Published online May 16, 2022. doi: 10.12998/wjcc.v10.i14.4480
Peer-review started: November 8, 2021
First decision: February 14, 2022
Revised: February 28, 2022
Accepted: March 25, 2022
Article in press: March 25, 2022
Published online: May 16, 2022
Processing time: 185 Days and 17.8 Hours
Hepatitis C virus (HCV) causes a large number of infections worldwide. New infections seem to be increasing according to a report of the World Health Organization in 2015. Although direct-acting antivirals are quite effective for most genotypes of the HCV, some genotypes fail to respond. Therefore, the trend of genotype distribution is vital to better control the development of this infection.
To analyze the distribution and trends of the HCV genotype before and after the emergence of direct-acting antivirals in China.
We searched all literature published in five electronic databases-China National Knowledge Infrastructure, Wan Fang Data, VIP Chinese Journal Database, Chinese Biomedical Literature Service System, and PubMed-from January 1, 2010 to December 31, 2020. The search strategy combined medical subject headings and free-text terms, including “hepatitis C virus” or “HCV” and “genotype” or “subtype” and ”China” or “Chinese”. Additional relevant articles were searched by manual selection. Data were extracted to build a database. All of the data were totaled according to regions, periods, routes of transmission, and sexes. The percentages in various stratifications were calculated.
There were 76110 samples from 30 provinces included in the study. Genotype 1 (G1) accounted for 58.2% of cases nationwide, followed by G2, G6, G3b, G3a, unclassified and mixed infections (17.5%, 7.8%, 6.4%, 4.9%, 1.8%, and 1.2%, respectively). The constitution of genotype varied among different regions, with G6 and G3b being more common in the south and southwest, respectively (28.1%, 15.4%). The past ten years have witnessed a decrease in G1 and G2 and an increase in G3 and G6 in almost all regions. The drug-use population had the most abundant genotypes, with G6 ranking first (33.3%), followed by G1 and G3b (23.4%, 18.5%).
G3 and G6 pose a new challenge for HCV infection. This study revealed the distribution of HCV genotypes in China over the past 10 years, providing information for HCV management strategies.
Core Tip: This article comprehensively included the literature published in recent years and reflects a picture of the genotype constitution of hepatitis C virus (HCV), showing increases in genotype 3 and genotype 6 owing to an increase in intravenous drug users, providing the prevention direction for future HCV control.
- Citation: Yang J, Liu HX, Su YY, Liang ZS, Rao HY. Distribution and changes in hepatitis C virus genotype in China from 2010 to 2020. World J Clin Cases 2022; 10(14): 4480-4493
- URL: https://www.wjgnet.com/2307-8960/full/v10/i14/4480.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i14.4480
According to the World Health Organization (WHO), there were 71 million people with chronic hepatitis C virus (HCV) infection in 2015 worldwide, and approximately 1.75 million new infections occurred in 2015. Unsafe health care procedures and injection drug use are responsible for most new HCV infections[1]. It is estimated that China has approximately 10 million HCV infections[2]. A report from the Ministry of Health of China found that new HCV infections rose from 70681 to 242897 between 2006 and 2017[3]. Left untreated, 20% or more of chronic HCV infections will develop into cirrhosis or hepatocellular carcinoma (HCC)[4]. As a result, in May 2016, the World Health Assembly endorsed the Global Health Sector Strategy (GHSS) for 2016-2021 on viral hepatitis, proposing to eliminate viral hepatitis as a public health threat by 2030[5].
HCV is a single-stranded RNA that can easily mutate. It is classified into seven confirmed genotypes and 67 subtypes[6]. Treatment is likely to be the most effective available option right now. The revolution in HCV therapy has been attributed to direct-acting antivirals (DAAs). The sustained virologic response rate has reached more than 90% of the main known genotypes and subtypes. However, the pangenotype regimen is not as effective in some of the genotypes, especially genotype 3b (G3b) with cirrhosis[2]. Limited data have shown that the SVR rate in G3b patients without cirrhosis is 96% and is 50% in cirrhosis patients. Genotype 1 (G1) and genotype 3 (G3) HCV infections are related to an increased risk for cirrhosis and HCC [7]. G3b also progresses more rapidly than G1[8]. Therefore, the Chinese Guidelines for HCV in 2019 recommend genotyping before treatment in regions with more than a 5% prevalence of G3b[2]. For treatment failure, genotyping is also needed to differentiate between relapse and reinfection[9]. HCV genotypes and subtypes remain the cornerstones in the management of HCV infection, even in the era of DAAs[10].
In addition, the geographic distribution of HCV genotypes offers information about the origin of infection and possible transmission routes[11]. The changes in distribution over time could provide information to guide monitoring and improve prevention for policy-makers. In this way, measures could be put into effect to avoid new and repeated infections.
However, there have been only six articles about China HCV distribution nationwide, four of which were cross-sectional studies[12-15] and two of which were literature reviews[3,16]. The most recent cross-sectional study and review were both from 2017. Most of these studies only included a few years or started in the 1990s. DAA officially came onto the market in 2017 in China. No studies of the HCV genotype distribution over the past 10 years have been published, although the first generation of DAA was approved by the FDA in 2011[5], swiftly evolving into the fourth generation currently. Our study included the genotype data of Chinese individuals from 2010 to 2020 and explored the evolution of HCV genotypes over time and regions in the different risk groups and both sexes.
We searched all of the literature published in five electronic databases-China National Knowledge Infrastructure (CNKI), Wanfang Data, VIP Chinese Journal Database (CQVIP), Chinese Biomedical Literature Service System (SinoMed), and PubMed-from January 1, 2010 to December 31, 2020. The search strategy combined medical subject headings and free-text terms including “hepatitis C virus” or “HCV” and “genotype” or “subtype” and “China” or “Chinese”. To acquire more comprehensive data, additional relevant articles were searched by manual selection from the references of published literature.
The inclusion criteria were as follows: (1) Studies reporting the distribution of HCV genotypes and with specific numbers of genotypes; (2) The sample sizes of genotypes equal to or greater than 50; (3) The population included being Chinese; and (4) Studies adopting the Simmond genotype nomenclature system. Studies were excluded if they met the following exclusion criteria: (1) Redundant publication; (2) Human immunodeficiency virus (HIV)/HCV or HBV/HCV coinfection; (3) Studies with no specific data of genotypes, no exact inclusion time or not in the required period; (4) Clinical trials; (5) Sample size of genotypes less than 50; and (6) Non-Chinese populations.
Data extraction from the included studies contained the following information: Year of publication, name of the first author, sampling time, risk group, province, route of transmission, HCV genotyping region, genotyping method and method of HCV RNA detection, ethnic group, sample size, numbers of subtypes in all populations and each sex population.
The database was built in Microsoft Excel software, version 2019, and the 30 provinces are divided into seven geographical regions according to the National Bureau of Statistics of China and other relevant studies[3]: North, South, Central, Northwest, Northeast, Southwest, and Southeast. The regions to which samples belong were determined by the areas where the infection was diagnosed. The periods were divided into two parts according to the sampling year: The 2010-2015 period and the 2016-2020 period. Although DAAs were not approved until 2017 in China, many patients had access to generic drugs from India before 2017. For the studies included, HCV genotypes were totaled by different classifications: Regions, periods, routes of transmission, and sexes. Then, we calculated the percentage in various stratifications; thus, the compositions of each genotype and subtype were obtained. Graphs were built using Excel software, version 2019, and R software, version 4.0.3.
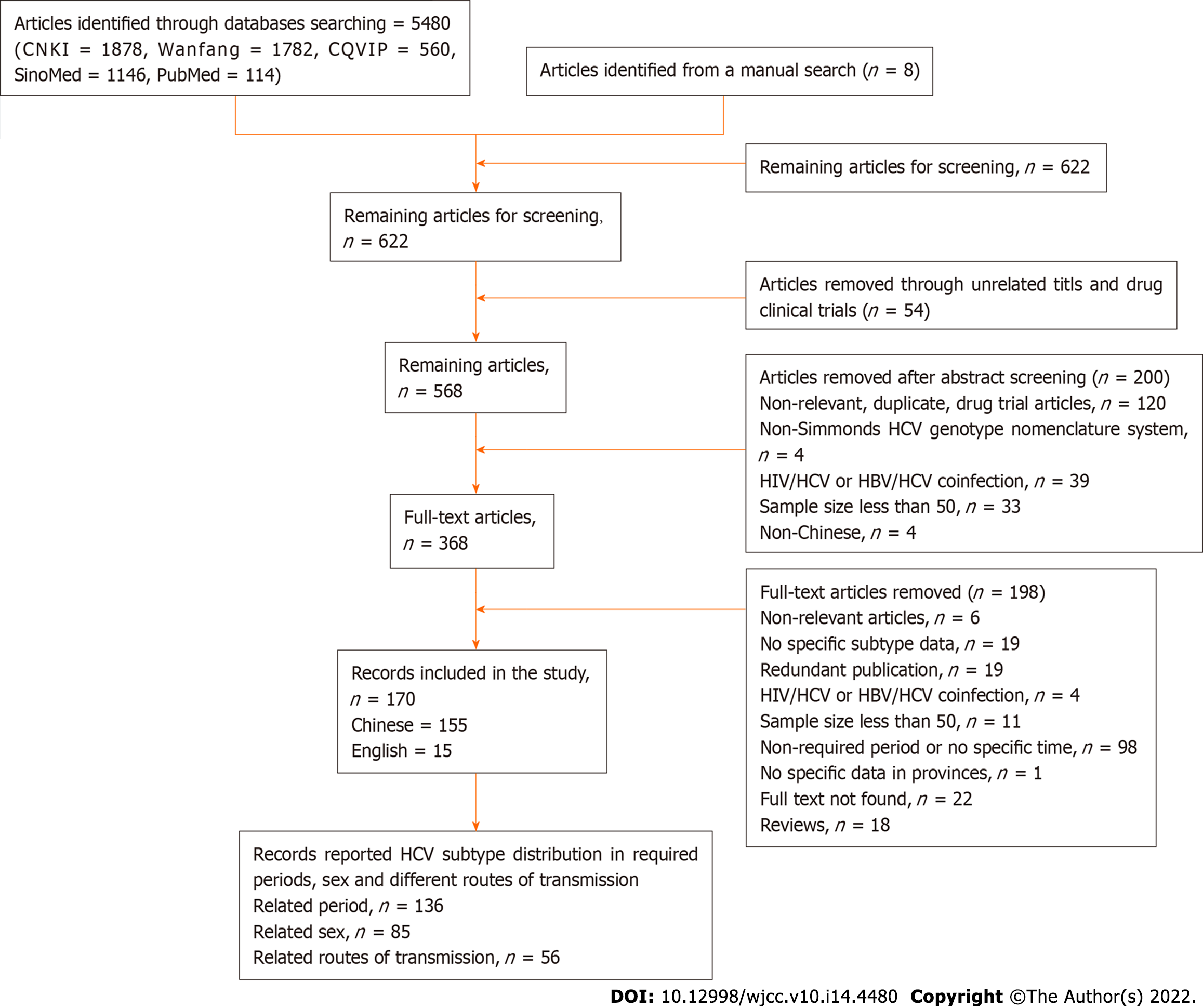
The criteria of screening identified 5480 articles from the five electronic databases (CNKI = 1878, Wanfang = 1782, CQVIP = 560, SinoMed = 1146, PubMed = 114), and another eight articles were identified from the references of the articles above. The literature screening procedure is shown in Figure 1.
The overall data are extracted from 170 articles from five databases from January 1, 2010 to December 31, 2020, involving 76110 samples from 30 provinces on the Chinese mainland. These samples included injecting drug users (IDUs), volunteer blood donors, formerly paid plasma donors (FDPs), sexually infected individuals, hemodialysis patients, a mother-to-child transmission group, an invasive operation group, and other unknown risk groups. The ten-year samples were classified into 2010 to 2015 and 2016 to 2020, comprising 30466 and 7607 samples in each period, respectively. The results are shown in Table 1. All of the studies included adopted the Simmond system. The methods used for genotyping varied from real-time fluorescent polymerase chain reaction (PCR), Tag Man probe hybridization, and Sanger sequencing to nested RT–PCR and pyrosequencing gene chip assays. The most common regions used for determining GT and subtypes were Core/E1 and NS5b, which are currently considered to be the gold standard for HCV genotyping[17,18].
| Region | Province | Number of articles included | Sample size | ||||
| 2010-2015 | 2016-2020 | Subtotal | 2010-2015 | 2016-2020 | Subtotal | ||
| Northwest | Shaanxi | 3 | 1 | 6 | 3799 | 268 | 4930 |
| Gansu | 5 | 1 | 6 | 298 | 112 | 699 | |
| Ningxia | 3 | 0 | 3 | 183 | 0 | 183 | |
| Qinghai | 2 | 0 | 4 | 150 | 0 | 562 | |
| Xinjiang | 5 | 2 | 12 | 1373 | 747 | 7694 | |
| Subtotal | 18 | 4 | 31 | 5803 | 1127 | 14068 | |
| Central | Hunan | 4 | 0 | 5 | 925 | 0 | 1877 |
| Hubei | 10 | 1 | 14 | 2602 | 84 | 2686 | |
| Henan | 7 | 0 | 8 | 840 | 0 | 1134 | |
| Subtotal | 21 | 1 | 27 | 4367 | 84 | 5697 | |
| North | Beijing | 5 | 0 | 7 | 2175 | 0 | 13222 |
| Hebei | 4 | 1 | 6 | 385 | 284 | 1427 | |
| Shanxi | 1 | 2 | 3 | 36 | 278 | 314 | |
| Tianjin | 3 | 0 | 3 | 777 | 0 | 777 | |
| Inner Mongolia | 4 | 0 | 6 | 1299 | 0 | 2203 | |
| subtotal | 17 | 3 | 25 | 4672 | 562 | 17943 | |
| South | Guangdong | 9 | 3 | 13 | 1553 | 946 | 5421 |
| Guangxi | 7 | 0 | 8 | 785 | 0 | 1629 | |
| Hainan | 2 | 1 | 5 | 283 | 128 | 1666 | |
| subtotal | 18 | 4 | 26 | 2621 | 1074 | 8716 | |
| Northeast | Liaoning | 3 | 1 | 5 | 436 | 100 | 627 |
| Jilin | 2 | 0 | 2 | 616 | 0 | 616 | |
| Heilongjiang | 3 | 0 | 3 | 0 | 360 | 360 | |
| subtotal | 8 | 1 | 10 | 1052 | 460 | 1603 | |
| Southwest | Yunnan | 7 | 3 | 13 | 538 | 2183 | 2861 |
| Guizhou | 6 | 1 | 9 | 926 | 359 | 2528 | |
| Sichuan | 10 | 5 | 18 | 3372 | 1664 | 11243 | |
| Chongqing | 3 | 0 | 3 | 993 | 0 | 993 | |
| subtotal | 26 | 9 | 43 | 5829 | 4206 | 17625 | |
| East | Shandong | 6 | 0 | 7 | 923 | 0 | 1371 |
| Jiangsu | 11 | 0 | 13 | 3805 | 0 | 4708 | |
| Anhui | 2 | 0 | 5 | 245 | 0 | 746 | |
| Zhejiang | 4 | 1 | 7 | 644 | 94 | 1619 | |
| Fujian | 2 | 0 | 5 | 147 | 0 | 638 | |
| Shanghai | 3 | 0 | 3 | 324 | 0 | 324 | |
| Jiangxi | 1 | 0 | 2 | 34 | 0 | 1052 | |
| subtotal | 29 | 1 | 42 | 6122 | 94 | 10458 | |
| Total1 | 137 | 23 | 204 | 30466 | 7607 | 76110 | |
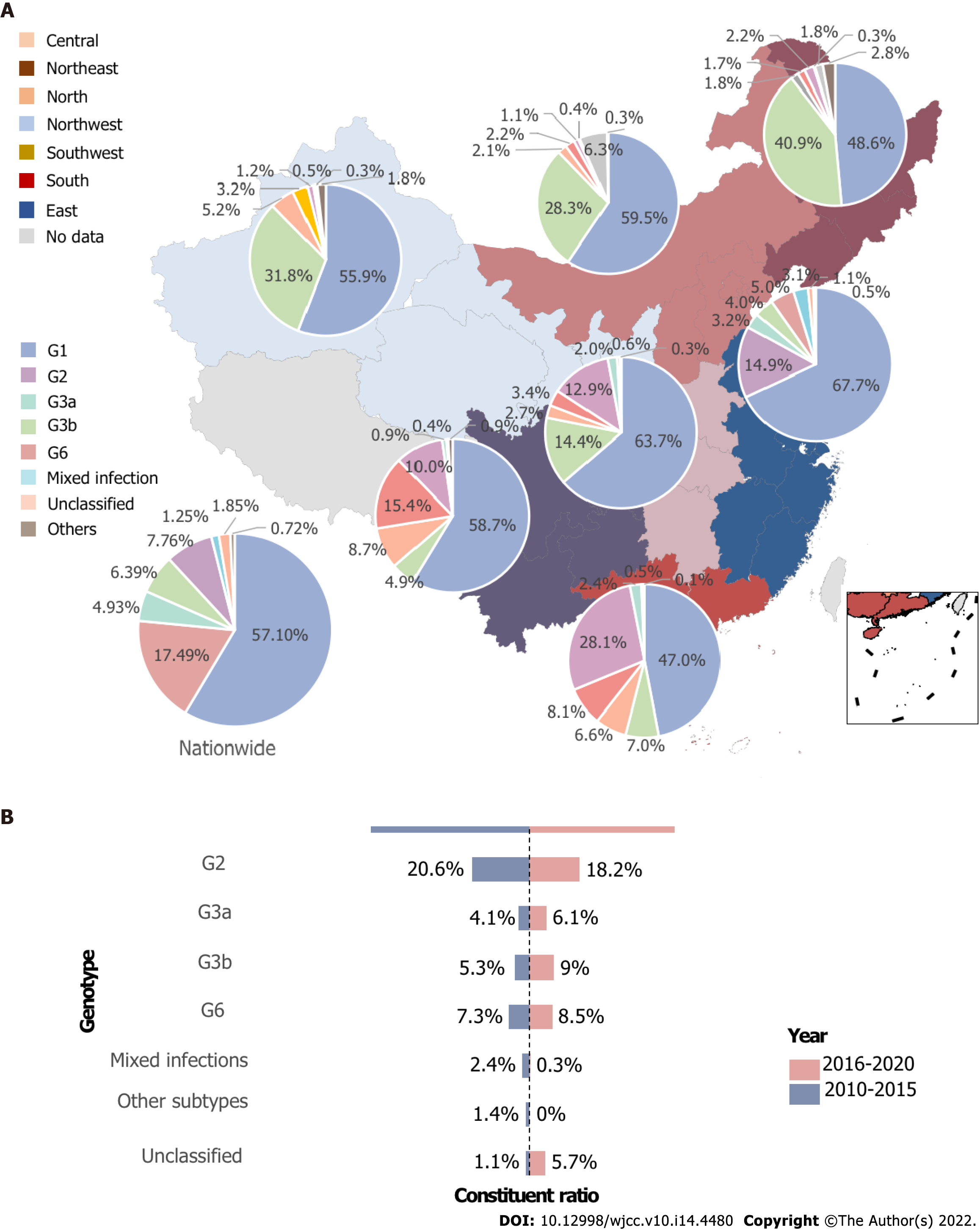
G1 accounted for 58.2% of all HCV infections, while G2 continued to follow G1 as the second major genotype nationwide, accounting for up to 18.4%. G3 altogether ranked as the third most predominant genotype, with 11.4%, and subtype 3b surpassed 3a (6.4% vs 4.9%). G6 was less common than G3 but exceeded 3a and 3b, causing 7.8% of the infections. Mixed infection, unclassified genotype, and other genotypes (for example, genotypes 4 and 5) accounted for 1.2%, 1.8%, and 0.7% of the total infections, respectively.
In the Northeast, G1 and G2 were the two most popular genotypes, with both of them accounting for more than eighty percent of all genotypes, and the proportion of G2 in this area was 40.8%, ranking the highest in seven regions, compared with 4.9% to 31.7% of other regions. The South is the only area where G1 constituted less than 50%, and the second-most common was G6 rather than G2, accounting for almost 30%, while G3b, G3a, G2 were less than 10%. The predominant genotype of the other five regions was G1, accounting for 55% to 62.5% in each area. In the north and northwest, apart from the unidentified ones and other genotypes, G3a, G3b, and G6 followed the majority G1 and G2 in order. However, in the central and eastern regions, G6 ranked third, G3b ranked fourth, and G3a ranked fifth. The structure of the genotype in South and Southwest China was quite different from that in other parts of China. The second most predominant genotype was G6 and G3b, with G3 and G6 altogether explaining 34.1% and 42.8% of all HCV infections, respectively. Mixed infection was more commonly seen in East, South, and Central China, with rates of 3.1%, 2.4%, and 2%, respectively, which are less than the 1% in other parts of the nation, as depicted in Figure 2A.
From 2010 to 2015, G1 accounted for 56.1% of the HCV infections on the Chinese mainland, while G2 and G6 followed as the second and third genotypes with 20.5% and 7.46%, respectively. G3a, G3b, and mixed genotype ranked as the fourth and fifth highest infections, and the unidentified type and other genotypes were the last two ones. However, from 2016 to 2020, as G1 remained the most popular genotype, G3a, G3b, G6, and unidentified genotypes increased, especially dramatically for G3b and the unclassified genotype. In this period, G3b rose to the third genotype with 9%. These changes are shown in Figure 2B.
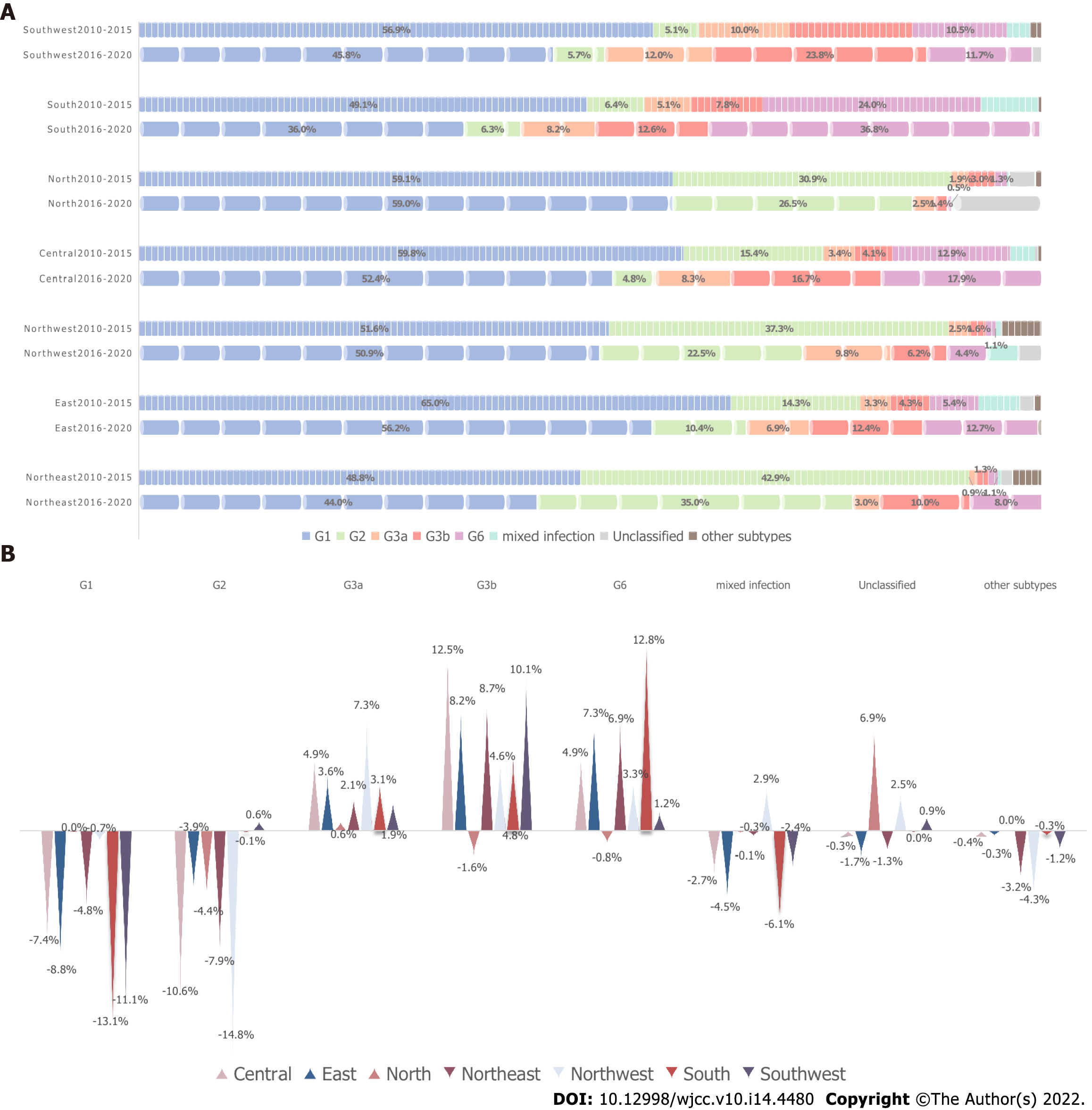
As the genotype distribution varied from area to area, the time trend of genotype distribution in the seven regions was also different. Except for the North, the proportion of genotype 1 in the other six areas all decreased from 2010-2015 to 2016-2020. The greatest decline was seen in the south at 13.1%, followed by the southwest at 11.1%. The proportion of G2 increased by 0.6% in the southwest, while other areas all decreased significantly. Mixed infections decreased in all regions other than the northwest. Compared with the mostly declining trend of G1 and G2, G3a, G3b, and G6 displayed the opposite tendency. G3b and G6 decreased only in the North, whereas G3a, G3b and G6 increased in all the other parts of China, with the greatest increases of G3a in the Northwest (7.3%), G3b in the Central (12.5%), and G6 in the South (12.8%), as shown in Figure 3.
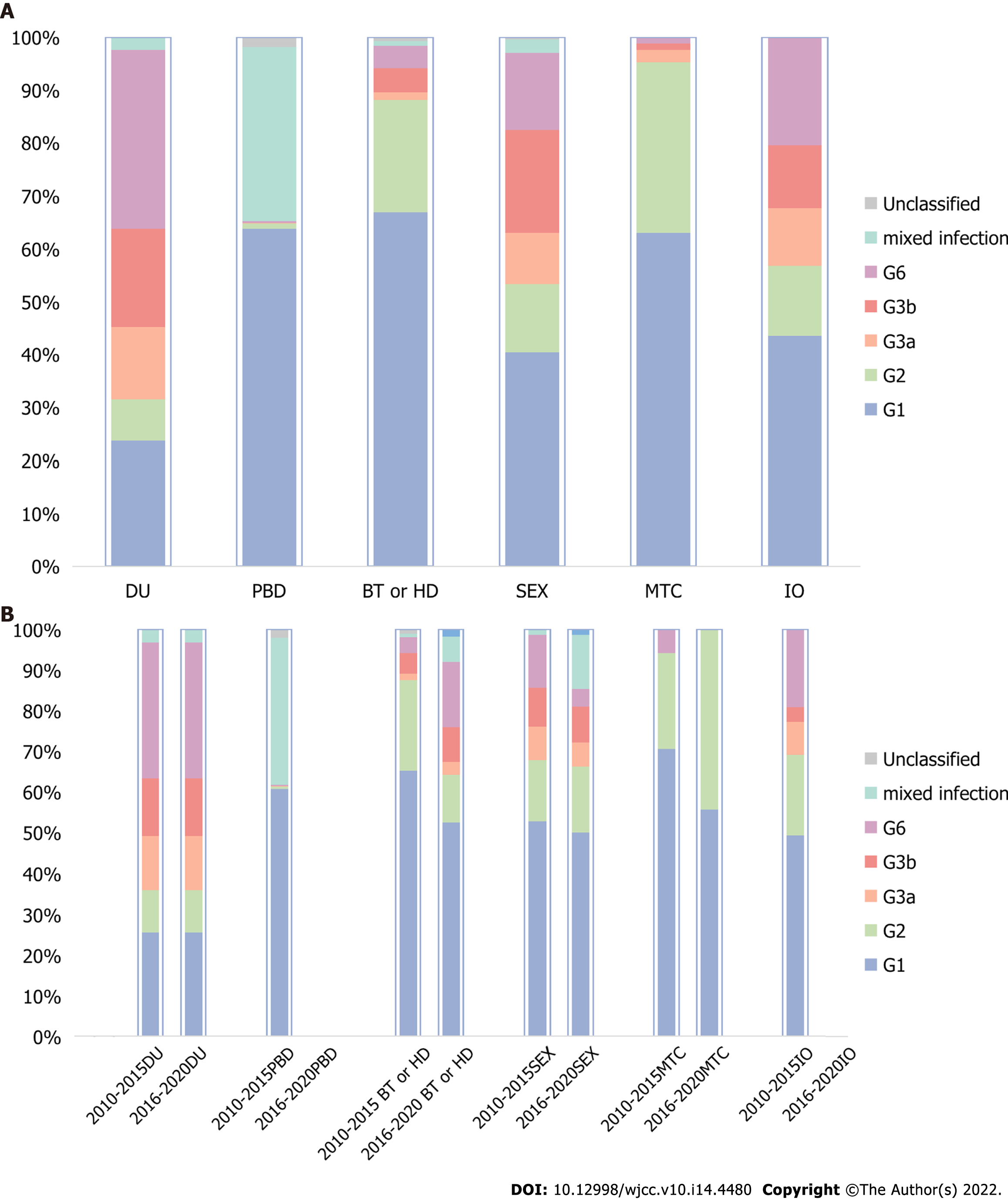
The composition ratio of HCV genotypes in different risk populations was distinctly different, as shown in Figure 4A. G1 accounted for the majority of the population of paid blood donor (PBD), blood transfusion or hemodialysis (BT or HD), and mother-to-child (MTC) transmissions. In these three populations, mixed infection was the second most predominant infection among paid blood donors, while G2 ranked second in the other two populations. The sex and invasive operation (IO) groups shared almost the same genotype composition except that G6 was higher and G3b was lower in the invasive operation group than in the sex group. The genotype distribution among drug users was quite different from all of the other risk groups. G6 ranked first, G1 and G3b ranked second and third, respectively, and G3a ranked fourth. G6, G3a, and G3b had larger proportions among drug users, sex, and invasive operation groups than other risk populations.
The time trend also varied in the various risk groups, as revealed by Figure 4B. There were no included studies from 2016-2020 in the PBD and IO groups. The distribution rarely changed between the two periods in the DU group. However, in the BT or HD populations, G1 and G2 showed obvious decreases, while G6 and 3b increased at the same time, especially G6. In contrast, in the sex group, G1, G2, and G6 all decreased, whereas the mixed infection rate increased dramatically. The MTC group had the simplest composition, with only three subtypes. As time went on, G2 increased while G1 and G6 decreased.
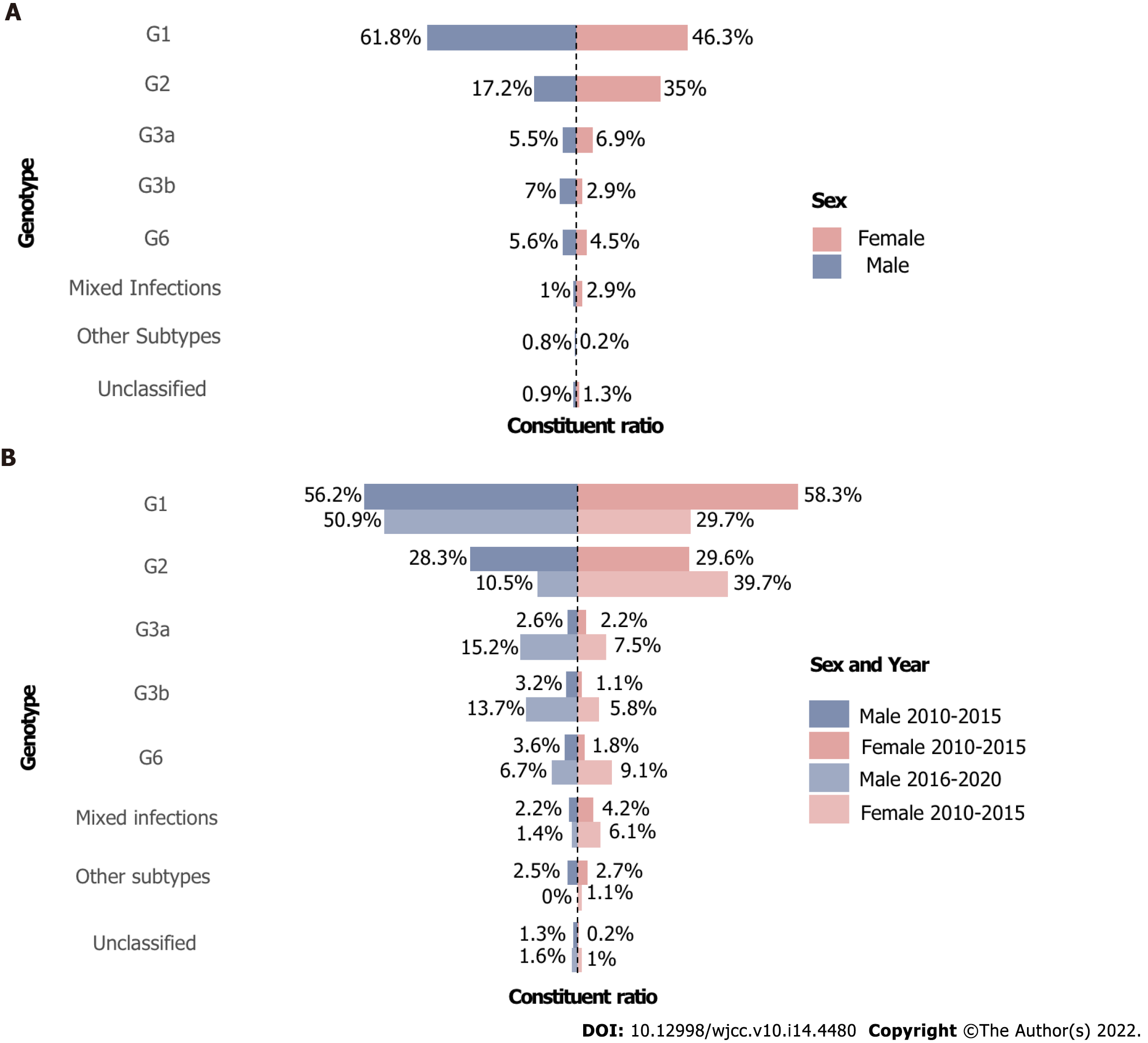
G1 and G2 were the two major genotypes in both sexes, except that G1 constituted a larger proportion in men than in women (61.8% vs 46.3%). In addition, women had more G2 and G3a and less G3b and G6 than men. Regarding the changes over time, the two populations also showed discrepancies. G1 decreased while G2 increased from 2010-2015 to 2016-2020 period and G2 became the majority of the female group in 2016-2020 with 39.7% instead of G1 with 29.7%. At the same time, G1 and G2 in the male group dropped. G3a, G3b, and G6 increased dramatically in both sexes, and the growth was even greater in men. In terms of mixed infections, they decreased in men and increased in women. The changes are demonstrated in Figure 5.
The WHO estimated that 399000 people had died from cirrhosis or HCC[6]. A clinical trial demo
At the whole-nation level, G1 remained the most common in the last ten years (the vast majority of cases were G1b), and G2 was the second most popular. G3 remained the third most prevalent, showing a tendency toward catching up to G2. G6 followed G3; the mixed infection reached 1.2%, and other genotypes were less than 1%, in line with previous studies[12,14,23-25].
G1 remained the preponderant strain in all regions, accounting for more than 60% in Central and East, although its proportion decreased significantly. G2 was more frequent in northern China. Similar observations were made in a 1540-patient study[12]. G2a was closely associated with blood transfusions and selling of blood before 1997[26]. HCV genotypes changed distinctly after 993 as the transmission of blood transfusions decreased[14]. G3a and G3b appeared to be concentrated in the southwest, and G6a was confined to the south in that study. However, G3a and G3b were also prevalent in the South, and G6 was not as common in the Central region, probably because of the economic development and migration flows in these three adjacent zones, driven by work, education, and transferring routes of transmission[27]. The unique composition of the southwest and south could be a result of their special geographic locations or caused by illegal drug transactions[14]. G3 and G6 spread from here to distant locations[3], resulting in the increases of G3 and G6 in other regions.
A multicenter, large-sample study reported that the Han Chinese population before 2012 had a relatively simple genotype constitution, with G1 dominating, G6 rarely seen, and G3 Less common[15]. Du et al[3] found that G1b decreased consistently from 72% through 63.8% to 54%, while G2a fell mildly from 16.2% to 15.4%. In contrast, G3a, G3b and G6a increased from 0.2%, 0.3% and 0.1%, respectively, to 5.4%, 7.1% and 7.5%, respectively, before and after 2000. Our study depicted the same trend over the past 10 years; G1 tended to continue to drop[3,16,25,28], and G2 decreased steadily. Simultaneously, G3a, G3b and G6 grew from 4.1%, 5.3% and 7.3% to 6.1%, 9.0% and 8.5%, respectively.
G1 declined in all regions except in the North. G2 decreased sharply in all regions but increased in the southwest, perhaps because selling blood was prevalent in central and northern areas in the 1990s[26]. From 1993, anti-HCV was screened in all blood donors, and HCV RNA was screened in them from 2015. Most of the remaining HCV infections with G1 and G2 had histories of blood transfusion before 1993 since they are older than G3 infected people[2,29]. G3 increased in all areas. G3b and G6 underwent the same increase apart from a slight decrease in the North. Interestingly, G6a replaced G2a as the second most prevalent subtype during 2001-2009 and continued to increase thereafter[3,30], reaching as high as 36.8% in the South during 2016-2020 in our study. G6 increased over time in Guangdong and spread from there to other parts of China[27]. Guangdong was responsible for one-sixth of all drug users in China at the time of 2015. New drug users are still increasing, with an estimated more than 75000 new drug users in 2018[31]. The other parts outside the south have also seen increases in G6. The South might be responsible for this increase with the origin of spreading owing to socioeconomic development. In addition, the higher viral load of G6a compared with other genotypes is also a cause of the higher infection rate[32]. The same story occurred in G3b in the southwest. Phylogeographic analysis indicated that G3b migrated from Yunnan to Guangdong and then was transmitted to other regions[31]. Furthermore, a poor response to DAAs also poses challenges to G3b elimination.
The mixed strain increased in the northwest and decreased in other regions. Mixed infections became more common, possibly owing to the increasing number of drug users, coverage of hemodialysis, population mobility, and advancement of detection methods[16].
Although G1 was the most prevalent in all risk groups, its share in the DU group was the lowest, in accordance with a previous study[3]. G2 is no longer the second most regular subtype in the various risk groups, except MTC and BT or HD populations, surpassed by G6 in DUs, as well as IO and mixed infection in PBD and by G3b in the sex group. The constitution of the PBD population in our study was quite different from that in previous studies, likely as a result of the Blood Donation Law in 1998. Illegal blood donation was forbidden, and voluntary blood donation was advocated for. The invasive operation group included higher proportions of G3 and G6, as observed in a multicenter study[14], and had similar distributions to the sex group. Transmission by IDU was more commonly seen in southern and western China, and G3 and G6 were more frequent in this group[27,33-35]. According to the phylogenetic analysis and risk factor study, G6a and G3b were more related to injecting drugs, and G3b could originate from Yunnan Province pertaining to the southwest[28,31,35]. G3a was more likely to spread by sex rather than by injecting drugs, according to an HIV/HCV coinfection study[36]. Although clean drug injection equipment is currently available, the proportion of genotypes barely changed over these ten years, and the share of G3 and G6 in the whole country continued to increase. A study regarding IDU in 2012 in Yunnan Province revealed that G6 increased while 3b and 1b decreased in the last five years before 2012[35]. A WHO desk review in 2019 found that less than half of countries valued the necessary interventions for people who inject drugs[37]. Nevertheless, in the whole population of south and southwest China, G6 and G3b rose dramatically, indicating spread from IDU to the general population. Therefore, the reuse of contaminated syringes should be prevented, especially in the south and southwest, to reduce the transmission of G3 and G6[14]. Moreover, screening and treating of patients with HCV infection in this population are also essential to preclude spread to the general population.
In contrast to a nationwide study showing that G1b and G2a constituted a larger proportion of women than men and that 3a, 3b, and 6a exhibited the opposite[13], G1b, G3b and G6 accounted for a larger proportion in men than women in our study. From 2010 to 2020, G6 accounted for 6% of men compared with 4.5% of women, in line with a blood volunteer study[30]. However, G6 in women surpassed that in men in 2016-2020, likely because of the decrease in G1. In addition, according to a meta-analysis, female drug addicts are increasing, and approximately 80% of them are engaging in sex work[37].
The main limitation of our study is that it was a literature review study based on published studies, and publication bias is inevitable. Nevertheless, our research contained as many as seventy thousand samples, well depicting a whole picture of the HCV distribution over the last ten years, the associated data of which have been lacking. Another limitation is that some of the samples were from patients in hospitals, which might not reflect the situation of the general population. However, HCV is a very silent disease without significant manifestations in the early phase of infection. It is theoretically impractical to detect all infections. Furthermore, our study merely totaled all the samples and calculated the proportions of the genotypes, and the exact prevalence of each genotype was unavailable. Hence, to acquire the actual prevalence of HCV genotypes, a cross-sectional study or survey consisting of samples from the whole nation is needed. However, the cost-effectiveness of this type of study is controversial.
In conclusion, over the past ten years, the construction of HCV genotypes has changed steadily. As various implementations were put into practice, G1 and G2 consistently decreased, and G3 and G6 became new challenges for the moment, particularly G3b. The two genotypes are mainly associated with IDUs. We should focus on the management of this population in the future regarding prevention and therapy to achieve the goal of WHO elimination in 2030.
Hepatitis C virus (HCV) infection remains a major problem worldwide since the infected population and new infections have not decreased much. Direct-acting antiviral, a revolutionary regimen to cure HCV, is highly effective except for some special genotypes. Genotype distribution has undergone great changes since the transformation of the route of transmission. Thus, being aware of the HCV genotype distribution is definitely helpful in HCV infection management.
HCV includes a variety of genotypes owing to different transmission routes in different risk populations. Although the pangenotype regimen has become the best solution to cure HCV currently, not all genotypes respond well. To better curb HCV infection and reduce the disease and economic burden of HCV infection, knowing the present genotype constitution would be quite helpful.
Our main objective was to describe the distribution of HCV genotypes in China over the past ten years and to determine the trends of distribution in the future to better manage HCV infection.
We searched all of the literature published in five electronic databases over the past 10 years. Then, we carefully selected literature that fulfilled our inclusion criteria and excluded literature that conformed to the exclusion criteria. After screening, the data of the remaining studies were extracted to build a database. Data were totaled and calculated according to different classifications: Regions, time periods, routes of transmission, and sexes.
A total of 76110 samples from 30 provinces were included in the study. Genotype 1 (G1) accounted for 58.2% nationwide, followed by G2, G6, G3b, and G3a. The constitution of genotypes varied among different regions. G6 and G3b were more common in the south and southwest (28.1%, 15.4%). The past ten years have witnessed a decrease in G1 and G2 and an increase in G3 and G6 in almost all regions. The drug-user population had the most abundant genotypes, with G6 ranking first (33.3%), followed by G1 and G3b (23.4%, 18.5%).
However, G1 and G2 accounted for the majority of HCV infections, and their decreasing tendency was clear and definite. G3 and G6 posed a new challenge in HCV infection. This study revealed the distribution of HCV genotypes in China over the past 10 years, providing information for HCV management strategies.
How to curb the development of new HCV infections and effectively cure already infected populations should be investigated in the future.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kao JT, Taiwan; Wang H, China S-Editor: Fan JR L-Editor: A P-Editor: Fan JR
| 1. | World Health Organization. Global Hepatitis Report 2017.2017. [cited 29 May 2021]. Available from: https://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/.pdf. |
| 2. | Chinese Society of Hepatology; Chinese Society of Infectious Diseases; Chinese Medical Association. [Guidelines for the prevention and treatment of hepatitis C (2019 version)]. Zhonghua Gan Zang Bing Za Zhi. 2019;27:962-979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 3. | Du G, Li X, Musa TH, Ji Y, Wu B, He Y, Ni Q, Su L, Li W, Ge Y. The nationwide distribution and trends of hepatitis C virus genotypes in mainland China. J Med Virol. 2019;91:401-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 4. | Sarin SK, Kumar M. Natural history of HCV infection. Hepatol Int. 2012;6:684-695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 5. | World Health Organization. Guidelines for the care and treatment of persons diagnosed with chronic hepatitis C virus infection.2018. [cited 29 May 2021]. Available from: https://www.who.int/publications/i/item/9789241550345.pdf. |
| 6. | Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59:318-327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 965] [Cited by in RCA: 982] [Article Influence: 89.3] [Reference Citation Analysis (1)] |
| 7. | Keikha M, Eslami M, Yousefi B, Ali-Hassanzadeh M, Kamali A, Yousefi M, Karbalaei M. HCV genotypes and their determinative role in hepatitis C treatment. Virusdisease. 2020;31:235-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 8. | Wu N, Rao HY, Yang WB, Gao ZL, Yang RF, Fei R, Gao YH, Jin Q, Wei L. Impact of hepatitis C virus genotype 3 on liver disease progression in a Chinese national cohort. Chin Med J (Engl). 2020;133:253-261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 9. | Namayandeh M, Jamalidoust M, Heydari Marandi N, Aliabadi N, Ziyaeyan A, Pouladfar G, Ziyaeyan M. Hepatitis C virus genotypes in patients with chronic hepatitis C infection in southern Iran from 2016 to 2019. Microbiol Immunol. 2020;64:762-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 10. | Grimaldi E, Della Pepa ME, Martora F, Magliocca P, Iovene MR, Coppola N, Donnarumma G, Galdiero M. Distribution of Hepatitis C Virus Genotypes and Subtypes in the Metropolitan Area of Naples, Italy, in the Era of Interferon-Free Regimens. Intervirology. 2017;60:82-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 11. | Zein NN. Clinical significance of hepatitis C virus genotypes. Clin Microbiol Rev. 2000;13:223-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 259] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 12. | Huang K, Chen J, Xu R, Jiang X, Ma X, Jia M, Wang M, Huang J, Liao Q, Shan Z, Dailey C, Song X, Lu L, Li C, Rong X, Zhang M, Fu Y. Molecular evolution of hepatitis C virus in China: A nationwide study. Virology. 2018;516:210-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 13. | Chen Y, Yu C, Yin X, Guo X, Wu S, Hou J. Hepatitis C virus genotypes and subtypes circulating in Mainland China. Emerg Microbes Infect. 2017;6:e95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 14. | Rao H, Wei L, Lopez-Talavera JC, Shang J, Chen H, Li J, Xie Q, Gao Z, Wang L, Wei J, Jiang J, Sun Y, Yang R, Li H, Zhang H, Gong Z, Zhang L, Zhao L, Dou X, Niu J, You H, Chen Z, Ning Q, Gong G, Wu S, Ji W, Mao Q, Tang H, Li S, Wei S, Sun J, Lu L, Jia J, Zhuang H. Distribution and clinical correlates of viral and host genotypes in Chinese patients with chronic hepatitis C virus infection. J Gastroenterol Hepatol. 2014;29:545-553. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 106] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 15. | Nie HM, Chen JJ, Wang R, Wang CB, Dong HL, Chen YY. [Genotypes distribution of hepatitis C virus through multi-center, large sample studies among chronic hepatitis C patients in Chinese Han population]. Zhonghua Liu Xing Bing Xue Za Zhi. 2012;33:501-504. [PubMed] |
| 16. | Zhang Y, Chen LM, He M. Hepatitis C Virus in mainland China with an emphasis on genotype and subtype distribution. Virol J. 2017;14:41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 64] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 17. | Cai Q, Zhao Z, Liu Y, Shao X, Gao Z. Comparison of three different HCV genotyping methods: core, NS5B sequence analysis and line probe assay. Int J Mol Med. 2013;31:347-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 61] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 18. | Simmonds P, Smith DB, McOmish F, Yap PL, Kolberg J, Urdea MS, Holmes EC. Identification of genotypes of hepatitis C virus by sequence comparisons in the core, E1 and NS-5 regions. J Gen Virol. 1994;75 ( Pt 5):1053-1061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 182] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 19. | Wei L, Lim SG, Xie Q, Văn KN, Piratvisuth T, Huang Y, Wu S, Xu M, Tang H, Cheng J, Le Manh H, Gao Y, Mou Z, Sobhonslidsuk A, Dou X, Thongsawat S, Nan Y, Tan CK, Ning Q, Tee HP, Mao Y, Stamm LM, Lu S, Dvory-Sobol H, Mo H, Brainard DM, Yang YF, Dao L, Wang GQ, Tanwandee T, Hu P, Tangkijvanich P, Zhang L, Gao ZL, Lin F, Le TTP, Shang J, Gong G, Li J, Su M, Duan Z, Mohamed R, Hou JL, Jia J. Sofosbuvir-velpatasvir for treatment of chronic hepatitis C virus infection in Asia: a single-arm, open-label, phase 3 trial. Lancet Gastroenterol Hepatol. 2019;4:127-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 89] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 20. | Grebely J, Page K, Sacks-Davis R, van der Loeff MS, Rice TM, Bruneau J, Morris MD, Hajarizadeh B, Amin J, Cox AL, Kim AY, McGovern BH, Schinkel J, George J, Shoukry NH, Lauer GM, Maher L, Lloyd AR, Hellard M, Dore GJ, Prins M; InC3 Study Group. The effects of female sex, viral genotype, and IL28B genotype on spontaneous clearance of acute hepatitis C virus infection. Hepatology. 2014;59:109-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 303] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 21. | Nkontchou G, Ziol M, Aout M, Lhabadie M, Baazia Y, Mahmoudi A, Roulot D, Ganne-Carrie N, Grando-Lemaire V, Trinchet JC, Gordien E, Vicaut E, Baghad I, Beaugrand M. HCV genotype 3 is associated with a higher hepatocellular carcinoma incidence in patients with ongoing viral C cirrhosis. J Viral Hepat. 2011;18:e516-e522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 128] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 22. | Bochud PY, Cai T, Overbeck K, Bochud M, Dufour JF, Müllhaupt B, Borovicka J, Heim M, Moradpour D, Cerny A, Malinverni R, Francioli P, Negro F; Swiss Hepatitis C Cohort Study Group. Genotype 3 is associated with accelerated fibrosis progression in chronic hepatitis C. J Hepatol. 2009;51:655-666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 203] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 23. | Su YY, Liu HX, Wang N. [Hepatitis C virus genotypes in China: a systematic review]. Zhonghua Liu Xing Bing Xue Za Zhi. 2013;34:80-84. [PubMed] |
| 24. | Jiang X, Lv X, Chang L, Yan Y, Ji H, Sun H, Guo F, Rodgers MA, Yin P, Wang L. Molecular characterization of hepatitis C virus for subtype determination and resistance-associated substitutions detection among Chinese voluntary blood donors. Antiviral Res. 2020;181:104871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 25. | Wang CB, Cheng ZX, Chen JJ, Chen YY, Nie HM, Ling QH, Dong YN. Epidemiological characteristics and risk factors of hepatitis C virus genotype 1 infection: a national epidemiological survey of Chinese Han population. Eur Rev Med Pharmacol Sci. 2016;20:1052-1056. [PubMed] |
| 26. | Peng J, Lu Y, Liu W, Zhu Y, Yan X, Xu J, Wang X, Wang Y, Sun Z. Genotype Distribution and Molecular Epidemiology of Hepatitis C Virus in Hubei, Central China. PLoS One. 2015;10:e0137059. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 27. | Yan J, Fu XB, Zhou PP, He X, Liu J, Huang XH, Yu GL, Yan XG, Li JR, Li Y, Lin P. Complicated HCV subtype expansion among drug users in Guangdong province, China. Infect Genet Evol. 2019;73:139-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 28. | Ju W, Yang S, Feng S, Wang Q, Liu S, Xing H, Xie W, Zhu L, Cheng J. Hepatitis C virus genotype and subtype distribution in Chinese chronic hepatitis C patients: nationwide spread of HCV genotypes 3 and 6. Virol J. 2015;12:109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 29. | Dong ZX, Zhou HJ, Wang JH, Xiang XG, Zhuang Y, Guo SM, Gui HL, Zhao GD, Tang WL, Wang H, Xie Q. Distribution of hepatitis C virus genotypes in Chinese patients with chronic hepatitis C: correlation with patients' characteristics and clinical parameters. J Dig Dis. 2012;13:564-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 30. | Fu Y, Wang Y, Xia W, Pybus OG, Qin W, Lu L, Nelson K. New trends of HCV infection in China revealed by genetic analysis of viral sequences determined from first-time volunteer blood donors. J Viral Hepat. 2011;18:42-52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 63] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 31. | Wang M, Liao Q, Xu R, Song D, Huang J, You Q, Shan Z, Huang K, Rong X, Fu Y. Hepatitis C virus 3b strains in injection drug users in Guangdong Province, China, may have originated in Yunnan Province. Arch Virol. 2019;164:1761-1770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 32. | Rong X, Xu R, Xiong H, Wang M, Huang K, Chen Q, Li C, Liao Q, Huang J, Xia W, Luo G, Ye X, Zhang M, Fu Y. Increased prevalence of hepatitis C virus subtype 6a in China: a comparison between 2004-2007 and 2008-2011. Arch Virol. 2014;159:3231-3237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 33. | Chen F, Zhang J, Guo F, Wen B, Luo S, Yuan D, Lin Y, Ou W, Tang P, Dai G, Li F, Liu W, Qu X. Hepatitis B, C, and D virus infection showing distinct patterns between injection drug users and the general population. J Gastroenterol Hepatol. 2017;32:515-520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 34. | Tao J, Liang J, Zhang H, Pei L, Qian HZ, Chambers MC, Jiang Y, Xiao Y. The Molecular Epidemiological Study of HCV Subtypes among Intravenous Drug Users and Non-Injection Drug Users in China. PLoS One. 2015;10:e0140263. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 35. | Tian D, Li L, Liu Y, Li H, Xu X, Li J. Different HCV genotype distributions of HIV-infected individuals in Henan and Guangxi, China. PLoS One. 2012;7:e50343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 36. | World Health Organization. Accelerating access to hepatitis C diagnostics and treatment.2021. [cited 29 May 2021]. Available from: https://www.who.int/publications/i/item/9789240027077.pdf. |
| 37. | Xia X, Luo J, Bai J, Yu R. Epidemiology of hepatitis C virus infection among injection drug users in China: systematic review and meta-analysis. Public Health. 2008;122:990-1003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 122] [Article Influence: 7.2] [Reference Citation Analysis (0)] |