Published online Apr 6, 2024. doi: 10.12998/wjcc.v12.i10.1750
Peer-review started: January 1, 2024
First decision: January 16, 2024
Revised: January 21, 2024
Accepted: February 29, 2024
Article in press: February 29, 2024
Published online: April 6, 2024
Processing time: 91 Days and 21.6 Hours
Both N6-methyladenosine (m6A) methylation and autophagy are considered relevant to the pathogenesis of ulcerative colitis (UC). However, a systematic exploration of the role of the com-bination of m6A methylation and autophagy in UC remains to be performed.
To elucidate the autophagy-related genes of m6A with a diagnostic value for UC.
The correlation between m6A-related genes and autophagy-related genes (ARGs) was analyzed. Finally, gene set enrichment analysis (GSEA) was performed on the characteristic genes. Additionally, the expression levels of four characteristic genes were verified in dextran sulfate sodium (DSS)-induced colitis in mice.
GSEA indicated that BAG3, P4HB and TP53INP2 were involved in the inflammatory response and TNF-α signalling via nuclear factor kappa-B. Furthermore, polymerase chain reaction results showed significantly higher mRNA levels of BAG3 and P4HB and lower mRNA levels of FMR1 and TP53INP2 in the DSS group compared to the control group.
This study identified four m6A-ARGs that predict the occurrence of UC, thus providing a scientific reference for further studies on the pathogenesis of UC.
Core Tip: Both N6-methyladenosine (m6A) methylation and autophagy are considered relevant to the pathogenesis of ulcerative colitis (UC). This study identified four m6A-related genes and autophagy-related genes that predict the occurrence of UC, thus providing a scientific reference for further studies on the pathogenesis of UC.
- Citation: Liu XY, Qiao D, Zhang YL, Liu ZX, Chen YL, Que RY, Cao HY, Dai YC. Identification of marker genes associated with N6-methyladenosine and autophagy in ulcerative colitis. World J Clin Cases 2024; 12(10): 1750-1765
- URL: https://www.wjgnet.com/2307-8960/full/v12/i10/1750.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i10.1750
Ulcerative colitis (UC) is a complex, chronic, immune-mediated, colitis disease. The UC lesions are mostly located in the sigmoid colon and rectum even the whole colon[1]. The clinical manifestations of UC can take many forms, with bloody diarrhoea as the most obvious early symptom. Other symptoms include abdominal pain, bloody stool, weight loss, tenesmus and vomiting. UC is characterised by the alternation of the active phase and remission phase; however, its clinical process is unclear[2]. The incidence rate of UC is on the rise worldwide, with treatment proving to be difficult. Approximately 15% of patients with UC experience an aggressive course, and some will even develop colorectal cancer. Current non-surgical treatment options include 5-aminosalicylic acid (5-ASA), glucocorticoids, immunosuppressants, biological agents and probiotics, which are limited by high recurrence rates and varying side effects. Current research on the pathogenesis of UC mainly focuses on microbiota, genetics, immunity and intestinal mucosal barrier. However, the exact pathogenesis of UC remains unclear. Therefore, exploring the aetiology of UC is of great significance for the diagnosis and treatment of UC[3].
N6-methyladenosine (m6A) methylation is one of the most common RNA modifications and plays a key role in the development and progression of various diseases. It has been reported that changes related to m6A are associated with intestinal microbiota changes and gastrointestinal cancer development[4]. Currently, studies have investigated m6A methylation in UC, which revealed the role of m6A methylation in the pathogenesis of UC[5,6].
Autophagy is a conservative degradation process, which is critical for regulating major cellular functions and biological metabolic processes. Thus, impaired autophagy could lead to many diseases, including cancer, cardiomyopathy, neurodegenerative diseases and ageing. Moreover, autophagy disorder can also lead to inflammation, intestinal barrier destruction and intestinal homeostasis imbalance, thus increasing the risk of colon disease[7]. Recently, autophagy-related gene polymorphisms have been reported to be strongly associated with an increased risk of UC. Additionally, the therapeutic effect of certain UC drugs is indicated to be mediated by regulating the autophagy pathway[8]. Recent studies have shown that the methylation of m6A RNA can regulate autophagy gene expression and affect autophagy function. Both m6A modification and autophagy play a key role in the occurrence and development of human diseases; however, the combined role of m6A and autophagy in UC remains unexplored[9].
In this study, we used publicly available data related to UC and comprehensive bioinformatics methods to elucidate the autophagy-related genes of m6A with a diagnostic value for UC. Additionally, animal models were also used to validate and explore the potential regulatory mechanism of m6A-autophagy in UC, thereby contributing to the deve
UC-related datasets (GSE8747 and GSE75214) were downloaded from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/). The training set was the GSE87473 dataset, which comprised 21 normal and 106 UC samples. The GSE75214 dataset, comprising 11 normal and 97 UC samples, was used as the external validation set. In total, 222 ARGs were acquired from the Human Autophagy database (http://www.autophagy.lu/index.html) (Supplementary Table 1). Furthermore, 23 m6A-related genes were obtained from the published literature (Supplementary Table 1)[10].
The mRNA expression levels between the UC and normal groups in the GSE87473 dataset were compared using the ‘limma’ package (version 3.52.4) with adj P < 0.05 and |log2FC| ≥ 0.5[11]. Subsequently, the ‘clusterProfiler’ R package (version 4.4.4) was used to perform the biological functional enrichment analysis of differentially expressed genes (DEGs) between the UC and normal groups with Gene Ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG )(P < 0.05)[12].
Correlations between the m6A-related genes and ARGs were calculated using the ‘rcorr’ function of the ‘Hmisc’ R package (version 4.7-1) (P < 0.05). The intersection of the m6A-autophagy-related genes (m6A-ARGs) and DEGs was performed using the Venn tool to acquire m6A-AR DEGs. STRING was adopted to explore the protein-protein interaction (PPI) of m6A-AR DEGs. Furthermore, Cytoscape (version 3.8.0) was utilised to visualise the co-expression network of m6A-ARGs and the PPI network of m6A-AR DEGs.
Univariate logistic regression was used to initially screen variables in the identified m6A-AR DEGs. Additionally, the least absolute shrinkage and selection operator (LASSO) and support vector machines (SVM) were further applied for screening characteristic genes. Moreover, the diagnostic value for UC of the characteristic gene was assessed using receiver operating characteristic (ROC) curves and the area under the curve (AUC).
The CIBERSORT algorithm (version 1.03) was utilised to assess the infiltrating abundance of 22 immune cells between the UC and normal groups in the training set[13]. Differences between UC and normal groups were analysed using a Wilcox test, and a boxplot was plotted using the ‘ggplot2’ R package (version 3.3.6). Additionally, the correlation between characteristic genes and differential immune cells was analysed using the cor function of R language.
Based on the median value of the expression of the characteristic genes, the samples in GSE87473 were grouped into high and low-expression groups, and differential analysis was performed. Hallmark gene sets were downloaded using the ‘msigdbr’ R package (version 7.5.1) as a reference. Sorted DEGs were subjected to enrichment analysis (adj. P < 0.05).
To further demonstrate the validity of our results, the expression levels of the characteristic genes were compared between the UC and normal groups in the GSE87473 and GSE75214 datasets for external validation.
C57BL/6 mice (6-8 wk old, Quality Certification of Laboratory Animals: SCXK (Shanghai) 2017-0012) were housed in the Animal Experiment Centre of Shanghai University of Traditional Chinese Medicine, at a temperature of 22℃ ± 2℃, alternating between light and dark (SPF class), 50% ± 10% relative humidity. All animals had unlimited access to standard diet and water and general health status was checked daily by veterinarians. All animals (n = 20) were divided randomly into two groups (five animals per cage): control group (n = 10) and dextran sulfate sodium (DSS) group (n = 10). The mice in the DSS group were given DSS solution (3.5%) for seven days. Mice in the control group drank water normally. All animal experiments conformed to the internationally accepted principles for the care and use of laboratory animals (No. PZSHUTCM210611001, the Animal Ethics Committee of the Shanghai University of Traditional Chinese Medicine).
Colitis was evaluated by disease activity index (DAI) score, including body weight, stool consistency, and fecal occult blood or gross bleeding[14]. DAI was calculated by grading on a scale of 0 to 4 using the following parameters: loss of body weight (0: normal; 1: 0%-5%; 2: 5%-10%; 3: 10%-15%; 4: > 15%); stool consistency (0: normal; 2: loose stools; 4: watery diarrhea); and occult blood (0: negative; 2: positive; 4: gross bleeding). The final result was expressed as the average of the three.
At the end of the experiments, all animals were euthanized by intraperitoneal injection of 80 mg/kg 1% sodium pentobarbital. After euthanasia, the whole colon was collected and the feces and surrounding connective tissue were removed. Freshly isolated organs were kept frozen at –80 °C for RNA isolation and analyses of gene expression.
After the experiment, colonic tissues were collected for quantitative real-time polymerase chain reaction (Table 1). Total RNA was extracted with TRIzol by homogenizing the tissue. Total RNA quality was assessed by measuring the absorbance at 260 and 280 nm using a NanoDrop-2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States) and the 260/280 ratio ranged between 1.8 and 2.0. Then samples were used to reverse transcription and synthesize cDNA by the Evo M-MLV RT Premix. The samples were diluted using the SYBR® Green Premix Pro Taq HS quantitative real-time polymerase chain reaction (qPCR) Kit (High Rox Plus) (Accurate Biology, China). qPCR and melting-curve analyses were performed using StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, United States). The PCR reactions were set for 40 cycles. Each cycle was fixed at 95 °C for 30 s, 95 °C for 3 s, and 60 °C for 30 s. These primers have been validated for specificity and efficiency using conventional RT-PCR. Relative gene expression was calculated based on 2–ΔΔCt method.
| Gene | Sequence (5’-3’) | PCR products (bp) | |
| Mus BAG3 | Forward | CACCTTTTCCATGCCTACTCCC | 108 |
| Reverse | TTCTGTCATGCCGCCACGTA | ||
| Mus P4HB | Forward | TCACTGAACAGACAGCTCCGAA | 144 |
| Reverse | ATAGGATCTTGCCCTTGAAGCC | ||
| Mus FMR1 | Forward | ATGTCATACAGGTTCCACGAAAC | 77 |
| Reverse | GTCCACGATCTCTTGAATCAGC | ||
| Mus TP53INP2 | Forward | CATTGAGCATCCCAGCATGTCC | 117 |
| Reverse | TCTCCATCGCTGAGGTCCTG | ||
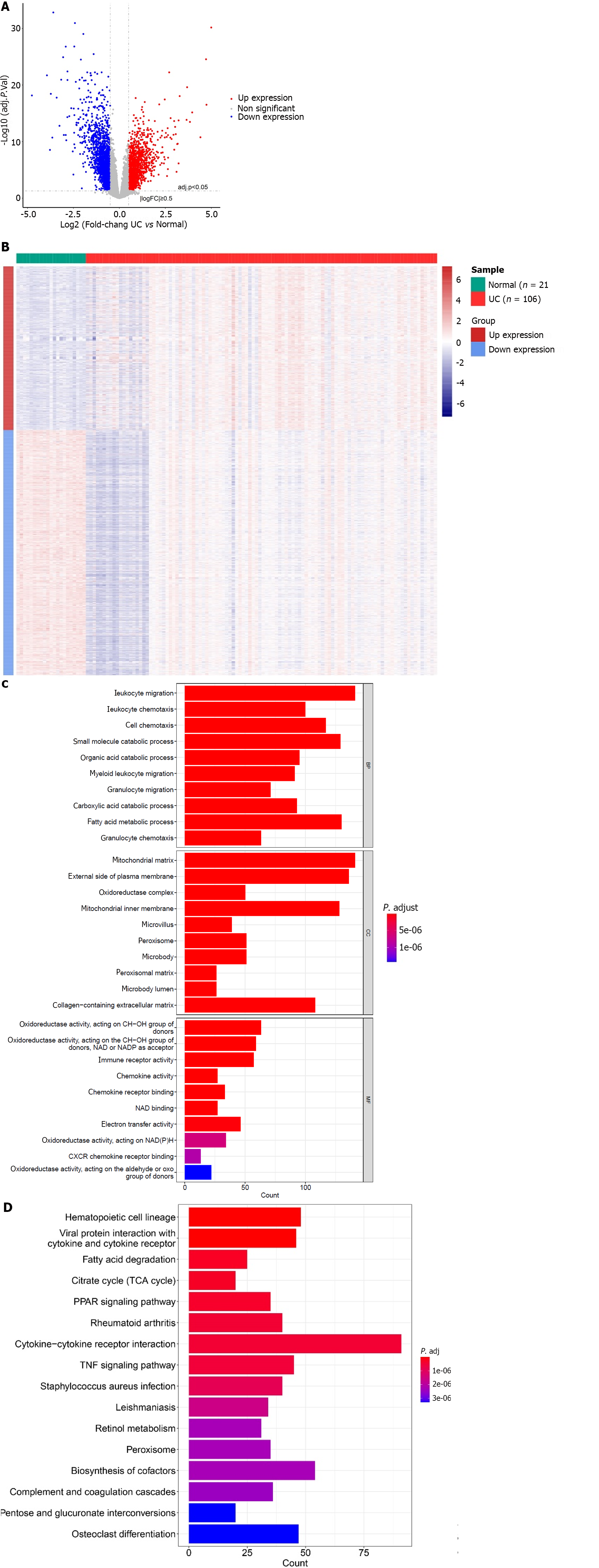
In total, 3512 DEGs were identified between the UC and normal groups in GSE87473 (Figure 1A and B). Enrichment analysis revealed a total of 1247 GO BP, 143 GO MF and 97 GO CC that were related to DEGs, such as small molecule catabolic process, oxidoreductase activity, catabolism of organic acids process, acting on CH-OH group of donors, mitochondrial matrix, peroxisomal matrix and microbody lumen (Figure 1C). Moreover, there were 67 KEGG pathways such as retinol metabolism, co-factors biosynthesis and cytokine-cytokine receptor interaction, that were related to DEGs (Figure 1D).
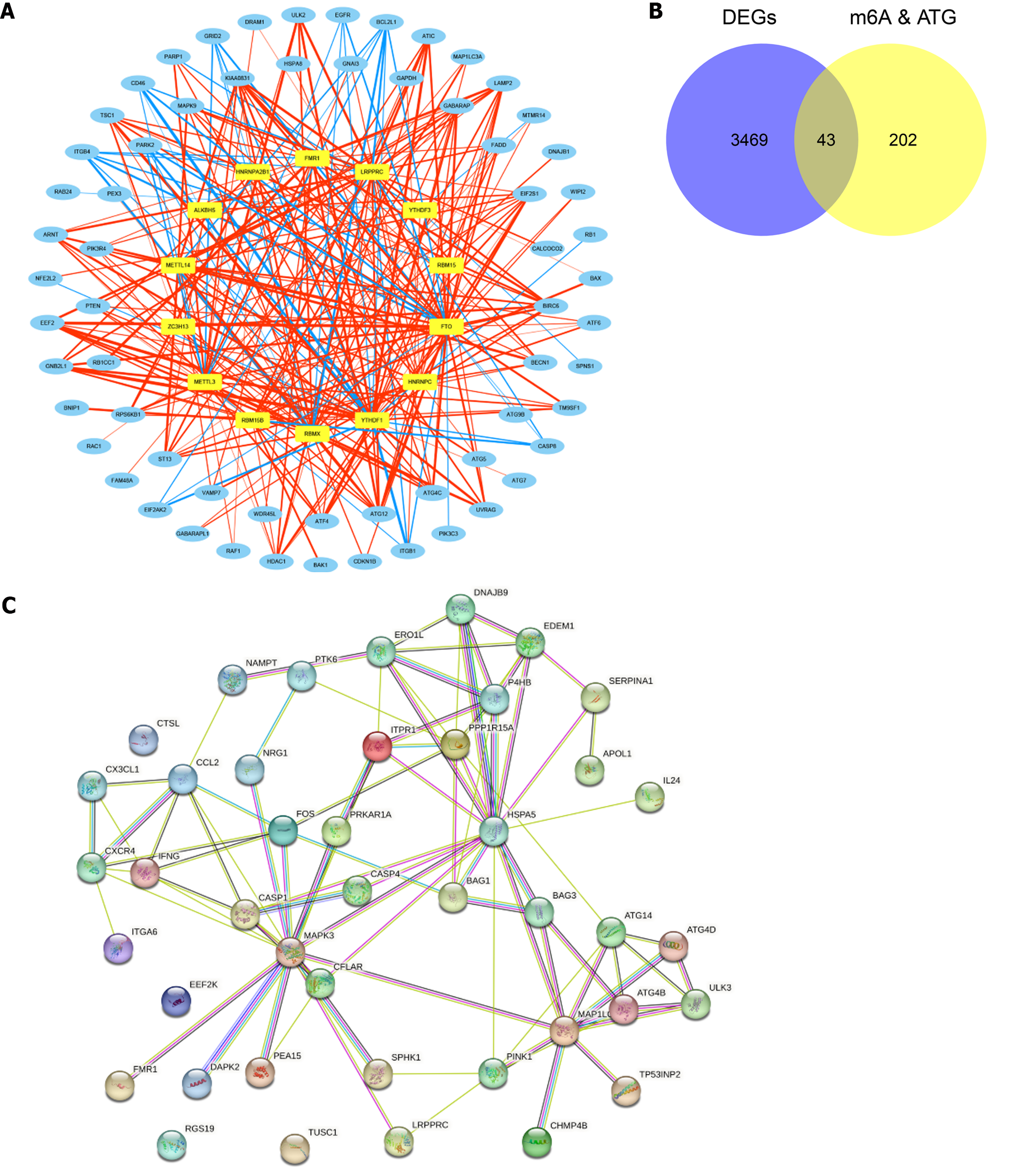
All 23 m6A-related genes and 222 ARGs were observed to be related to each other. Figure 2A presents the co-expression network of m6A-autophagy-related genes that contains 14 m6A-related genes and 64 ARGs (|r| > 0.8 and P < 0.05) (Supplementary Table 2). A total of 43 m6A-AR DEGs were obtained by the intersection of m6A-autophagy-related genes and DEGs between the UC and normal groups (Figure 2B). Moreover, using the PPI network of m6A-AR DEGs, a strong reciprocal relationship was observed between heat shock protein family A (Hsp70) member 5 (HSPA5) and dnaJ heat shock protein family member B9 (DNAJB9), HSPA5 and tumour protein P53 inducible nuclear protein 2 (TP53INP2), death-associated protein kinase 2 (DAPK2) and mitogen-activated protein kinase 3 (MAPK3) (Figure 2C). However, four proteins did not interact with other proteins, namely Cathepsin L, eukaryotic elongation factor 2 kinase, a regulator of G protein signalling 19 and tumour suppressor candidate 1 (TUSC1) (Figure 2C).
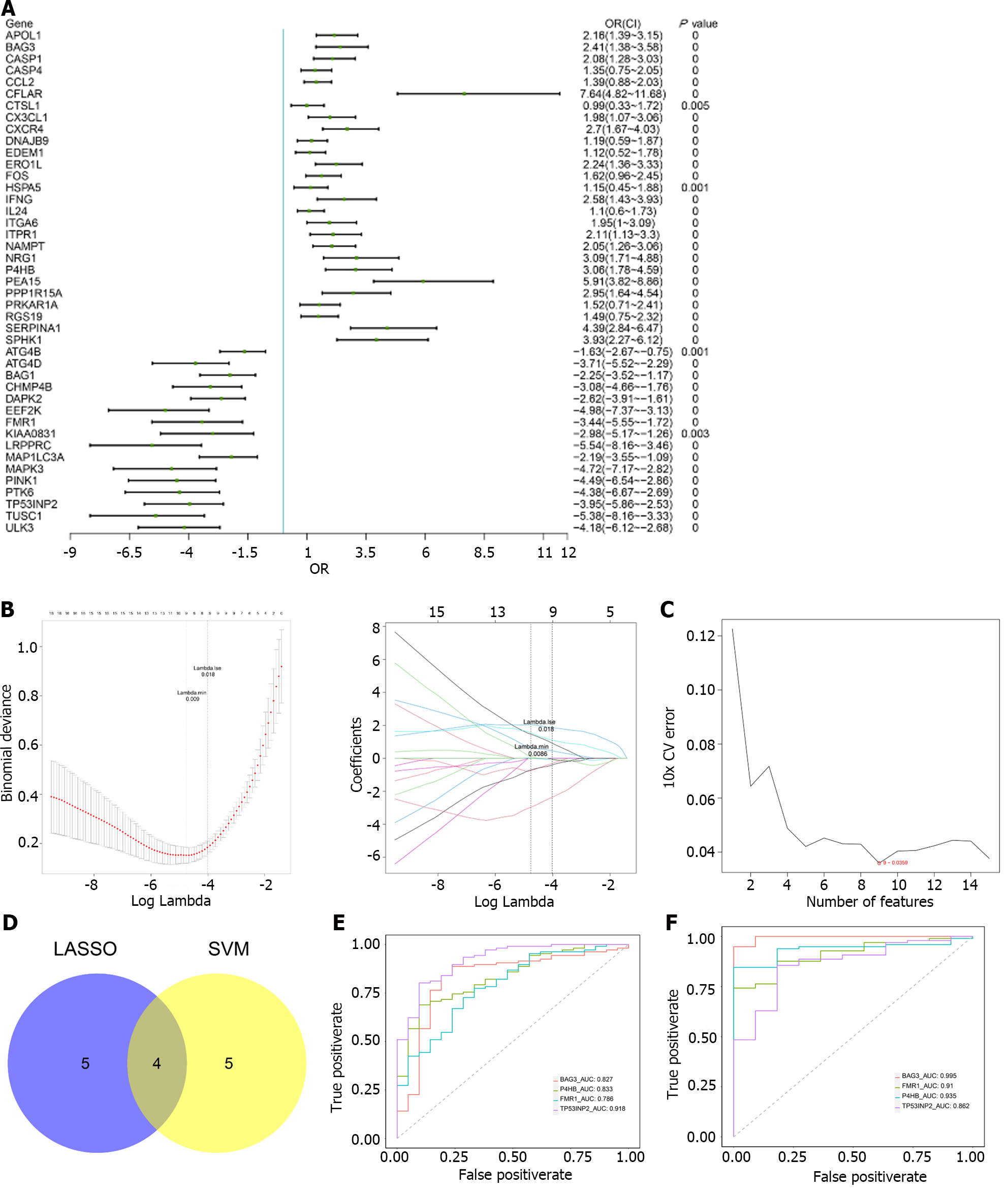
Univariate logistic regression revealed that all 43 m6A-AR DEGs were associated with the occurrence of UC (Figure 3A). Nine characteristic genes were further identified using LASSO, namely BLC2-associated athanogene 3 (BAG3), CC Chemokine Ligand 2, prolyl 4-hydroxylase subunit beta (P4HB), proliferation and apoptosis adaptor protein 15, serpin family a member 1, fragile X mental retardation 1 (FMR1), MAPK3, TP53INP2 and TUSC1 (Figure 3B). From the SVM algorithm, we obtained nine eigengenes, namely DNAJB9, BAG3, TP53IMP2, recombinant human caspase-1, P4HB, breast tumour kinase/protein-tyrosine kinase 6, DAPK2, FMR1, leucine-rich pentatricopeptide repeat containing (Figure 3C). Finally, four characteristic genes (FMR1, BAG3, P4HB and TP53IMP2) were selected through the cross-talk between LASSO and SVM (Figure 3D). The diagnostic accuracy of the four characteristic genes was evaluated using ROC curve analysis in GSE87466 and GSE75214. The AUC values of the four genes were greater than 0.7 in both datasets, indicating their high predictive accuracy for the occurrence of UC (Figure 3E and F).
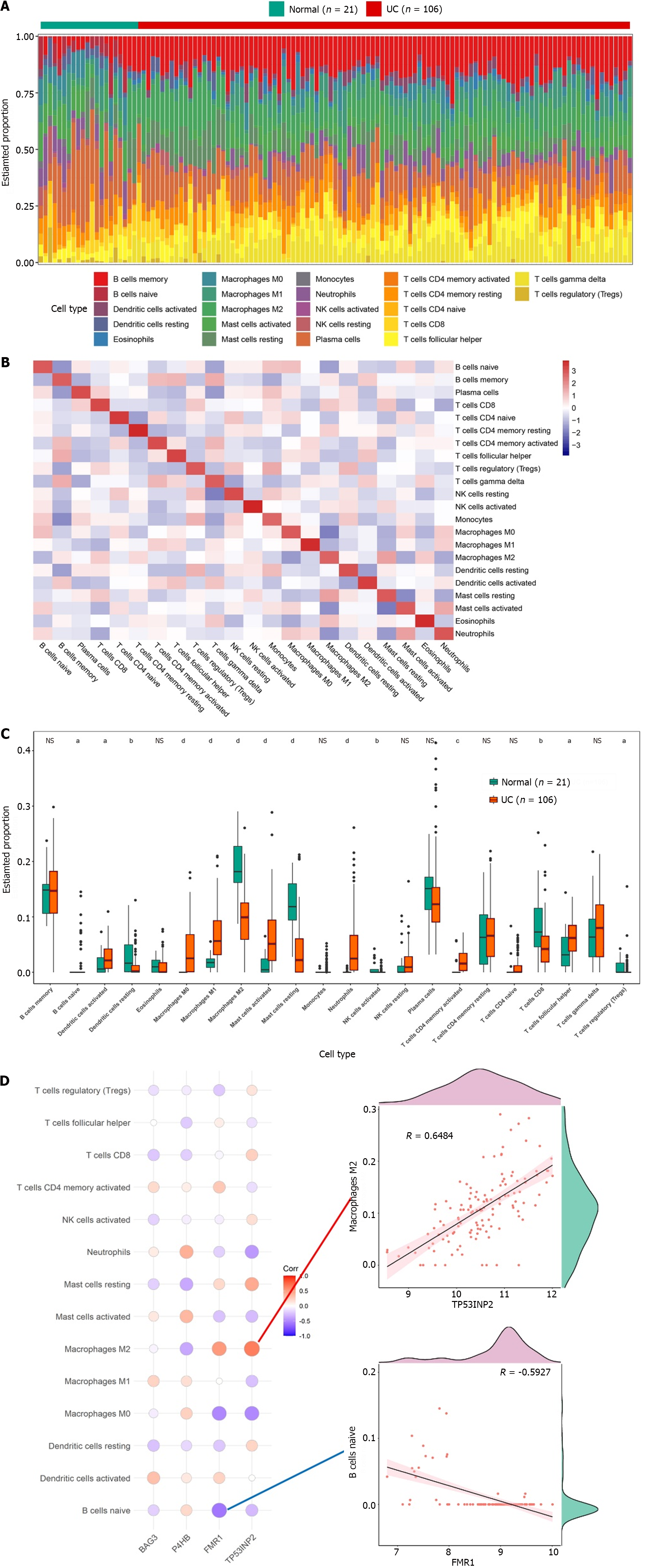
The infiltrating abundance of the 22 immune cells between the UC and normal groups was demonstrated using a bar chart, which showed that the content of B cells and T cells was higher in the UC group (Figure 4A). The correlation of infiltration levels of the 22 immune cells with each other was demonstrated using a heat map (Figure 4B). Moreover, 14 immune cells significantly differed between the UC and normal groups (Figure 4C), such as M0 macrophages, neutrophils and activated NK cells. Finally, correlation analysis between characteristic genes and differential immune cells showed that TP53INP2 had the strongest positive correlation with M2 macrophages, while FMR1 had the strongest negative correlation with naive B cells (Figure 4D).
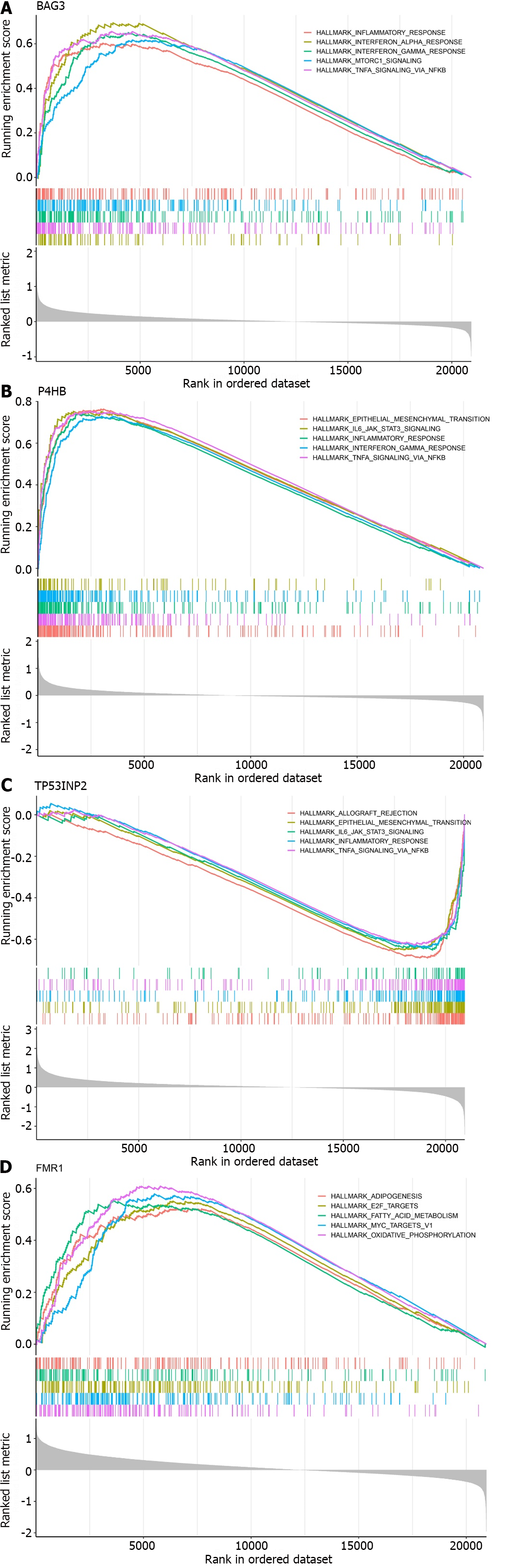
To further understand the impact of the characteristic genes on the development of UC, we performed gene set enrichment analysis (GSEA) of the characteristic genes. BAG3, P4HB and TP53INP2 were found to be involved in the inflammatory response and tumour necrosis factor-α (TNF-α) signalling through nuclear factor kappa-B (NF-κB) (Figure 5A-C). Notably, BAG3 and P4HB were positively associated with the two signalling pathways, whereas TP53INP2 was negatively correlated. FMR1 was mainly enriched in adipogenesis, MYC targets V1, E2F targets, oxidative phosphorylation and fatty acid metabolism (Figure 5D).
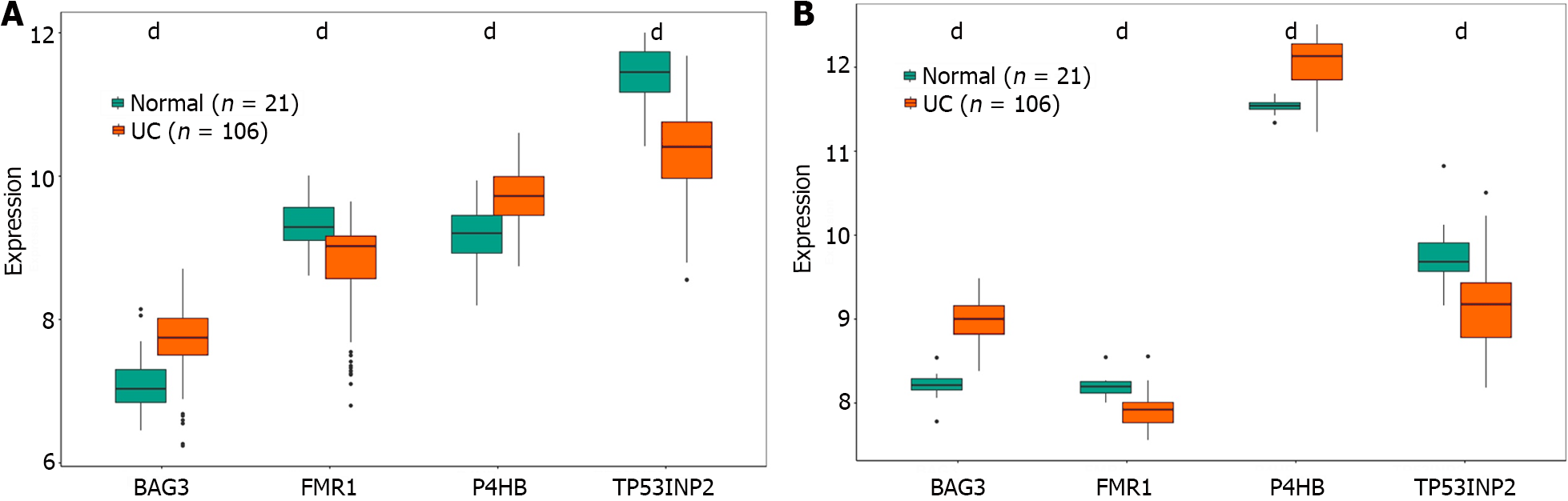
The visualised data exhibited the expressions of BAG3, FMR1, P4HB and TP53IMP2. The expression of four characteristic genes were shown in the GSE87473 and GSE75214 datasets (Figure 6A and B). The expression of the four characteristic genes between the normal and UC groups was significantly different. Moreover, the expression trends of the four genes were consistent in both datasets, with BAG3 and P4HB expressions elevated in the UC group whereas TP53INP2 and FMR1 expressions were lowered. The results suggested that these four genes had great diagnostic value in predicting the occurrence of UC.
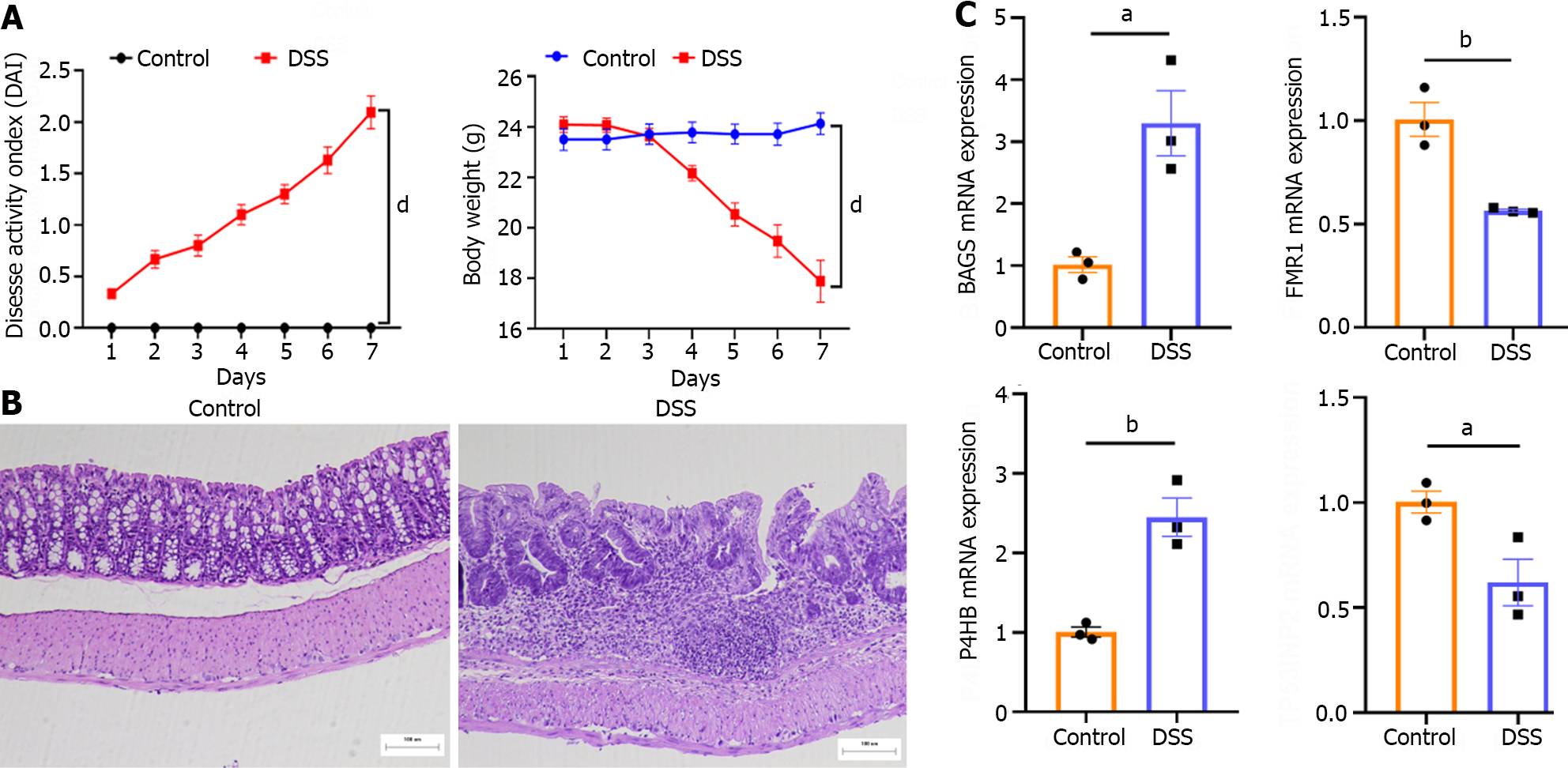
The control group mice had a good mental state, sensitive reaction, shiny hair, gradually increased body mass and formed faeces; The mice in the DSS group had a poor mental state, slow movement, matted hair, significantly reduced body weight and mucus purulent stool. Compared with the control group, the DAI of the DSS group mice increased significantly on the 7th day of administration (Figure 7A). HE staining showed that the colon tissue structure in the control group mice was complete and orderly arranged, the goblet cells and crypt structure were normal, and there was no congestion, oedema or ulcer. Meanwhile, in the DSS group, the colonic tissue was destroyed, goblet cells and crypts disappeared, a large number of inflammatory cells infiltrated the tissue and large ulcerative lesions were observed (Figure 7B). PCR also revealed significantly higher mRNA levels of BAG3 and P4HB and lower mRNA levels of FMR1 and TP53INP2 in the DSS group compared to the control group (Figure 7C). The flowchart systematically describes our study (Figure 8).
The role of m6A modification, especially concerning autophagy regulation, in human diseases such as obesity, heart disease, azoospermia or oligozoospermia, intervertebral disc degeneration and cancer has been extensively studied by scholars. Such studies contribute to the optimisation of treatment strategies for various diseases[9,15,16]. However, research on the mechanism of m6A autophagy interaction in colitis is still lacking.
In our study, four characteristic genes (FMR1, BAG3, P4HB and TP53INP2) were acquired using the machine learning algorithms. FMR1 is related to m6A, while BAG3, P4HB and TP53INP2 are related to autophagy. FMR1, the gene responsible for fragile X syndrome, encodes the fragile X mental retardation protein[17]. BAG proteins compete with Hip for binding to the Hsc70/Hsp70 ATPase domain and promote substrate release. Additionally, diseases associated with BAG3 include myopathy, myofibrillar and cardiomyopathy[18]. P4HB is an important endoplasmic reticulum (ER) molecular chaperone. Traditionally, it regulates the post-translational modification of proteins in the ER, which in turn is crucial for cell proliferation, apoptosis and autophagy regulation[19]. The protein encoded by TP53INP2 promotes autophagy and is essential for proper autophagosome formation and processing[20]. Recent studies report that the relationship between these four genes and intestinal diseases is mainly studied in colorectal cancer, with very little attention being paid to colitis.
In this study, through machine learning and animal model verification, the expression of Fmr1 and TP53INP2 was observed to be reduced in colitis. Researchers report that the relative abundance of Proteus, Deironobacteria and Bacteroides in Fmr1 knockout mice was higher than that in the wild-type (WT) mice, whereas that of Firmicutes and Tenericutes was lower in Fmr1 knockout mice than in the WT mice[21]. Therefore, we speculate that Fmr1 could affect the occurrence and development of colitis through the intestinal flora and its metabolites. Studies also report that the downregulation of TP53INP2 inhibits epithelial-to-mesenchymal transition (EMT) via the GSK-3β/β-catenin/Snail1 pathway in bladder cancer[22]. We hypothesised that TP53INP2 could inhibit the occurrence of EMT by regulating EMT-related transcription factors, thus blocking the key link required for UC-associated colorectal cancer (CAC) trans
GSEA revealed that BAG3, P4HB and TP53INP2 were involved in the NF-κB/TNF-α signalling pathway. The NF-κB signalling pathway plays an important role in the development of UC. The loss of the NF-κB signalling pathway and its regulatory factors lead to pathological changes in the UC intestinal tract, which, acting as positive feedback, further activates NF-κB and aggravates inflammation. Cytokines such as TNF-α, interleukin (IL)-6 and IL-1β are influenced by NF-κB and regulate immunity and inflammation in different ways[25]. Therefore, BAG3, P4HB and TP53INP2-related NF- κB/TNF-α signalling pathways have curative potential in clinical UC treatment[26].
Immuno-infiltration analysis showed that TP53INP2 had the highest positive correlation with M2 macrophages, while FMR1 had the highest negative correlation with naive B cells. Intestinal macrophages are involved in intestinal immune homeostasis and intestinal inflammation. The imbalance of the classical activated pro-inflammatory phenotype (M1)/alternative activated anti-inflammatory phenotype (M2) macrophage polarization can lead to intestinal inflammation. In UC patients, intestinal inflammation is closely related to the imbalance of intestinal M1/M2 macrophages polarization[27]. Therefore, targeted therapy that promotes macrophage polarisation by regulating TP53INP2 can reconstruct the homeostasis of intestinal immune microenvironment and restore post-inflammatory tissue homeostasis, which is a new focus of UC therapy. Through single-cell RNA sequencing of B cells from three cohorts of patients with UC, researchers have drawn the composition, transcription and clonal map of the intestinal mucosa and circulating B cells and found major perturbations within the mucosal B cell compartment, including an expansion of naive B cells and IgG+ plasma cells with curtailed diversity and maturation. These findings suggest that B cells play an important role in the pathogenesis of UC[28]. Nevertheless, the mechanism of how FMR1 regulates naive B cells requires further study.
In conclusion, our study analysed the diagnostic value of m6A and ARGs in predicting the occurrence of UC. A total of four key genes (FMR1, BAG3, P4HB and TP53INP2) were identified and verified using animal models, providing a foundation for the clinical diagnosis and treatment of UC. However, m6A and ARGs how to participate in the occurrence and development of UC, as well as the genes identification are as possible markers for assessing UC severity and developing innovative UC targeted therapeutic approaches. The specific regulatory mechanisms of these genes need further experimental research and clinical application research.
Both N6-methyladenosine (m6A) methylation and autophagy are considered relevant to the pathogenesis of ulcerative colitis (UC). However, a systematic exploration of the role of the combination of m6A methylation and autophagy in UC remains to be performed.
In this study, we used publicly available data related to UC and comprehensive bioinformatics methods to elucidate the autophagy-related genes of m6A with a diagnostic value for UC, thereby contributing to the development of treatment options for patients with UC.
To elucidate the autophagy-related genes of m6A with a diagnostic value for UC.
The correlation between m6A-related genes and autophagy-related genes (ARGs) was analysed. Finally, gene set enrichment analysis (GSEA) was performed on the characteristic genes. Additionally, the expression levels of four characteristic genes were verified in DSS-induced colitis in mice.
GSEA indicated that BAG3, P4HB and TP53INP2 were involved in the inflammatory response and TNF-α signalling via NF- κB. Furthermore, polymerase chain reaction results showed significantly higher mRNA levels of BAG3 and P4HB and lower mRNA levels of FMR1 and TP53INP2 in the DSS group compared to the control group.
This study identified four m6A-ARGs that predict the occurrence of UC, thus providing a scientific reference for further studies on the pathogenesis of UC.
The specific regulatory mechanisms of these genes need further experimental research and clinical application research.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medical informatics
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Nakaji K, Japan S-Editor: Liu JH L-Editor: A P-Editor: Cai YX
| 1. | Du L, Ha C. Epidemiology and Pathogenesis of Ulcerative Colitis. Gastroenterol Clin North Am. 2020;49:643-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 359] [Article Influence: 71.8] [Reference Citation Analysis (0)] |
| 2. | Ordás I, Eckmann L, Talamini M, Baumgart DC, Sandborn WJ. Ulcerative colitis. Lancet. 2012;380:1606-1619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1151] [Cited by in RCA: 1547] [Article Influence: 119.0] [Reference Citation Analysis (5)] |
| 3. | Cui DJ, Chen C, Yuan WQ, Yang YH, Han L. Integrative analysis of ferroptosis-related genes in ulcerative colitis. J Int Med Res. 2021;49:3000605211042975. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 4. | Sebastian-delaCruz M, Olazagoitia-Garmendia A, Gonzalez-Moro I, Santin I, Garcia-Etxebarria K, Castellanos-Rubio A. Implication of m6A mRNA Methylation in Susceptibility to Inflammatory Bowel Disease. Epigenomes. 2020;4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 5. | Gu C, Wu J, Zhang W, Yao Y, Yan W, Yuan Y, Wang W, Shang A. Immune Infiltration of Ulcerative Colitis and Detection of the m6A Subtype. J Immunol Res. 2022;2022:7280977. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 6. | Lu TX, Zheng Z, Zhang L, Sun HL, Bissonnette M, Huang H, He C. A New Model of Spontaneous Colitis in Mice Induced by Deletion of an RNA m(6)A Methyltransferase Component METTL14 in T Cells. Cell Mol Gastroenterol Hepatol. 2020;10:747-761. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 99] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 7. | Qiao D, Zhang Z, Zhang Y, Chen Q, Chen Y, Tang Y, Sun X, Tang Z, Dai Y. Regulation of Endoplasmic Reticulum Stress-Autophagy: A Potential Therapeutic Target for Ulcerative Colitis. Front Pharmacol. 2021;12:697360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 8. | Qiu P, Liu L, Fang J, Zhang M, Wang H, Peng Y, Chen M, Liu J, Wang F, Zhao Q. Identification of Pharmacological Autophagy Regulators of Active Ulcerative Colitis. Front Pharmacol. 2021;12:769718. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 9. | Chen X, Wang J, Tahir M, Zhang F, Ran Y, Liu Z. Current insights into the implications of m6A RNA methylation and autophagy interaction in human diseases. Cell Biosci. 2021;11:147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 44] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 10. | Wu X, Bai Z. Multi-omics analysis of m(6)A modification-related patterns based on m(6)A regulators and tumor microenvironment infiltration in lung adenocarcinoma. Sci Rep. 2021;11:20921. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 11. | Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16184] [Cited by in RCA: 25721] [Article Influence: 2572.1] [Reference Citation Analysis (0)] |
| 12. | Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb). 2021;2:100141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 522] [Cited by in RCA: 4778] [Article Influence: 1194.5] [Reference Citation Analysis (0)] |
| 13. | Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol. 2018;1711:243-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 993] [Cited by in RCA: 2458] [Article Influence: 351.1] [Reference Citation Analysis (0)] |
| 14. | Zhang Z, Qiao D, Zhang Y, Chen Q, Chen Y, Tang Y, Que R, Zheng L, Dai Y, Tang Z. Portulaca oleracea L. Extract Ameliorates Intestinal Inflammation by Regulating Endoplasmic Reticulum Stress and Autophagy. Mol Nutr Food Res. 2022;66:e2100791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 15. | Peng J, Zheng H, Liu F, Wu Q, Liu S. The m6A methyltransferase METTL3 affects autophagy and progression of nasopharyngeal carcinoma by regulating the stability of lncRNA ZFAS1. Infect Agent Cancer. 2022;17:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 36] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 16. | Cui YH, Yang S, Wei J, Shea CR, Zhong W, Wang F, Shah P, Kibriya MG, Cui X, Ahsan H, He C, He YY. Autophagy of the m(6)A mRNA demethylase FTO is impaired by low-level arsenic exposure to promote tumorigenesis. Nat Commun. 2021;12:2183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 101] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 17. | Zhang G, Xu Y, Wang X, Zhu Y, Wang L, Zhang W, Wang Y, Gao Y, Wu X, Cheng Y, Sun Q, Chen D. Dynamic FMR1 granule phase switch instructed by m6A modification contributes to maternal RNA decay. Nat Commun. 2022;13:859. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 50] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 18. | Jiang H, Ji Y, Sheng J, Wang Y, Liu X, Xiao P, Ding H. Genome-Wide Identification of the Bcl-2 Associated Athanogene (BAG) Gene Family in Solanum lycopersicum and the Functional Role of SlBAG9 in Response to Osmotic Stress. Antioxidants (Basel). 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 19. | Xie L, Li H, Zhang L, Ma X, Dang Y, Guo J, Liu J, Ge L, Nan F, Dong H, Yan Z, Guo X. Autophagy-related gene P4HB: a novel diagnosis and prognosis marker for kidney renal clear cell carcinoma. Aging (Albany NY). 2020;12:1828-1842. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 20. | You Z, Xu Y, Wan W, Zhou L, Li J, Zhou T, Shi Y, Liu W. TP53INP2 contributes to autophagosome formation by promoting LC3-ATG7 interaction. Autophagy. 2019;15:1309-1321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 21. | Ouyang YY, Chen SQ, Tian LR, Yang XY, Xing XM, Zhuang HM, Huang XF, Deng XY. Analysis of intestinal flora composition in Fmr1 knockout mice. Zhonghua Linchuang Shiyanshi Guanli (Electronic Edition). 2020;8:158-165. [DOI] [Full Text] |
| 22. | Zhou Z, Liu X, Li Y, Li J, Deng W, Zhong J, Chen L, Zeng X, Wang G, Zhu J, Fu B. TP53INP2 Modulates Epithelial-to-Mesenchymal Transition via the GSK-3β/β-Catenin/Snail1 Pathway in Bladder Cancer Cells. Onco Targets Ther. 2020;13:9587-9597. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 23. | Li N, Chen M, Cao Y, Li H, Zhao J, Zhai Z, Ren F, Li K. Bcl-2-associated athanogene 3(BAG3) is associated with tumor cell proliferation, migration, invasion and chemoresistance in colorectal cancer. BMC Cancer. 2018;18:793. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 24. | Zhou Y, Lu LH, Xu QH, Liao BL. Expression and significance of P4HB in colorectal cancer. Beichuan Yixueyuan Xuebao. 2019;34:343-346. [DOI] [Full Text] |
| 25. | Wang Y, Tang Q, Duan P, Yang L. Curcumin as a therapeutic agent for blocking NF-κB activation in ulcerative colitis. Immunopharmacol Immunotoxicol. 2018;40:476-482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 78] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 26. | Lu PD, Zhao YH. Targeting NF-κB pathway for treating ulcerative colitis: comprehensive regulatory characteristics of Chinese medicines. Chin Med. 2020;15:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 102] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 27. | Yang Z, Lin S, Feng W, Liu Y, Song Z, Pan G, Zhang Y, Dai X, Ding X, Chen L, Wang Y. A potential therapeutic target in traditional Chinese medicine for ulcerative colitis: Macrophage polarization. Front Pharmacol. 2022;13:999179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 37] [Reference Citation Analysis (0)] |
| 28. | Uzzan M, Martin JC, Mesin L, Livanos AE, Castro-Dopico T, Huang R, Petralia F, Magri G, Kumar S, Zhao Q, Rosenstein AK, Tokuyama M, Sharma K, Ungaro R, Kosoy R, Jha D, Fischer J, Singh H, Keir ME, Ramamoorthi N, O'Gorman WE, Cohen BL, Rahman A, Cossarini F, Seki A, Leyre L, Vaquero ST, Gurunathan S, Grasset EK, Losic B, Dubinsky M, Greenstein AJ, Gottlieb Z, Legnani P, George J, Irizar H, Stojmirovic A, Brodmerkel C, Kasarkis A, Sands BE, Furtado G, Lira SA, Tuong ZK, Ko HM, Cerutti A, Elson CO, Clatworthy MR, Merad M, Suárez-Fariñas M, Argmann C, Hackney JA, Victora GD, Randolph GJ, Kenigsberg E, Colombel JF, Mehandru S. Ulcerative colitis is characterized by a plasmablast-skewed humoral response associated with disease activity. Nat Med. 2022;28:766-779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 133] [Article Influence: 44.3] [Reference Citation Analysis (0)] |