Published online Oct 6, 2019. doi: 10.12998/wjcc.v7.i19.2916
Peer-review started: March 22, 2019
First decision: August 1, 2019
Revised: August 22, 2019
Accepted: September 9, 2019
Article in press: September 9, 2019
Published online: October 6, 2019
Processing time: 191 Days and 8.4 Hours
Colorectal cancer (CRC) remains a major contributor to the number of cancer-related deaths that occur annually worldwide. With the development of molecular biology methods, an increasing number of molecular biomarkers have been identified and investigated. CRC is believed to result from an accumulation of epigenetic changes, and detecting aberrant DNA methylation patterns is useful for both the early diagnosis and prognosis of CRC. Numerous studies are focusing on the development of DNA methylation detection methods or DNA methylation panels. Thus, this review will discuss the commonly used techniques and technologies to evaluate DNA methylation, their merits and deficiencies as well as the prospects for new methods.
Core tip: We screened the literature on the application of DNA methylation detection technologies as well as colorectal cancer (CRC) associated DNA methylation markers in the diagnosis or prognosis evaluation of CRC. Apart from introducing each method in detail and describing the methylation status of several candidate genes being assayed, we also evaluated the advantages or disadvantages of each method. More importantly, we discuss the new DNA methylation detection methods and their potential use. At last, we prospect on how these methylation markers could be used and what kind of methylation detection techniques might be more practical in clinical work.
- Citation: Zhan YX, Luo GH. DNA methylation detection methods used in colorectal cancer. World J Clin Cases 2019; 7(19): 2916-2929
- URL: https://www.wjgnet.com/2307-8960/full/v7/i19/2916.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v7.i19.2916
Colorectal cancer (CRC) is a major contributor to cancer-related deaths worldwide[1]. According to an epidemiological study, the number of newly emerged CRC cases worldwide is predicted to rise to 2.2 million and more and will cause approximately 1.1 million deaths by the year of 2030[2]. Furthermore, an article published in 2012 reported that there are more than 1 million CRC cases diagnosed per year, and nearly 700000 annual deaths result from this disease[3]. Genetic analyses of CRC have shown that CRC arises from an accumulation of genetic mutations and epigenetic changes[4], with CRC typically having 3 to 6 driver mutations and 33 to 66 hitchhiker or passenger mutations that lead to protein expression silencing[5]. In these instances, molecular data are often used as evidence to support the clinical and pathological changes[6]. Changes in the methylation levels of specific genes have been shown to be associated with the early stages of CRC or with good or bad patient prognoses[7]. Epigenetic modifications can be affected by environmental factors, and the identification of site-specific changes in DNA methylation may offer insights into carcinogenetic mechanisms and ultimately aid in uncovering the role of environmental influences in the development of CRC[8]. Furthermore, colonoscopies and polypectomies are complicated technical procedures that require training and experience to maximize accuracy and safety[9]. In addition, the procedures for tests such as colonoscopies are not acceptable to all individuals and can have complications[10,11]. Future research and further investigations aiming to identify predictive biomarkers can be generally useful in developing available, noninvasive, cost-effective diagnostic and prognostic panels that would have immense societal benefits[12].
For the reasons described above, we believe that further investigations on gene methylation related to CRC and the development of rapid and reliable methylation detection methods for CRC patients are worthwhile. It seems like that CRC epigenetics will be the epitome for multistep tumorigenesis, because new high-throughput methods will make detailed mapping of the CRC chromatin landscape accessible, which is able to offer new vision of CRC etiology and biology, discover more genetic variations in epigenetic regulators linked to CRC, and uncover potential epigenetic biomarkers for CRC diagnosis or prediction of prognosis and response to treatment[13]. Numerous studies have been undertaken to identify epigenetic biomarkers for diagnosing CRC in early stage or evaluating disease prognosis or to develop brand-new technologies or techniques to assess DNA methylation in CRC, all the results of which may promote a decrease in CRC-associated morbidity and mortality. Dozens of methylation detection methods have been developed, which utilize highly different approaches. Therefore, the intent of this review is to discuss the most widely used methods as well as newly investigated or invented technologies. This review may be useful to researchers or scholars who are interested in the relationship between aberrant DNA methylation and CRC or those who want to investigate new methods for the detection of CRC. We will summarize the techniques used for CRC DNA methylation level analysis, compare different methods, and analyze both the advantages and disadvantages of the most widely used and newly developed methods. Finally, we will make conclusions regarding the preferred techniques and evaluate the value of novel methods that are under development.
In this review, we focus on the methylation detection methods developed for or implemented in the diagnosis CRC. The advantages and disadvantages of each method are summarized and compared to offer valuable information to researchers who want to undertake investigations in this field.
From January 15 to February 7, 2019, we searched for articles involving the detection of DNA methylation in CRC patients. The key words we used were “gene” OR “DNA” AND “methylation” AND “detection” AND “colon” OR “intestine” OR “rectal” OR “colorectal” OR “CRC” AND “cancer”.
We performed literature searches using six online databases, including PubMed, MEDLINE, Ovid, EMBASE, Science Direct, and Web of Science. We only included original research written in English and reported within the past decade.
The selection criteria for our review were as follows: (1) Studies involving DNA methylation detection for CRC; and (2) Studies that implemented or developed a novel method to detect the level of DNA methylation in CRC. The rejection criteria for our review included studies that did not mention the sensitivity of the method or did not report the methods.
To evaluate the quality of the included studies, two reviewers were independently chosen to extract the data and assess quality of each study that included DNA methylation detection in CRC patients.
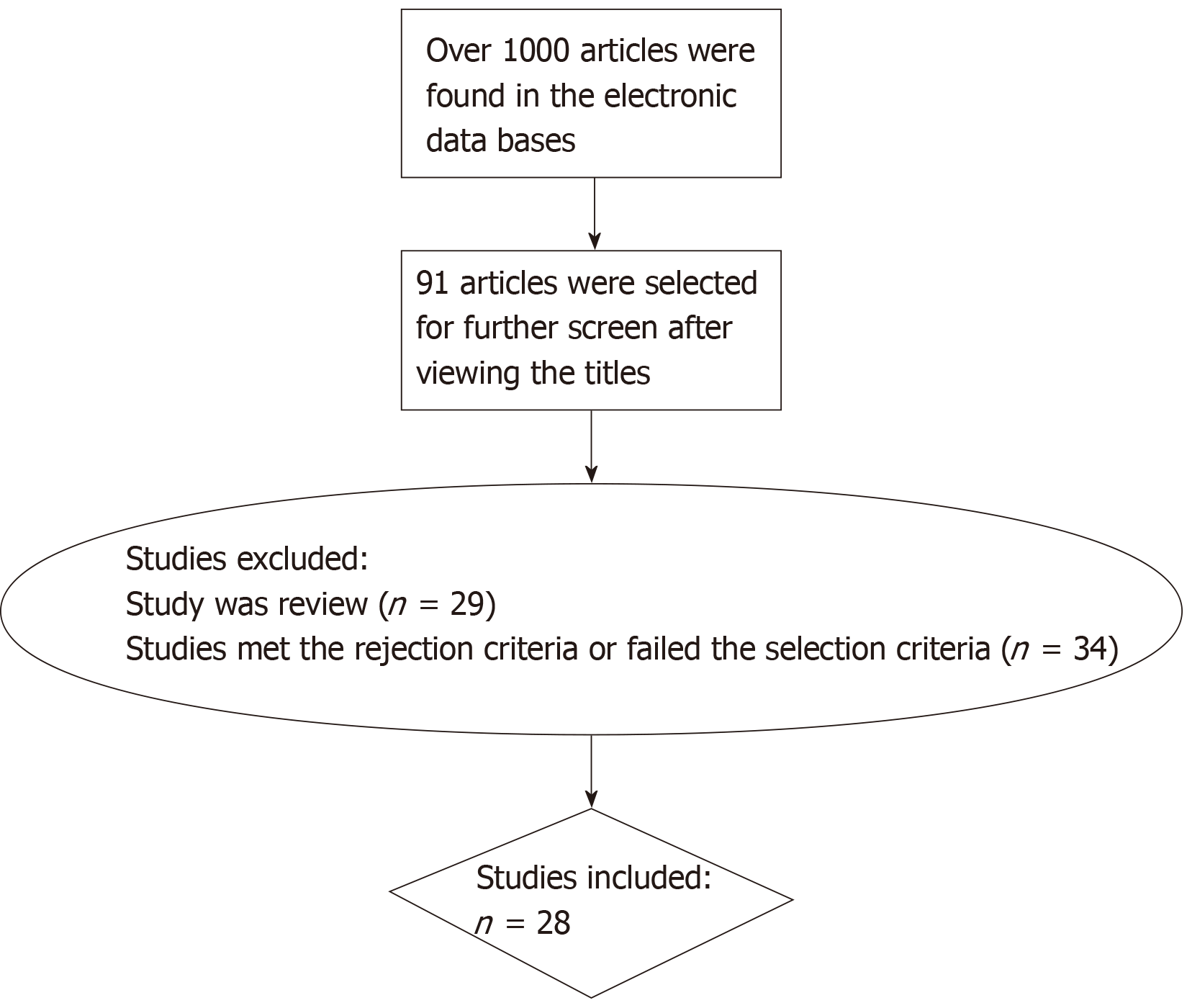
All of the data were extracted from 28 carefully selected articles. The selection process used is shown in Figure 1. We focused on the methods and candidate genes the researchers chose, and we collected or calculated the diagnostic sensitivity and specificity of each reported marker in these articles. For those not mentioning the sensitivity and specificity, we used the statistics in the articles for calculation. Generally, the sensitivity for DNA methylation in CRC diagnosis is the proportion of those CRC patients with a positive test. Specificity is negative test results among persons without CRC.
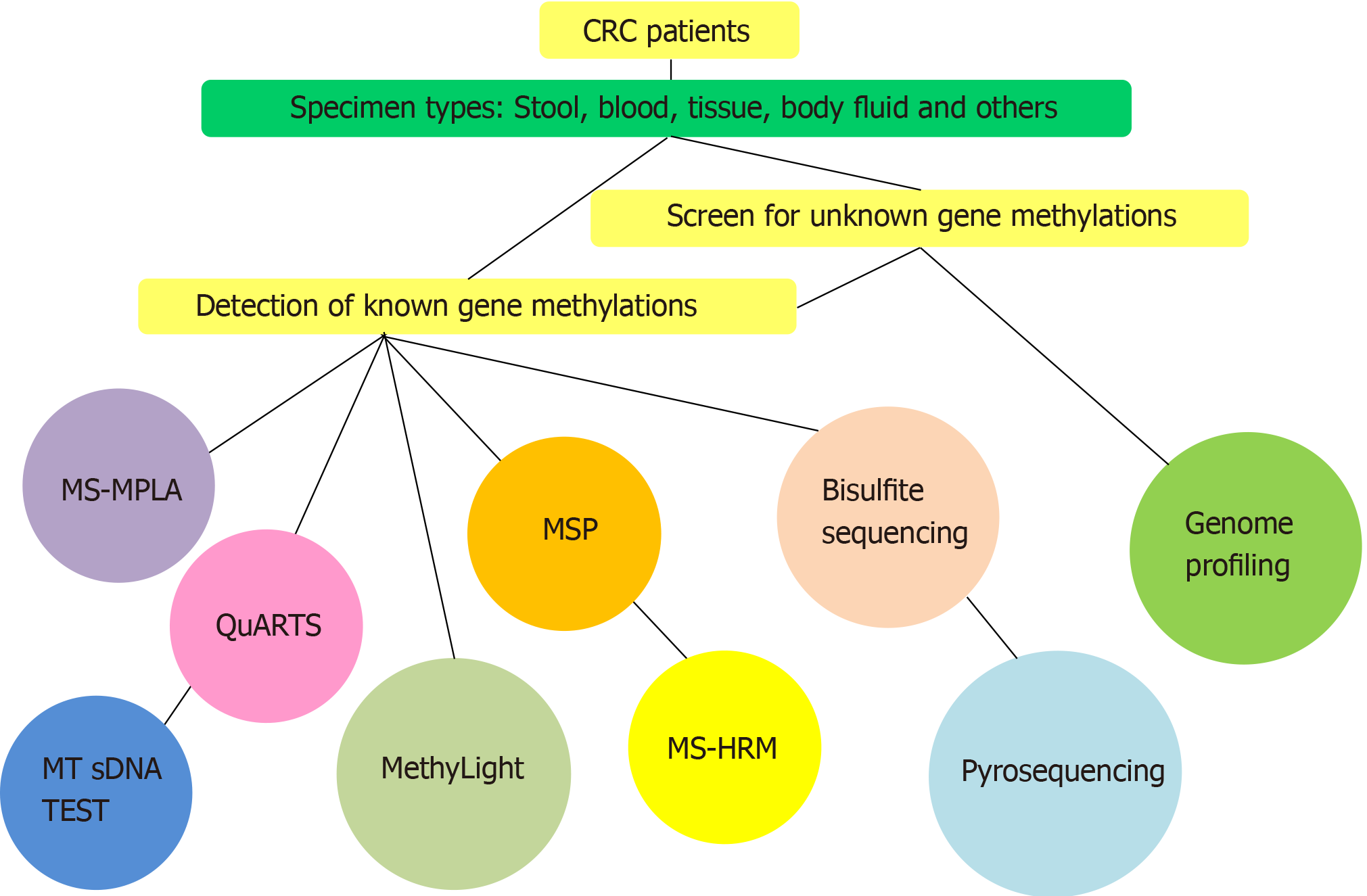
We searched more than 100 articles and summarized all commonly used DNA methylation methods used in these studies (Figure 2). Among the different studies that focused on DNA methylation in CRC, various types of techniques were used (Table 1). The diagnostic sensitivity and specificity of the molecular markers obtained using specific methylation detection methods are also provided in Table 1. We observed that most of the methods are PCR-based techniques or technologies, such as methylation-specific multiple ligation-dependent probe amplification (MS-MLPA), direct or nested methylation-specific polymerase chain reaction (MSP), multitarget stool DNA (MT-sDNA) test, methylation-specific high resolution melting curve analysis (MS-HRM), and MethyLight assay. DNA sequencing is also a commonly used technique, including bisulfite sequencing, pyrosequencing, and quantitative allele-specific real-time target and signal amplification (QuARTS) assays. Other methods involving Illumina technologies were typically used for genome profiling.
| No.[Ref.] | Year | Specimen | Gene | Sample number | Method | Sensitivity (95%CI) | Specificity (95%CI) | Conclusion and comments | |
| Case | Control | ||||||||
| 1[77] | 2014 | Tissue | 18 genes1 | 20 | 20 | Illumina2 | 96% (87%-105%) | 100% | These results suggest that methylation biomarkers in this study may be developed that will, at minimum, serve as useful objective and quantitative diagnostic complements to colonoscopy as a cancer-screening tool |
| 2[78] | 2015 | Tissue | FAM78A, FSTL1, KCNC1, MYOCD, SLC6A4 | 28 | 97 | Illumina3 | 100.00% | 100% | Four distinct DNA methylation subgroups of CRC identified can provide novel insight regarding the role of CIMP-specific DNA hypermethylation in gene silencing |
| 3[20] | 2013 | Stool | NDRG4, BMP3 | 86 | 794 | Automated MT sDNA assay | 98% (95%-101%)4 | 90% (88%-92%) | This automated high-throughput system could be a widely accessible noninvasive approach to general CRC screening |
| 92% (86%-98%)5 | |||||||||
| 4[4] | 2014 | Stool | BMP3, NDRG4 | 65 | 9167 | MT sDNA test | 92.3% (83.0%-97.5%) | 87% (86%-87%) | This stool test shows a higher single-application sensitivity than a commercial FIT for both colorectal cancer and advanced precancerous lesions, although with lower specificity |
| 5[21] | 2012 | Stool | Vimentin, NDRG4, BMP3, TFPI2 | 252 | 293 | QuARTS | 89% (83%-93%) | 90% (85%-94%) | Next-generation sDNA tests appear capable of detecting early-stage CRC and large adenomas with a high sensitivity and specificity at all sites throughout the colon; Validation of optimized next-generation sDNA tests in the screening setting is now needed |
| 6[19] | 2018 | Tissues | BMP3 | 421 | 44 | QuARTS | 58% (53%-63%) | 100% (90%-102%) | This novel panel is highly specific for colitis-associated neoplasia across geographically diverse patient subsets and among those with varying risk; The marker panel has been optimized by the discovery and incorporation of methylated ZDHHC1, an epithelium-specific marker, as a normalizer for the neoplasm-specific markers, in stool. Further studies are needed to corroborate and extend this finding. Except for DTX1, each of these novel markers is individually more sensitive than the most sensitive marker in the MT-sDNA panel |
| NDRG4 | 78% (74%-82%) | 100% (90%-102%) | |||||||
| SFMBT_895 | 90% (86%-92%) | 100% (90%-102%) | |||||||
| SFMBT_896 | 89% (86%-92%) | 100% (90%-102%) | |||||||
| SFMBT_897 | 89% (85%-92%) | 100% (90%-102%) | |||||||
| CHST2_7890 | 82% (77%-85%) | 100% (90%-102%) | |||||||
| PDGFD | 83% (79%-87%) | 100% (90%-102%) | |||||||
| CHST2_7889 | 82% (78%-86%) | 100% (90%-102%) | |||||||
| VAV3 | 82% (78%-85%) | 100% (90%-102%) | |||||||
| DTX1 | 71% (66%-75% | 100% (90%-102%) | |||||||
| Stool | BMP3, VAV3, ZDHHC1 | 12 | 291 | MSP | 92% (60%-100%) | 90% (86%-93%) | |||
| 7[79] | 2018 | Tissue | BCAT1 | 36 | 36 | MSP | 92% (83% -101%) | 69% (54%-85%) | This study has shown that highly methylated BCAT1 and IKZF1 exist in all stages of CRC; A positive correlation was observed between hypermethylated IKZF1 and gene activity silencing |
| IKZF1 | 72% (58%-87%) | 97% (92%-103%) | |||||||
| 8[80] | 2016 | Plasma | BCAT1/IKZF | 28 | 94 | MSP | 68% (48%-84%) | 87% (79%-95%) | Methylated BCAT1/IKZF1 blood test has a significantly higher sensitivity for recurrent CRC than the CEA test |
| 9[81] | 2015 | Plasma | BCAT1 | 129 | 450 | MSP | 57% (48%-66%) | 95% (93%-97%) | Accuracy of this two-marker blood test approximates that of gFOBT; This two-marker blood test for screening is likely a rescue strategy for those refusing more sensitive RCT-proven methods such as FIT, flexible sigmoidoscopy, or colonoscopy |
| IKZF | 48% (39%-57%) | 99% (98.1%-100%) | |||||||
| BCAT1/IKZF | 66% (57%-74%) | 95% (93%-97%) | |||||||
| 10[82] | 2014 | Plasma | BCAT1 | 74 | 144 | MSP | 65% (54%-76%) | 96% (93%-99%) | Detection of methylated BCAT1 and/or IKZF1 DNA in plasma may have clinical application as a novel blood test for CRC; Combining the results from the two methylation-specific PCR assays improves CRC detection with a minimal change in specificity |
| IKZF | 68% (57% -79%) | 95% (91%-99%) | |||||||
| BCAT1/IKZF | 77% (64 %-84%) | 92% (88%-96%) | |||||||
| 11[6] | 2018 | Tissue | SEPT9 | 145 | 50 | qMSP | 86% (80%-91%) | 94% (87%-101%) | Results indicate that MGMT/RASSF1A/SEPT9 gene promoter methylation panel accurately identifies CRC, irrespective of molecular subtype and may have a better performance than currently available epigenetic based biomarkers but requiring assessment of its performance in liquid biopsies |
| MGMT/SEPT9 | 145 | 50 | 94% (90%-98%) | 82% (71%-93%) | |||||
| MGMT/RASSF1A/SEPT9 | 145 | 50 | 97% (94%-100%) | 74% (62%-86%) | |||||
| 12[35] | 2013 | Serum | NPY, PENK, WIF1 | 32 | 161 | qMSP | 94% (86%-102%)6 | 34% (27%-41%) | This test can be a cost-effective screening tool for detection of asymptomatic cancer patients for colonoscopy, who are at CRC risk that is hard to access and does not necessarily need further examination |
| 59% (42%-76%)7 | 95% (86%-98%) | ||||||||
| 13[36] | 2018 | Bowel lavage fluid | SDC2 | 10 | 54 | qMSP | 80% (44%-97%) | 89% (77%-96%) | The SDC2 DNA methylation in BLF shows a high sensitivity and specificity in patients with CRC and precancerous lesions |
| 14[39] | 2017 | Stool | SDC2 | 50 | 22 | qMSP | 90% (78%-97%) | 91% (71%-99%) | Abnormal SDC2 methylation can be a new potential diagnostic biomarker for noninvasive screening of CRC |
| 15[27] | 2017 | Tissue | UNC5C | 73 | 28 | MSP | 78% (69%-88%) | 100% | This study found that aberrant methylation of UNC5C gene exists in CRCs and APs; UNC5C protein expression is negatively correlated with methylation status |
| 16[83] | 2017 | Serum | MGMT | 30 | 40 | MSP | 90% (79%-101%) | 100% | MGMT hypermethylation can be used “as a clinical biomarker” for early diagnosis and prognostic assessment of the disease |
| 17[37] | 2017 | Plasma | RASSF1A | 108 | 78 | MSP | 44% (35%-54%)8 | 95% (90%-100%) | Methylation of RASSF1A in the promoter region is independently associated with prognosis in CRC patients treated with oxaliplatin-based chemotherapy, and might be a promising target for improving chemotherapeutic effects |
| 21% (14%-29%)9 | |||||||||
| 18[84] | 2015 | Tissue | hMLH1 | 61 | 14 | MSP | 25% (14%-36%) | 57% (31%-83%) | The findings suggest that DNA methylation is a useful marker and that promoter methylation in certain genes is associated with more advanced tumor stages, poor differentiation, and metastasis, which can have an application as a risk assessment tool or as a marker of recurrence to help decide on the aggressiveness of the treatment |
| MGMT | 61 | 27 | 66% (54%-78%) | 19% (4%-33%) | |||||
| APC | 61 | 14 | 30% (19%-42%) | 14% (-4%-32%) | |||||
| CDH1 | 61 | 31 | 87% (79%-95%) | 25.8% (10%-41%) | |||||
| 2 OR MORE | 61 | N/A | 71% (60%-82%) | N/A | |||||
| ALL | 61 | N/A | 10% (3%-18%) | N/A | |||||
| 19[85] | 2014 | Tissue | CDKN2A | 44 | 155 | MSP | 82% (71%-93%) | 32% (25%-39%) | The presence of CDKN2A methylation is associated with poorer overall survival in stages B and C combined |
| 20[86] | 2017 | Tissue | DIRAS1 | 146 | 50 | MSP, BS-Seq | 47% (39%-55%) | 100% | Methylation of DIRAS1 is a marker of poor prognosis in human colorectal cancer; Methylation of DIRAS1 may promote colorectal carcinogenesis and progression |
| 21[87] | 2017 | Tissue | ZNF331 | 146 | 10 | MSP, BS-Seq | 67% (60%-75%) | 100 % | Methylation of ZNF331 is a poor prognostic marker in human colorectal cancer; ZNF331 may serve as a tumor suppressor in human colorectal cancer |
| 22[34] | 2016 | Tissue | RET10 | 25 | 184 | Direct MSP | 11% (-1%-23%) | 87% (82%-91%) | The data also show that qualitative techniques such as direct MSP and nested MSP (although showing different methylation frequencies) can, when carefully developed, optimized, and interpreted, yield comparable clinical results as pyrosequencing and MS-HRM and could therefore be used for biomarker detection/validation |
| 71 | 169 | Nested MSP | 33% (22%-44%) | 74% (67%-80%) | |||||
| 62 | 178 | Pyrosequencing | 23% (12%- 33%) | 73% (67%-80%) | |||||
| 41 | 199 | Pyrosequencing | 18% (6%-30%) | 84% (79%-89%) | |||||
| 37 | 203 | Pyrosequencing | 16% (4%-28%) | 85% (80%-90%) | |||||
| 32 | 208 | Pyrosequencing | 14% (2%-27%) | 88% (83%-92%) | |||||
| 25 | 215 | Pyrosequencing | 11% (-1%-23 %) | 90% (86%-94%) | |||||
| 33 | 206 | MS-HRM | 15% (3%-28%) | 87.7% (83.2%-92.2%) | |||||
| 23[7] | 2017 | Tissue | EDNRB location 1 | 45 | 45 | MS-HRM | 91% (83%-99%) | 88.9% (80%-98%) | The combination of EDNRB locations 1 and 2 can be used as a powerful biomarker, in which the first location is significantly correlated with tumor stage and grade while the second one is aberrantly methylated independent of any clinicopathological features |
| EDNRB location 2 | 91% (83%-99%) | 80% (68%-92%) | |||||||
| EDNRB location 3 | 76% (63%-88%) | 78% (66%-90%) | |||||||
| EDNRB location 4 | 80% (68%-92%) | 84% (74%-95%) | |||||||
| KISS1 | 58% (43%-72%) | 71% (58%-84%) | |||||||
| 24[88] | 2013 | Tissue | AGTR1, WNT2, SLIT2 | 60 | 38 | Pyrosequencing | 95% (89%-101%) | 89% (79%-99%) | This study offers a novel panel of specific methylation markers that can be assessed in stools and may complement currently applied protocols for the early detection of sporadic CRC, which may contribute to improving the follow-up and early diagnosis of high-risk patients with IBD when assessed in non-neoplastic tissues obtained by surveillance colonoscopy |
| SEPT9 | 14 | 26 | 93% (50%-100%) | 100.00% | |||||
| VIM | 12 | 26 | 83% (33%-92%) | 86.00% (73%-99%) | |||||
| Stool | AGTR1, WNT2, SLIT4 | 64 | N/A | 78% (68%-88%) | N/A | ||||
| SEPT9 | 35 | N/A | 20% (6%-31%) | N/A | |||||
| VIM | 33 | N/A | 55% (33%-70%). | N/A | |||||
| 25[60] | 2016 | Tissue | CMTM3 + SSTR2 + MDF | 42 | 42 | Pyrosequencing | 81% (69%-93%) | 91% (82%-100%) | This study has shown that the combination of CMTM3, SSTR2, and MDFI gene methylation may be an epigenetic biomarker for early stages of CRC but should be studied further to determine their potential role as non-invasive diagnostic biomarkers for CRC |
| 26[89] | 2013 | Tissue | O6-MGMT | 111 | 53 | Pyrosequencing | 34% (25%-43%) | 100% | p14arf, RASSF1A, APC1A, and O6-MGMT methylation as biomarkers of prognosis in CRC could be utilized as a relevant stratification factor in future prospective and interventional studies on CRC, and might serve as a tool in tailoring treatment for individual patients |
| p14ARF | 29% (21%-37%) | 100% | |||||||
| p16INK4a | 28% (20%-36%) | 100% | |||||||
| RASSF1A | 14% (8%-21%) | 100% | |||||||
| APC1A | 27% (19%-35%) | 100% | |||||||
| 27[90] | 2016 | Plasma | PDX1 | 20 | 20 | SYBR Green detection | 45% (23%-67%) | 70% (50%-90%) | These data demonstrate the utility of close consideration of the background levels of DNA methylation in WBC DNA as an important step in the selection of biomarkers suitable for development as plasma-based assays |
| SDC2 | 44 | 44 | 59% (45-74%) | 84.1% (73%-95%) | |||||
| IKZF1 | 74 | 144 | 68% (57-78%) | 95% (92%-99%) | |||||
| BCAT1 | 74 | 144 | MethyLight | 65% (54%-76%) | 97% (94%-100%) | ||||
| FGF5 | 20 | 40 | 85% (69%-101%) | 83% (71%-94%) | |||||
| GRASP | 44 | 44 | 55% (40%-69%) | 93%(86%-101%%) | |||||
| IRF4 | 22 | 24 | 59% (39%-80%) | 96% (88%-104%) | |||||
| SEPT9 | 44 | 44 | 59.1% (45%-74%) | 96% (89 %-102%) | |||||
| SOX21 | 20 | 20 | 85% (69%-101%) | 50% (28%-72%) | |||||
| 28[46] | 2017 | Plasma | SFRP1 | 47 | 37 | MethyLight | 85% (75%-95%) | 81% (69%-94%) | The present study offers the possibility to measure the hypermethylation of the marker panel in cell-free plasma DNA and provides a potential non-invasive, epigenetic diagnostic test. It also shows that the altered methylation pattern might serve as a key for early diagnosis of precancerous stages |
| SFRP2 | 47 | 37 | 72% (60%-85%) | 89% (79%-99%) | |||||
| SDC2 | 47 | 37 | 89% (81%-98%) | 97% (92%-103%) | |||||
| PRIMA1 | 47 | 37 | 81% (70%-92%) | 73% (59%-87%) | |||||
| 4 genes as a panel | 47 | 37 | 92% (84%-100%) | 97% (92%-102%) | |||||
The development of MT-sDNA test was a remarkable progress that was made on solving problems of CRC screening while keeping high accuracy. It is independent of the CRC lesion site and is believed to be beneficial for ameliorating both the diagnostic yield and quality of the colonoscopy examination[14]. The MT-sDNA test is a noninvasive technology by detecting CRC-related DNA markers and occult hemoglobin in the feces, including aberrantly methylated promoter regions of BMP3 and NDRG4[4,15].
Accordingly, recent advances have allowed a combined fecal immunochemical test (FIT) and multitarget DNA stool test to become commercially available. There is high possibility that MT-sDNA test could be an optimal screening choice for asymptomatic people with a moderate risk for CRC, and the early adoption of this assay is recommended[15,16]. Despite being the most sensitive noninvasive CRC screening test currently available, further investigations on this sDNA test seems to be needed[17]. What’s more, it is reported that the MT-sDNA test is not as effective as FIT and colonoscopy and even more expensive when participation rates of all strategies were identical[15].
The QuARTS is a technology combing polymerase-based target amplification with signal amplification based on invasive cleavage, with the fluorescence-based signal detection resembling real-time PCR. To the best of our knowledge, QuARTS was first used to detect DNA methylation by Zou et al[18]. Kisiel et al[19] measured candidate markers for CRC through QuARTS assays using DNA extracted from normal colon, adenoma, and CRC frozen tissues. An automated sDNA assay developed by Lidgard et al[20] uses the QuARTS method and has been reported to have a 90% specificity and 98% sensitivity when used to test the DNA methylation of stool samples from individuals with CRC[20]. Ahlquist et al[21] demonstrated that QuARTS is a very sensitive and specific method for detecting CRC in early stage and large adenomas in the colon regardless of the sites, although optimization and standardization of feces collecting methods as well as stool storing protocols are of great importance in improving the performance of QuARTS.
MS-MLPA is a quick, reliable, and cost-effective method derived from MLPA that does not require sodium bisulfite treatment and instead uses the methylation-sensitive endonuclease HhaI. If the target DNA is methylated, the HhaI enzyme cannot cut its recognition sequence, and the target sequence will then be amplified by PCR. When analyzing the data of MS-MLPA, the percentage of methylated DNA is calculated by comparing the peak patterns of HhaI-digested and -undigested products[22-24]. To perform MS-MLPA, genomic DNA must first be denatured, after which the MS-MLPA probes are added and a 16 h hybridization step is performed[25]. Furlan et al[26] used this technique to assess CpG methylation of tumor suppressor genes in CRC samples using a well-characterized series of sporadic colorectal adenocarcinoma and neuroendocrine carcinomas with known clinicopathologic and molecular profiles. The results of this study showed that an outstanding merit of MS-MLPA is being able to screen multiple cancer genes in only an assay with better sensitivity and specificity compared with the rest of the DNA methylation detection assays. Their results indicate that the MS-MLPA assay is a simple, cost-effective, reliable method for epigenetic analysis of tumor tissues and can provide innovative aspects for clinical diagnosis and treatment[27]. In a study by Mäki-Nevala et al[28] focusing on Lynch syndrome-associated adenomas, they employed MS-MLPA to evaluate DNA methylation conditions in the promoters of 8 CIMP-related genes and 7 chosen candidate genes such as TSGs and the gene LINE-1. In an investigation of the use of constitutive MLH1 methylation to diagnose Lynch syndrome in patients with tumor MLH1 downregulation, MS-MPLA was used to analyze MLH1 promoter methylation in peripheral blood leukocyte DNA from the index patients[29].
MSP is probably the most ubiquitously used technique to study DNA methylation and can rapidly detect the methylation status of DNA without the use of methylation-sensitive restriction enzymes[30]. Despite only requiring minute amounts of DNA, MSP can be utilized to detect less than 0.1% of allele methylation in a specific locus and could be applied to analyzing DNA methylation in various types of specimens like body fluids and paraffin-embedded specimens[31]. To perform MSP, primers are needed for amplifying the genomic DNA that has been treated with sodium bisulfite[27,31,32]. Normally, based on the amplification results, MSP can produce quantitative or qualitative (using agarose gels) results to assess changes in DNA methylation[33]. Draht et al[34] tested a number of tissue samples using variations of this method, including MSP, direct MSP, and nested MSP. As expected, the results showed that direct MSP is less sensitive than nested MSP. The reason is that the sensitivity of nested MSP in detecting decreased CpG island methylation of the RET promoter was higher compared with direct MSP. In a study in which a panel of tumor-specific methylation genes (NPY, PENK, and WIF1) was evaluated, a quantitative multiplex methylation-specific PCR (QM-MSP) assay was shown to be not as sensitive and specific as a direct assay of the tissue samples, but it was suggested to be an efficient test to assess serum DNA methylation[35]. To assess SDC2 methylation in DNA from bowel lavage fluid, Park et al[36] used a two-step fluorescence-based quantitative methylation-specific PCR (qMSP) method to measure SDC2 methylation of DNA from tissues and bowel lavage fluid. Sun et al[37] used MSP to detect the methylation status of RASSF1A in blood samples from patients before and after chemotherapy. Exner et al[38] used a microfluidic high-throughput and methylation-sensitive qPCR approach to assess methylated sites of DNA in freshly frozen and formalin-fixed paraffin-embedded samples. With the intension of determining whether SDC2 methylation detection in stool DNA could be used to screen CRC and adenoma, Oh et al[39] performed qMSP to analyze stool DNA.
As an endpoint analysis technique[40], the primary limitation of MSP is that it yields qualitative results, meaning that well-standardized MSP assays provide information that is restricted to the presence of methylated, unmethylated, or both methylated and unmethylated alleles[41].
MethyLight is a quantitative, fluorescence-based, real-time PCR method[42] that is capable of detecting and quantifying DNA. It is especially suitable for detecting hypomethylated DNA regions when the ratio of unmethylated DNA is relatively high because of the combined use of methylation-specific priming and methylation-specific fluorescent probing[43]. The remarkable advantages of this technique, which was proposed by Eads et al[44], are its high throughput and great sensitivity other than the ability of yielding high-resolution information of methylation. The MethyLight technique is able to show the methylation level of a specific DNA region without requiring a significant amount of DNA, and it differs from MSP by being able to quantify the relative amounts of a particular methylation pattern. The way of MethyLight quantifying products is through recording the cycle number right when the fluorescent signal goes across a threshold during the exponential amplification phase of PCR[44,45]. Barták et al[46] successfully measured the methylation level of four chosen DNA patterns in two types of specimens, colon tissues and plasma, via MethyLight assay. As reported by Li et al[47], the detection limit for SFRP2 methylation using the MethyLight assay was approximately 200 pg (approximately 60 copies of human genome) per reaction. In addition, as a PCR-based method, the throughput of MethyLight can be of great use. For example, He et al[48] reported on a multiplex MethyLight assay for the detection of methylated genes in CRC.
MS-HRM can detect DNA methylation status with a relatively good sensitivity and specificity. In MS-HRM, the difference between methylated and unmethylated DNA PCR products will be shown as different high-resolution melting curves. By comparing the results of samples with reference amplicons, the extent of methylation of the unknown samples can be estimated[49,50]. Compared to MSP and pyrosequencing, it has been reported that the MS-HRM method is an inexpensive and simple technique with high accuracy and its methylation level quantification function works as well as or somehow even better than pyrosequencing[51]. MS-HRM has a higher sensitivity than MSP, is high throughput, and is performed in a closed tube[52]. Kidambi et al[53] applied MS-HRM to detecting MLH1 intron 1 methylation not only in cancer samples, but also in peripheral white blood cells as well as buccal cells. However, the accuracy of the HRM method can be affected by factors such as the location or length of the amplicon and the DNA extraction method used[54,55]. Thus, the use of MS-HRM is typically restricted due to the fact that it could only provide the approximate range of methylation level[56].
Bisulfite sequencing (BS-Seq) is an available option for profiling methylated cytosine in DNA at the genome-wide level at single nucleotide resolution. BS-Seq is regarded as the “gold standard” for single-base resolution measurements of DNA methylation levels[57]. The basic methylation detection principle of BS-Seq is that unmethylated cytosine can be deaminated to uracil under sodium bisulfite treatment but methylated cytosine cannot. The converted DNA can be amplified using a gene-specific primer[58], and methylation status of mappable cytosine in genome was measured by means of deep sequencing of the bisulfite-treated genomic DNA[59,60]. Like all sequencing technologies, BS-Seq involves sophisticated procedures, and problems remain in the sequencing of short amplicons amplified from a bisulfite-modified template[61].
Pyrosequencing is a DNA sequencing technique that is based on the “sequencing by synthesis” method. By examining the activity of both DNA polymerase and another chemiluminescent enzyme, a single chain of DNA is sequenced using this technique by synthesizing the complementary strand and detecting which base is added at each step[62]. This method has high throughput after years of investigations[63,64]. When using this technique, DNA samples are typically treated with bisulfite to detect DNA methylation. Pyrosequencing can be employed to directly detect DNA methylation or to identify the products from MSP.
When screening for abnormal methylation patterns of genes, genome profiling is the first option used, and the Illumina BeadChip is a popular platform for profiling DNA methylation. The Illumina HumanMethylation450 BeadChip can profile more than 450000 CpGs across the human genome in a cost-effective, comprehensive manner[65,66]. Lin et al[67] used an Illumina HumanMethylation450 array while studying the clinical relevance of plasma DNA methylation in CRC patients. Sung-Eun Kim and his team used the Illumina Infinium HumanMethylation27 (HM27) BeadChip to analyze the methylation status of more than 27 thousand individual CpG sites located at promoter regions of over 14 thousand genes in cultivated cells genes in cultivated cells[68]. Pekow et al[8] utilized an Illumina Infinium HumanMethylation450 array for the analysis of around half a million methylation sites in each specimen at single-nucleotide resolution. Despite its popularity, this array-based measurement of DNA methylation status has issues with measurement variation. For instance, the negative values or truncation of low intensity signals produced by background subtraction methods can potentially introduce additional bias[69]. One study showed that part of the probes used in genome profiling could potentially cause false signals due to unspecific hybridization with unintended genomic sequences that are highly homologous to the target genes. Additionally, probes targeting polymorphism of CpGs which have overlapping SNPs have been discovered[70]. Furthermore, this technique requires the use of complicated procedures to perform DNA profiling[66,71].
In this review, we summarize and discuss all of the techniques or technologies used in scientific studies or in clinical settings. Despite being time-consuming and involving the use of sophisticated procedures, the use of DNA sequencing techniques is needed to identify genes that are aberrantly methylated for use as disease markers. When the candidate genes are identified, it is obviously unwise to use DNA sequencing to screen for them in many types of specimens. In these cases, PCR-based methods have the advantages of being generally more sensitive, scalable, specific, reliable, time-saving, and cost-effective than other methods[72]. For this reason, we believe that MS-PCR is a preferred technology, as methylation-specific PCR alone can be used to determine the methylation status of a gene. Developing methods for only one specific gene[73], in our point of view, will limit the use of PCR technology. Combined with various types of methods, the use of methylation-specific PCR can be promising to assess the methylation status of multiple genes, which can be genuinely valuable. Furthermore, not all PCR-based methods require extra equipment to further confirm multiple products. Techniques such as MS-HRM have been successfully developed for the closed-tube screening of aberrant DNA methylation with satisfactory throughput, although we have concerns regarding the deficiencies of this technique. For example, ambiguous results obtained for several samples in an HRM-based screening experiment did not allow for the straightforward classification of the methylation status of the sample[61]. Thus, we believe that the development of new high-throughput technologies is still urgently needed to allow for faster and cheaper DNA methylation detection in CRC and other diseases.
As shown in Table 1, although only a portion of the investigated markers of CRC are presented, we can see that the number of useful methylation markers is considerable, which we should make good use of. We think individuals with positive FIT results should first have the CRC DNA methylation marker screening test done instead of going straight for tests like colonoscopy. If the DNA markers show that the individual is at the average or high risk of CRC, colonoscopy and other invasive tests are recommended. However, we believe that more studies are needed for selecting the methylation markers that could indicate the precancerous changes in colon. We also suggest that combined methylation marker screening should be a routine test of regular physical examination. What’s more, in terms of detection methods, if 10 or more of these markers could be simultaneously assayed in just one closed tube using PCR-based method, the societal benefits would be immense. If the SNPs of multiple genes could be detected and identified in just one tube[74,75], there could be a means of developing a PCR-based method to detect the methylation status of multiple genes. With the identification of Hachimoji DNA[76], faster, simpler, and more cost-effective technologies for methylation analysis are not far off.
Manuscript source: Invited manuscript
Specialty type: Medicine, research and experimental
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Miyoshi E S-Editor: Ma RY L-Editor: Wang TQ E-Editor: Wu YXJ
| 1. | Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330-337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6773] [Cited by in RCA: 6645] [Article Influence: 511.2] [Reference Citation Analysis (0)] |
| 2. | Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3058] [Cited by in RCA: 3287] [Article Influence: 410.9] [Reference Citation Analysis (3)] |
| 3. | Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20108] [Cited by in RCA: 20495] [Article Influence: 2049.5] [Reference Citation Analysis (20)] |
| 4. | Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, Ahlquist DA, Berger BM. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287-1297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 1231] [Article Influence: 111.9] [Reference Citation Analysis (1)] |
| 5. | Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546-1558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5906] [Cited by in RCA: 5586] [Article Influence: 465.5] [Reference Citation Analysis (0)] |
| 6. | Freitas M, Ferreira F, Carvalho S, Silva F, Lopes P, Antunes L, Salta S, Diniz F, Santos LL, Videira JF, Henrique R, Jerónimo C. A novel DNA methylation panel accurately detects colorectal cancer independently of molecular pathway. J Transl Med. 2018;16:45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 7. | Mousavi Ardehaie R, Hashemzadeh S, Behrouz Sharif S, Ghojazadeh M, Teimoori-Toolabi L, Sakhinia E. Aberrant methylated EDNRB can act as a potential diagnostic biomarker in sporadic colorectal cancer while KISS1 is controversial. Bioengineered. 2017;8:555-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 8. | Pekow J, Hernandez K, Meckel K, Deng Z, Haider HI, Khalil A, Zhang C, Talisila N, Siva S, Jasmine F, Li YC, Rubin DT, Hyman N, Bissonnette M, Weber C, Kibriya MG. IBD-associated Colon Cancers Differ in DNA Methylation and Gene Expression Profiles Compared With Sporadic Colon Cancers. J Crohns Colitis. 2019;13:884-893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 9. | Rex DK, Bond JH, Winawer S, Levin TR, Burt RW, Johnson DA, Kirk LM, Litlin S, Lieberman DA, Waye JD, Church J, Marshall JB, Riddell RH; U. S. Multi-Society Task Force on Colorectal Cancer. Quality in the technical performance of colonoscopy and the continuous quality improvement process for colonoscopy: recommendations of the U.S. Multi-Society Task Force on Colorectal Cancer. Am J Gastroenterol. 2002;97:1296-1308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 691] [Cited by in RCA: 721] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 10. | Keating NL, James O'Malley A, Onnela JP, Landon BE. Assessing the impact of colonoscopy complications on use of colonoscopy among primary care physicians and other connected physicians: an observational study of older Americans. BMJ Open. 2017;7:e014239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 11. | McCarty TR, Liu A, Njei B. Splenic Injury and Hemoperitoneum as a Complication of Colonoscopy: A Case Report and Literature Review. Conn Med. 2016;80:217-221. [PubMed] |
| 12. | Lech G, Słotwiński R, Słodkowski M, Krasnodębski IW. Colorectal cancer tumour markers and biomarkers: Recent therapeutic advances. World J Gastroenterol. 2016;22:1745-1755. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 234] [Cited by in RCA: 285] [Article Influence: 31.7] [Reference Citation Analysis (8)] |
| 13. | van Engeland M, Derks S, Smits KM, Meijer GA, Herman JG. Colorectal cancer epigenetics: complex simplicity. J Clin Oncol. 2011;29:1382-1391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 139] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 14. | Johnson DH, Kisiel JB, Burger KN, Mahoney DW, Devens ME, Ahlquist DA, Sweetser S. Multitarget stool DNA test: clinical performance and impact on yield and quality of colonoscopy for colorectal cancer screening. Gastrointest Endosc. 2017;85:657-665.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 15. | Malik P. A novel multitarget stool DNA test for colorectal cancer screening. Postgrad Med. 2016;128:268-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 16. | Finney Rutten LJ, Jacobson RM, Wilson PM, Jacobson DJ, Fan C, Kisiel JB, Sweetser S, Tulledge-Scheitel SM, St Sauver JL. Early Adoption of a Multitarget Stool DNA Test for Colorectal Cancer Screening. Mayo Clin Proc. 2017;92:726-733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 17. | Kadiyska T, Nossikoff A. Stool DNA methylation assays in colorectal cancer screening. World J Gastroenterol. 2015;21:10057-10061. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 22] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 18. | Zou H, Allawi H, Cao X, Domanico M, Harrington J, Taylor WR, Yab T, Ahlquist DA, Lidgard G. Quantification of methylated markers with a multiplex methylation-specific technology. Clin Chem. 2012;58:375-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 67] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 19. | Kisiel JB, Klepp P, Allawi HT, Taylor WR, Giakoumopoulos M, Sander T, Yab TC, Moum BA, Lidgard GP, Brackmann S, Mahoney DW, Roseth A, Ahlquist DA. Analysis of DNA Methylation at Specific Loci in Stool Samples Detects Colorectal Cancer and High-Grade Dysplasia in Patients With Inflammatory Bowel Disease. Clin Gastroenterol Hepatol. 2019;17:914-921.e5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 20. | Lidgard GP, Domanico MJ, Bruinsma JJ, Light J, Gagrat ZD, Oldham-Haltom RL, Fourrier KD, Allawi H, Yab TC, Taylor WR, Simonson JA, Devens M, Heigh RI, Ahlquist DA, Berger BM. Clinical performance of an automated stool DNA assay for detection of colorectal neoplasia. Clin Gastroenterol Hepatol. 2013;11:1313-1318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 114] [Article Influence: 9.5] [Reference Citation Analysis (1)] |
| 21. | Ahlquist DA, Zou H, Domanico M, Mahoney DW, Yab TC, Taylor WR, Butz ML, Thibodeau SN, Rabeneck L, Paszat LF, Kinzler KW, Vogelstein B, Bjerregaard NC, Laurberg S, Sørensen HT, Berger BM, Lidgard GP. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology. 2012;142:248-256; quiz e25-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 232] [Article Influence: 17.8] [Reference Citation Analysis (1)] |
| 22. | Kim MJ, Cho SI, Chae JH, Lim BC, Lee JS, Lee SJ, Seo SH, Park H, Cho A, Kim SY, Kim JY, Park SS, Seong MW. Pitfalls of Multiple Ligation-Dependent Probe Amplifications in Detecting DMD Exon Deletions or Duplications. J Mol Diagn. 2016;18:253-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 23. | Moelans CB, Atanesyan L, Savola SP, van Diest PJ. Methylation-Specific Multiplex Ligation-Dependent Probe Amplification (MS-MLPA). Methods Mol Biol. 2018;1708:537-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 24. | Alkorta-Aranburu G, Sukhanova M, Carmody D, Hoffman T, Wysinger L, Keller-Ramey J, Li Z, Johnson AK, Kobiernicki F, Botes S, Fitzpatrick C, Das S, Del Gaudio D. Improved molecular diagnosis of patients with neonatal diabetes using a combined next-generation sequencing and MS-MLPA approach. J Pediatr Endocrinol Metab. 2016;29:523-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 25. | Nygren AO, Ameziane N, Duarte HM, Vijzelaar RN, Waisfisz Q, Hess CJ, Schouten JP, Errami A. Methylation-specific MLPA (MS-MLPA): simultaneous detection of CpG methylation and copy number changes of up to 40 sequences. Nucleic Acids Res. 2005;33:e128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 275] [Cited by in RCA: 284] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 26. | Furlan D, Sahnane N, Mazzoni M, Pastorino R, Carnevali I, Stefanoli M, Ferretti A, Chiaravalli AM, La Rosa S, Capella C. Diagnostic utility of MS-MLPA in DNA methylation profiling of adenocarcinomas and neuroendocrine carcinomas of the colon-rectum. Virchows Arch. 2013;462:47-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 27. | Wu J, Wang G, He B, Chen X, An Y. Methylation of the UNC5C gene and its protein expression in colorectal cancer. Tumour Biol. 2017;39:1010428317697564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 28. | Mäki-Nevala S, Valo S, Ristimäki A, Sarhadi V, Knuutila S, Nyström M, Renkonen-Sinisalo L, Lepistö A, Mecklin JP, Peltomäki P. DNA methylation changes and somatic mutations as tumorigenic events in Lynch syndrome-associated adenomas retaining mismatch repair protein expression. EBioMedicine. 2019;39:280-291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 29. | Pinto D, Pinto C, Guerra J, Pinheiro M, Santos R, Vedeld HM, Yohannes Z, Peixoto A, Santos C, Pinto P, Lopes P, Lothe R, Lind GE, Henrique R, Teixeira MR. Contribution of MLH1 constitutional methylation for Lynch syndrome diagnosis in patients with tumor MLH1 downregulation. Cancer Med. 2018;7:433-444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 30. | Ramalho-Carvalho J, Henrique R, Jerónimo C. Methylation-Specific PCR. Methods Mol Biol. 2018;1708:447-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 31. | McCormick TM, Carvalho MDGDC. Using Methylation-Specific PCR to Study RB1 Promoter Hypermethylation. Methods Mol Biol. 2018;1726:29-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 32. | Wani K, Aldape KD. PCR Techniques in Characterizing DNA Methylation. Methods Mol Biol. 2016;1392:177-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 33. | Lizardi PM, Yan Q, Wajapeyee N. Methylation-Specific Polymerase Chain Reaction (PCR) for Gene-Specific DNA Methylation Detection. Cold Spring Harb Protoc. 2017;2017:pdb.prot094847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 34. | Draht MX, Smits KM, Jooste V, Tournier B, Vervoort M, Ramaekers C, Chapusot C, Weijenberg MP, van Engeland M, Melotte V. Analysis of RET promoter CpG island methylation using methylation-specific PCR (MSP), pyrosequencing, and methylation-sensitive high-resolution melting (MS-HRM): impact on stage II colon cancer patient outcome. Clin Epigenetics. 2016;8:44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 35. | Roperch JP, Incitti R, Forbin S, Bard F, Mansour H, Mesli F, Baumgaertner I, Brunetti F, Sobhani I. Aberrant methylation of NPY, PENK, and WIF1 as a promising marker for blood-based diagnosis of colorectal cancer. BMC Cancer. 2013;13:566. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 114] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 36. | Park YS, Kim DS, Cho SW, Park JW, Jeon SJ, Moon TJ, Kim SH, Son BK, Oh TJ, An S, Kim JH, Chae JD. Analysis of Syndecan-2 Methylation in Bowel Lavage Fluid for the Detection of Colorectal Neoplasm. Gut Liver. 2018;12:508-515. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 37. | Sun X, Yuan W, Hao F, Zhuang W. Promoter Methylation of RASSF1A indicates Prognosis for Patients with Stage II and III Colorectal Cancer Treated with Oxaliplatin-Based Chemotherapy. Med Sci Monit. 2017;23:5389-5395. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 38. | Exner R, Pulverer W, Diem M, Spaller L, Woltering L, Schreiber M, Wolf B, Sonntagbauer M, Schröder F, Stift J, Wrba F, Bergmann M, Weinhäusel A, Egger G. Potential of DNA methylation in rectal cancer as diagnostic and prognostic biomarkers. Br J Cancer. 2015;113:1035-1045. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 39. | Oh TJ, Oh HI, Seo YY, Jeong D, Kim C, Kang HW, Han YD, Chung HC, Kim NK, An S. Feasibility of quantifying SDC2 methylation in stool DNA for early detection of colorectal cancer. Clin Epigenetics. 2017;9:126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 40. | Choudhury JH, Das R, Laskar S, Kundu S, Kumar M, Das PP, Choudhury Y, Mondal R, Ghosh SK. Detection of p16 Promoter Hypermethylation by Methylation-Specific PCR. Methods Mol Biol. 2018;1726:111-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 41. | Hernández HG, Tse MY, Pang SC, Arboleda H, Forero DA. Optimizing methodologies for PCR-based DNA methylation analysis. Biotechniques. 2013;55:181-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 120] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 42. | Campan M, Weisenberger DJ, Trinh B, Laird PW. MethyLight. Methods Mol Biol. 2009;507:325-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 95] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 43. | Campan M, Weisenberger DJ, Trinh B, Laird PW. MethyLight and Digital MethyLight. Methods Mol Biol. 2018;1708:497-513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 44. | Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1017] [Cited by in RCA: 1054] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 45. | Dallol A, Al-Ali W, Al-Shaibani A, Al-Mulla F. Analysis of DNA methylation in FFPE tissues using the MethyLight technology. Methods Mol Biol. 2011;724:191-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 46. | Barták BK, Kalmár A, Péterfia B, Patai ÁV, Galamb O, Valcz G, Spisák S, Wichmann B, Nagy ZB, Tóth K, Tulassay Z, Igaz P, Molnár B. Colorectal adenoma and cancer detection based on altered methylation pattern of SFRP1, SFRP2, SDC2, and PRIMA1 in plasma samples. Epigenetics. 2017;12:751-763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 97] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 47. | Li H, Wang Z, Zhao G, Ma Y, Chen Y, Xue Q, Zheng M, Fei S. Performance of a MethyLight assay for methylated SFRP2 DNA detection in colorectal cancer tissue and serum. Int J Biol Markers. 2019;34:54-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 48. | He Q, Chen HY, Bai EQ, Luo YX, Fu RJ, He YS, Jiang J, Wang HQ. Development of a multiplex MethyLight assay for the detection of multigene methylation in human colorectal cancer. Cancer Genet Cytogenet. 2010;202:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 91] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 49. | Wojdacz TK, Dobrovic A. Methylation-sensitive high resolution melting (MS-HRM): a new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res. 2007;35:e41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 377] [Cited by in RCA: 406] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 50. | Wojdacz TK, Dobrovic A, Hansen LL. Methylation-sensitive high-resolution melting. Nat Protoc. 2008;3:1903-1908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 224] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 51. | Switzeny OJ, Christmann M, Renovanz M, Giese A, Sommer C, Kaina B. MGMT promoter methylation determined by HRM in comparison to MSP and pyrosequencing for predicting high-grade glioma response. Clin Epigenetics. 2016;8:49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 57] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 52. | Xiao Z, Li B, Wang G, Zhu W, Wang Z, Lin J, Xu A, Wang X. Validation of methylation-sensitive high-resolution melting (MS-HRM) for the detection of stool DNA methylation in colorectal neoplasms. Clin Chim Acta. 2014;431:154-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 53. | Kidambi TD, Blanco A, Van Ziffle J, Terdiman JP. Constitutional MLH1 methylation presenting with colonic polyposis syndrome and not Lynch syndrome. Fam Cancer. 2016;15:275-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 54. | Martín-Núñez GM, Gómez-Zumaquero JM, Soriguer F, Morcillo S. High resolution melting curve analysis of DNA samples isolated by different DNA extraction methods. Clin Chim Acta. 2012;413:331-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 55. | Słomka M, Sobalska-Kwapis M, Wachulec M, Bartosz G, Strapagiel D. High Resolution Melting (HRM) for High-Throughput Genotyping-Limitations and Caveats in Practical Case Studies. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 80] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 56. | Migheli F, Stoccoro A, Coppedè F, Wan Omar WA, Failli A, Consolini R, Seccia M, Spisni R, Miccoli P, Mathers JC, Migliore L. Comparison study of MS-HRM and pyrosequencing techniques for quantification of APC and CDKN2A gene methylation. PLoS One. 2013;8:e52501. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 57. | Li Q, Hermanson PJ, Springer NM. Detection of DNA Methylation by Whole-Genome Bisulfite Sequencing. Methods Mol Biol. 2018;1676:185-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 58. | Lizardi PM, Yan Q, Wajapeyee N. DNA Bisulfite Sequencing for Single-Nucleotide-Resolution DNA Methylation Detection. Cold Spring Harb Protoc. 2017;2017:pdb.prot094839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 59. | Chen YR, Yu S, Zhong S. Profiling DNA Methylation Using Bisulfite Sequencing (BS-Seq). Methods Mol Biol. 2018;1675:31-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 60. | Li J, Chen C, Bi X, Zhou C, Huang T, Ni C, Yang P, Chen S, Ye M, Duan S. DNA methylation of CMTM3, SSTR2, and MDFI genes in colorectal cancer. Gene. 2017;630:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 61. | Wojdacz TK, Møller TH, Thestrup BB, Kristensen LS, Hansen LL. Limitations and advantages of MS-HRM and bisulfite sequencing for single locus methylation studies. Expert Rev Mol Diagn. 2010;10:575-580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 56] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 62. | Uhlen M. Magnetic separation of DNA. Nature. 1989;340:733-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 138] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 63. | Ronaghi M, Uhlén M, Nyrén P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363, 365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1055] [Cited by in RCA: 898] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 64. | Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5541] [Cited by in RCA: 5065] [Article Influence: 253.3] [Reference Citation Analysis (0)] |
| 65. | Phipson B, Maksimovic J, Oshlack A. missMethyl: an R package for analyzing data from Illumina's HumanMethylation450 platform. Bioinformatics. 2016;32:286-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 550] [Article Influence: 55.0] [Reference Citation Analysis (0)] |
| 66. | Carless MA. Determination of DNA methylation levels using Illumina HumanMethylation450 BeadChips. Methods Mol Biol. 2015;1288:143-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 67. | Lin PC, Lin JK, Lin CH, Lin HH, Yang SH, Jiang JK, Chen WS, Chou CC, Tsai SF, Chang SC. Clinical Relevance of Plasma DNA Methylation in Colorectal Cancer Patients Identified by Using a Genome-Wide High-Resolution Array. Ann Surg Oncol. 2015;22 Suppl 3:S1419-S1427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 68. | Kim SE, Hinoue T, Kim MS, Sohn KJ, Cho RC, Weisenberger DJ, Laird PW, Kim YI. Effects of folylpolyglutamate synthase modulation on global and gene-specific DNA methylation and gene expression in human colon and breast cancer cells. J Nutr Biochem. 2016;29:27-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 69. | Xu Z, Niu L, Li L, Taylor JA. ENmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. Nucleic Acids Res. 2016;44:e20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 183] [Cited by in RCA: 278] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 70. | Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ, Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1050] [Cited by in RCA: 1132] [Article Influence: 94.3] [Reference Citation Analysis (0)] |
| 71. | Lehne B, Drong AW, Loh M, Zhang W, Scott WR, Tan ST, Afzal U, Scott J, Jarvelin MR, Elliott P, McCarthy MI, Kooner JS, Chambers JC. A coherent approach for analysis of the Illumina HumanMethylation450 BeadChip improves data quality and performance in epigenome-wide association studies. Genome Biol. 2015;16:37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 299] [Cited by in RCA: 278] [Article Influence: 27.8] [Reference Citation Analysis (1)] |
| 72. | Chen JJ, Wang AQ, Chen QQ. DNA methylation assay for colorectal carcinoma. Cancer Biol Med. 2017;14:42-49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 73. | Wang Y, Chen PM, Liu RB. Advance in plasma SEPT9 gene methylation assay for colorectal cancer early detection. World J Gastrointest Oncol. 2018;10:15-22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 38] [Cited by in RCA: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 74. | Luo G, Zheng L, Zhang X, Zhang J, Nilsson-Ehle P, Xu N. Genotyping of single nucleotide polymorphisms using base-quenched probe: a method does not invariably depend on the deoxyguanosine nucleotide. Anal Biochem. 2009;386:161-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 75. | Mao H, Luo G, Zhang J, Xu N. Detection of simultaneous multi-mutations using base-quenched probe. Anal Biochem. 2018;543:79-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 76. | Hoshika S, Leal NA, Kim MJ, Kim MS, Karalkar NB, Kim HJ, Bates AM, Watkins NE, SantaLucia HA, Meyer AJ, DasGupta S, Piccirilli JA, Ellington AD, SantaLucia J, Georgiadis MM, Benner SA. Hachimoji DNA and RNA: A genetic system with eight building blocks. Science. 2019;363:884-887. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 307] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 77. | Cesaroni M, Powell J, Sapienza C. Validation of methylation biomarkers that distinguish normal colon mucosa of cancer patients from normal colon mucosa of patients without cancer. Cancer Prev Res (Phila). 2014;7:717-726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 78. | Hinoue T, Weisenberger DJ, Lange CP, Shen H, Byun HM, Van Den Berg D, Malik S, Pan F, Noushmehr H, van Dijk CM, Tollenaar RA, Laird PW. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22:271-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 455] [Cited by in RCA: 486] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 79. | Jedi M, Young GP, Pedersen SK, Symonds EL. Methylation and Gene Expression of BCAT1 and IKZF1 in Colorectal Cancer Tissues. Clin Med Insights Oncol. 2018;12:1179554918775064. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 80. | Young GP, Pedersen SK, Mansfield S, Murray DH, Baker RT, Rabbitt P, Byrne S, Bambacas L, Hollington P, Symonds EL. A cross-sectional study comparing a blood test for methylated BCAT1 and IKZF1 tumor-derived DNA with CEA for detection of recurrent colorectal cancer. Cancer Med. 2016;5:2763-2772. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 81] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 81. | Pedersen SK, Symonds EL, Baker RT, Murray DH, McEvoy A, Van Doorn SC, Mundt MW, Cole SR, Gopalsamy G, Mangira D, LaPointe LC, Dekker E, Young GP. Evaluation of an assay for methylated BCAT1 and IKZF1 in plasma for detection of colorectal neoplasia. BMC Cancer. 2015;15:654. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 98] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 82. | Pedersen SK, Baker RT, McEvoy A, Murray DH, Thomas M, Molloy PL, Mitchell S, Lockett T, Young GP, LaPointe LC. A two-gene blood test for methylated DNA sensitive for colorectal cancer. PLoS One. 2015;10:e0125041. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 83. | Alizadeh Naini M, Kavousipour S, Hasanzarini M, Nasrollah A, Monabati A, Mokarram P. O6-Methyguanine-DNA Methyl Transferase (MGMT) Promoter Methylation in Serum DNA of Iranian Patients with Colorectal Cancer. Asian Pac J Cancer Prev. 2018;19:1223-1227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 84. | Michailidi C, Theocharis S, Tsourouflis G, Pletsa V, Kouraklis G, Patsouris E, Papavassiliou AG, Troungos C. Expression and promoter methylation status of hMLH1, MGMT, APC, and CDH1 genes in patients with colon adenocarcinoma. Exp Biol Med (Maywood). 2015;240:1599-1605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 85. | Kohonen-Corish MR, Tseung J, Chan C, Currey N, Dent OF, Clarke S, Bokey L, Chapuis PH. KRAS mutations and CDKN2A promoter methylation show an interactive adverse effect on survival and predict recurrence of rectal cancer. Int J Cancer. 2014;134:2820-2828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 86. | Zheng R, Gao D, He T, Zhang M, Zhang X, Linghu E, Wei L, Guo M. Methylation of DIRAS1 promotes colorectal cancer progression and may serve as a marker for poor prognosis. Clin Epigenetics. 2017;9:50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 87. | Wang Y, He T, Herman JG, Linghu E, Yang Y, Fuks F, Zhou F, Song L, Guo M. Methylation of ZNF331 is an independent prognostic marker of colorectal cancer and promotes colorectal cancer growth. Clin Epigenetics. 2017;9:115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 88. | Carmona FJ, Azuara D, Berenguer-Llergo A, Fernández AF, Biondo S, de Oca J, Rodriguez-Moranta F, Salazar R, Villanueva A, Fraga MF, Guardiola J, Capellá G, Esteller M, Moreno V. DNA methylation biomarkers for noninvasive diagnosis of colorectal cancer. Cancer Prev Res (Phila). 2013;6:656-665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 102] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 89. | Nilsson TK, Löf-Öhlin ZM, Sun XF. DNA methylation of the p14ARF, RASSF1A and APC1A genes as an independent prognostic factor in colorectal cancer patients. Int J Oncol. 2013;42:127-133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 90. | Mitchell SM, Ho T, Brown GS, Baker RT, Thomas ML, McEvoy A, Xu ZZ, Ross JP, Lockett TJ, Young GP, LaPointe LC, Pedersen SK, Molloy PL. Evaluation of Methylation Biomarkers for Detection of Circulating Tumor DNA and Application to Colorectal Cancer. Genes (Basel). 2016;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 44] [Article Influence: 4.9] [Reference Citation Analysis (0)] |