarithromycin resistance in stool samples.
We searched PubMed, Medline, Embase, and other databases for articles that evaluated the value of the PCR analysis of stool samples for detecting the resistance of H. pylori to clarithromycin. We collected cross-sectional studies that met the inclusion criteria. Diagnostic accuracy measures were pooled using a random-effects model. The risk of bias was assessed using the Quality Assessment of Diagnostic Accuracy Studies-2 tool. Subgroup analysis was also conducted according to PCR type, purification technique, reference standard, mutation site, sample weight, number of patients, and age group, and the clinical utility of diagnostic tests was evaluated using the Likelihood Ratio Scatter Graph.
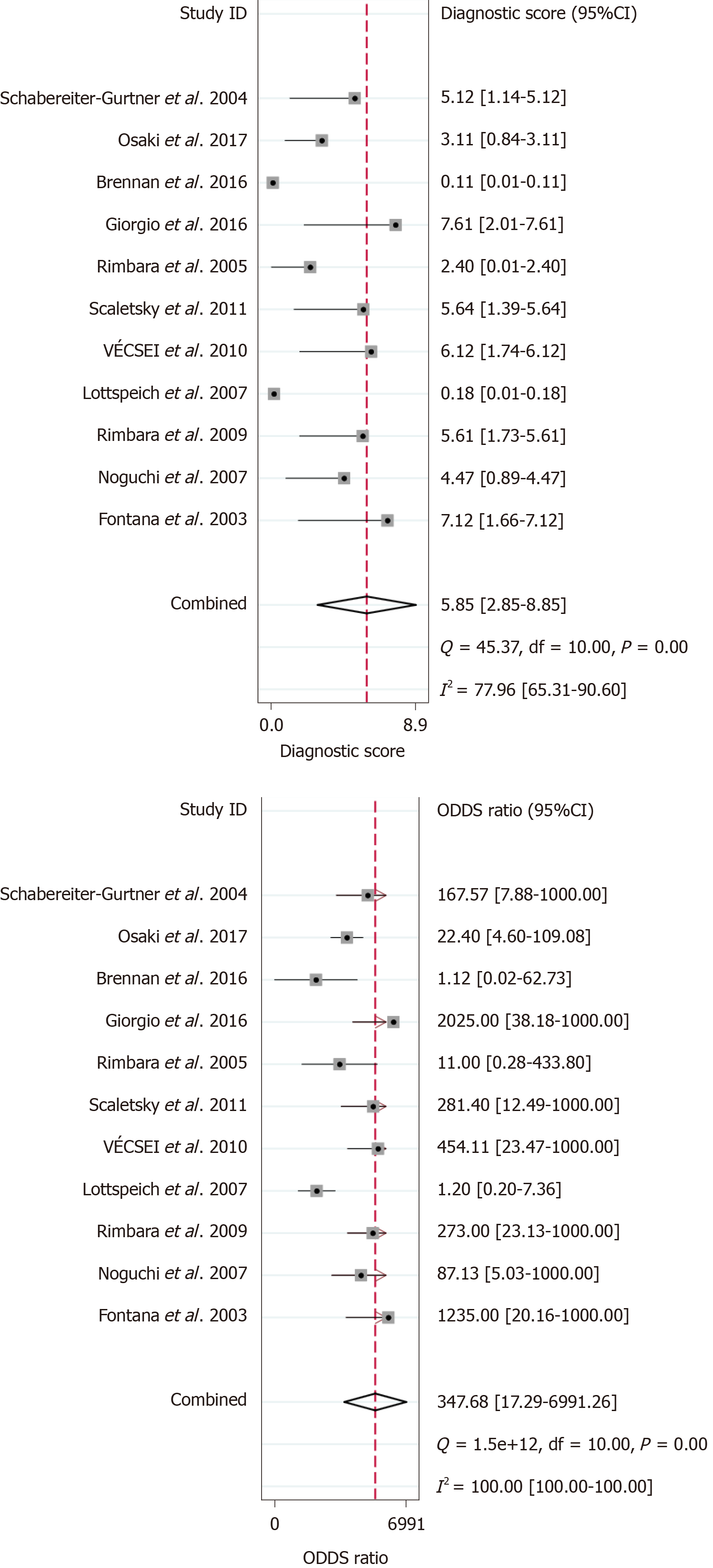
Out of the 1818 identified studies, only 11 met the eligibility criteria, with a total of 592 patients assessed. A meta-analysis of the random-effect model showed that PCR-based analysis of stool samples had high diagnostic accuracy for detecting clarithromycin resistance in patients infected with H. pylori. The combined sensitivity was 0.91 [95% confidence interval (CI): 0.83-0.95], Q = 30.34, and I2 = 67.04, and the combined specificity was 0.97 (95%CI: 0.62-1.00), Q = 279.54, and I2 = 96.42. The likelihood ratio for a positive test was 33.25 (95%CI: 1.69-652.77), and that for a negative test was 0.10 (95%CI: 0.05-0.18), with an area under the curve of 0.94. The diagnostic odds ratio was 347.68 (95%CI: 17.29-6991.26). There was significant statistical heterogeneity, and the sub-analyses showed significant differences in the number of patients, sample weight, purification methods, PCR types, mutation points, and reference standards. The included studies showed no risk of publication bias.
PCR-based tests on stool samples have high diagnostic accuracy for detecting H. pylori clarithromycin resistance.
Core Tip: No consensus is available in the literature about the reliability of polymerase chain reaction (PCR)-based tests for detecting Helicobacter pylori (H. pylori) clarithromycin resistance in stool samples. This is the first meta-analysis deciphering these methods based on the numbers of true-positive, false-positive, false-negative, and true-negative test results. Our results show that PCR-based approaches on stool samples have high diagnostic accuracy for detecting H. pylori clarithromycin resistance.
- Citation: Gong RJ, Xu CX, Li H, Liu XM. Polymerase chain reaction-based tests for detecting Helicobacter pylori clarithromycin resistance in stool samples: A meta-analysis. World J Clin Cases 2021; 9(1): 133-147
- URL: https://www.wjgnet.com/2307-8960/full/v9/i1/133.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i1.133
Helicobacter pylori (H. pylori) is a gram-negative bacterium firstly identified from antral mucosa in 1984[1]. H. pylori is closely related to digestive diseases such as chronic gastritis, peptic ulcers, mucosa associated lymphoid tissue lymphoma and gastric carcinoma. The World Health Organization classified H. pylori as a group I carcinogen for stomach cancer, and its eradication therapy is highly recommended by the Kyoto global consensus[2]. Clarithromycin-based triple therapy encompassing a proton pump inhibitor and another antibiotic (amoxicillin or metronidazole) is generally conducted as the first-line treatment[3]. The eradication rate of H. pylori, however, has gradually decreased due to antibiotic resistance worldwide. In particular, clarithromycin resistance significantly increased from 13% in 2006-2008 to 21% in 2012–2016[4]. The latest clinical guidelines point out that clarithromycin triple therapy should be limited to patients that reside in areas with low H. pylori clarithromycin resistance[5,6]. Compared with previous empirical treatments for H. pylori eradication, tailored therapy including an antimicrobial susceptibility test leads to better outcomes[7]. The Maastricht IV/Florence Consensus Report suggests that susceptibility testing should be performed in regions where the clarithromycin resistance rate greater than 20%[8]. In addition, the Toronto Consensus recommends that susceptibility testing should be encouraged when patients undergo endoscopy[9]. Therefore, the detection of clarithromycin resistance is necessary prior to the treatment of H. pylori, especially in cases of refractory H. pylori infection. However, conducting susceptibility tests in all patients is currently impractical or impossible.
In the past, phenotypic methods have been widely used, including the broth dilution method, agar dilution method, disk diffusion tests, and E-test, all of which were conducted using H. pylori isolated from patient stomach biopsy samples[10]. These methods have high accuracy, but the time-consuming nature, harsh conditions, and vulnerability to certain medicines caused by these methods limit their wide clinical application. The need for repeated endoscopy to obtain biopsies after failed therapy may render this approach cost-prohibitive. The emerging roles of genotypic methods have been recognized. For example, point mutations at specific loci of the 23S ribosomal ribonucleic acid (rRNA) were found to explain clarithromycin resistance in 1996[11]. Point mutations launch decreased affinity between ribosomes and clarithromycin so that the antibiotic is unable to interfere with bacterial protein biosynthesis[12]. Polymerase chain reaction (PCR)-based tests have been gradually acknowledged to evaluate the clarithromycin resistance by detecting 23S rRNA in H. pylori strains or biopsies[13-15]. However, this technique requires the use of a gastroscopy, which is particularly difficult for elderly and pediatric patients. PCR-based analysis of stool samples has received increasing attention for its noninva-siveness, convenience, and low cost. Studies have shown that the detection of clarithromycin resistance in feces by PCR has high accuracy[16,17], but some studies have suggested otherwise[18].
Although many studies have been conducted on the efficacy of PCR-based tests to detect clarithromycin resistance in stool, there are no data available on the reliability of these tests. Here, we performed a meta-analysis by collecting all useful data to assess the reliability of these noninvasive tests. This was the first meta-analysis to evaluate the reliability of PCR-based tests for detecting H. pylori clarithromycin resistance in stool samples.
Our research was registered and approved on the International Prospective Register of Systematic Reviews, with the registration number CRD42019142429. We conducted this study following the recommendations of the Preferred Reporting Items for Systematic reviews and Meta-analyses[19].
We searched PubMed, Medline, Embase, and other databases for related articles from January 1, 1987 through January 31, 2019 using the search terms “Helicobacter pylori”, “H. pylori”, “Helicobacter infection”, “clarithromycin resistance”, “antibiotic resistance”, “feces”, “polymerase chain reaction”, and “PCR” with its Medical Subject Headings terms and keywords. Letters were also included if they could provide additional data. There were no restrictions on language and age in the literature search.
Studies were eligible when they met the following criteria: (1) Observational studies about the detection of human clarithromycin resistance by PCR in feces without restrictions of language and age; (2) Patients infected with H. pylori; (3) Gold standard was provided; (4) Data could be extracted; (5) Full text was available; and (6) Letters could be included if they could provide the information required for studies to be included.
Studies were excluded if they were abstracts, reviews, case reports, or animal experiments or if they could not provide useful data.
All the data were extracted from eligible full-text studies by two investigators (RJG and HL) independently. The data included the first author, year of publication, number of patients, country, age groups, gender, method used to diagnose H. pylori, sample weight, purification methods, PCR types, point mutations, reference standard, and diagnostic study data.
Two investigators (RJG and HL) independently assessed the quality of each study using the QUADAS-2 tool, which was designed to assess the quality of primary diagnostic accuracy studies included in the article.
The tool includes four important areas: Patient selection, index tests, reference standard, flow, and timing[20]. Using this tool, the risk of bias was judged as “low”, “high”, and “unclear”. If all the signaling questions for an area were answered with “yes”, then the risk of bias was considered “low”. If any signaling questions were answered with “no”, then this could indicate potential bias. The level of agreement on article quality was generally high (> 80% crude agreement, kappa = 0.65). Funnel plots were used to evaluate the risk of publication bias. Any divergence was resolved by a third reviewer (XML).
The meta-analysis was conducted using stata15.0 software (College Station, TX, United States). A random-effect model was used in all the analyses due to heterogeneity. The diagnostic accuracy indexes used in the analysis were pooled sensitivity, pooled specificity, positive likelihood ratio, negative likelihood ratio, diagnostic odds ratio, and summary receiver operating characteristics curve. I-squared statistic and Q test were used to assess heterogeneity. Deeks’ funnel plot asymmetry test was used for publication bias, and slope coefficient P values < 0.05 indicated significant asymmetry. We also performed a subgroup analysis to assess whether PCR type, purification technique, reference criteria, mutation site, sample weight, number of patients, and age group affected the pooled estimates.
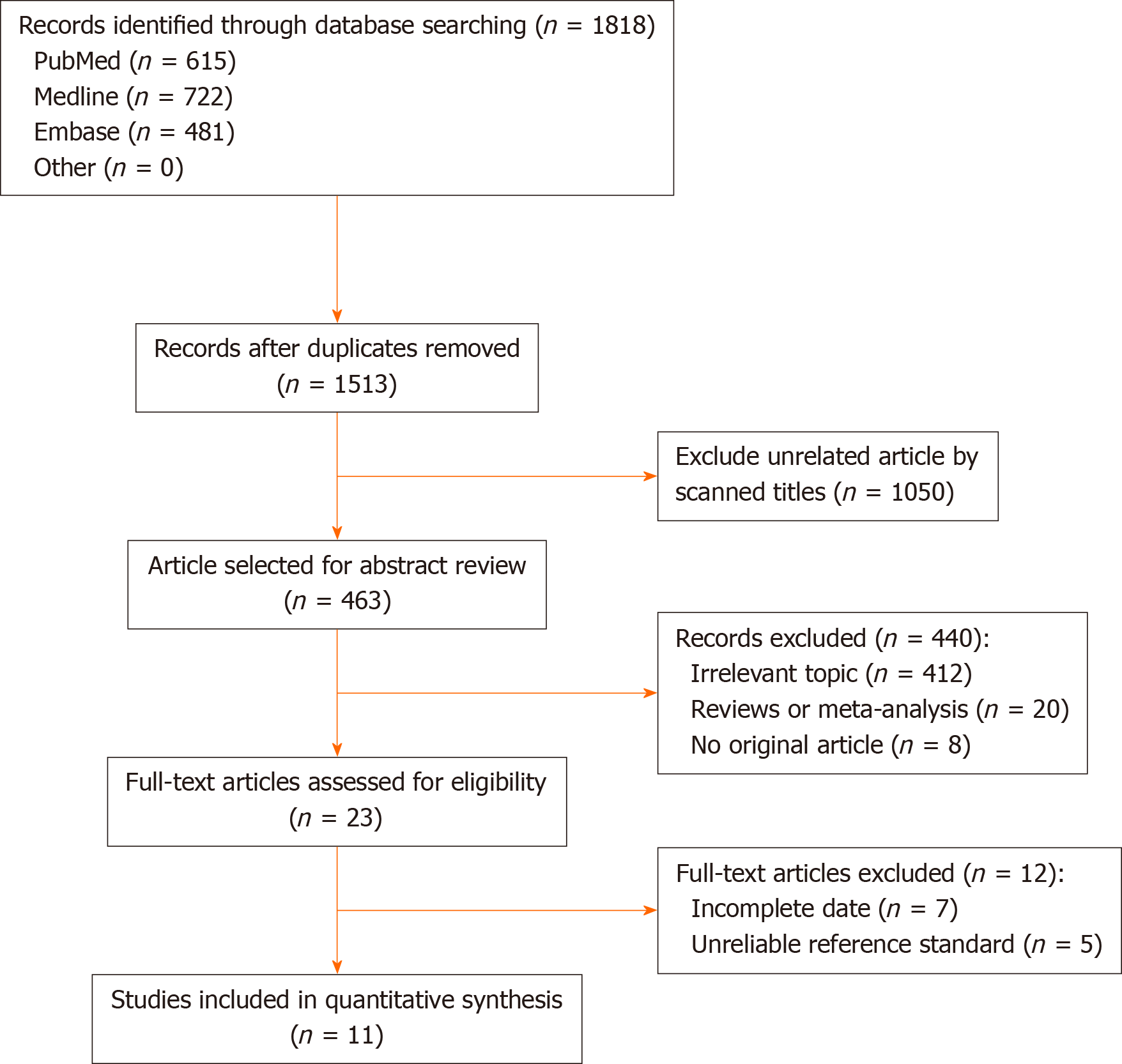
From the 1818 initial articles, 11 studies enrolling a total of 592 participants were finally included. The study selection process is shown in Figure 1.
The characteristics of all the included studies are shown in Table 1; the data extracted by the two investigators (RJG and HL) were identical. A total of 11 studies with 592 participants were included in the meta-analysis. These studies were conducted in seven countries, and all the studies were published in English. All the eligible studies were published between 2004 and 2017. The sample sizes ranged from seven to 125. The patients enrolled included adults and children. Most of the studies did not provide accurate data about the male-female ratios and age groups. There was variation in the type of PCR used in the different studies. Nested PCR was used in five studies (45%)[16,21-24]; real-time PCR and Genotype were used in five studies[17,25-28] and one study (10%)[18], respectively. The reference standards for clarithromycin resistance were different: Minimum inhibitory concentration in nine studies (82%) and PCR-based test of biopsy samples in two studies (18%). The gene mutation loci detected were A2142 and A2143 in 10 studies (91%) and A2143, A2142 and A2717 for one study (9%). Key data were successfully extracted from all the studies, including the number of true positives, false positives, false negatives, and true negatives.
| Ref. | Country | Diagnose H.pylori | Patient number | Source | Gender, M/F | Sample weight, mg | Purification | PCR type | Point mutations | Gold standard |
| Fontana et al[21], 2003 | Italy | Culture | 125 | Adult + child | NA | 220 | Qiagen | Nested | A2143, A2142, A2717 | MIC > 1 μg/mL |
| Noguchi et al[22], 2007 | Japan | UBT | 98 | Adult | NA | 50 | Promega | Nested | A2142, A2143 | MIC > 1 μg/mL |
| Rimbara et al[23], 2009 | Japan | NA | 50 | Adult | 24/26 | 50 | Promega | Nested | A2142, A2143 | MIC > 1 μg/mL |
| Lottspeich et al[25], 2007 | Germany | Histo; Culture; UBT; HpSA | 46 | Child | NA | 200 | Qiagen | RT | A2142, A2143 | MIC > 1 μg/mL |
| VÉCSEI et al[26], 2010 | Austria | RUT; Histo; Culture | 67 | Child | NA | 200 | Qiagen | RT | A2142, A2143 | MIC > 1 μg/mL |
| Scaletsky et al[27], 2011 | Brazil | Culture; Histo; RUT | 45 | Child | NA | 200 | Qiagen | RT | A2142, A2143 | MIC > 1 mg/L |
| Rimbara et al[24], 2005 | Japan | HpSA; Culture | 7 | NA | NA | NA | Q-BIOgene | Nested | A2142, A2143 | MIC > 1 μg/mL |
| Giorgio et al[28], 2016 | Italy | UBT | 52 | Adult | 23/29 | 300 | THD fecal test | RT | A2142, A2143 | PCR in biopsy |
| Brennan et al[18], 2016 | Ireland | UBT; RUT | 17 | Adult | NA | NA | PSP spin stool | Genotype | A2146, A2147 | PCR in biopsy |
| Osaki et al[16], 2017 | Japan | HpSA | 40 | Adult | NA | 200 | DNA Plus Kit | Nested | A2142, A2143 | MIC > 0.5 mg/L |
| Schabereiter-Gurtner et al[17], 2004 | Austria | Histo; RUT;Culture | 45 | Adult | NA | 200 | Qiagen | RT | A2142, A2143 | MIC ≥ 1 μg/mL |
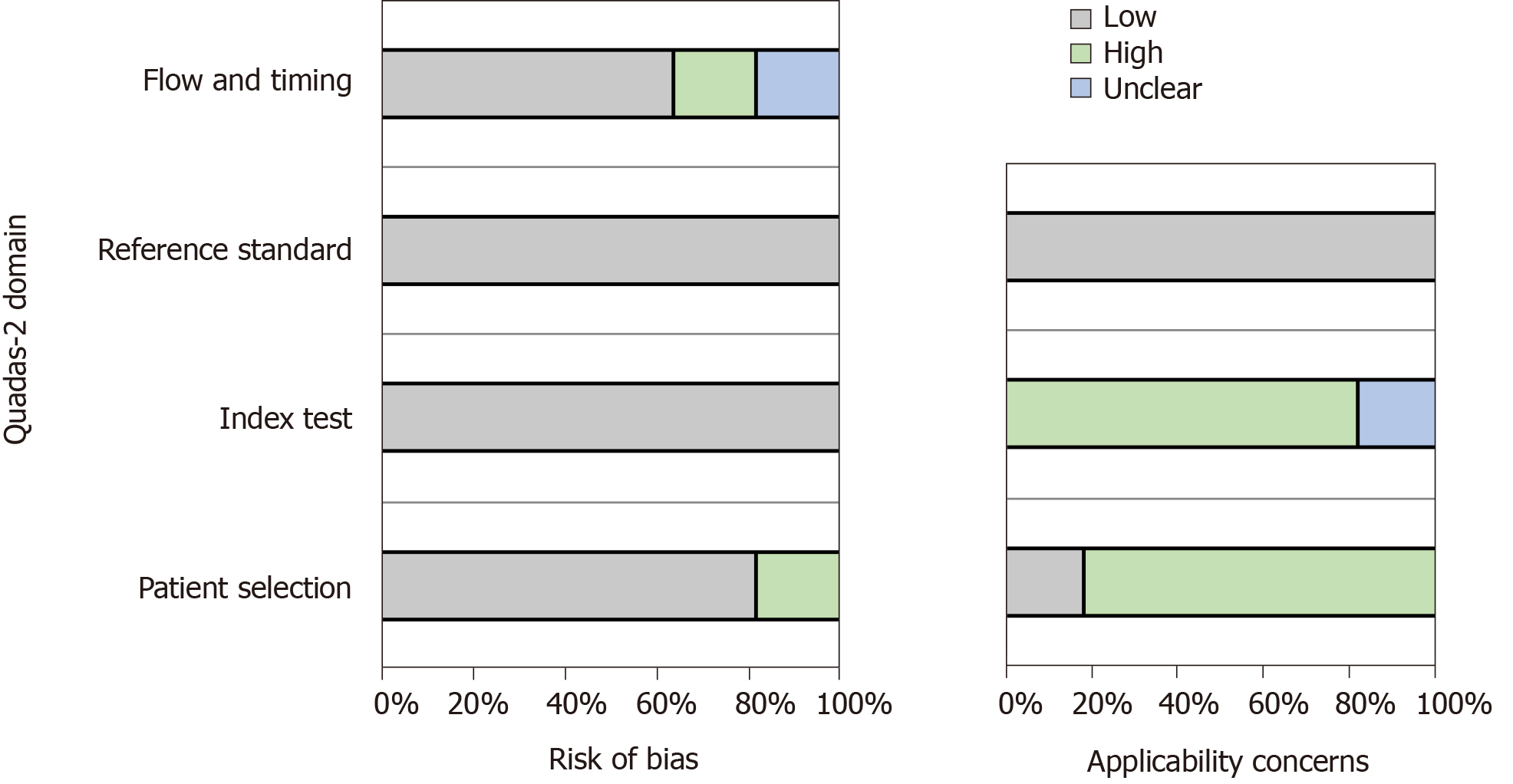
The quality assessment scores of more than 50% of the articles indicated that the risk of bias was low. Figure 2 visually shows the risk of bias estimated for all of the included studies.
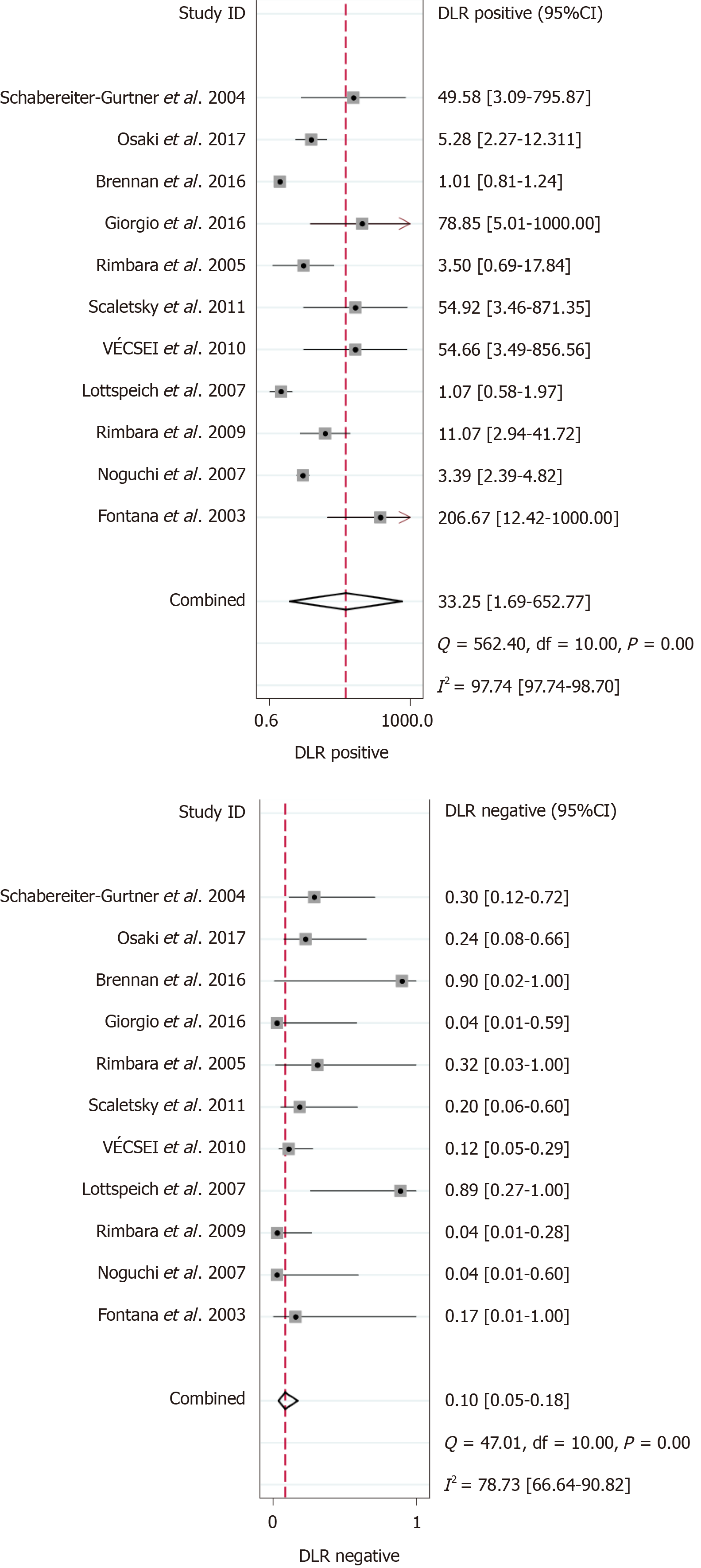
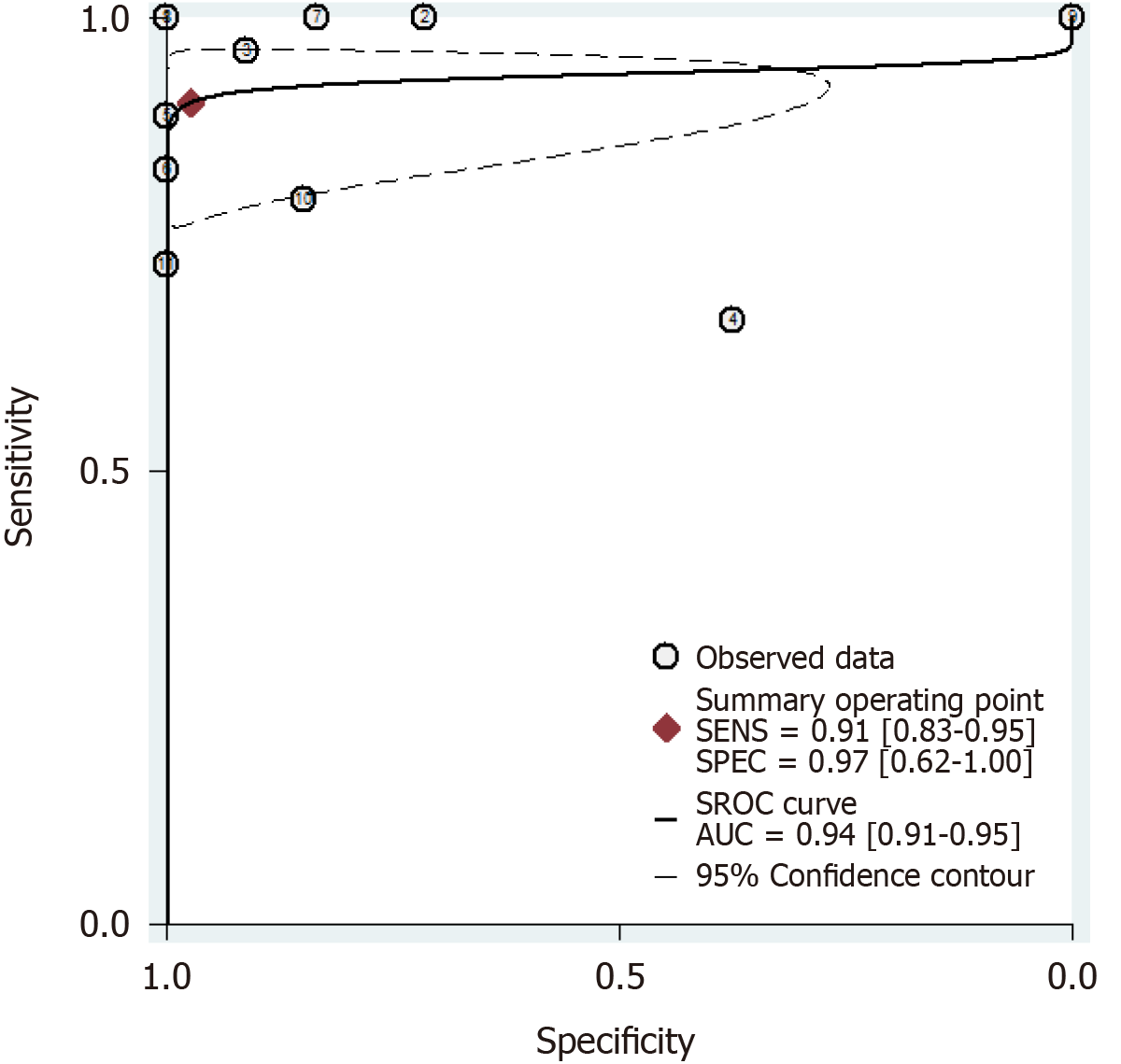
The results of the analysis are shown in Figure 3-6. The pooled sensitivity was 0.91 [95% confidence interval (CI): 0.83-0.95], and the pooled specificity was 0.97 (95%CI: 0.62-1.00) (Figure 3). The positive likelihood ratio was 33.25 (95%CI: 1.69-652.77), and the negative likelihood ratio was 0.10 (95%CI: 0.05-0.18) (Figure 4) with an area under the curve (Figure 5) of 0.94. The diagnostic odds ratio was 347.68 (95%CI: 17.29-6991.26) (Figure 6). There was significant statistical heterogeneity; the I2 values for sensitivity and specificity were 67.04 and 96.42, respectively, with a statistically significant Q test result (P < 0.05).
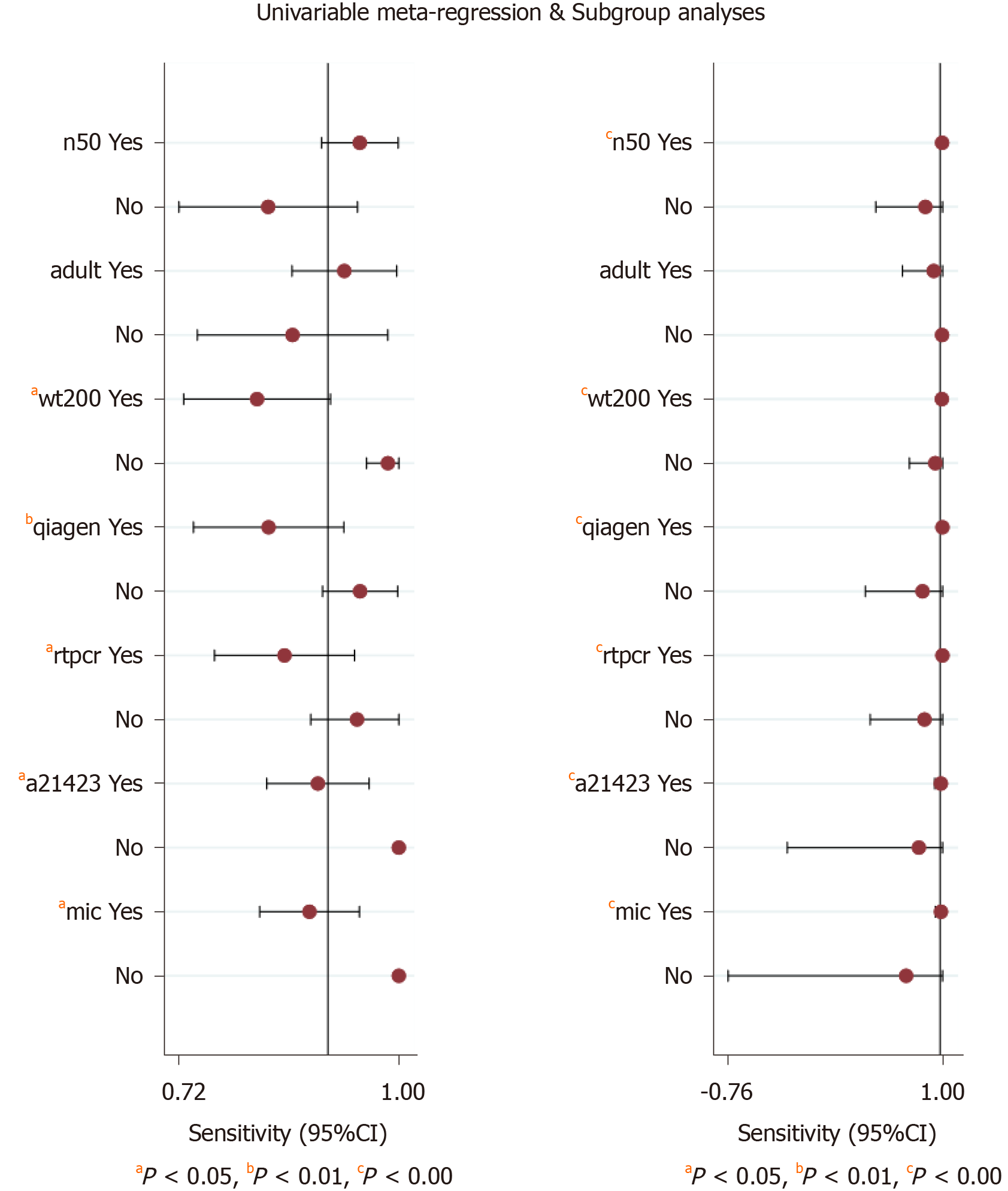
The significant heterogeneity could be explained by clinical and methodological variation. We used stata15.0 software to conduct regression and subgroup analyses to determine the source of heterogeneity based on these variables. The results indicated that sample weight, purification methods, PCR types, mutation points, and reference standards accounted for the heterogeneity in the pooled sensitivity and specificity, and the number of patients only explained the heterogeneity of the pooled sensitivity (Figure 7).
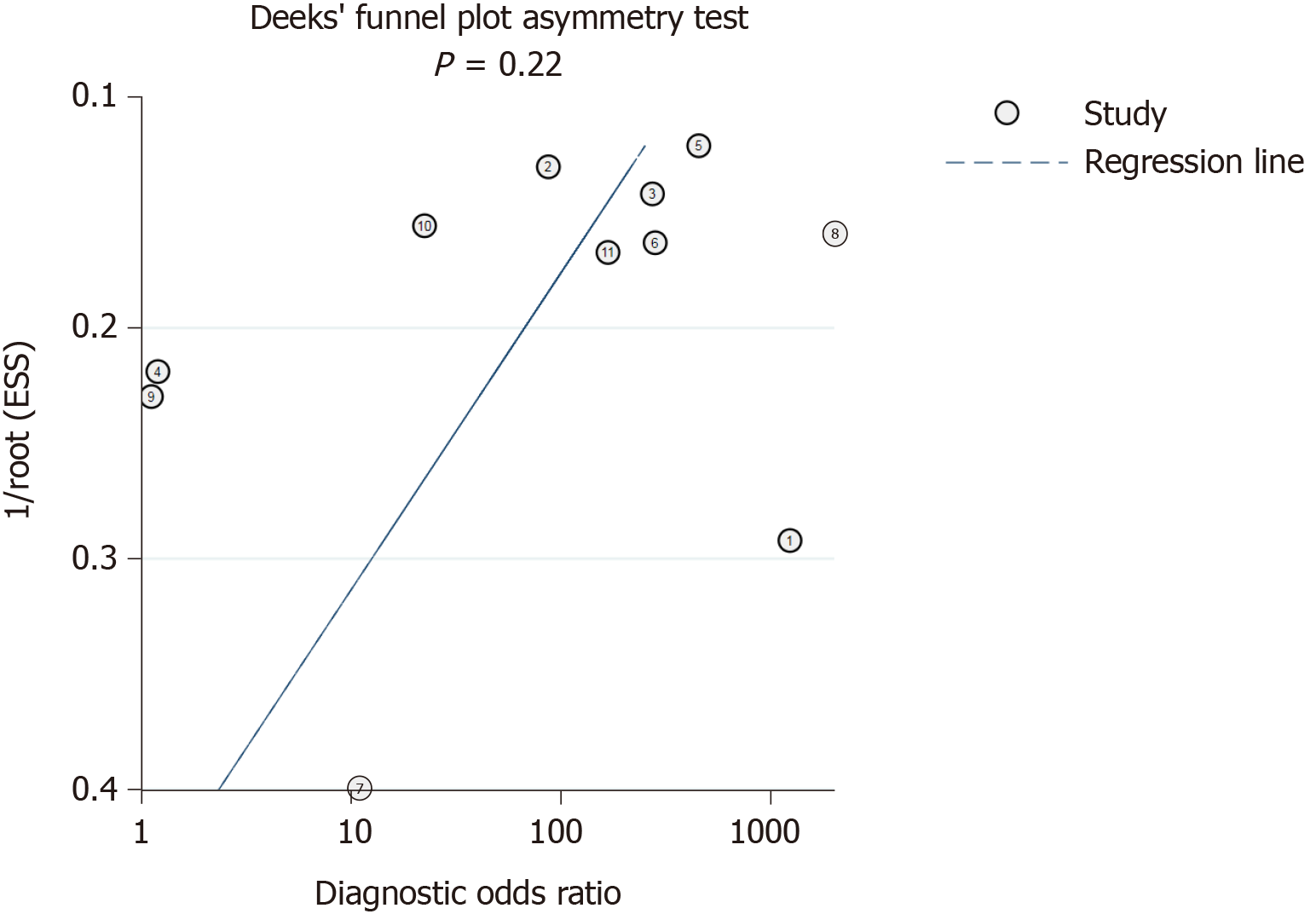
Deeks’ funnel plot test (Figure 8) indicated that the risk of publication bias was not significant (P = 0.22).
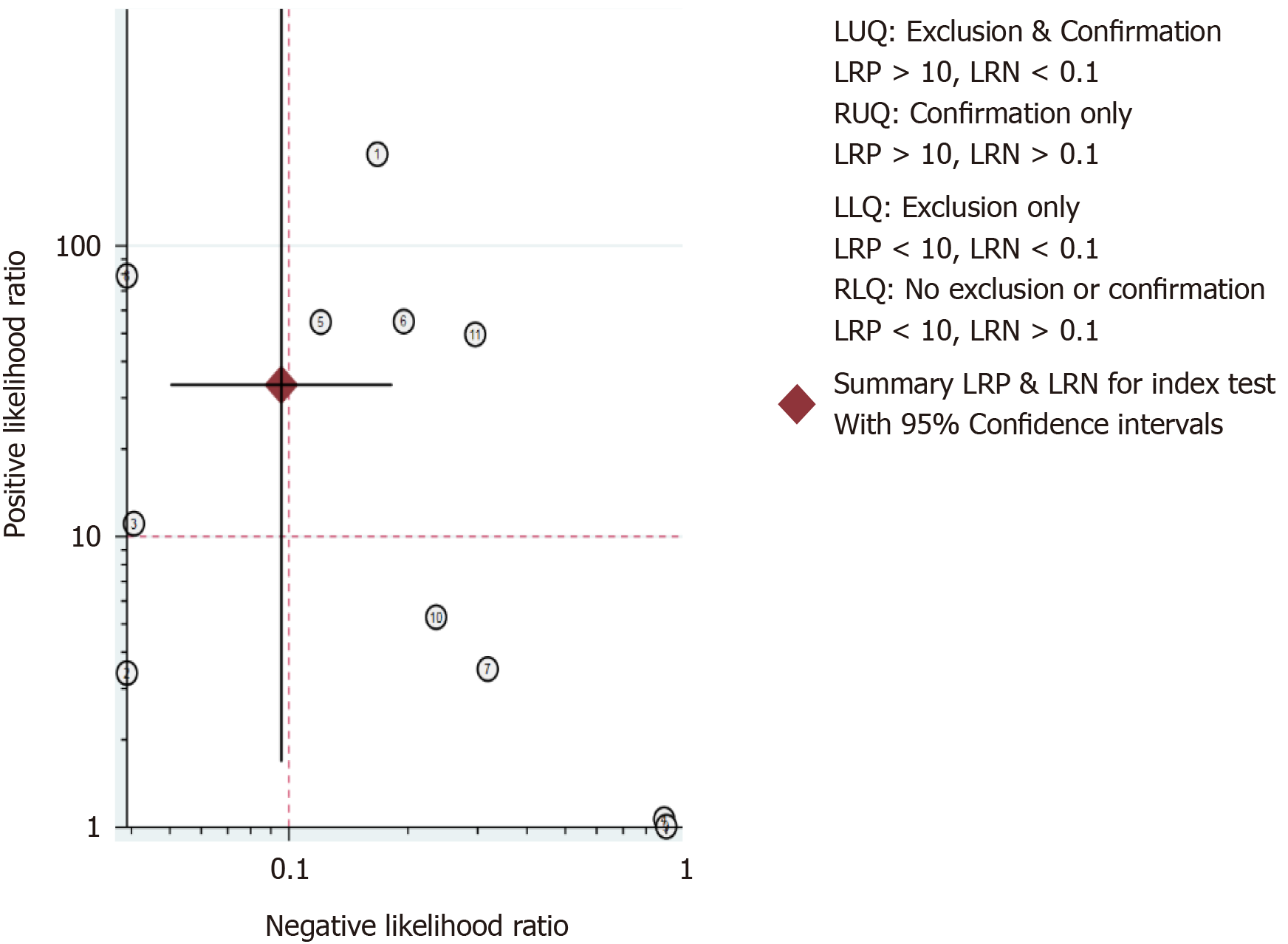
The clinical utility of a diagnostic test was evaluated using the Likelihood Ratio Scatter Graph (Figure 9). The likelihood ratio summation points in the upper left quadrant were a function of mean sensitivity and specificity, indicating that the test was useful for confirming the presence and absence of disease. Specifically, this figure shows the summary points of the likelihood ratio in the upper left quadrant, which was a function of the mean sensitivity and specificity.
This study is the first meta-analysis to assess the reliability of detecting clarithromycin resistance in the feces of patients with H. pylori by PCR-based tests. Eleven studies were included to evaluate the clinical application value of the test. The meta-analysis results showed that the technique has a high diagnostic accuracy for detecting clarithromycin resistance in patients infected with H. pylori. The heterogeneity can be explained by the number of patients, sample weight, purification method, PCR type, mutation point, and reference standard, and the variation in the methodological quality of the included studies may also lead to heterogeneity. The included studies had no risk of publication bias. A likelihood ratio scatter graph revealed that the test was useful for confirming the presence and absence of the disease. A previous study[18] reached a different conclusion, namely, that the Genotype HelicoDR assay was unsuitable for the accurate detection of clarithromycin resistance in stool specimens. This considerable discrepancy may arise for several reasons in pre-analysis and laboratory practice. Improper storage/transportation and repeated freezing/thawing of stool samples may impair test sensitivity due to enzymatic or mechanical degradation of DNA, and the presence of a large number of diverse symbiotic bacteria in feces may hinder the specificity of detection. Due to our overall data analysis, we think that the PCR-based tests still have high diagnostic accuracy.
H. pylori has been reported to be closely related not only to diseases of the digestive system but also to cardiovascular, vascular, and autoimmune diseases. The Kyoto global consensus recommends that H. pylori infection should be efficiently treated to prevent more severe complications. The clarithromycin-based standard triple therapy has been recommended by the Maastricht Treaty consensus to eradicate H. pylori in children and adults[8]. However, the rate of H. pylori eradication has decreased annually, primarily as a result of the development of resistance to antibiotics, such as clarithromycin[11]. Furthermore, H. pylori infection is frequently acquired during childhood[29]. Some studies have shown that the eradication of H. pylori may benefit children with nonnuclear dyspepsia[30,31], and that there are unexpectedly higher rates of clarithromycin-resistant H. pylori in younger age groups[32,33]. However, gastro-duodenal endoscopy is not generally advisable for the pediatric population. Thus, developing simple and noninvasive means to diagnose antibiotic susceptibility will greatly facilitate antibiotic therapy[34]. Susceptibility testing should be carried out in regions with high clarithromycin resistance[8] or in patients who are not suitable for gastroscopy or have refractory infections, as susceptibility testing can avoid the hazards associated with the abuse of antibiotics and prevent increases in the development of secondary or multiple antibiotics resistance.
The application of phenotypic detection technology is limited by its time-consuming nature, demanding conditions, and vulnerability to certain drugs. In addition, traditional PCR tests specimens, including biopsies, isolates, and gastric juices, that require invasive testing. PCR-based tests usually allow for both the detection of H. pylori and clarithromycin susceptibility simultaneously. The former detects specific genes, such as 23S rRNA, to confirm H. pylori infection, which has been proved to have high diagnostic accuracy[35,36]. Specific point mutation functional domains of the 23S rRNA gene usually lead to clarithromycin resistance, which is most frequently located in domain V and domain VI.
In a real clinical setting, the novel fecal PCR-based test has many advantages compared with traditional methods. First, patients only need to provide a small amount (nearly 200 mg) of fresh stool samples rather than undergo gastroscopy. Although special storage containers may be required, the results will be communicated to patients within hours, which certainly facilitates treatment decisions. Furthermore, the cost of fecal PCR is clearly lower than conventional methods for assessing antimicrobial resistance, and no additional expenses of general anesthesia and hospital admission for gastroscopy are needed. This approach could also be used with a wider range of people, including elderly and pediatric patients, and would thus be feasible for clinical application in small- and medium-scale hospitals in developing countries. In addition, this technique can not only provide accurate gene-level information before eradication but also meets the requirements for noninvasive testing for posttreatment follow-up examinations. Resistance to other antibiotics can also be predicted. For example, the rdxA and frxA genes are associated with metronidazole resistance[37,38], 16S ribosomal DNA mutations participate in tetracycline resistance[39], and the role of the gyrA gene in fluoroquinolone-resistant strains has been determined[40].
Overall, PCR-based analysis of stool samples has high diagnostic accuracy for detecting clarithromycin resistance in patients infected with H. pylori. The advantages of these tests include its noninvasiveness, convenience, and low cost compared with traditional detection methods. Therefore, this method could be help improve the eradication rate of H. pylori, especially in regions showing high resistance to clarithromycin.
The eradication rate of Helicobacter pylori (H. pylori) is gradually decreasing due to antibiotic resistance worldwide, in particular clarithromycin resistance.
The detection of clarithromycin resistance is necessary prior to the treatment of H. pylori, accurate data on the feasibility of stool polymerase chain reaction (PCR)-based tests are not available.
We performed a meta-analysis to assess the feasibility of PCR-based tests for detecting H. pylori clarithromycin resistance in stool samples.
We collected cross-sectional studies that met the inclusion criteria. This is the first meta-analysis based on true-positive, false-positive, false-negative, and true-negative test results.
A meta-analysis of the random-effect model showed that PCR-based analysis of stool samples had high diagnostic accuracy for detecting clarithromycin resistance in patients infected with H. pylori.
PCR-based tests on stool samples have high diagnostic accuracy for detecting H. pylori clarithromycin resistance.
This non-invasive, convenient, and inexpensive method can increase the eradication rate of H. pylori, especially in areas with high clarithromycin resistance.
Manuscript source: Unsolicited manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Slomiany BL S-Editor: Gao CC L-Editor: Filipodia P-Editor: Li JH
| 1. | Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311-1315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3302] [Cited by in RCA: 3263] [Article Influence: 79.6] [Reference Citation Analysis (1)] |
| 2. | Sugano K, Tack J, Kuipers EJ, Graham DY, El-Omar EM, Miura S, Haruma K, Asaka M, Uemura N, Malfertheiner P; faculty members of Kyoto Global Consensus Conference. Kyoto global consensus report on Helicobacter pylori gastritis. Gut. 2015;64:1353-1367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1322] [Cited by in RCA: 1183] [Article Influence: 118.3] [Reference Citation Analysis (0)] |
| 3. | McNicholl AG, Bordin DS, Lucendo A, Fadeenko G, Fernandez MC, Voynovan I, Zakharova NV, Sarsenbaeva AS, Bujanda L, Perez-Aisa Á, Vologzhanina L, Zaytsev O, Ilchishina T, Coba C, Lasala JP, Alekseenko S, Modolell I, Molina-Infante J, Ruiz-Zorrilla Lopez R, Alonso-Galan H, Moreno NF, Hinojosa J, Santaella I, Varela P, Gonzalez-Cordero PL, Barrio J, Dominguez-Jimenez JL, Nuñez O, Alcedo J, Nyssen OP, Caldas M, Donday MG, Shvetz O, Megraud F, O'Morain C, Gisbert JP. Combination of Bismuth and Standard Triple Therapy Eradicates Helicobacter pylori Infection in More than 90% of Patients. Clin Gastroenterol Hepatol. 2020;18:89-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 61] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 4. | Savoldi A, Carrara E, Graham DY, Conti M, Tacconelli E. Prevalence of Antibiotic Resistance in Helicobacter pylori: A Systematic Review and Meta-analysis in World Health Organization Regions. Gastroenterology 2018; 155: 1372-1382. e17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 617] [Cited by in RCA: 822] [Article Influence: 117.4] [Reference Citation Analysis (0)] |
| 5. | Chey WD, Leontiadis GI, Howden CW, Moss SF. ACG Clinical Guideline: Treatment of Helicobacter pylori Infection. Am J Gastroenterol. 2017;112:212-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 744] [Cited by in RCA: 1018] [Article Influence: 127.3] [Reference Citation Analysis (1)] |
| 6. | Randel A. H. pylori Infection: ACG Updates Treatment Recommendations. Am Fam Physician. 2018;97:135-137. [PubMed] |
| 7. | Chen H, Dang Y, Zhou X, Liu B, Liu S, Zhang G. Tailored Therapy Versus Empiric Chosen Treatment for Helicobacter pylori Eradication: A Meta-Analysis. Medicine (Baltimore). 2016;95:e2750. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 46] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 8. | Malfertheiner P, Megraud F, O'Morain CA, Atherton J, Axon AT, Bazzoli F, Gensini GF, Gisbert JP, Graham DY, Rokkas T, El-Omar EM, Kuipers EJ; European Helicobacter Study Group. Management of Helicobacter pylori infection--the Maastricht IV/ Florence Consensus Report. Gut. 2012;61:646-664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1719] [Cited by in RCA: 1589] [Article Influence: 122.2] [Reference Citation Analysis (5)] |
| 9. | Fallone CA, Chiba N, van Zanten SV, Fischbach L, Gisbert JP, Hunt RH, Jones NL, Render C, Leontiadis GI, Moayyedi P, Marshall JK. The Toronto Consensus for the Treatment of Helicobacter pylori Infection in Adults. Gastroenterology 2016; 151: 51-69. e14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 695] [Cited by in RCA: 635] [Article Influence: 70.6] [Reference Citation Analysis (0)] |
| 10. | Mégraud F, Lehours P. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin Microbiol Rev. 2007;20:280-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 476] [Cited by in RCA: 488] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 11. | Versalovic J, Shortridge D, Kibler K, Griffy MV, Beyer J, Flamm RK, Tanaka SK, Graham DY, Go MF. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 340] [Cited by in RCA: 331] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 12. | Vester B, Douthwaite S. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob Agents Chemother. 2001;45:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 401] [Cited by in RCA: 394] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 13. | Marais A, Monteiro L, Occhialini A, Pina M, Lamouliatte H, Mégraud F. Direct detection of Helicobacter pylori resistance to macrolides by a polymerase chain reaction/DNA enzyme immunoassay in gastric biopsy specimens. Gut. 1999;44:463-467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 79] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 14. | Maeda S, Yoshida H, Ogura K, Kanai F, Shiratori Y, Omata M. Helicobacter pylori specific nested PCR assay for the detection of 23S rRNA mutation associated with clarithromycin resistance. Gut. 1998;43:317-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 45] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 15. | van Doorn LJ, Glupczynski Y, Kusters JG, Mégraud F, Midolo P, Maggi-Solcà N, Queiroz DM, Nouhan N, Stet E, Quint WG. Accurate prediction of macrolide resistance in Helicobacter pylori by a PCR line probe assay for detection of mutations in the 23S rRNA gene: multicenter validation study. Antimicrob Agents Chemother. 2001;45:1500-1504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 120] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 16. | Osaki T, Mabe K, Zaman C, Yonezawa H, Okuda M, Amagai K, Fujieda S, Goto M, Shibata W, Kato M, Kamiya S. Usefulness of detection of clarithromycin-resistant Helicobacter pylori from fecal specimens for young adults treated with eradication therapy. Helicobacter. 2017;22 . [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 17. | Schabereiter-Gurtner C, Hirschl AM, Dragosics B, Hufnagl P, Puz S, Kovách Z, Rotter M, Makristathis A. Novel real-time PCR assay for detection of Helicobacter pylori infection and simultaneous clarithromycin susceptibility testing of stool and biopsy specimens. J Clin Microbiol. 2004;42:4512-4518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 147] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 18. | Brennan DE, Omorogbe J, Hussey M, Tighe D, Holleran G, O'Morain C, Smith SM, McNamara D. Molecular detection of Helicobacter pylori antibiotic resistance in stool vs biopsy samples. World J Gastroenterol. 2016;22:9214-9221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 38] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (2)] |
| 19. | Yoshitoku Y, Toyonori O. Practice guideline of evidence-based medicine: Preferred Reporting Items for Systematic Reviews and Meta-analyses (the PRISMA statement). Inf Process Manag. 2011;54:254-266. [RCA] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 20. | Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA, Bossuyt PM; QUADAS-2 Group. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6953] [Cited by in RCA: 9580] [Article Influence: 684.3] [Reference Citation Analysis (0)] |
| 21. | Fontana C, Favaro M, Pietroiusti A, Pistoia ES, Galante A, Favalli C. Detection of clarithromycin-resistant Helicobacter pylori in stool samples. J Clin Microbiol. 2003;41:3636-3640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 22. | Noguchi N, Rimbara E, Kato A, Tanaka A, Tokunaga K, Kawai T, Takahashi S, Sasatsu M. Detection of mixed clarithromycin-resistant and -susceptible Helicobacter pylori using nested PCR and direct sequencing of DNA extracted from faeces. J Med Microbiol. 2007;56:1174-1180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 58] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 23. | Rimbara E, Tamura R, Tanuma M, Noguchi N, Kawai T, Sasatsu M. Evaluation of clarithromycin resistance in Helicobacter pylori obtained from culture isolates, gastric juice, and feces. Helicobacter. 2009;14:156-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 24. | Rimbara E, Noguchi N, Yamaguchi T, Narui K, Kawai T, Sasatsu M. Development of a highly sensitive method for detection of clarithromycin-resistant Helicobacter pylori from human feces. Curr Microbiol. 2005;51:1-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 25. | Lottspeich C, Schwarzer A, Panthel K, Koletzko S, Rüssmann H. Evaluation of the novel Helicobacter pylori ClariRes real-time PCR assay for detection and clarithromycin susceptibility testing of H. pylori in stool specimens from symptomatic children. J Clin Microbiol. 2007;45:1718-1722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 57] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 26. | Vécsei A, Innerhofer A, Binder C, Gizci H, Hammer K, Bruckdorfer A, Riedl S, Gadner H, Hirschl AM, Makristathis A. Stool polymerase chain reaction for Helicobacter pylori detection and clarithromycin susceptibility testing in children. Clin Gastroenterol Hepatol. 2010;8:309-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 27. | Scaletsky IC, Aranda KR, Garcia GT, Gonçalves ME, Cardoso SR, Iriya K, Silva NP. Application of real-time PCR stool assay for Helicobacter pylori detection and clarithromycin susceptibility testing in Brazilian children. Helicobacter. 2011;16:311-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 28. | Giorgio F, Ierardi E, Sorrentino C, Principi M, Barone M, Losurdo G, Iannone A, Giangaspero A, Monno R, Di Leo A. Helicobacter pylori DNA isolation in the stool: an essential pre-requisite for bacterial noninvasive molecular analysis. Scand J Gastroenterol. 2016;51:1429-1432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 29. | Banatvala N, Mayo K, Megraud F, Jennings R, Deeks JJ, Feldman RA. The cohort effect and Helicobacter pylori. J Infect Dis. 1993;168:219-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 334] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 30. | Uc A, Chong SK. Treatment of Helicobacter pylori gastritis improves dyspeptic symptoms in children. J Pediatr Gastroenterol Nutr. 2002;34:281-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 32] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 31. | Farrell S, Milliken I, Murphy JL, Wootton SA, McCallion WA. Nonulcer dyspepsia and Helicobacter pylori eradication in children. J Pediatr Surg. 2005;40:1547-1550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 32. | Okamura T, Suga T, Nagaya T, Arakura N, Matsumoto T, Nakayama Y, Tanaka E. Antimicrobial resistance and characteristics of eradication therapy of Helicobacter pylori in Japan: a multi-generational comparison. Helicobacter. 2014;19:214-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 33. | Taneike I, Goshi S, Tamura Y, Wakisaka-Saito N, Matsumori N, Yanase A, Shimizu T, Yamashiro Y, Toyoda S, Yamamoto T. Emergence of clarithromycin-resistant Helicobacter pylori (CRHP) with a high prevalence in children compared with their parents. Helicobacter. 2002;7:297-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 34. | Ierardi E, Giorgio F, Iannone A, Losurdo G, Principi M, Barone M, Pisani A, Di Leo A. Noninvasive molecular analysis of Helicobacter pylori: Is it time for tailored first-line therapy? World J Gastroenterol. 2017;23:2453-2458. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 34] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 35. | Khadangi F, Yassi M, Kerachian MA. Review: Diagnostic accuracy of PCR-based detection tests for Helicobacter Pylori in stool samples. Helicobacter. 2017;22 . [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 36. | Iannone A, Giorgio F, Russo F, Riezzo G, Girardi B, Pricci M, Palmer SC, Barone M, Principi M, Strippoli GF, Di Leo A, Ierardi E. New fecal test for non-invasive Helicobacter pylori detection: A diagnostic accuracy study. World J Gastroenterol. 2018;24:3021-3029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 36] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 37. | Kwon DH, Kato M, El-Zaatari FA, Osato MS, Graham DY. Frame-shift mutations in NAD(P)H flavin oxidoreductase encoding gene (frxA) from metronidazole resistant Helicobacter pylori ATCC43504 and its involvement in metronidazole resistance. FEMS Microbiol Lett. 2000;188:197-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 42] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 38. | Jenks PJ, Ferrero RL, Labigne A. The role of the rdxA gene in the evolution of metronidazole resistance in Helicobacter pylori. J Antimicrob Chemother. 1999;43:753-758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 68] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 39. | Dailidiene D, Bertoli MT, Miciuleviciene J, Mukhopadhyay AK, Dailide G, Pascasio MA, Kupcinskas L, Berg DE. Emergence of tetracycline resistance in Helicobacter pylori: multiple mutational changes in 16S ribosomal DNA and other genetic loci. Antimicrob Agents Chemother. 2002;46:3940-3946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 68] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 40. | Moore RA, Beckthold B, Wong S, Kureishi A, Bryan LE. Nucleotide sequence of the gyrA gene and characterization of ciprofloxacin-resistant mutants of Helicobacter pylori. Antimicrob Agents Chemother. 1995;39:107-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 122] [Article Influence: 4.1] [Reference Citation Analysis (0)] |