Published online Nov 26, 2019. doi: 10.12998/wjcc.v7.i22.3683
Peer-review started: April 1, 2019
First decision: May 30, 2019
Revised: October 23, 2019
Accepted: October 30, 2019
Article in press: October 29, 2019
Published online: November 26, 2019
Processing time: 239 Days and 24 Hours
Colorectal cancer (CRC) is the third most common cancer in men (746000 cases per year) and the second most common cancer in women globally (614000 cases per year). The incidence rate of CRC in developed countries (737000 cases per year) is higher than that in less developed countries (624000 cases per year). CRC can arise from genetic causes such as chromosomal instability and microsatellite instability. Several etiologic factors underlie CRC including age, diet, and lifestyle. Gut microbiota represent a proven cause of the disease, where they play pivotal roles in modulating and reshaping the host epigenome. Several active microbial metabolites have been found to drive carcinogenesis, invasion, and metastasis via modifying both the methylation landscape along with histone structure in intestinal cells. Gut microbiota, in response to diet, can exert both beneficial and harmful functions in humans, according to the intestinal balance of number and types of these bacteria. Although the intestinal microbial community is diverse among individuals, these microbes cumulatively produce 100-fold more proteins than the human genome itself, which calls for further studies to elaborate on the complicated interaction between these microorganisms and intestinal cells. Therefore, understanding the exact role that gut microbiota play in inducing CRC will help attain reliable strategies to precisely diagnose and treat this fatal disease.
Core tip: Colorectal cancer is serious disease that affects males and females late in their lives. Several etiologic factors trigger colorectal cancer; however, the gut microbiome is responsible of most of the cases. Gut bacteria can produce a variety of chemical compounds that affect intestinal cells and might transform them into malignant ones. In this review, we describe the main mechanisms by which gut microbiota exert these functions.
- Citation: Sabit H, Cevik E, Tombuloglu H. Colorectal cancer: The epigenetic role of microbiome. World J Clin Cases 2019; 7(22): 3683-3697
- URL: https://www.wjgnet.com/2307-8960/full/v7/i22/3683.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v7.i22.3683
Colorectal cancer (CRC) is one of the primary causes of cancer-related deaths globally[1]. It occurs as a result of complicated sequences involving mutation accumulation that is either genetic or epigenetic[2]. The process of CRC carcinogenesis is a quite slow, starting with minor inflammation followed by adenomatous polyps in the epithelium, and finally adenocarcinoma[3]. Given the crucial role of epigenetic changes in developing CRC, 95% of cases are sporadic i.e. appear in patients with no family history for the disease. Meanwhile, minor ratio (3%) is attributed to hereditary nonpolyposis CRC, and 2% of cases are caused by other hereditary disorders such as MYH-associated polyposis and familial adenomatous polyposis[2,4].
Microorganisms occupy almost every part of the human body, armed with a huge number of genes, where it could interact, modulate, or disrupt a wide array of human genes especially in colonic cells[5]. Interestingly, the human microbiome encodes for approximately 100-fold more proteins than the human genome itself. This microbiota comprise 1000 to 1500 bacterial species, and the composition of the microbiome is significantly diverse among individuals[6]. These species belong to just a few phyla: Firmicutes, Bacteroidetes (most prevalent), Proteobacteria, Verrucomicrobia, Actinobacteria, Fusobacteria, and Cyanobacteria[7]. Although distribution of the microbiota in terms of types and number is common in healthy individuals, it differs significantly in diseased persons. In addition to bacteria that compose the gut microbiome, eukaryotic fungal species have recently been found to co-exist with bacterial species, the major component of the microbiome[8].
It is well established that gut microbiota play critical roles in the progression of CRC either via their metabolites or interaction with their host intestinal epithelial cells[9]. Imbalance of this microbiota has been associated with several disorders including inflammatory bowel disease and CRC[10]. Nonetheless, several studies have related the changes in microbiota to a cause of disease, while others have indicated that these changes are merely a result; however, this issue demands further investigation[11]. In this review, we highlight the recent studies that addresses the causal link between gut microbiota and CRC onset and progression. Meanwhile, the epigenetic changes underlying CRC and its microbial root will also be described.
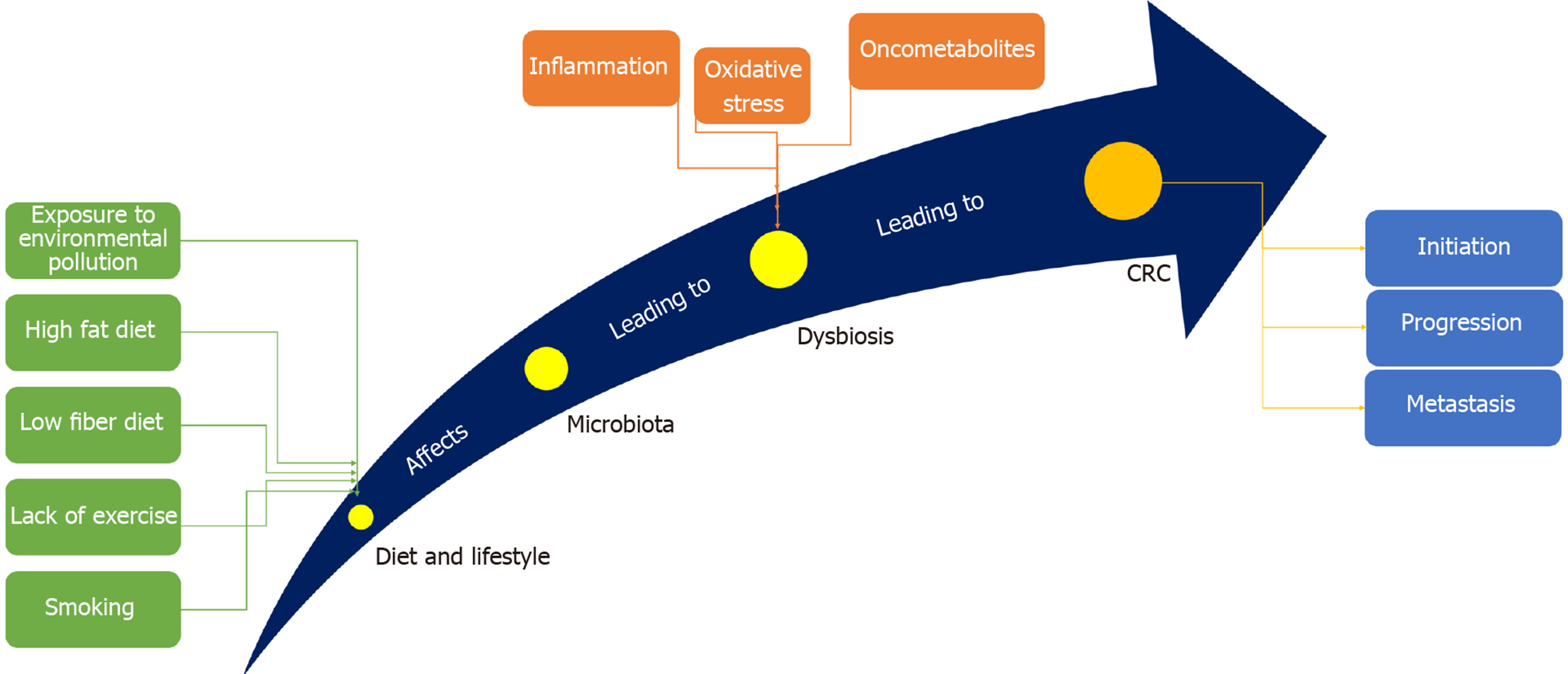
CRC is one of the most prevalent malignant tumors and the third most common cause of cancer-related death worldwide[12]. It is the third most common malignancy in males and the second in females, with a lifetime risk of about 6%[13]. Being well-developed, CRC can metastasize -even after operation- to distant organs such as the liver and lungs, forming secondary CRC[14]. The common risk factors underlying CRC involve genetics[15], environmental pollution[16], diet[17], age[18], alcohol consumption[19], smoking[20], obesity[21], and physical inactivity[22], with gut microbiota standing alone as a potent risk factor[23] (Figure 1). It is well established that CRC arises due to accumulation of genetic mutations. Large studies showed that approximately 13000 mutations in 67 genes correlate with CRC. Among them, only 12 genes were found to be closely related to CRC[24]. Different types of genomic instability predispose patients to CRC including microsatellite instability (MSI), in which frequent insertions and deletions are prevalent, and chromosomal instability (CIN), in which gain or loss of chromosomes prevail[25]. CIN is responsible for about 85% of CRC cases, where loss of chromosomal segment/arm includes 15q11-q21, 17p12-13, and18q12-21 and gain of chromosomal segment/arm includes 1q32, 7p, 7q, 8q, 13q, 20p, and 20q[26,27].
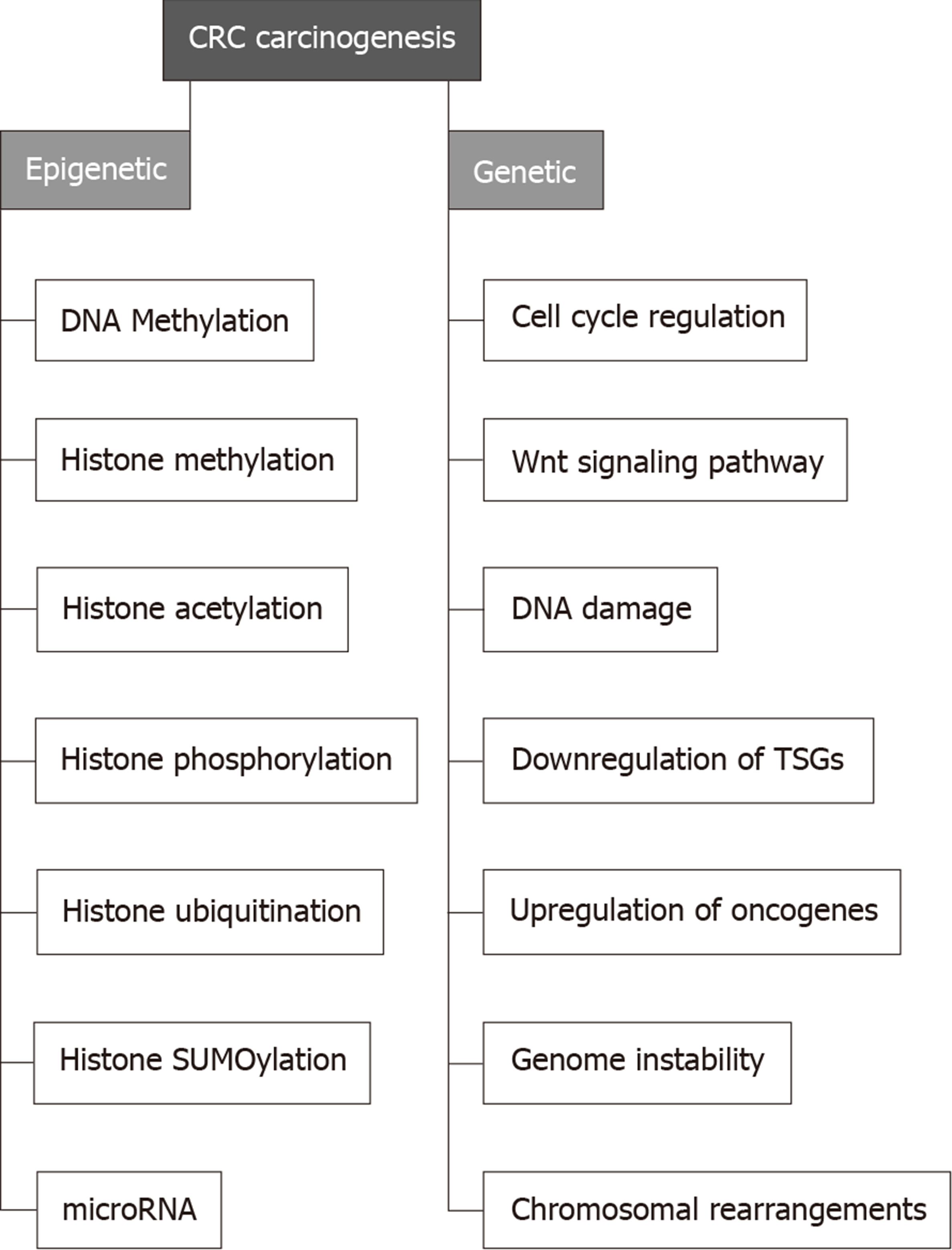
Several genes are directly correlated with CRC. Examples include APC in which inactivation leads to triggering the Wnt signaling pathway to initiate colon polyps which can be benign (e.g., hyperplastic polyp), pre-malignant (e.g., tubular adenoma) or malignant (e.g., colorectal adenocarcinoma)[28]. Furthermore, transforming growth factor receptor 2 TGFBR2 is involved in almost 30% of CRC cases. The downstream effector of this genes, i.e. KRAS was found to be activated in 55%-60% of CRC cases. Mismatch repair genes such as MLH1, MSH6, MSH2, and PMS2 causing frameshift mutations were found to induce MSI, triggering the development of CRC[29] (Figure 2).
Epigenetic regulation of gene expression analysis is a validated tool to correlate gene expression changes with cancer development[2,30]. Through the last three decades, solid common knowledge has been established to indicate that the perturbation of epigenetic mechanisms leads to cancer initiation and progression[29,31]. Identification of CRC epigenetic changes has revealed that almost all CRCs have abnormally methylated genes. Although rare data have been provided to highlight the pattern of specific histone modifications in CRC, certain histone modifications (such as acetylation, methylation, and phosphorylation) have been found to work in harmony with DNA methylation to regulate CRC-related gene expression that is involved in cancer initiation and progression[1,32]. Therefore, a deep understanding of epigenetic changes related to CRC pathogenesis might help develop epigenetic-based biomarkers for CRC diagnosis and prognosis, and hence, epigenetic-based therapy[29].
In the normal adult person, the gut microbiota comprise approximately 1014 bacterial cells that live in commensalism with the host, where they substantially facilitate various aspects in the host health and disease[33]. The normal gut microbiota are rich in anaerobic bacteria, which are 100- to 1000-times more than aerobic and facultative anaerobic bacteria, respectively[34]. The colon has a reductive environment devoid of oxygen, which allows Bacteroidetes and Firmicutes to be the dominant phyla followed by Actinobacteria and Verrucomicrobia[35]. For bacteria, the colon represents a suitable niche as it provides them with elevated pH, nutrients, and low concentration of bile salts and pancreatic secretions. These conditions, indeed, are favored by bacteria to flourish and proliferate[10]. Commensal bacteria start colonization of the host during birth and continue to variate in number and type along with the host development[36]. It is well established that our gut microbiome is responding to any dietary shift, where switching from polysaccharides-rich diets to that high in animal fat eventually leads to a hasty shift in the gut microbiome[37]. Commonly in the gut, the prevailed microbial product is lipopolysaccharide (LPS), produced by Gram-negative bacteria, function to stimulate the innate immunity, thus, protecting against inflammation that leads to cancer[38] (Table 1).
| Microorganism | Role in CRC initiation/progression | Ref. |
| Lactobacillus casei BL23 | Immunomodulatory effect via downregulation of the IL-22, and an antiproliferative effect, via upregulation of caspase-7 and caspase-9 | [109] |
| Escherichia coli NC101 | Production of colibactin that induces CRC | [110] |
| Fusobacterium nucleatum | Activation of β-catenin signaling and induction of oncogenic gene expression that promotes growth of CRC cells via the FadA adhesion virulence facto. It produces also the autotransporter protein, Fap2, that has been shown to potentiate the progress of CRC via inhibiting immune cell activity | [111] |
| Eubacterium rectale | Production of butyrate to induce IL-10, the anti-inflammatory cytokine | [112] |
| Bacteroides fragilis | Production of Enterotoxigenic Bacteroides fragilis (ETBF) toxin that promotes CRC by modulating the mucosal immune response and inducing epithelial cell changes. ETBF stimulates E-cadherin cleavage and facilitates cell tumor metastasis | [113] |
| Streptococcus bovis | Triggering of inflammations, bacteremia, and endocarditis, that leads ultimately to colorectal cancer | [114] |
| Clostridium septicum | Production of alpha toxin that binds GPI-anchored cell surface receptors including the human folate receptor as well as the neuronal molecules contactin and Thy-1 (CD90) | [115] |
| Enterococcus faecalis | Damaging the colonic epithelial cell DNA | [116] |
| Bifidobacterium | Production of β-galactosidases, which has antitumor activity | [117] |
| Helicobacter pylori | Induction of inflammatory responses, alteration of gut microflora and release of gastrin, which may contribute to tumor formation | [118] |
| Faecalibacterium prausnitzii | Production of butyrate to induce IL-10, the anti-inflammatory cytokine that protects against cancer formation | [119] |
| Enterotoxigenic bacteroides | Induction of early-stage carcinogenic, that might lead to early colorectal carcinogenesis | [113] |
| Clostridium nexile | Contribution to the anticancer effect of Pseudomonas aeruginosa. It improves also malnutrition in infants | [120] |
| Fusobacterium varium | Activate the E-cadherin/β-catenin signaling pathway and association with epigenetic phenotype, such as microsatellite instability and hypermethylation, via its strong adhesive and invasive abilities resulting in malignant transformation of epithelial cells | [121] |
| Actinomyces odontolyticus | Causes colon actinomycosis only when the epithelial barrier was perished | [122] |
| Veillonella dispar | Might be able to enhance the dosage response to CRC chemotherapeutic agents or reduce the side effects of these drugs | [123] |
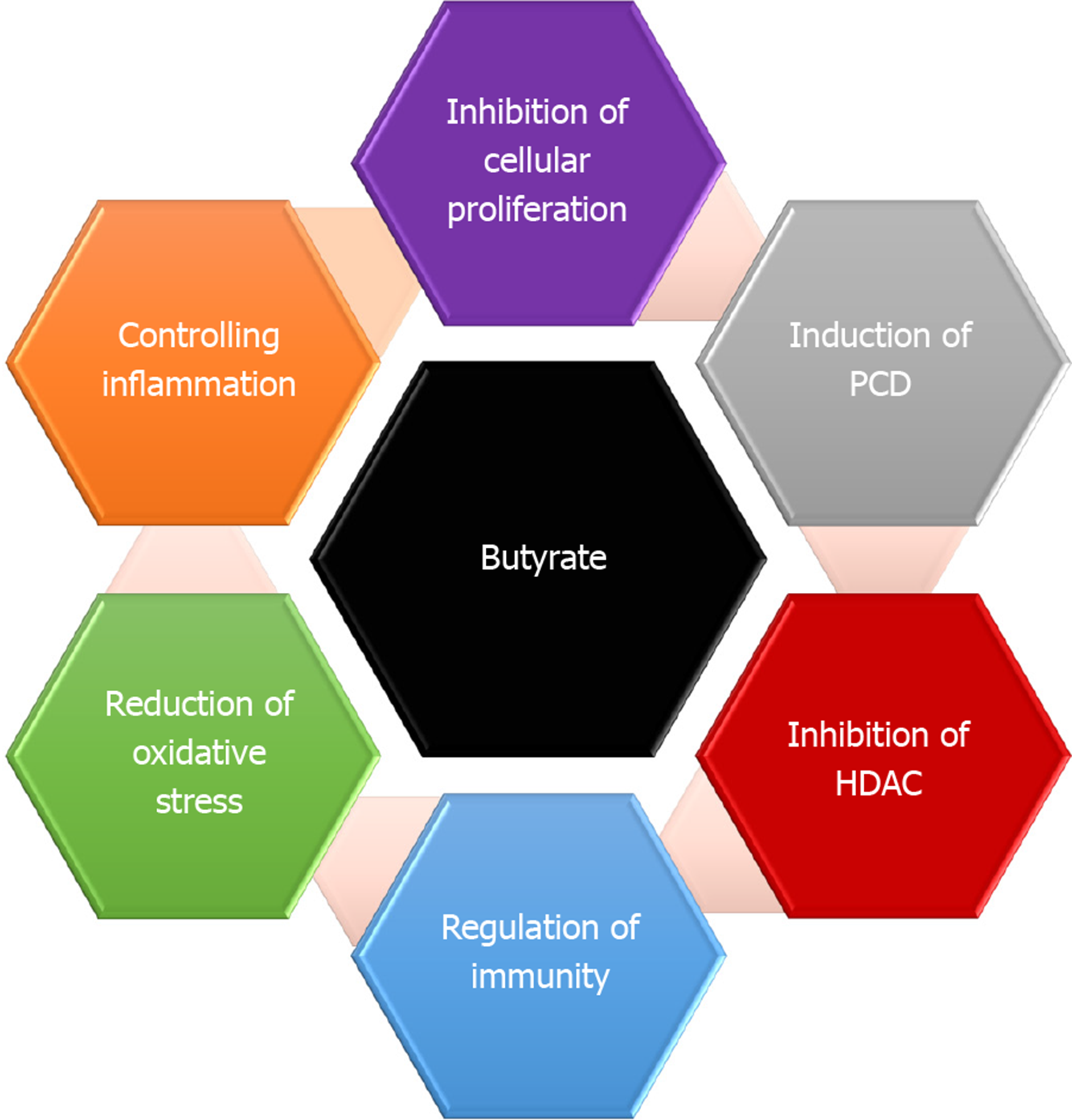
Gut microbiota is crucial for numerous characteristics of host biology[10,39]. They enable the host to digest and metabolize indigestible polysaccharides[40]. The gut microbiota plays an important role in maintaining gut homeostasis[41]. Furthermore, gut microbial community also participates effectively in the normal cellular proliferation. To keep its habitat for millions of years, several gut microbiota essentially protect the host against CRC[42]. Reports have indicated that enterotoxigenic Bacteroides fragilis is capable of induce apoptosis in CRC cells[42]. Generally, diet is metabolized by microbiota into potent oncometabolites and tumor-suppressive metabolites. Whereas, the same microbiota can digest fiber into short-chain fatty acids (SCFAs)[43]. It is well known that SCFAs have anti-inflammatory properties and can increase the level of colonic regulatory T cells (Tregs) and protect the host against colitis[43,44]. The most common SCFAs produced in the gut are acetate, propionate, and butyrate[45]. Butyrate is one of the important sources of energy, where it provides the cells with 5%-15% of its caloric requirements. Faecalibacterium prausnitzii and Eubacterium rectale are the main gut bacterial species that produce butyrate[44,46]. Butyrate controls cell proliferation, differentiation, and apoptosis among other functions in colon cells[23,47]. It exerts also a preventive role where it ameliorates the harmful effects of N-nitroso compounds, a product that accumulates in the colon upon intake of heat-treated and processed meat[48] (Figure 3). It has been indicated that Clostridium species enhances Treg cell abundance by increasing the production of potent anti-inflammatory molecules such as cytokine interleukin 10 (IL-10)[49]. Lactic acid bacteria have also shown protective properties against CRC via different mechanisms that include strengthening the mucosal barrier and altering luminal secretions, resulting in underpinning of the host immune system. Ursodeoxycholic acid (UDCA, ursodiol) is a metabolic byproduct of intestinal bacteria, with a chemical structure that resembles deoxycholic acid (DCA)[50]. While DCA promotes the initiation of CRC, UDCA function to prevent the disease. It was reported that UDCA inhibits the expression of cyclooxygenase-2 (COX-2) by Ras-dependent and RAS-independent mechanisms in CRC cells[51]. UDCA prevents colon cells from the harmful effect of DCA via inhibiting the DCA-induced extracellular signal-regulated kinase and Raf-1 kinase activity and the activation of epidermal growth factor receptor (EGFR)[52].
Microbiota-mediated carcinogenesis is a complex process that takes place through changing host cell proliferation, influencing the host cell immune system, and metabolizing dietary factors[53]. A plethora of research has suggested that an imbalance in normal intestinal microbiota can trigger inflammatory conditions by producing carcinogenic metabolites that lead to cancer formation, and about 16% of human cancers are triggered by bacteria[36,53]. Gut bacteria can attack intestinal epithelial cells to cause inflammation, that in turn, increase the risk of developing CRC[54]. For CRC to occur, the microbiota-host interaction must be dysregulated, resulting in disruption of cellular homoeostasis[55]. The major component of the gut immune system, Peyer’s patch, is robustly influenced by gut microbiota[56]. The host diet can trigger gut microbiota to be involved in the early stages of CRC carcinogenesis[57]. Upon metabolism of saturated fatty acid- and sugar-rich diets, gut bacteria produce harmful procarcinogenic products including polyamines hydrogen sulfide, secondary bile acids such as DCA and lithocholic acid (LCA), and reactive oxygen species (ROS), which induce chronic inflammation, and hence elevate the susceptibility of cells to develop CRC[58]. DCA is a metabolite of the gut microbiota that induces cancer stemness by regulating the muscarinic 3 receptor/Wnt intracellular signaling pathway[59]. It can also trigger the production of Nur77 protein, which is positively correlated with CRC when upregulated[60]. Meanwhile, DCA induces CRC via downregulation of miR-199a-5p that degrades CAC1, the tumor suppressor gene plays a role in CRC[61]. LCA (aka 3α-hydroxy-5β-cholan-24-oic acid), a secondary bile acid synthesized by gut microbiota, is verified to a promoter of CRC[62]. Gut bacteria produce LCA by utilizing DCA[63]. Both LCA and DCA can enhance cancer stemness[64]. Furthermore, LCA and DCA activate the EGFR signaling pathway, inducing DNA damage, and causing oxidative stress, apoptosis, mutation, and activation of the protein kinase C pathway[59].
Trimethylamine (TMA) is solely synthesized by gut microbiota (in humans) from various dietary nutrients including choline and carnitine (found in red meat)[65]. It reacts with flavin monooxygenase to produce trimethylamine-N-oxide (TMAO), a microbial metabolite involved in CRC progression[66]. A high incidence rate of CRC was noticed in omnivorous individuals, as they produce more TMAO compared to vegans and vegetarians who show low incidence rate[67]. The genetic pathway by which TMAO triggers CRC remains unclear.
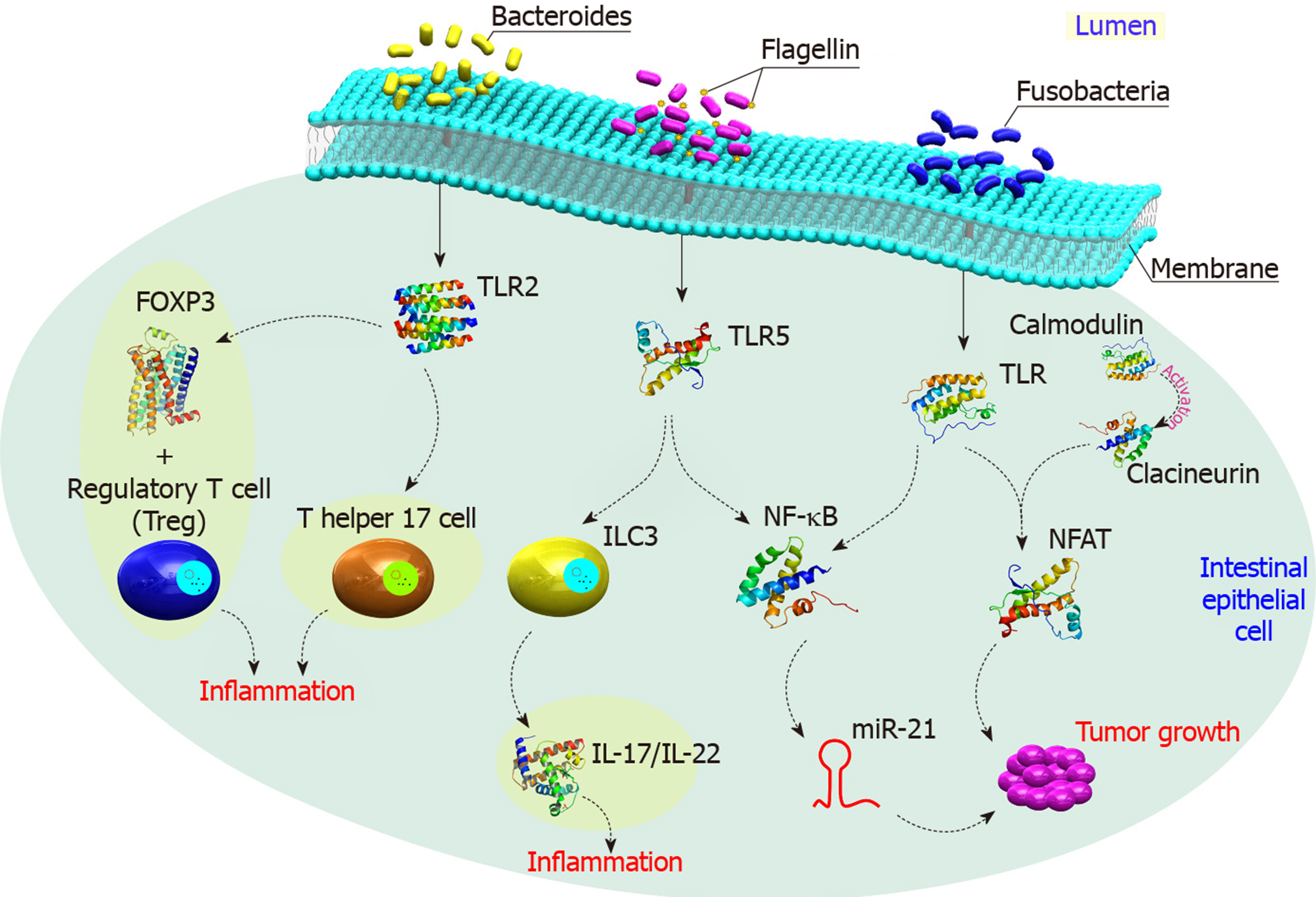
Furthermore, specific gut bacteria such as Enterococcus faecalis was found to induce COX-2, that generates pro-proliferative signals through prostaglandin E(2) (PGE2)[68]. Several Gram-negative bacteria produce LPS that activates TLR4, COX-2, and then PGE2 leading to inhibition of programmed cell death and increase cell proliferation[69]. Moreover, there is an increased resistance to macrophage killing and MAPK activation in those who have the pks (polyketide synthase) island in E. coli isolated from CRC patients[70]. Activated TLR also enhances angiogenesis through MAPK and NF-κB signaling networks[71] (Figure 4).
Other CRC-related bacterial metabolites were highlighted including fragilysin[72]. This extracellular 20 kDa zinc-dependent metalloproteinase metabolite, produced by B. fragilis, could hydrolyze the extracellular domain of E-cadherin and activate the β-catenin nuclear signaling, leading to induction of CRC[73]. Meanwhile fragilysin can damage the tight junction of the intestines, increases intestinal permeability[74]. Colibactin is a bacterial-derived genotoxin first reported in 2006 by Nougayrede and colleagues. It is a hybrid polyketide-non ribosomal peptide produced through an intricate biosynthetic mechanism and encoded by the pks pathogenicity island[75]. E. coli strains harboring this pks island were found to be associated with CRC[76]. Moreover, colibactin, a kind of bacterial toxin synthetized by the pks genomic island can trigger chromosomic instability and DNA damage that might lead eventually to CRC[77].
Several epigenetic changes are common in CRC including DNA methylation, histone modification, and miRNA-mediated post-transcriptional regulation[28,78]. Abnormal epigenetic modifications (AKA epimutations) occur in the promoter regions of tumor-suppressor genes and proto-oncogenes. These epimutations were reported in several malignancies including CRC, where many genes such as MLH1, LKB1, APC, p16INK4a, and GATA4 represent common targets[2,40,79]. Microbial community in our guts are armed with an arsenal of genes that produces millions of proteins, let alone their outpouring of metabolites[67,80]. This microbiota produces low-molecular-weight substances that interact within the human cells with different targets to trigger genomic and epigenomic changes[81]. Research teams everywhere highlight the association between gut microbiota and human diseases; thus, all these diseases should be revisited once again to elucidate the actual role played by microbial community. Being very stable, DNA might not be affected by microbial metabolites, and this is pointing to a more fragile component in our cells; epigenome.
Linking diseases to epigenetic changes was first addressed in 1983[82]. Based on that first note, numerous researches indicated that cancer cells undergo global hypomethylation along with site-specific hypermethylation in the promotors of cancer-related genes[83]. A bunch of reports have indicated that the microbial metabolites can modulate epigenetic landscape of the host gut’s cells via modifying the methylation pattern of cancer-related genes, as they represent a validated source of microbial-induced epigenetic change. Thus, the deep understanding of how epigenetic modifications influenced by the gut microbiota take place could offer possible therapeutic targets to prevent and treat CRC[10,80].
In DNA methylation, DNMTs add methyl group (CH3) to the fifth carbon atom in the cytosine residue using the intracellular methyl substrate S-Adenosyl methionine (SAM) as a methyl donor to convert the normal cytosine into 5-methylcytosine (5-mC)[84]. Meanwhile, ten-eleven translocation enzyme can reverse this process via the oxygenation of 5-mC to produce 5-hydroxymethylcytosine[85]. It is well known that folate is the main source of SAM, and this vitamin could be produced by Bifidobacterium and Lactobacillus, common probiotic bacteria[86]. Folate is required for DNA methylation (5-methyltetrahydrofolate) or DNA synthesis (5-formyltetrahydrofolate and 5, 10-methenyltetrahydrofolate)[87]. A study indicated that volunteers administered Bifidobacterium showed a high concentration of folate in their feces, meaning that these probiotic bacteria were capable to generate it and hence, affect DNA methylation pattern[88]. On the other hand, deficiency of folate synthesis might contribute to DNA hypomethylation, which is an established phenomenon in CRC[89]. Meanwhile, pathogenic bacteria such as Helicobacter pylori that infects the stomach and causes gastritis or gastric ulcers or in severe infection gastric cancer can induce several epigenetic changes[90]. A comparison between gastric biopsies excised from patients with gastritis (upon infection with Helicobacter pylori) and healthy individuals showed that chronic gastritis was associated with promoter hypermethylation of E-cadherin (CDH1), MGMT, WIF1, and MLH1[91]. Although studies that address the relationship between the microbiome and epigenetic changes in CRC are rare, a population-based study reported that Fusobacterium nucleatum was associated with genomic hypermutation independent of CIMP and BRAF mutations[92]. Other study indicated that Fusobacterium was correlated with the CIMP phenotype, wild-type TP53, hMLH1 methylation, genomic hypermutation, and CHD7/8 mutation[93]. These studies strongly suggest the contribution of F. nucleatum to the epigenetic changes.
In addition to DNA methylation changes, histone modification patterns are also altered in human cancers[84,94]. Some bacterial metabolites such as short chain fatty acids (butyrate and acetate) can induce epigenetic changes in colonic cells[23,45]. Butyrate, a byproduct of the fermentation process of undigested dietary carbohydrates and proteins carried out by Firmicutes, has been shown to regulate over 4000 genes including many involved in apoptosis and cell cycle regulation[95]. It is known also to inhibit histone deacetylases and induce hyperacetylation of histones, that lead to changes in the expression of critical cell cycle regulatory genes such as CCND3 and CDKN1A in intestinal cells. Butyrate triggers epithelial generation of ROS and function also to suppress NF-kB, the protein complex controls DNA transcription[44,96]. Furthermore, Bacteroides thetaiotaomicron stimuli the inflammatory signaling by inhibiting NF-κB pathway through binding to IkB (inhibitor of κB), inhibitory component of the NF-κB pathway[97]. It was reported that infection with Listeria monocytogenes (L. monocytogenes) can cause deacetylation of histone H3K18 in many genes in colonic cells such as SMAD1, IRF2, SMARCA2 and CXCL12[98]. L. monocytogenes execute the deacetylation process by translocating NAD-dependent deacetylase sirtuin 2 to the host nucleus. By doing so, L. monocytogenes epigenetically regulates cell cycle-related genes and modulate the host immune response to enable its invasion[99].
MiRNAs are a class of small single-stranded non-coding RNA molecules that are evolutionarily conserved and encoded by nearly 1% of the genome in most species[100]. MiRNAs were found to be involved in initiation, progression, and metastasis of CRC, where it regulate of various cancer-related gene expression at the post-transcriptional level[101]. Deregulated miRNAs identified in different types of cancers might put us a step forward towards understanding the tumor microenvironment, which necessitate deep investigation of their actual role in cancer progression and spreading[102]. Numerous miRNAs were found to be associated with CRC, such as miR-21, Let-7, miR-145, miR-221, miR-17-19 cluster, and miR-143[103]. Table 2 highlights some of miRNAs related to CRC development, progression, and metastasis. Studies addressed the expression levels of different miRNA in CRC, reported that miR-31, miR-20, miR-25, miR-223, miR-133b, miR-92, miR-93, miR-135a, miR-203, miR-183, and miR-17 were upregulated in CRC, while miR-26b, miR-192, miR-145, let-7a, miR-143, miR-215, miR-16, and miR-191 were downregulated in patients with CRC[13,104]. Some miRNAs were suggested to serve as diagnostic markers for CRC, including miR-133a, miR-145, miR-484-5p, miR-139, miR-143, and miR-106a[105], while another study indicated different set of miRNAs that could be used as biomarkers, including miR-125b, miR-125a, miR-143miR-30a-3p, and miR-145[106]. However, this variation in miRNA list might be attributed to the samples used in the identification process (cell line, tissue, blood or stool) and to the techniques employed. Reports also highlight the role of human diet in modulating the expression of miRNA[107]. For example, butyrate was found to regulate the expression of Let-7, miR-18-106a, miR-25-106b, and miR-17-92a in CRC[96]. The later miRNA cluster (miR-17-92a) was found to be associated with c-Myc to inhibit the activity of PTEN and promotes PI3K-Akt-mTOR axis raising the cell survival in early stage adenoma in CRC[108].
| miRNA | Role in CRC | Ref. |
| miR-15a/miR-16 | Their low expression levels were associated with poor disease-free survival and overall survival | [124] |
| miR-17-5p | Its high expression was associated with disease-free survival | [125] |
| miR-21 | Its high level of expression was associated with poor survival and poor therapeutic outcomes | [126] |
| miR-29a | Its elevated level of expression was associated with a longer disease-free survival in stage II CRC patients | [127] |
| miR-34a-5p | Its high expression was correlated with disease-free survival | [128] |
| miR-106a | Its downregulation was associated with shortened overall survival | [125] |
| miR-132 | Its decreased expression level was associated with poorer overall survival and occurrence of distant metastasis especially in liver | [129] |
| miR-150 | Its elevated expression level was associated with longer overall survival. While its low level of expression was associated with poor therapeutic outcome in patients treated with 5-Fluro uracil | [130] |
| miR-195 | Its low expression rate was associated with occurrence of lymph node metastasis and advanced tumor grade/stage | [131] |
| miR-199b | Increased in metastatic CRC tissue compared with non-metastatic CRC tissue. Furthermore, its low expression was associated with longer overall survival | [132] |
| miR-203 | Its elevated expression level was associated with advanced TNM staging and poorer overall survival | [130] |
| miR-320e | Its high expression was associated with poorer overall survival in stage III colon cancer patients | [133] |
| miR-429 | Its overexpression was associated with overall survival; low level of expression was associated with response to 5-Fluro uracil-based chemotherapy | [134] |
| miR-494 | Its elevated expression was associated with shorter DFS and overall survival | [135] |
| miR-625-3p | High expressions were associated with higher overall survival and enhanced response to therapy | [136] |
Gut microbiota is an enhancer to our second brain; the intestine. With millions of proteins expressed by the microbiota’s arsenal, human could make use of various kinds of dietary ingredients, that otherwise will be rubbish-in/rubbish-out. Although genetic factors and age play a role in the pathogenesis of CRC, still gut microbiota has the lion’s share in this complicated process. Armed with an enormous number of identified and yet-to-be-identified metabolites, this population of bacteria can modify the gut’s cells methylation pattern and histone structure causing inflammation, that lead eventually to cancer. It is quite important to keep these microorganisms under focus by deeply investigate their intricate communications with our cells. By doing so, we would be able to avoid at least life-threatening diseases such as CRC[1].
Manuscript source: Invited manuscript
Specialty type: Medicine, Research and Experimental
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): E
P-Reviewer: Demetrashvili Z, Garg P, Ng QX S-Editor: Dou Y L-Editor: Filipodia E-Editor: Liu JH
| 1. | Kalady MF, Boland CR, Church JM. Chapter 165 - Inherited Colorectal Cancer and the Genetics of Colorectal Cancer. In: Yeo CJ. Shackelford's Surgery of the Alimentary Tract, 2 Volume Set (Eighth Edition). Philadelphia: Content Repository Only 2019; 1959-1980. [DOI] [Full Text] |
| 2. | Baretti M, Azad NS. The role of epigenetic therapies in colorectal cancer. Curr Probl Cancer. 2018;42:530-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 3. | Wang X, Yu H, Sun W, Kong J, Zhang L, Tang J, Wang J, Xu E, Lai M, Zhang H. The long non-coding RNA CYTOR drives colorectal cancer progression by interacting with NCL and Sam68. Mol Cancer. 2018;17:110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 112] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 4. | Lei Z, Ma X, Li H, Zhang Y, Gao Y, Fan Y, Li X, Chen L, Xie Y, Chen J, Wu S, Tang L, Zhang X. Up-regulation of miR-181a in clear cell renal cell carcinoma is associated with lower KLF6 expression, enhanced cell proliferation, accelerated cell cycle transition, and diminished apoptosis. Urol Oncol. 2018;36:93.e23-93.e37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 5. | Serino M. Molecular Paths Linking Metabolic Diseases, Gut Microbiota Dysbiosis and Enterobacteria Infections. J Mol Biol. 2018;430:581-590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 6. | Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768-1773. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 875] [Cited by in RCA: 767] [Article Influence: 51.1] [Reference Citation Analysis (0)] |
| 7. | Sommer F, Bäckhed F. The gut microbiota--masters of host development and physiology. Nat Rev Microbiol. 2013;11:227-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2011] [Cited by in RCA: 2407] [Article Influence: 200.6] [Reference Citation Analysis (0)] |
| 8. | Górska A, Peter S, Willmann M, Autenrieth I, Schlaberg R, Huson DH. Dynamics of the human gut phageome during antibiotic treatment. Comput Biol Chem. 2018;74:420-427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 9. | Wang J, Lu R, Fu X, Dan Z, Zhang YG, Chang X, Liu Q, Xia Y, Liu X, Sun J. Novel Regulatory Roles of Wnt1 in Infection-Associated Colorectal Cancer. Neoplasia. 2018;20:499-509. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 10. | Kahouli I, Tomaro-Duchesneau C, Prakash S. Probiotics in colorectal cancer (CRC) with emphasis on mechanisms of action and current perspectives. J Med Microbiol. 2013;62:1107-1123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 89] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 11. | Harris RA, Shah R, Hollister EB, Tronstad RR, Hovdenak N, Szigeti R, Versalovic J, Kellermayer R. Colonic Mucosal Epigenome and Microbiome Development in Children and Adolescents. J Immunol Res. 2016;2016:9170162. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 12. | Herbst CL, Miot JK, Moch SL, Ruff P. Access to colorectal cancer (CRC) chemotherapy and the associated costs in a South African public healthcare patient cohort. J Cancer Policy. 2018;15:18-24. [RCA] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 13. | Zullig LL, Smith VA, Jackson GL, Danus S, Schnell M, Lindquist J, Provenzale D, Weinberger M, Kelley MJ, Bosworth HB. Colorectal Cancer Statistics From the Veterans Affairs Central Cancer Registry. Clin Colorectal Cancer. 2016;15:e199-e204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 14. | Jacobsen A, Bosch LJW, Martens-de Kemp SR, Carvalho B, Sillars-Hardebol AH, Dobson RJ, de Rinaldis E, Meijer GA, Abeln S, Heringa J, Fijneman RJA, Feenstra KA. Aurora kinase A (AURKA) interaction with Wnt and Ras-MAPK signalling pathways in colorectal cancer. Sci Rep. 2018;8:7522. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 15. | Brosens LA, Offerhaus GJ, Giardiello FM. Hereditary Colorectal Cancer: Genetics and Screening. Surg Clin North Am. 2015;95:1067-1080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 16. | López-Abente G, García-Pérez J, Fernández-Navarro P, Boldo E, Ramis R. Colorectal cancer mortality and industrial pollution in Spain. BMC Public Health. 2012;12:589. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 17. | Baena R, Salinas P. Diet and colorectal cancer. Maturitas. 2015;80:258-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 138] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 18. | Marshall ML, Roberts M, Susswein LR, Mcgill AK, Xu Z, Klein RT, Hruska KS. High prevalence of pathogenic variants in individuals with colorectal cancer ≤ age 35. J Clin Oncol. 2018;36:576-576. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 19. | Phipps AI, Baron J, Newcomb PA. Prediagnostic smoking history, alcohol consumption, and colorectal cancer survival: the Seattle Colon Cancer Family Registry. Cancer. 2011;117:4948-4957. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 87] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 20. | Botteri E, Iodice S, Bagnardi V, Raimondi S, Lowenfels AB, Maisonneuve P. Smoking and colorectal cancer: a meta-analysis. JAMA. 2008;300:2765-2778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 510] [Cited by in RCA: 577] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 21. | Bardou M, Barkun AN, Martel M. Obesity and colorectal cancer. Gut. 2013;62:933-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 474] [Cited by in RCA: 587] [Article Influence: 48.9] [Reference Citation Analysis (0)] |
| 22. | Silva DAS, Tremblay MS, Souza MFM, Mooney M, Naghavi M, Malta DC. Mortality and years of life lost by colorectal cancer attributable to physical inactivity in Brazil (1990-2015): Findings from the Global Burden of Disease Study. PLoS One. 2018;13:e0190943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 23. | Bishop KS, Xu H, Marlow G. Epigenetic Regulation of Gene Expression Induced by Butyrate in Colorectal Cancer: Involvement of MicroRNA. Genet Epigenet. 2017;9:1179237X17729900. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 24. | Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, Raymond CK, Roberts BS, Juhl H, Kinzler KW, Vogelstein B, Velculescu VE. The colorectal microRNAome. Proc Natl Acad Sci USA. 2006;103:3687-3692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 710] [Cited by in RCA: 735] [Article Influence: 38.7] [Reference Citation Analysis (0)] |
| 25. | Jass JR. Molecular heterogeneity of colorectal cancer: Implications for cancer control. Surg Oncol. 2007;16 Suppl 1:S7-S9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 70] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 26. | Hermsen M, Postma C, Baak J, Weiss M, Rapallo A, Sciutto A, Roemen G, Arends JW, Williams R, Giaretti W, De Goeij A, Meijer G. Colorectal adenoma to carcinoma progression follows multiple pathways of chromosomal instability. Gastroenterology. 2002;123:1109-1119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 233] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 27. | Douglas EJ, Fiegler H, Rowan A, Halford S, Bicknell DC, Bodmer W, Tomlinson IP, Carter NP. Array comparative genomic hybridization analysis of colorectal cancer cell lines and primary carcinomas. Cancer Res. 2004;64:4817-4825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 144] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 28. | Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4616] [Cited by in RCA: 4464] [Article Influence: 120.6] [Reference Citation Analysis (0)] |
| 29. | Söreide K, Janssen EA, Söiland H, Körner H, Baak JP. Microsatellite instability in colorectal cancer. Br J Surg. 2006;93:395-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 174] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 30. | Nguyen HT, Duong HQ. The molecular characteristics of colorectal cancer: Implications for diagnosis and therapy (review). Oncol Lett. 2018;16:9-18. [RCA] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 128] [Article Influence: 18.3] [Reference Citation Analysis (1)] |
| 31. | Luo N, Nixon MJ, Gonzalez-Ericsson PI, Sanchez V, Opalenik SR, Li H, Zahnow CA, Nickels ML, Liu F, Tantawy MN, Sanders ME, Manning HC, Balko JM. DNA methyltransferase inhibition upregulates MHC-I to potentiate cytotoxic T lymphocyte responses in breast cancer. Nat Commun. 2018;9:248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 188] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 32. | Druliner BR, Wang P, Bae T, Baheti S, Slettedahl S, Mahoney D, Vasmatzis N, Xu H, Kim M, Bockol M, O'Brien D, Grill D, Warner N, Munoz-Gomez M, Kossick K, Johnson R, Mouchli M, Felmlee-Devine D, Washechek-Aletto J, Smyrk T, Oberg A, Wang J, Chia N, Abyzov A, Ahlquist D, Boardman LA. Molecular characterization of colorectal adenomas with and without malignancy reveals distinguishing genome, transcriptome and methylome alterations. Sci Rep. 2018;8:3161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 33. | Shenderov BA. Gut indigenous microbiota and epigenetics. Microb Ecol Health Dis. 2012;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 34. | Li DY, Tang WHW. Contributory Role of Gut Microbiota and Their Metabolites Toward Cardiovascular Complications in Chronic Kidney Disease. Semin Nephrol. 2018;38:193-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 35. | Tomazetto G, Hahnke S, Wibberg D, Pühler A, Klocke M, Schlüter A. Proteiniphilum saccharofermentans str. M3/6T isolated from a laboratory biogas reactor is versatile in polysaccharide and oligopeptide utilization as deduced from genome-based metabolic reconstructions. Biotechnol Rep (Amst). 2018;18:e00254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 36. | Haag LM, Fischer A, Otto B, Plickert R, Kühl AA, Göbel UB, Bereswill S, Heimesaat MM. Intestinal microbiota shifts towards elevated commensal Escherichia coli loads abrogate colonization resistance against Campylobacter jejuni in mice. PLoS One. 2012;7:e35988. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 105] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 37. | Lovegrove A, Edwards CH, De Noni I, Patel H, El SN, Grassby T, Zielke C, Ulmius M, Nilsson L, Butterworth PJ, Ellis PR, Shewry PR. Role of polysaccharides in food, digestion, and health. Crit Rev Food Sci Nutr. 2017;57:237-253. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 342] [Cited by in RCA: 334] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 38. | Liang D, Leung RK, Guan W, Au WW. Involvement of gut microbiome in human health and disease: brief overview, knowledge gaps and research opportunities. Gut Pathog. 2018;10:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 132] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 39. | Sun J, Kato I. Gut microbiota, inflammation and colorectal cancer. Genes Dis. 2016;3:130-143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 167] [Cited by in RCA: 192] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 40. | Yadav R, Kumar V, Baweja M, Shukla P. Gene editing and genetic engineering approaches for advanced probiotics: A review. Crit Rev Food Sci Nutr. 2018;58:1735-1746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 41. | Mori G, Rampelli S, Orena BS, Rengucci C, De Maio G, Barbieri G, Passardi A, Casadei Gardini A, Frassineti GL, Gaiarsa S, Albertini AM, Ranzani GN, Calistri D, Pasca MR. Shifts of Faecal Microbiota During Sporadic Colorectal Carcinogenesis. Sci Rep. 2018;8:10329. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 66] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 42. | Bober JR, Beisel CL, Nair NU. Synthetic Biology Approaches to Engineer Probiotics and Members of the Human Microbiota for Biomedical Applications. Annu Rev Biomed Eng. 2018;20:277-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 81] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 43. | Nagpal R, Wang S, Ahmadi S, Hayes J, Gagliano J, Subashchandrabose S, Kitzman DW, Becton T, Read R, Yadav H. Human-origin probiotic cocktail increases short-chain fatty acid production via modulation of mice and human gut microbiome. Sci Rep. 2018;8:12649. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 237] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 44. | Bultman SJ. Interplay between diet, gut microbiota, epigenetic events, and colorectal cancer. Mol Nutr Food Res. 2017;61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 185] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 45. | Bourassa MW, Alim I, Bultman SJ, Ratan RR. Butyrate, neuroepigenetics and the gut microbiome: Can a high fiber diet improve brain health? Neurosci Lett. 2016;625:56-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 315] [Cited by in RCA: 417] [Article Influence: 46.3] [Reference Citation Analysis (0)] |
| 46. | Wang Q, Li L, Xu R. A systems biology approach to predict and characterize human gut microbial metabolites in colorectal cancer. Sci Rep. 2018;8:6225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 47. | Landi D, Moreno V, Guino E, Vodicka P, Pardini B, Naccarati A, Canzian F, Barale R, Gemignani F, Landi S. Polymorphisms affecting micro-RNA regulation and associated with the risk of dietary-related cancers: a review from the literature and new evidence for a functional role of rs17281995 (CD86) and rs1051690 (INSR), previously associated with colorectal cancer. Mutat Res. 2011;717:109-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 48. | Ferguson LR. Meat and cancer. Meat Sci. 2010;84:308-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 166] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 49. | Cui H, Cai Y, Wang L, Jia B, Li J, Zhao S, Chu X, Lin J, Zhang X, Bian Y, Zhuang P. Berberine Regulates Treg/Th17 Balance to Treat Ulcerative Colitis Through Modulating the Gut Microbiota in the Colon. Front Pharmacol. 2018;9:571. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 189] [Cited by in RCA: 179] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 50. | Pearson T, Caporaso JG, Yellowhair M, Bokulich NA, Padi M, Roe DJ, Wertheim BC, Linhart M, Martinez JA, Bilagody C, Hornstra H, Alberts DS, Lance P, Thompson PA. Effects of ursodeoxycholic acid on the gut microbiome and colorectal adenoma development. Cancer Med. 2019;8:617-628. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 73] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 51. | Wang D, Dubois RN. The role of COX-2 in intestinal inflammation and colorectal cancer. Oncogene. 2010;29:781-788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 569] [Cited by in RCA: 657] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 52. | Khare S, Cerda S, Wali RK, Von Lintig FC, Tretiakova M, Joseph L, Stoiber D, Cohen G, Nimmagadda K, Hart J, Sitrin MD, Boss GR, Bissonnette M. Ursodeoxycholic acid inhibits Ras mutations, wild-type Ras activation, and cyclooxygenase-2 expression in colon cancer. Cancer Res. 2003;63:3517-3523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 53. | Cani PD, Jordan BF. Gut microbiota-mediated inflammation in obesity: a link with gastrointestinal cancer. Nat Rev Gastroenterol Hepatol. 2018;15:671-682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 277] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 54. | Zhang Y, Yu X, Yu E, Wang N, Cai Q, Shuai Q, Yan F, Jiang L, Wang H, Liu J, Chen Y, Li Z, Jiang Q. Changes in gut microbiota and plasma inflammatory factors across the stages of colorectal tumorigenesis: a case-control study. BMC Microbiol. 2018;18:92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 55. | Rajilić-Stojanović M. Function of the microbiota. Best Pract Res Clin Gastroenterol. 2013;27:5-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 67] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 56. | Shi N, Li N, Duan X, Niu H. Interaction between the gut microbiome and mucosal immune system. Mil Med Res. 2017;4:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 223] [Cited by in RCA: 406] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 57. | Culpepper T, Ukhanova M, Wang X, Sun Y, Mai V. Associations between diet, gut microbiota and markers of CRC risk. Cancer & Metabolism. 2014;2:2049-3002. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 58. | Yang Y, Nirmagustina DE, Kumrungsee T, Okazaki Y, Tomotake H, Kato N. Feeding of the water extract from Ganoderma lingzhi to rats modulates secondary bile acids, intestinal microflora, mucins, and propionate important to colon cancer. Biosci Biotechnol Biochem. 2017;81:1796-1804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 59. | Centuori SM, Martinez JD. Differential regulation of EGFR-MAPK signaling by deoxycholic acid (DCA) and ursodeoxycholic acid (UDCA) in colon cancer. Dig Dis Sci. 2014;59:2367-2380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 60. | Wang JR, Gan WJ, Li XM, Zhao YY, Li Y, Lu XX, Li JM, Wu H. Orphan nuclear receptor Nur77 promotes colorectal cancer invasion and metastasis by regulating MMP-9 and E-cadherin. Carcinogenesis. 2014;35:2474-2484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 61. | Kong Y, Bai PS, Sun H, Nan KJ, Chen NZ, Qi XG. The deoxycholic acid targets miRNA-dependent CAC1 gene expression in multidrug resistance of human colorectal cancer. Int J Biochem Cell Biol. 2012;44:2321-2332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 62. | Ocvirk S, O'Keefe SJ. Influence of Bile Acids on Colorectal Cancer Risk: Potential Mechanisms Mediated by Diet - Gut Microbiota Interactions. Curr Nutr Rep. 2017;6:315-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 110] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 63. | Mikó E, Vida A, Kovács T, Ujlaki G, Trencsényi G, Márton J, Sári Z, Kovács P, Boratkó A, Hujber Z, Csonka T, Antal-Szalmás P, Watanabe M, Gombos I, Csoka B, Kiss B, Vígh L, Szabó J, Méhes G, Sebestyén A, Goedert JJ, Bai P. Lithocholic acid, a bacterial metabolite reduces breast cancer cell proliferation and aggressiveness. Biochim Biophys Acta Bioenerg. 2018;1859:958-974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 140] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 64. | Farhana L, Nangia-Makker P, Arbit E, Shango K, Sarkar S, Mahmud H, Hadden T, Yu Y, Majumdar AP. Bile acid: a potential inducer of colon cancer stem cells. Stem Cell Res Ther. 2016;7:181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 132] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 65. | Landfald B, Valeur J, Berstad A, Raa J. Microbial trimethylamine-N-oxide as a disease marker: something fishy? Microb Ecol Health Dis. 2017;28:1327309. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 66. | Liu ZY, Tan XY, Li QJ, Liao GC, Fang AP, Zhang DM, Chen PY, Wang XY, Luo Y, Long JA, Zhong RH, Zhu HL. Trimethylamine N-oxide, a gut microbiota-dependent metabolite of choline, is positively associated with the risk of primary liver cancer: a case-control study. Nutr Metab (Lond). 2018;15:81. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 67. | Zou S, Fang L, Lee MH. Dysbiosis of gut microbiota in promoting the development of colorectal cancer. Gastroenterol Rep (Oxf). 2018;6:1-12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 187] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 68. | Wang X, Allen TD, Yang Y, Moore DR, Huycke MM. Cyclooxygenase-2 generates the endogenous mutagen trans-4-hydroxy-2-nonenal in Enterococcus faecalis-infected macrophages. Cancer Prev Res (Phila). 2013;6:206-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 69. | Floch MH, Ringel Y, Walker WA. The Microbiota in Gastrointestinal Pathophysiology: Implications for Human Health, Prebiotics, Probiotics, and Dysbiosis. US: Academic Press. 2016;. |
| 70. | Veziant J, Gagnière J, Jouberton E, Bonnin V, Sauvanet P, Pezet D, Barnich N, Miot-Noirault E, Bonnet M. Association of colorectal cancer with pathogenic Escherichia coli: Focus on mechanisms using optical imaging. World J Clin Oncol. 2016;7:293-301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 52] [Cited by in RCA: 42] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 71. | Zou J, Shankar N. Roles of TLR/MyD88/MAPK/NF-κB Signaling Pathways in the Regulation of Phagocytosis and Proinflammatory Cytokine Expression in Response to E. faecalis Infection. PLoS One. 2015;10:e0136947. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 72. | Boleij A, Hechenbleikner EM, Goodwin AC, Badani R, Stein EM, Lazarev MG, Ellis B, Carroll KC, Albesiano E, Wick EC, Platz EA, Pardoll DM, Sears CL. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis. 2015;60:208-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 313] [Cited by in RCA: 451] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 73. | Shiryaev SA, Remacle AG, Chernov AV, Golubkov VS, Motamedchaboki K, Muranaka N, Dambacher CM, Capek P, Kukreja M, Kozlov IA, Perucho M, Cieplak P, Strongin AY. Substrate cleavage profiling suggests a distinct function of Bacteroides fragilis metalloproteinases (fragilysin and metalloproteinase II) at the microbiome-inflammation-cancer interface. J Biol Chem. 2013;288:34956-34967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 74. | Lv Y, Ye T, Wang HP, Zhao JY, Chen WJ, Wang X, Shen CX, Wu YB, Cai YK. Suppression of colorectal tumorigenesis by recombinant Bacteroides fragilis enterotoxin-2 in vivo. World J Gastroenterol. 2017;23:603-613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 11] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 75. | Nougayrède JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313:848-851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 688] [Cited by in RCA: 833] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 76. | Buc E, Dubois D, Sauvanet P, Raisch J, Delmas J, Darfeuille-Michaud A, Pezet D, Bonnet R. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One. 2013;8:e56964. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 401] [Cited by in RCA: 406] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 77. | Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède JP. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci USA. 2010;107:11537-11542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 613] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 78. | Wang Y, Zhang R, Wu D, Lu Z, Sun W, Cai Y, Wang C, Jin J. Epigenetic change in kidney tumor: downregulation of histone acetyltransferase MYST1 in human renal cell carcinoma. J Exp Clin Cancer Res. 2013;32:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 79. | Lannagan TRM, Lee YK, Wang T, Roper J, Bettington ML, Fennell L, Vrbanac L, Jonavicius L, Somashekar R, Gieniec K, Yang M, Ng JQ, Suzuki N, Ichinose M, Wright JA, Kobayashi H, Putoczki TL, Hayakawa Y, Leedham SJ, Abud HE, Yilmaz ÖH, Marker J, Klebe S, Wirapati P, Mukherjee S, Tejpar S, Leggett BA, Whitehall VLJ, Worthley DL, Woods SL. Genetic editing of colonic organoids provides a molecularly distinct and orthotopic preclinical model of serrated carcinogenesis. Gut. 2019;68:684-692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 94] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 80. | Raskov H, Burcharth J, Pommergaard HC. Linking Gut Microbiota to Colorectal Cancer. J Cancer. 2017;8:3378-3395. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 101] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 81. | Gerhauser C. Impact of dietary gut microbial metabolites on the epigenome. Philos Trans R Soc Lond B Biol Sci. 2018;373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 82. | Bruserud Ø, Stapnes C, Ersvaer E, Gjertsen BT, Ryningen A. Histone deacetylase inhibitors in cancer treatment: a review of the clinical toxicity and the modulation of gene expression in cancer cell. Curr Pharm Biotechnol. 2007;8:388-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 124] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 83. | Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, Shu J, Chen X, Waterland RA, Issa JP. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:2023-2036. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 251] [Cited by in RCA: 274] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 84. | Sandoval-Basilio J, González-González R, Bologna-Molina R, Isiordia-Espinoza M, Leija-Montoya G, Alcaraz-Estrada SL, Serafín-Higuera I, González-Ramírez J, Serafín-Higuera N. Epigenetic mechanisms in odontogenic tumors: A literature review. Arch Oral Biol. 2018;87:211-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 85. | Cheng YW, Chou CJ, Yang PM. Ten-eleven translocation 1 (TET1) gene is a potential target of miR-21-5p in human colorectal cancer. Surg Oncol. 2018;27:76-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 86. | Kopp M, Dürr K, Steigleder M, Clavel T, Rychlik M. Development of stable isotope dilution assays for the quantitation of intra- and extracellular folate patterns of Bifidobacterium adolescentis. J Chromatogr A. 2016;1469:48-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 87. | Zhou HR, Zhang FF, Ma ZY, Huang HW, Jiang L, Cai T, Zhu JK, Zhang C, He XJ. Folate polyglutamylation is involved in chromatin silencing by maintaining global DNA methylation and histone H3K9 dimethylation in Arabidopsis. Plant Cell. 2013;25:2545-2559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 88. | Pompei A, Cordisco L, Amaretti A, Zanoni S, Matteuzzi D, Rossi M. Folate production by bifidobacteria as a potential probiotic property. Appl Environ Microbiol. 2007;73:179-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 209] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 89. | Wasson GR, McGlynn AP, McNulty H, O'Reilly SL, McKelvey-Martin VJ, McKerr G, Strain JJ, Scott J, Downes CS. Global DNA and p53 region-specific hypomethylation in human colonic cells is induced by folate depletion and reversed by folate supplementation. J Nutr. 2006;136:2748-2753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 83] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 90. | Cheng AS, Li MS, Kang W, Cheng VY, Chou JL, Lau SS, Go MY, Lee CC, Ling TK, Ng EK, Yu J, Huang TH, To KF, Chan MW, Sung JJ, Chan FK. Helicobacter pylori causes epigenetic dysregulation of FOXD3 to promote gastric carcinogenesis. Gastroenterology. 2013;144:122-133.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 112] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 91. | Kawanaka M, Watari J, Kamiya N, Yamasaki T, Kondo T, Toyoshima F, Ikehara H, Tomita T, Oshima T, Fukui H, Daimon T, Das KM, Miwa H. Effects of Helicobacter pylori eradication on the development of metachronous gastric cancer after endoscopic treatment: analysis of molecular alterations by a randomised controlled trial. Br J Cancer. 2016;114:21-29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 92. | Koi M, Okita Y, Carethers JM. Fusobacterium nucleatum Infection in Colorectal Cancer: Linking Inflammation, DNA Mismatch Repair and Genetic and Epigenetic Alterations. J Anus Rectum Colon. 2018;2:37-46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 93. | Inamura K. Colorectal Cancers: An Update on Their Molecular Pathology. Cancers (Basel). 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 122] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 94. | Supic G, Zeljic K, Magic Z. Chapter 15 - Epigenetic Nutraceuticals in Cancer Treatment, In: Holban AM, Grumezescu AM. Therapeutic Foods. Academic Press. 2018;449-493. [DOI] [Full Text] |
| 95. | Kurita-Ochiai T, Hashizume T, Yonezawa H, Ochiai K, Yamamoto M. Characterization of the effects of butyric acid on cell proliferation, cell cycle distribution and apoptosis. FEMS Immunol Med Microbiol. 2006;47:67-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 96. | Hu S, Liu L, Chang EB, Wang JY, Raufman JP. Butyrate inhibits pro-proliferative miR-92a by diminishing c-Myc-induced miR-17-92a cluster transcription in human colon cancer cells. Mol Cancer. 2015;14:180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 141] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 97. | Jung J, Ko SH, Yoo do Y, Lee JY, Kim YJ, Choi SM, Kang KK, Yoon HJ, Kim H, Youn J, Kim JM. 5,7-Dihydroxy-3,4,6-trimethoxyflavone inhibits intercellular adhesion molecule 1 and vascular cell adhesion molecule 1 via the Akt and nuclear factor-κB-dependent pathway, leading to suppression of adhesion of monocytes and eosinophils to bronchial epithelial cells. Immunology. 2012;137:98-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 98. | Minarovits J, Niller HH. Patho-Epigenetics of Infectious Disease. Part of the Advances in Experimental Medicine and Biology book series (AEMB, volume 879). Cham: Springer 2015; . [RCA] [DOI] [Full Text] [Cited by in Crossref: 6] [Reference Citation Analysis (0)] |
| 99. | Jing H, Lin H. Sirtuins in epigenetic regulation. Chem Rev. 2015;115:2350-2375. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 202] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 100. | Migault M, Donnou-Fournet E, Galibert MD, Gilot D. Definition and identification of small RNA sponges: Focus on miRNA sequestration. Methods. 2017;117:35-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 101. | Balmayor ER, Font Tellado S, Van Griensven M, 2.26 MicroRNA as Biomaterial. In: Ducheyne P. Comprehensive Biomaterials II. 2nd Ed. Elsevier: Oxford 2017; 558-570. [DOI] [Full Text] |
| 102. | Kai K, Dittmar RL, Sen S. Secretory microRNAs as biomarkers of cancer. Semin Cell Dev Biol. 2018;78:22-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 80] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 103. | Strubberg AM, Madison BB. MicroRNAs in the etiology of colorectal cancer: pathways and clinical implications. Dis Model Mech. 2017;10:197-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 112] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 104. | Kim SW. [The Role of MicroRNAs in Colorectal Cancer]. Korean J Gastroenterol. 2017;69:206-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 105. | Schee K, Boye K, Abrahamsen TW, Fodstad Ø, Flatmark K. Clinical relevance of microRNA miR-21, miR-31, miR-92a, miR-101, miR-106a and miR-145 in colorectal cancer. BMC Cancer. 2012;12:505. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 138] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 106. | Cekaite L, Eide PW, Lind GE, Skotheim RI, Lothe RA. MicroRNAs as growth regulators, their function and biomarker status in colorectal cancer. Oncotarget. 2016;7:6476-6505. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 77] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 107. | Karius T, Schnekenburger M, Dicato M, Diederich M. MicroRNAs in cancer management and their modulation by dietary agents. Biochem Pharmacol. 2012;83:1591-1601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 108. | Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762-765. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1736] [Cited by in RCA: 1696] [Article Influence: 106.0] [Reference Citation Analysis (0)] |
| 109. | Jacouton E, Chain F, Sokol H, Langella P, Bermúdez-Humarán LG. Probiotic Strain Lactobacillus casei BL23 Prevents Colitis-Associated Colorectal Cancer. Front Immunol. 2017;8:1553. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 156] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 110. | Kåhrström CT. Bacterial pathogenesis: E. coli claims the driving seat for cancer. Nat Rev Cancer. 2012;12:658-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 111. | Shang FM, Liu HL. Fusobacterium nucleatum and colorectal cancer: A review. World J Gastrointest Oncol. 2018;10:71-81. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 192] [Cited by in RCA: 197] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 112. | Rivière A, Selak M, Lantin D, Leroy F, De Vuyst L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut. Front Microbiol. 2016;7:979. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 979] [Cited by in RCA: 1103] [Article Influence: 122.6] [Reference Citation Analysis (0)] |
| 113. | Purcell RV, Pearson J, Aitchison A, Dixon L, Frizelle FA, Keenan JI. Colonization with enterotoxigenic Bacteroides fragilis is associated with early-stage colorectal neoplasia. PLoS One. 2017;12:e0171602. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 163] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 114. | Tsai CE, Chiu CT, Rayner CK, Wu KL, Chiu YC, Hu ML, Chuah SK, Tai WC, Liang CM, Wang HM. Associated factors in Streptococcus bovis bacteremia and colorectal cancer. Kaohsiung J Med Sci. 2016;32:196-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 115. | Melton-Witt JA, Bentsen LM, Tweten RK. Identification of functional domains of Clostridium septicum alpha toxin. Biochemistry. 2006;45:14347-14354. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 33] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 116. | Huycke MM, Abrams V, Moore DR. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis. 2002;23:529-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 327] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 117. | Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, Kumagai H, Ashida H, Hirose J, Kitaoka M. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem. 2011;286:34583-34592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 332] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 118. | Papastergiou V, Karatapanis S, Georgopoulos SD. Helicobacter pylori and colorectal neoplasia: Is there a causal link? World J Gastroenterol. 2016;22:649-658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 36] [Cited by in RCA: 42] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 119. | Martín R, Miquel S, Benevides L, Bridonneau C, Robert V, Hudault S, Chain F, Berteau O, Azevedo V, Chatel JM, Sokol H, Bermúdez-Humarán LG, Thomas M, Langella P. Functional Characterization of Novel Faecalibacterium prausnitzii Strains Isolated from Healthy Volunteers: A Step Forward in the Use of F. prausnitzii as a Next-Generation Probiotic. Front Microbiol. 2017;8:1226. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 211] [Cited by in RCA: 269] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 120. | Chen L, Tai WC, Brar MS, Leung FC, Hsiao WL. Tumor grafting induces changes of gut microbiota in athymic nude mice in the presence and absence of medicinal Gynostemma saponins. PLoS One. 2015;10:e0126807. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 121. | Zhou Z, Chen J, Yao H, Hu H. Fusobacterium and Colorectal Cancer. Front Oncol. 2018;8:371. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 93] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 122. | McFarlane ME, Coard KC. Actinomycosis of the colon with invasion of the abdominal wall: An uncommon presentation of a colonic tumour. Int J Surg Case Rep. 2010;1:9-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 123. | Deng X, Li Z, Li G, Li B, Jin X, Lyu G. Comparison of Microbiota in Patients Treated by Surgery or Chemotherapy by 16S rRNA Sequencing Reveals Potential Biomarkers for Colorectal Cancer Therapy. Front Microbiol. 2018;9:1607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 81] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 124. | Xiao G, Tang H, Wei W, Li J, Ji L, Ge J. Aberrant Expression of MicroRNA-15a and MicroRNA-16 Synergistically Associates with Tumor Progression and Prognosis in Patients with Colorectal Cancer. Gastroenterol Res Pract. 2014;2014:364549. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 125. | Díaz R, Silva J, García JM, Lorenzo Y, García V, Peña C, Rodríguez R, Muñoz C, García F, Bonilla F, Domínguez G. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosomes Cancer. 2008;47:794-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 183] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 126. | Karaayvaz M, Pal T, Song B, Zhang C, Georgakopoulos P, Mehmood S, Burke S, Shroyer K, Ju J. Prognostic significance of miR-215 in colon cancer. Clin Colorectal Cancer. 2011;10:340-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 114] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 127. | Weissmann-Brenner A, Kushnir M, Lithwick Yanai G, Aharonov R, Gibori H, Purim O, Kundel Y, Morgenstern S, Halperin M, Niv Y, Brenner B. Tumor microRNA-29a expression and the risk of recurrence in stage II colon cancer. Int J Oncol. 2012;40:2097-2103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 128. | Gao J, Li N, Dong Y, Li S, Xu L, Li X, Li Y, Li Z, Ng SS, Sung JJ, Shen L, Yu J. miR-34a-5p suppresses colorectal cancer metastasis and predicts recurrence in patients with stage II/III colorectal cancer. Oncogene. 2015;34:4142-4152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 134] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 129. | Mokutani Y, Uemura M, Munakata K, Okuzaki D, Haraguchi N, Takahashi H, Nishimura J, Hata T, Murata K, Takemasa I, Mizushima T, Doki Y, Mori M, Yamamoto H. Down-Regulation of microRNA-132 is Associated with Poor Prognosis of Colorectal Cancer. Ann Surg Oncol. 2016;23:599-608. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 69] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 130. | Ma Y, Zhang P, Wang F, Zhang H, Yang J, Peng J, Liu W, Qin H. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut. 2012;61:1447-1453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 153] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 131. | Wang X, Wang J, Ma H, Zhang J, Zhou X. Downregulation of miR-195 correlates with lymph node metastasis and poor prognosis in colorectal cancer. Med Oncol. 2012;29:919-927. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 100] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 132. | Shen ZL, Wang B, Jiang KW, Ye CX, Cheng C, Yan YC, Zhang JZ, Yang Y, Gao ZD, Ye YJ, Wang S. Downregulation of miR-199b is associated with distant metastasis in colorectal cancer via activation of SIRT1 and inhibition of CREB/KISS1 signaling. Oncotarget. 2016;7:35092-35105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 133. | Perez-Carbonell L, Sinicrope FA, Alberts SR, Oberg AL, Balaguer F, Castells A, Boland CR, Goel A. MiR-320e is a novel prognostic biomarker in colorectal cancer. Br J Cancer. 2015;113:83-90. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 134. | Dong SJ, Cai XJ, Li SJ. The Clinical Significance of MiR-429 as a Predictive Biomarker in Colorectal Cancer Patients Receiving 5-Fluorouracil Treatment. Med Sci Monit. 2016;22:3352-3361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 135. | Yang IP, Tsai HL, Miao ZF, Huang CW, Kuo CH, Wu JY, Wang WM, Juo SH, Wang JY. Development of a deregulating microRNA panel for the detection of early relapse in postoperative colorectal cancer patients. J Transl Med. 2016;14:108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 136. | Rasmussen MH, Jensen NF, Tarpgaard LS, Qvortrup C, Rømer MU, Stenvang J, Hansen TP, Christensen LL, Lindebjerg J, Hansen F, Jensen BV, Hansen TF, Pfeiffer P, Brünner N, Ørntoft TF, Andersen CL. High expression of microRNA-625-3p is associated with poor response to first-line oxaliplatin based treatment of metastatic colorectal cancer. Mol Oncol. 2013;7:637-646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 80] [Article Influence: 6.7] [Reference Citation Analysis (0)] |