Published online Aug 6, 2022. doi: 10.12998/wjcc.v10.i22.7760
Peer-review started: February 11, 2022
First decision: April 13, 2022
Revised: April 27, 2022
Accepted: June 13, 2022
Article in press: June 13, 2022
Published online: August 6, 2022
Processing time: 160 Days and 22.5 Hours
The diagnostic value of metagenomic next-generation sequencing (mNGS) in central nervous system (CNS) infectious diseases after empirical treatment has not been reported.
To investigate the diagnostic value of mNGS of cerebrospinal fluid (CSF) in the empirically treated CNS infectious diseases.
A total of 262 CSF samples from patients with suspected CNS infections were collected between August 2020 and December 2021. Both mNGS and conventional methods were used for testing. The conventional methods included microbial culture, smear, polymerase chain reaction, etc.
Among 262 suspected cases, 183 cases (69.84%) were diagnosed as CNS infection, including 86 cases of virus infection (47.00%), 70 cases of bacterial infection (38.25%) and 27 cases of fungal infection (14.76%). The sensitivity and specificity of mNGS were 65.6% (95%CI: 58.2%-72.3%) and 89.6% (95%CI: 79.1%-95.3%), respectively. The PPV of mNGS was 94.5% (95%CI: 88.6%-97.6%), and the NPV was 48.8% (95%CI: 39.7%–57.9%). The pathogen detective sensitivity and accuracy of mNGS were higher than those of conventional methods (Sensitivity: 65.6% vs 37.2%; P < 0.001; Accuracy: 72.0% vs 50%, P < 0.001). The results showed that compared with conventional methods, mNGS technology was a more sensitive method for the diagnosis of CNS infection after empirical treatment.
mNGS can be a better method applied in the diagnosis of CNS infection after empirical treatment.
Core Tip: The study found that metagenomic next-generation sequencing (mNGS) had a higher sensitivity than the conventional methods in the diagnosis of infectious diseases of the central nervous system after empirical treatment. mNGS has significant clinical value and should be used mainly in early pathogen diagnosis in the future.
- Citation: Chen YY, Guo Y, Xue XH, Pang F. Application of metagenomic next-generation sequencing in the diagnosis of infectious diseases of the central nervous system after empirical treatment. World J Clin Cases 2022; 10(22): 7760-7771
- URL: https://www.wjgnet.com/2307-8960/full/v10/i22/7760.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i22.7760
Infectious diseases of the central nervous system (CNS) are acute or chronic inflammatory (or noninflammatory) diseases caused by pathogenic microorganisms that invade the parenchyma, meninges and blood vessels of the CNS[1]. The pathogenic microorganisms include viruses, bacteria, fungi, mycobacterium tuberculosis, spirochetes, parasites, etc. The infections are characterized by high morbidity, rapid disease progression, and high rates of disability and death. With the exception of stroke, meningitis ranks first in terms of disability and second in terms of mortality among neurological diseases. The pathogenic diagnosis of CNS infections mainly relies on cerebrospinal fluid (CSF) smear microscopy, pathogen culture, antigen-antibody tests, polymerase chain reaction (PCR) tests, etc. Early diagnosis of this disease is difficult to realize and it always followed by poor curative effects and prognosis[2,3]. In view of this, it is important for us to find a way to diagnose, identify and treat CNS infections at an early stage, which is also the topic of our research.
As an emerging pathogen detection method, metagenomic next-generation sequencing (mNGS) is of increasing interest to researchers, as it is able to detect all potential pathogens in a single test[4,5]. In recent years, mNGS has been proven to be outstanding in the diagnosis of infectious diseases, especially in the infections of blood, respiratory tract and CNS. Studies have reported that mNGS can increase the diagnosis rate of CNS infections by 25%[6-8]. The application of mNGS can improve and optimize the diagnostic strategy for infectious encephalitis/meningitis. However, the clinical value of mNGS in diagnosing CNS infection after empirical treatment remains to be discussed. In this study, we compared the differences of mNGS and conventional methods in the detection of pathogens including viruses, bacteria, and fungi in CNS infections after empirical therapy.
According to standard procedures, we recruited 262 patients with suspected CNS infections who visited our hospital from August 2020 to December 2021 as study subjects. The inclusion criteria were high-level clinical suspicion of CNS infectious diseases (e.g., fever, impaired consciousness, manifestations of increased intracranial pressure, signs of meningeal irritation, etc.). Exclusion criteria were refusal of lumbar puncture to obtain CSF or bloody CSF. Specimens from all patients were routinely tested for pathogens, including culture of bacteria and fungi from CSF, detection of bacteria on the stained CSF smear, serology test and PCR. CSF specimens were collected simultaneously for mNGS, and the clinical and laboratory data of the patients were recorded. The 262 patients enrolled in this study were diagnosed by three chief neurologists based on their clinical presentation, imaging findings, response to antibiotic treatment, follow-up results, and pathogenic evidence. Among them, 195 patients were diagnosed with CNS infections (183 infections with viruses, bacteria, and fungi and 12 other infections). The remaining 67 patients were diagnosed with noninfectious diseases, including autoimmune encephalitis, malignancy and venous sinus thrombosis. The study conformed to the principles of the Declaration of Helsinki and received ethical approval from the Ethics Committee of Liaocheng People's Hospital. Patients or their families gave informed consent to the diagnosis and treatment and signed the informed consent form.
CSF was extracted from each patient and tested in our laboratory (provided by Shenzhen UW Medical Laboratory Co., Ltd.) under aseptic conditions. The procedures of nucleic acid extraction and second-generation sequencing were as follows: Add 0.3 mL of 0.5 mm diameter glass beads to the broken wall tube, and then add 0.6 mL of specimen. Shake the tube at 2800-3200 r/min for 20 min at high speed, and then the 300 µl of nucleic acid is extracted. The DNA was extracted using the TINAamp Micro DNA Kit (DP316, Tiangen Biochemical Technology Co., Ltd., China) according to the standard operation.
The DNA libraries were constructed according to the following steps: Firstly, randomly cut the DNA into fragments, then perform the end-repair and adaptor ligation. Finally, the DNA fragments after ligation were amplified by PCR. The libraries were quality-controlled using Agilent 2100 (Agilent Technologies, United States) and Qubit 2.0 (Invitrogen, United States). The double-stranded DNA was converted to single-stranded circular DNA by DNA degradation and cyclization, and the DNA nanoballs (DNBs) were generated by rolling circle amplification (RCA) technology. The Qubit 2.0 was used for quality control of the DNBs. The qualified DNBs were loaded onto chips and sequenced on the the BGISEQ-50 platform (BGI Genetics Ltd., China) with 20 mol/L of sequencing data.
After removing low-quality short reads (length < 35 bp) to obtain sequencing data with high-quality, the high-quality reads were aligned to the human reference genome (fg19) sequence by BWA. Then the low-complexity reads were removed. The remaining reads were simultaneously aligned with four microbial genomic databases, which mainly composed of viruses, bacteria, fungi, and parasites. These database were downloaded from the National Center for Biotechnology Information (ftp://ftp.ncbi.nlm.nih.gov/genomes/), which mainly contain 1979 DNA viral whole-genome sequences, 6350 bacterial genomes, 1064 fungal genomes associated with human infections, and 234 parasite genome sequences associated with human diseases.
The final sequencing data were obtained by removing common background microorganisms and suspected pathogenic microorganisms that appeared in > 50% of the samples in the past 3 mo.
For viruses and bacteria, the positive criteria were mapped when the reads of one microorganism (species level) were 10 times greater than those of any other microorganism. For fungi, the coverage (species level) of one fungus was 5 times wider than that of any other fungus. The positive criteria in this study were as follows: (1) After excluding the microorganisms from normal skin and other flora, the top 5 genera with a relative high abundance were selected, and the top 2 species in each genus were selected; (2) Viruses, bacteria and fungi were considered positive when their number of specific sequences was ≥ 3 reads; (3) The clinical manifestations and imaging changes of the patients were discussed by experienced physicians and excluded the possible colonization and contamination cases; and (4) The condition of the patient improved after a targeted treatment. Consistent with clinical judgment or targeted treatment from three experienced senior physicians.
Results of mNGS or conventional methods were considered positive only if the pathogen tested corresponding to the final clinical diagnosis. If the patient's final clinical diagnosis was a non-CNS infection, a positive test result was considered a false positive, and a negative result was considered a true negative. If the patient's final clinical diagnosis was a CNS infectious disease, a positive result was considered a true positive, and a negative test was considered a false negative.
Using the final clinical diagnosis as the gold standard (the final clinical diagnosis was determined by three chief physicians based on the imaging, clinical manifestation, response to the medical therapy, and follow-up of the patient) to divide the subject patients into CNS infection and non-CNS infection groups. Differences in continuous variables between the groups were calculated using t tests and χ2 tests. P values <0.05 was considered as statistically significant. Sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy (ACC) were calculated, and the χ2 test was used to compare the sensitivity and specificity of mNGS with conventional pathogen detection methods. All statistics were reported as absolute values with 95% confidence intervals (CIs), and the SPSS 25.0 was used for statistics analysis.
A total of 262 patients were involved in this study, only 195 cases of them were diagnosed with CNS infections and the remaining 67 cases were diagnosed with non-CNS infectious diseases. The patients with CNS infections included 183 cases of viral, bacterial, and fungal infections and 12 cases of parasitic infections with 95 males and 97 females and an average age of 41.2 (14-91) years. All patients had received empirical treatment prior to CSF collections. The final CNS infections were classified as viral, bacterial, fungal, other infections, and noninfectious diseases. The non-CNS infections included autoimmune encephalitis, malignancy, and venous sinus thrombosis.
Comparison of the diagnostic performance of mNGS and conventional methods: Based on the final clinical diagnosis, 86 of the 121 patients with suspected viral infections were diagnosed with viral encephalitis and/or meningitis, and all 121 patients were given empirical antiviral therapy. A total of 135 CSF specimens were tested by mNGS and conventional methods respectively (serology and PCR) (5 of these patients underwent multiple mNGS tests). Compared with the conventional methods, the mNGS had a subtle advantage of detective sensitivity (66.3% vs 53.5%, P = 0.087), specificity (88.6% vs 85.7%, P = 1.00), accuracy (72.7% vs 62.8%, P = 0.009), positive predictive value (93.4% vs 90.2%, P = 0.779), and negative predictive value (51.7% vs 42.9%, P = 0.316) (Table 1).
| Sensitivity | Specificity | Accuracy | Positive predictive value | Negative predictive value | |
| Conventional methods+ | 53.5% (42.7%, 64.2%) | 85.7% (69.0%, 94.6%) | 62.8% (53.5%, 71.3%) | 90.2% (77.8%, 96.3%) | 42.9% (31.3%, 55.2%) |
| mNGS+ | 66.3% (55.2%, 75.9%) | 88.6% (72.3%, 96.3%) | 72.7% (63.7%, 80.2%) | 93.4% (83.3%, 97.9%) | 51.7% (38.5%, 64.6%) |
| P value | 0.087 | 1.000 | 0.099 | 0.779 | 0.316 |
Inconsistency between mNGS and conventional methods for virus detection: Among 86 specimens of CSF from patients with confirmed viral infection, 29 specimens were tested negative by mNGS while 40 were tested negative by conventional methods. In addition, mNGS confirmed the diagnosis of viral infection in 19 of the specimens that tested negative by conventional methods. Among these specimens that tested negative by mNGS, only 7 cases were confirmed by conventional methods (Table 2).
| mNGS | Conventional methods (+, -) | Total |
| + | 38, 19 | 57 |
| _ | 7, 22 | 29 |
| Total | 45, 41 | 86 |
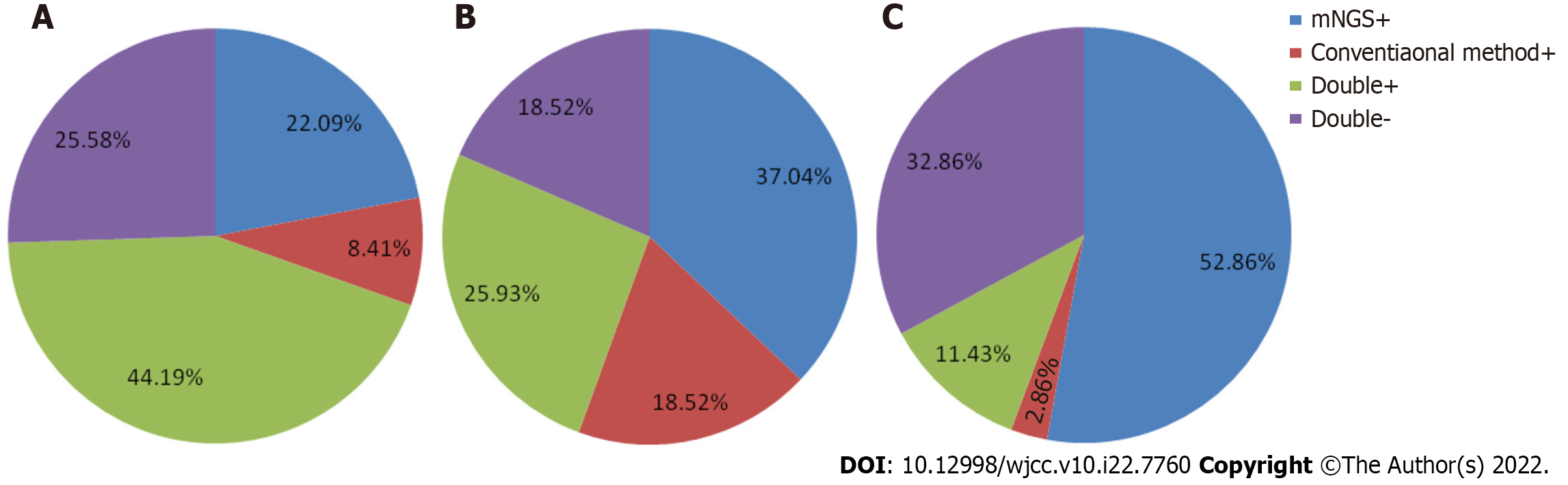
Consistency between mNGS and conventional methods for virus detection: Thirty-eight of 86 (11.43%) cases were tested both positive by mNGS and conventional methods and 22 of 86 (32.86%) cases were tested both negative by these two methods respectively. Moreover, 19 cases (52.86%) were tested positive only by mNGS and 7 (32.86%) were tested positive only by conventional methods (Figure 1A).
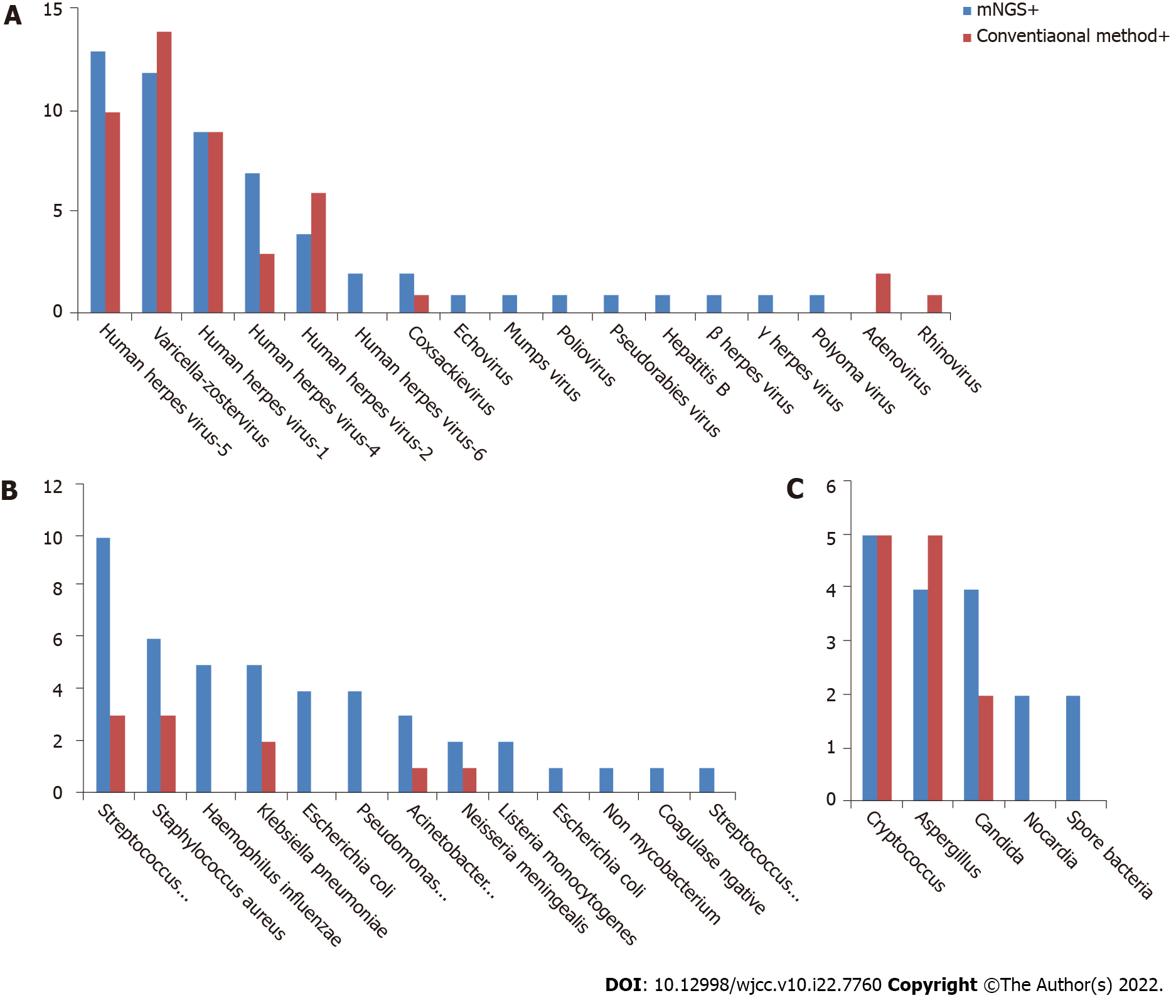
Comparison of pathogenic detection between mNGS and conventional methods: The positive detection rate of mNGS in CNS viral infections was 57/86 (66.28%), and the top 3 pathogens with the highest detection rate were herpes simplex virus type 3, cytomegalovirus, and herpes simplex virus type 1. The positive detection rate of conventional methods was 46/86 (53.49%), and the top 3 pathogens with the highest detection rate were cytomegalovirus, herpes simplex virus type 3, and herpes simplex virus type. A total of 17 pathogenic viruses were detected by mNGS, and 8 by conventional methods (Figure 2A). That means the mNGS can detect a wider range of pathogens than conventional methods.
Comparison of the diagnostic performance of mNGS and conventional methods: Based on the final clinical diagnosis, 96 patients with suspected bacterial infections were given empirical antibiotics, and finally, 70 patients were diagnosed with bacterial meningitis or encephalitis. A total of 110 specimens were prepared for mNGS and bacterial smear-culture respectively (6 of these patients underwent multiple mNGS tests). Compared with the conventional methods, the mNGS had a distinct advantage of detective sensitivity (65.7% vs 14.3%, P < 0.001), specificity (88.5% vs 84.6%, P = 1.000), accuracy (71.9% vs 33.3%, P < 0.001), positive predictive value (93.9% vs 71.4.%, P = 0.061), and negative predictive value (48.9% vs 26.8%, P = 0.011) (Table 3).
| Sensitivity | Specificity | Accuracy | Positive predictive value | Negative predictive value | |
| Conventional methods+ | 14.3% (8.5%, 23.6%) | 84.6% (64.3%, 95.0%) | 33.3% (24.2%, 43.8%) | 71.4% (42.0%, 90.4%) | 26.8% (17.9%, 37.9%) |
| mNGS+ | 65.7% (53.3%, 76.4%) | 88.5% (68.7%, 97.0%) | 71.9% (61.6%, 80.3%) | 93.9% (82.1%, 98.4%) | 48.9% (34.3%, 63.7%) |
| P value | < 0.001 | 1.000 | < 0.001 | 0.061 | 0.011 |
Inconsistency between mNGS and conventional methods for bacterial detection: Among the 70 confirmed CSF specimens, 24 of them were tested negative by mNGS and 60 were tested negative by conventional methods. Moreover, mNGS confirmed the bacterial infections in 37 specimens that were tested negative by conventional methods. Among these specimens that tested negative by mNGS, only 2 cases were confirmed by conventional methods (Table 4).
| mNGS | Conventional test (+, -) | Total |
| + | 8, 37 | 45 |
| _ | 2, 23 | 25 |
| Total | 10, 60 | 70 |
Consistency between mNGS and conventional methods for bacterial detection: Eight of 70 (11.43%) cases were tested both positive by mNGS and conventional methods and 23 of 70 (32.86%) cases were tested both negative by these two methods respectively. Moreover, 37 cases (52.86%) were tested positive only by mNGS and 2 (2.86%) were tested positive only by conventional methods (Figure 1B).
Comparison of pathogenic detection between mNGS and conventional methods: The mNGS positive detection rate of mNGS in CNS bacterial infections was 46/70 (65.71%) and the top 3 pathogens with the highest detection rate were Streptococcus pneumoniae, Haemophilus influenzae, and Klebsiella pneumoniae. The positive detection rate of conventional methods was 10/70 (14.29%), and the top 3 pathogens with the highest detection rate were S. pneumoniae, Staphylococcus aureus, and K. pneumoniae. A total of 13 pathogenic bacteria were detected by mNGS, and 5 by conventional methods (Figure 2B). That means the mNGS can detect a wider range of pathogens than conventional methods.
Comparison of the diagnostic performance of mNGS and conventional methods: Based on the final clinical diagnosis, 33 patients with suspected fungal infections were given antifungal treatment, and finally, 27 patients were diagnosed with fungal infections. A total of 27 CSF specimens were prepared for mNGS and fungal culture respectively. Compared with the conventional methods, the mNGS had a slight advantage of detective sensitivity (63.0% vs 44.4%, P = 0.127), specificity (100% vs 83.3%, P = 0.01), accuracy (69.7% vs 51.5%, P = 0.131), positive predictive value (100% vs 92.3%, P = 0.433), and negative predictive value (37.5% vs 25.0%, P = 0.656) (Table 5).
| Sensitivity | Specificity | Accuracy | Positive predictive value | Negative predictive value | |
| Conventional methods | 44.4% (26.0%, 64.4%) | 83.3% (36.5%, 99.1%) | 51.5% (33.9%, 68.8%) | 92.3% (62.1%, 99.6%) | 25.0% (9.6%, 49.4%) |
| mNGS | 63.0% (42.5%, 79.9%) | 100.0% (51.7%, 100.0%) | 69.7% (51.1%, 83.8%) | 100.0% (77.1%, 100.0%) | 37.5% (16.3%, 64.1%) |
| P value | 0.172 | 1.000 | 0.131 | 0.433 | 0.656 |
Inconsistency between mNGS and conventional methods for fungal detection: Among the 27 confirmed CSF specimens, 10 of them were tested negative by mNGS and 15 were tested negative by conventional methods. Moreover, mNGS confirmed the fungal infections in 10 specimens that were tested negative by conventional methods. Among these specimens that tested negative by mNGS, only 5 cases were confirmed by conventional methods (Table 6).
| mNGS | Conventional test (+, -) | Total |
| + | 7, 10 | 17 |
| _ | 5, 5 | 10 |
| Total | 12, 15 | 27 |
Consistency between mNGS and conventional methods for fungal detection: Seven of 27 (25.93%) cases were tested both positive by mNGS and conventional methods and 5 of 27 (18.52%) cases were tested both negative by these two methods respectively. Moreover, 10 cases (37.04%) were tested positive only by mNGS and 5 (18.52%) were tested positive only by conventional methods (Figure 1C).
Comparison of pathogenic detection between mNGS and conventional methods: The mNGS positive detection rate of mNGS in CNS fungal infections was 17/27 (62.96%) and the top 3 pathogens with the highest detection rate were Cryptococcus, Aspergillus, and Candida. The positive detection rate of conventional methods was 12/27 (44.44%), and the top 3 pathogens with the highest detection rate were Cryptococcus, Aspergillus, and Candida. A total of 5 pathogenic fungi were detected by mNGS, and 3 detected by conventional methods (Figure 2C). Among the negative specimens detected by mNGS, only 5 of them were confirmed by conventional methods.
Of the 262 CSF samples, 49.23% (129/262) of the samples were positively detected by conventional methods, while 26.34% (69/262) of the samples were positively detected by mNGS.
The sensitivity and specificity of mNGS were 65.6% (95%CI: 58.2%-72.3%) and 89.6% (95%CI: 79.1%-95.3%), respectively. The PPV of mNGS was 94.5% (95%CI: 88.6%-97.6%), and the NPV was 48.8% (95%CI: 39.7%-57.9%). The pathogen detective sensitivity and accuracy of mNGS were higher than those of conventional methods (sensitivity: 65.6% vs 37.2%; P < 0.001; accuracy: 72.0% vs 50%, P < 0.001). The results showed that compared with conventional methods, mNGS technology was a more sensitive method for the diagnosis of CNS infection after empirical treatmen (Table 7).
| Sensitivity | Specificity | Accuracy | Positive predictive value | Negative predictive value | |
| Conventional methods | 37.2% (30.2%, 44.6%) | 85.1% (73.8%, 92.2%) | 50.0% (43.7%, 56.3%) | 87.2% (77.2%, 83.3%) | 33.1% (26.3%, 40.8%) |
| mNGS | 65.6% (58.2%, 72.3%) | 89.6% (79.1%, 95.3%) | 72.0% (65.9%, 77.4%) | 94.5% (88.6%, 97.6%) | 48.8% (39.7%, 57.9%) |
| P value | < 0.001 | 0.436 | < 0.001 | 0.065 | 0.007 |
The accurate and rapid pathogen detection is essential for the diagnosis of CNS infections[9]. Despite previous studies have already reported the use of mNGS in the diagnosis of CNS infection[10], few studies have comprehensively evaluated the overall diagnostic performance of it in those patients already receiving the empiric treatment before. This cross-sectional study evaluated the diagnostic rate and additional diagnostic value of mNGS in patients with CNS infection after empirical treatment.
The aim of this study was to apply mNGS directly to clinical specimens in order to evaluate its diagnostic efficacy on CNS infections. We employed the result of clinical diagnosis rather than that of conventional methods as a reference standard. All CSF samples were collected after medical therapy, which would reduce the diagnostic efficacy of the conventional method. In addition, the advantages of other testing methods will not be revealed if the conventional methods are used as the reference standard. Although the clinical diagnosis lacks evidence of pathological findings, a follow-up period longer than 1 mo could greatly reduce the possibility of misdiagnosis. Therefore, we considered that using clinical diagnosis as the reference standard could be an appropriate choice when comparing the two diagnostic methods.
In this study, a systematic comparison of CSF tested by mNGS and conventional methods was performed, and the results showed several advantages of mNGS. First, mNGS is faster, costing an average of 2 d from sample collection to report. In contrast, the conventional methods cost at least 2-5 d. The mNGS is characterized by a short detection time and unbiased detection, which could facilitate the detection of clinical specimens without prior suspicion of certain pathogens. Second, the overall positive detection rate of mNGS (91.1%) was significantly higher than that of the conventional methods (62.2%). The results suggest that the detective sensitivity of mNGS is superior to that of conventional methods, especially in bacterial and fungal infections. In this study, 6 rare pathogens causing CNS infections and 3 mixed infections of CNS were identified, which means that the mNGS has the potential in detecting mixed infections, rare and unanticipated pathogens.
Virus infection is one of the most common types of CNS infections. At present, the cause of more than half of the CNS infections can not be confirmed. The pathogen detection methods of viral encephalitis/meningitis mainly include CSF staining, antigen antibody detection and PCR technology. These methods are often characterized by low sensitivity and time-consuming, resulting in delayed treatments. A significant merit of mNGS compared with traditional viral etiology test is that it can detect thousands of viruses at the same time within one test. It is no need to rely on clinical prediction, which means the detection range is wide, especially when extracting the CSF is difficult with limited resources[11,12].
The results showed that the detective sensitivity of mNGS and conventional methods for the diagnosis of CNS virus infection was 66.3% and 53.5% respectively, while the detective specificity of mNGS and conventional methods was 88.6% and 85.7% respectively. Compared with conventional methods, mNGS had high sensitivity and similar specificity in identifying the pathogens which cause the virus infection in CNS. The mNGS detected 9 additional pathogenic viruses compared with conventional methods. Eight of these viruses were rare pathogens, suggesting the latent capacity of mNGS in detecting rare and unanticipated viruses[13]. Most of the viruses detected by mNGS in patients with viral encephalitis or meningitis in this study were DNA viruses, with the most common viruses as the herpes simplex virus type 1, 2 type 5, varicella-zoster virus, and cytomegalovirus. RNA viruses were detected in 3 cases including the case 22 with influenza virus, the case 68 with enterovirus, and the case 139 with rhinovirus, which gave a fact that the DNA viruses were more common in CSF in patients with acute or subacute encephalitis or meningitis. And the mNGS was helpful in their detection and diagnosis.
Bacterial meningitis is a serious disease that can be fatal to both children and adults. The incidence rate and mortality rate of bacterial meningitis vary from different pathogen types[13]. In order to effectively treat bacterial meningitis, it is necessary to determine the microorganisms and their antibiotic sensitivity patterns as soon as possible. At present, CSF culture is the gold standard for the diagnosis of bacterial meningitis. However, the lower bacterial proliferation rates could lead to results with higher false negative. Therefore, new test methods are urgently needed. The mGNS is a rapid and high-throughput pathogen detection method, which has been applied to CSF samples in many studies. Miao et al[8] had systematically compared mGNS and CSF culture, and found that the mGNS had advantages in several aspects. The mGNS is highly sensitive to pathogen identification and is less affected by the previous application of antibiotics[14,15].
In this study, we used CSF samples to compare the differences between conventional methods and mGNS in the diagnosis of patients with bacterial meningitis. The sensitivity and accuracy of mNGS in detecting and diagnosing bacterial infections in CNS were 65.7% and 71.9% respectively, significantly higher than those of conventional methods as 14.3% and 33.3%. All patients received antibiotics before the extraction of CSF. This study suggests that mNGS could have a diagnostic advantage over conventional methods in patients who received empiric antimicrobial therapy before sample collection. The use of empirical antibiotic can reduce the detection rate of conventional methods by approximately 20% without affecting the detection rate of mNGS[16], as the microbial culture is susceptible to antimicrobial therapy. The mNGS only requires the DNA fragments of microorganisms for identification, which may explain its relatively high detection rate after treatment[17]. In our study, mNGS identified a total of 37 culture-negative pathogens. Among these pathogens, the non-mycobacterium may require a relative long incubation time, and some other pathogens can’t be cultured under standard conditions, such as Streptococcus haemolyticus. While the mNGS has a short TAT and is non-targeted, enabling rapid detection of these pathogens[18]. Considering these merits, mNGS could be an important complement to conventional culture which can improve the pathogen detection rate and the disease management in patients with complex infectious diseases. Similar conclusions have been reached in previous studies. When used as a complement of conventional methods, mNGS could improve the diagnosis of focal and CNS infections.
In recent years, with the increasing using of immunosuppressants, the prevalence of CNS mycosis has increased significantly. The main pathogens causing fungal infection in human CNS are opportunistic fungi, such as aspergillus, cryptococcus, pneumocystis girovii and endemic fungi. The conventional methods for the diagnosis of CNS fungal infection mainly include culture, histopathology, antigen detection, serology, imaging and molecular diagnosis. The culture method is often regarded as the gold standard of fungal infection, but most fungi should be cultured a relative long time, some of them even need a culture time as long as one month. The histopathological diagnosis is an invasive examination, whose sensitivity and specificity largely depend on the experience of pathologists, and there often exists a certain misdiagnosis rate[19,20]. This study showed that the positive rate of the fungal CNS infection detected by mNGS was 17/27(62.96%), while that of the conventional methods was only 12/17(44.4%). The diagnostic value of mNGS in fungal infection of CNS is worth more concern from the researchers and physicians. However, the mNGS has no obvious diagnostic advantage in cryptococcal infection in the CNS. In 5 cases of cryptococcal infection, capsular polysaccharide antigen was positive. For cryptococcal infection of CNS, it is recommended to detect the capsular polysaccharide antigen. If the patient's medical history, clinical manifestations and imaging findings are highly suspected of cryptococcal infection, mNGS is not recommended. The reason may be that the thick cryptococcus capsule is difficult to fully destroyed and the DNA used for mNGS can hardly released, which would reduce the diagnostic efficiency of mNGS. Although the positive detection rate of fungal culture is low, the combination of culture, GM test and mNGS is of great significance for the diagnosis of fungal CNS infections to avoid the omissions.
Fungal infections usually occur in immunocompromised individuals. However, in our study, a large proportion 15/27 (55.6%) of patients with CNS fungal infections had normal immune function[21]. There are increasing reports of immunocompetent patients with fungal infections, possibly due to the exposures to environmental genetic factors. We found that mNGS has a higher diagnostic value for CNS fungal infections than traditional methods (ink blot staining, culture, and antigen-antibody testing). mNGS is suitable for the detection of pathogens that cannot be identified by other available detection technologies and in situations when patients fail to respond to standardized drug therapy. For rare and slow-proliferating pathogens, mNGS shows considerable advantages in reducing the time required to diagnose and confirm the type of pathogen, and further facilitating the targeted drug therapy, and improving patient prognosis
We performed multiple mNGS tests in 11 patients to observe the dynamic changes of mNGS in CNS infections. In all cases, the positive detection rate of mNGS decreased within weeks when the patients received effective drug therapy. mNGS not only has a confirmatory value but also evaluates the efficiency of treatment to some extent.
The mNGS findings in 15 patients led to a change in treatment strategy. mNGS diagnosed rare pathogen infections in 6 cases, as the case 4 with porcine streptococcal meningitis, the case 6 with feline rickettsial meningitis, the case 12 with porcine cysticercosis, the case 82 with porcine pseudorabies infection, the case 111 with Listeria monocytogenes infection, and the case 124 with Mycoplasma pneumoniae encephalitis. While the conventional methods identified only 1 case, which is the case 111 with Listeria monocytogenes infection, and the mNGS positively identified this case earlier than the conventional methods, leading to a rapid change in anti-infective therapies. However, mNGS findings must be combined with epidemiological and clinical features to identify the pathogens.
As a revolutionary diagnostic tool, mNGS can detect all pathogens simultaneously. However, there are some inherent defects of mNGS. For example, microbial contaminants may interfere the interpretation of mNGS results, leading to unnecessary testing and inappropriate processing. One hindrance is the genome background of the human host with high abundance, which may limit the extraction of pathogenic sequences and lead to an insufficient sensitivity of mNGS. Other hindrances such as different registration cohorts and reference standards, as well as different types of infectious diseases still exist. Furthermore, we used the results of clinical diagnosis as a reference standard rather than that of conventional methods, which may be incorrect in some cases. These results need to be further explored with studies of more samples.
The mNGS method is a useful complement to conventional methods. It has higher positive rate, higher sensitivity and wider pathogen spectrum, especially for rare pathogens and pathogens that are difficult to culture. mNGS showed a good diagnostic efficiency on CNS infection after empirical treatment, which is superior to conventional methods, and can be used to detect special pathogens and mixed infections. All in all, the mNGS technology has great potential in the diagnosis of CNS infection.
The value of metagenomic next-generation sequencing (mNGS) in central nervous system infectious diseases after empirical treatment has not been reported.
The authors evaluated the value of mNGS in cerebrospinal fluid in the diagnosis of empirically treated central nervous system (CNS) infectious diseases.
This study evaluated the value of mNGS in central nervous system infection and whether mNGS can be used to diagnose the pathogen of central nervous system infection
A total of 262 empirically treated central nervous system-infected samples were analyzed by mNGS Confirmed pathogen. Using the final clinical diagnosis as the gold standard (the final cpatients were divided into CNS infection and non-CNS infection groups. Differences in continuous variables between groups were calculated using tests and χ2 tests.
mNGS is potentially advantageous in terms of speed and sensitivity. mNGS detected six rare pathogens.
mNGS has a better diagnosis of CNS infection after empirical treatment, and the overall detection rate is better than that of conventional assays.
mNGS has a better diagnosis of CNS infection after empirical treatment, and the overall detection rate is better than that of conventional assays. mNGS has diagnostic advantages.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Clinical neurology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Alkhatib AJ, Jordan; Novita BD, Indonesia A-Editor: Zhu JQ, China S-Editor: Ma YJ L-Editor: A P-Editor: Ma YJ
| 1. | Varıcı Balcı FK, Sayıner AA. [A Seven-Year Evaluation of Viral Central Nervous System Infections]. Mikrobiyol Bul. 2019;53:434-441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 2. | Stahl JP, Azouvi P, Bruneel F, De Broucker T, Duval X, Fantin B, Girard N, Herrmann JL, Honnorat J, Lecuit M, Mailles A, Martinez-Almoyna L, Morand P, Piroth L, Tattevin P; reviewing group. Guidelines on the management of infectious encephalitis in adults. Med Mal Infect. 2017;47:179-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 3. | He T, Kaplan S, Kamboj M, Tang YW. Laboratory Diagnosis of Central Nervous System Infection. Curr Infect Dis Rep. 2016;18:35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 44] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 4. | Hu B, Tao Y, Shao Z, Zheng Y, Zhang R, Yang X, Liu J, Li X, Sun R. A Comparison of Blood Pathogen Detection Among Droplet Digital PCR, Metagenomic Next-Generation Sequencing, and Blood Culture in Critically Ill Patients With Suspected Bloodstream Infections. Front Microbiol. 2021;12:641202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 50] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 5. | Perlejewski K, Bukowska-Ośko I, Rydzanicz M, Pawełczyk A, Caraballo Cortѐs K, Osuch S, Paciorek M, Dzieciątkowski T, Radkowski M, Laskus T. Next-generation sequencing in the diagnosis of viral encephalitis: sensitivity and clinical limitations. Sci Rep. 2020;10:16173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 6. | Miller S, Naccache SN, Samayoa E, Messacar K, Arevalo S, Federman S, Stryke D, Pham E, Fung B, Bolosky WJ, Ingebrigtsen D, Lorizio W, Paff SM, Leake JA, Pesano R, DeBiasi R, Dominguez S, Chiu CY. Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Res. 2019;29:831-842. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 362] [Cited by in RCA: 394] [Article Influence: 65.7] [Reference Citation Analysis (0)] |
| 7. | Gu W, Miller S, Chiu CY. Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection. Annu Rev Pathol. 2019;14:319-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 389] [Cited by in RCA: 839] [Article Influence: 119.9] [Reference Citation Analysis (0)] |
| 8. | Miao Q, Ma Y, Wang Q, Pan J, Zhang Y, Jin W, Yao Y, Su Y, Huang Y, Wang M, Li B, Li H, Zhou C, Li C, Ye M, Xu X, Li Y, Hu B. Microbiological Diagnostic Performance of Metagenomic Next-generation Sequencing When Applied to Clinical Practice. Clin Infect Dis. 2018;67:S231-S240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 596] [Article Influence: 99.3] [Reference Citation Analysis (0)] |
| 9. | Wilson MR, O'Donovan BD, Gelfand JM, Sample HA, Chow FC, Betjemann JP, Shah MP, Richie MB, Gorman MP, Hajj-Ali RA, Calabrese LH, Zorn KC, Chow ED, Greenlee JE, Blum JH, Green G, Khan LM, Banerji D, Langelier C, Bryson-Cahn C, Harrington W, Lingappa JR, Shanbhag NM, Green AJ, Brew BJ, Soldatos A, Strnad L, Doernberg SB, Jay CA, Douglas V, Josephson SA, DeRisi JL. Chronic Meningitis Investigated via Metagenomic Next-Generation Sequencing. JAMA Neurol. 2018;75:947-955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 213] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 10. | Simner PJ, Miller S, Carroll KC. Understanding the Promises and Hurdles of Metagenomic Next-Generation Sequencing as a Diagnostic Tool for Infectious Diseases. Clin Infect Dis. 2018;66:778-788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 242] [Cited by in RCA: 508] [Article Influence: 84.7] [Reference Citation Analysis (0)] |
| 11. | Guan H, Shen A, Lv X, Yang X, Ren H, Zhao Y, Zhang Y, Gong Y, Ni P, Wu H, Zhu Y, Cui L. Detection of virus in CSF from the cases with meningoencephalitis by next-generation sequencing. J Neurovirol. 2016;22:240-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 92] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 12. | Hong NTT, Anh NT, Mai NTH, Nghia HDT, Nhu LNT, Thanh TT, Phu NH, Deng X, van Doorn HR, Chau NVV, Delwart E, Thwaites G, Tan LV. Performance of Metagenomic Next-Generation Sequencing for the Diagnosis of Viral Meningoencephalitis in a Resource-Limited Setting. Open Forum Infect Dis. 2020;7:ofaa046. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 13. | van de Beek D, Cabellos C, Dzupova O, Esposito S, Klein M, Kloek AT, Leib SL, Mourvillier B, Ostergaard C, Pagliano P, Pfister HW, Read RC, Sipahi OR, Brouwer MC; ESCMID Study Group for Infections of the Brain (ESGIB). ESCMID guideline: diagnosis and treatment of acute bacterial meningitis. Clin Microbiol Infect. 2016;22 Suppl 3:S37-S62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 350] [Cited by in RCA: 502] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 14. | Xie YP, Hua CZ, Wang HJ, Sun AN, Shen J. Diagnostic Yield of Pneumococcal Antigen Detection in Cerebrospinal Fluid for Diagnosis of Pneumococcal Meningitis Among Children in China. Indian Pediatr. 2020;57:39-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 15. | Bagheri-Nesami M, Babamahmoodi F, Nikkhah A. Types, Risk Factors, Clinical symptoms and Diagnostic Tests of Acute Adult Meningitis in Northern Iran During 2006-2012. J Clin Diagn Res. 2015;9:IC01-IC05. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 16. | Zhang Y, Cui P, Zhang HC, Wu HL, Ye MZ, Zhu YM, Ai JW, Zhang WH. Clinical application and evaluation of metagenomic next-generation sequencing in suspected adult central nervous system infection. J Transl Med. 2020;18:199. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 107] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 17. | Guo LY, Li YJ, Liu LL, Wu HL, Zhou JL, Zhang Y, Feng WY, Zhu L, Hu B, Hu HL, Chen TM, Guo X, Chen HY, Yang YH, Liu G. Detection of pediatric bacterial meningitis pathogens from cerebrospinal fluid by next-generation sequencing technology. J Infect. 2019;78:323-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 18. | Bronska E, Kalmusova J, Dzupova O, Maresova V, Kriz P, Benes J. Dynamics of PCR-based diagnosis in patients with invasive meningococcal disease. Clin Microbiol Infect. 2006;12:137-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 19. | Nigrovic LE, Malley R, Macias CG, Kanegaye JT, Moro-Sutherland DM, Schremmer RD, Schwab SH, Agrawal D, Mansour KM, Bennett JE, Katsogridakis YL, Mohseni MM, Bulloch B, Steele DW, Kaplan RL, Herman MI, Bandyopadhyay S, Dayan P, Truong UT, Wang VJ, Bonsu BK, Chapman JL, Kuppermann N; American Academy of Pediatrics, Pediatric Emergency Medicine Collaborative Research Committee. Effect of antibiotic pretreatment on cerebrospinal fluid profiles of children with bacterial meningitis. Pediatrics. 2008;122:726-730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 142] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 20. | Chen J, Zhang R, Liu L, Qi T, Wang Z, Song W, Tang Y, Sun J, Liu D, Lin Y, Xu S, Yang J, Shen Y, Lu H Dr. Clinical usefulness of metagenomic next-generation sequencing for the diagnosis of central nervous system infection in people living with HIV. Int J Infect Dis. 2021;107:139-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 21. | McHugh KE, Gersey M, Rhoads DD, Procop GW, Zhang Y, Booth CN, Sturgis CD. Sensitivity of Cerebrospinal Fluid Cytology for the Diagnosis of Cryptococcal Infections: A 21-Year Single-Institution Retrospective Review. Am J Clin Pathol. 2019;151:198-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |