Published online Aug 6, 2022. doi: 10.12998/wjcc.v10.i22.7772
Peer-review started: February 28, 2022
First decision: May 11, 2022
Revised: May 19, 2022
Accepted: June 17, 2022
Article in press: June 17, 2022
Published online: August 6, 2022
Processing time: 143 Days and 23.2 Hours
Non-small-cell lung cancer (NSCLC) has the highest morbidity and mortality rates among all malignant tumor types. Although therapies targeting the mutated genes such as KRAS have been used in the clinic for many years, the prognosis remains poor. Therefore, it is necessary to further study the aberrant expression or mutation of non-target genes affecting the survival and prognosis.
To explore the impact of simultaneous abnormalities of multiple genes on the prognosis and survival of patients.
We used R packages to analyze gene expression data and clinical data down
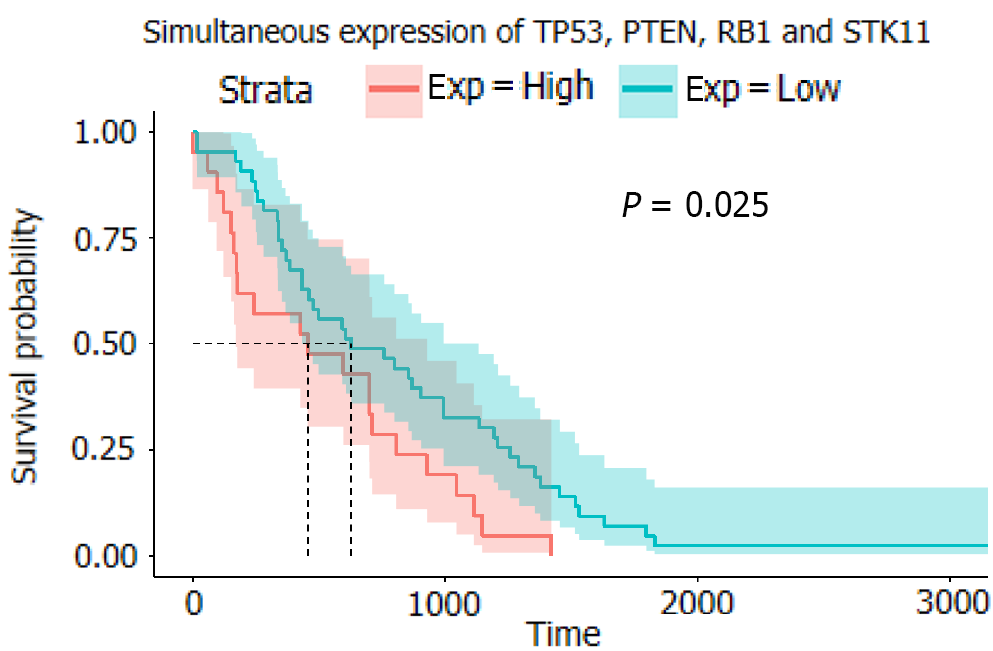
Analysis of gene expression data from TCGA revealed that the overexpression of the following single genes affected overall survival: TP53 (P = 0.79), PTEN (P = 0.94), RB1 (P = 0.49), CTNNB1 (P = 0.24), STK11 (P = 0.32), and PIK3CA (P = 0.013). However, the probability of multiple genes (TP53, PTEN, RB1, and STK11) affecting survival was 0.025. Retrospective analysis of clinical data revealed that sex (hazard ratio [HR] = 1.29; [95%CI: 0.64-2.62]), age (HR = 1.05; [95%CI: 1.02-1.07]), smoking status (HR = 2.26; [95%CI: 1.16-4.39]), tumor histology (HR = 0.58; [95%CI: 0.30-1.11]), cancer stage (HR = 16.63; [95%CI: 4.8-57.63]), epidermal growth factor receptor (EGFR) mutation (HR = 1.82; [95%CI: 1.05-3.16]), abundance (HR = 4.95; [95%CI: 0.78-31.36]), and treatment with tyrosine kinase inhibitors (TKIs) (HR = 0.58; [95%CI: 0.43-0.78]) affected patient survival. Co-occurring mutations of TP53, PTEN, RB1, and STK11 did not significantly affect the overall survival of patients receiving chemotherapy (P = 0.96) but significantly affected the overall survival of patients receiving TKIs (P = 0.045).
Co-occurring mutation or overexpression of different genes has different effects on the overall survival and prognosis of NSCLC patients. Combined with TKI treatment, the co-occurring mutation of some genes may have a synergistic effect on the survival and prognosis of NSCLC patients.
Core Tip: Non-small-cell lung cancer (NSCLC) has the highest morbidity and mortality rates among all malignant tumors. To explore the impact of simultaneous abnormalities of multiple genes on the prognosis and survival of patients. We used R packages to analyze gene expression data and clinical data downloaded from The Cancer Genome Atlas database. We also collected samples from 85 NSCLC patients from the First People's Hospital of Jingzhou City and retrospectively followed the patients for multivariate Cox regression analysis and survival analysis. Co-occurring mutation or overexpression of different genes has different effects on the overall survival and prognosis of patients. Combined with TKI treatment, the co-occurring mutation of some genes may have a synergistic effect on the survival and prognosis of NSCLC patients.
- Citation: Yan LD, Yang L, Li N, Wang M, Zhang YH, Zhou W, Yu ZQ, Peng XC, Cai J. Prognostic role of multiple abnormal genes in non-small-cell lung cancer. World J Clin Cases 2022; 10(22): 7772-7784
- URL: https://www.wjgnet.com/2307-8960/full/v10/i22/7772.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i22.7772
The prognosis of lung cancer is poor, and the associated mortality rate is among the highest due to the tumor’s highly invasive and metastatic nature[1]. Non-small-cell lung cancer (NSCLC) accounts for 85% of lung malignancy cases[2]. Despite advances in gene-targeted therapy and immunotherapy, the long-term survival benefits of NSCLC patients are still limited[3]. Cancer patients receiving molecularly targeted therapies have clinically different survival prognoses mainly because currently used targeted therapies primarily target single-gene mutations, while tumor tissues are highly heterogeneous. Moreover, the complex tumor microenvironment plays a crucial role in the survival, ability to evade immune surveillance, and drug resistance of cancer cells[4].
Single-gene-targeted therapy has provided a survival benefit to patients over conventional che
Next-generation sequencing analyses have revealed significant differences in gene mutation sites among patients, with the differences also being apparent between early and late stages of cancer and in the mutation frequency of each site[10]. The differences in gene mutations may also be responsible for differences in the risk of drug resistance and differences in individual treatment responses.
The co-occurrence of TP53 and EGFR mutations is often associated with a worse prognosis and accelerated proliferation and invasion of cancer cells[11,12]. In this study, we collected clinical data from The Cancer Genome Atlas (TCGA) and analyzed the impact of common mutations in NSCLC patients on targeted therapy and survival prognosis.
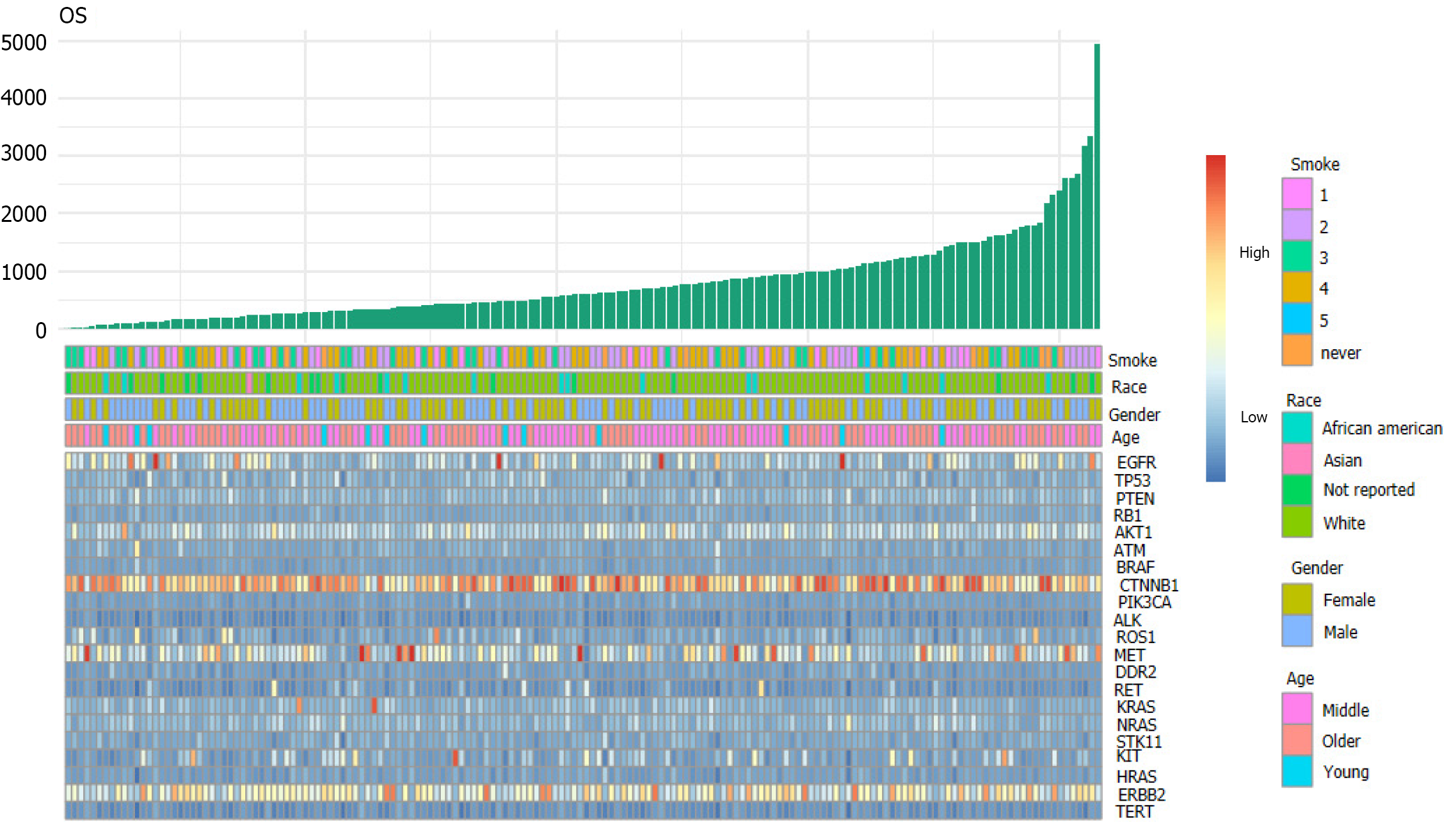
First, we downloaded the clinical and original gene expression data (counts) of lung adenocarcinoma (LUAD) patients from the TCGA public database. Next, we screened 21 target genes detected by second-generation sequencing during clinical treatment and observed the effect of multiple gene expression on the survival and prognosis of patients using the pheatmap package of R software to plot a heat map for visual analysis.
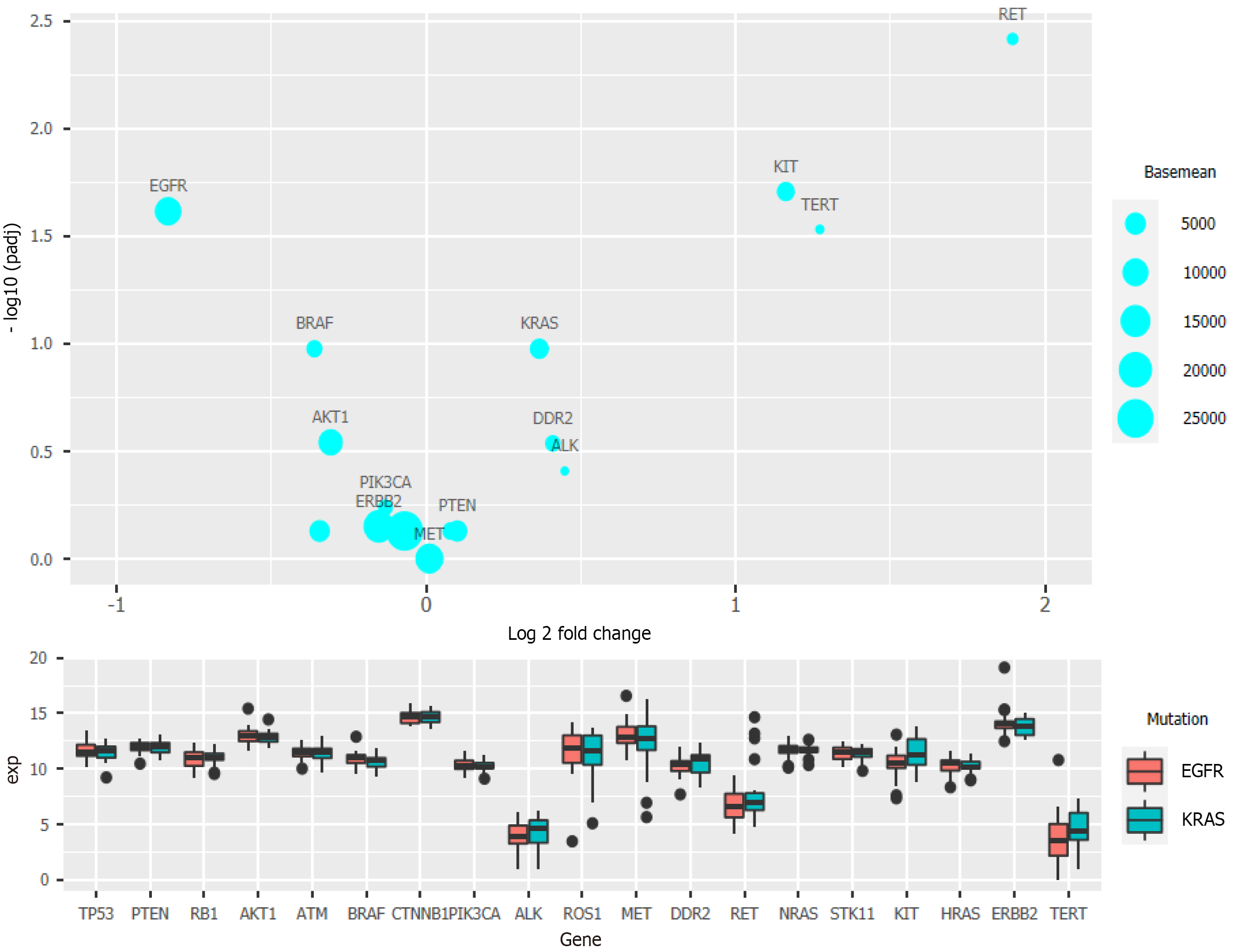
Then, we used the patient mutation information provided in the database and divided all patient samples into EGFR and KRAS mutation groups to study the gene expression differences and explore the correlation between mutations at other loci and these two most common mutations.
Finally, we analyzed the overall survival (OS) of patients whose samples showed different gene expression profiles. The significance of single-gene analysis was improved by considering an expression Z score of more than 1 as high expression and an expression Z score less than -1 as low expression. Then, considering 0 as the critical value, we analyzed the co-expression of multiple genes.
We collected the next-generation sequencing results of more than 300 NSCLC patients from the First People’s Hospital of Jingzhou City from 2017 to 2020. After follow-up, clear OS and detailed data of 85 patients were obtained. If some patients were examined many times during the treatment, we took the first detection results as the basis for analysis.
We collected the medical history and general clinical data of the subjects through the hospital information system and telephone return visit in the hospital's oncology department. We collected the patient’s sex, age, smoking history, pathological type, cancer stage, next-generation sequencing results, treatment with TKIs, OS, and other results and divided the patient population based on the gene mutation status for multivariate Cox regression analysis. Finally, the patients were divided into two groups based on whether they received targeted TKI therapy or chemotherapy to explore the effect of co-occurrence of gene mutations on the OS of patients. All clinical data were collected after being submitted to the ethics committee of Jingzhou First People’s Hospital for approval. All patients provided informed consent before the next-generation sequencing analysis.
All patients were followed until December 31, 2020. We screened patients with complete basic information and definite OS data. The results of all patients were obtained using the same high-throughput sequencing equipment. All statistical analyses were performed using several R packages such as edgeR, DESeq, TCGAbiolinks, and ggplot2. We used the Kaplan-Meier method to analyze the differences in OS and compared the effects of gene mutations using the log-rank test. We also investigated the influence of various factors on the total survival of patients using multivariate Cox regression analysis. The Fisher exact test was used for comparing different groups. All P values are based on a two-tail hypothesis with statistical significance defined as P < 0.05.
The volcano map (Figure 1) of gene expression data of 533 cancer tissues and 53 normal tissues from TCGA reveals a large number of upregulated (red) and downregulated (green) genes[13]. Differences in gene expression were also seen among different subtypes of cancer tissues. In this study, 21 genes detected by next-generation sequencing were selected to explore the effect of gene mutation and overexpression on OS. A two-dimensional heat map of various parameters was plotted to intuitively observe patients' basic indicators and gene expression (Figure 2, which shows apparent differences in the expression of different genes, but it is necessary to clarify which indicators impact the OS of patients.
Clinical data regarding the EGFR and KRAS mutation status were also analyzed and compared with data on mutations of other genes[14,15]. The National Comprehensive Cancer Network guidelines have pointed out that KRAS mutation can reduce sensitivity toward EGFR inhibitors. Clinically, the probability of simultaneous occurrence of KRAS mutation and EGFR mutation is very low, and a mutually exclusive relationship between them is also reported[16]. After grouping the samples based on the EGFR and KRAS mutation status with more than 20 samples in each group, the prepared bubble chart and box chart (Figure 3) show that RET, KIT, and TERT exhibited significantly different expression levels between the two groups. These three genes were upregulated in patients with KRAS mutation, while EGFR and BRAF were downregulated (Figure 3).
In the analysis of the survival of patients with single-gene mutations, to amplify the single-gene effect, we considered genes with a Z score greater than 1 to be highly expressed and those with a Z score less than -1 to have a low expression level. After calculating the P value, it was found that except for PIK3CA (P < 0.05), there was no statistical significance in the high expression of other single genes: TP53 (P = 0.79), PTEN (P = 0.94), RB1 (0.49), CTNNB1 (P = 0.24), and STK11 (P = 0.32) (Figure 4). We speculated that significant PIK3CA overexpression indicates a poor prognosis and OS[17-19]. Given that the expression of other single genes did not seem to affect prognosis significantly, we suspected that the simultaneous overexpression of multiple genes, especially the four tumor suppressor genes (TP53, PTEN, RB1, and STK11), would have some clinical implication. Therefore, we used the Z score of 0 as the critical value and divided the four genes into two groups in which all had a high expression or a low expression level at the same time (Figure 5). A P value of 0.025 showed that when TP53, PTEN, RB1, and STK11 were highly expressed simultaneously, the OS was significantly different from that when these genes showed a low expression level. Moreover, the high expression group had a significantly shorter OS.
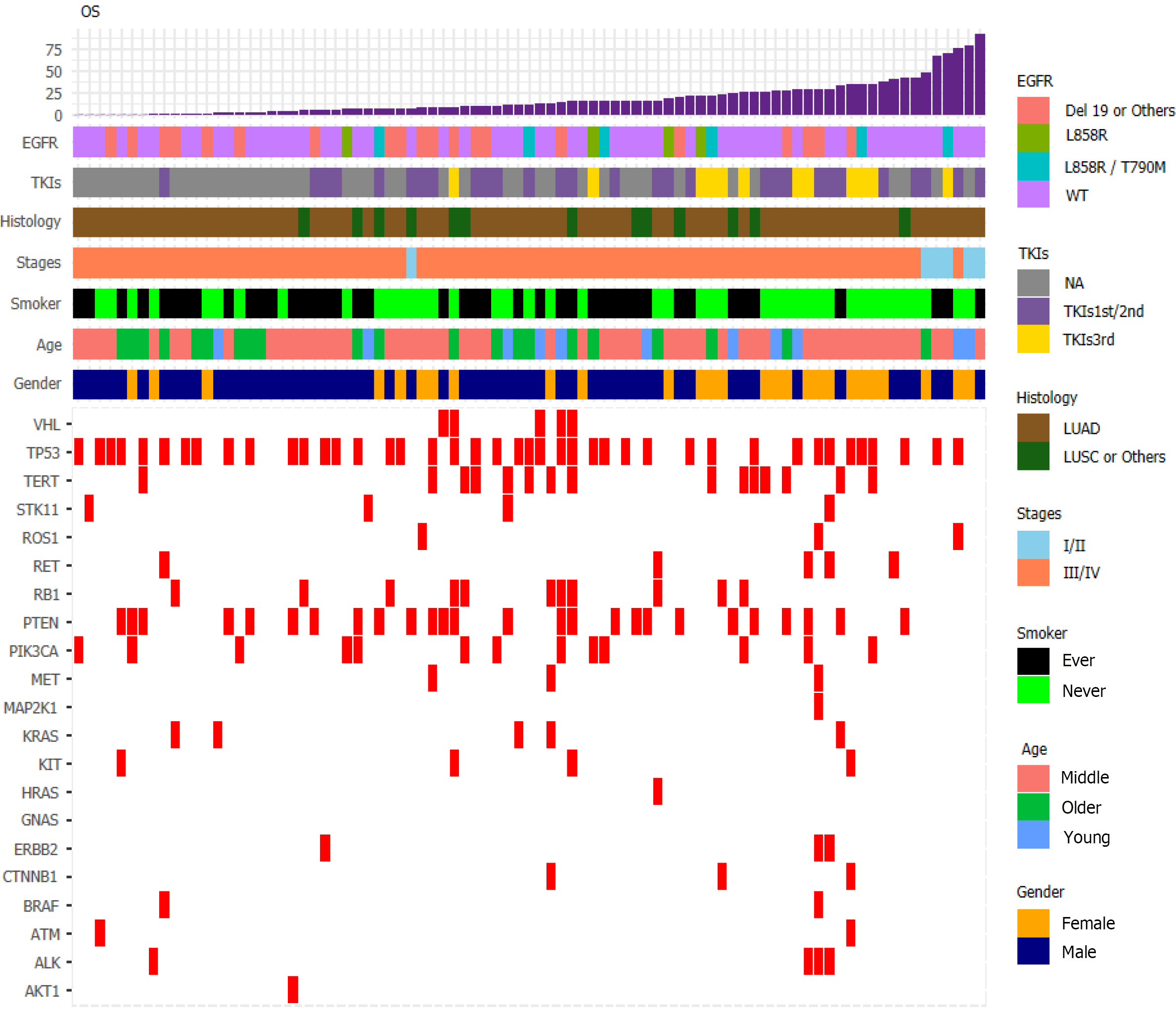
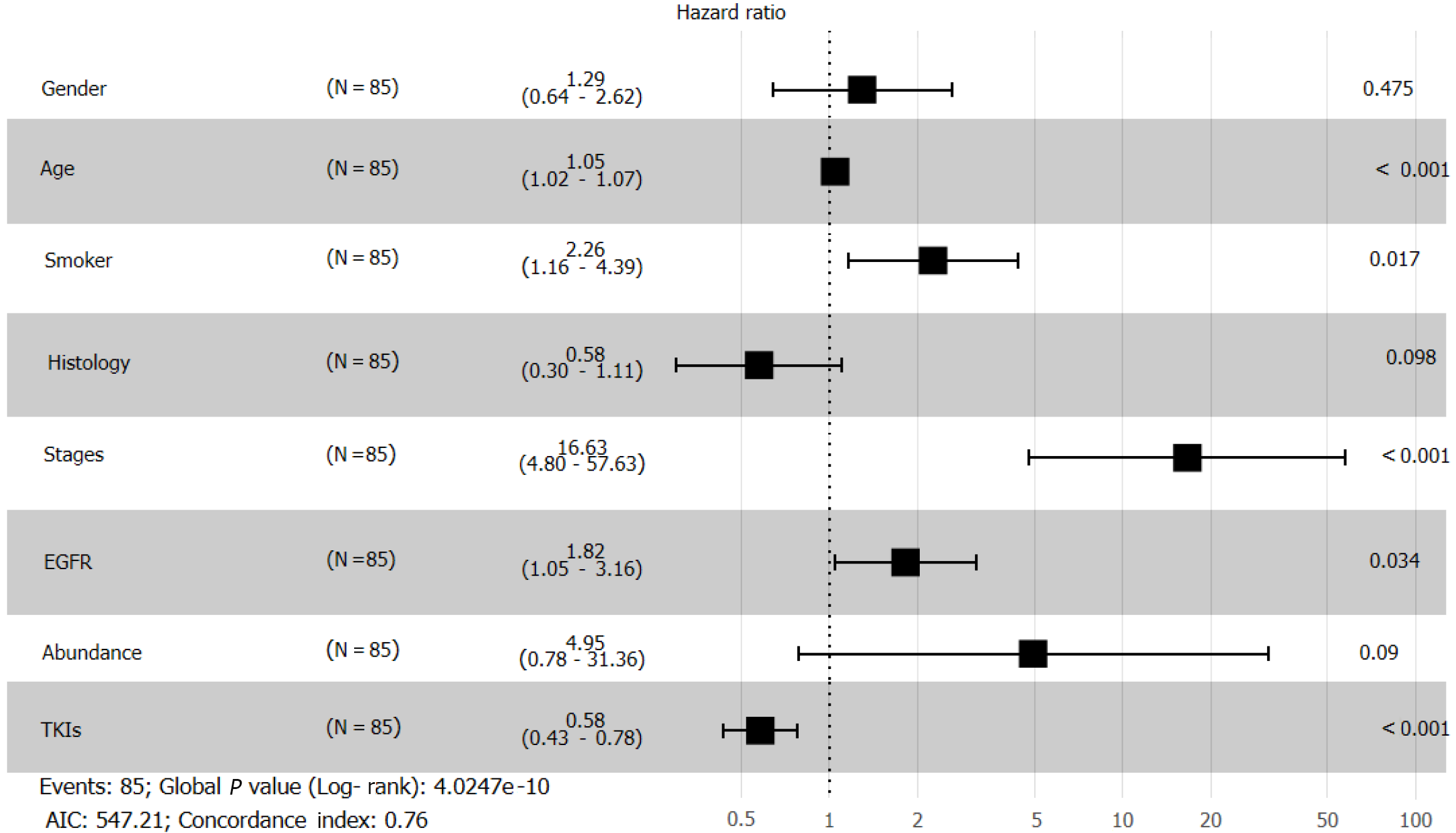
We visualized data regarding gene mutations and basic clinical information collected from patients by plotting a heat map (Figure 6). First, we divided the patients based on whether they received chemotherapy or TKI therapy and then carried out Fisher's exact test (Table 1). We found that sex (P = 0.0021), smoking history (P = 0.0302), and OS (P = 0.0022) differed significantly based on the treatment. To understand the impact of various factors on patients, we separately analyzed the impact of basic indicators and gene mutations by multivariate Cox regression (Figure 7), which demonstrated that sex (hazard ratio [HR] = 1.29; [95%CI: 0.64-2.62]; P = 0.475), age (HR = 1.05; [95%CI: 1.02-1.07]; P < 0.001), smoking history (HR = 2.26; [95%CI: 1.16-4.39]; P = 0.017), tumor histology (HR = 0.58; [95%CI: 0.30-1.11]; P = 0.098), cancer stage (HR = 16.63; [95%CI: 4.8-57.63]; P < 0.001), EGFR mutation (HR = 1.82; [95%CI: 1.05-3.16]; P = 0.034), abundance (HR = 4.95; [95%CI: 0.78-31.36]; P = 0.09), and TKI treatment (HR = 0.58; [95%CI: 0.43-0.78]; P < 0.001) affected patient survival.
| Therapy type | Chemotherapy | TKI therapy | P value |
| Sex | |||
| Female | 7 (8.2) | 20 (23.5) | 0.0021 |
| Male | 37 (43.5) | 21 (23.7) | |
| Age | |||
| Less than 50 yr | 7 (8.2) | 4 (4.7) | 0.7334 |
| 50-70 yr | 27 (31.8) | 26 (30.6) | |
| Greater than 70 yr | 10 (11.8) | 11 (12.9) | |
| Smoking history | |||
| Ever | 27 (31.7) | 15 (17.6) | 0.0302 |
| Never | 17 (20.0) | 26 (30.6) | |
| Tumor histology | |||
| LUAD | 35 (41.1) | 37 (43.5) | 0.2317 |
| LUSC & Others | 9 (10.6) | 4 (4.7) | |
| Cancer stage | |||
| I and II | 2 (2.4) | 4 (4.7) | 0.4227 |
| III and IV | 42 (49.4) | 37 (43.5) | |
| OS | |||
| < 12 mo | 28 (32.9) | 12 (14.1) | 0.0022 |
| ≥ 12 mo | 16 (18.8) | 29 (34.1) |
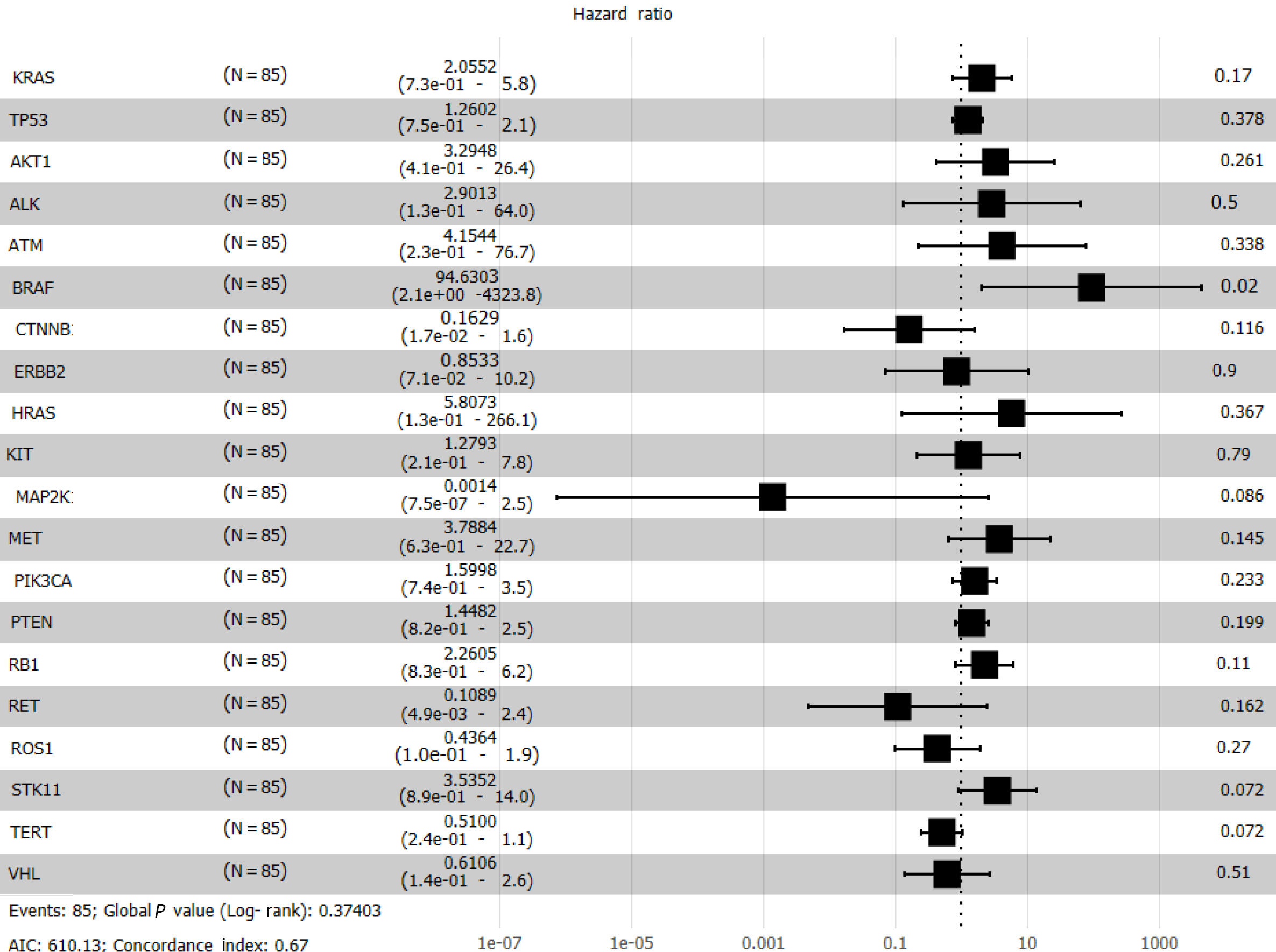
Cox regression analysis (Figure 8) revealed a significant effect of all gene mutations except for BRAF (P = 0.02), which indicates that the OS of patients is under the combined influence of multiple-locus genes. The HR values of some gene mutations were positive, such as MAP2K1 (HR = 0.0014), CTNNB1 (HR = 0.1629), and RET (HR = 0.1089), indicating that mutations of these genes may benefit the patients, but further confirmation using more samples is necessary. We selected TP53, PTEN, Rb1, and STK11, whose mutation frequency is high in NSCLC patients, to study if the co-occurrence of mutations of these genes has a similar superposition effect[20-22]. Patients with mutations in these tumor suppressor genes have a worse survival prognosis[23,24]. These co-occurring mutations can allow cancer cells to escape immune surveillance, proliferate aberrantly to malignancy, and develop resistance to targeted therapy. After identifying the effect of TP53 (HR = 1.2602), PTEN (HR = 1.4428), Rb1 (HR = 2.2605), and STK11 (HR = 3.5352), we sought to determine whether either of them could be used individually as an indicator of survival and prognosis of patients. However, the effect of age (P < 0.001) was significant even if its HR value was only 1.05 (Figure 7).
The patients were divided into two groups based on whether they received chemotherapy (n = 41) or TKI treatment (n = 44). According to the number of mutations in the four tumor suppressor genes (TP53, PTEN, Rb1, and STK11), we classified those with more than one mutation into the greater than (GT) 1 group (Figure 9). The number of mutations of these tumor suppressor genes in the chemotherapy group did not bear significance between the GT1 and non-GT1 groups (P = 0.96). Still, it was significant in the TKI treatment group (P = 0.045). Co-occurrence of mutations in these genes worsened the prognosis similarly in both groups. We found that some patients discontinued targeted therapy not because of disease progression but because of economic reasons; this could have affected the results. The complexity of the tumor genome determines that cancer treatment cannot target a single gene. Co-mutation is likely to completely change the biological characteristics of the original tumor through synergy, endow the tumor with new biological characteristics, and make the tumor tolerant to targeted therapy. This co-mutation may occur gradually in the process of targeted therapy.
In this study, we focused on the effect of the co-occurrence of gene mutations using TCGA database and clinical patient data. Our analyses revealed significant differences in the gene expression profiles between adenocarcinoma and normal tissues in NSCLC. The upregulated and downregulated genes also differed in different patients and between LUAD patients with EGFR and KRAS mutations. Most tumor-derived TP53 mutations occur in the region encoding the DNA binding domain of p53. The TP53 mutation significantly impacts the progression of various types of cancer[25,26]. While the overexpression of some genes was significantly associated with good OS and prognosis, such as PIK3CA, the single expression of most genes did not have a significant effect. The simultaneous overexpression of multiple tumor suppressor genes (TP53, PTEN, RB1, and STK11) was associated with a poor OS. Cox multivariate regression analysis revealed that for NSCLC patients, the most critical factor affecting OS was not the type of treatment or gene mutation but the disease stage, which underscores the importance of early diagnosis of solid tumors. The effects of recognized risk factors such as smoking history were also confirmed in the analysis. Finally, after grouping based on treatment, we found that in patients receiving traditional chemotherapy, mutations of TP53, PTEN, RB1, and STK11 had no significant influence on the OS; however, these mutations had a significant effect in patients receiving TKIs. Simultaneous mutations in multiple tumor suppressor genes resulted in a risk superposition effect[24].
These genes have been studied in the context of non-target therapy. While the frequency of mutations in these genes is high in NSCLC patients, the problem of tumor heterogeneity and the possibility of personalized medicine need to be further explored. The influence of these gene mutations on the OS and prognosis of patients receiving immunotherapy has also received attention[27-29]. In the future, the development of new molecular targeted drugs will help deal with the heterogeneity of different mutant subtypes.
In conclusion, this study summarizes the impact of the co-occurrence of mutations or overexpression of multiple genes on the OS and prognosis of NSCLC patients. The results indicate that the co-occurrence of mutations results in a risk superposition effect, and such genes must be studied further when predicting patients' disease progression.
Among all malignant tumor types, non-small cell lung cancer (NSCLC) has the highest incidence rate and mortality.
To investigate the effect of simultaneous polygenic abnormalities on the prognosis and survival of NSCLC patients.
To study the effect of polygene mutation and abnormal expression on the prognosis and survival of patients with non-small cell lung cancer.
We used R packages to analyze gene expression data and clinical data downloaded from The Cancer Genome Atlas (TCGA) database. We also collected samples from 85 NSCLC patients from the First People’s Hospital of Jingzhou City and retrospectively followed the patients. Multivariate Cox regression analysis and survival analysis were performed.
The probability of multiple genes (TP53, PTEN, RB1, and STK11) affecting survival was 0.025. Retrospective analysis of clinical data revealed that sex (hazard ratio [HR] = 1.29), age (HR = 1.05), smoking status (HR = 2.26), tumor histology (HR = 0.58), cancer stage (HR = 16.63), epidermal growth factor receptor (EGFR) mutation (HR = 1.82), abundance (HR = 4.95), and treatment with tyrosine kinase inhibitors (TKIs) (HR = 0.58) affected patient survival. Co-occurring mutation of TP53, PTEN, RB1, and STK11 did not significantly affect the overall survival of patients receiving chemotherapy (P = 0.96) but significantly affected the overall survival of patients receiving TKIs (P = 0.045).
Co-mutation or overexpression of different genes has different effects on the overall survival and prognosis of NSCLC patients. Combined with TKI treatment, the co-mutations of some genes may have a synergistic effect on the survival and prognosis of NSCLC patients.
In the future, the development of new molecular targeted drugs will help deal with the heterogeneity of different mutant subtypes.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cho SG, South Korea; Sezer HF, Turkey A-Editor: Anaya-Prado R, Mexico S-Editor: Wang LL L-Editor: Wang TQ P-Editor: Qi WW
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12667] [Cited by in RCA: 15295] [Article Influence: 3059.0] [Reference Citation Analysis (4)] |
| 2. | Arbour KC, Riely GJ. Systemic Therapy for Locally Advanced and Metastatic Non-Small Cell Lung Cancer: A Review. JAMA. 2019;322:764-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 505] [Cited by in RCA: 761] [Article Influence: 126.8] [Reference Citation Analysis (0)] |
| 3. | Hastings K, Yu HA, Wei W, Sanchez-Vega F, DeVeaux M, Choi J, Rizvi H, Lisberg A, Truini A, Lydon CA, Liu Z, Henick BS, Wurtz A, Cai G, Plodkowski AJ, Long NM, Halpenny DF, Killam J, Oliva I, Schultz N, Riely GJ, Arcila ME, Ladanyi M, Zelterman D, Herbst RS, Goldberg SB, Awad MM, Garon EB, Gettinger S, Hellmann MD, Politi K. EGFR mutation subtypes and response to immune checkpoint blockade treatment in non-small-cell lung cancer. Ann Oncol. 2019;30:1311-1320. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 242] [Cited by in RCA: 282] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 4. | Kim J, DeBerardinis RJ. Mechanisms and Implications of Metabolic Heterogeneity in Cancer. Cell Metab. 2019;30:434-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 398] [Article Influence: 66.3] [Reference Citation Analysis (0)] |
| 5. | Zhang L, Ma S, Song X, Han B, Cheng Y, Huang C, Yang S, Liu X, Liu Y, Lu S, Wang J, Zhang S, Zhou C, Zhang X, Hayashi N, Wang M; INFORM investigators. Gefitinib vs placebo as maintenance therapy in patients with locally advanced or metastatic non-small-cell lung cancer (INFORM; C-TONG 0804): a multicentre, double-blind randomised phase 3 trial. Lancet Oncol. 2012;13:466-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 191] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 6. | Herbst RS, Ansari R, Bustin F, Flynn P, Hart L, Otterson GA, Vlahovic G, Soh CH, O'Connor P, Hainsworth J. Efficacy of bevacizumab plus erlotinib vs erlotinib alone in advanced non-small-cell lung cancer after failure of standard first-line chemotherapy (BeTa): a double-blind, placebo-controlled, phase 3 trial. Lancet. 2011;377:1846-1854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 324] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 7. | Shaw AT, Bauer TM, de Marinis F, Felip E, Goto Y, Liu G, Mazieres J, Kim DW, Mok T, Polli A, Thurm H, Calella AM, Peltz G, Solomon BJ; CROWN Trial Investigators. First-Line Lorlatinib or Crizotinib in Advanced ALK-Positive Lung Cancer. N Engl J Med. 2020;383:2018-2029. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 529] [Cited by in RCA: 686] [Article Influence: 137.2] [Reference Citation Analysis (0)] |
| 8. | Wu YL, Tsuboi M, He J, John T, Grohe C, Majem M, Goldman JW, Laktionov K, Kim SW, Kato T, Vu HV, Lu S, Lee KY, Akewanlop C, Yu CJ, de Marinis F, Bonanno L, Domine M, Shepherd FA, Zeng L, Hodge R, Atasoy A, Rukazenkov Y, Herbst RS; ADAURA Investigators. Osimertinib in Resected EGFR-Mutated Non-Small-Cell Lung Cancer. N Engl J Med. 2020;383:1711-1723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 671] [Cited by in RCA: 1129] [Article Influence: 225.8] [Reference Citation Analysis (0)] |
| 9. | Ramalingam SS, Vansteenkiste J, Planchard D, Cho BC, Gray JE, Ohe Y, Zhou C, Reungwetwattana T, Cheng Y, Chewaskulyong B, Shah R, Cobo M, Lee KH, Cheema P, Tiseo M, John T, Lin MC, Imamura F, Kurata T, Todd A, Hodge R, Saggese M, Rukazenkov Y, Soria JC; FLAURA Investigators. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N Engl J Med. 2020;382:41-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1120] [Cited by in RCA: 1900] [Article Influence: 380.0] [Reference Citation Analysis (0)] |
| 10. | Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543-550. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3395] [Cited by in RCA: 4302] [Article Influence: 391.1] [Reference Citation Analysis (0)] |
| 11. | VanderLaan PA, Rangachari D, Mockus SM, Spotlow V, Reddi HV, Malcolm J, Huberman MS, Joseph LJ, Kobayashi SS, Costa DB. Mutations in TP53, PIK3CA, PTEN and other genes in EGFR mutated lung cancers: Correlation with clinical outcomes. Lung Cancer. 2017;106:17-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 150] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 12. | Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer. 2019;19:495-509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 583] [Cited by in RCA: 672] [Article Influence: 112.0] [Reference Citation Analysis (0)] |
| 13. | Zhang B, Zhang L, Yue D, Li C, Zhang H, Ye J, Gao L, Zhao X, Chen C, Huo Y, Pang C, Li Y, Chen Y, Chuai S, Zhang Z, Giaccone G, Wang C. Genomic characteristics in Chinese non-small cell lung cancer patients and its value in prediction of postoperative prognosis. Transl Lung Cancer Res. 2020;9:1187-1201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 14. | Roberts PJ, Stinchcombe TE, Der CJ, Socinski MA. Personalized medicine in non-small-cell lung cancer: is KRAS a useful marker in selecting patients for epidermal growth factor receptor-targeted therapy? J Clin Oncol. 2010;28:4769-4777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 214] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 15. | Kim D, Xue JY, Lito P. Targeting KRAS(G12C): From Inhibitory Mechanism to Modulation of Antitumor Effects in Patients. Cell. 2020;183:850-859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 161] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 16. | Amodio V, Yaeger R, Arcella P, Cancelliere C, Lamba S, Lorenzato A, Arena S, Montone M, Mussolin B, Bian Y, Whaley A, Pinnelli M, Murciano-Goroff YR, Vakiani E, Valeri N, Liao WL, Bhalkikar A, Thyparambil S, Zhao HY, de Stanchina E, Marsoni S, Siena S, Bertotti A, Trusolino L, Li BT, Rosen N, Di Nicolantonio F, Bardelli A, Misale S. EGFR Blockade Reverts Resistance to KRASG12C Inhibition in Colorectal Cancer. Cancer Discov. 2020;10:1129-1139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 331] [Article Influence: 66.2] [Reference Citation Analysis (0)] |
| 17. | Aldea M, Hendriks L, Mezquita L, Jovelet C, Planchard D, Auclin E, Remon J, Howarth K, Benitez JC, Gazzah A, Lavaud P, Naltet C, Lacroix L, de Kievit F, Morris C, Green E, Ngo-Camus M, Rouleau E, Massard C, Caramella C, Friboulet L, Besse B. Circulating Tumor DNA Analysis for Patients with Oncogene-Addicted NSCLC With Isolated Central Nervous System Progression. J Thorac Oncol. 2020;15:383-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 18. | Fang W, Huang Y, Gu W, Gan J, Wang W, Zhang S, Wang K, Zhan J, Yang Y, Zhao H, Zhang L. PI3K-AKT-mTOR pathway alterations in advanced NSCLC patients after progression on EGFR-TKI and clinical response to EGFR-TKI plus everolimus combination therapy. Transl Lung Cancer Res. 2020;9:1258-1267. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 19. | Jamme P, Fernandes M, Copin MC, Descarpentries C, Escande F, Morabito A, Grégoire V, Jamme M, Baldacci S, Tulasne D, Kherrouche Z, Cortot AB. Alterations in the PI3K Pathway Drive Resistance to MET Inhibitors in NSCLC Harboring MET Exon 14 Skipping Mutations. J Thorac Oncol. 2020;15:741-751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 56] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 20. | Lopez-Chavez A, Thomas A, Rajan A, Raffeld M, Morrow B, Kelly R, Carter CA, Guha U, Killian K, Lau CC, Abdullaev Z, Xi L, Pack S, Meltzer PS, Corless CL, Sandler A, Beadling C, Warrick A, Liewehr DJ, Steinberg SM, Berman A, Doyle A, Szabo E, Wang Y, Giaccone G. Molecular profiling and targeted therapy for advanced thoracic malignancies: a biomarker-derived, multiarm, multihistology phase II basket trial. J Clin Oncol. 2015;33:1000-1007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 188] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 21. | Mazieres J, Drilon A, Lusque A, Mhanna L, Cortot AB, Mezquita L, Thai AA, Mascaux C, Couraud S, Veillon R, Van den Heuvel M, Neal J, Peled N, Früh M, Ng TL, Gounant V, Popat S, Diebold J, Sabari J, Zhu VW, Rothschild SI, Bironzo P, Martinez-Marti A, Curioni-Fontecedro A, Rosell R, Lattuca-Truc M, Wiesweg M, Besse B, Solomon B, Barlesi F, Schouten RD, Wakelee H, Camidge DR, Zalcman G, Novello S, Ou SI, Milia J, Gautschi O. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann Oncol. 2019;30:1321-1328. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 433] [Cited by in RCA: 965] [Article Influence: 193.0] [Reference Citation Analysis (0)] |
| 22. | Passaro A, Attili I, Rappa A, Vacirca D, Ranghiero A, Fumagalli C, Guarize J, Spaggiari L, de Marinis F, Barberis M, Guerini-Rocco E. Genomic Characterization of Concurrent Alterations in Non-Small Cell Lung Cancer (NSCLC) Harboring Actionable Mutations. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 23. | Wang F, Zhao N, Gao G, Deng HB, Wang ZH, Deng LL, Yang Y, Lu C. Prognostic value of TP53 co-mutation status combined with EGFR mutation in patients with lung adenocarcinoma. J Cancer Res Clin Oncol. 2020;146:2851-2859. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 24. | Chevallier M, Tsantoulis P, Addeo A, Friedlaender A. Influence of Concurrent Mutations on Overall Survival in EGFR-mutated Non-small Cell Lung Cancer. Cancer Genomics Proteomics. 2020;17:597-603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 25. | El-Arabey AA, Abdalla M, Abd-Allah AR. SnapShot: TP53 status and macrophages infiltration in TCGA-analyzed tumors. Int Immunopharmacol. 2020;86:106758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 26. | El-Arabey AA, Denizli M, Kanlikilicer P, Bayraktar R, Ivan C, Rashed M, Kabil N, Ozpolat B, Calin GA, Salama SA, Abd-Allah AR, Sood AK, Lopez-Berestein G. CORRIGENDUM: GATA3 as a master regulator for interactions of tumor-associated macrophages with high-grade serous ovarian carcinoma. Cell Signal. 2022;89:110147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 27. | Alos L, Fuster C, Castillo P, Jares P, Garcia-Herrera A, Marginet M, Agreda F, Arance A, Gonzalvo E, Garcia M, Puig S, Teixido C. TP53 mutation and tumoral PD-L1 expression are associated with depth of invasion in desmoplastic melanomas. Ann Transl Med. 2020;8:1218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 28. | Fang C, Zhang C, Zhao WQ, Hu WW, Wu J, Ji M. Co-mutations of TP53 and KRAS serve as potential biomarkers for immune checkpoint blockade in squamous-cell non-small cell lung cancer: a case report. BMC Med Genomics. 2019;12:136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 29. | Biton J, Mansuet-Lupo A, Pécuchet N, Alifano M, Ouakrim H, Arrondeau J, Boudou-Rouquette P, Goldwasser F, Leroy K, Goc J, Wislez M, Germain C, Laurent-Puig P, Dieu-Nosjean MC, Cremer I, Herbst R, Blons H, Damotte D. TP53, STK11, and EGFR Mutations Predict Tumor Immune Profile and the Response to Anti-PD-1 in Lung Adenocarcinoma. Clin Cancer Res. 2018;24:5710-5723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 261] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 30. | Bersani F, Morena D, Picca F, Morotti A, Tabbò F, Bironzo P, Righi L, Taulli R. Future perspectives from lung cancer pre-clinical models: new treatments are coming? Transl Lung Cancer Res. 2020;9:2629-2644. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 31. | Kuang PP, Li N, Liu Z, Sun TY, Wang SQ, Hu J, Ou W, Wang SY. Circulating Tumor DNA Analyses as a Potential Marker of Recurrence and Effectiveness of Adjuvant Chemotherapy for Resected Non-Small-Cell Lung Cancer. Front Oncol. 2020;10:595650. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 32. | Page K, Martinson LJ, Hastings RK, Fernandez-Garcia D, Gleason KLT, Gray MC, Rushton AJ, Goddard K, Guttery DS, Stebbing J, Coombes RC, Shaw JA. Prevalence of ctDNA in early screen-detected breast cancers using highly sensitive and specific dual molecular barcoded personalised mutation assays. Ann Oncol. 2021;32:1057-1060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |