Published online Dec 6, 2022. doi: 10.12998/wjcc.v10.i34.12578
Peer-review started: August 1, 2022
First decision: August 21, 2022
Revised: September 1, 2022
Accepted: November 14, 2022
Article in press: November 14, 2022
Published online: December 6, 2022
Processing time: 123 Days and 2.9 Hours
The vaginal microbiome plays a critical role in the health of pregnant women and their newborns. Group B Streptococcus (GBS) and vaginal cleanliness significantly affect the vaginal microecosystem and are closely associated with vaginal diseases.
To explore the effects of GBS status and vaginal cleanliness on vaginal microecosystems.
We collected 160 vaginal swabs from pregnant women and divided them into the following four groups based on GBS status and vaginal cleanliness: GBS-positive + vaginal cleanliness I–II degree, GBS-negative + vaginal cleanliness I–II degree, GBS-positive + vaginal cleanliness III–IV degree, and GBS-negative + vaginal cleanliness III–IV degree. Samples were subjected to 16S rRNA gene amplicon sequencing.
Alpha diversity analysis showed that the Shannon index did not significantly differ between the four groups. We identified significant variation in taxa abundance between the GBS-positive and GBS-negative groups and between the vaginal cleanliness I–II degree and III–IV degree groups. Principal coordinate analysis and non-metric multidimensional scaling analysis further confirmed the microbial diversity of the four groups. Moreover, the linear discriminant analysis demonstrated that Lactobacillus jensenii and Actinobacteria were strongly associated with GBS-positive status, and Lactobacillus iners, Lactobacillaceae, Lactobacillus, Lactobacillales, Bacilli and Firmicutes were closely correlated with GBS-negative status.
GBS status and vaginal cleanliness significantly affect vaginal microbiome differences in pregnant women. Our findings provide instructional information for clinical antibiotic treatment in pregnant women with different GBS statuses and vaginal cleanliness degrees.
Core Tip: To explore the correlation between Group B Streptococcus (GBS) status and vaginal cleanliness with the vaginal microbiome, we collected 160 vaginal swabs from pregnant women and the samples were subjected to 16S rRNA gene amplicon sequencing. We identified significant variation in taxa abundance between the GBS-positive and GBS-negative groups and between the vaginal cleanliness I–II degree and III–IV degree groups. Our findings provide new insights into understanding the vaginal microenvironment.
- Citation: Liao Q, Zhang XF, Mi X, Jin F, Sun HM, Wang QX. Influence of group B streptococcus and vaginal cleanliness on the vaginal microbiome of pregnant women. World J Clin Cases 2022; 10(34): 12578-12586
- URL: https://www.wjgnet.com/2307-8960/full/v10/i34/12578.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i34.12578
The vagina is a complex and sensitive microecosystem controlled by the vaginal anatomy, endocrine regulation, microbial composition, and the local immune system[1]. In dynamic equilibrium, the vaginal microbiome species are mutually interdependent, antagonistic, and controlled by the local immune system, endocrine system, and internal environment[2]. Vaginal pH, estrogen levels, local immunity, Lactobacillus species, and vaginal cleanliness play essential roles in maintaining the microecological balance of the vagina[2]. The vaginal microbiome significantly affects vaginal homeostasis. Hence, understanding the vaginal microbiome is essential for vaginal health.
Group B Streptococcus (GBS) is a gram-positive bacterium that transiently and asymptomatically colonizes the vagina and gastrointestinal tracts of healthy women. Thus, it is the principal reason for invasive bacterial disorders in newborns and lethal diseases in infants[3]. Globally, more than three million annual neonatal deaths are caused by GBS infections[4]. The features of GBS have been primarily studied in 4025 women, and a significantly low likelihood of detecting coagulase-negative Lactobacillus, Prevotella, and Staphylococcus has been observed in GBS-positive patients[5,6]. Vaginal cleanliness significantly affects vaginal health. However, the correlation between GBS status and vaginal cleanliness with the vaginal microbiome is still elusive.
In this study, we aimed to investigate the effects of GBS status and vaginal cleanliness on the vaginal microbiome of pregnant women. We successfully identified a novel landscape in which GBS status and vaginal cleanliness significantly affected vaginal microbiome differences in pregnant women.
A total of 160 vaginal swab samples from pregnant women were collected at our hospital between June 2018 and January 2019. The samples were divided based on GBS status and vaginal cleanliness into the following four groups: GBS-positive + vaginal cleanliness I–II degree (group A, n = 24), GBS-negative + vaginal cleanliness I–II degree (group B, n = 53), GBS-positive + vaginal cleanliness III–IV degree (group C, n = 35), and GBS-negative + vaginal cleanliness III–IV degree (group D, n = 48). Samples were acquired from the patients and healthy participants after obtaining written informed consent. This study was approved by the Ethics Committee of the Shunyi Women and Children’s Hospital of Beijing Children’s Hospital.
DNA samples were obtained from vaginal swabs using a DNA isolation kit (Omega, USA). DNA was quantified using NanoDrop ND-2000 (Thermo Fisher Scientific, USA). The V1–V2 hypervariable regions were also measured. Two standard bacterial 16S rRNA amplicon polymerase chain reaction (PCR) primers were used. A QIAquick PCR Purification Kit (Qiagen, USA) was used to purify the amplicons, followed by quantification using NanoDrop ND-2000 (Thermo Fisher Scientific, USA). Further, 16S rRNA sequencing was performed using HiSeq 2500 (Illumina, USA).
The 16S rRNA sequencing datasets were filtered and merged using the FLASH method [7]. Sequencing was performed using Quantitative Insights into Microbial Ecology (QIIME, version 1.9.1) software (http://qiime.org/)[8]. Chimeric sequences were deleted applying usearch61 using de novo methods[9]. Sequencing was clustered on the 2013 Greengenes (13_8 release) ribosomal database 97% reference dataset (http://greengenes.secondgenome.com/?prefix=downloads/greengenes_database/). Taxono
Data are presented as mean ± standard deviation. The Wilcoxon rank-sum test was used to evaluate alpha diversity. Analysis of similarities (ANOSIM) of beta diversity matrices was performed to analyze significant differences in microbial communities using QIIME. The microbial biomarkers were analyzed using linear discriminant analysis effect size with the web-based Galaxy interface (http://hutten
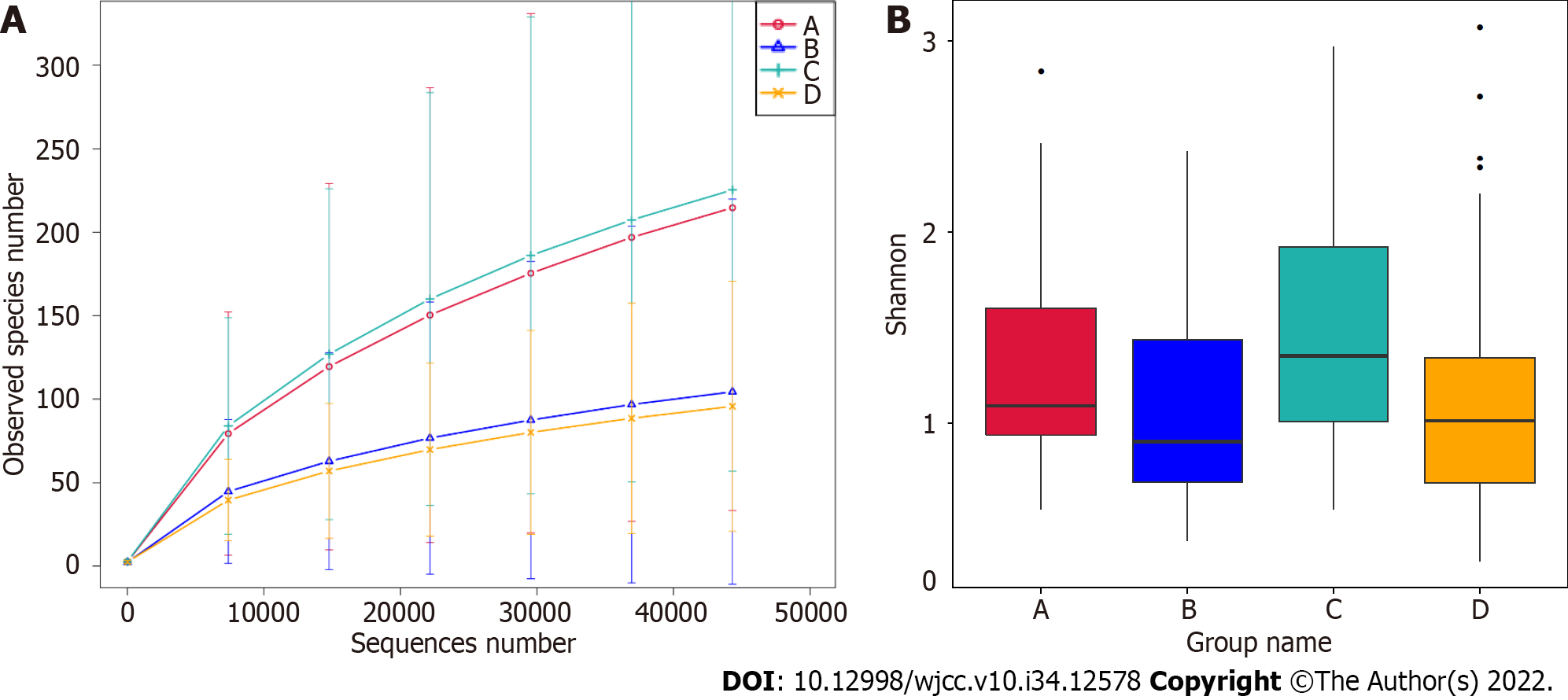
The effect of GBS status and vaginal cleanliness on the vaginal microbiome was determined using 16S rRNA gene amplicon sequencing. Significantly, the rarefaction curve of the observed species revealed that the depth of 16S rRNA gene amplicon sequencing satisfactorily demonstrated sequencing diversity among the four groups (Figure 1A). In addition, alpha diversity analysis revealed that the Shannon index failed to exhibit significant differences among the four groups (Figure 1B, Wilcoxon rank-sum test, P > 0.05).
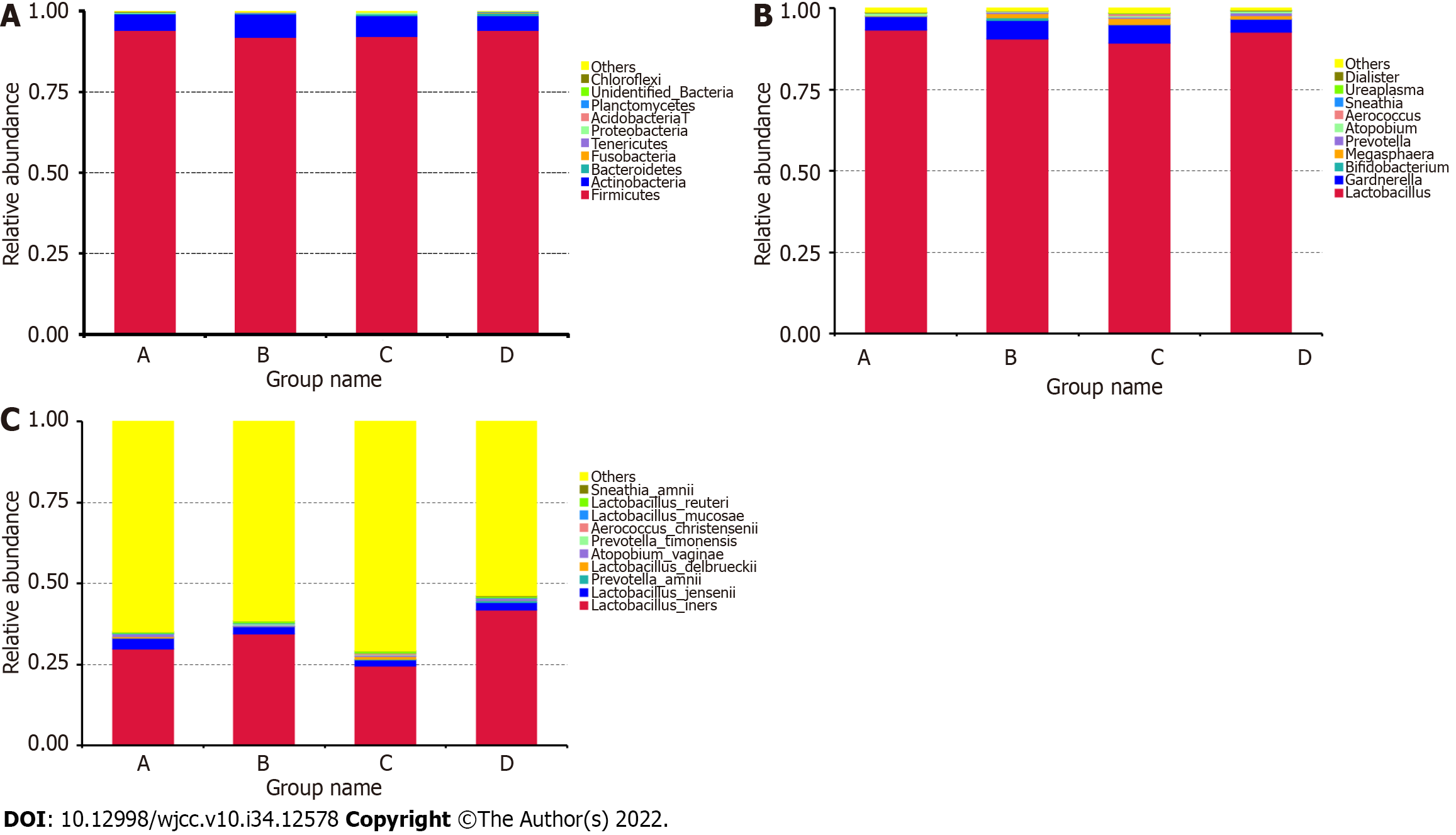
We then compared the taxa abundance and identified the top ten significant taxa at the phylum, genus, and species levels. At the phylum level, the abundance of Firmicutes, Actinobacteria, Bacteroidetes, Fusobacteria, Tenericutes, Proteobacteria, Acidobacteria, Planctomycetes, Chloroflexi, and some unidentified bacteria differed between the GBS-positive and GBS-negative groups, as well as between the vaginal cleanliness I–II and III–IV degree groups (Figure 2A). At the genus level, the abundance of Lactobacillus, Gardnerella, Bifidobacterium, Megasphaera, Prevotella, Atopobium, Aerococcus, Sneathia, Ureaplasma, and Dialister were different between the GBS-positive and GBS-negative groups and between the vaginal cleanliness degrees I–II and III–IV (Figure 2B). At the species level, the abundance of Lactobacillus iners, Lactobacillus jensenii, Prevotella amnii, Lactobacillus delbrueckii, Atopobium vaginae, Prevotella timonensis, Aerococcus christensenii, Lactobacillus mucosae, Lactobacillus reuteri, and Sneathia amnii were different between the GBS-positive and GBS-negative groups and between the vaginal cleanliness I–II and III–IV degree groups (Figure 2C).
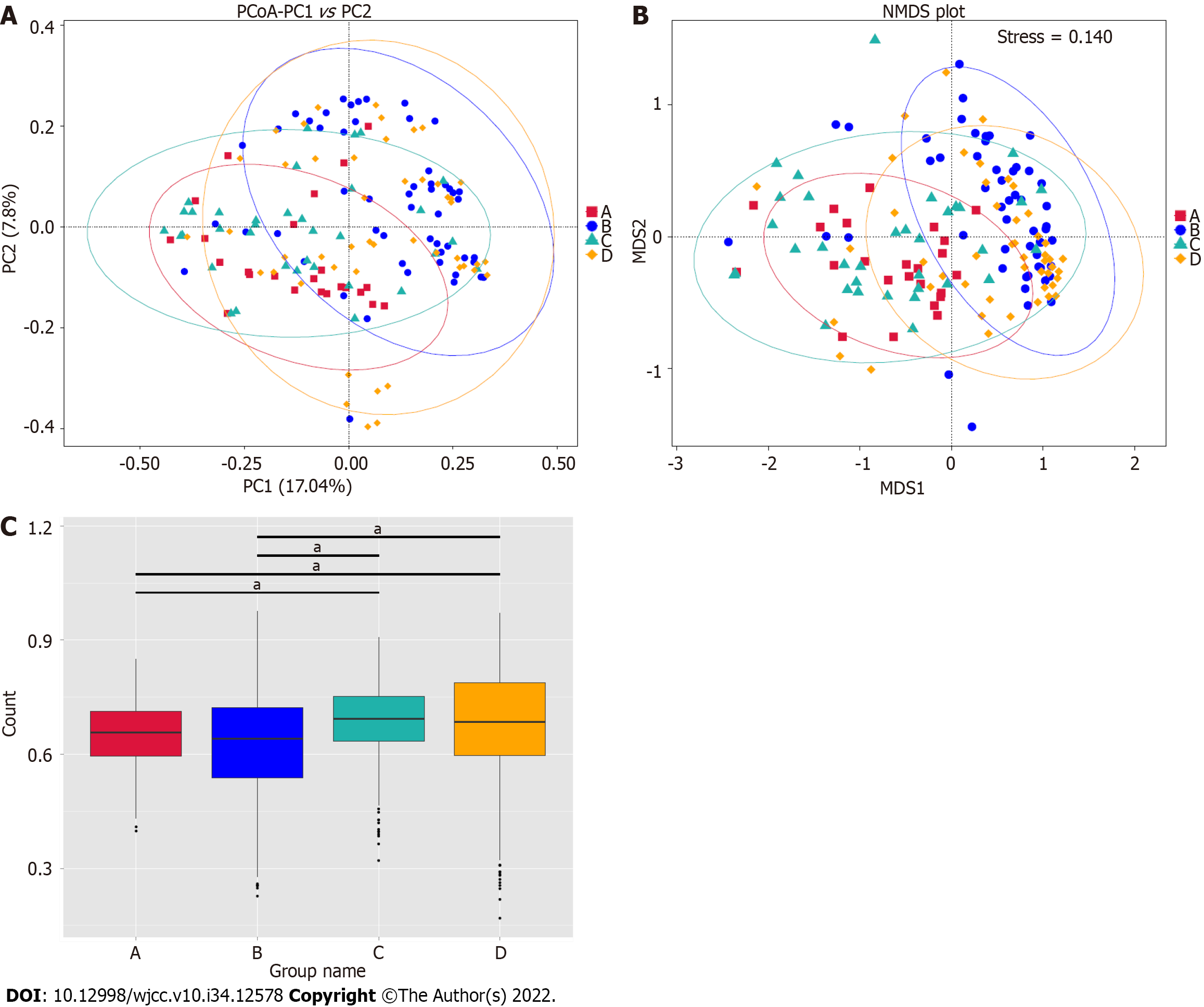
We further explored the microbiome differences between the GBS-positive and GBS-negative groups and between the vaginal cleanliness degrees groups I–II and III–IV using beta diversity analyses, including principal coordinates analysis (PCoA) and non-metric multidimensional scaling (NMDS). The PCoA (Figure 3A), unweighted Unifrac Distance, ANOSIM), NMDS analysis (Figure 3B), and unwei
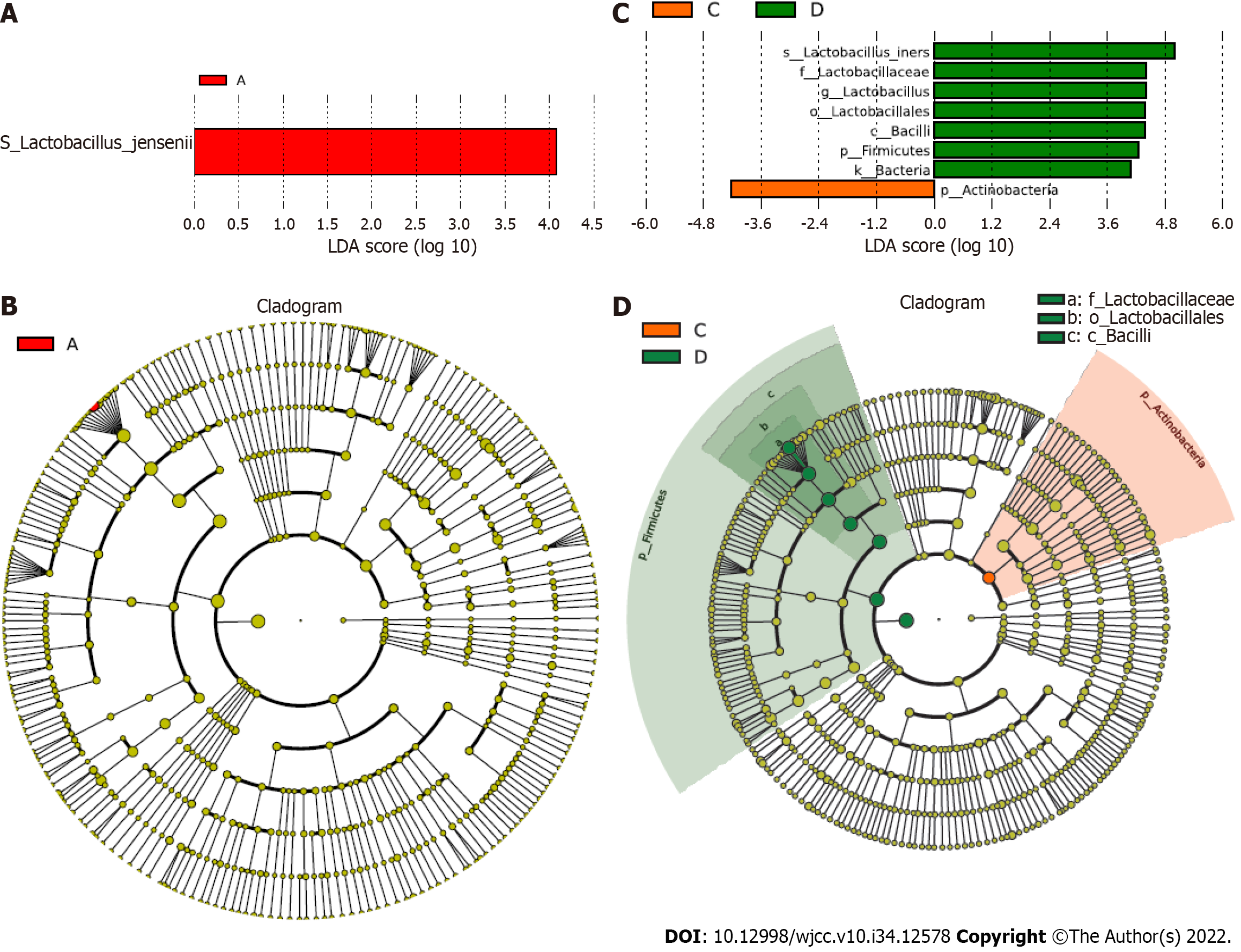
Additionally, we investigated the association of specific taxa with GBS status and vaginal cleanliness based on LDA. Significantly, we observed several specific taxa between the GBS-positive and GBS-negative groups based on groups A and B and groups C and D (Figure 4). Lactobacillus jensenii and Actinobacteria were closely correlated with GBS-positive status. Lactobacillus iners, Lactobacillaceae, Lactobacillus, Lactobacillales, Bacilli, Firmicutes, and Bacteria were strongly associated with GBS-negative status.
Diseases associated with vaginal infections cause significant burden to society[12]. Clinical data plus microbiome may be a good model to predict the personalized health status[13]. The vaginal microbiome plays an essential role in women’s reproductive health, and GBS status and vaginal cleanliness are crucial in maintaining the vaginal microenvironment[14,15]. However, the impact of GBS status and vaginal cleanliness on the vaginal microbiome remains unclear. In this study, we demonstrated the influence of GBS status and vaginal cleanliness on the vaginal microbiome of 160 vaginal swab samples using 16S rRNA gene amplicon sequencing.
Several 16S rRNA gene amplicon sequencing studies have characterized the vaginal microbiome, including the relationship between the vaginal microbiome and polycystic ovary syndrome[16]. Two 16S rRNA gene investigations have also reported the role of the vaginal microbiome in reproductive-aged women and preterm newborns[17,18]. Another 16S rRNA gene amplicon sequencing identified the vagina-uterine microbiome features and showed that elucidating the vaginal microbiome may help detect prevalent diseases in the upper reproductive tract[19]. Although both GBS and vaginal cleanliness are critical factors for the vaginal microbiome of pregnant women, the combined analysis of GBS and vaginal cleanliness using 16S rRNA gene amplicon sequencing remains limited. Here, we focused on the correlation of GBS status and vaginal cleanliness with the vaginal microbiome. Our data showed no alpha diversity between the GBS-positive and GBS-negative groups and between the vaginal cleanliness I–II degree and III–IV degree groups. However, the PCoA and NMDS analysis showed significant microbiome differences between the GBS-positive and GBS-negative groups and between the vaginal cleanliness I–II degree and III–IV degree groups, respectively. These data suggest that GBS status and vaginal cleanliness degree can affect the vaginal microbiome, providing new evidence of the association between GBS status and vaginal cleanliness in the vaginal microenvironment. Moreover, our study provides an example of a comprehensive combined analysis of the GBS status and vaginal cleanliness in pregnant women.
Numerous studies have shown that the dominance of a single OTU mainly characterizes the normal vaginal microbiome, most closely related to Lactobacillus species[20-22]. The Lactobacillus species repress pathogenic microorganisms by maintaining an acidic vaginal pH[23,24]. Lactobacillus dominates the healthiest vaginal microbiota, and Prevotella, generally identified in the vagina, is associated with bacterial vaginosis and has been correlated with GBS-positive status[25-28]. Megasphaera is also associated with a GBS-positive status, which is closely related to bacterial vaginosis[25-27,29]. Nevertheless, investigation of the correlation between vaginal cleanliness and the vaginal microbiome is remarkably limited. In this study, we identified that Lactobacillus iners, Lactobacillus jensenii, Prevotella amnii, Lactobacillus delbrueckii, Atopobium vaginae, Prevotella timonensis, Aerococcus christensenii, Lactobacillus mucosae, Lactobacillus reuteri, and Sneathia amnii differed between the GBS-positive and GBS-negative groups, and between the vaginal cleanliness I–II and III–IV degree groups at the species level. Moreover, we found that Lactobacillus jensenii and Actinobacteria were strongly correlated with GBS-positive status, and Lactobacillus iners, Lactobacillaceae, Lactobacillus, Lactobacillales, Bacilli, Firmicutes, and Bacteria were strongly associated with the GBS-negative status. In Pace et al study, they assessed positive GBS clinical cultivation, and found a limited number of differentially abundant taxa, including an increased enrichment of Ureaplasma urealyticum, Corynebacterium glucuronolyticum, Propionibacterium acnes, and Haemophilus haemolyticus[30]. These data indicated a correlation between GBS status and vaginal cleanliness in the vaginal microenvironment. Importantly, we present a landscape of the specific vaginal microbiome, such as Lactobacillus iners, Prevotella timonensis, and Sneathia amnii, associated with the GBS status and vaginal cleanliness, and demonstrate the precise vaginal microbiome associated with GBS-positive or -negative status, providing instructional information for clinical antibiotic treatment of pregnant women with different GBS status and vaginal cleanliness degrees.
In summary, we discovered that GBS status and vaginal cleanliness significantly affect vaginal microbiota differences in pregnant women. We identified several specific vaginal microbiomes, including Lactobacillus iners, Prevotella timonensis, and Sneathia amnii, in patients with varying GBS statuses. We also found that Lactobacillus jensenii and Actinobacteria were particularly associated with GBS-positive status, and Lactobacillus iners, Lactobacillaceae, Lactobacillus, Lactobacillales, Bacilli, Firmicutes, and Bacteria strongly correlated with GBS-negative status. Our findings provide new insights into understanding the vaginal microenvironment, presenting a landscape of the association of GBS status and vaginal cleanliness with the vaginal microbiome of pregnant women. Our results provide instructional information for clinical antibiotic treatment in pregnant women with different GBS statuses and vaginal cleanliness degrees.
The vaginal microbiome significantly affects vaginal homeostasis. Hence, understanding the vaginal microbiome is essential for vaginal health. Group B Streptococcus (GBS) is a gram-positive bacterium that transiently and asymptomatically colonizes the vagina and gastrointestinal tracts of healthy women. However, the correlation between GBS status and vaginal cleanliness with the vaginal microbiome is still elusive.
This study explored the effects of GBS status and vaginal cleanliness on vaginal microecosystems. This study would provide instructional information for clinical antibiotic treatment in pregnant women with different GBS statuses and vaginal cleanliness degrees.
We aimed to investigate the effects of GBS status and vaginal cleanliness on the vaginal microbiome of pregnant women.
We collected 160 vaginal swabs from pregnant women and divided them into the following four groups based on GBS status and vaginal cleanliness: GBS-positive + vaginal cleanliness I–II degree, GBS-negative + vaginal cleanliness I–II degree, GBS-positive + vaginal cleanliness III–IV degree, and GBS-negative + vaginal cleanliness III–IV degree. Samples were subjected to 16S rRNA gene amplicon sequencing.
Alpha diversity analysis showed that the Shannon index did not significantly differ between the four groups. We identified significant variation in taxa abundance between the GBS-positive and GBS-negative groups and between the vaginal cleanliness I–II degree and III–IV degree groups. Principal coordinate analysis and non-metric multidimensional scaling analysis further confirmed the microbial diversity of the four groups. Moreover, the linear discriminant analysis demonstrated that Lactobacillus jensenii and Actinobacteria were strongly associated with GBS-positive status, and Lactobacillus iners, Lactobacillaceae, Lactobacillus, Lactobacillales, Bacilli and Firmicutes were closely correlated with GBS-negative status.
We identified several specific vaginal microbiomes, including Lactobacillus iners, Prevotella timonensis, and Sneathia amnii, in patients with varying GBS statuses. We also found that Lactobacillus jensenii and Actinobacteria were particularly associated with GBS-positive status, and Lactobacillus iners, Lactobacillaceae, Lactobacillus, Lactobacillales, Bacilli, Firmicutes, and Bacteria strongly correlated with GBS-negative status.
Our findings provide new insights into understanding the vaginal microenvironment, presenting a landscape of the association of GBS status and vaginal cleanliness with the vaginal microbiome of pregnant women. Our results provide instructional information for clinical antibiotic treatment in pregnant women with different GBS statuses and vaginal cleanliness degrees.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Microbiology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Colpaert C, Belgium; Dobrocky T, Switzerland S-Editor: Wang JL L-Editor: A P-Editor: Zhao S
| 1. | Yu F, Tang YT, Hu ZQ, Lin XN. Analysis of the Vaginal Microecological Status and Genital Tract Infection Characteristics of 751 Pregnant Women. Med Sci Monit. 2018;24:5338-5345. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 2. | Ma B, Forney LJ, Ravel J. Vaginal microbiome: rethinking health and disease. Annu Rev Microbiol. 2012;66:371-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 409] [Cited by in RCA: 472] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 3. | Korir ML, Manning SD, Davies HD. Intrinsic Maturational Neonatal Immune Deficiencies and Susceptibility to Group B Streptococcus Infection. Clin Microbiol Rev. 2017;30:973-989. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 4. | Rick AM, Aguilar A, Cortes R, Gordillo R, Melgar M, Samayoa-Reyes G, Frank DN, Asturias EJ. Group B Streptococci Colonization in Pregnant Guatemalan Women: Prevalence, Risk Factors, and Vaginal Microbiome. Open Forum Infect Dis. 2017;4:ofx020. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 5. | Kubota T, Nojima M, Itoh S. Vaginal bacterial flora of pregnant women colonized with group B streptococcus. J Infect Chemother. 2002;8:326-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 6. | Altoparlak U, Kadanali A, Kadanali S. Genital flora in pregnancy and its association with group B streptococcal colonization. Int J Gynaecol Obstet. 2004;87:245-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 26] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 7. | Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957-2963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7679] [Cited by in RCA: 9237] [Article Influence: 659.8] [Reference Citation Analysis (0)] |
| 8. | Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335-336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29318] [Cited by in RCA: 23744] [Article Influence: 1582.9] [Reference Citation Analysis (0)] |
| 9. | Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460-2461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14305] [Cited by in RCA: 13984] [Article Influence: 932.3] [Reference Citation Analysis (0)] |
| 10. | Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141-D145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3794] [Cited by in RCA: 3483] [Article Influence: 217.7] [Reference Citation Analysis (0)] |
| 11. | Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11842] [Cited by in RCA: 10319] [Article Influence: 737.1] [Reference Citation Analysis (0)] |
| 12. | Ding W, Ma Y, Ma C, Malone DC, Ma A, Tang W, Si L. The Lifetime Cost Estimation of Human Papillomavirus-related Diseases in China: A Modeling Study. J Transl Int Med. 2021;9:200-211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 13. | Huang Q, Fang Q, Hu Z. A P4 Medicine Perspective of Gut Microbiota and Prediabetes: Systems Analysis and Personalized Intervention. J Transl Int Med. 2020;8:119-130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 14. | Martín V, Cárdenas N, Ocaña S, Marín M, Arroyo R, Beltrán D, Badiola C, Fernández L, Rodríguez JM. Rectal and Vaginal Eradication of Streptococcus agalactiae (GBS) in Pregnant Women by Using Lactobacillus salivarius CECT 9145, A Target-specific Probiotic Strain. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 15. | Zheng N, Guo R, Yao Y, Jin M, Cheng Y, Ling Z. Lactobacillus iners Is Associated with Vaginal Dysbiosis in Healthy Pregnant Women: A Preliminary Study. Biomed Res Int. 2019;2019:6079734. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 16. | Hong X, Qin P, Huang K, Ding X, Ma J, Xuan Y, Zhu X, Peng D, Wang B. Association between polycystic ovary syndrome and the vaginal microbiome: A case-control study. Clin Endocrinol (Oxf). 2020;93:52-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 17. | Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4680-4687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2090] [Cited by in RCA: 2694] [Article Influence: 179.6] [Reference Citation Analysis (0)] |
| 18. | Fettweis JM, Serrano MG, Brooks JP, Edwards DJ, Girerd PH, Parikh HI, Huang B, Arodz TJ, Edupuganti L, Glascock AL, Xu J, Jimenez NR, Vivadelli SC, Fong SS, Sheth NU, Jean S, Lee V, Bokhari YA, Lara AM, Mistry SD, Duckworth RA 3rd, Bradley SP, Koparde VN, Orenda XV, Milton SH, Rozycki SK, Matveyev AV, Wright ML, Huzurbazar SV, Jackson EM, Smirnova E, Korlach J, Tsai YC, Dickinson MR, Brooks JL, Drake JI, Chaffin DO, Sexton AL, Gravett MG, Rubens CE, Wijesooriya NR, Hendricks-Muñoz KD, Jefferson KK, Strauss JF 3rd, Buck GA. The vaginal microbiome and preterm birth. Nat Med. 2019;25:1012-1021. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 589] [Reference Citation Analysis (0)] |
| 19. | Chen C, Song X, Wei W, Zhong H, Dai J, Lan Z, Li F, Yu X, Feng Q, Wang Z, Xie H, Chen X, Zeng C, Wen B, Zeng L, Du H, Tang H, Xu C, Xia Y, Xia H, Yang H, Wang J, Madsen L, Brix S, Kristiansen K, Xu X, Li J, Wu R, Jia H. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat Commun. 2017;8:875. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 330] [Cited by in RCA: 569] [Article Influence: 71.1] [Reference Citation Analysis (0)] |
| 20. | Ahmed A, Earl J, Retchless A, Hillier SL, Rabe LK, Cherpes TL, Powell E, Janto B, Eutsey R, Hiller NL, Boissy R, Dahlgren ME, Hall BG, Costerton JW, Post JC, Hu FZ, Ehrlich GD. Comparative genomic analyses of 17 clinical isolates of Gardnerella vaginalis provide evidence of multiple genetically isolated clades consistent with subspeciation into genovars. J Bacteriol. 2012;194:3922-3937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 123] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 21. | Albert AY, Chaban B, Wagner EC, Schellenberg JJ, Links MG, van Schalkwyk J, Reid G, Hemmingsen SM, Hill JE, Money D; VOGUE Research Group. A Study of the Vaginal Microbiome in Healthy Canadian Women Utilizing cpn60-Based Molecular Profiling Reveals Distinct Gardnerella Subgroup Community State Types. PLoS One. 2015;10:e0135620. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 83] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 22. | Paramel Jayaprakash T, Schellenberg JJ, Hill JE. Resolution and characterization of distinct cpn60-based subgroups of Gardnerella vaginalis in the vaginal microbiota. PLoS One. 2012;7:e43009. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 70] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 23. | Rosen GH, Randis TM, Desai PV, Sapra KJ, Ma B, Gajer P, Humphrys MS, Ravel J, Gelber SE, Ratner AJ. Group B Streptococcus and the Vaginal Microbiota. J Infect Dis. 2017;216:744-751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 24. | Bhandari P, Prabha V. Evaluation of profertility effect of probiotic Lactobacillus plantarum 2621 in a murine model. Indian J Med Res. 2015;142:79-84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 25. | Ling Z, Kong J, Liu F, Zhu H, Chen X, Wang Y, Li L, Nelson KE, Xia Y, Xiang C. Molecular analysis of the diversity of vaginal microbiota associated with bacterial vaginosis. BMC Genomics. 2010;11:488. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 262] [Cited by in RCA: 237] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 26. | Dols JA, Smit PW, Kort R, Reid G, Schuren FH, Tempelman H, Bontekoe TR, Korporaal H, Boon ME. Microarray-based identification of clinically relevant vaginal bacteria in relation to bacterial vaginosis. Am J Obstet Gynecol. 2011;204:305.e1-305.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 27. | Datcu R. Characterization of the vaginal microflora in health and disease. Dan Med J. 2014;61:B4830. [PubMed] |
| 28. | O'Hanlon DE, Moench TR, Cone RA. Vaginal pH and microbicidal lactic acid when lactobacilli dominate the microbiota. PLoS One. 2013;8:e80074. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 304] [Cited by in RCA: 335] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 29. | Balkus JE, Srinivasan S, Anzala O, Kimani J, Andac C, Schwebke J, Fredricks DN, McClelland RS. Impact of Periodic Presumptive Treatment for Bacterial Vaginosis on the Vaginal Microbiome among Women Participating in the Preventing Vaginal Infections Trial. J Infect Dis. 2017;215:723-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 30. | Pace RM, Chu DM, Prince AL, Ma J, Seferovic MD, Aagaard KM. Complex species and strain ecology of the vaginal microbiome from pregnancy to postpartum and association with preterm birth. Med (N Y). 2021;2:1027-1049. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |