Published online May 26, 2022. doi: 10.12998/wjcc.v10.i15.4923
Peer-review started: September 22, 2021
First decision: January 10, 2022
Revised: January 23, 2022
Accepted: April 3, 2022
Article in press: April 3, 2022
Published online: May 26, 2022
Processing time: 244 Days and 2.3 Hours
Hereditary spherocytosis (HS) is characterized by anemia, jaundice, splenomegaly, and cholelithiasis, and is caused by abnormal genes encoding red blood cell membrane components. The most common mutations found in HS are in the ANK1 gene.
A 4-mo-old girl was admitted to our hospital with pallor that had lasted for more than 2 mo. She presented with jaundice, anemia and splenomegaly. A heterozygous mutation of ANK1 (exon23: c.G2467T:p.E823X) was identified, and the mutation was determined to be autosomal dominant. This mutation is linked to the relatively serious anemia she had after birth; this anemia improved with age.
The utilization of next-generation sequencing may assist with the accurate diagnosis of HS, especially in atypical cases.
Core Tip: We report a case of hereditary spherocytosis. The clinical manifestation and a mutation of ANK1 gene (exon23: c.G2467T:p.E823X) were assessed and related literature was reviewed. The information in this report may help with the diagnosis of hereditary spherocytosis.
- Citation: Fu P, Jiao YY, Chen K, Shao JB, Liao XL, Yang JW, Jiang SY. Targeted next-generation sequencing identifies a novel nonsense mutation in ANK1 for hereditary spherocytosis: A case report. World J Clin Cases 2022; 10(15): 4923-4928
- URL: https://www.wjgnet.com/2307-8960/full/v10/i15/4923.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i15.4923
Hereditary spherocytosis (MIM: 182900) is the third most common hemolytic disease after glucose-6-phosphate dehydrogenase deficiency and ABO hemolytic disease[1]. HS is a common, inherited, red cell membrane disorder with an incidence of 1/2000 in Northern Europe and Northern America[2], whereas in China it is considered less common and affects approximately 1.27 cases per 100000 people in males and 1.49 cases per 100000 people in females[3]; however, these discrepancies may reflect a considerable number of undiagnosed, asymptomatic cases. HS is fundamentally characterized by a mechanical abnormality of erythrocytes, along with osmotic changes, and can result in kernicterus in newborn babies. The main biochemical defects of HS lie in the proteins of the erythrocyte membrane, including ankyrin, band 3, α-spectrin, β-spectrin, and protein 4.2, which are encoded by the ANK1, SLC4A1, SPTA1, SPTB and EPB42 genes, respectively[4]. Approximately 50% of patients with HS present with anemia and 10 to 15% with jaundice or splenomegaly[5]. The symptoms of HS can vary widely from asymptomatic patients to those that require blood transfusions or even life-threatening anemia. An initial symptom of HS in neonates may be hyperbilirubinemia that is unrelated to blood incompatibility. Here, we present a HS case with a novel mutation of ANK1 present within a Chinese family.
A pale complexion for more than 2 mo.
A female neonate displayed splenomegaly and moderate anemia with a Hb level of 60 g/L at examination 1 mo after birth. The girl was then taken to Shanghai Children’s Hospital for further treatment 3 mo later.
The child was pale at the age of 1 mo. Anemia was detected in the blood after a routine examination at the local hospital, but no treatment was given.
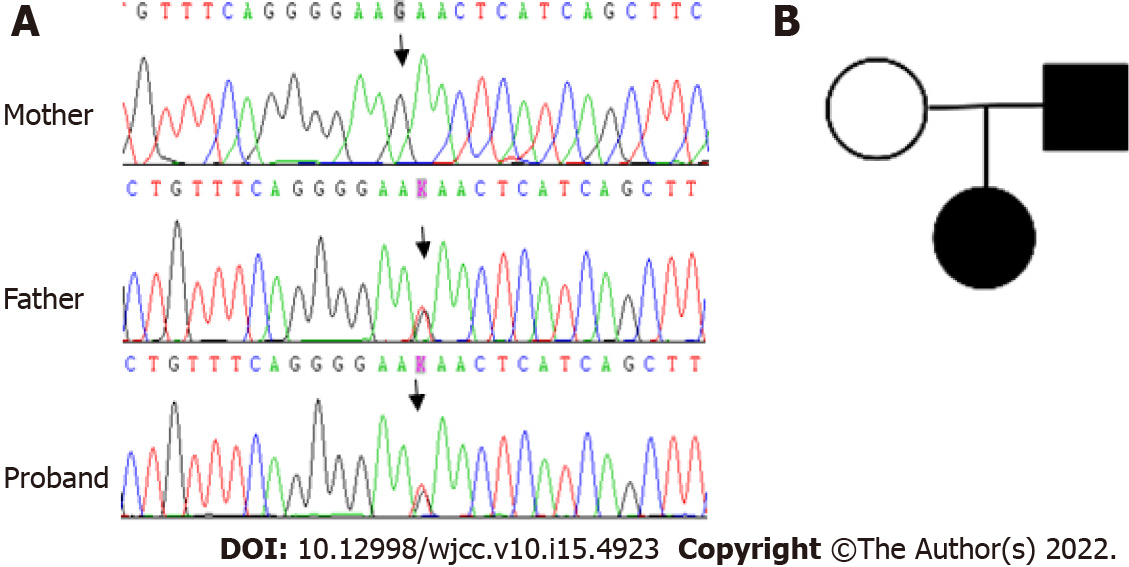
G1P1, gestational age 39 wk, birth weight 3110 g, she was the first child of the family, and her father currently has mild splenomegaly but no anemia and no history of blood transfusion.
A further physical examination confirmed pallor and splenomegaly.
Blood tests showed moderate anemia, reticulocytosis, and hyperbilirubinemia that was mainly unconjugated bilirubin (Table 1). An osmotic fragility test showed that a concentration of NaCl solution of 0.52% (ref: 0.38%-0.41%) initiated hemolysis and at a concentration of 0.44% (ref: 0.30%-0.34%) there was complete hemolysis; indicative of increased erythrocyte osmotic fragility. The activity of glucose-6-phosphate dehydrogenase (G-6-PD) was normal and a Coomb’s test was negative. Hemoglobin electrophoresis, and α- and β-globin genetic analysis excluded α- and β-thalassemia. A significant increase of spherocytes after a peripheral blood smear was absent.
| Test | Results | Reference |
| Hb (g/L) | 62 | 110-160 |
| MCV (fL) | 77.4 | 73-100 |
| MCH (pg) | 27.4 | 27-34 |
| MCHC (g/L) | 354 | 320-410 |
| RBC (x 1012/L) | 2.26 | 4-5.5 |
| RET (%) | 11.2 | 0.5-1.5 |
| TB (μmol/L) | 22.67 | 3.4-17.1 |
| DB (μmol/L) | 9.28 | 0-6.8 |
| ALT (U/L) | 21 | 5-40 |
| FER (ng/ml) | 154.4 | 11-306.8 |
| LDH (U/L) | 300 | 180-430 |
| Serum iron (μmol/L) | 21.68 | 9.00-32.00 |
| Unsaturatediron (μmol/L) | 31.08 | 34.20-48.20 |
| TIBC (μmol/L) | 52.76 | 45.00-72.00 |
A chest X-ray showed no active lesions in the lungs. Echocardiography showed an atrial septal defect (1.2 mm) and the oval foramen was not closed.
To identify the cause of the unexplained hemolysis, genetic analysis by next-generation sequencing (NGS) was performed. DNA was extracted from peripheral blood collected before transfusion, and genetic analysis was conducted on 700 genes associated with hematological diseases. Genomic DNA was extracted using the QIAamp DNA Blood Mini Kit according to the manufacturer’s instructions. The DNA sample was quantified by Qubit dsDNA BR Assay kit. All libraries were prepared using the KAPA HTP Library Preparation Kit according to the manufacturer’s instructions. Fragmented DNA was repaired, 3’ dA-tailed, ligated with Illumina adapters, size selected, amplified, and assessed using the Agilent 2100 Bioanalyzer. The captured DNA library was finally amplified and sequenced on an Illumina Novoseq 6000 sequencer for paired reads at 150 bp. The original data were converted from bcl files to fastq format files by Illumina CASAVA1.8 (Illumina, San Diego, CA, United States), and reads were compared to the GRCh37/hg19 human genome reference using BWA, samtools, picard, and GATK to remove repeated sequences and identify genetic variants. All of the identified variants were evaluated by browsing through databases, including NCBI dbSNP, OMIM, HGMD and NCBI ClinVar. The putative effects on the ANK1 protein of all the identified variants were explored using a variety of prediction algorithms, including PolyPhen2, SIFT and Mutation Taster. Finally, a heterozygous mutation in ANK1 (exon23:c.G2467T:p.E823X) was detected in the girl and her father, whereas her mother was wild type (Figure 1).
Hereditary spherocytosis.
She received suspension RBCs transfusion during her hospitalization.
To date, and 4 mo after her last blood transfusion, the child’s hemoglobin levels have fluctuated between 90 g/L and 100 g/L and a further blood transfusion was not required.
According to the HGMD, ExAC, gnomAD, and 1000 genomes databases, the detected mutation (ANK1:exon23:c.G2467T:p.E823X) in this Chinese family led to premature termination of the protein, thereby forming a truncated protein without normal function. Since the mutation is novel, the clinical manifestations associated with it are unclear. At the time of diagnosis in early infancy, the proband had splenomegaly and moderate anemia that required a blood transfusion. However, at present, the degree of anemia is reduced, and the patient is not dependent upon a blood transfusion. Similarly, her father displayed a history of anemia in his childhood, but his current symptoms are now only mild. Therefore, it is speculated that this mutation leads to relatively serious anemia after birth, but that anemia improves with age. The diagnostics for HS in this family were based upon laboratory findings and clinical examinations, as well as a positive family history, and we found a novel nonsense mutation of ANK1, detected by targeted next generation sequencing and identified by Sanger sequencing in this study.
Mutations within the ANK1 gene are the most common cause of HS (up to 50% of cases), followed by mutations in the spectrin gene (SPTB, up to 20%; SPTA1, up to 5%), SLC4A1 (up to 15%), and EPB42 (up to 10%)[6]. It has been reported that heterozygous ANK1 mutations account for 52% of all Korean HS patients and approximately 31% of all Japanese HS patients[7,8]. The ANK1 gene is located at 8p11.21[9], contains 42 exons, and with a cDNA of 8300 bp in length. The ANK1 gene encodes a critical component of the erythrocyte membrane skeleton, the ankyrin 1 protein, which is composed of 1881 amino acids and possesses three main structural domains[10]. Each erythrocyte contains approximately 1.24 × 106 ankyrins[8]. ANK1 mutations are primarily frameshift, nonsense, and splice site mutations[7,11], and it is frequently mutated de novo. Most of the mutations occur in the coding sequence of genes and can result in functional deficiencies of the protein. Autosomal recessive inheritance has occasionally been recorded in a few patients[12]. For recessive HS, the common mutational types are missense mutations and with a mutation in the putative ankyrin-1 promoter[13].
The complexity of gene mutations and gene regulation may contribute to the heterogeneity of clinical manifestations. Patients with ANK1 gene mutations tend to be more anemic and with a higher level of reticulocytosis than those without[8]. However, even if the mutation site is the same, disease severity can greatly differ[14], and furthermore, for the same patient, the degree of anemia at any given period can also vary extensively. We report here a new mutation in a family with HS in which the child showed initial transfusion dependence during the early infantile period; however, the anemia improved with age.
In general, children with Coomb’s negative hemolytic disease can be diagnosed as HS according to a family history of HS, positive osmotic fragility test, spherocytosis on a blood smear, and the exclusion of possible causes of secondary spherocytosis. Yet, 20% of HS patients do not display typical spherocytosis; hence, for such patients without spherocytosis, genetic testing is very important to make a diagnosis of HS. It has been reported that splenectomy was required for a girl with highly suspected HS and with an anemia recovery, and then decades later, her son was subsequently diagnosed with HS by genetic testing[12]. With the rapid development and wide clinical application of gene technologies, a growing number of HS cases have been identified by genetic tests, especially asymptomatic patients. This has led to the identification of several new mutants in HS genes in our center through this gene testing[14]. Guideline recommendations are that further molecular testing is not a requirement when a family history, clinical manifestations, and laboratory tests all support the diagnosis of HS[15]. However, in view of the fact that there are no hotspot mutations in HS and most mutations are sporadic and specific to individual patients or their families[16], it is necessary to detect and characterize gene mutations in patients with atypical clinical presentations.
When compared directly with Sanger sequencing, NGS has higher diagnostic efficiency for suspected red blood cell membrane disorders in patients[17,18]. In addition to helping to confirm a diagnosis, genetic technology also helps to assess the risk of HS for descendants of a family and provide information for potential genetic counselling and future research and understanding of HS.
Regarding treatment, splenectomy, as the standard surgical treatment in moderate and severe patients with HS[19], is efficient but does have drawbacks, of which the most recognized is a risk of infection. In addition, the heterogeneity of clinical manifestations requires close observation of the disease progression to appropriately determine the timing of surgery. Before undertaking a splenectomy, the diagnosis should be confirmed by clinical data and a genetic examination.
A novel ANK1 mutation considered causative of HS was identified in a Chinese family and its clinical features were elucidated and documented. This novel study expands the current spectrum of ANK1 mutations. Moreover, the analysis of pathogenic gene mutations via NGS and Sanger sequencing can provide a powerful tool to support HS diagnosis and the associated genetic consultation of HS patients. However, the pathogenic mechanism of the ANK1 mutation is unclear and needs to be explored further to help with the diagnosis of HS.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Hematology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Malik S, Pakistan; Sanal MG, India; Ramasamy I, United Kingdom S-Editor: Ma YJ L-Editor: Filipodia P-Editor: Ma YJ
| 1. | Christensen RD, Yaish HM, Gallagher PG. A pediatrician's practical guide to diagnosing and treating hereditary spherocytosis in neonates. Pediatrics. 2015;135:1107-1114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 2. | Da Costa L, Galimand J, Fenneteau O, Mohandas N. Hereditary spherocytosis, elliptocytosis, and other red cell membrane disorders. Blood Rev. 2013;27:167-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 224] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 3. | Wang C, Cui Y, Li Y, Liu X, Han J. A systematic review of hereditary spherocytosis reported in Chinese biomedical journals from 1978 to 2013 and estimation of the prevalence of the disease using a disease model. Intractable Rare Dis Res. 2015;4:76-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 4. | Perrotta S, Gallagher PG, Mohandas N. Hereditary spherocytosis. Lancet. 2008;372:1411-1426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 401] [Cited by in RCA: 420] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 5. | Krueger HC, Burgert EO Jr. Hereditary spherocytosis in 100 children. Mayo Clin Proc. 1966;41:821-830. [PubMed] |
| 6. | An X, Mohandas N. Disorders of red cell membrane. Br J Haematol. 2008;141:367-375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 125] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 7. | Park J, Jeong DC, Yoo J, Jang W, Chae H, Kim J, Kwon A, Choi H, Lee JW, Chung NG, Kim M, Kim Y. Mutational characteristics of ANK1 and SPTB genes in hereditary spherocytosis. Clin Genet. 2016;90:69-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 65] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 8. | Nakanishi H, Kanzaki A, Yawata A, Yamada O, Yawata Y. Ankyrin gene mutations in japanese patients with hereditary spherocytosis. Int J Hematol. 2001;73:54-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 9. | Lambert S, Yu H, Prchal JT, Lawler J, Ruff P, Speicher D, Cheung MC, Kan YW, Palek J. cDNA sequence for human erythrocyte ankyrin. Proc Natl Acad Sci U S A. 1990;87:1730-1734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 113] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 10. | Lux SE, John KM, Bennett V. Analysis of cDNA for human erythrocyte ankyrin indicates a repeated structure with homology to tissue-differentiation and cell-cycle control proteins. Nature. 1990;344:36-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 418] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 11. | Eber S, Lux SE. Hereditary spherocytosis--defects in proteins that connect the membrane skeleton to the lipid bilayer. Semin Hematol. 2004;41:118-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 155] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 12. | Han JH, Kim S, Jang H, Kim SW, Lee MG, Koh H, Lee JH. Identification of a novel p.Q1772X ANK1 mutation in a Korean family with hereditary spherocytosis. PLoS One. 2015;10:e0131251. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 13. | Eber SW, Gonzalez JM, Lux ML, Scarpa AL, Tse WT, Dornwell M, Herbers J, Kugler W, Ozcan R, Pekrun A, Gallagher PG, Schröter W, Forget BG, Lux SE. Ankyrin-1 mutations are a major cause of dominant and recessive hereditary spherocytosis. Nat Genet. 1996;13:214-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 142] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 14. | Hao L, Li S, Ma D, Chen S, Zhang B, Xiao D, Zhang J, Jiang N, Jiang S, Ma J. Two novel ANK1 loss-of-function mutations in Chinese families with hereditary spherocytosis. J Cell Mol Med. 2019;23:4454-4463. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 15. | Bolton-Maggs PH, Langer JC, Iolascon A, Tittensor P, King MJ; General Haematology Task Force of the British Committee for Standards in Haematology. Guidelines for the diagnosis and management of hereditary spherocytosis--2011 update. Br J Haematol. 2012;156:37-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 242] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 16. | He BJ, Liao L, Deng ZF, Tao YF, Xu YC, Lin FQ. Molecular Genetic Mechanisms of Hereditary Spherocytosis: Current Perspectives. Acta Haematol. 2018;139:60-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 17. | van Dijk EL, Jaszczyszyn Y, Naquin D, Thermes C. The Third Revolution in Sequencing Technology. Trends Genet. 2018;34:666-681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 656] [Article Influence: 93.7] [Reference Citation Analysis (0)] |
| 18. | van Vuren A, van der Zwaag B, Huisjes R, Lak N, Bierings M, Gerritsen E, van Beers E, Bartels M, van Wijk R. The Complexity of Genotype-Phenotype Correlations in Hereditary Spherocytosis: A Cohort of 95 Patients: Genotype-Phenotype Correlation in Hereditary Spherocytosis. Hemasphere. 2019;3:e276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 19. | Manciu S, Matei E, Trandafir B. Hereditary Spherocytosis - Diagnosis, Surgical Treatment and Outcomes. A Literature Review. Chirurgia (Bucur). 2017;112:110-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (1)] |