Published online Jan 21, 2022. doi: 10.12998/wjcc.v10.i3.1067
Peer-review started: June 22, 2021
First decision: July 26, 2021
Revised: August 9, 2021
Accepted: December 23, 2021
Article in press: December 23, 2021
Published online: January 21, 2022
Fabry disease (FD) is a rare X-linked lysosomal storage disease caused by a deficiency of the enzyme α-galactosidase A.
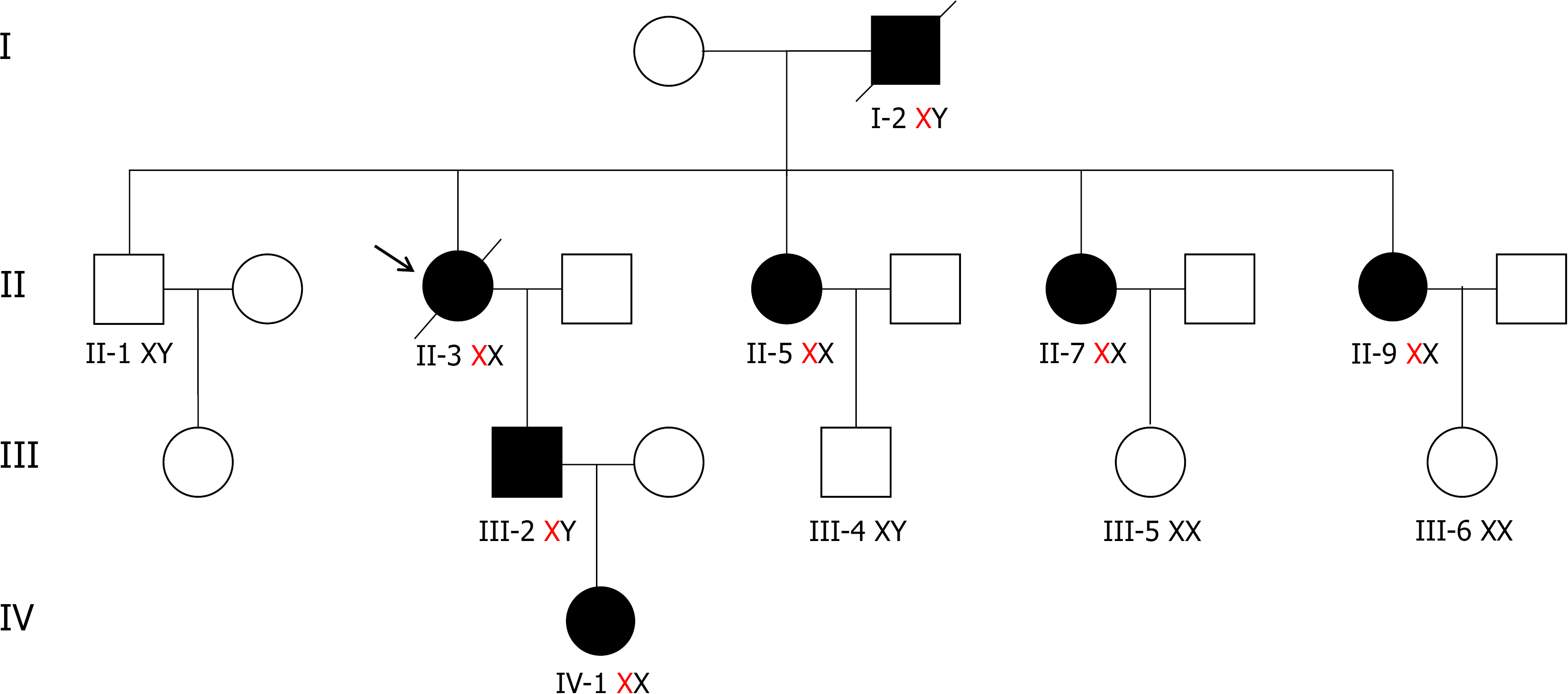
Herein, we analyzed a four-generation Chinese family. The proband is a 57-year-old woman who was diagnosed with left ventricular hypertrophy and atrial fibrillation 7 years ago. Echocardiography showed an end-diastolic diameter of the interventricular septum of 19.9 mm, left ventricular end-diastolic diameter of 63.1 mm, and moderate-to-severe mitral regurgitation. Cardiac magnetic resonance indicated an enlarged left heart and right atrium, decreased left ventricular systolic and diastolic function, a left ventricular ejection fraction of 20%, and thickening of the left ventricular septum. In March 2019, gene and enzyme activity tests confirmed the diagnosis of FD. Her son was diagnosed with FD after gene and enzyme activity assay, and was prescribed agalsidase-β for enzyme replacement therapy in July 2020. Two sisters of the proband were also diagnosed with FD by genetic testing. Both of them had a history of atrial fibrillation.
A novel mutation was identified in a Chinese family with FD, in which the male patient had a low level of enzyme activity, early-onset, and severe organ involvement. Comprehensive analysis of clinical phenotype genetic testing and enzyme activity testing helped in the diagnosis and treatment of this FD family.
Core Tip: Fabry disease (FD) is a rare X-linked lysosomal storage disorder caused by a deficiency of α-galactosidase A. We present herein a case of novel mutation (348delG:p.G116fs) in exon 2 in a Chinese FD family. Time delay in the diagnosis was 6 years. The proband died of respiratory circulatory failure. The son of the proband had a low level of enzyme activity, early-onset, and severe organ involvement. He was prescribed agalsidase-β for enzyme replacement therapy to delay progression of the disease. This case highlights that clinical phenotype, gene detection, and enzyme activity results should be analyzed comprehensively for patients suspected of having FD.
- Citation: Fu AY, Jin QZ, Sun YX. Novel α-galactosidase A gene mutation in a Chinese Fabry disease family: A case report. World J Clin Cases 2022; 10(3): 1067-1076
- URL: https://www.wjgnet.com/2307-8960/full/v10/i3/1067.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i3.1067
Fabry disease (FD, OMIM 301500) is a progressive, X-linked inherited disorder of glycosphingolipid metabolism caused by a deficient or absent lysosomal α-galactosidase A (α-GLA) activity[1]. The prevalence of FD was once believed to be very rare, occurring approximately in 1:50000 patients[2]. Substrates of this lysosomal enzyme accumulate, resulting in cellular dysfunction in multiple organs. FD is commonly known as a silent disease that appears later in life and could be easily misdiagnosed. Patients lacking α-GLA activity exhibit a 10-20 year shortened life span: Male patients with FD have a median survival of 57 years, and the median female survival is 72 years[3].
Classically affected FD males with no residual α-GLA activity, may display neurological (pain and acroparesthesia), cutaneous (angiokeratoma), renal (proteinuria and kidney failure), cardiovascular (cardiomyopathy, arrhythmia, and valvulopathy), cochleovestibular, and cerebrovascular (transient ischemic attacks and strokes) signs while heterozygous females have symptoms ranging from mild to severe[1]. Male patients are usually severely affected, while the clinical presentation in female patients may be more variable[4].
There are currently 967 known GLA mutations, including 671 missense/nonsense mutations, listed in the Human Gene Mutation Database[5]. The type of amino acid exchange domain in the α-GLA 3D-structure determines the disease severity and temporal course of clinical presentation. Patients with active site or buried mutations showed a severe phenotype with multi-organ involvement and early disease manifestation. Patients with certain mutations showed a milder phenotype with less organ impairment and later disease onset[6]. In male patients, the α-GLA enzyme activity is often significantly decreased, while about a third of female patients have enzymes within the normal range.
Enzyme replacement therapy (ERT) and chaperone therapy are currently considered the main targeted treatments for FD. As two representative drugs of enzyme replacement therapy, agalsidase-α and agalsidase-β have been shown to be clinically effective for patients with FD; yet, these are very expensive (approximately $200000 per patient annually in China). Some patients with amenable GLA mutations have residual activity in α-GLA. In these patients, small molecular chaperones could promote enzyme stability and are clinically effective.
In the present study, we describe a novel frameshift mutation in GLA and different α-GLA enzymatic activity in a Chinese family in which both male and female members presented with left ventricular hypertrophy and atrial fibrillation.
The proband was a 57-year-old woman who has experienced paroxysmal chest tightness and shortness of breath for 7 years.
A 57-year-old woman was diagnosed with left ventricular hypertrophy and atrial fibrillation 7 years ago. Echocardiography showed an end-diastolic diameter of the interventricular septum of 19.9 mm, left ventricular end-diastolic diameter of 63.1 mm, moderate-to-severe mitral regurgitation, and a left ventricular ejection fraction (LVEF) of 45%. Cardiac magnetic resonance (CMR) indicated an enlarged left heart and right atrium, decreased left ventricular systolic and diastolic function, an LVEF of about 20%, and thickening of the left ventricular septum. In March 2019, gene and enzyme activity tests confirmed the diagnosis of FD. Her son was diagnosed with FD after gene and enzyme activity assay, and was prescribed agalsidase-β for enzyme replacement therapy in July 2020. Two sisters of the proband were also diagnosed with FD by genetic testing. Both of them had a history of atrial fibrillation.
The proband had a history of hypertension for more than 20 years.
The proband did not have any significant personal history. Her father died of cardiomyopathy, while her mother died of colon cancer. Her son and two sisters were diagnosed with FD.
At the last admission, the proband’s blood pressure was 99/65 mmHg. She was conscious, but presented with an appearance of weakness. Her tongue stuck out to the right. Her jugular vein was filling. Rales could be heard widely over both lung fields. The heart rate was 96 bpm. The intensity of the first heart sound was unequal. Systolic murmur (III/6) was identified in the apex of the heart. There was no edema in her lower limbs.
Enzymatic measurement of α-GLA: The enzyme activity of α-GLA was reduced to only 1.0 nmol/h/mg protein in the son of the proband, while a normal range was observed in all other family members (Table 1).
| Variables | II-3 | II-5 | II-7 | II-1 | III-2 |
| Sex | Female | Female | Female | Male | Male |
| Age at diagnosis | 56 | 52 | 47 | 63 | 22 |
| Mutation | NM_000169.2:c.348delG:p.G116fs | NM_000169.2:c.348delG:p.G116fs | NM_000169.2:c.348delG:p.G116fs | - | NM_000169.2:c.348delG:p.G116fs |
| Genotype | Heterozygote | Heterozygote | Heterozygote | Wild Type | Hemizygote |
| Enzymatic activity (nmol/h/mg protein) | N/A | 42 | 34 | 60 | 1 |
| Cardiovascular symptoms | |||||
| Heart rhythm | AF | AF | AF | SR | SR |
| LVPWD (mm) | 9.6 | 9.9 | 13.4 | 9.8 | 14.5 |
| IVST (mm) | 16 | 15.8 | 14.5 | 9.8 | 17.4 |
| EF (%) | 28.1 | 58.3 | 55.6 | 67.3 | 61.5 |
| hsTNI (ng/mL) | 0.21 | 0.59 | 0.11 | < 0.01 | < 0.110 |
| NT-proBNP (pg/mL) | > 25000 | 1124 | 3242 | < 10 | 586 |
| Kidney symptoms | |||||
| Serum creatinine(μmol/L) | 258 | 57 | 63 | N/A | 344 |
| Neuralgia | + | - | - | - | + |
| Neurological symptoms | |||||
| Neuropathic pain | - | - | - | - | + |
| Cerebrovascular involvement | + | + | - | - | - |
| Gastrointestinal symptoms | |||||
| Nausea | + | - | - | - | - |
| Abdominal Pain | + | - | - | - | - |
| Chronic diarrhea | + | - | - | - | - |
| Skin | |||||
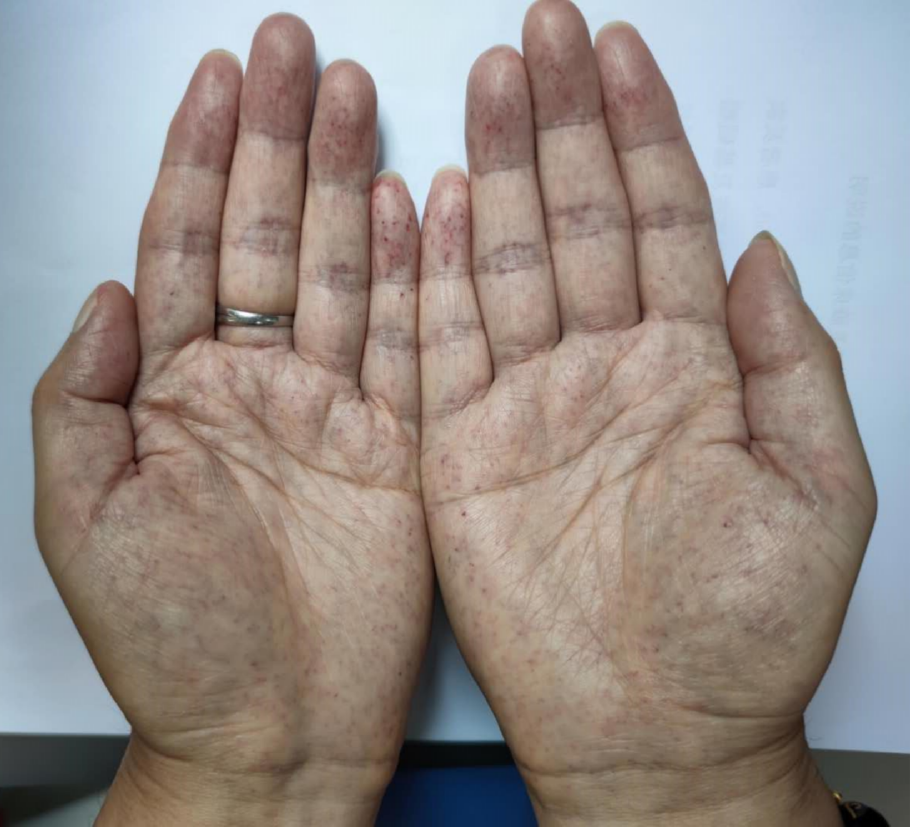
| Angiokeratoma | + | - | - | - | + |
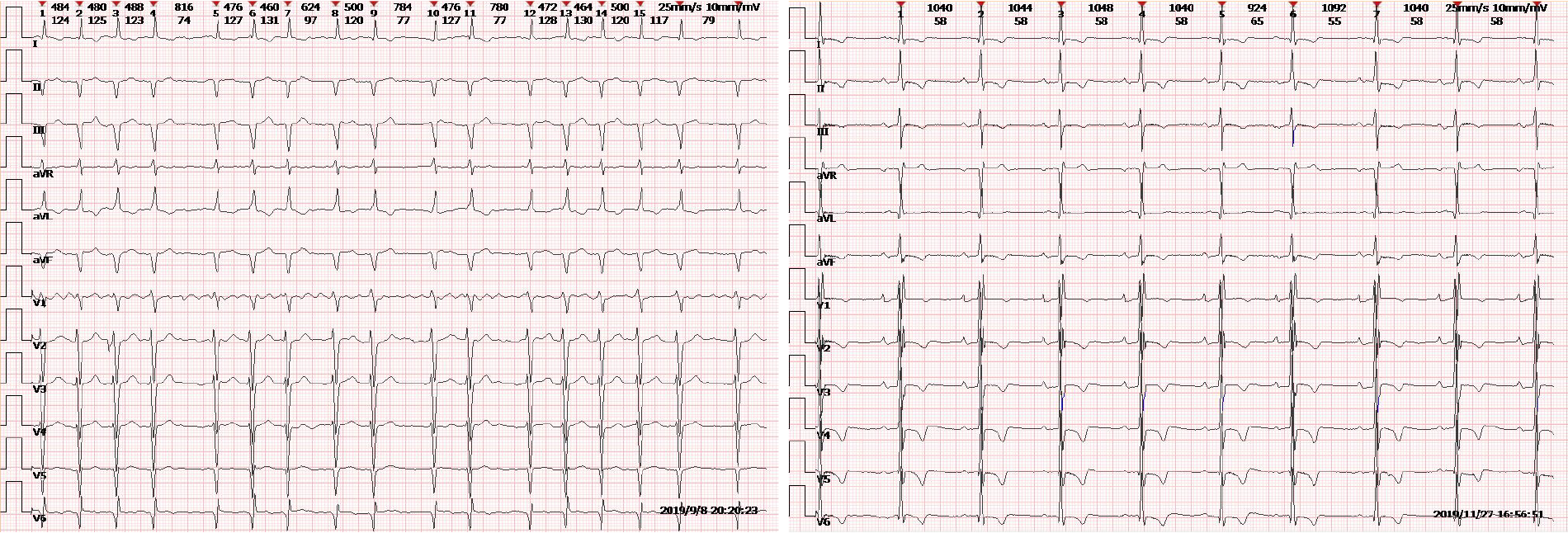
Clinical and biochemical studies: All of the patients in this family, whether hemizygous or heterozygote, had left ventricular hypertrophy. All female family members (II3, II5, and II7) had atrial fibrillation, except the propositus granddaughter (IV-1), who did not undergo inspection due to being only 2 years old. The levels of troponin I were all increased, and the ejection fraction was generally lower in female than male patients (III-2). Female heterozygotes suffered more severe cardiovascular damage while the kidney damage occurred earlier in males than in female family members. Stroke was more common in women, possibly due to atrial fibrillation and older age. At the same time, cutaneous and neuralgia manifestations were present in males of the same lineage, suggesting a wider range of glycosphingolipid deposition in the hemizygote and more involved organs (Table 1, Figure 1-5).
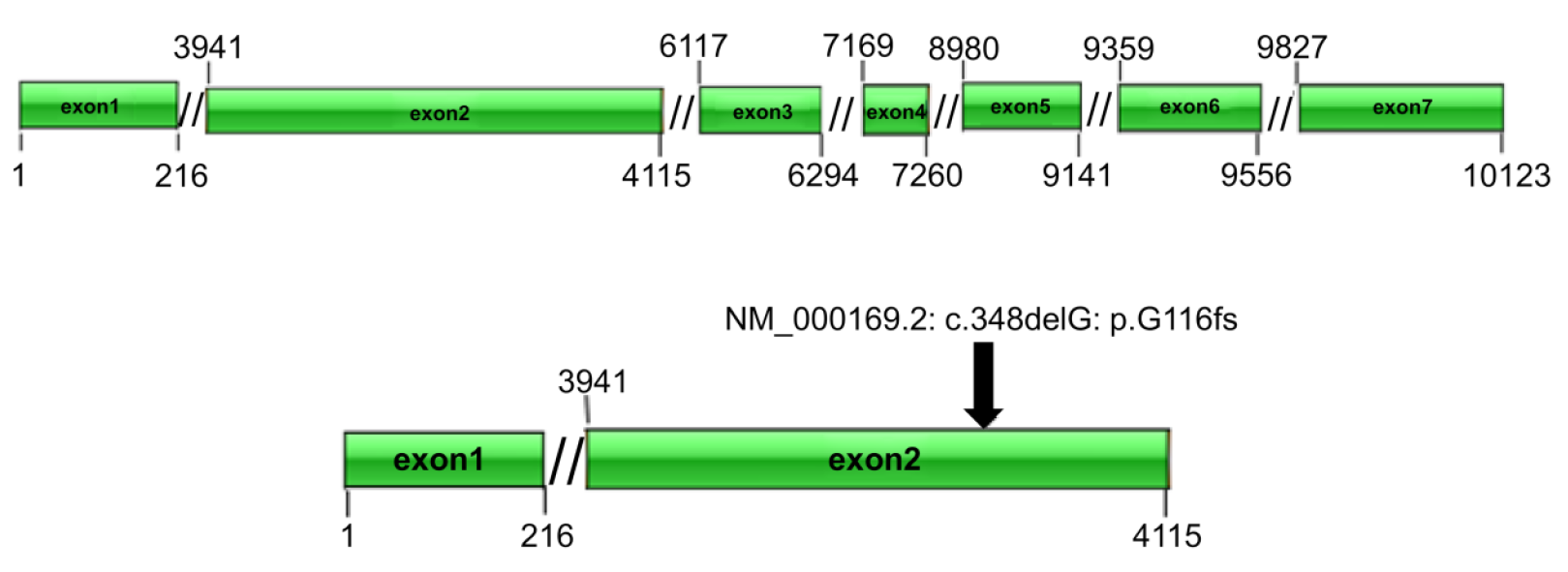
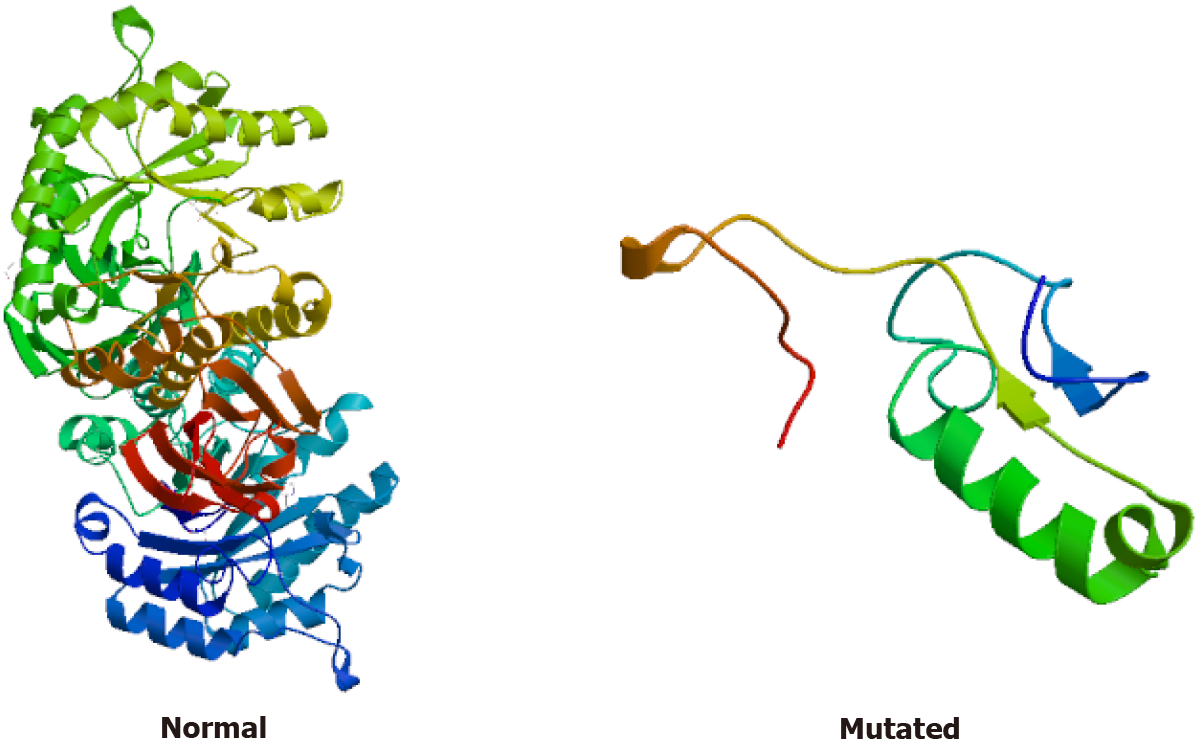
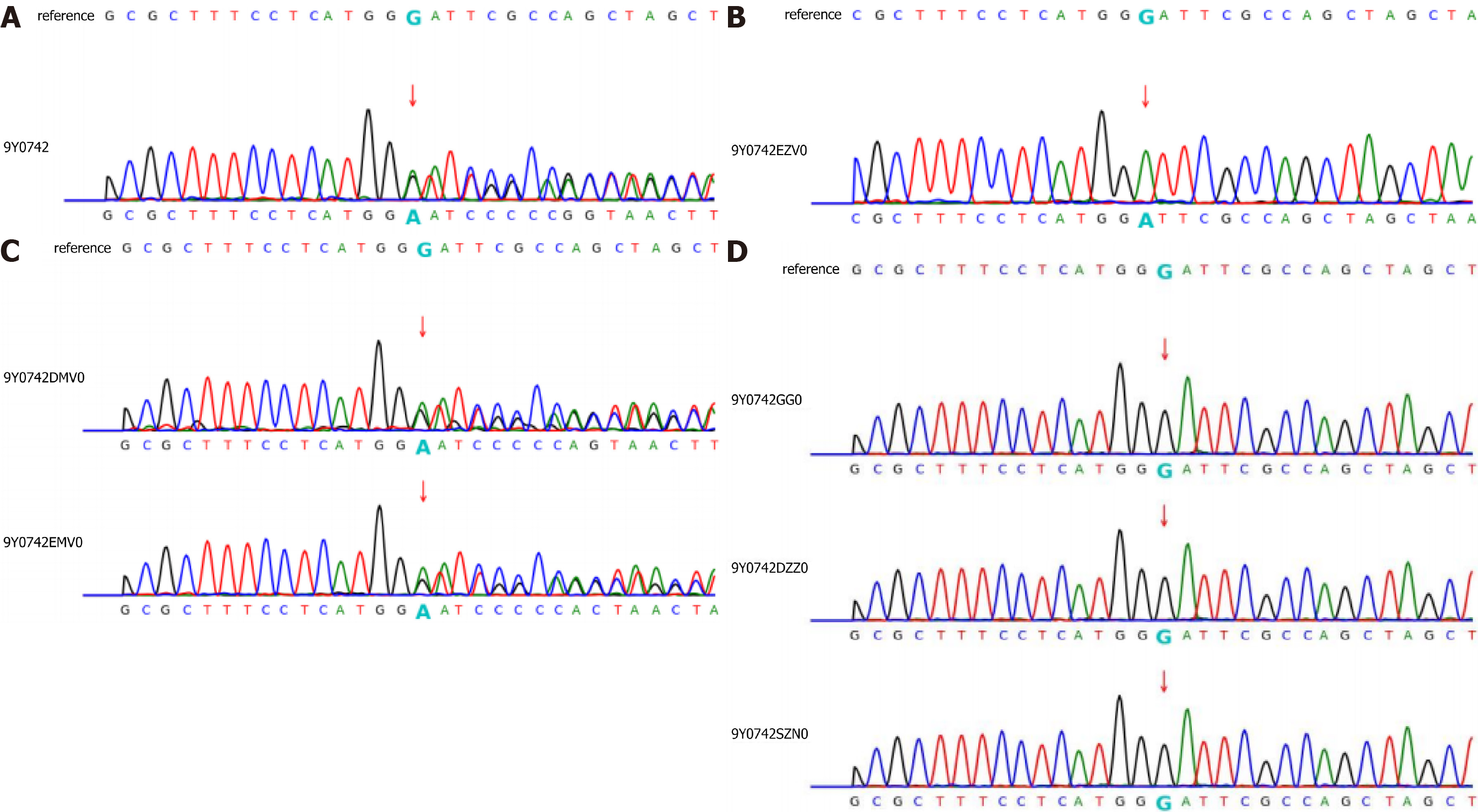
Gene expression: (1) Whole exome sequencing results: In this family, we found a frameshift mutation (348delG:p.G116fs) according to the guidelines for mutation nomenclature recommended by the Human Genome Variation Society (www.hgvs.org/mutnomen). The mutation occurred in the guanine deletion at position 348 of the GLA gene, resulting in a series of changes in the code of the 116th amino acid and its downstream (Figure 6). This mutation causes a change in the GLA protein domain (Figure 7)[7,8]; and (2) Conventional sequencing results: Sanger sequencing confirmed that the mutation occurred due to the guanine deletion in exon 2. Figure 8A shows the gene sequencing results of the proband (II-3). Adenine takes the place of guanine, thus causing a rearrangement of the subsequent amino acid sequence. The son of the proband (III-2) and the two sisters showed the same mutation (Figure 8B and C).
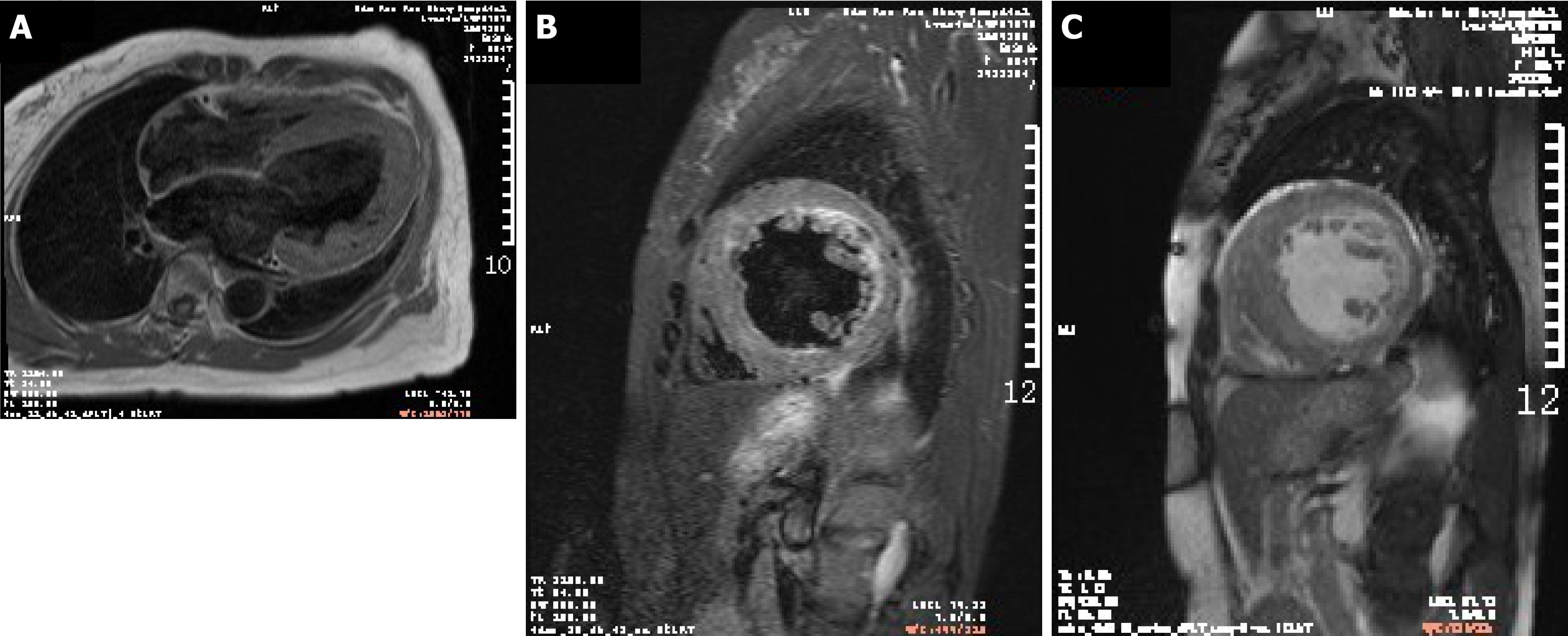
CMR showed patchy enhancement of interventricular septum and left ventricular anterior wall hypertrophy. Delayed enhancement suggested the formation of a large number of fibrous scars in left ventricular hypertrophy (Figure 3).
The final diagnosis of the presented case was FD.
The proband underwent atrial fibrillation radiofrequency ablation 7 years ago. Three years ago, she was given prednisone and tacrolimus because of edema and proteinuria. The son of the proband was given enzyme replacement therapy with agalsidase-β 65 mg (biweekly, intramuscularly) starting from July, 2020.
After 6 mo of ERT, serum creatinine in the son of the proband had no significant decrease. The pain in his skin was markedly relieved.
FD (MIM301500) was first described in 1898 by William Anderson and Johannes Fabry. After 65 years, Sweeley and Klionsky found an accumulation of a glycosphingolipid, globotriaosylceramide (Gb3), in patients with FD[9]. The incidence of FD in male newborns is 1/110000 to 1/4 million[10]. The main international databases for FD is the Fabry Outcome Survey in Europe, which currently lists 967 different GLA mutations in the human gene mutation database, including 671 missense/nonsense mutations[5].
The GLA gene encoding α-Gal A is located on Xq22.1, with 7 exons and 12 kb in length[11]. Herein, we report a novel frameshift mutation in the GLA gene in four members of a family with classical FD phenotype, with early-onset signs in affected men. Genotype-phenotype correlation in FD is challenging. Many GLA mutations are family-specific; in some families, there are quite marked phenotype variations. In contrast, the disease manifestation may vary within patients carrying the same mutation[12]. Garman et al[13] discovered two types of α-GLA mutations that are responsible for the disease progression: Mutations near the active sites and mutations of buried residues far from the active sites. Mutations near the active sites have a higher pathogenic frequency and severe clinical phenotype, while mutations far from the enzyme active sites are relatively mild[13]. The structure of α-GLA is a homodimeric glycoprotein with each monomer composed of two domains. The first domain contains the active site and extends from residues 32 to 330, and the second domain is comprised of residues 331-429, burying much surface area within one monomer. Rickert et al[6] found that patients with active site or buried mutations showed a severe phenotype with multi-organ involvement and early disease manifestation. Patients with other mutations had a milder phenotype with less organ impairment and later disease onset. In addition, the α-GalA activity was lower in patients with active site or buried mutations than in those with other mutations while lyso-Gb3 levels were higher.
In the proband of our study, a frameshift mutation (348delG:p.G116fs) occurred due to the guanine deletion at position 348 of the GLA gene, resulting in a series of changes in the code of the 116th amino acid and its downstream, so that the GLA peptide chain was transformed into a completely different peptide sequence. Enzyme activity tests confirmed that the enzyme activity of the female members of the family was moderately decreased and that of the male members was extremely decreased.
The α-Gal activity in female subjects who carry a heterozygous pathogenic variant in the GLA gene, is subject to X chromosome inactivation, typically random, cell-dependent, often nonuniform across the silenced chromosome[14]. Likewise, it complicates correlations among the genetic variants, functional data, and organ involvement. Nevertheless, as a group, α-Gal activity is higher in female subjects with pathogenic GLA variants than in male subjects' corresponding values. Consequently, up to one-third of X-chromosomal genes are expressed from both the active and inactive X chromosomes (Xa and Xi, respectively) in female cells, with the degree of “escape” from inactivation varying between genes and individuals[15], posing significant diagnostic challenges. In this study, the proband was a heterozygote but had classical characteristics such as heart failure and renal failure. Her sisters present nonclassical characteristics, whose manifestations are limited to cardiac involvement.
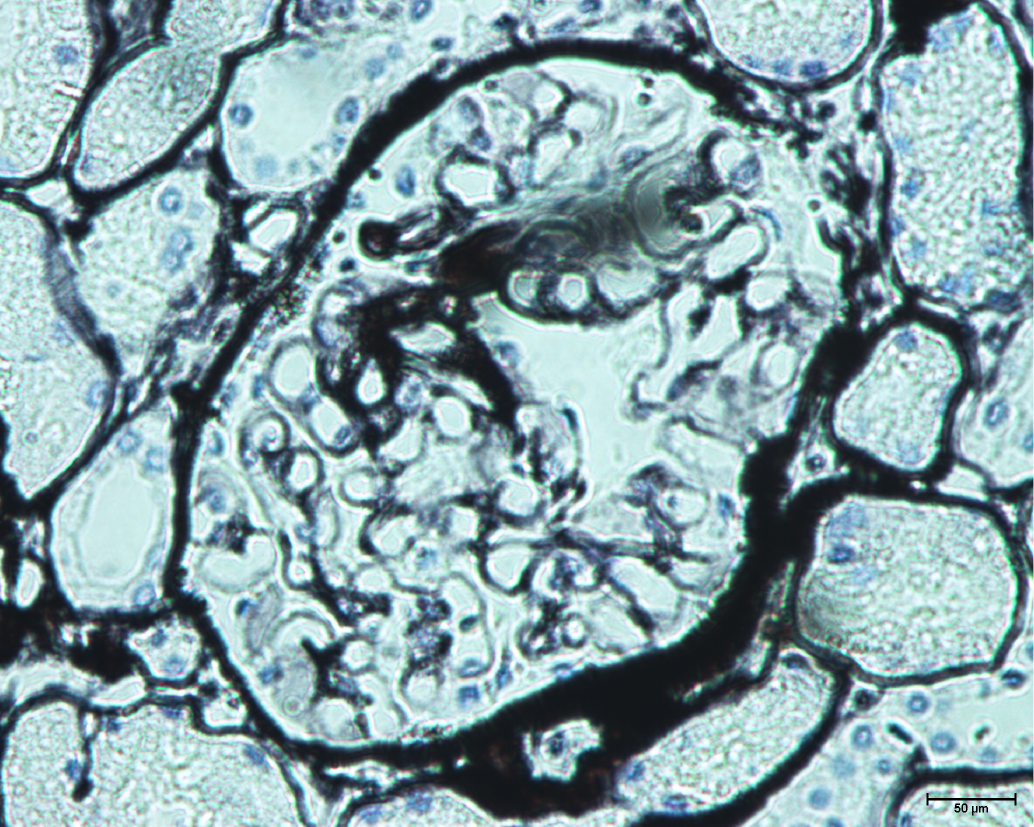
Clinically, FD diagnosis is primarily based on the clinical manifestation of multiple systems involving the brain, kidney, heart, and peripheral nerves, and also based on the comprehensive interrogation of family history. Patients may seek help from multiple medical specialists before a correct diagnosis is made, resulting in delayed treatment initiation[16]. Cardiac involvement is characterized by progressive cardiac hypertrophy, fibrosis, arrhythmias, heart failure, and sudden cardiac death. As myocardial fibrosis develops, the posterior and inferior LV wall can thin and become hypokinetic or akinetic[17]. Thinning of the LV posterior wall is a feature of FD related cardiomyopathy in the late stage. Further laboratory tests may include GLA activity test, pathological biopsy, and gene test, which are also considered the gold standard for diagnosis. Also, microscopic formation of typical onion-like osmiophilic inclusion bodies (such as myeloid corpuscle and zebra-corpuscle) in glomerular and tubular epithelial cell lysosomes is a typical pathological feature of lysosomal glycolipid aggregation, which is of great value in disease diagnosis. Early detection and treatment are crucial for achieving the best outcome.
Genetic testing, performed by whole-exome sequencing, and targeted analysis of the GLA gene could confirm clinical diagnosis. Nevertheless, the findings of a missense variant should not be considered an unequivocal validation of the diagnosis. Recently, a study examined 115 Japanese families with FD. No pathogenic mutations were identified in six families (5.2%). In total, 73 different disease-causing mutations were identified: 41 missense (56.2%), 11 nonsense (15.1%), four in-frame deletion (5.5%), 10 frameshift (13.7%), six splice site (8.2%), and one intronic (1.4%)[18]. As a result, many GLA variants of unknown significance (VOS) were identified. Therefore, the diagnosis of FD should not over-rely on genetic testing, and both clinical manifestations and family history should be considered comprehensively[19].
The treatment of FD relies on specific and non-specific treatments. Non-specific treatment is used to deal with the involvement of various organs. In this family, all the women (II3, II5, and II7) developed atrial fibrillation and underwent radical ablation, and in one case (II5) left atrial appendage occlusion was performed. Specific treatments include ERT, which is currently approved to be marketed as a galactosidase-α and a galactosidase-β. In this study, the son of the proband had started ERT treatment, and the effect will be followed closely. The European Union also approved the molecular chaperone migalastat in 2016 for the long-term treatment of specific mutated FD in patients over the age of 16 years, which could increase endogenous α-Gal A activity in a prospective observational multicenter study[20]. Pre-treatment clinical assessment, continuous clinical monitoring, and establishment and improvement of disease database should be made during treatment.
Since FD is an X-linked genetic disorder, genetic counseling and prenatal diagnosis should also be performed for all patients. Here we report a female patient who had a son who was also diagnosed with FD. The son had a daughter and he definitely passed the abnormal X chromosome to her (with 1 abnormal X chromosome and 1 normal X chromosome). However, the daughter has a heterozygous GLA allele, which may have relatively mild clinical manifestations and still need to be followed closely.
In summary, our findings suggest that the novel mutation 348delG:p.G116fs may be associated with classical manifestations of FD. These new data can be helpful in the diagnosis of FD and increase clinical and molecular knowledge about the correlations between mutations in the GLA gene, enzyme activity, and clinical phenotype of FD.
The authors would like to acknowledge the technical staff from the Berry Genomics for providing the genetic tests. We would also like to thank the Shanghai Jiaotong University School of Medicine, Shanghai Institute for Pediatric Research, and the Key Laboratory of Cardiovascular Intervention and Regenerative Medicine of Zhejiang Province for their help with enzyme activity testing.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Cardiac and Cardiovascular Systems
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Spinelli L S-Editor: Chang KL L-Editor: Wang TQ P-Editor: Chang KL
| 1. | Germain DP. Fabry disease. Orphanet J Rare Dis. 2010;5:30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 782] [Cited by in F6Publishing: 751] [Article Influence: 53.6] [Reference Citation Analysis (1)] |
| 2. | GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–. [PubMed] [Cited in This Article: ] |
| 3. | Vedder AC, Linthorst GE, van Breemen MJ, Groener JE, Bemelman FJ, Strijland A, Mannens MM, Aerts JM, Hollak CE. The Dutch Fabry cohort: diversity of clinical manifestations and Gb3 levels. J Inherit Metab Dis. 2007;30:68-78. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 132] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 4. | Basta M, Pandya AM. Genetics, X-Linked Inheritance. 2021 May 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2021 Jan-. [PubMed] [Cited in This Article: ] |
| 5. | Cooper DN BE, Stenson PD, Phillips AD, Evans K, Heywood S, Hayden MJ, Azevedo L, Mort ME, Hussain M. The Human Gene Mutation Database (2017) [cited 2019 Sep 8]. Available from: http://www.hgmd.cf.ac.uk/ac/index.php. [Cited in This Article: ] |
| 6. | Rickert V, Wagenhäuser L, Nordbeck P, Wanner C, Sommer C, Rost S, Üçeyler N. Stratification of Fabry mutations in clinical practice: a closer look at α-galactosidase A-3D structure. J Intern Med. 2020;288:593-604. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 7. | Studer G, Rempfer C, Waterhouse AM, Gumienny R, Haas J, Schwede T. QMEANDisCo-distance constraints applied on model quality estimation. Bioinformatics. 2020;36:1765-1771. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 257] [Cited by in F6Publishing: 396] [Article Influence: 99.0] [Reference Citation Analysis (0)] |
| 8. | Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296-W303. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5454] [Cited by in F6Publishing: 6693] [Article Influence: 1338.6] [Reference Citation Analysis (0)] |
| 9. | Miller JJ, Kanack AJ, Dahms NM. Progress in the understanding and treatment of Fabry disease. Biochim Biophys Acta Gen Subj. 2020;1864:129437. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 60] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 10. | Shi Q, Chen J, Pongmoragot J, Lanthier S, Saposnik G. Prevalence of Fabry disease in stroke patients--a systematic review and meta-analysis. J Stroke Cerebrovasc Dis. 2014;23:985-992. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 11. | Kornreich R, Desnick RJ, Bishop DF. Nucleotide sequence of the human alpha-galactosidase A gene. Nucleic Acids Res. 1989;17:3301-3302. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 118] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 12. | Verovnik F, Benko D, Vujkovac B, Linthorst GE. Remarkable variability in renal disease in a large Slovenian family with Fabry disease. Eur J Hum Genet. 2004;12:678-681. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 25] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 13. | Garman SC, Garboczi DN. The molecular defect leading to Fabry disease: structure of human alpha-galactosidase. J Mol Biol. 2004;337:319-335. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 272] [Cited by in F6Publishing: 284] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 14. | Berletch JB, Yang F, Disteche CM. Escape from X inactivation in mice and humans. Genome Biol. 2010;11:213. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 129] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 15. | Tukiainen T, Villani AC, Yen A, Rivas MA, Marshall JL, Satija R, Aguirre M, Gauthier L, Fleharty M, Kirby A, Cummings BB, Castel SE, Karczewski KJ, Aguet F, Byrnes A; GTEx Consortium; Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration &Visualization—EBI; Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz, Lappalainen T, Regev A, Ardlie KG, Hacohen N, MacArthur DG. Landscape of X chromosome inactivation across human tissues. Nature. 2017;550:244-248. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 507] [Cited by in F6Publishing: 580] [Article Influence: 96.7] [Reference Citation Analysis (0)] |
| 16. | Reisin R, Perrin A, García-Pavía P. Time delays in the diagnosis and treatment of Fabry disease. Int J Clin Pract. 2017;71. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 17. | Linhart A, Germain DP, Olivotto I, Akhtar MM, Anastasakis A, Hughes D, Namdar M, Pieroni M, Hagège A, Cecchi F, Gimeno JR, Limongelli G, Elliott P. An expert consensus document on the management of cardiovascular manifestations of Fabry disease. Eur J Heart Fail. 2020;22:1076-1096. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 78] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 18. | Marian AJ. Challenges in the Diagnosis of Anderson-Fabry Disease: A Deceptively Simple and Yet Complicated Genetic Disease. J Am Coll Cardiol. 2016;68:1051-1053. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 19. | Kobayashi M, Ohashi T, Kaneshiro E, Higuchi T, Ida H. Mutation spectrum of α-Galactosidase gene in Japanese patients with Fabry disease. J Hum Genet. 2019;64:695-699. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 20. | Lenders M, Nordbeck P, Kurschat C, Karabul N, Kaufeld J, Hennermann JB, Patten M, Cybulla M, Müntze J, Üçeyler N, Liu D, Das AM, Sommer C, Pogoda C, Reiermann S, Duning T, Gaedeke J, Stumpfe K, Blaschke D, Brand SM, Mann WA, Kampmann C, Muschol N, Canaan-Kühl S, Brand E. Treatment of Fabry's Disease With Migalastat: Outcome From a Prospective Observational Multicenter Study (FAMOUS). Clin Pharmacol Ther. 2020;108:326-337. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |