Copyright
©The Author(s) 2025.
World J Clin Oncol. Aug 24, 2025; 16(8): 107208
Published online Aug 24, 2025. doi: 10.5306/wjco.v16.i8.107208
Published online Aug 24, 2025. doi: 10.5306/wjco.v16.i8.107208
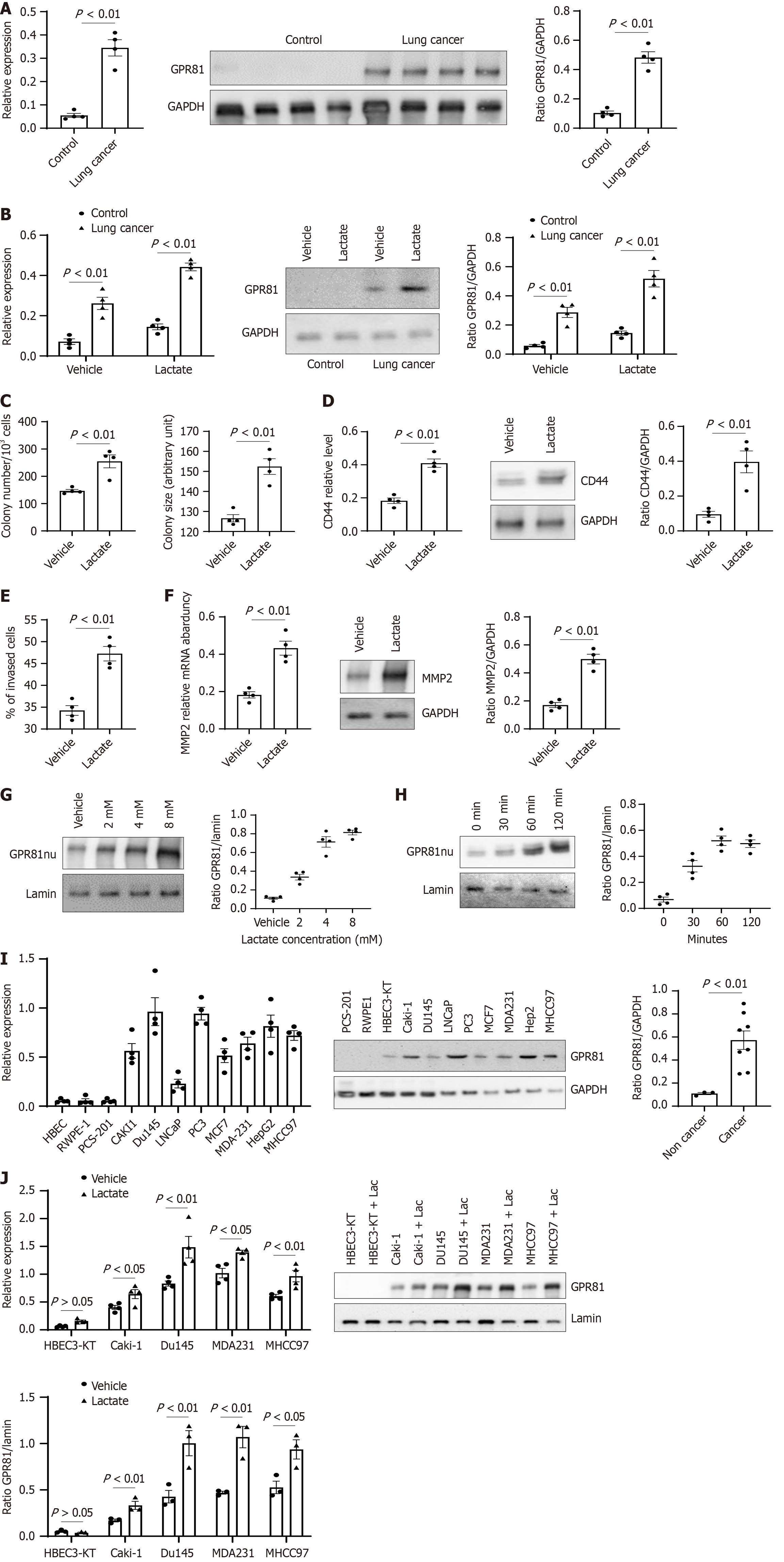
Figure 1 Lactate promotes GPR81 expression and GPR81 nuclear translocation in cancer cells.
A: GPR81 expression level in lung cancer and control cells were measured with reverse transcriptase-PCR (RT-PCR) (left panel) and western blot (middle panel). Densitometry values summarizing western blot data are shown in the right panel. GAPDH served as a loading control; B: GPR81 expression level in lung cancer and control cells cultured under vehicle/Lactate (5 mmol/L) conditions were measured with RT-PCR (left panel) and western blot (middle panel). Densitometry values summarizing western blot data are shown in the right panel. GAPDH served as a loading control; C: Lung cancer cells self-renewal was assessed in the colony forming assay; D: Cancer stemness marker CD44 and cell proliferation marker Ki67 were quantified with RT-PCR and western blot analysis; E: Lung cancer cell invasion was assessed; F: Cell motion marker MMP2 was quantified in RT-PCR and Western blot analyses. A-F: n = 4, each of control and lung cancer cells (Control 202, 205, 272, 392; lung cancer A549, 661, 858, 645); G: Lung cancer cells were cultured with varying concentration of lactate or vehicle and the cells were collected at 4 hours and nuclear fractions isolated for Western blot analysis. Lamin served as a loading control; H: Lung cancer cells were cultured with 5 mmol/L of lactate and the cells were collected at the indicated time and nuclear fractions were isolated for Western blot analysis. G and H: n = 4 Lung cancer cell lines (A549, 645, 661, 858); I: GPR81 expression level in cancer and control cells was measured with RT-PCR (left panel) and western blot (middle panel). Densitometry values summarizing western blot data are shown in the right panel. GAPDH served as a loading control; J: GPR81 Level in cancer and control cells cultured under vehicle/Lactate (5 mmol/L) conditions were measured with RT-PCR (left panel) and the nuclear fraction was analysed with western blot (middle panel). Densitometry values summarizing western blot data are shown in the right panel. Lamin served as a loading control.
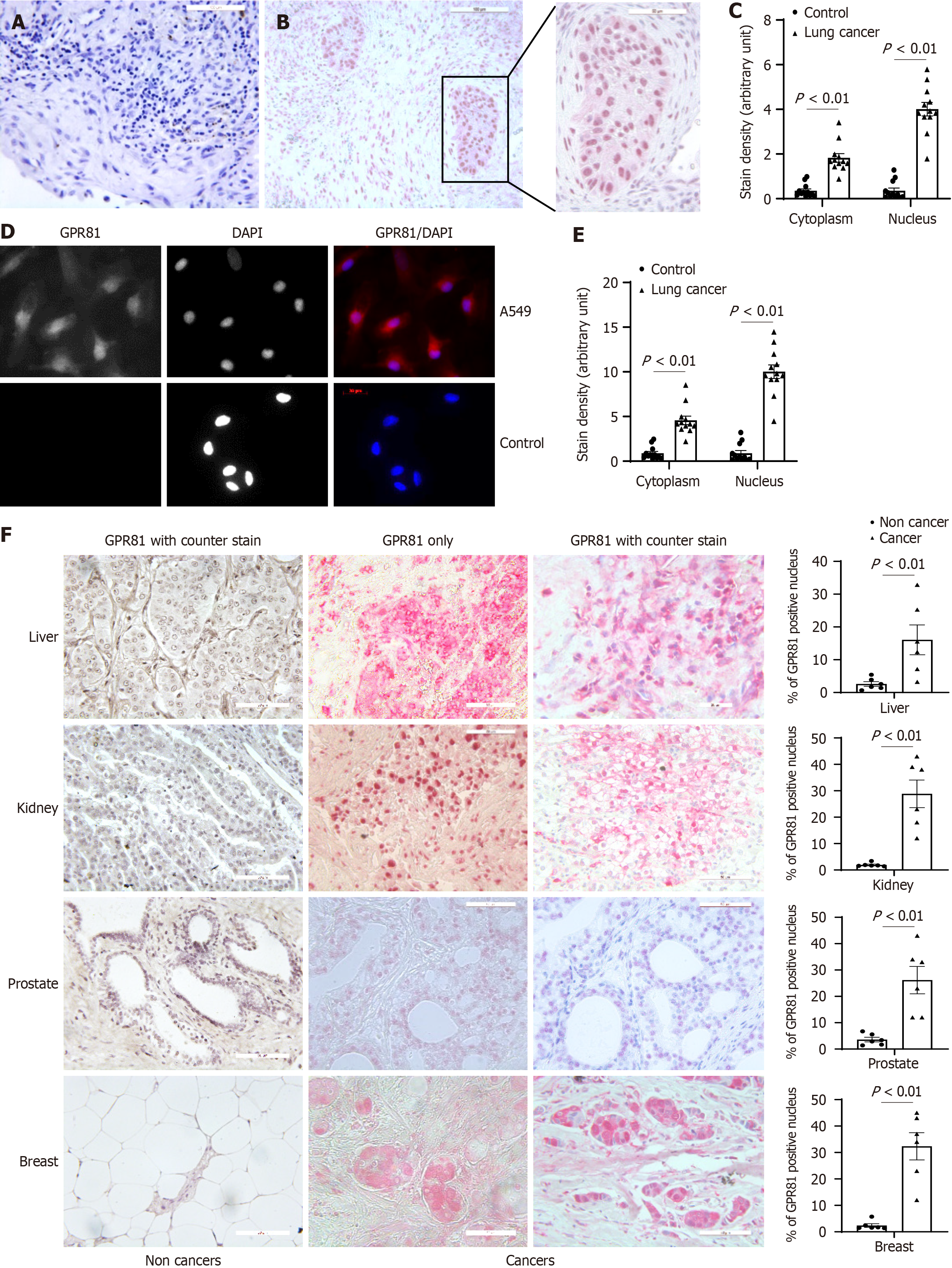
Figure 2 Nuclear GPR81 is present in non-small cell lung cancer cells and in non-small cell lung cancer tissue sections.
Immuno
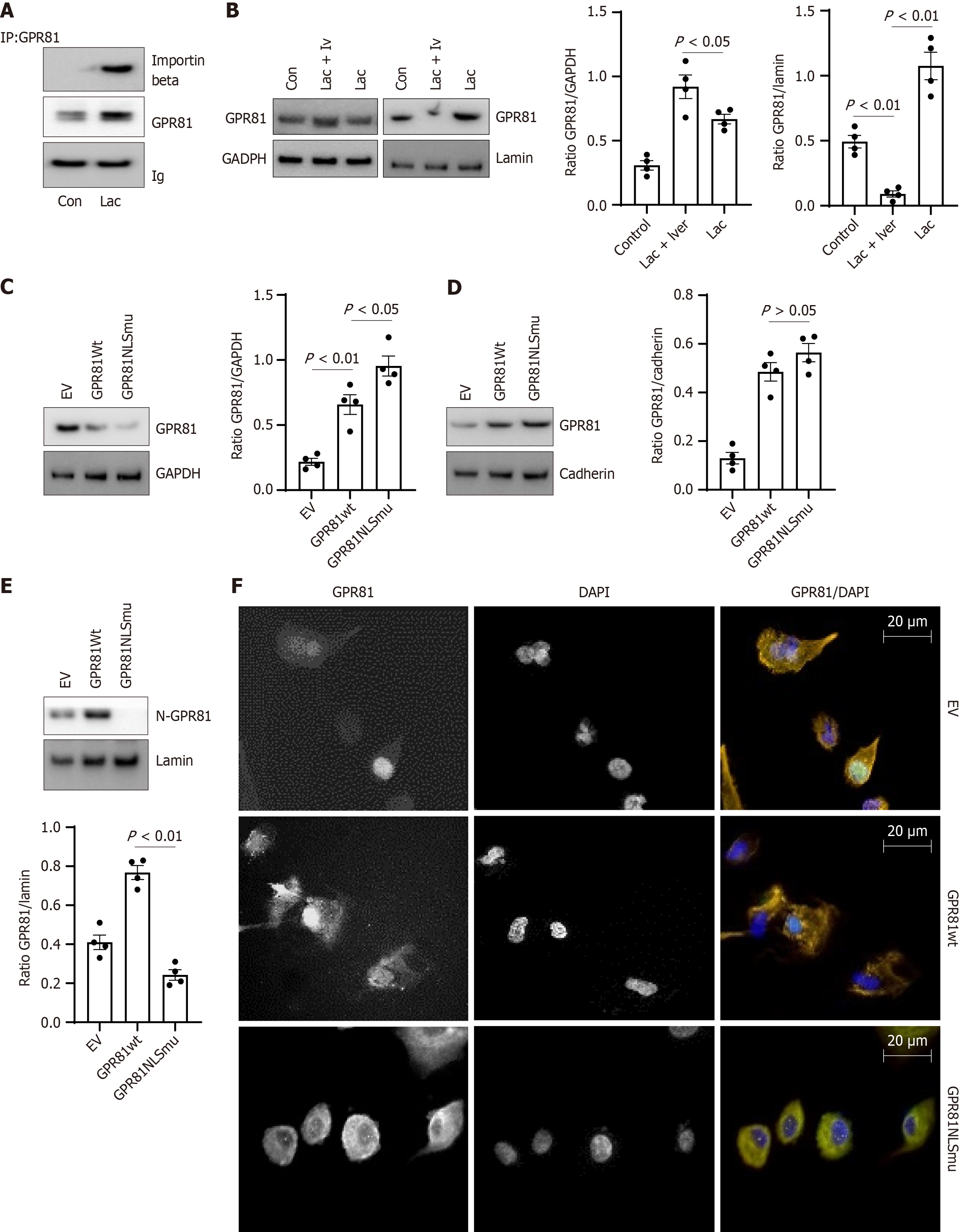
Figure 3 GPR81 is translocated to cell nucleus with importin beta.
A: Anti-importin beta was used to immunoprecipitate proteins from lung cancer cell with lactate or vehicle overnight treatment and the precipitated material was used in Western blot analysis. Densitometry values summarizing western blot data are shown in the right panel. Ig served as a loading control. Cell H661 was used; B: GPR81 expression levels were quantified in lung cancer cell exposed to 5 mmol/L lactate, with or without ivermectin (10 μmol/L) vs vehicle control in western blot (middle panel). Densitometry values summarizing western blot data are shown in the right panel. GAPDH served as a loading control. A549, H661, H858, H645 were used; C-E: GPR81 expression level in cell fractions of GPR81 NLS mutant transduced lung cancer cells, GPR81 wild-type transduced and empty vector transduced cells cultured under lactate or vehicle conditions were measured with western blot; D and E: Loading markers were GAPDH for cytoplasm, cadherin for cell membrane and lamin for nucleus. Densitometry values summarizing western blot data are shown in the right panel. GAPDH served as a loading control. n = 4 Lung cancer cells (A549, H661, H858, H645); F: Immunoflurescent stain was performed on human lung cancer A549 cells (transduced with Empty vector, GPR81 wild-type and GPR81 NLSmutant as marked) with GPR81 antibody. Scale bar = 20 μmol/L.
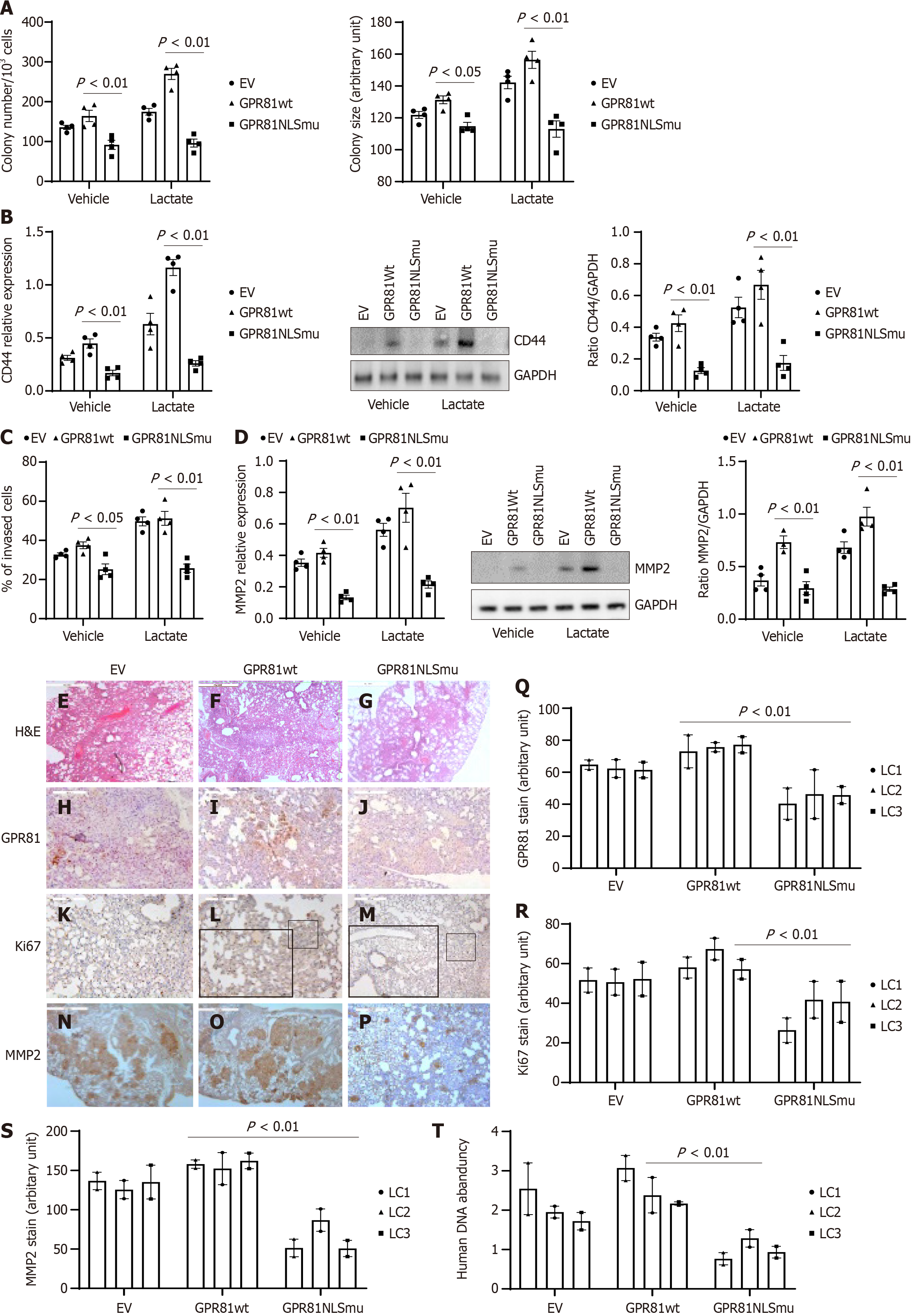
Figure 4 Inhibition of GPR81 nuclear translocation reduces cancer cell self-renewal and invasion.
Lung cancer cells were transduced with empty vector, GPR81 wild-type or NLS mutant and cultured under vehicle or 5 mmol/L lactate conditions. A: Lung cancer cell self-renewal was assessed in the colony forming assay; B: Cancer stemness marker CD44 and proliferation marker Ki67 were quantified with reverse transcriptase-PCR (RT-PCR) and western blot analysis; C: Lung cancer cells invasion was assessed; D: Cell motion marker MMP2 was quantified in RT-PCR and western blot analyses using cell lines H549, H661, H848 and H448. Data are shown as mean ± SE. 3 technical replicates were performed for all experiments. GPR81 translocation affects lung cancer progression in vivo. NSG mice implanted with lung cancer cells (A549, H848 and H661) stably transduced with either empty vector, GPR81 NLS mutant, or wild-type GPR81 via IV (3 × 105 cells/50µl); 6 mice/group; E-P: Serial 4 µm sections of right lung tissue from lung cancer tumors transduced with GPR81NLS mutant or wild-type GPR81. Representative H&E (E-G; scale bar: 500 µm) and immunohistochemistry (IHC) stains assessing GPR81, Ki67 and MMP2 positivity. IHC identifies GPR81, Ki67 and MMP2 and determines the distribution of GPR81 expression cells (H-J), Ki67 expressing cells (K-M) and MMP2 expressing cells (N-P). IHC for GPR81 and Ki67 was conducted to assess the distribution of human cells expressing GPR81, Ki67 and MMP2 protein expressing cells from mice from tumors of lung cancer cells transduced with empty vector, GPR81 wild-type (GPR81wt) or GPR81 NLS mutant (GPR81NLSmu). Inner frame shows an enlarged image from the indicated area. Scale bar = 200 μmol/L; Q-S: GPR81, Ki67 and positive cells in lung cancer cell explanted mice lung tissue sections; T: Cell quantification was conducted with quantitative PCR.
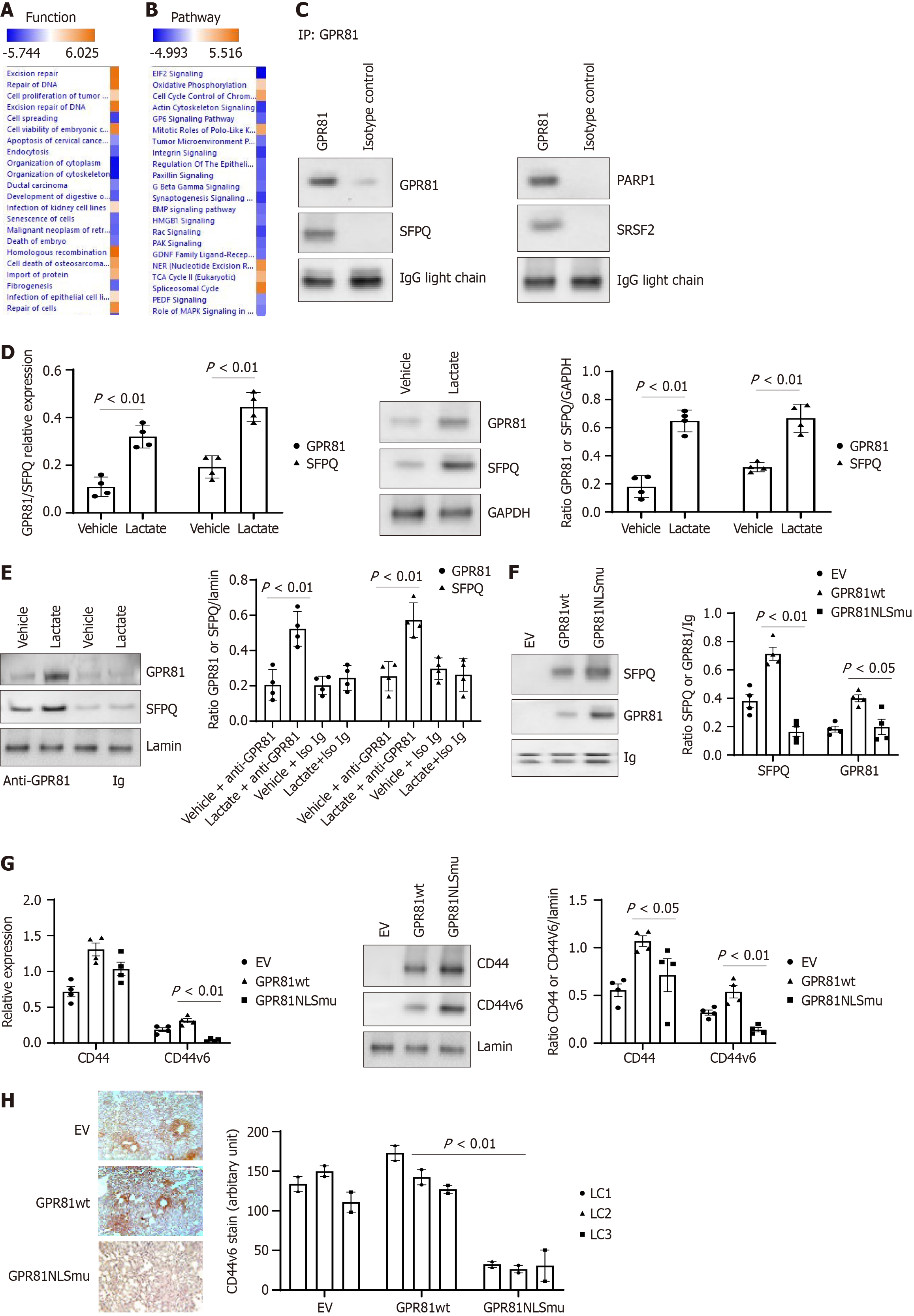
Figure 5 The interaction of GPR81 with other nuclear proteins affects lung cancer cell function.
Proteins identified from GPR81 immunoprecipitated proteins in lung cancer cells in Proteomics analysis were applied to Ingenuity Pathway Analysis (IPA). A: Top functions associated with the lung cancer cell dataset as shown by IPA pathway analysis; B: Top canonical pathways associated with proteins from the lung cancer cell dataset as shown by IPA pathway analysis. Cell functions, or pathways identified are represented on the y-axis. The x-axis corresponds to the –log of the P-value (Fisher’s exact test) and the orange points on each pathway bar represent the ratio of the number of proteins in a given pathway that meet the cutoff criteria, divided by the total number of proteins that map to that pathway; C: Immunoprecipitation and western blot analysis with GPR81 bound proteins. The nuclear fraction from A549 was used in immunoprecipitation with the GPR81 antibody. Then SFPQ, PARP1 and SRSF2 antibodies were used in western blot analysis with the GPR81 pull down portion of the lung cancer nuclear fraction. Lactate promotes SFPQ expression and formation of GPR81/SFPQ complex in lung cancer cells; D: GPR81 reverse transcriptase-PCR (RT-PCR) was conducted with lung cancer cells cultured under vehicle/Lactate (5 mmol/L) conditions. (left panel). GPR81 antibody was used in immunoprecipitation with lung cancer cells cultured under vehicle/Lactate (5 mmol/L) conditions and the SFPQ levels were measured with western blot (middle panel). Densitometry values summarizing western blot data are shown in the right panel. GAPDH served as a loading control; E: GPR81 immunoprecipitation was conducted with the cell nuclear fraction from lung cancer cell cultured under vehicle/Lactate (5 mmol/L) conditions. Then the protein bands were detected with anti-GPR81 and SFPQ antibodies. Western blot was shown in the left panel. Densitometry values summarizing western blot data are shown in the right panel. Lamin served as a loading control; F: Lung cancer cells transduced with Lenti virus with empty vector, GPR81 wild-type or GPR81 NLS mutant, antibody was used in immunoprecipitation with lung cancer cell nuclear fraction; G: The same set of cells was used to analyze CD44 and CD44v6 expression. The CD44 and CD44v6 Levels were measured with RT-PCR (left panel) and western blot (middle panel). Densitometry values summarizing western blot data are shown in the right panel. GAPDH served as a loading control. Lung cancer cell A549, H661, H858, H645 were used in figure D-G; H: Anti-CD44v6 was used to assess the distribution of human cells expressing CD44v6 protein expressing cells from mice lung regions from mice receiving lung cancer cells transduced with empty vector, GPR81 wild-type (GPR81wt) (middle panel) or GPR81 NLS mutant (GPR81NLSmu) (right panel). Scale bar = 200 μmol/L. Quantification was summed up in the bottom figure. Image J was used to quantify CD44v6 protein positive cells in lung cancer cell explanted mice lung tissue sections.
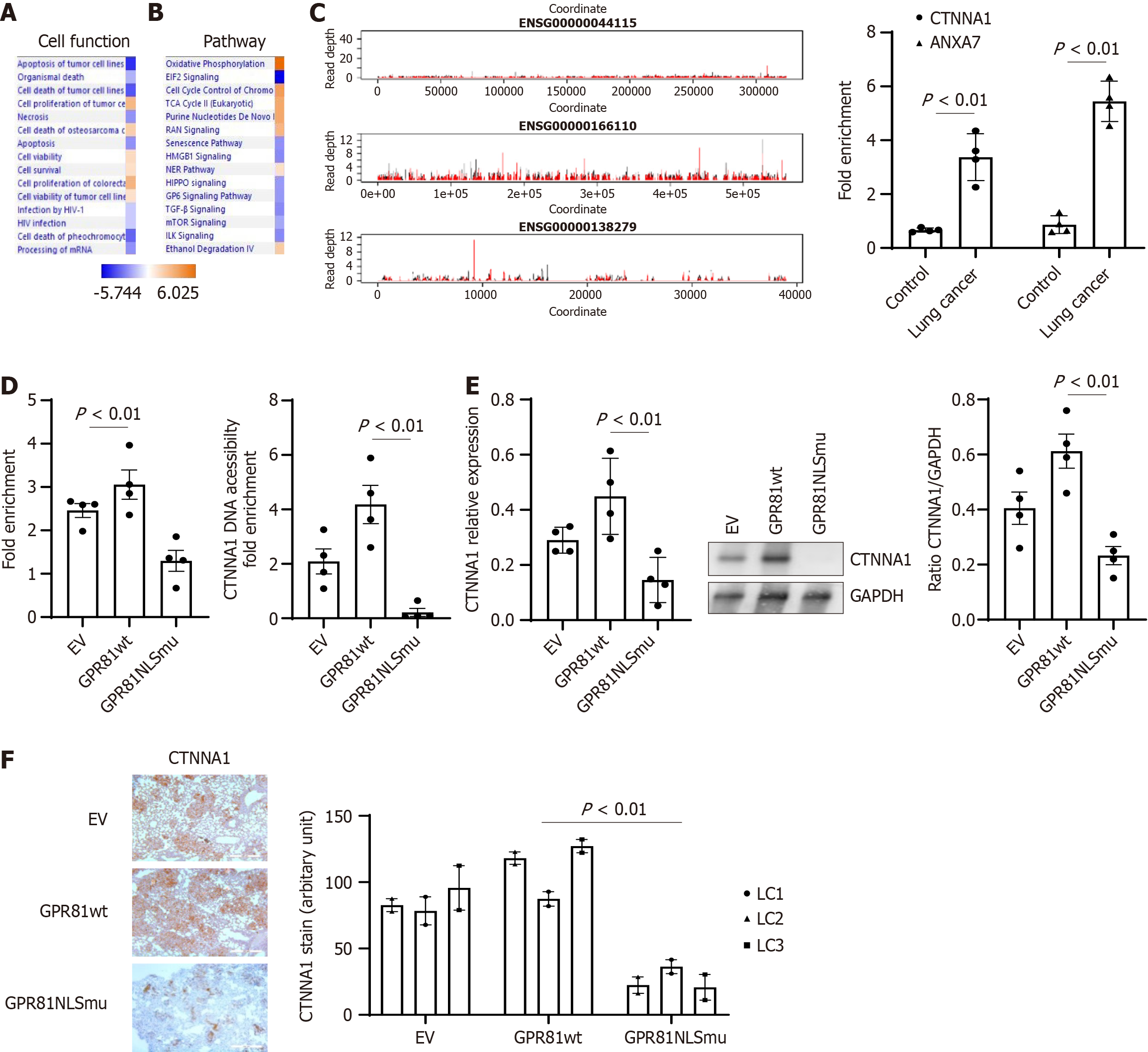
Figure 6 GPR81 interacts with genes and change gene expression.
Genes identified from GPR81 immunoprecipitated chromatin in lung cancer cells in Chromatin immunoprecipitation (ChIP) sequencing analysis were applied to Ingenuity Pathway Analysis (IPA). A: Top cell functions associated with the lung cancer cell dataset as shown by IPA pathway analysis; B: Top canonical pathways associated with proteins from the lung cancer cell dataset as shown by IPA pathway analysis. Cell functions, or pathways identified are represented on the Y-axis. The X-axis corresponds to the –log of the P-value (Fisher’s exact test) and the orange points on each pathway bar represent the ratio of the number of proteins in a given pathway that meet the cutoff criteria, divided by the total number of proteins that map to that pathway; C: The read depth plots for the GPR81 ChIP peaks of gene interest. Each plot shows the gene body. The red line shows the mean read depth across the +lactate samples and the black line shows the mean across the -lactate samples. The vertical blue lines show the boundaries of the gene body. Right panel: ChIP assays were performed on cross linked chromatin from nuclear fractions from control and lung cancer cells with CTNNA1 and ANXA7 primers; D: ChIP assays were performed on cross linked chromatin from nuclear fractions from GPR81 wt and GPR81 NLSmutant cells (n = 4 cell lines each) using immune GPR81 antibody (Isotype IgG as blank control) (left panel). Right panel: DNA Accessibility Assays were performed on cross linked chromatin from nuclear fractions from GPR81 wt and GPR81 NLSmutant cells (n = 4 cell lines A549, H661, H858, H645); E: CTNNA1 expression level was quantified with reverse transcriptase-PCR and western blot analysis in above set of cells; F: Anti-CTNNA1 was used to assess the distribution of human cells expressing CTNNA1 protein expressing cells from mice lung regions from mice receiving lung cancer cells transduced with empty vector, GPR81 wild-type (GPR81wt) (left panel) or GPR81 NLS mutant (GPR81NLSmu) (right panel). Scale bar = 200 μmol/L. Quantification was summed up in the bottom figure. Image J was used to quantify CTNNA1 protein positive cells in lung cancer cell explanted mice lung tissue sections.
- Citation: Yang L, Kono T, Gilbertsen A, Li Y, Sun B, Jacobson BA, Karam S, Dehm SM, Henke CA, Kratzke RA. GPR81 nuclear transportation is critical for cancer growth and progression in lung and other solid cancers. World J Clin Oncol 2025; 16(8): 107208
- URL: https://www.wjgnet.com/2218-4333/full/v16/i8/107208.htm
- DOI: https://dx.doi.org/10.5306/wjco.v16.i8.107208