Published online Dec 6, 2023. doi: 10.12998/wjcc.v11.i34.8111
Peer-review started: July 27, 2023
First decision: September 5, 2023
Revised: September 26, 2023
Accepted: November 27, 2023
Article in press: November 27, 2023
Published online: December 6, 2023
Processing time: 131 Days and 21.1 Hours
Inflammatory bowel disease (IBD) is a disorder of the immune system and intestinal microecosystem caused by environmental factors in genetically susceptible people. Paneth cells (PCs) play a central role in IBD pathogenesis, especially in Crohn's disease development, and their morphology, number and function are regulated by susceptibility genes. In the intestine, PCs participate in the formation of the stem cell microenvironment by secreting antibacterial particles and play a role in helping maintain the intestinal microecology and intestinal mucosal homeostasis. Moreover, PC proliferation and maturation depend on symbiotic flora in the intestine. This paper describes the interactions among susceptibility genes, PCs and intestinal microecology and their effects on IBD occurrence and development.
Core Tip: Inflammatory bowel disease (IBD) is a disorder of the immune system and intestinal microecosystem caused by environmental factors in genetically susceptible people. Paneth cells (PCs) play a central role in IBD pathogenesis, especially in Crohn's disease development, and their morphology, number and function are regulated by susceptibility genes. In the intestine, PCs participate in the formation of the stem cell microenvironment by secreting antibacterial particles and play a role in helping maintain the intestinal microecology and intestinal mucosal homeostasis. Moreover, PC proliferation and maturation depend on symbiotic flora in the intestine. This paper describes the interactions among susceptibility genes, PCs and intestinal microecology and their effects on IBD occurrence and development.
- Citation: Zhou QM, Zheng L. Research progress on the relationship between Paneth cells-susceptibility genes, intestinal microecology and inflammatory bowel disease. World J Clin Cases 2023; 11(34): 8111-8125
- URL: https://www.wjgnet.com/2307-8960/full/v11/i34/8111.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i34.8111
Inflammatory bowel disease (IBD) is a disorder of the immune system and intestinal microecosystem caused by environmental factors in genetically susceptible people. Paneth cells (PCs) play a central role in IBD pathogenesis, especially in Crohn's disease (CD) development, and their morphology, number and function are regulated by susceptibility genes. In the intestine, PCs participate in the formation of the stem cell microenvironment by secreting antibacterial particles and play a role in helping maintain the intestinal microecology and intestinal mucosal homeostasis. Moreover, PC proliferation and maturation depend on symbiotic flora in the intestine. This paper describes the interactions among susceptibility genes, PCs and intestinal microecology and their effects on IBD occurrence and development.
IBD is a group of chronic, nonspecific inflammatory diseases that include CD and ulcerative colitis (UC). PCs, derived from intestinal pluripotent stem cells, are gradually growing columnar epithelial cells located at the junction of villi and crypts[1]. During differentiation and maturation, PCs migrate to the base of crypts and spread throughout the small intestine. PCs are pyramidal, and the cytoplasm at the “top” of the pyramid is full of coarse and large eosinophilic secretory granules. The main components are α-defensin, lysozyme, phospholipase A2 and other antibacterial substances that can be released into the intestinal cavity and play an important role in the natural defense of the small intestinal mucosa[2]. Due to the high secretion capacity of PCs and their close relationship with intestinal microecology, PC abnormalities and flora disorders often occur in the intestinal inflammatory response[3]. In recent years, PCs have been found throughout the gastrointestinal tract, including the stomach and colon. However, the distribution of PCs throughout the gastrointestinal tract occurs mainly as a response to mucosal inflammation, and PC presence in abnormal areas is called metaplasia. Colorectal PCs are widely found in UC and inflammatory enteritis[4].
In the past 20 years, the results of whole-genome scanning have revealed that IBD susceptibility genes are distributed on chromosomes 1, 3, 4, 5, 6, 7, 10, 12, 14, 16, 19 and X, among which 9 susceptibility genes associated with IBD were named IBD1-9: IBD1 on chromosome 16q, IBD2 on 12p13.2-q24.1, IBD3 on 6p, IBD4 on 14q11-q12, IBD5 on 5q31, IBD6 on 19p13, IBD7 on 1p36, IBD8 on 16p and IBD9 on 3p26[5]. Studies have shown that more than 200 gene loci are associated with IBD susceptibility, including more than 150 that increase the risk of CD[6]. Studies have shown that IBD susceptibility genes can affect the important physiological processes of PC, leading to abnormal PCs and promoting the occurrence and development of intestinal mucosal inflammation[7].
Changes in the intestinal microecology are involved in IBD pathogenesis, which mainly manifests as a flora imbalance, including flora diversity, species and abundance changes. Studies have shown that the abundance of Firmicutes and Bacteroidetes, which dominate the intestinal flora of IBD patients, decreases, while the proportion of Proteobacteria and actinomycetes increases[8]. Therefore, based on the above knowledge, this paper reviews the relationship among PCs, susceptibility genes, intestinal microecology and IBD.
In 1745, German anatomist Johann Nathanael Lieberkuhn first described intestinal glands, or crypts, present in the intestines. In 1872, Gustav Schwalbe observed PCs in the crypt of the small intestine. In 1888, the Austrian physician Joseph Paneth described PCs graphically as a group of specialized cylindrical cells in the crypts of the small intestine epithelium, whose cytoplasm is filled with granular matter. The cells have been named PCs in honor of Dr. Paneth. PCs are rare cells in the small intestine that provide the host with protection against microbial invasion. Their function is the secretion of antibacterial proteins.
When bacteria or bacterial antigens invade the body, PCs secrete antibacterial molecules such as defensins between the villi of the bowel to help maintain the gastrointestinal barrier[9]. PCs are characteristic cells of the small intestinal gland, located at the bottom of the gland, the cells are pyramidal, and the top cytoplasm is full of coarse and large eosinophilic secretory particles. Under the electron microscope, the cytoplasm contains a large number of rough endoplasmic reticulums and developed Golgi complexes, and the secretory particles contain defensin and lysozyme, which can kill intestinal microorganisms. Most of the substances secreted by PCs are antibacterial proteins, which are expelled from the recess of the small intestine and dispersed into the mucosal layer to assist the mucosal immune barrier in its function. Later, PCs were found in the gastrointestinal tract, including the stomach, small intestineand colon[10]. However, PCs in the gastrointestinal tract are mainly found in response to mucous membrane inflammation, which is called metaplasia in abnormal colorectal regions. Paneth cell metaplasia is widespread in inflammatory enteritis.
In adult mammalian tissues, the small intestine epithelium has a remarkable capacity for self-renewal. The renewal of the small intestinal epithelium depends on stem cells in the small intestinal crypt. Progenitor cells differentiated from small intestinal stem cells migrate from the bottom of the crypt to the small intestinal villi and continue to differentiate into goblet cells, plexus cells, neuroendocrine cells, intestinal epithelial cells and other cells[11]. After differentiation, these cells migrate from the crypt to the apex of the villi, where they gradually die and are replaced by new cells that migrate from the lower end. Intestinal epithelial cells have a life cycle of only four to five days, and this rapid self-renewal is thought to be essential for intestinal integrity. PCs are also derived from small intestine stem cells, but unlike other cells, PCs do not migrate upward after they are produced[12]. They always reside in the crypt, and the lifespan of this group of cells is more than 1 mo.
Intestinal stem cells are located at the base of the crypt, and recent research suggests that there may be two types of stem cells present. One type is crypt base columnar cells (CBC cells), which are spaced apart from PCs at the base of the crypt[13]. The target gene of Wnt, Lgr5, is the most representative marker of CBC cells and is expressed on the surface of CBC cells. The other type is static +4 cells, which are located above PCs, and the main markers include Bmi-1, Hopx, mTert and Lrig1. Little is known about +4 cells, but studies have shown that there is a close relationship between CBC cells and PCs[14].
The close spatial relationship between PCs and CBC cells has prompted speculation that PCs provide an important niche for stem cells[15]. However, one laboratory refuted this hypothesis. They created a transgenic mouse model in which PCs specifically expressed diphtheria toxin, causing most PCs to be knocked out, but this did not affect the proliferation of CBC cells in the crypt[16]. Later, with the identification of the Lgr5 marker and the establishment of a crypt system in vitro, the hypothesis that PCs provide a niche for stem cells was re-established. In vitro, isolated Lgr5hi CBC cells hardly grew into crypt bodies[17]. However, when PCs and stem cells were cultured together, the stem cells could differentiate and develop into crypt bodies. Further studies in in vivo mouse models, including the Gordon model mentioned earlier, showed that knocking out PCs resulted in the loss of Lgr5 stem cells[18]. In terms of gene expression, PCs produce not only germicidal substances but also epidermal growth factor, Wnt3 and Notch ligand Dll4 in large quantities, providing necessary conditions for PCs to become a niche[19]. In conclusion, PCs provide the necessary niche signal for Lgr5hi CBC stem cells (Figure 1).
PCs contain a large number of endoplasmic reticulum and Golgi complexes, which have a major role in protein secretion. The main secretions of PCs are protein polypeptides with bactericidal ability, such as the cryptdin-related sequence peptide, lysozyme, IIA secretory phospholipase A2 (secretory group IIA phospholipase A2), regenerated insulin-derived proteins REG3β and REG3γ, angiogenin 4, and ANG4[20].
Antimicrobial peptides are important effectors in the innate immune response against pathogenic microbial infection. α-Defensins are one of the earliest recognized antimicrobial peptide families. They are the main components of secretory granules in phagocytes[21]. In addition to phagocytes, epithelial cells in various mucous membranes also secrete α-defensin. Mouse PCs secrete several subtypes of defensins, which can be divided into 6 subtypes from 1 to 6 by purification analysis in vitro. Immunohistochemical analysis showed that α-defensin was specifically expressed in PCs and was secreted into the intestinal cavity in a polar manner[22]. These α-defensins, which are secreted extracellularly, are thought to have important host defense functions.
Defensins are a class of small (15-20 residues) cationic proteins rich in cysteine. They are amphoteric molecules that can bind to bacterial cell membranes and form transmembrane ion channels, destroying the integrity of the membranes and causing cell contents to leak, thus killing bacteria[23]. At the same time, defensins can inactivate a variety of bacterial toxins by combining with them in order to denature them. However, how intestinal symbiotic bacteria coexist with the abundant bactericidal defensins in the gut has always been a perplexing problem. Recent studies have shown that intestinal symbiotic bacteria usually express dephosphatase (LpxF) to remove negative charges on the surface of bacteria to resist killing by cationic antibacterial peptides such as defensins[24].
Studies have shown that α-defensin secreted by PCs plays an important role in the response to pathogen infection. Gram-positive bacteria, gram-negative bacteria, lipopolysaccharide, muramic acid, muramyl dipeptides, and lipid A all stimulate defensin secretion by PCs in the small intestine[25]. Live fungi and protozoa do not stimulate PC degranulation. When PCs in the mouse small intestine encounter pathogens or pathogen antigens, they secrete granules rich in antimicrobial peptides within a few minutes to kill pathogenic microorganisms[26]. This secretion activity is dose-dependent for the pathogens or pathogen antigens. α-Defensins account for 70% of the total antimicrobial peptide killing activity[27].
It has been found that α-defensin derived from PCs plays an important role in establishing and maintaining the balance of intestinal microecology[28]. Mouse α-defensin is synthesized as a nonactivated precursor peptide and must be cleaved by matrix metalloproteinase 7 (MMP7) to be activated[29]. Two animal models, DEFA5 transgenic mice and MMP7-deficient mice, have been studied. DEFA5 transgenic mice express α-defensin 5 (also known as HD-5), which is an α-defensin overexpression model[30]. MMP7-deficient mice do not produce active α-defensin and are a model of α-defensin deficiency in the small intestine. In both mouse models, the mRNA expression levels of PC effector factors, such as lysozyme, Defa1 and Defcr4 encoding α-defcr4, and the total number of intestinal bacteria did not change significantly, but the composition of intestinal bacteria did[31]. The proportion of Firmicutes in DEFA5 transgenic mice was significantly reduced, while the proportion of Bacteroides was significantly increased, but the opposite result was obtained in MMP7-deficient mice[32]. This suggests that changes in intestinal bacterial composition are dependent on α-defensin but α-defensin does not affect the total number of intestinal bacteria[33]. Moreover, segmented filamentous bacteria, important members of Firmicutes, were barely detected in DEFA5 transgenic mice, and the proportion and number of Th17 cells in the lamina proper were also affected. These results indicate that α-defensins derived from PCs affect the intestinal symbiotic bacterial composition and intestinal homeostasis[34].
PCs not only secrete antimicrobial peptides stored in vesicles under microbial stimulation but also regulate the production of some antimicrobial peptides upon sensing microorganisms[35]. It has been found that the presence of enterobacteria can greatly enhance REG3γ expression in PCs, the upregulation of REG3γ expression depends on the MyD88 signaling pathway of the downstream adaptor molecule of the Toll-like receptor (TLR), and the REG3γ expression is necessary to prevent microbial invasion into the host tissue. Using a model with MyD88 overexpression in PCs, researchers found that PCs directly sense microorganisms through the TLR-MyD88 pathway and activate the expression of MYD88-dependent antimicrobial peptides, such as Reg3γ[36]. These results demonstrated that MyD88 signaling pathway activation in PCs is sufficient to prevent microbial invasion of the host and does not require MyD88 signaling from other sources, such as bone marrow cells[37]. This study further employed a mouse model with a defensin promoter regulating the expression of diphtheria toxin (CR2-tox176) to deplete PCs and showed that mice with PC depletion did not effectively control intestinal symbiotic and pathogenic bacterial invasion into the spleen and mucosa-associated lymph nodes. Thus, the antibacterial substances derived from PCs are very important for controlling the invasion and diffusion of microorganisms in vivo[38].
Nucleotide-binding oligomeric domain protein 2 (NOD2)/CARD15, the first discovered C and D susceptibility gene, is located around the centromeres of chromosome 16 (16q12) and is mainly expressed in macrophages and PCs specific to the small intestinal gland. It encodes 2 CARD domains and 6 LRR (1 eukaryotic repeat) proteins. CARD15 protein activates NF-κB by recognizing the muramyl dipeptide (MDP) of foreign bacteria and plays a role in the immune response to bacterial LPS[39]. In addition, CARD15 can induce the expression of human β-defensin-2 (HBD-2) in epithelial cells when encountering invading microorganisms, which constitutes the first line of rapid defense of epithelial cells against foreign microorganisms[40]. Therefore, mutation of the CARD15 gene and subsequent alteration of the structure of the encoded protein is a risk factor for CD[41]. Most studies suggest that NOD2/CARD15 is closely related to genetic susceptibility to CD but not to UC. However, the presence of TL4 or CD14 gene mutations in conjunction with NOD2/CARD15 mutations increases UC susceptibility. The genes in IBD1 near D16s408 are also associated with the incidence of UC. For example, single allelic mutations increase the incidence of UC, while double allelic mutations can lead to severe CD[42]. Therefore, although not as significantly as with C and D, the NOD2 gene is also associated with UC.
The human leucine-rich repetitive kinase 2 (LRRK2) gene consists of 51 exons located on chromosome 12q12 and encodes the LRRK2 protein, a multidomain protein composed of 2527 amino acids. LRRK2 is a multidomain protein kinase with a wide range of functions, including vesicle transport and entosis, protein synthesis, immune response regulation, and inflammation[43]. The LRRK2 protein consists of an ARM repeat, ankyrin repeat (ANK), leucine-rich repeat region (LRR), Ras protein complex (ROC), Ras protein C-terminal repeat (COR), kinase domain (MAPKKK) and tryptophan aspartic acid repeat region (WD40)[44]. Three scaffold domains, ANK, LRR and WD40, are involved in interactions with other proteins that can maintain the conformation and stability of those proteins. The ROC domain and MAPKKK domain are related to the GTPase activity and kinase activity of LRRK2, respectively, while the functions of the ARM repeat sequence and COR domain are not clear. Y1699C and R1628P mutations in the ROC domain have been found to be associated with Parkinson disease (PD) and leprosy, respectively[45]. Pathological and physiological studies of LRRK2 have indicated that the LRRK2 domain is significantly related to its various cellular functions, suggesting that LRRK2 is a pivotal protein with a wide range of functions. Gain-of-function mutations in the LRRK2 kinase domain lead to increased LRRK2 kinase activity and play an important role in disease pathogenesis[46].
Most members of the tumor necrosis factor (TNF)/tumor necrosis factor receptor superfamily proteins (TNFR SFP) are expressed in immune cells and play a key role in the immune response[47]. Tumor necrosis factor ligand 1A (TL1A), a member of the TNFSF family, is the encoding product of the TNFSF 15 gene, and its expression is increased in the intestinal inflammatory region of patients with IBD[48]. TL1A protein has been found to be expressed in mononuclear macrophages and CD4+/CD8+ lymphocytes in the intestinal lamina propria in patients with CD, and the expression level and the number of positive cells were positively correlated with the severity of intestinal lesions[49]. Furthermore, the number of DR3-positive T lymphocytes increased in the intestinal lamina propria of CD patients. The uniform upregulation of TL1A and DR3 expression indicates that downstream cytokines after TL1A and DR3 binding play an important role in CD[50]. In addition, TL1A helps balance promotion and inhibition of the inflammatory response in the intestinal mucosa in CD. At the initial stage of inflammation, when T cells are recruited to the inflammatory site of intestinal mucosa, TL1A interacts with DR3 to enhance inflammatory cytokine secretion, and these cytokines cause the recruitment and activation of macrophages and neutrophils, stimulating further inflammation[51].
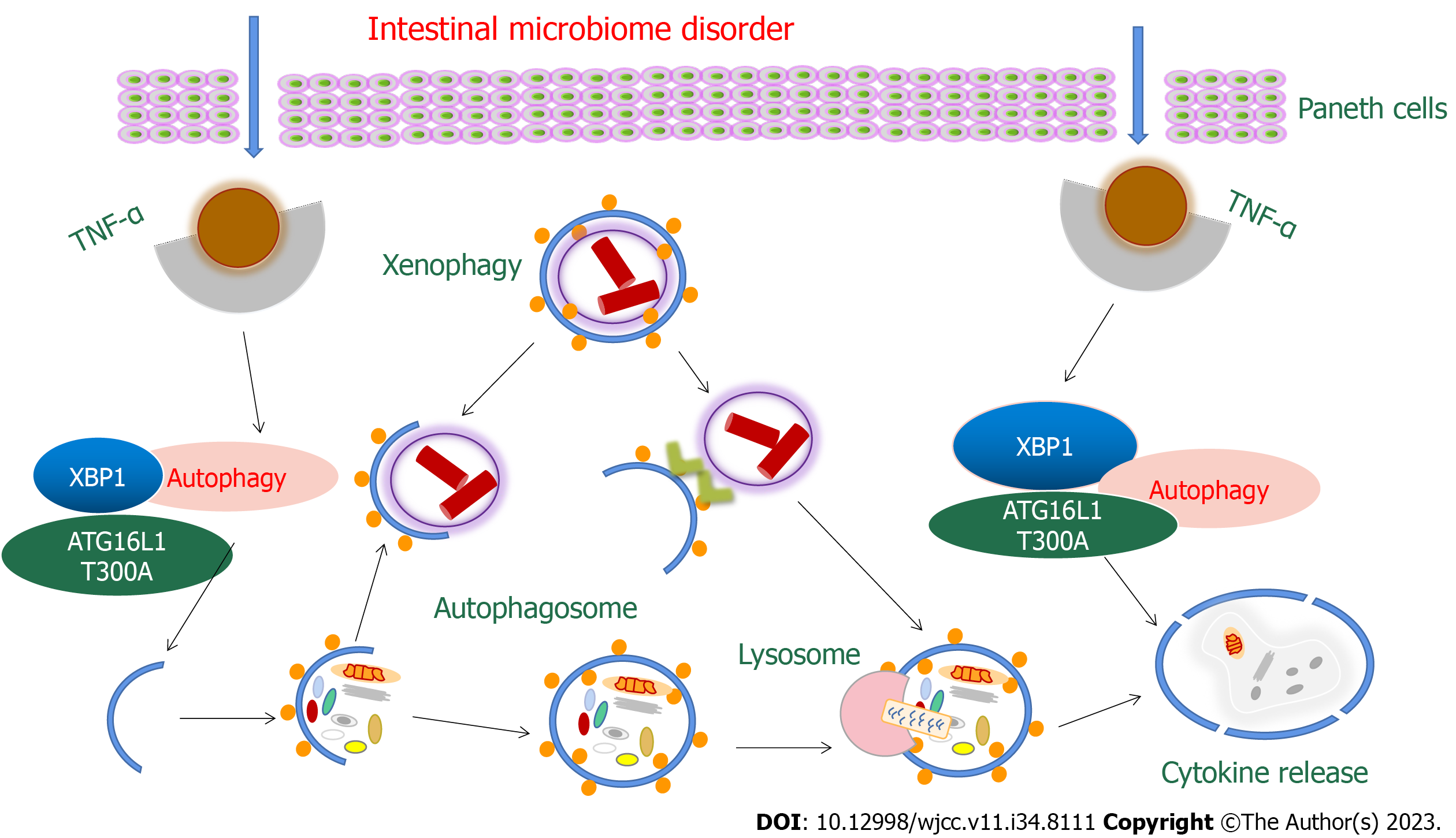
The Atg16L1 gene, which is involved in autophagy, is related to CD development and plays an important role in PCs, suggesting the importance of autophagy to the normal physiological function of PCs. CD patients with Atg16L1 mutations have an altered gut microbiota and abnormal PC granules[52]. A similar phenomenon was observed in mice with low expression of Atg16L1 protein. Notably, Zhang et al[53] found that autophagy of PCs was specifically activated in some CD patients, and this state was not related to mucosal inflammation and Atg16L1 mutation. This result suggested that in addition to Atg16L1, more autophagy-related genes might be involved in the pathological mechanism of CD[54]. Moreover, we do not currently know which genes are involved. Additionally, autophagy and endoplasmic reticulum stress have compensatory effects in PCs. When Atg16L1 and Xbp1 were knocked out simultaneously in the intestinal epithelium, mice developed more severe idiopathic enteritis than when either gene was knocked out alone[55].
Changes in intestinal microecology are involved in IBD pathogenesis and development. Intestinal microecology includes intestinal microbes, intestinal epithelial cells and immune cells, among which intestinal microbes play the most important role in intestinal microecology[56]. Intestinal microorganisms are distributed on the surface of the intestinal cavity and are mainly composed of bacteria, viruses, fungi and parasites, among which the number of bacteria is approximately 1014, approximately 10 times the number of human cells[57]. The total weight of intestinal bacteria is approximately 0.2 kg, accounting for 60% of the dry weight of stool. There are more than 50 bacterial groups and approximately 1100 species, most of which are Bacteroides and Firmicutes (90%), while a small portion are Actinobacteria and Proteus[58]. Many factors have been found to influence the composition of gut microbes. At birth, the environment can directly affect the intestinal microflora, including the birth canal, early diet, antibiotic use, pet contact, sex, and the mother's health, all of which are related to the intestinal microflora composition of infants in the early period[59]. The intestinal microbial diversity of infants under 1 year of age increases rapidly and tends to stabilize at 3 years of age, the intestinal microbial composition becomes more stable at 5 years of age, and Bacteroides dominates. Adult exposure to various environmental factors (such as smoking, air pollution, hygiene habits, stress, diet, drugs, etc.) can change the intestinal microbial composition[60].
Evidence suggests that the intestinal barrier plays an important role in intestinal microbial maintenance. The intestinal barrier is composed of intestinal symbiotic bacteria, the intestinal mucous layer, the intestinal epithelium and various lymphocytes in the lamina propria[61]. The intestinal mucus layer covers the intestinal epithelium, and its components are secreted by intestinal epithelial cells; this layer act as a physical barrier between the flora and the intestinal epithelium and provides nutrients and a living environment for the intestinal flora[62]. The mucus layer is rich in mucus secreted by goblet cells, a variety of antibacterial substances secreted by PCs and ordinary epithelial cells, and IgA secreted by B cells, which are difficult obstacles for intestinal bacteria to cross, effectively preventing contact with and invasion of intestinal epithelium by intestinal bacteria and preventing inflammation[63]. Intestinal epithelial cells include goblet cells, PCs, M cells, neuroendocrine cells and absorptive intestinal epithelial cells, and intestinal epithelial cells are mainly connected by tight junctions, which can prevent the invasion of bacteria and their derivatives[64]. Moreover, there are a variety of gut-associated lymphoid tissues in the intestinal epithelium and lamina propria, such as Peyer's patches in the small intestine and lymphatic follicles and colonic patches in the large intestine[65]. Many immune cells, such as dendritic cells, T lymphocytes and B lymphocytes, exist in these enteric-associated lymphoid tissues. These lymphocytes cooperate with each other to promote immune tolerance and participate in host defense. Among them, M cells and dendritic cells directly sense intestinal contents and transmit information about the intestinal flora to other immune cells, inducing an immune response or tolerance[66].
Normally, PC secretions are slowly released, and degranulation can be caused by feeding, microbial stimulation, and M receptor agonists[67]. There are many bacteria or foreign bacteria in the lumen. PCs can directly detect bacteria and express large quantities of particulate matter containing antibacterial factors through TLRs, which induces degranulation to increase the concentration of antibacterial factors in the intestinal cavity, inhibit the invasion of exogenous microorganisms, and control the microbial population in the small intestine[68]. This is an important reason why the microbial colonization density in the small intestine is lower than that in the large intestine. Therefore, PCs play important roles in controlling the passage of symbiotic bacteria and pathogenic bacteria through the intestinal barrier and in maintaining host and microbiological stability on the mucosal surface[69].
PCs and crypt basal columnar stem cells together constitute the stem cell microenvironment. The epithelial growth factors Wnt3 and Notch act on intestinal stem cells, which can promote self-renewal of intestinal stem cells and immune differentiation of different intestinal cell lines[70]. Therefore, PCs can regulate intestinal microecology and maintain intestinal mucosal homeostasis through mucosal defense and regulation of intestinal epithelial differentiation[71].
Studies have shown that the distribution of intestinal bacteria in ordinary wild mice, laboratory mice and specific pathogen-free mice decreased successively, and the distribution of PCs in the same part of the small intestine of mice among these three populations also decreased successively, suggesting that the reduction in the number of bacteria in a certain part of the small intestine could lead to a decrease in the number of PCs in that part of the small intestine[72]. After consumption of amoxicillin for 3 d, the number of PCs in each segment of the small intestine decreased significantly, which may be because amoxicillin, as a broad-spectrum antibacterial, can kill a large number of gram-positive and gram-negative bacilli in the intestine, resulting in a decrease in the total number of bacteria in each segment of the small intestine and then the number of PCs in each segment of the small intestine[73]. However, after 1 d of amoxicillin consumption, the number of PCs in the jejunum and ileum increased significantly, which may be related to the sudden disturbance of intestinal microecology leading to a temporary increase in the number of PCs in a certain period of time, which improves the body's defense function[74].
In normal mice at 4 to 6 wk of age, there is a certain degree of particle abnormality in PCs, but with increasing age, the number of normal PCs gradually increases, while the number of abnormal PCs gradually decreases[75]. However, there is no obvious PC proliferation and maturation with age in germ-free mice. Compared with that in conditions without specific pathogens, the average number of PCs per crypt is lower under completely sterile conditions, and the proportion of the normal form of PCs was almost zero in young and old mice[76]. This finding suggests that the number, morphology, and maturation of normal PCs are dependent on the intestinal flora[77].
Changes in intestinal microecology have an obvious effect on the number of PCs in the small intestine of mice[78]. After Escherichia coli (E. coli) infection, the number of PCs in each segment of the small intestine of mice is significantly reduced, which may be related to intestinal damage caused by E. coli infection[79]. Studies have shown that the intestinal villi of mice infected with E. coli have varying degrees of damage, and the longer the infection extends, the more serious the intestinal villus damage, especially duodenum damage, which is the most serious, with thinning of some parts of the intestinal wall and intestinal inflammatory cell infiltration[80]. In addition, the decrease in the number of PCs after E. coli infection may be related to the large number of released particles participating in the intestinal inflammatory response[81]. A significant decrease in the number of PCs will lead to a decrease in the barrier defense function of the intestinal tract, resulting in further damage to the small intestine of mice by opportunistic pathogens or infecting E. coli[82]. Therefore, with the extension of infection time, the number of PCs is significantly reduced. Thus, the number of PCs is closely related to the intestinal microecological balance.
PCs are an important component of the intestinal epithelial barrier, so it is not surprising that functional abnormalities of PCs play a role in CD development[83]. The first pathological analysis of small intestine samples from CD patients revealed the presence of intracellular vesicle abnormalities in PCs in these patients[84]. A better understanding of the role of PCs in CD development resulted from the discovery of CD susceptibility genes. A study showed that a CD susceptibility gene was highly expressed in PCs, which supported PCs as the origin of the disease[85]. Recently, it has been found that many CD susceptibility genes are involved in the important physiological activities of PCs, and research on the pathways regulated by these genes has revealed the significance of these pathways in regulating the physiological activities of PCs[86].
The abnormalities of PCs in IBD are mainly reflected in the abnormal quantity, morphology and function of PCs and their secretory granules[87]. Upon morphological examination of PCs from CD patients, abnormal PCs were found in 20%-50% of patients. Changes in susceptibility genes or risk-associated polymorphic sites can also cause PC abnorma
Many studies have shown that microecological disorders exist in the intestinal tract of both IBD patients and mouse models[90]. Obvious PC defects can be seen in children with CD, and abnormal PCs can cause an increased abundance of inflammatory bacteria (Corynebacterium, Erysipelotrichaceae, etc.) and reduced abundance of barrier bacteria (Faecalibacterium, Blautia, etc.). Prevotella was found to be significantly enriched in IBD patients with susceptibility genes, which may lead to a loss of intestinal barrier function and in turn to increased epithelial cell penetration and chronic inflammation[91].
Nod2 is the most significant CD susceptibility gene. In macrophages, NOD2 recognizes bacterial-derived muramyl dipeptides and activates the immune response[92]. Three major Nod2 mutants (R702W, G908R and L1007insC) have been found to be associated with the development of CD, resulting in the inability of NOD2 to effectively activate the downstream immune response[93]. In NOD2-deficient mice, α-defensin expression in PCs is decreased, terminal ileal symbiosis was increased, and pathogenic bacteria were enriched. In particular, granulomatous inflammation characterized by increased expression of Th1-related genes and inflammatory cytokines was observed in the ileum of NOD2-deficient mice inoculated with Helicobacter hepatis[94]. However, the overexpression of α-defensin in mouse PCs via transgenic technology resulted in decreased Th1 inflammation. Therefore, NOD2 can effectively inhibit the development of Th1-induced ileal granulomatous inflammation in mice. It is currently believed that lysozyme sorting in PCs is carried out through a NOD2-LRRK2-RIP2-Rab2A pathway dependent on intestinal symbiotic bacterial stimulation[95]. The absence of any one of NOD2, LRRK2, RIP2, Rab2A or symbiotic bacteria will cause lysozyme in PCs to be degraded by lysosomes, resulting in the breakdown of the balance between the organism and symbiotic bacteria, thus promoting the occurrence of CD[96].
However, the specific role of Nod2 mutations in the development of inflammatory enteritis is still under investigation. Because all three Nod2 mutations reduce the ability of Nod2 to activate the immune response, Nod2-/-mice are widely used to study the role of NOD2 in inflammatory enteritis[97]. Nod2 is mainly expressed in PCs and bone marrow-derived lymphocytes in the small intestine. The downregulation of α-defensin expression in Nod2-/-mouse PCs was reported to result in a decreased immune response to listeria[98]. It has been found that α-defensin expression in PCs is significantly reduced in patients with ileum CD compared with that in healthy individuals or patients with other types of IBD[99]. The decrease in α-defensin levels in PCs is not affected by intestinal inflammation, suggesting that the decrease in α-defensin levels is not caused by inflammation but is probably an inherent phenomenon that occurs early[100]. There was also a decrease in α-defensin levels in CD patients with 1007fs (SNP13) NOD2 mutations. However, some studies have not been able to identify defects in α-defensin production in Nod2-/- mice, which may be related to the genetic background of the mice[101]. Additional studies have shown that Nod2 functions by regulating the intestinal flora, as Nod2-/- mice have an altered intestinal flora, and Nod2-/- mice have granulomatous lesions in the ileum after infection with Helicobacter pylori, consistent with the pathological state of CD[102]. This granulomatous damage is relieved when PCs of Nod2-/- mice are transferred to α-defensin HD5 gene knockout mice.
TLRs in PCs can activate and promote the formation and secretion of antibacterial particles such as lysozyme after directly sensing intestinal bacteria and their metabolites. NOD2 secreted by PCs senses the presence of symbiotic bacteria by detecting the cell wall acyl dipeptides of symbiotic bacteria, and symbiotic microbiota-derived signals trigger NOD2 binding to receptor interaction protein 2 (RIP2)[103]. LRRK2 and Rab2a are recruited into dense core vesicles (DCVs) containing lysozyme to regulate the sorting of lysozyme in DCVs[104]. In this process, on the one hand, LRRK2 can affect the composition of intestinal microorganisms by regulating lysozyme sorting. On the other hand, symbiotic flora constituents can not only guide lysozyme sorting in PCs but also promote symbiosis between the symbiotic flora and host through the NOD2-LRRK2-Rab2a axis[105].
Cellular life activities depend on the precise transfer and directional transport and secretion of intracellular substance transport systems. If the regulation of the vesicle transport system is abnormal, the normal life activities of cells will be affected[106]. Abnormal PC vesicle transport, characterized by a reduced number of DCVs containing lysozyme, occurs in CD patients[107]. A similar phenomenon was also observed in LRRK2-/- mice. Although the expression level of lysozyme mRNA in the PCs of LRRK2-/- mice was normal, lysozyme deficiency was also found in the intestinal cavity because lysozyme was degraded by intracellular lysosomes[108]. However, other antibacterial substances, such as defensin and islet RegIIIγ, in PCs were not affected by LRRK2 knockout, suggesting that LRRK2 can specifically regulate lysozyme transport and secretion in PCs[109].
CD patients often exhibit significant dysregulation of innate immunity in the intestine. In CD pathogenesis, cytokines such as interferon-γ (IFN-γ), IFN-β, TNF-α and IL-6 can induce and upregulate LRRK2 expression[109]. Additional studies have shown that LRRK2 is involved in inflammatory cytokine production and macrophage chemotaxis in the innate immune response. LRRK2 mutation may regulate the innate immune response in CD14+ monocytes[110]. Therefore, abnormal LRRK2 expression may aggravate the innate immune disorder and further damage the tissue.
The NOD(1/2)/RIP2 signaling pathway is an important signaling pathway for the innate immune response to bacterial infection and endoplasmic reticulum stress[111]. LRRK2 enhances the activity of RIP2 by promoting the phosphorylation of RIP2 at Ser176, thus enhancing NOD(1/2)/RIP2 signaling and promoting the production of inflammatory cytokines[112]. Because LRRK2 overexpression activates NF-κB and promotes inflammatory cytokine secretion in lamina propria dendritic cells in mice, an LRRK2 overexpression group exhibited more severe colitis symptoms in DSS-induced colitis mice than a control group (wild-type mice)[113]. LRRK2 inhibitors can reduce LP-induced TNF receptor-related factor 6 (TRAF6) interaction with LRRK2 and inhibit MAPK and NF-κB suppressor protein α (Ik Bα) phosphorylation by inhibiting LLP-induced kinase activity of the LRRK2 protein[114]. Thus, the production of the inflammatory cytokine TNF-α in the dendritic cells of CD patients is reduced, which exerts an anti-inflammatory activity and ameliorates the symptoms of DSS-induced colitis[115]. In addition, a similar phenomenon has been found in macrophages, where LRRK2 defects significantly inhibit the secretion of inflammatory cytokines by macrophages when NOD2 is activated by muramyl dipeptide and NOD1 is activated by γ-D-glutamine-racemo-disaminophenic acid (I-e-DAP) or endoplasmic reticulum stress. In conclusion, LRRK2 can positively regulate the secretion of inflammatory cytokines[116].
However, other studies have shown that LRRK2 has the opposite regulatory effect. Activated T nuclear factor (NFAT) is an important mediator in the immune response, while LRRK2 is a major component of the NRON complex, an inhibitor of NFAT[117]. Normally, LRRK2 can trap NFAT by forming NRON complexes in the cytoplasm, thus sequestering NFAT in the cytoplasm[118]. However, in LRRK2-/- mice, LRRK2 deficiency leads to the failure of NFAT sequestration in the cytoplasm, thus causing NFAT translocation to the nucleus and inducing the transcription of inflammatory cytokines, increasing the level of inflammatory cytokines, increasing susceptibility to DSS-induced colitis and aggravating colitis symptoms[119]. This finding suggests that LRRK2 may also negatively regulate the secretion of inflammatory mediators and cytokines by sequestering NFAT in the cytoplasm.
When LRRK2 is defective or mutated, NFAT regulation is altered, which activates the expression of inflammatory genes in macrophages, exacerbating intestinal inflammation in colitis mouse models and leading to the development of CD[120]. It has also been found that the lysozyme mRNA expression level in the PCs of LRRK2-/- mice was normal, but lysozyme is readily degraded by intracellular lysosomes, while other antibacterial substances in PCs are not affected by LRRK2 deletion, suggesting that LRRK2 regulates lysozyme transport and secretion in PCs[121].
Another remarkable genetic factor of the CD unfolded protein response (UPR) is the transcription factor X-box binding protein-1 (XBP-1), which is a key transcription factor in the endoplasmic reticulum stress response and is involved in UPR regulation, endoplasmic reticulum amplification, and the development of hypersecretory cells (such as PCs)[122]. XBP-1 can regulate the number of PCs by preventing apoptosis and mediating cell renewal. Furthermore, mucosal defense function and susceptibility to IBD are affected[123]. The specific clearance of XBP1 from IECs induces endoplasmic reticulum stress, PC loss, reduced lysozyme and defensin expression, increased IEC death and idiopathic enteritis. Similarly, PC-specific clearance of XBP1 can produce similar symptoms of spontaneous ileitis, suggesting that the PC-specific UPR plays an important role in maintaining ileal mucosal homeostasis in mice[124]. Xbp1 knockout in the mouse epithelium resulted in spontaneous enteritis in mice[125]. Pathological analysis showed that the deletion of Xbp1 resulted in endoplasmic reticulum stress and a lack of lysozyme and α-defensin expression in PCs[125]. Therefore, XBP1 may play a partially compensatory role in inhibiting proinflammatory signals, maintaining mucosal homeostasis and assisting PC function in mice, thus supporting the function of PCs[126].
TL1A/DR3 (TL1A functional receptor) signal transduction can not only promote the proliferation of T-effector cell subsets but also promote the production of cytokines and accelerate the progression of inflammatory diseases[127]. Different levels of TL1A expression in mice have different effects on PCs and intestinal inflammation. With increasing age, the number of PCs in wild-type mice gradually increased, lysozyme particles gradually matured, and there was no spontaneous ileitis[128]. Although the number of PCs in TL1A-/- mice was reduced, normal lysozyme particle morphology was maintained, so spontaneous ileitis did not occur in older TL1A-/- mice[129]. However, in the TL1A-overexpressing mouse model, the number of PCs increased significantly, and the granules did not mature with age, accompanied by spontaneous ileitis and long-distance intestinal stenosis[130]. However, an anti-TL1A antibody could block TL1A function, reverse colonic fibrosis, and reduce existing colonic inflammation. Therefore, overexpression of TL1A can aggravate proximal intestinal inflammation and fibrous stenosis and promote disease progression[131].
ATG16L1 is an autophagy-related protein that protects against PC necrosis by participating in autophagosome formation, maintaining autophagy and mitochondrial homeostasis, and preventing PC necrotic apoptosis mediated by TNF-α[132]. In addition, ATG16L1 can also play an important role in the pathogenesis of CD by affecting the extracellular secretion of PC particles in patients or activating the transcription factor XBP1 in the endoplasmic reticulum stress response[133]. In IEC-specific ATG16L1 knockout mice, loss of autophagy resulted in increased IEC sensitivity to TNF-induced cell death. Moreover, since defensins and antimicrobial peptides are mainly secreted by PCs, changes in the abundance distribution of protein components in PCs caused by ATG16L1 defects may also trigger CD[134].
ATG16L1 T300A is the most important risk-associated polymorphic site of ATG16L1. Abnormal particle morphology and antibacterial protein packaging in PCs can be seen in mice injected with ATG16L1 T300A or in CD patients carrying ATG16L1 T300A[133]. Lysozyme in PCs is packaged and secreted in secretory autophagy during bacterial infection, and secretory autophagy was inhibited in mice carrying ATG16L1 T300A in PCs[133]. In addition, ATG16L1 T300A can also reduce selective autophagy, shorten the remission interval, increase cytokine release and reduce intracellular bacterial clearance, leading to abnormal PCs, early immune infiltration and intestinal ecological disorders[134] (Figure 2).
IBD pathogenesis is influenced by genetics, the environment, the intestinal flora and immunity, among which abnormal PCs play a central role. Both susceptibility genes and their risk-associated polymorphic loci can cause the development of abnormal PCs, and the more susceptibility genes a patient carries, the higher the proportion of abnormal PCs. The interaction between abnormal PCs and intestinal microecology is reflected in two aspects. On the one hand, the stability of the intestinal microecology needs to be maintained by the physiological function of PCs, and abnormal PCs can also cause an imbalance in intestinal microecology. On the other hand, the proliferation and maturation of PCs depend on intestinal microecology, and environmental conditions can also aggravate the influence of susceptibility genes on PCs by changing the intestinal microecology. Intestinal microecology and susceptibility genes interact with each other. On the one hand, intestinal microecology can enhance the effect of susceptibility gene expression products and promote the occurrence of IBD. If there is no intestinal flora, abnormal susceptibility genes cannot cause idiopathic enteritis. On the other hand, when IBD-related genes are abnormal, the abundance of specific bacteria in the gut is altered, which can disrupt the intestinal barrier and promote chronic inflammation. In conclusion, susceptibility genes may cause PC abnormalities and intestinal microecological disorders, and the interaction among the three can lead to potential diseases or aggravate existing diseases when they reach a specific functional threshold. In the future, more clinical and disease mechanism studies are needed to identify other genes associated with PC abnormalities and explore the cellular and molecular mechanisms of PC abnormalities caused by susceptibility genes. In addition, exploring ways to restore intestinal homeostasis by regulating the intestinal microecosystem, improving diseases by utilizing the mutual benefit between the host and intestinal flora, and identifying peripheral markers related to CD development and PC activity changes can also provide new methods for the diagnosis, treatment and prognosis of CD.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Infectious diseases
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D, D
Grade E (Poor): 0
P-Reviewer: Maslennikov R, Russia; Nikolić M, Croatia S-Editor: Yan JP L-Editor: A P-Editor: Yan JP
| 1. | Yang E, Shen J. The roles and functions of Paneth cells in Crohn's disease: A critical review. Cell Prolif. 2021;54:e12958. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 2. | Wahida A, Müller M, Hiergeist A, Popper B, Steiger K, Branca C, Tschurtschenthaler M, Engleitner T, Donakonda S, De Coninck J, Öllinger R, Pfautsch MK, Müller N, Silva M, Usluer S, Thiele Orberg E, Böttcher JP, Pfarr N, Anton M, Slotta-Huspenina JB, Nerlich AG, Madl T, Basic M, Bleich A, Berx G, Ruland J, Knolle PA, Rad R, Adolph TE, Vandenabeele P, Kanegane H, Gessner A, Jost PJ, Yabal M. XIAP restrains TNF-driven intestinal inflammation and dysbiosis by promoting innate immune responses of Paneth and dendritic cells. Sci Immunol. 2021;6:eabf7235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 3. | Mei X, Gu M, Li M. Plasticity of Paneth cells and their ability to regulate intestinal stem cells. Stem Cell Res Ther. 2020;11:349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 4. | Hassan M, Juanola O, Keller I, Nanni P, Wolski W, Martínez-López S, Caparrós E, Francés R, Moghadamrad S. Paneth Cells Regulate Lymphangiogenesis under Control of Microbial Signals during Experimental Portal Hypertension. Biomedicines. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 5. | Guo F, Cai D, Li Y, Gu H, Qu H, Zong Q, Bao W, Chen A, Liu HY. How Early-Life Gut Microbiota Alteration Sets Trajectories for Health and Inflammatory Bowel Disease? Front Nutr. 2021;8:690073. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 6. | Zhao H, Zhang W, Cheng D, You L, Huang Y, Lu Y. Investigating dysbiosis and microbial treatment strategies in inflammatory bowel disease based on two modified Koch's postulates. Front Med (Lausanne). 2022;9:1023896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 7. | De Bruyn M, Ceuleers H, Hanning N, Berg M, De Man JG, Hulpiau P, Hermans C, Stenman UH, Koistinen H, Lambeir AM, De Winter BY, De Meester I. Proteolytic Cleavage of Bioactive Peptides and Protease-Activated Receptors in Acute and Post-Colitis. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 8. | Ganna A, Satterstrom FK, Zekavat SM, Das I, Kurki MI, Churchhouse C, Alfoldi J, Martin AR, Havulinna AS, Byrnes A, Thompson WK, Nielsen PR, Karczewski KJ, Saarentaus E, Rivas MA, Gupta N, Pietiläinen O, Emdin CA, Lescai F, Bybjerg-Grauholm J, Flannick J; GoT2D/T2D-GENES Consortium, Mercader JM, Udler M; SIGMA Consortium Helmsley IBD Exome Sequencing Project; FinMetSeq Consortium; iPSYCH-Broad Consortium, Laakso M, Salomaa V, Hultman C, Ripatti S, Hämäläinen E, Moilanen JS, Körkkö J, Kuismin O, Nordentoft M, Hougaard DM, Mors O, Werge T, Mortensen PB, MacArthur D, Daly MJ, Sullivan PF, Locke AE, Palotie A, Børglum AD, Kathiresan S, Neale BM. Quantifying the Impact of Rare and Ultra-rare Coding Variation across the Phenotypic Spectrum. Am J Hum Genet. 2018;102:1204-1211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 82] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 9. | Fu Q, Song T, Ma X, Cui J. Research progress on the relationship between intestinal microecology and intestinal bowel disease. Animal Model Exp Med. 2022;5:297-310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 29] [Reference Citation Analysis (0)] |
| 10. | Sun T, Xue M, Yang J, Pei Z, Zhang N, Qin K, Liang H. Metabolic regulation mechanism of fucoidan via intestinal microecology in diseases. J Sci Food Agric. 2021;101:4456-4463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 11. | Zhu G, Hu J, Xi R. The cellular niche for intestinal stem cells: a team effort. Cell Regen. 2021;10:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 50] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 12. | Won JH, Choi JS, Jun JI. CCN1 interacts with integrins to regulate intestinal stem cell proliferation and differentiation. Nat Commun. 2022;13:3117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 32] [Reference Citation Analysis (0)] |
| 13. | Wallaeys C, Garcia-Gonzalez N, Libert C. Paneth cells as the cornerstones of intestinal and organismal health: a primer. EMBO Mol Med. 2023;15:e16427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 48] [Reference Citation Analysis (0)] |
| 14. | Verdile N, Mirmahmoudi R, Brevini TAL, Gandolfi F. Evolution of pig intestinal stem cells from birth to weaning. Animal. 2019;13:2830-2839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 15. | Vishy CE, Swietlicki EA, Gazit V, Amara S, Heslop G, Lu J, Levin MS, Rubin DC. Epimorphin regulates the intestinal stem cell niche via effects on the stromal microenvironment. Am J Physiol Gastrointest Liver Physiol. 2018;315:G185-G194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 16. | Naser AN, Lu Q, Chen YH. Three-Dimensional Culture of Murine Colonic Crypts to Study Intestinal Stem Cell Function Ex Vivo. J Vis Exp. 2022;. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 17. | Lanik WE, Xu L, Luke CJ, Hu EZ, Agrawal P, Liu VS, Kumar R, Bolock AM, Ma C, Good M. Breast Milk Enhances Growth of Enteroids: An Ex Vivo Model of Cell Proliferation. J Vis Exp. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 18. | Glal D, Sudhakar JN, Lu HH, Liu MC, Chiang HY, Liu YC, Cheng CF, Shui JW. ATF3 Sustains IL-22-Induced STAT3 Phosphorylation to Maintain Mucosal Immunity Through Inhibiting Phosphatases. Front Immunol. 2018;9:2522. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 19. | Wehkamp J, Stange EF. An Update Review on the Paneth Cell as Key to Ileal Crohn's Disease. Front Immunol. 2020;11:646. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 20. | Ma Q, Li Y, Li P, Wang M, Wang J, Tang Z, Wang T, Luo L, Wang C, Zhao B. Research progress in the relationship between type 2 diabetes mellitus and intestinal flora. Biomed Pharmacother. 2019;117:109138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 261] [Article Influence: 43.5] [Reference Citation Analysis (0)] |
| 21. | Yang W, Yu T, Huang X, Bilotta AJ, Xu L, Lu Y, Sun J, Pan F, Zhou J, Zhang W, Yao S, Maynard CL, Singh N, Dann SM, Liu Z, Cong Y. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat Commun. 2020;11:4457. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 415] [Cited by in RCA: 649] [Article Influence: 129.8] [Reference Citation Analysis (0)] |
| 22. | Liu Z, Dai X, Zhang H, Shi R, Hui Y, Jin X, Zhang W, Wang L, Wang Q, Wang D, Wang J, Tan X, Ren B, Liu X, Zhao T, Pan J, Yuan T, Chu C, Lan L, Yin F, Cadenas E, Shi L, Zhao S. Gut microbiota mediates intermittent-fasting alleviation of diabetes-induced cognitive impairment. Nat Commun. 2020;11:855. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 344] [Article Influence: 68.8] [Reference Citation Analysis (0)] |
| 23. | Song X, Sun X, Oh SF, Wu M, Zhang Y, Zheng W, Geva-Zatorsky N, Jupp R, Mathis D, Benoist C, Kasper DL. Microbial bile acid metabolites modulate gut RORγ(+) regulatory T cell homeostasis. Nature. 2020;577:410-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 321] [Cited by in RCA: 702] [Article Influence: 117.0] [Reference Citation Analysis (0)] |
| 24. | Li G, Lin J, Zhang C, Gao H, Lu H, Gao X, Zhu R, Li Z, Li M, Liu Z. Microbiota metabolite butyrate constrains neutrophil functions and ameliorates mucosal inflammation in inflammatory bowel disease. Gut Microbes. 2021;13:1968257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 261] [Article Influence: 87.0] [Reference Citation Analysis (0)] |
| 25. | Li H, Xiang Y, Zhu Z, Wang W, Jiang Z, Zhao M, Cheng S, Pan F, Liu D, Ho RCM, Ho CSH. Rifaximin-mediated gut microbiota regulation modulates the function of microglia and protects against CUMS-induced depression-like behaviors in adolescent rat. J Neuroinflammation. 2021;18:254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 148] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 26. | Lin S, Mukherjee S, Li J, Hou W, Pan C, Liu J. Mucosal immunity-mediated modulation of the gut microbiome by oral delivery of probiotics into Peyer's patches. Sci Adv. 2021;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 135] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 27. | Zhu Y, Wang X, Zhu L, Tu Y, Chen W, Gong L, Pan T, Lin H, Lin J, Sun H, Ge Y, Wei L, Guo Y, Lu C, Chen Y, Xu L. Lactobacillus rhamnosus GG combined with inosine ameliorates alcohol-induced liver injury through regulation of intestinal barrier and Treg/Th1 cells. Toxicol Appl Pharmacol. 2022;439:115923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 29] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 28. | Li R, Zhou R, Wang H, Li W, Pan M, Yao X, Zhan W, Yang S, Xu L, Ding Y, Zhao L. Gut microbiota-stimulated cathepsin K secretion mediates TLR4-dependent M2 macrophage polarization and promotes tumor metastasis in colorectal cancer. Cell Death Differ. 2019;26:2447-2463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 253] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 29. | Shil A, Chichger H. Artificial Sweeteners Negatively Regulate Pathogenic Characteristics of Two Model Gut Bacteria, E. coli and E. faecalis. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 30. | Hu J, Wang C, Huang X, Yi S, Pan S, Zhang Y, Yuan G, Cao Q, Ye X, Li H. Gut microbiota-mediated secondary bile acids regulate dendritic cells to attenuate autoimmune uveitis through TGR5 signaling. Cell Rep. 2021;36:109726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 124] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 31. | Xu Y, Zhu J, Feng B, Lin F, Zhou J, Liu J, Shi X, Lu X, Pan Q, Yu J, Zhang Y, Li L, Cao H. Immunosuppressive effect of mesenchymal stem cells on lung and gut CD8(+) T cells in lipopolysaccharide-induced acute lung injury in mice. Cell Prolif. 2021;54:e13028. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 32. | Luck H, Khan S, Kim JH, Copeland JK, Revelo XS, Tsai S, Chakraborty M, Cheng K, Tao Chan Y, Nøhr MK, Clemente-Casares X, Perry MC, Ghazarian M, Lei H, Lin YH, Coburn B, Okrainec A, Jackson T, Poutanen S, Gaisano H, Allard JP, Guttman DS, Conner ME, Winer S, Winer DA. Gut-associated IgA(+) immune cells regulate obesity-related insulin resistance. Nat Commun. 2019;10:3650. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 142] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 33. | Tyler CJ, Guzman M, Lundborg LR, Yeasmin S, Zgajnar N, Jedlicka P, Bamias G, Rivera-Nieves J. Antibody secreting cells are critically dependent on integrin α4β7/MAdCAM-1 for intestinal recruitment and control of the microbiota during chronic colitis. Mucosal Immunol. 2022;15:109-119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 28] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 34. | Lyu S, Pan F, Ge H, Yang Q, Duan X, Feng M, Liu X, Zhang T, Liu J. Fermented egg-milk beverage alleviates dextran sulfate sodium-induced colitis in mice through the modulation of intestinal flora and short-chain fatty acids. Food Funct. 2022;13:702-715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 35. | Pan LJ, Ma SY, Wen J, Zhang XQ, Xing HJ, Jia CS. Direct contact moxibustion promotes apoptosis of gastric cancer cells in rats by regulating intestinal flora. J Tradit Chin Med. 2021;41:943-952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 36. | Mao Q, Pan H, Zhang Y, Zhu Q, Hong Y, Huang Z, Li Y, Feng X, Fang Y, Chen W, Chen P, Shen B, Ouyang H, Liang Y. GelNB molecular coating as a biophysical barrier to isolate intestinal irritating metabolites and regulate intestinal microbial homeostasis in the treatment of inflammatory bowel disease. Bioact Mater. 2023;19:251-267. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 9.0] [Reference Citation Analysis (1)] |
| 37. | Yu J, He X, Wei A, Liu T, Zhang Q, Pan Y, Hao Z, Yang L, Yuan Y, Zhang Z, Zhang C, Hao C, Liu Z, Li W. HPS1 Regulates the Maturation of Large Dense Core Vesicles and Lysozyme Secretion in Paneth Cells. Front Immunol. 2020;11:560110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 38. | Li Y, Li H, Tong H, Maegelev M, Gu Z. Outer membrane vesicles derived from heatstroke-associated intestinal microbiota promote multiple organ injury in mice. Microb Pathog. 2022;170:105653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 39. | Schmitt H, Neurath MF, Atreya R. Role of the IL23/IL17 Pathway in Crohn's Disease. Front Immunol. 2021;12:622934. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 125] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 40. | Ruan J, Schlüter D, Naumann M, Waisman A, Wang X. Ubiquitin-modifying enzymes as regulators of colitis. Trends Mol Med. 2022;28:304-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 41. | Wu Y, Murray GK, Byrne EM, Sidorenko J, Visscher PM, Wray NR. GWAS of peptic ulcer disease implicates Helicobacter pylori infection, other gastrointestinal disorders and depression. Nat Commun. 2021;12:1146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 111] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 42. | Kumar A, Priyamvada S, Ge Y, Jayawardena D, Singhal M, Anbazhagan AN, Chatterjee I, Dayal A, Patel M, Zadeh K, Saksena S, Alrefai WA, Gill RK, Zadeh M, Zhao N, Mohamadzadeh M, Dudeja PK. A Novel Role of SLC26A3 in the Maintenance of Intestinal Epithelial Barrier Integrity. Gastroenterology. 2021;160:1240-1255.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 96] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 43. | Kim S, Lee JY, Shin SG, Kim JK, Silwal P, Kim YJ, Shin NR, Kim PS, Won M, Lee SH, Kim SY, Sasai M, Yamamoto M, Kim JM, Bae JW, Jo EK. ESRRA (estrogen related receptor alpha) is a critical regulator of intestinal homeostasis through activation of autophagic flux via gut microbiota. Autophagy. 2021;17:2856-2875. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 44. | Sazonovs A, Stevens CR, Venkataraman GR, Yuan K, Avila B, Abreu MT, Ahmad T, Allez M, Ananthakrishnan AN, Atzmon G, Baras A, Barrett JC, Barzilai N, Beaugerie L, Beecham A, Bernstein CN, Bitton A, Bokemeyer B, Chan A, Chung D, Cleynen I, Cosnes J, Cutler DJ, Daly A, Damas OM, Datta LW, Dawany N, Devoto M, Dodge S, Ellinghaus E, Fachal L, Farkkila M, Faubion W, Ferreira M, Franchimont D, Gabriel SB, Ge T, Georges M, Gettler K, Giri M, Glaser B, Goerg S, Goyette P, Graham D, Hämäläinen E, Haritunians T, Heap GA, Hiltunen M, Hoeppner M, Horowitz JE, Irving P, Iyer V, Jalas C, Kelsen J, Khalili H, Kirschner BS, Kontula K, Koskela JT, Kugathasan S, Kupcinskas J, Lamb CA, Laudes M, Lévesque C, Levine AP, Lewis JD, Liefferinckx C, Loescher BS, Louis E, Mansfield J, May S, McCauley JL, Mengesha E, Mni M, Moayyedi P, Moran CJ, Newberry RD, O'Charoen S, Okou DT, Oldenburg B, Ostrer H, Palotie A, Paquette J, Pekow J, Peter I, Pierik MJ, Ponsioen CY, Pontikos N, Prescott N, Pulver AE, Rahmouni S, Rice DL, Saavalainen P, Sands B, Sartor RB, Schiff ER, Schreiber S, Schumm LP, Segal AW, Seksik P, Shawky R, Sheikh SZ, Silverberg MS, Simmons A, Skeiceviciene J, Sokol H, Solomonson M, Somineni H, Sun D, Targan S, Turner D, Uhlig HH, van der Meulen AE, Vermeire S, Verstockt S, Voskuil MD, Winter HS, Young J; Belgium IBD Consortium; Cedars-Sinai IBD; International IBD Genetics Consortium; NIDDK IBD Genetics Consortium; NIHR IBD BioResource; Regeneron Genetics Center; SHARE Consortium; SPARC IBD Network; UK IBD Genetics Consortium, Duerr RH, Franke A, Brant SR, Cho J, Weersma RK, Parkes M, Xavier RJ, Rivas MA, Rioux JD, McGovern DPB, Huang H, Anderson CA, Daly MJ. Large-scale sequencing identifies multiple genes and rare variants associated with Crohn's disease susceptibility. Nat Genet. 2022;54:1275-1283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 112] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 45. | Nasser J, Bergman DT, Fulco CP, Guckelberger P, Doughty BR, Patwardhan TA, Jones TR, Nguyen TH, Ulirsch JC, Lekschas F, Mualim K, Natri HM, Weeks EM, Munson G, Kane M, Kang HY, Cui A, Ray JP, Eisenhaure TM, Collins RL, Dey K, Pfister H, Price AL, Epstein CB, Kundaje A, Xavier RJ, Daly MJ, Huang H, Finucane HK, Hacohen N, Lander ES, Engreitz JM. Genome-wide enhancer maps link risk variants to disease genes. Nature. 2021;593:238-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 389] [Cited by in RCA: 395] [Article Influence: 98.8] [Reference Citation Analysis (0)] |
| 46. | Lin R, Wu W, Chen H, Gao H, Wu X, Li G, He Q, Lu H, Sun M, Liu Z. GPR65 promotes intestinal mucosal Th1 and Th17 cell differentiation and gut inflammation through downregulating NUAK2. Clin Transl Med. 2022;12:e771. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 38] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 47. | Zheng HB, de la Morena MT, Suskind DL. The Growing Need to Understand Very Early Onset Inflammatory Bowel Disease. Front Immunol. 2021;12:675186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 48. | Gettler K, Levantovsky R, Moscati A, Giri M, Wu Y, Hsu NY, Chuang LS, Sazonovs A, Venkateswaran S, Korie U, Chasteau C; UK IBD Genetics Consortium, National Institute of Diabetes, Digestive and Kidney Diseases Inflammatory Bowel Disease Genetics Consortium, Duerr RH, Silverberg MS, Snapper SB, Daly MJ, McGovern DP, Brant SR, Rioux JD, Kugathasan S, Anderson CA, Itan Y, Cho JH. Common and Rare Variant Prediction and Penetrance of IBD in a Large, Multi-ethnic, Health System-based Biobank Cohort. Gastroenterology. 2021;160:1546-1557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 58] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 49. | Tuganbaev T, Mor U, Bashiardes S, Liwinski T, Nobs SP, Leshem A, Dori-Bachash M, Thaiss CA, Pinker EY, Ratiner K, Adlung L, Federici S, Kleimeyer C, Moresi C, Yamada T, Cohen Y, Zhang X, Massalha H, Massasa E, Kuperman Y, Koni PA, Harmelin A, Gao N, Itzkovitz S, Honda K, Shapiro H, Elinav E. Diet Diurnally Regulates Small Intestinal Microbiome-Epithelial-Immune Homeostasis and Enteritis. Cell. 2020;182:1441-1459.e21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 121] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 50. | Sharma D, Malik A, Guy CS, Karki R, Vogel P, Kanneganti TD. Pyrin Inflammasome Regulates Tight Junction Integrity to Restrict Colitis and Tumorigenesis. Gastroenterology. 2018;154:948-964.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 118] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 51. | Djouina M, Waxin C, Leprêtre F, Tardivel M, Tillement O, Vasseur F, Figeac M, Bongiovanni A, Sebda S, Desreumaux P, Launay D, Dubuquoy L, Body-Malapel M, Vignal C. Gene/environment interaction in the susceptibility of Crohn's disease patients to aluminum. Sci Total Environ. 2022;850:158017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 52. | Garcia-Carbonell R, Yao SJ, Das S, Guma M. Dysregulation of Intestinal Epithelial Cell RIPK Pathways Promotes Chronic Inflammation in the IBD Gut. Front Immunol. 2019;10:1094. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 53. | Zhang H, Cui Z, Cheng D, Du Y, Guo X, Gao R, Chen J, Sun W, He R, Ma X, Peng Q, Martin BN, Yan W, Rong Y, Wang C. RNF186 regulates EFNB1 (ephrin B1)-EPHB2-induced autophagy in the colonic epithelial cells for the maintenance of intestinal homeostasis. Autophagy. 2021;17:3030-3047. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 54. | Saul D, Leite Barros L, Wixom AQ, Gellhaus B, Gibbons HR, Faubion WA, Kosinsky RL. Cell Type-Specific Induction of Inflammation-Associated Genes in Crohn's Disease and Colorectal Cancer. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 55. | Rusu I, Mennillo E, Bain JL, Li Z, Sun X, Ly KM, Rosli YY, Naser M, Wang Z, Advincula R, Achacoso P, Shao L, Razani B, Klein OD, Marson A, Turnbaugh JA, Turnbaugh PJ, Malynn BA, Ma A, Kattah MG. Microbial signals, MyD88, and lymphotoxin drive TNF-independent intestinal epithelial tissue damage. J Clin Invest. 2022;132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 56. | Lin S, Wang Z, Lam KL, Zeng S, Tan BK, Hu J. Role of intestinal microecology in the regulation of energy metabolism by dietary polyphenols and their metabolites. Food Nutr Res. 2019;63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 57. | Cui S, Jiang J, Li B, Ross RP, Stanton C, Zhao J, Zhang H, Yang B, Chen W. Effects of the short-term administration of Pediococcus pentosaceus on physiological characteristics, inflammation, and intestinal microecology in mice. Food Funct. 2021;12:1695-1707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 58. | Mu J, Lin Q, Liang Y. An update on the effects of food-derived active peptides on the intestinal microecology. Crit Rev Food Sci Nutr. 2022;1-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 13] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 59. | Guo K, Yan Y, Zeng C, Shen L, He Y, Tan Z. Study on Baohe Pills Regulating Intestinal Microecology and Treating Diarrhea of High-Fat and High-Protein Diet Mice. Biomed Res Int. 2022;2022:6891179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 60. | Wang J, Liang J, He M, Xie Q, Wu Q, Shen G, Zhu B, Yu J, Yu L, Tan X, Wei L, Ren J, Lv Y, Deng L, Yin Q, Zhou H, Wu W, Zhang M, Yang W, Qiao M, Shu R, Xia Z, Li Z, Huang Z, Hu W, Wang L, Liu Z, Pi G, Ren H, Ji Y, Qi X, Chen P, Shao L, Chen F, Xu X, Chen W, Wang Q, Guo Z; Tumor and Microecology Committee of China Anti-Cancer Association. Chinese expert consensus on intestinal microecology and management of digestive tract complications related to tumor treatment (version 2022). J Cancer Res Ther. 2022;18:1835-1844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 61. | Zheng L, Ji YY, Wen XL, Duan SL. Fecal microbiota transplantation in the metabolic diseases: Current status and perspectives. World J Gastroenterol. 2022;28:2546-2560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 31] [Cited by in RCA: 34] [Article Influence: 11.3] [Reference Citation Analysis (4)] |
| 62. | Zheng L, Wen XL, Duan SL. Role of metabolites derived from gut microbiota in inflammatory bowel disease. World J Clin Cases. 2022;10:2660-2677. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 30] [Cited by in RCA: 25] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 63. | Zheng L, Duan SL, Dai YC, Wu SC. Role of adherent invasive Escherichia coli in pathogenesis of inflammatory bowel disease. World J Clin Cases. 2022;10:11671-11689. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 15] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 64. | Zheng L, Duan SL, Wen XL, Dai YC. Molecular regulation after mucosal injury and regeneration in ulcerative colitis. Front Mol Biosci. 2022;9:996057. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 65. | Zhang P, Jia J, Jiang P, Zheng W, Li X, Song S, Ai C. Polysaccharides from edible brown seaweed Undaria pinnatifida are effective against high-fat diet-induced obesity in mice through the modulation of intestinal microecology. Food Funct. 2022;13:2581-2593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 66. | Pan ST, Wang RC, Su YZ, Hsieh YC, Chuang SS. Lymphomatous effusion of monomorphic epitheliotropic intestinal T-cell lymphoma is characterized by azurophilic granules and is a dismal sign: Report of two new cases with literature review. Diagn Cytopathol. 2021;49:E247-E252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 67. | Hang JF, Yuan CT, Chang KC, Wang RC, Chen BJ, Hsieh PP, Huang WT, Chuang WY, Chen TW, Yeh YC, Lin SY, Hsiao CH, Chou SC, Tseng CE, Pan ST, Chang SL, Chuang SS. Targeted Next-generation Sequencing Reveals a Wide Morphologic and Immunophenotypic Spectrum of Monomorphic Epitheliotropic Intestinal T-Cell Lymphoma. Am J Surg Pathol. 2022;46:1207-1218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 68. | Pan S, Hong F, Li L, Guo Y, Qiao X, Zhang J, Xu P, Zhai Y. Melatonin Attenuates Dextran Sodium Sulfate Induced Colitis in Obese Mice. Pharmaceuticals (Basel). 2021;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 69. | Gu M, Pan S, Deng W, Li Q, Qi Z, Chen C, Bai N. Effects of glutamine on the IKK/IκB/NF-кB system in the enterocytes of turbot Scophthalmus maximus L. stimulated with soya-saponins. Fish Shellfish Immunol. 2021;119:373-378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 70. | Zhang S, Hu H, He W, Muhammad Z, Wang L, Liu F, Pan S. Regulatory Roles of Pectin Oligosaccharides on Immunoglobulin Production in Healthy Mice Mediated by Gut Microbiota. Mol Nutr Food Res. 2019;63:e1801363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 71. | Gu M, Pan S, Li Q, Qi Z, Deng W, Bai N. Protective effects of glutamine against soy saponins-induced enteritis, tight junction disruption, oxidative damage and autophagy in the intestine of Scophthalmus maximus L. Fish Shellfish Immunol. 2021;114:49-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 72. | Tan Y, Chen Q, Pan S, An W, Xu H, Xing Y, Zhang J. LMOD1, an oncogene associated with Lauren classification, regulates the metastasis of gastric cancer cells through the FAK-AKT/mTOR pathway. BMC Cancer. 2022;22:474. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 73. | Tao M, Li R, Zhang Z, Wu T, Xu T, Zogona D, Huang Y, Pan S, Xu X. Vitexin and Isovitexin Act through Inhibition of Insulin Receptor to Promote Longevity and Fitness in Caenorhabditis elegans. Mol Nutr Food Res. 2022;66:e2100845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 74. | Hong F, Pan S, Xu P, Xue T, Wang J, Guo Y, Jia L, Qiao X, Li L, Zhai Y. Melatonin Orchestrates Lipid Homeostasis through the Hepatointestinal Circadian Clock and Microbiota during Constant Light Exposure. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 72] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 75. | Pan ST, Ko YH, Tan SY, Chuang SS. Primary cutaneous peripheral T-cell lymphoma with a late relapse solely in the ileum mimicking monomorphic epitheliotropic intestinal T-cell lymphoma. Pathol Res Pract. 2018;214:2106-2109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 76. | Peng H, Bai H, Pan Y, Li J, Pei Z, Liao Y, Wu C, Li C, Tao L, Zhong S, Ma C, Chen Z, Li X, Gong Y, Wang L, Li F. Immunological pathogenesis of Bovine E. coli infection in a model of C. elegans. BMC Microbiol. 2022;22:311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 77. | Asadi Karam MR, Habibi M, Bouzari S. Urinary tract infection: Pathogenicity, antibiotic resistance and development of effective vaccines against Uropathogenic Escherichia coli. Mol Immunol. 2019;108:56-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 105] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 78. | Oberc AM, Fiebig-Comyn AA, Tsai CN, Elhenawy W, Coombes BK. Antibiotics Potentiate Adherent-Invasive E. coli Infection and Expansion. Inflamm Bowel Dis. 2019;25:711-721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 79. | Manges AR, Geum HM, Guo A, Edens TJ, Fibke CD, Pitout JDD. Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages. Clin Microbiol Rev. 2019;32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 343] [Article Influence: 57.2] [Reference Citation Analysis (0)] |
| 80. | Lee JB, Kim SK, Yoon JW. Pathophysiology of enteropathogenic Escherichia coli during a host infection. J Vet Sci. 2022;23:e28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 81. | Liu H, Xu M, He Q, Wei P, Ke M, Liu S. Untargeted serum metabolomics reveals specific metabolite abnormalities in patients with Crohn's disease. Front Med (Lausanne). 2022;9:814839. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 82. | Kattah MG, Milush JM, Burt T, McCabe RP Jr, Whang MI, Ma A, Mahadevan U. Anti-TNF and thiopurine therapy in pregnant IBD patients does not significantly alter a panel of B-cell and T-cell subsets in 1-year-old infants. Clin Transl Gastroenterol. 2018;9:143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 83. | Neubauer K, Woźniak-Stolarska B, Krzystek-Korpacka M. Peripheral Lymphocytes of Patients with Inflammatory Bowel Disease Have Altered Concentrations of Key Apoptosis Players: Preliminary Results. Biomed Res Int. 2018;2018:4961753. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 84. | Almazni I, Stapley RJ, Khan AO, Morgan NV. A comprehensive bioinformatic analysis of 126 patients with an inherited platelet disorder to identify both sequence and copy number genetic variants. Hum Mutat. 2020;41:1848-1865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 85. | Wu J, Lv X, Zhu S, Li T, Cheng H, Chen J. Critical Roles of Balanced Innate Lymphoid Cell Subsets in Intestinal Homeostasis, Chronic Inflammation, and Cancer. J Immunol Res. 2019;2019:1325181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 86. | Lu X, Wang H, Zhang J, Jin K, Ma L, Wang Y, Yang S, Wang X, Shen Q, Zhou T, Xu H, Zhang W. Comparison of Gut Viral Communities in Atopic Dermatitis and Healthy Children. Front Med (Lausanne). 2022;9:835467. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 87. | Belibasakis GN, Bostanci N, Marsh PD, Zaura E. Applications of the oral microbiome in personalized dentistry. Arch Oral Biol. 2019;104:7-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 82] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 88. | Sun Y, Liu X, Sun L, Men M, Wang B, Deng L, Zhao L, Han Y, Jong C, Bi R, Zhao M, Li X, Liu W, Shi S, Gai Z, Xu X. Microecological insight to fungal structure and key fungal communities regulating nitrogen transformation based on spatial heterogeneity during cow manure composting by multi-angle and multi-aspect analyses. Waste Manag. 2022;142:132-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 89. | Kang J, Sun M, Chang Y, Chen H, Zhang J, Liang X, Xiao T. Butyrate ameliorates colorectal cancer through regulating intestinal microecological disorders. Anticancer Drugs. 2023;34:227-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 90. | Tian Z, Zhang Y, Zheng Z, Zhang M, Zhang T, Jin J, Zhang X, Yao G, Kong D, Zhang C, Wang Z, Zhang Q. Gut microbiome dysbiosis contributes to abdominal aortic aneurysm by promoting neutrophil extracellular trap formation. Cell Host Microbe. 2022;30:1450-1463.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 106] [Reference Citation Analysis (0)] |
| 91. | Huang X, Li M, Huang Y, Yang H, Geng Y, Ouyang P, Chen D, Yin L, Yang S, Jiang J, Luo W, He Z. Microbiome analysis reveals microecological advantages of emerging ditchless rice-crayfish co-culture mode. Front Microbiol. 2022;13:892026. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 92. | Liu J, Xiao L, Nie H, Pan Y, Liu Y, Zhang Z, Lin X, Zhang Y, Cai J, Yang M, Zhang L, Xu A, Zhu C. Microecological preparation combined with an modified low-carbon diet improves glucolipid metabolism and cardiovascular complication in obese patients. Diabetol Metab Syndr. 2021;13:77. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 93. | Zhang H, Jin S, Ji A, Zhang C, Shi S. Correlation between Vaginal Microecological Status and Prognosis of CIN Patients with High-Risk HPV Infection. Biomed Res Int. 2022;2022:3620232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 94. | Du H, Ding S, Gao L, Zeng J, Lu J. Microecological investigation and comparison of two clinical methods to evaluate axillary osmidrosis. Mol Med Rep. 2020;22:4207-4212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 95. | Yang L, Fan W, Xu Y. Chameleon-like microbes promote microecological differentiation of Daqu. Food Microbiol. 2023;109:104144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 96. | Xiang Y, Zhang S, Cui Z, Yang Y. Exploring the effect of microecological agents on postoperative immune function in patients undergoing liver cancer surgery: a systematic review and meta-analysis. Ann Palliat Med. 2021;10:11615-11627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 97. | Ma Y, Chen X, Khan MZ, Xiao J, Cao Z. A Combination of Novel Microecological Agents and Molasses Role in Digestibility and Fermentation of Rice Straw by Facilitating the Ruminal Microbial Colonization. Front Microbiol. 2022;13:948049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 98. | Tan J, Zhou H, Deng J, Sun J, Zhou X, Tang Y, Qin W. Effectiveness of Microecological Preparations for Improving Renal Function and Metabolic Profiles in Patients With Chronic Kidney Disease. Front Nutr. 2022;9:850014. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 99. | Gurses SA, Banskar S, Stewart C, Trimoski B, Dziarski R, Gupta D. Nod2 protects mice from inflammation and obesity-dependent liver cancer. Sci Rep. 2020;10:20519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 100. | Takada R, Watanabe T, Hara A, Sekai I, Kurimoto M, Otsuka Y, Masuta Y, Yoshikawa T, Kamata K, Minaga K, Kudo M. NOD2 deficiency protects mice from the development of adoptive transfer colitis through the induction of regulatory T cells expressing forkhead box P3. Biochem Biophys Res Commun. 2021;568:55-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 101. | Butera A, Di Paola M, Pavarini L, Strati F, Pindo M, Sanchez M, Cavalieri D, Boirivant M, De Filippo C. Nod2 Deficiency in mice is Associated with Microbiota Variation Favouring the Expansion of mucosal CD4+ LAP+ Regulatory Cells. Sci Rep. 2018;8:14241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 102. | Wong SY, Coffre M, Ramanan D, Hines MJ, Gomez LE, Peters LA, Schadt EE, Koralov SB, Cadwell K. B Cell Defects Observed in Nod2 Knockout Mice Are a Consequence of a Dock2 Mutation Frequently Found in Inbred Strains. J Immunol. 2018;201:1442-1451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 103. | Goethel A, Turpin W, Rouquier S, Zanello G, Robertson SJ, Streutker CJ, Philpott DJ, Croitoru K. Nod2 influences microbial resilience and susceptibility to colitis following antibiotic exposure. Mucosal Immunol. 2019;12:720-732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 104. | Lee C, Choi C, Kang HS, Shin SW, Kim SY, Park HC, Hong SN. NOD2 Supports Crypt Survival and Epithelial Regeneration after Radiation-Induced Injury. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 105. | Umiker B, Lee HH, Cope J, Ajami NJ, Laine JP, Fregeau C, Ferguson H, Alves SE, Sciammetta N, Kleinschek M, Salmon M. The NLRP3 inflammasome mediates DSS-induced intestinal inflammation in Nod2 knockout mice. Innate Immun. 2019;25:132-143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 106. | Arbez N, He X, Huang Y, Ren M, Liang Y, Nucifora FC, Wang X, Pei Z, Tessarolo L, Smith WW, Ross CA. G2019S-LRRK2 mutation enhances MPTP-linked Parkinsonism in mice. Hum Mol Genet. 2020;29:580-590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 107. | Cheng X, Wu X, Zhang Y, Li W, Feng L, You H, Yang S, Yang D, Chen X, Pan X. LRRK2 Deficiency Aggravates Sleep Deprivation-Induced Cognitive Loss by Perturbing Synaptic Pruning in Mice. Brain Sci. 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 108. | Albanese F, Domenicale C, Volta M, Morari M. Modeling Parkinson's disease in LRRK2 mice: focus on synaptic dysfunction and the autophagy-lysosomal pathway. Biochem Soc Trans. 2022;50:621-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 109. | Yan J, Yu W, Wang G, Lu C, Liu C, Jiang L, Jiang Z, Liang Z, Liu D. LRRK2 deficiency mitigates colitis progression by favoring resolution of inflammation and restoring homeostasis of gut microbiota. Genomics. 2022;114:110527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 110. | Kozina E, Byrne M, Smeyne RJ. Mutant LRRK2 in lymphocytes regulates neurodegeneration via IL-6 in an inflammatory model of Parkinson's disease. NPJ Parkinsons Dis. 2022;8:24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 22] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 111. | Ning B, Guo G, Gu C, Xu J, Bibic A, He X, Liu H, Chen L, Wei Z, Duan W, Liu P, Lu H, van Zijl PCM, Ross CA, Smith W, Hua J. Mutant G2019S-LRRK2 Induces Abnormalities in Arteriolar Cerebral Blood Volume in Mouse Brains: An MRI Study. Neurodegener Dis. 2020;20:65-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 112. | Shani V, Safory H, Szargel R, Wang N, Cohen T, Elghani FA, Hamza H, Savyon M, Radzishevsky I, Shaulov L, Rott R, Lim KL, Ross CA, Bandopadhyay R, Zhang H, Engelender S. Physiological and pathological roles of LRRK2 in the nuclear envelope integrity. Hum Mol Genet. 2019;28:3982-3996. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 113. | Lara Ordónez AJ, Fernández B, Fdez E, Romo-Lozano M, Madero-Pérez J, Lobbestael E, Baekelandt V, Aiastui A, López de Munaín A, Melrose HL, Civiero L, Hilfiker S. RAB8, RAB10 and RILPL1 contribute to both LRRK2 kinase-mediated centrosomal cohesion and ciliogenesis deficits. Hum Mol Genet. 2019;28:3552-3568. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 114. | Chen X, Xie C, Tian W, Sun L, Zheng W, Hawes S, Chang L, Kung J, Ding J, Chen S, Le W, Cai H. Parkinson's disease-related Leucine-rich repeat kinase 2 modulates nuclear morphology and genomic stability in striatal projection neurons during aging. Mol Neurodegener. 2020;15:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 115. | Wallings RL, Tansey MG. LRRK2 regulation of immune-pathways and inflammatory disease. Biochem Soc Trans. 2019;47:1581-1595. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 107] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 116. | Tian Y, Lv J, Su Z, Wu T, Li X, Hu X, Zhang J, Wu L. LRRK2 plays essential roles in maintaining lung homeostasis and preventing the development of pulmonary fibrosis. Proc Natl Acad Sci U S A. 2021;118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (1)] |
| 117. | Marchand A, Drouyer M, Sarchione A, Chartier-Harlin MC, Taymans JM. LRRK2 Phosphorylation, More Than an Epiphenomenon. Front Neurosci. 2020;14:527. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 118. | Liu Z, Xu E, Zhao HT, Cole T, West AB. LRRK2 and Rab10 coordinate macropinocytosis to mediate immunological responses in phagocytes. EMBO J. 2020;39:e104862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 72] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 119. | Zhao Y, Zhang W, Huo M, Wang P, Liu X, Wang Y, Li Y, Zhou Z, Xu N, Zhu H. XBP1 regulates the protumoral function of tumor-associated macrophages in human colorectal cancer. Signal Transduct Target Ther. 2021;6:357. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 57] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 120. | Wang Q, Bu Q, Liu M, Zhang R, Gu J, Li L, Zhou J, Liang Y, Su W, Liu Z, Wang M, Lian Z, Lu L, Zhou H. XBP1-mediated activation of the STING signalling pathway in macrophages contributes to liver fibrosis progression. JHEP Rep. 2022;4:100555. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 46] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 121. | Song M, Sandoval TA, Chae CS, Chopra S, Tan C, Rutkowski MR, Raundhal M, Chaurio RA, Payne KK, Konrad C, Bettigole SE, Shin HR, Crowley MJP, Cerliani JP, Kossenkov AV, Motorykin I, Zhang S, Manfredi G, Zamarin D, Holcomb K, Rodriguez PC, Rabinovich GA, Conejo-Garcia JR, Glimcher LH, Cubillos-Ruiz JR. IRE1α-XBP1 controls T cell function in ovarian cancer by regulating mitochondrial activity. Nature. 2018;562:423-428. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 305] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 122. | Zhao N, Cao J, Xu L, Tang Q, Dobrolecki LE, Lv X, Talukdar M, Lu Y, Wang X, Hu DZ, Shi Q, Xiang Y, Wang Y, Liu X, Bu W, Jiang Y, Li M, Gong Y, Sun Z, Ying H, Yuan B, Lin X, Feng XH, Hartig SM, Li F, Shen H, Chen Y, Han L, Zeng Q, Patterson JB, Kaipparettu BA, Putluri N, Sicheri F, Rosen JM, Lewis MT, Chen X. Pharmacological targeting of MYC-regulated IRE1/XBP1 pathway suppresses MYC-driven breast cancer. J Clin Invest. 2018;128:1283-1299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 167] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 123. | Zhang Z, Qian Q, Li M, Shao F, Ding WX, Lira VA, Chen SX, Sebag SC, Hotamisligil GS, Cao H, Yang L. The unfolded protein response regulates hepatic autophagy by sXBP1-mediated activation of TFEB. Autophagy. 2021;17:1841-1855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 67] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 124. | Valatas V, Kolios G, Bamias G. TL1A (TNFSF15) and DR3 (TNFRSF25): A Co-stimulatory System of Cytokines With Diverse Functions in Gut Mucosal Immunity. Front Immunol. 2019;10:583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 125. | Zhang J, Zhang D, Pan Y, Liu X, Xu J, Qiao X, Cui W, Dong L. The TL1A-DR3 Axis in Asthma: Membrane-Bound and Secreted TL1A Co-Determined the Development of Airway Remodeling. Allergy Asthma Immunol Res. 2022;14:233-253. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 126. | Jacob N, Kumagai K, Abraham JP, Shimodaira Y, Ye Y, Luu J, Blackwood AY, Castanon SL, Stamps DT, Thomas LS, Gonsky R, Shih DQ, Michelsen KS, Targan SR. Direct signaling of TL1A-DR3 on fibroblasts induces intestinal fibrosis in vivo. Sci Rep. 2020;10:18189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 127. | Steele H, Sachen K, McKnight AJ, Soloff R, Herro R. Targeting TL1A/DR3 Signaling Offers a Therapeutic Advantage to Neutralizing IL13/IL4Rα in Muco-Secretory Fibrotic Disorders. Front Immunol. 2021;12:692127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 128. | Clarke AW, Poulton L, Shim D, Mabon D, Butt D, Pollard M, Pande V, Husten J, Lyons J, Tian C, Doyle AG. An anti-TL1A antibody for the treatment of asthma and inflammatory bowel disease. MAbs. 2018;10:664-677. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 129. | Machida K, Aw M, Salter BMA, Ju X, Mukherjee M, Gauvreau GM, O'Byrne PM, Nair P, Sehmi R. The Role of the TL1A/DR3 Axis in the Activation of Group 2 Innate Lymphoid Cells in Subjects with Eosinophilic Asthma. Am J Respir Crit Care Med. 2020;202:1105-1114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 130. | Aden K, Tran F, Ito G, Sheibani-Tezerji R, Lipinski S, Kuiper JW, Tschurtschenthaler M, Saveljeva S, Bhattacharyya J, Häsler R, Bartsch K, Luzius A, Jentzsch M, Falk-Paulsen M, Stengel ST, Welz L, Schwarzer R, Rabe B, Barchet W, Krautwald S, Hartmann G, Pasparakis M, Blumberg RS, Schreiber S, Kaser A, Rosenstiel P. ATG16L1 orchestrates interleukin-22 signaling in the intestinal epithelium via cGAS-STING. J Exp Med. 2018;215:2868-2886. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 130] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 131. | Chesney KL, Men H, Hankins MA, Bryda EC. The Atg16 L1 gene: characterization of wild type, knock-in, and knock-out phenotypes in rats. Physiol Genomics. 2021;53:269-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 132. | Zhang Z, Guo M, Li Y, Shen M, Kong D, Shao J, Ding H, Tan S, Chen A, Zhang F, Zheng S. RNA-binding protein ZFP36/TTP protects against ferroptosis by regulating autophagy signaling pathway in hepatic stellate cells. Autophagy. 2020;16:1482-1505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 333] [Article Influence: 66.6] [Reference Citation Analysis (0)] |
| 133. | Xu Y, Zhou P, Cheng S, Lu Q, Nowak K, Hopp AK, Li L, Shi X, Zhou Z, Gao W, Li D, He H, Liu X, Ding J, Hottiger MO, Shao F. A Bacterial Effector Reveals the V-ATPase-ATG16L1 Axis that Initiates Xenophagy. Cell. 2019;178:552-566.e20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 258] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 134. | Oestreich AK, Chadchan SB, Popli P, Medvedeva A, Rowen MN, Stephens CS, Xu R, Lydon JP, Demayo FJ, Jungheim ES, Moley KH, Kommagani R. The Autophagy Gene Atg16L1 is Necessary for Endometrial Decidualization. Endocrinology. 2020;161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |