Published online Jul 22, 2021. doi: 10.4291/wjgp.v12.i4.59
Peer-review started: March 17, 2021
First decision: April 15, 2021
Revised: April 24, 2021
Accepted: May 19, 2021
Article in press: May 19, 2021
Published online: July 22, 2021
Processing time: 120 Days and 15.9 Hours
Despite advances in antiretroviral treatment (ART), human immunodeficiency virus (HIV) continues to be a major global public health issue owing to the increased mortality rates related to the prevalent oncogenic viruses among people living with HIV (PLWH). Human papillomavirus (HPV) is the most common sexually transmitted viral disease in both men and women worldwide. High-risk or oncogenic HPV types are associated with the development of HPV-related malignancies, including cervical, penile, and anal cancer, in addition to oral cancers. The incidence of anal squamous cell cancers is increasing among PLWH, necessitating the need for reliable screening methods in this population at risk. In fact, the currently used screening methods, including the Pap smear, are invasive and are neither sensitive nor specific. Investigators are interested in circulatory and tissue micro ribonucleic acids (miRNAs), as these small non-coding RNAs are ideal biomarkers for early detection and prognosis of cancer. Multiple miRNAs are deregulated during HIV and HPV infection and their deregulation contributes to the pathogenesis of disease. Here, we will review the molecular basis of HIV and HPV co-infections and focus on the pathogenesis and epidemiology of anal cancer in PLWH. The limitations of screening for anal cancer and the need for a reliable screening program that involves specific miRNAs with diagnostic and therapeutic values is also discussed.
Core Tip: Human papillomavirus (HPV) is the most common sexually transmitted infection worldwide. People living with human immunodeficiency virus (HIV) are at high risk of acquiring HPV infection and developing HPV-associated malignancies, including anal cancer, independent of acquired immune deficiency syndrome. This high risk is associated with several factors including the dysregulation of cellular micro ribonucleic acids (miRNAs) and the direct interaction between HIV and HPV. Dysregulated miRNAs are known to play a role in HIV, HPV infections, and HPV-related cancers. Here, we discuss the role of HIV in HPV-associated pathogenesis and important implications of miRNAs on current screening for and early detection of anal cancer.
- Citation: Al Bitar S, Ballouz T, Doughan S, Gali-Muhtasib H, Rizk N. Potential role of micro ribonucleic acids in screening for anal cancer in human papilloma virus and human immunodeficiency virus related malignancies. World J Gastrointest Pathophysiol 2021; 12(4): 59-83
- URL: https://www.wjgnet.com/2150-5330/full/v12/i4/59.htm
- DOI: https://dx.doi.org/10.4291/wjgp.v12.i4.59
At the end of 2019, approximately 36900000 people were estimated to be living with human immunodeficiency virus (HIV)[1]. Despite the advances in antiretroviral treatment (ART) and the increase in number of patients accessing ART since 2010, cancer mortality in people living with HIV (PLWH) remains high[2]. Mortality from HIV associated illnesses decreased substantially since its peak in 2004 mainly due to a decrease in the incidence of opportunistic infections. With the introduction of highly active ART in 1996, there has been a substantial improvement of clinical outcomes in PLWH[3-5]. This has brought an increase in life expectancy and a change in the age distribution of PLWH[6,7]. The risk of developing cancer increases with age; and as PLWH are now aging, the burden of cancer has substantially increased in this population. Since the beginning of the epidemic, HIV was associated with Kaposi’s sarcoma, aggressive B-cell lymphomas, and invasive cervical cancer. Diagnosis of these cancers in PLWH confers the diagnosis of acquired immune deficiency syndrome (AIDS) and are thus termed as AIDS-defining cancers. Other types of cancers are non-AIDS defining, such as anal carcinoma, Hodgkin lymphoma, hepatocellular carcinoma, and lung cancer. These have been increasingly recognized to occur in PLWH and have become a leading cause of death[8-11]. One reason behind the increase in the rate of non-AIDS defining cancers in PLWH is increased prevalence of oncogenic viruses in this population, one of which is human papilloma virus (HPV)[12].
HPV is the most common sexually transmitted viral disease in both men and women worldwide[13]. HPV targets epithelial cells and includes more than 200 types that exist with genomic differences. About 40 types specifically infect the anogenital epithelium and upper digestive tract, among which 15-20 types are considered as high-risk HPV (HR-HPV), including HPV16 and HPV18[14]. Oncogenic or HR-HPV types are associated with the development of high-grade intraepithelial lesions and consequently, cancers of the anogenital region and oropharynx. About 99.9% of cervical cancers and 80%-90% of anal squamous cell cancers (ASCC) are associated with infection with HR-HPV[15]. While the incidence of cervical cancer has remained stable over the years, the incidence of ASCC has increased, particularly in PLWH[16]. With these increasing trends, it is imperative to screen for anal cancer in this high-risk population. However, many of the currently used screening methods, including the Pap smear, are invasive and require specialized equipment. In addition, the Pap smear is neither specific (specificity is approximately 75%) nor sensitive (approximately 55%)[17]. Thus, identification of non-invasive and more effective methods is crucial.
Micro ribonucleic acids (miRNAs) have emerged as clinically useful molecular biomarkers for better management and treatment of many types of cancers. In HPV-associated cancers, miRNAs have been shown to be deregulated and involved in the pathogenesis of the disease. Given that the molecular mechanisms involved in anal cancer development during HIV infection are still unclear, characterization of miRNA expression in the context of HIV infection and anal cancer and the identification of relevant biomarkers could help elucidate the potential role of HIV and HPV in the progression of ASCC, as well as help prevent and treat anal cancer.
In this review, we will focus on the mechanisms and pathogenesis underlying HIV and HPV infections and the epidemiology and risk factors of anal cancer. We will also discuss the need for anal cancer screening, especially in HIV-infected individuals and the potential implementation of miRNAs as screening and therapeutic tools in high-risk populations.
HIV-1 is the causative agent of AIDS. HIV-1 is a retrovirus whose genome is composed of 2 copies of single-stranded RNA molecules. HIV genome has 9 open reading frames and encodes for precursor proteins that give rise to 15 viral proteins. These proteins can be classified into structural and regulatory. The structural proteins include Gag, Env, and Pol. The matrix, capsid (CA), nucleocapsid, and p6 proteins are generated from Gag precursor and make up the core of the virus particle. The Env polyprotein is subsequently processed to generate the envelope proteins, gp120 and gp41. The pol gene encodes viral enzymes: Protease (PR), reverse transcriptase, and integrase. The HIV genome also encodes essential regulatory elements, Tat and Rev, and accessory regulatory proteins: Vif, Vpr and Nef[18].
HIV envelope glycoprotein mediates HIV cell entry by binding to its primary receptor, CD4 molecule, expressed on target cells, such as CD4+ T cells, monocytes, and macrophages. HIV entry also requires binding a chemokine coreceptor, CCR5 or CXCR4[19]. Viral entry is followed by reverse transcription of the viral RNA genome, integration of the provirus into cellular genome, synthesis of viral genome, and assembly and budding of the newly formed virions. When no new viral proteins are produced, infected cells can revert to latency[20].
HIV targets and kills CD4+ T cells, monocytes, macrophages, and microglial cells, however the main targets of HIV infection and subsequent destruction are the CD4+ T cells[21,22].
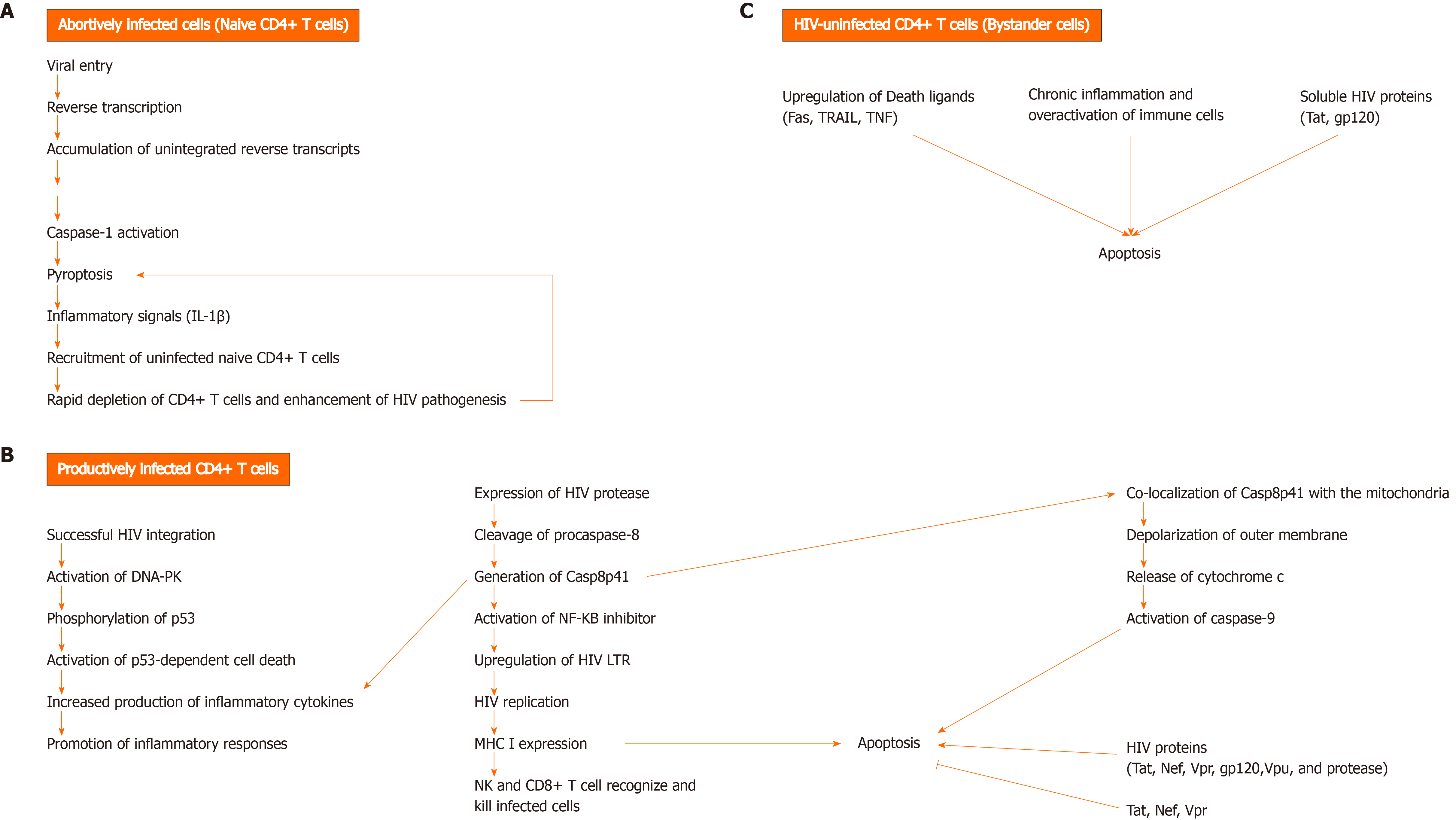
The mechanisms underlying CD4+ T cell death are still not well defined. The permissivity status of CD4+ T cells during HIV infection determines the pathway by which these cells die (Figure 1). Abortively-infected[23,24], productively infected[25-28], and HIV-uninfected (bystander) CD4+ T cells undergo cell death through different mechanisms[27].
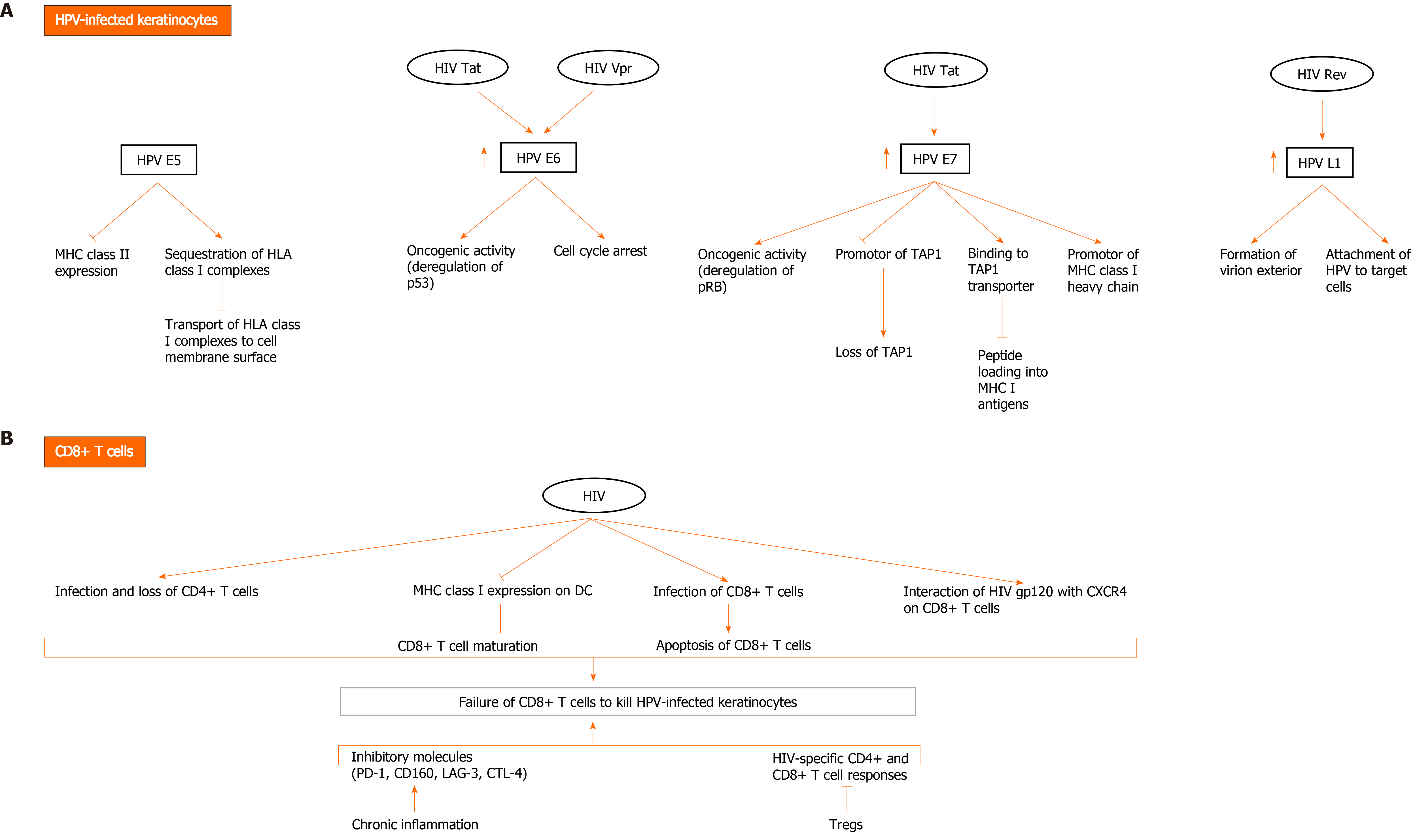
In addition to progressive CD4 lymphopenia, HIV infection is also associated with impaired HIV-specific CD8+ T cell responses. CD8+ T cells play an important role in eliminating viruses. Recognition of infected cells occurs through T cell receptor that binds processed viral antigen expressed by major histocompatibility complex (MHC) I molecules on the surface of infected cells. Recognition is followed by a cascade of activation events leading to the release of granzymes and perforin and killing of infected cell. Activated CD8+ T cells also release anti-viral cytokines that act to control viral replication[29]. Despite the over activation of the immune system during HIV infection, it seems that HIV-specific CD8+ T cell responses fail to clear viral infection[30-32] and this can be attributed to several factors. HIV-infected cells sometimes revert to latency and are known to act as viral reservoirs. In this case, the absence of HIV protein expression on the surface of infected cells hinders recognition by CD8+ T cells[33]. Interestingly, several studies have shown that HIV proteins are capable of escaping CD8+ T cell recognition by modulating the expression of MHC I on surface of infected cells (Figure 2)[34]. Andrieu et al[35] showed that the Nef protein can down-regulate surface MHC I expression on DC, thereby impairing CD8+ T-cell maturation. In addition, HIV viruses are prone to rapid mutations which enables them to escape immune surveillance[36,37]. Chronic immune stimulation can have adverse effects on CD8+ T cell function. Several inhibitory molecules (Figure 2) are expressed by CD8+ T cells during chronic inflammation, and therefore impair the function of HIV-specific CD8+ T cell response[38]. Importantly, a small fraction of CD8+ T cells become infected with HIV and are susceptible to the direct cytotoxic effects of the virus[39,40]. It has also been shown that CD8+ T cell counts begin to decline during late stages of infection[41]. The pro-apoptotic properties of HIV gp120 protein may contribute to this decline[42,43]. Several studies showed that CD4+ T cell loss also impacts the function of CD8+ T cell, whereby CD4+ T cells are required to maintain cell-mediated immune responses against HIV[44,45]. Tregs, a subpopulation of CD4+ T cells that have a regulatory and suppressive role in autoimmune diseases and cancer, have been shown to contribute to the progression of AIDS disease by inhibiting HIV-specific CD4+ and CD8+ T cell responses[46].
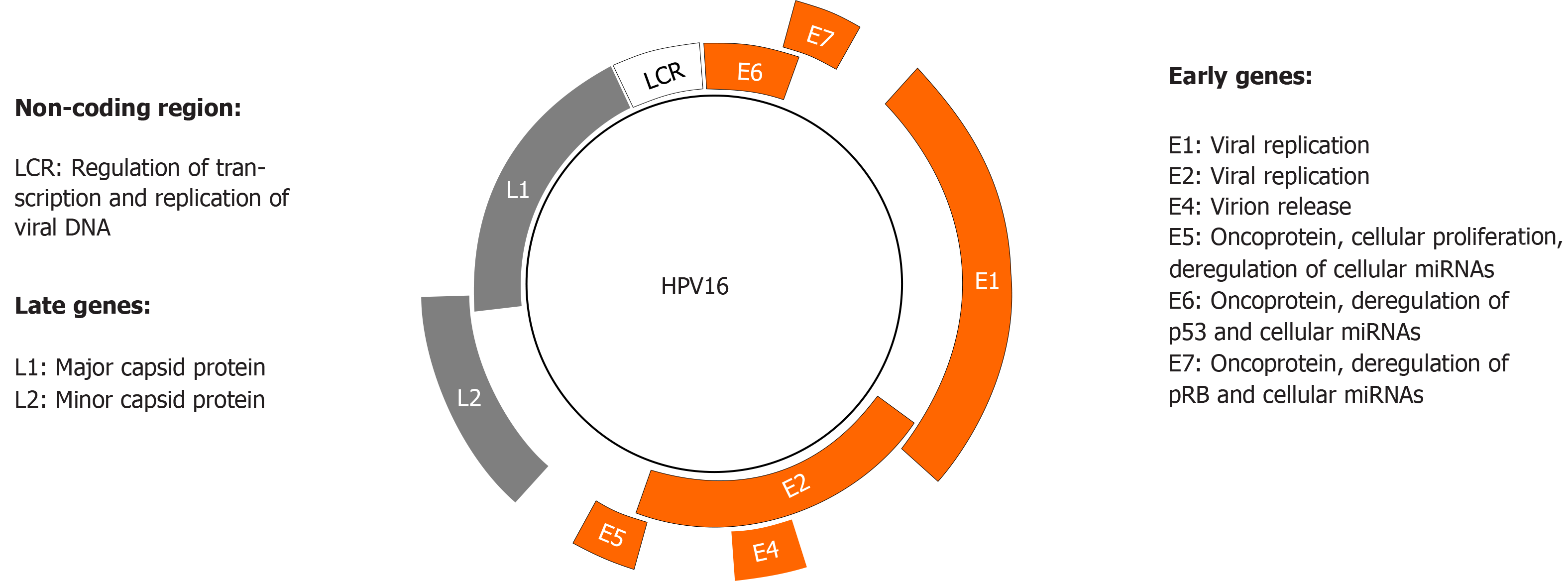
HPV can deregulate cellular proteins, including p53 and Retinoblastoma protein (pRb), thus mediating epithelial transformation and malignancy. HPV genome consists of a circular DNA that encodes the early proteins E1, E2, E4, E5, E6, and E7, and the late proteins L1 and L2 (Figure 3). E1 and E2 play an important role during HPV replication by binding to the viral replication origin, whereas E4 proteins are involved in virion release. E5, E6, and E7 are viral oncoproteins whose increased expression and activity is associated with enhanced proliferation of HPV-infected epithelial cells. L1 and L2 are structural proteins that form the viral capsid[47].
CD8+ T cells play a key role in the immune responses against HPV. In vivo studies using mouse models have shown that cells expressing HPV-16 E6 and E7 antigens are recognized and killed by cytotoxic T lymphocyte (CTL) cells[48,49]. In fact, E7-specific CTLs were detected in lesions containing tumor cells[50]. CD8+ T cells recognize viral antigens presented by MHC I/peptide complexes expressed on the surface of infected cells. However, this interaction is not sufficient to induce the killing of the infected cell. Signaling from activated dendritic cells and virus specific CD4+ T cells is highly important for stimulating and maintaining an efficient CTL activation[51]. It has been reported that peripheral blood mononuclear cell (PBMC) cultures from healthy individuals showed HPV16-specific CD4+ T-cell and CTL responses directed against HPV16 E2, E6 and/or E7[52-55]. Activated circulatory CD4+ and CD8+ T cells migrate from peripheral blood to infected tissues in healthy individuals[56]. Interestingly, these responses are mostly detected in women without cervical intraepithelial neoplasia (CIN)[57], and less commonly in women with CIN[54]. Nakagawa et al[55] also showed that the absence of CTL response to E6 proteins is associated with persistence of HPV16 infection in HPV-infected women without squamous intraepithelial lesions. HPV deregulates MHC I expression during infection (Figure 2). Multiple studies have reported the down regulation of MHC I expression in cervical cancer cells[57] and laryngeal papilloma[58]. This may be due to the loss of the peptide transporter 1 associated with antigen processing (TAP1), whose promotor appears to be downregulated by HPV 16 and 18 E7. The latter proteins also downregulate the promotor of MHC class I heavy chain[59]. A study has documented that HPV 11 E7 binds to the TAP transporter protein, thereby blocking peptide loading into MHC I antigens[60]. Other studies have reported that HPV 16 E5 plays a role in sequestering human leukocyte antigen class I complexes in the Golgi apparatus, which thus prevents their transport to the cell membrane surface[61]. The expression of MHC II is also modulated during HPV infection and carcinogenesis. MHC II are usually expressed by antigen presenting cells only however, it has been shown that keratinocytes, in cervical premalignant lesions and cancer, upregulate the expression of MHC II, because of the production of pro-inflammatory cytokines. On the other hand, HPV 16 E5 can block the expression of these molecules[62].
Nevertheless, the induction of a systemic T cell-mediated response against HPV proteins (E6, E7, and others) results in successful viral clearance in healthy individuals. In contrast, HIV infection leads to a progressive loss of CD4+ T cells[63]. Thus, even though antigen presenting cells express and present HPV peptides on their cell surface, in the absence of CD4+ T cell, CD8+ T cells fail to maintain their activity and thus, fail to kill HPV-infected cells.
HPV and HIV have developed a wide spectrum of mechanisms to evade immune responses. Given the ability of both viruses to modulate cellular pathways in infected and uninfected cells, and thus immune surveillance and responses[64], many mechanisms of immune evasion may be possible (Figure 2). HIV infection may directly or indirectly result in protecting HPV-infected keratinocytes from CTL-mediated killing. Therefore, HIV may affect both keratinocytes and CD8+ T cells, and thus favor HPV pathogenesis. Importantly, HIV proteins have been shown to interact with HPV proteins directly and indirectly by enhancing their expression and/or activation, promoting cancer[65-68]. HIV Tat increases the expression of HPV16 E6 and E7, enhancing their oncogenic effects. It also increases the expression of HPV L1, which forms the exterior of the virion and mediates initial attachment to target cells[65,66]. Rev indirectly upregulates HPV L1 expression[67]. Vpr interacts with HPV E6 protein to induce cell cycle arrest in cervical cancer cells[68]. However, evidence of interaction between the two viruses remains scarce and needs further investigation.
ASCC are cancers that arise in the transitional or squamous zone of the anal canal and are mostly caused by HPV16 and 18. It is believed that the basal layer cells in the epithelium of this transitional zone can become infected with HPV after the occurrence of micro-abrasions. Most individuals who acquire HPV mount the appropriate immune response and clear HPV infection within a year. However, HPV may persist in others and could lead to either low-grade or high-grad (HSIL) squamous intraepithelial lesions , and can be further classified into anal intraepithelial neoplasia (AIN) 1, 2 or 3[69,70].
Anal cancer is uncommon with 48541 new cases reported worldwide in 2018 as by the GLOBOCAN estimates[71]. However, its epidemiology has changed over the past 2 decades. A steady increase in the incidence and prevalence rates of ASCC has been reported. In the United Kingdom, a 70% increase in its incidence rates has been noted since the early 1990s[72]. The United States has reported similar trends with a 2.9% increase in incident rates each year since 1975[73]. In 2021, there will be an estimated 9090 new anal cancer cases and 1430 new anal cancer deaths[74]. The increase in incidence has been associated with multiple factors that include lifetime number of sexual partners, smoking, receptive anal intercourse, genital warts, and infection with HIV[75-77]. More than 90% of ASCCs have been found to be related to HPV, mainly HPV 16 and 18. Among men, the highest proportion of HPV is in men who have sex with men (MSM) and ranges between 50%-60%[76,78]. This proportion is even higher in HIV-infected MSM and reaches 90% in some studies[79,80]. Additionally, this population is infected with multiple HR-HPV types[81,82]. Not surprisingly, the prevalence of HSIL and anal cancer mirrors that of anal HPV in these populations where the incidence of neoplasia is higher than that of the general population. Compared to heterosexual men, MSMs have a 20 times increased risk of developing ASCC. HIV-positive MSMs have an even greater risk[76].
An obvious relationship between HIV, HPV, and anal cancer was illustrated in a population study in the United States between 1980-2005[73]. Authors found that HIV infection had a strong impact on the trends of anal cancer among males where incidence rate increased by 3.4% annually overall and by 1.7% in those without HIV-infection. A meta-analysis of 53 studies by Machalek et al[76] assessed the prevalence and incidence of HPV, AIN, and anal cancer in MSM and reported a substantial difference between HIV-positive and HIV-negative men for prevalence of any type of HPV (P = 0.005), including any HR-HPV (P = 0.01), prevalence of any anal cytological abnormality (P = 0.005), and low-grade anal lesions (P = 0.01). Analysis of recent studies reporting on histological abnormalities, high-grade AIN, and anal cancer revealed a significant difference between HIV-positive MSM vs HIV-negative MSM[76]. In another study, the incident rate for anal cancer was reported to be 69 per 100,000 person-years (PY) in HIV-positive MSMs vs 14 per 100,000 PY in HIV-negative MSMs[83]. Contrary to AIDS defining cancers, whose rates have decreased after the introduction of ART, the incidence rates of ASCC have shown an increase by 3%[8,76,84]. This may be attributable to a longer lifespan of PLWH allowing them to live longer with oncogenic HPV giving time for the development of HSIL and ASCC. In addition, PLWH have been found to have multiple types of HPV with Müller et al[82] reporting PLWH having a 7 times higher risk of having multiple types of HPV as compared to HIV-negative individuals.
The link between a lower rate of HR-HPV clearance and development of ASCC in PLWH is still being investigated. Studies have shown that in HIV-positive individuals, HR-HPV infection is cleared at a slower rate than HIV-negative individuals. Geskus et al[85] observed that HPV16 had the lowest clearance for both prevalent positive and incident positive infection. Additionally, authors reported a decreasing clearance rate with increasing HIV viral load. Results from a recently updated meta-analysis showed that clearance rate of HPV infection among PLWH was approximately half compared to that of HIV-negative individuals, with similar findings reported for HR-HPV[86]. Whether CD4 count affects the clearance rate or not is not well established. In the same meta-analysis above, Looker et al[86] reported a possible, but non-significant, reduction in clearance of HPV with lower CD4 counts. In a nested case-control study from the Swiss HIV Cohort Study, lower CD4 counts in PLWH were correlated with the development of ASCC. Authors reported that the best predictor was a CD4 count 6-7 years prior to ASCC diagnosis. Beyond that point, authors found that the ASCC risk was less sensitive to CD4 counts, highlighting the importance of starting ART early before the establishment of precancerous lesions[87].
There are no formal guidelines on anal cancer screening due to the lack of trials assessing the effectiveness of such screening practices. However, with the accumulating evidence of an increasing incidence of anal cancer in PLWH, there is increased advocacy for screening in these high-risk populations, drawing on the proven value of cervical cytology in reducing cervical cancer. Additionally, cost-effective models of screening MSM for AIN every 2-3 years have shown possible gains in life-expectancy and quality of life[88,89].
Screening consists of detection and treatment of anal HSIL. Detection can be done through anal cytology, digital rectal examination, high resolution anoscopy (HRA), and/or biopsy. While some experts have advocated the use of HRA for initial screening because of the high prevalence of AIN in PLWH and MSM, anal cytology remains a preferred initial method due to limited availability of HRA especially in developing countries[64]. Yet, all the previously mentioned tools have several limitations and disadvantages. For example, HRA is invasive, and cytology is neither sensitive nor specific. Therefore, a non-invasive method with high sensitivity and specificity for detection of precancerous and cancerous anal lesions is needed.
Circulatory and tissue miRNAs have become of interest to investigators, as these small non-coding RNAs possess distinctive properties that make them ideal biomarkers for detection and prognosis of cancer. They play an important role in gene regulation by inducing the degradation and inhibiting the translation of the corresponding mRNAs[90,91]. They can also activate the expression of genes by targeting their promotors[92,93]. They are well known for their pleiotropic effects in many important cellular processes, such as apoptosis, proliferation, and differentiation[94]. They are signi
Globular profiling of miRNAs in cancer and normal tissues has been established in different types of cancers, including breast[102], lung[103], colon, liver, and pancreatic[104] cancers, which have allowed for the identification of a series of miRNAs that are deregulated in these cancers. However, an invasive method, such as surgery and biopsy collection, is needed to analyze the tissues. Thus, researchers are investigating the use of plasma and serum miRNAs as potential circulatory biomarkers for different purposes. This would allow for non-invasive quantification of these biomarkers and potentially for detection of premalignant lesions and screening of early tumorigenesis. In the context of HPV-associated cancer, miRNAs have been studied and documented as mediators or suppressors of pathogenesis[96,97,105]. Some of these miRNAs have been shown to be deregulated by HPV E5, E6, and E7 oncoproteins in different cells and tissues (Table 1). By downregulating p53, E6 alters the expression of many miRNAs that are transcribed by p53. On the other hand, E7 releases E2F transcription factor from pRB-E2F complex by degrading pRB. As a result, E2F becomes free to activate the transcription of many miRNAs. The mechanism by which E5 deregulates cellular miRNAs is still unclear[106]. The deregulation of many of these miRNAs was shown to affect several hallmarks of cancer, including enhanced proliferation, inhibition of apoptosis, invasion, and metastasis. A recent study showed that miR-129 was significantly upregulated in the serum and cervical cancer tissues collected from 72 patients, suggesting the possibility of using this miRNA as a biomarker for the detection of cervical cancer. Interestingly, HPV typing detected HPV16 in all cancer samples studied[107]. Another study identified a miRNA signature panel consisting of 9 miRNAs (miR-9, miR-15b, miR-20a, miR-31, miR-93, miR-183, miR-184, miR-222, and let-7b) with a combined area under the curve of 0.89 for CIN3 detection in HPV-positive self-samples of women with CIN3[108]. Recently, Shi et al[109] identified an optimal subset of 7 signature miRNAs, including miR144, miR147b, miR2182, miR425, miR451, miR483, and miR486 in cervical cancer. Functional enrichment analysis showed that the latter miRNAs are involved in carcinogenic pathways, such as Wnt signaling pathway and transforming growth factor-β signaling pathway. Importantly, altered miRNAs have been investigated mainly in cervical cancer cell lines[96] and cervical carcinoma samples[110]. However, miRNAs have been less studied in anal cancer and a single study showed that HPV16-E7 protein is capable of inducing miR-15b in anal carcinoma biopsies[111].
| HPV protein | miRNA target | miRNA expression level | Sample type | Biological effect(s) | Ref. |
| E5 | miR-146a | + | E5-expressing HaCat cells | Promoted cell proliferation | [106] |
| miR-203 | - | E5-expressing HaCat cells | Increased expression of p63 | [106] | |
| miR-324-5p | - | E5-expressing HaCat cells | Contributed to cervical carcinogenesis | [106] | |
| E6 | miR-20a | + | CaSki and SiHa (HPV16+) human cervical cell lines | Promoted cell growth through downregulating PDCD6 and activating Akt and p38 | [156] |
| miR-20b | + | HeLa (HPV18+), SiHa and Caski human cervical cancer cell lines; Cervical carcinoma tissues | Reduced TIMP2 expression and induced EMT, migration, and invasion | [157] | |
| miR-23b | - | SiHa and CaSki cell lines | Increased expression of uPA and induction of migration in human cervical cancer cells | [158] | |
| miR-30c-2* | - | HPV-infected NSCLC; TL1 cell line | Correlated with tumor stage and lymph node metastasis | [159] | |
| miR-34a | - | CaSki and SiHa cell lines, HPV18-positive cell lines HeLa and C411, HPV68-positive cell line ME180Cervical cancer tissues | Inhibited cell proliferation; Increased LDHA expression levels, inhibited Warburg effect and reprogrammed glycolysis through targeting LDHA | [160,161] | |
| miR-145 | - | Hela, SiHa, and CaSki cell lines; Cervical cancer tissues | Modulated invasion and therapy resistance of cervical cancer cells | [162] | |
| miR-195 | - | HeLa and SiHa cell line cervical cancer tissue samples | Promoted cell proliferation, invasion, and metastasis | [163] | |
| miR-218 | - | HPV16 positive cervical cell lines and tissues; Cervical cancer tissues | Increased expression of LAMB3, SFMFBT1, and DCUN1D1, promoted EMT, migration, and invasion in cervical cancer associated with clinicopathological characteristics of patients | [164,165] | |
| miR-375 | - | SiHa and CaSki cell lines; Cervical tissue samples | Modulated EMT in cervical cancer; Enhanced invasion and metastasis of cervical carcinoma cells through targeting SP1 | [166,167] | |
| miR-2861 | - | SiHa and CaSki cell lines; Cervical cancer tissues | Enhanced cell proliferation and invasion, and inhibited apoptosis in cervical cancer cells; Negatively associated with advanced tumor stage and lymph node metastasis | [168] | |
| E7 | miR-15b | + | HPV16 E7-expressing tumors from anal carcinoma patients; CaSki cell line | Downregulated cyclin E1; Increased expression of several E2F-regulated genes | [111] |
| miR-20a | + | OSCC tissues | Inhibited cell proliferation, invasion, and migration | [169] | |
| miR-21 | + | HPV16 E7-transfected Hela cells; Cervical cancer tissue | Enhanced cervical carcinoma cell proliferation, growth, and invasion; Involved in cervicitis and cervical cancer progression | [170,171] | |
| miR-25 | + | HVK-derived raft tissues infected with either HPV16 or HPV18 | Increased expression correlated with the progression of the cervical lesions, making it a potential biomarker for CINs and cervical cancer | [96] | |
| miR-27b | + | HPV 16-positive human cervical carcinoma tissues; SiHa and CaSki cell lines | Reduced PLK2 expression; Promoted cell proliferation and inhibited paclitaxel-induced cell apoptosis; Inhibited PPARγ expression and promoted proliferation and invasion | [172,173] | |
| miR-205 | + | HPV-positive keratinocytes | Activated Akt pathway and upregulated cyclin D1 levels, resulting in increased proliferation | [174] | |
| miR-323 | + | Cervical cancer cell lines transfected with HPV 16 E7 and SiHa cervical cancer cells | --- | [175] | |
| E6/E7 | miR-16 | + | HFK-derived raft cultures with HPV16 infection; HVK-derived raft tissues infected with either HPV16 or HPV18; CIN3 and Cervical carcinoma tissues with HR HPV infection | --- | [96] |
| miR-22 | - | HVK-derived raft tissues infected with either HPV16 or HPV18 | Suppressed tumor growth and metastasis | [96,176] | |
| miR-24 | + | HPV-positive keratinocytes | Reduced p27 expression level and enhanced proliferation | [174] | |
| miR-29a | - | HVK-derived raft tissues infected with either HPV16 or HPV18 | --- | [96] | |
| miR-92a | + | CIN and cervical carcinoma tissues with HR HPV infection, and raft tissues with HPV16 or HPV18 infection | Increased expression correlated with the progression of the cervical lesions, and may serve as a biomarker for CINs and cervical cancer | [96] | |
| miR-100 | - | HFK-derived raft cultures with HPV18 infection | --- | [96] | |
| miR-125a | - | Cervical carcinoma tissuesSiHa and HeLa cell lines | Increased STAT3 expression and enhanced tumorigenesis and metastasis | [177] | |
| miR-146a-5p | - | HPV16 E6/E7-positive keratinocytes; Hela, SiHa, and CaSki cell lines | Enhanced expression of KDM2B; Promoted proliferation and migration | [178] | |
| miR-203 | - | NHKs and NFKs expressing E6, E7, or combination | Increased expression of p63 and promotion of cell proliferation | [179,180] | |
| miR-378 | + | CIN3 and cervical carcinoma tissues with HR HPV infection, and raft tissues with HPV16 or HPV18 infection | --- | [96] |
HIV infection also dysregulates cellular miRNA biogenesis and expression profiles[112-114]. For example, HIV Tat and Vpr affect miRNA biogenesis by binding Dicer or Drosha[115-117], while trans-activation response modulates TRBP, an important component of the miRNA generation complex. HIV infection is known to both upregulate and downregulate several cellular miRNAs in HIV-infected human PBMC, T cells, monocyte-derived macrophages (MDMs), latently infected CD4+ T cells, plasma samples, HUT78 cells, and CD4+ T cells from either acute or chronic HIV-infected individuals. Few studies determined the expression of cellular miRNAs in HIV-infected cell lines (Table 2). Recently, Biswas et al[118] established a comparative global miRNA expression profile in human PBMC and MDMs infected with HIV-1/HIV-2. Differentially expressed miRNAs were identified in these cells. Pathway analysis using Kyoto Encyclopedia of Genes and Genomes database showed that the deregulated miRNAs are likely to be involved in p53 signaling pathway, PI3K-Akt signaling pathways, Mitogen-activated protein kinase signaling pathways, FoxO signaling pathway, and NF-kappaB inhibitor signaling pathway, all of which play a role in carcinogenesis[118,119]. HIV Tat, Nef, and Vpr have been reported to alter the expression levels of many miRNAs and contribute to HIV pathogenesis (Table 3). On the other hand, cellular miRNAs also target HIV genome, but it is still unclear whether these miRNAs are effective during HIV infection[112].
| Sample type | miRNAs deregulated | Ref. |
| PBMCs from HIV-infected patients | ↑ miR-9; ↓ miR-29c, miR-31, miR-125b, miR-146b-5p, miR-150, Let-7g | [181] |
| T cells from HIV-infected individuals | ↓ miR-16, miR-146b, miR-150, miR-223 | [182] |
| HIV-1 infected PBMCs | ↑ miR-223; ↓ miR-21, miR-26a, miR-29a, miR-29b, miR-29c, miR-155 | [183] |
| HIV-1 infected PBMCs | ↑ miR-3195, miR-3656, miR-4492, and miR-6087; ↓ miR-1273h-3p, miR-1273h-5p, miR-671-5p, and miR-7-5p | [118] |
| HIV-2-infected PBMCs | ↑ miR-18a-3p and hsa-miR-320b | [118] |
| HIV-2-infected MDMs | ↑ miR-542-3p, miR-375, miR-195-5p, miR-30c-2-3p, miR-4802-3p, and miR-26b-5p | [118] |
| HIV-1- and HIV-2-infected MDMs | ↓ miR-148b-5p, hsa-miR-26a-2-3p, miR-199a-1, miR-199a-2, and miR-874-5p | [118] |
| HIV-1-transfected HeLa cells | ↓ miR-16, miR-93, miR-148b, miR-221 | [184] |
| HIV-1 infected Jurkat cells. | ↑ miR-122, miR-297, miR-370, and miR-373; ↓ miR-17-5p and miR-20a | [185] |
| latently infected CD4+ T cells | ↑ miR-196b and miR-1290 | [186] |
| PBMCs obtained from HIV-1 positive individuals with high viral load | ↑ miR19b, miR-34a, miR-144, miR-146a, miR-155, miR-382, miR-615-3p | [187] |
| Plasma obtained from patients with HIV infection | ↓ miR-3162-3p | [188] |
| HIV-1-infected HUT78 cells and CD4+ T cells from chronic HIV-1 infected individuals | ↓ Let-7 miRs | [189] |
| HIV-1 positive plasma samples in the acute stage infection | ↑ miR-16-5p, miR-20b-5p, miR-24-3p, miR-142-5p, miR-195-5p, miR-206, miR-223-3p, miR-885-5p, and let-7 g-3p; ↓ miR-34c-3p, miR-181c-3p, miR-202-3p, and miR-409-3p | [190] |
| HIV-1 infected CD4+ T cells | ↓ miR-20a and miR-106b | [191] |
| HIV protein | miRNA target | Effect | Sample type | Biological effect | Ref. |
| Tat | miR-21, miR-29a, miR-222, miR-1290 | + | Tat101-expressing Jurkat cells; Resting PBMCs from healthy donors were transiently transfected with Tat101-expressing vector | Targeted mRNAs of genes involved in apoptosis, T cell migration, and proliferation | [192] |
| miR-128a, and miR-3182 | - | Tat101-expressing Jurkat cells | ---- | [192] | |
| miR-132 | + | Tat-transfected astrocytes and neurons, astrocytes from Tat-transgenic mice, and HIV-infected astrocytes | Involved in the direct neurotoxicity of Tat | [193] | |
| miR-129, miR-135a, miR-181a, miR-495, miR-523, miR-524, miR-539, let-7 | - | U-87MG (astrocyte cell line), HEK 293T, and HeLa cells transfected with wild-type Tat | Downregulation of β-catenin activity | [194] | |
| miR-101 | + | BMVECs exposed to Tat C | Decreased the expression of VE-cadherin | [195] | |
| miR-34a and miR-138 | + | Astrocytoma cell line A172 and rat primary astrocytes exposed to Tat | Upregulated NF-κB and promoted activation of astrocytes | [196] | |
| Nef | miR-573 and miR-638 | + | Human monocytic U937 cells that stably expressed HIV-1 Nef | Altered several pathways involved in HIV pathogenesis | [197] |
| miR16-1, miR-18, miR-19a, miR-20a, miR-21, miR-27a, miR-29b, miR-125b, miR-146a, miR-146b-3p, miR-181a, miR-223, miR-570, miR-610 and miR-624 | - | Human monocytic U937 cells that stably expressed HIV-1 Nef | Altered several pathways involved in HIV pathogenesis | [197] | |
| miR-17, miR-19a, miR-19b, miR-20a, miR26a, miR-28, miR-29a, miR-29b, miR-29c, miR-92a, miR-125b, miR-149, miR-150, miR-223, miR-324-5p, miR-378 and miR-382 | + | Nef exosomes | Inhibited HIV replication | [197] | |
| Vpr | miR-942-5p | + | PEL cells | Targeted IκBα and activation of NF-κB signalling | [198] |
| miR-711 | + | PEL cells | Directly targeted Notch1 and reduced levels of IκBα transcript | [198] |
Overall, molecular mechanisms that contribute to anal cancer pathogenesis and progression are still elusive. One of the reasons that little progress has been made in understanding the mechanisms of carcinogenesis in this type of cancer is the scarcity of in vitro and in vivo model systems for investigating anal cancer. Thus, further studies are required to gain insight into the mechanisms involved in anal caner. This is particularly important as these mechanisms may involve miRNAs, which may be further investigated as potential targets for cancer therapy. The use of miRNA-based therapeutics has been investigated in clinical trials in several countries. MicroRNA mimics and anti-miRNAs (antagomirs) are now under investigation as potential therapeutic agents for multiple cancers. miRNA mimics may be administered to replace downregulated miRNAs, which usually act as tumor suppressors in cancers. On the other hand, many miRNAs have been targeted for inhibition in the treatment of several cancers. These miRNAs are referred to as oncomiRs and their overexpression in cancer contributes to pathogenesis. In the context of cervical cancer, Lee et al[110] showed that treatment with anti-miR-199a suppressed cervical cell growth in vitro. Additionally, a study has shown a promising role for the tumor suppressor miR-34a, which is downregulated in HPV-positive cancers, in repressing oncogenic transformations. Both miR-34a and miR-125 are downregulated in cervical cancer samples and correlate with cervical cancer invasiveness[120]. Interestingly, a recent phase 1 study of MRX34, a liposomal miR-34a mimic, was conducted with patients having advanced solid tumors[121]. Thereby, this miRNA may be a good candidate for treatment of HPV-related cancers, including anal cancer[122]. In addition, anti-miRs targeted at miR-122, which has been shown to be upregulated in HIV-1 infected Jurkat cells, reached clinical phase II trials and were investigated for treating hepatitis C infection[123]. Other candidate miRNAs are being tested in clinical trials, paving the way for developing miRNA-based drugs for treating several illnesses and cancer diseases[122,124].
In the case of anal cancer, where HIV and HPV pathogenesis play a role in the development of the disease in PLWH, a major challenge is to distinguish HIV-specific miRNAs, HPV-specific miRNAs, and HIV and HPV co-infection-specific miRNAs. Major limitations include the absence of studies implementing computational models to identify these miRNAs, technical issues associated with conventional miRNA extraction and detection tools, and scarcity of anal cancer in vitro and in vivo models. Ongoing studies are still being conducted to study miRNA profiles during HIV[125-127] and HPV[128-130] infections. With the appropriate application of advanced bioinformatic analysis tools and computational models, the identification of the most predictive miRNAs, even from complex datasets would be possible. These tools are becoming widespread and have already been used to identify potential miRNA biomarkers for Ebola[131] and severe acute respiratory syndrome coronavirus 2[132], in addition, these tools have been used to decipher potential miRNA biomarkers in a wide variety of cancers, including melanoma[133,134], breast[135], colon[136], and lung cancer[137].
In addition to the conventional miRNA detection platforms which include Northern blotting, in situ hybridization, next generation sequencing, reverse transcription qPCR, and microarrays[138], new miRNA extraction and detection platforms have emerged to compensate for the limitations of conventional assays[139]. These technologies are referred to as point-of-care (PoC) technologies and include isothermal amplification-based assays[140], lateral flow assay-based systems[141], nanobead-based[142], electro-chemical-based[143], and microfluidic chip-based[144] strategies. The latter, which is also known as Lab-on-a-chip or microchip, is highly specific, cost-effective, and a quick approach for the multiplexed detection of miRNAs[139]. It has been used to test miRNAs in several biological samples, including blood of breast cancer patients[145]. Importantly, this system has been also used to quantify miRNAs in plasma extracellular vesicles (EVs), including exosomes. EVs are secreted by body cells and are found in body fluids including plasma, urine, and synovial fluid[146]. They have been shown to carry and stabilize miRNAs in the blood[147]. A unique feature of exosomes is the presence of cell-specific proteins[148], which enables identification of exosomes released from cancer cells. Examining specific miRNAs released from tumors and tumor niche, instead of whole blood miRNA profiling would provide a more accurate way of distinguishing HIV-specific and HPV-specific miRNAs, given the unique viral tropism of each. Exosomal miRNAs would enable the identification of the cell origin and might be a better source when compared to non-exosomal, cell-free miRNAs. Recently, studies that profiled and analyzed miRNAs from different sources were reviewed[149]. Authors concluded that 71% of the studies stated that exosomes are the best source of miRNAs as biomarkers. Detecting EVs miRNA signature has already been proven to be a good prognostic tool in several cancers including colorectal[150] and pancreatic cancer[151].
Interestingly, organ-on-chip and organoids are being used to study infectious diseases and cancer. These models can be used to assess HPV virus-Langerhans cells interactions[152] and HPV-oral mucosa epithelia interactions[153]. Cell-to-cell communication can be also studied by co-culturing cancer cells with immune cells, and thus allows the study of cancer-immune interaction. Organoids can be used to model tumor-derived EVs, also known as oncosomes, in addition to EVs released by stromal cells in tumor microenvironment[154]. Very recently, researchers established organoid cultures from human ecto-and endocervix. Cells collected using Pap brush method were used to derive organoids from cervical tissue. The established patient-derived model system resembled causative HPV infection[155], and thus could be used for modeling HPV-related pathogenesis, in addition to exploring the role of HPV and HIV in deregulating miRNAs. The same derivation method can be used to derive organoids from healthy or tumor anal tissue to assess miRNA deregulation by HIV and/or HPV. These model systems could be used to test the efficacy of engineered miRNA-loaded EVs in targeting anal cancer cells to deliver potential miRNA thera
It is important to note that although extensive research has been conducted to identify candidate miRNA biomarkers for cancer screening, the development of new techniques, such as PoC for miRNA detection is still at the very early stage and a work on progress. Further progress is required to achieve the desired goal of using PoC testing for detecting and distinguishing miRNAs deregulated by oncogenic viral infections, including HPV. Therefore, the identification of miRNAs deregulated by HIV, HPV, and HIV-HPV co-infection warrants further research. More accurate and standardized methods are required for implementation of miRNAs as biomarkers for anal cancer diagnosis[181-198]. Importantly, the widespread use of high-throughput sequencing, PoC technologies, and advanced computational analysis tools may facilitate discovering and distinguishing these miRNAs.
HPV is the most common sexually transmitted infection worldwide. PLWH are at high risk of acquiring HPV infection and developing HPV-associated malignancies, independent of AIDS. Anal cancer incidence, though rare in the general population, has been rising significantly in PLWH. The lack of standard screening programs contributes to the increased incidence of anal cancer, and thus, there is a need for anal dysplasia screening and treatment in PLWH. The discovery of highly sensitive and specific biomarkers would enable the early detection of anal cancer and the improved survival of HIV-infected patients. There is a need for relevant biomarkers that could be integrated into clinical practice and thus, aid in the detection, diagnosis, and treatment of high-risk patients. miRNAs have become valuable tools for detection and treatment of many types of cancer. Given their deregulation and potentially significant role in HPV-related pathogenesis and in HIV infections, miRNAs may serve as diagnostic and prognostic biomarkers that can enhance HIV patients’ outcomes and provide better management of the disease. Genome-wide profiling of miRNAs and validation of miRNA targets in tissue and blood samples of people infected with HIV and HR-HPV is important to establish miRNA expression signatures in this population and would help develop non-invasive miRNA therapeutic strategies for treatment of anal cancer.
The authors would like to thank Mr. Rawad Abdul Salam for his valued assistance in drawing the illustrations.
Manuscript source: Invited manuscript
Specialty type: Infectious diseases
Country/Territory of origin: Lebanon
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Gao F, Serban D S-Editor: Zhang L L-Editor: Filipodia P-Editor: Li JH
| 1. | GBD 2017 Disease and Injury Incidence and Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1789-1858. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9354] [Cited by in RCA: 8390] [Article Influence: 1198.6] [Reference Citation Analysis (4)] |
| 2. | Agace WW, Roberts AI, Wu L, Greineder C, Ebert EC, Parker CM. Human intestinal lamina propria and intraepithelial lymphocytes express receptors specific for chemokines induced by inflammation. Eur J Immunol. 2000;30:819-826. [PubMed] |
| 3. | May MT, Sterne JA, Costagliola D, Sabin CA, Phillips AN, Justice AC, Dabis F, Gill J, Lundgren J, Hogg RS, de Wolf F, Fätkenheuer G, Staszewski S, d'Arminio Monforte A, Egger M; Antiretroviral Therapy (ART) Cohort Collaboration. HIV treatment response and prognosis in Europe and North America in the first decade of highly active antiretroviral therapy: a collaborative analysis. Lancet. 2006;368:451-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 179] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 4. | Mocroft A, Brettle R, Kirk O, Blaxhult A, Parkin JM, Antunes F, Francioli P, D'Arminio Monforte A, Fox Z, Lundgren JD; EuroSIDA study group. Changes in the cause of death among HIV positive subjects across Europe: results from the EuroSIDA study. AIDS. 2002;16:1663-1671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 217] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 5. | Palella FJ Jr, Baker RK, Moorman AC, Chmiel JS, Wood KC, Brooks JT, Holmberg SD; HIV Outpatient Study Investigators. Mortality in the highly active antiretroviral therapy era: changing causes of death and disease in the HIV outpatient study. J Acquir Immune Defic Syndr. 2006;43:27-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1002] [Cited by in RCA: 1080] [Article Influence: 56.8] [Reference Citation Analysis (0)] |
| 6. | Gueler A, Moser A, Calmy A, Günthard HF, Bernasconi E, Furrer H, Fux CA, Battegay M, Cavassini M, Vernazza P, Zwahlen M, Egger M; Swiss HIV Cohort Study; Swiss National Cohort. Life expectancy in HIV-positive persons in Switzerland: matched comparison with general population. AIDS. 2017;31:427-436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 185] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 7. | Samji H, Cescon A, Hogg RS, Modur SP, Althoff KN, Buchacz K, Burchell AN, Cohen M, Gebo KA, Gill MJ, Justice A, Kirk G, Klein MB, Korthuis PT, Martin J, Napravnik S, Rourke SB, Sterling TR, Silverberg MJ, Deeks S, Jacobson LP, Bosch RJ, Kitahata MM, Goedert JJ, Moore R, Gange SJ; North American AIDS Cohort Collaboration on Research and Design (NA-ACCORD) of IeDEA. Closing the gap: increases in life expectancy among treated HIV-positive individuals in the United States and Canada. PLoS One. 2013;8:e81355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1010] [Cited by in RCA: 1097] [Article Influence: 91.4] [Reference Citation Analysis (0)] |
| 8. | Shiels MS, Engels EA. Evolving epidemiology of HIV-associated malignancies. Curr Opin HIV AIDS. 2017;12:6-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 161] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 9. | Smith CJ, Ryom L, Weber R, Morlat P, Pradier C, Reiss P, Kowalska JD, de Wit S, Law M, el Sadr W, Kirk O, Friis-Moller N, Monforte Ad, Phillips AN, Sabin CA, Lundgren JD; D:A:D Study Group. Trends in underlying causes of death in people with HIV from 1999 to 2011 (D:A:D): a multicohort collaboration. Lancet. 2014;384:241-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 667] [Cited by in RCA: 735] [Article Influence: 66.8] [Reference Citation Analysis (0)] |
| 10. | Yarchoan R, Uldrick TS. HIV-Associated Cancers and Related Diseases. N Engl J Med. 2018;378:2145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 11. | Abbar B, Veyri M, Solas C, Poizot-Martin I, Spano JP. [HIV and cancer: Update 2020]. Bull Cancer. 2020;107:21-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 12. | de Martel C, Shiels MS, Franceschi S, Simard EP, Vignat J, Hall HI, Engels EA, Plummer M. Cancers attributable to infections among adults with HIV in the United States. AIDS. 2015;29:2173-2181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 84] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 13. | Giuliano AR, Tortolero-Luna G, Ferrer E, Burchell AN, de Sanjose S, Kjaer SK, Muñoz N, Schiffman M, Bosch FX. Epidemiology of human papillomavirus infection in men, cancers other than cervical and benign conditions. Vaccine. 2008;26 Suppl 10:K17-K28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 245] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 14. | Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370:890-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1829] [Cited by in RCA: 1883] [Article Influence: 104.6] [Reference Citation Analysis (0)] |
| 15. | Centers for Disease Control and Prevention (CDC). Human papillomavirus-associated cancers - United States, 2004-2008. MMWR Morb Mortal Wkly Rep. 2012;61:258-261. [PubMed] |
| 16. | Shiels MS, Pfeiffer RM, Gail MH, Hall HI, Li J, Chaturvedi AK, Bhatia K, Uldrick TS, Yarchoan R, Goedert JJ, Engels EA. Cancer burden in the HIV-infected population in the United States. J Natl Cancer Inst. 2011;103:753-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 552] [Cited by in RCA: 563] [Article Influence: 40.2] [Reference Citation Analysis (0)] |
| 17. | Nkwabong E, Laure Bessi Badjan I, Sando Z. Pap smear accuracy for the diagnosis of cervical precancerous lesions. Trop Doct. 2019;49:34-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 18. | Frankel AD, Young JA. HIV-1: fifteen proteins and an RNA. Annu Rev Biochem. 1998;67:1-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 559] [Cited by in RCA: 549] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 19. | Dragic T. An overview of the determinants of CCR5 and CXCR4 co-receptor function. J Gen Virol. 2001;82:1807-1814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 97] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 20. | Craigie R, Bushman FD. HIV DNA integration. Cold Spring Harb Perspect Med. 2012;2:a006890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 239] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 21. | Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2622] [Cited by in RCA: 2578] [Article Influence: 62.9] [Reference Citation Analysis (0)] |
| 22. | Lifson JD, Reyes GR, McGrath MS, Stein BS, Engleman EG. AIDS retrovirus induced cytopathology: giant cell formation and involvement of CD4 antigen. Science. 1986;232:1123-1127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 405] [Cited by in RCA: 399] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 23. | Doitsh G, Cavrois M, Lassen KG, Zepeda O, Yang Z, Santiago ML, Hebbeler AM, Greene WC. Abortive HIV infection mediates CD4 T cell depletion and inflammation in human lymphoid tissue. Cell. 2010;143:789-801. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 377] [Cited by in RCA: 346] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 24. | Doitsh G, Galloway NL, Geng X, Yang Z, Monroe KM, Zepeda O, Hunt PW, Hatano H, Sowinski S, Muñoz-Arias I, Greene WC. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505:509-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 887] [Cited by in RCA: 875] [Article Influence: 79.5] [Reference Citation Analysis (0)] |
| 25. | Cooper A, García M, Petrovas C, Yamamoto T, Koup RA, Nabel GJ. HIV-1 causes CD4 cell death through DNA-dependent protein kinase during viral integration. Nature. 2013;498:376-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 177] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 26. | Cossarizza A. Apoptosis and HIV infection: about molecules and genes. Curr Pharm Des. 2008;14:237-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 27. | Cummins NW, Badley AD. Mechanisms of HIV-associated lymphocyte apoptosis: 2010. Cell Death Dis. 2010;1:e99. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 138] [Cited by in RCA: 149] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 28. | Chávez-Galán L, Arenas-Del Angel MC, Zenteno E, Chávez R, Lascurain R. Cell death mechanisms induced by cytotoxic lymphocytes. Cell Mol Immunol. 2009;6:15-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 226] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 29. | Yang OO, Kalams SA, Trocha A, Cao H, Luster A, Johnson RP, Walker BD. Suppression of human immunodeficiency virus type 1 replication by CD8+ cells: evidence for HLA class I-restricted triggering of cytolytic and noncytolytic mechanisms. J Virol. 1997;71:3120-3128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 273] [Cited by in RCA: 266] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 30. | Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68:6103-6110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1489] [Cited by in RCA: 1452] [Article Influence: 46.8] [Reference Citation Analysis (0)] |
| 31. | Ndhlovu ZM, Kamya P, Mewalal N, Kløverpris HN, Nkosi T, Pretorius K, Laher F, Ogunshola F, Chopera D, Shekhar K, Ghebremichael M, Ismail N, Moodley A, Malik A, Leslie A, Goulder PJ, Buus S, Chakraborty A, Dong K, Ndung'u T, Walker BD. Magnitude and Kinetics of CD8+ T Cell Activation during Hyperacute HIV Infection Impact Viral Set Point. Immunity. 2015;43:591-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 215] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 32. | Dalod M, Dupuis M, Deschemin JC, Goujard C, Deveau C, Meyer L, Ngo N, Rouzioux C, Guillet JG, Delfraissy JF, Sinet M, Venet A. Weak anti-HIV CD8(+) T-cell effector activity in HIV primary infection. J Clin Invest. 1999;104:1431-1439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 114] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 33. | Huang SH, Ren Y, Thomas AS, Chan D, Mueller S, Ward AR, Patel S, Bollard CM, Cruz CR, Karandish S, Truong R, Macedo AB, Bosque A, Kovacs C, Benko E, Piechocka-Trocha A, Wong H, Jeng E, Nixon DF, Ho YC, Siliciano RF, Walker BD, Jones RB. Latent HIV reservoirs exhibit inherent resistance to elimination by CD8+ T cells. J Clin Invest. 2018;128:876-889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 152] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 34. | Collins KL, Chen BK, Kalams SA, Walker BD, Baltimore D. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature. 1998;391:397-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 796] [Cited by in RCA: 797] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 35. | Andrieu M, Chassin D, Desoutter JF, Bouchaert I, Baillet M, Hanau D, Guillet JG, Hosmalin A. Downregulation of major histocompatibility class I on human dendritic cells by HIV Nef impairs antigen presentation to HIV-specific CD8+ T lymphocytes. AIDS Res Hum Retroviruses. 2001;17:1365-1370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 38] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 36. | Phillips RE, Rowland-Jones S, Nixon DF, Gotch FM, Edwards JP, Ogunlesi AO, Elvin JG, Rothbard JA, Bangham CR, Rizza CR. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature. 1991;354:453-459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 823] [Cited by in RCA: 811] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 37. | Koup RA. Virus escape from CTL recognition. J Exp Med. 1994;180:779-782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 115] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 38. | Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, Betts MR, Freeman GJ, Vignali DA, Wherry EJ. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. 2009;10:29-37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1703] [Cited by in RCA: 1643] [Article Influence: 102.7] [Reference Citation Analysis (0)] |
| 39. | De Maria A, Pantaleo G, Schnittman SM, Greenhouse JJ, Baseler M, Orenstein JM, Fauci AS. Infection of CD8+ T lymphocytes with HIV. Requirement for interaction with infected CD4+ cells and induction of infectious virus from chronically infected CD8+ cells. J Immunol. 1991;146:2220-2226. [PubMed] |
| 40. | Brenchley JM, Hill BJ, Ambrozak DR, Price DA, Guenaga FJ, Casazza JP, Kuruppu J, Yazdani J, Migueles SA, Connors M, Roederer M, Douek DC, Koup RA. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J Virol. 2004;78:1160-1168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 317] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 41. | Margolick JB, Muñoz A, Donnenberg AD, Park LP, Galai N, Giorgi JV, O'Gorman MR, Ferbas J. Failure of T-cell homeostasis preceding AIDS in HIV-1 infection. The Multicenter AIDS Cohort Study. Nat Med. 1995;1:674-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 174] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 42. | Herbein G, Mahlknecht U, Batliwalla F, Gregersen P, Pappas T, Butler J, O'Brien WA, Verdin E. Apoptosis of CD8+ T cells is mediated by macrophages through interaction of HIV gp120 with chemokine receptor CXCR4. Nature. 1998;395:189-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 314] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 43. | Blanco J, Barretina J, Cabrera C, Gutiérrez A, Clotet B, Esté JA. CD4(+) and CD8(+) T cell death during human immunodeficiency virus infection in vitro. Virology. 2001;285:356-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 44. | Kalams SA, Buchbinder SP, Rosenberg ES, Billingsley JM, Colbert DS, Jones NG, Shea AK, Trocha AK, Walker BD. Association between virus-specific cytotoxic T-lymphocyte and helper responses in human immunodeficiency virus type 1 infection. J Virol. 1999;73:6715-6720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 275] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 45. | Matloubian M, Concepcion RJ, Ahmed R. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J Virol. 1994;68:8056-8063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 885] [Cited by in RCA: 910] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 46. | Chevalier MF, Weiss L. The split personality of regulatory T cells in HIV infection. Blood. 2013;121:29-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 168] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 47. | Alp Avcı G. [Genomic organization and proteins of human papillomavirus]. Mikrobiyol Bul. 2012;46:507-515. [PubMed] |
| 48. | Chen LP, Thomas EK, Hu SL, Hellström I, Hellström KE. Human papillomavirus type 16 nucleoprotein E7 is a tumor rejection antigen. Proc Natl Acad Sci USA. 1991;88:110-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 158] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 49. | Feltkamp MC, Smits HL, Vierboom MP, Minnaar RP, de Jongh BM, Drijfhout JW, ter Schegget J, Melief CJ, Kast WM. Vaccination with cytotoxic T lymphocyte epitope-containing peptide protects against a tumor induced by human papillomavirus type 16-transformed cells. Eur J Immunol. 1993;23:2242-2249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 607] [Cited by in RCA: 603] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 50. | Evans EM, Man S, Evans AS, Borysiewicz LK. Infiltration of cervical cancer tissue with human papillomavirus-specific cytotoxic T-lymphocytes. Cancer Res. 1997;57:2943-2950. [PubMed] |
| 51. | Melief CJ, Van Der Burg SH, Toes RE, Ossendorp F, Offringa R. Effective therapeutic anticancer vaccines based on precision guiding of cytolytic T lymphocytes. Immunol Rev. 2002;188:177-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 83] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 52. | de Jong A, van der Burg SH, Kwappenberg KM, van der Hulst JM, Franken KL, Geluk A, van Meijgaarden KE, Drijfhout JW, Kenter G, Vermeij P, Melief CJ, Offringa R. Frequent detection of human papillomavirus 16 E2-specific T-helper immunity in healthy subjects. Cancer Res. 2002;62:472-479. [PubMed] |
| 53. | de Jong A, van Poelgeest MI, van der Hulst JM, Drijfhout JW, Fleuren GJ, Melief CJ, Kenter G, Offringa R, van der Burg SH. Human papillomavirus type 16-positive cervical cancer is associated with impaired CD4+ T-cell immunity against early antigens E2 and E6. Cancer Res. 2004;64:5449-5455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 236] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 54. | Nakagawa M, Stites DP, Farhat S, Sisler JR, Moss B, Kong F, Moscicki AB, Palefsky JM. Cytotoxic T lymphocyte responses to E6 and E7 proteins of human papillomavirus type 16: relationship to cervical intraepithelial neoplasia. J Infect Dis. 1997;175:927-931. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 110] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 55. | Nakagawa M, Stites DP, Patel S, Farhat S, Scott M, Hills NK, Palefsky JM, Moscicki AB. Persistence of human papillomavirus type 16 infection is associated with lack of cytotoxic T lymphocyte response to the E6 antigens. J Infect Dis. 2000;182:595-598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 115] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 56. | van den Hende M, van Poelgeest MI, van der Hulst JM, de Jong J, Drijfhout JW, Fleuren GJ, Valentijn AR, Wafelman AR, Slappendel GM, Melief CJ, Offringa R, van der Burg SH, Kenter GG. Skin reactions to human papillomavirus (HPV) 16 specific antigens intradermally injected in healthy subjects and patients with cervical neoplasia. Int J Cancer. 2008;123:146-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 57. | IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Human papillomaviruses. IARC Monogr Eval Carcinog Risks Hum. 2007;90:1-636. [PubMed] |
| 58. | Vambutas A, Bonagura VR, Steinberg BM. Altered expression of TAP-1 and major histocompatibility complex class I in laryngeal papillomatosis: correlation of TAP-1 with disease. Clin Diagn Lab Immunol. 2000;7:79-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 59. | Georgopoulos NT, Proffitt JL, Blair GE. Transcriptional regulation of the major histocompatibility complex (MHC) class I heavy chain, TAP1 and LMP2 genes by the human papillomavirus (HPV) type 6b, 16 and 18 E7 oncoproteins. Oncogene. 2000;19:4930-4935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 118] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 60. | Vambutas A, DeVoti J, Pinn W, Steinberg BM, Bonagura VR. Interaction of human papillomavirus type 11 E7 protein with TAP-1 results in the reduction of ATP-dependent peptide transport. Clin Immunol. 2001;101:94-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 52] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 61. | Ashrafi GH, Haghshenas MR, Marchetti B, O'Brien PM, Campo MS. E5 protein of human papillomavirus type 16 selectively downregulates surface HLA class I. Int J Cancer. 2005;113:276-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 170] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 62. | Zhang B, Li P, Wang E, Brahmi Z, Dunn KW, Blum JS, Roman A. The E5 protein of human papillomavirus type 16 perturbs MHC class II antigen maturation in human foreskin keratinocytes treated with interferon-gamma. Virology. 2003;310:100-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 86] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 63. | Ahdieh L, Klein RS, Burk R, Cu-Uvin S, Schuman P, Duerr A, Safaeian M, Astemborski J, Daniel R, Shah K. Prevalence, incidence, and type-specific persistence of human papillomavirus in human immunodeficiency virus (HIV)-positive and HIV-negative women. J Infect Dis. 2001;184:682-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 228] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 64. | Brickman C, Palefsky JM. Human papillomavirus in the HIV-infected host: epidemiology and pathogenesis in the antiretroviral era. Curr HIV/AIDS Rep. 2015;12:6-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 80] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 65. | Kim RH, Yochim JM, Kang MK, Shin KH, Christensen R, Park NH. HIV-1 Tat enhances replicative potential of human oral keratinocytes harboring HPV-16 genome. Int J Oncol. 2008;33:777-782. [PubMed] |
| 66. | Tornesello ML, Buonaguro FM, Beth-Giraldo E, Giraldo G. Human immunodeficiency virus type 1 tat gene enhances human papillomavirus early gene expression. Intervirology. 1993;36:57-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 50] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 67. | Tan W, Felber BK, Zolotukhin AS, Pavlakis GN, Schwartz S. Efficient expression of the human papillomavirus type 16 L1 protein in epithelial cells by using Rev and the Rev-responsive element of human immunodeficiency virus or the cis-acting transactivation element of simian retrovirus type 1. J Virol. 1995;69:5607-5620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 77] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 68. | Toy EP, Rodríguez-Rodríguez L, McCance D, Ludlow J, Planelles V. Induction of cell-cycle arrest in cervical cancer cells by the human immunodeficiency virus type 1 viral protein R. Obstet Gynecol. 2000;95:141-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 69. | Darragh TM, Colgan TJ, Thomas Cox J, Heller DS, Henry MR, Luff RD, McCalmont T, Nayar R, Palefsky JM, Stoler MH, Wilkinson EJ, Zaino RJ, Wilbur DC; Members of the LAST Project Work Groups. The Lower Anogenital Squamous Terminology Standardization project for HPV-associated lesions: background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology. Int J Gynecol Pathol. 2013;32:76-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 407] [Cited by in RCA: 387] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 70. | Leeds IL, Fang SH. Anal cancer and intraepithelial neoplasia screening: A review. World J Gastrointest Surg. 2016;8:41-51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 113] [Cited by in RCA: 116] [Article Influence: 12.9] [Reference Citation Analysis (1)] |
| 71. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55853] [Article Influence: 7979.0] [Reference Citation Analysis (132)] |
| 72. | Wilkinson JR, Morris EJ, Downing A, Finan PJ, Aravani A, Thomas JD, Sebag-Montefiore D. The rising incidence of anal cancer in England 1990-2010: a population-based study. Colorectal Dis. 2014;16:O234-O239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 69] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 73. | Shiels MS, Kreimer AR, Coghill AE, Darragh TM, Devesa SS. Anal Cancer Incidence in the United States, 1977-2011: Distinct Patterns by Histology and Behavior. Cancer Epidemiol Biomarkers Prev. 2015;24:1548-1556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 124] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 74. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8287] [Cited by in RCA: 11943] [Article Influence: 2985.8] [Reference Citation Analysis (4)] |
| 75. | Daling JR, Madeleine MM, Johnson LG, Schwartz SM, Shera KA, Wurscher MA, Carter JJ, Porter PL, Galloway DA, McDougall JK. Human papillomavirus, smoking, and sexual practices in the etiology of anal cancer. Cancer. 2004;101:270-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 565] [Cited by in RCA: 570] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 76. | Machalek DA, Poynten M, Jin F, Fairley CK, Farnsworth A, Garland SM, Hillman RJ, Petoumenos K, Roberts J, Tabrizi SN, Templeton DJ, Grulich AE. Anal human papillomavirus infection and associated neoplastic lesions in men who have sex with men: a systematic review and meta-analysis. Lancet Oncol. 2012;13:487-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 796] [Cited by in RCA: 719] [Article Influence: 55.3] [Reference Citation Analysis (0)] |
| 77. | Frisch M, Glimelius B, van den Brule AJ, Wohlfahrt J, Meijer CJ, Walboomers JM, Goldman S, Svensson C, Adami HO, Melbye M. Sexually transmitted infection as a cause of anal cancer. N Engl J Med. 1997;337:1350-1358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 425] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 78. | Chin-Hong PV, Husnik M, Cranston RD, Colfax G, Buchbinder S, Da Costa M, Darragh T, Jones D, Judson F, Koblin B, Mayer KH, Palefsky JM. Anal human papillomavirus infection is associated with HIV acquisition in men who have sex with men. AIDS. 2009;23:1135-1142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 110] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 79. | Darwich L, Cañadas MP, Videla S, Coll J, Molina-López RA, Sirera G, Clotet B; Can Ruti HIV-HPV Team. Prevalence, clearance, and incidence of human papillomavirus type-specific infection at the anal and penile site of HIV-infected men. Sex Transm Dis. 2013;40:611-618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 49] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 80. | Vajdic CM, van Leeuwen MT, Jin F, Prestage G, Medley G, Hillman RJ, Stevens MP, Botes LP, Zablotska I, Tabrizi SN, Grulich AE. Anal human papillomavirus genotype diversity and co-infection in a community-based sample of homosexual men. Sex Transm Infect. 2009;85:330-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 75] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 81. | Hernandez AL, Efird JT, Holly EA, Berry JM, Jay N, Palefsky JM. Incidence of and risk factors for type-specific anal human papillomavirus infection among HIV-positive MSM. AIDS. 2014;28:1341-1349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 82. | Müller EE, Rebe K, Chirwa TF, Struthers H, McIntyre J, Lewis DA. The prevalence of human papillomavirus infections and associated risk factors in men-who-have-sex-with-men in Cape Town, South Africa. BMC Infect Dis. 2016;16:440. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 83. | D'Souza G, Wiley DJ, Li X, Chmiel JS, Margolick JB, Cranston RD, Jacobson LP. Incidence and epidemiology of anal cancer in the multicenter AIDS cohort study. J Acquir Immune Defic Syndr. 2008;48:491-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 249] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 84. | Robbins HA, Shiels MS, Pfeiffer RM, Engels EA. Epidemiologic contributions to recent cancer trends among HIV-infected people in the United States. AIDS. 2014;28:881-890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 165] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 85. | Geskus RB, González C, Torres M, Del Romero J, Viciana P, Masiá M, Blanco JR, Iribarren M, De Sanjosé S, Hernández-Novoa B, Ortiz M, Del Amo J; CoRIS-HPV Study Group. Incidence and clearance of anal high-risk human papillomavirus in HIV-positive men who have sex with men: estimates and risk factors. AIDS. 2016;30:37-44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 46] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 86. | Looker KJ, Rönn MM, Brock PM, Brisson M, Drolet M, Mayaud P, Boily MC. Evidence of synergistic relationships between HIV and Human Papillomavirus (HPV): systematic reviews and meta-analyses of longitudinal studies of HPV acquisition and clearance by HIV status, and of HIV acquisition by HPV status. J Int AIDS Soc. 2018;21:e25110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 115] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 87. | Bertisch B, Franceschi S, Lise M, Vernazza P, Keiser O, Schöni-Affolter F, Bouchardy C, Dehler S, Levi F, Jundt G, Ess S, Pawlita M, Kovari H, Wandeler G, Calmy A, Cavassini M, Stöckle M, Clifford G; Swiss HIV Cohort Study Investigators. Risk factors for anal cancer in persons infected with HIV: a nested case-control study in the Swiss HIV Cohort Study. Am J Epidemiol. 2013;178:877-884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 103] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 88. | Goldie SJ, Kuntz KM, Weinstein MC, Freedberg KA, Palefsky JM. Cost-effectiveness of screening for anal squamous intraepithelial lesions and anal cancer in human immunodeficiency virus-negative homosexual and bisexual men. Am J Med. 2000;108:634-641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 145] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 89. | Goldie SJ, Kuntz KM, Weinstein MC, Freedberg KA, Welton ML, Palefsky JM. The clinical effectiveness and cost-effectiveness of screening for anal squamous intraepithelial lesions in homosexual and bisexual HIV-positive men. JAMA. 1999;281:1822-1829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 307] [Cited by in RCA: 288] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 90. | Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835-840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3232] [Cited by in RCA: 3113] [Article Influence: 207.5] [Reference Citation Analysis (0)] |
| 91. | Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 842] [Cited by in RCA: 827] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 92. | Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA. 2008;105:1608-1613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 860] [Cited by in RCA: 918] [Article Influence: 54.0] [Reference Citation Analysis (0)] |
| 93. | Huang V, Place RF, Portnoy V, Wang J, Qi Z, Jia Z, Yu A, Shuman M, Yu J, Li LC. Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res. 2012;40:1695-1707. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 235] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 94. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14460] [Cited by in RCA: 16089] [Article Influence: 1005.6] [Reference Citation Analysis (2)] |
| 95. | Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Zen K, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997-1006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3218] [Cited by in RCA: 3554] [Article Influence: 209.1] [Reference Citation Analysis (0)] |
| 96. | Wang X, Wang HK, Li Y, Hafner M, Banerjee NS, Tang S, Briskin D, Meyers C, Chow LT, Xie X, Tuschl T, Zheng ZM. microRNAs are biomarkers of oncogenic human papillomavirus infections. Proc Natl Acad Sci USA. 2014;111:4262-4267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 160] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 97. | Lajer CB, Garnæs E, Friis-Hansen L, Norrild B, Therkildsen MH, Glud M, Rossing M, Lajer H, Svane D, Skotte L, Specht L, Buchwald C, Nielsen FC. The role of miRNAs in human papilloma virus (HPV)-associated cancers: bridging between HPV-related head and neck cancer and cervical cancer. Br J Cancer. 2012;106:1526-1534. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 186] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 98. | Harden ME, Prasad N, Griffiths A, Munger K. Modulation of microRNA-mRNA Target Pairs by Human Papillomavirus 16 Oncoproteins. mBio. 2017;8:e02170-16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 59] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 99. | Mar-Aguilar F, Mendoza-Ramírez JA, Malagón-Santiago I, Espino-Silva PK, Santuario-Facio SK, Ruiz-Flores P, Rodríguez-Padilla C, Reséndez-Pérez D. Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis Markers. 2013;34:163-169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 126] [Reference Citation Analysis (0)] |
| 100. | Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7:e30679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 712] [Cited by in RCA: 862] [Article Influence: 66.3] [Reference Citation Analysis (2)] |
| 101. | Nedaeinia R, Manian M, Jazayeri MH, Ranjbar M, Salehi R, Sharifi M, Mohaghegh F, Goli M, Jahednia SH, Avan A, Ghayour-Mobarhan M. Circulating exosomes and exosomal microRNAs as biomarkers in gastrointestinal cancer. Cancer Gene Ther. 2017;24:48-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 164] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 102. | Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Ménard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065-7070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2930] [Cited by in RCA: 3055] [Article Influence: 152.8] [Reference Citation Analysis (0)] |
| 103. | Tian F, Li R, Chen Z, Shen Y, Lu J, Xie X, Ge Q. Differentially Expressed miRNAs in Tumor, Adjacent, and Normal Tissues of Lung Adenocarcinoma. Biomed Res Int. 2016;2016:1428271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 104. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7723] [Cited by in RCA: 7370] [Article Influence: 368.5] [Reference Citation Analysis (0)] |
| 105. | Gocze K, Gombos K, Kovacs K, Juhasz K, Gocze P, Kiss I. MicroRNA expressions in HPV-induced cervical dysplasia and cancer. Anticancer Res. 2015;35:523-530. [PubMed] |
| 106. | Louten J, Beach M, Palermino K, Weeks M, Holenstein G. MicroRNAs Expressed during Viral Infection: Biomarker Potential and Therapeutic Considerations. Biomark Insights. 2015;10:25-52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 107. | Farzanehpour M, Mozhgani SH, Jalilvand S, Faghihloo E, Akhavan S, Salimi V, Azad TM. Serum and tissue miRNAs: potential biomarkers for the diagnosis of cervical cancer. Virol J. 2019;16:116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 108. | Snoek BC, Verlaat W, Babion I, Novianti PW, van de Wiel MA, Wilting SM, van Trommel NE, Bleeker MCG, Massuger LFAG, Melchers WJG, Sie D, Heideman DAM, Snijders PJF, Meijer CJLM, Steenbergen RDM. Genome-wide microRNA analysis of HPV-positive self-samples yields novel triage markers for early detection of cervical cancer. Int J Cancer. 2019;144:372-379. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 109. | Shi C, Yang Y, Zhang L, Zhang T, Yu J, Qin S, Gao Y. Optimal subset of signature miRNAs consisting of 7 miRNAs that can serve as a novel diagnostic and prognostic predictor for the progression of cervical cancer. Oncol Rep. 2019;41:3167-3178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 110. | Lee JW, Choi CH, Choi JJ, Park YA, Kim SJ, Hwang SY, Kim WY, Kim TJ, Lee JH, Kim BG, Bae DS. Altered MicroRNA expression in cervical carcinomas. Clin Cancer Res. 2008;14:2535-2542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 239] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 111. | Myklebust MP, Bruland O, Fluge Ø, Skarstein A, Balteskard L, Dahl O. MicroRNA-15b is induced with E2F-controlled genes in HPV-related cancer. Br J Cancer. 2011;105:1719-1725. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 112. | Sun G, Rossi JJ. MicroRNAs and their potential involvement in HIV infection. Trends Pharmacol Sci. 2011;32:675-681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 113. | Tan Gana NH, Onuki T, Victoriano AF, Okamoto T. MicroRNAs in HIV-1 infection: an integration of viral and cellular interaction at the genomic level. Front Microbiol. 2012;3:306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 114. | Swaminathan G, Navas-Martín S, Martín-García J. MicroRNAs and HIV-1 infection: antiviral activities and beyond. J Mol Biol. 2014;426:1178-1197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 92] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 115. | Bennasser Y, Le SY, Benkirane M, Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22:607-619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 343] [Cited by in RCA: 344] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 116. | Bennasser Y, Yeung ML, Jeang KT. HIV-1 TAR RNA subverts RNA interference in transfected cells through sequestration of TAR RNA-binding protein, TRBP. J Biol Chem. 2006;281:27674-27678. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 90] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 117. | Casey Klockow L, Sharifi HJ, Wen X, Flagg M, Furuya AK, Nekorchuk M, de Noronha CM. The HIV-1 protein Vpr targets the endoribonuclease Dicer for proteasomal degradation to boost macrophage infection. Virology. 2013;444:191-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 118. | Biswas S, Chen E, Haleyurgirisetty M, Lee S, Hewlett I, Devadas K. Comparison of miRNA Expression Profiles between HIV-1 and HIV-2 Infected Monocyte-Derived Macrophages (MDMs) and Peripheral Blood Mononuclear Cells (PBMCs). Int J Mol Sci. 2020;21:6970. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 119. | Barbu MG, Condrat CE, Thompson DC, Bugnar OL, Cretoiu D, Toader OD, Suciu N, Voinea SC. MicroRNA Involvement in Signaling Pathways During Viral Infection. Front Cell Dev Biol. 2020;8:143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 106] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 120. | Ribeiro J, Marinho-Dias J, Monteiro P, Loureiro J, Baldaque I, Medeiros R, Sousa H. miR-34a and miR-125b Expression in HPV Infection and Cervical Cancer Development. Biomed Res Int. 2015;2015:304584. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 121. | Hong DS, Kang YK, Borad M, Sachdev J, Ejadi S, Lim HY, Brenner AJ, Park K, Lee JL, Kim TY, Shin S, Becerra CR, Falchook G, Stoudemire J, Martin D, Kelnar K, Peltier H, Bonato V, Bader AG, Smith S, Kim S, O'Neill V, Beg MS. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br J Cancer. 2020;122:1630-1637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 409] [Cited by in RCA: 594] [Article Influence: 118.8] [Reference Citation Analysis (0)] |
| 122. | Chakraborty C, Sharma AR, Sharma G, Lee SS. Therapeutic advances of miRNAs: A preclinical and clinical update. J Adv Res. 2021;28:127-138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 264] [Cited by in RCA: 281] [Article Influence: 70.3] [Reference Citation Analysis (0)] |
| 123. | Ottosen S, Parsley TB, Yang L, Zeh K, van Doorn LJ, van der Veer E, Raney AK, Hodges MR, Patick AK. In vitro antiviral activity and preclinical and clinical resistance profile of miravirsen, a novel anti-hepatitis C virus therapeutic targeting the human factor miR-122. Antimicrob Agents Chemother. 2015;59:599-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 184] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 124. | Hanna J, Hossain GS, Kocerha J. The Potential for microRNA Therapeutics and Clinical Research. Front Genet. 2019;10:478. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 366] [Cited by in RCA: 559] [Article Influence: 93.2] [Reference Citation Analysis (0)] |
| 125. | Moghoofei M, Najafipour S, Mostafaei S, Tavakoli A, Bokharaei-Salim F, Ghorbani S, Javanmard D, Ghaffari H, Monavari SH. MicroRNAs Profiling in HIV, HCV, and HIV/HCV Co-Infected Patients. Curr HIV Res. 2021;19:27-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 126. | Ma L, Zhang H, Zhang Y, Li H, An M, Zhao B, Ding H, Xu J, Shang H, Han X. Integrated analysis of lncRNA, miRNA and mRNA profiles reveals potential lncRNA functions during early HIV infection. J Transl Med. 2021;19:135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 127. | Heinson AI, Woo J, Mukim A, White CH, Moesker B, Bosque A, Spina CA, Woelk CH, Macarthur BD, Beliakova-Bethell N. Micro RNA Targets in HIV Latency: Insights into Novel Layers of Latency Control. AIDS Res Hum Retroviruses. 2021;37:109-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 128. | Shah UJ, Nasiruddin M, Dar SA, Khan MKA, Akhter MR, Singh N, Rabaan AA, Haque S. Emerging biomarkers and clinical significance of HPV genotyping in prevention and management of cervical cancer. Microb Pathog. 2020;143:104131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 129. | Sadri Nahand J, Moghoofei M, Salmaninejad A, Bahmanpour Z, Karimzadeh M, Nasiri M, Mirzaei HR, Pourhanifeh MH, Bokharaei-Salim F, Mirzaei H, Hamblin MR. Pathogenic role of exosomes and microRNAs in HPV-mediated inflammation and cervical cancer: A review. Int J Cancer. 2020;146:305-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 182] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 130. | Chen Y, Song Y, Mi Y, Jin H, Cao J, Li H, Han L, Huang T, Zhang X, Ren S, Ma Q, Zou Z. microRNA-499a promotes the progression and chemoresistance of cervical cancer cells by targeting SOX6. Apoptosis. 2020;25:205-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 131. | Teng Y, Wang Y, Zhang X, Liu W, Fan H, Yao H, Lin B, Zhu P, Yuan W, Tong Y, Cao W. Systematic Genome-wide Screening and Prediction of microRNAs in EBOV During the 2014 Ebolavirus Outbreak. Sci Rep. 2015;5:9912. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 132. | Saini S, Saini A, Thakur CJ, Kumar V, Gupta RD, Sharma JK. Genome-wide computational prediction of miRNAs in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) revealed target genes involved in pulmonary vasculature and antiviral innate immunity. Mol Biol Res Commun. 2020;9:83-91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 133. | Mumford SL, Towler BP, Pashler AL, Gilleard O, Martin Y, Newbury SF. Circulating MicroRNA Biomarkers in Melanoma: Tools and Challenges in Personalised Medicine. Biomolecules. 2018;8:21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 134. | Shams R, Saberi S, Zali M, Sadeghi A, Ghafouri-Fard S, Aghdaei HA. Identification of potential microRNA panels for pancreatic cancer diagnosis using microarray datasets and bioinformatics methods. Sci Rep. 2020;10:7559. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 135. | Rehman O, Zhuang H, Muhamed Ali A, Ibrahim A, Li Z. Validation of miRNAs as Breast Cancer Biomarkers with a Machine Learning Approach. Cancers (Basel). 2019;11:431. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 136. | Zhu J, Xu Y, Liu S, Qiao L, Sun J, Zhao Q. MicroRNAs Associated With Colon Cancer: New Potential Prognostic Markers and Targets for Therapy. Front Bioeng Biotechnol. 2020;8:176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 137. | Kang HW, Crawford M, Fabbri M, Nuovo G, Garofalo M, Nana-Sinkam SP, Friedman A. A mathematical model for microRNA in lung cancer. PLoS One. 2013;8:e53663. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 138. | Zhu CS, Zhu L, Tan DA, Qiu XY, Liu CY, Xie SS, Zhu LY. Avenues Toward microRNA Detection In Vitro: A Review of Technical Advances and Challenges. Comput Struct Biotechnol J. 2019;17:904-916. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 139. | Dave VP, Ngo TA, Pernestig AK, Tilevik D, Kant K, Nguyen T, Wolff A, Bang DD. MicroRNA amplification and detection technologies: opportunities and challenges for point of care diagnostics. Lab Invest. 2019;99:452-469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 155] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 140. | Jia H, Li Z, Liu C, Cheng Y. Ultrasensitive detection of microRNAs by exponential isothermal amplification. Angew Chem Int Ed Engl. 2010;49:5498-5501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 407] [Cited by in RCA: 413] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 141. | Ying N, Ju C, Sun X, Li L, Chang H, Song G, Li Z, Wan J, Dai E. Lateral flow nucleic acid biosensor for sensitive detection of microRNAs based on the dual amplification strategy of duplex-specific nuclease and hybridization chain reaction. PLoS One. 2017;12:e0185091. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 142. | Degliangeli F, Pompa PP, Fiammengo R. Nanotechnology-based strategies for the detection and quantification of microRNA. Chemistry. 2014;20:9476-9492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 143. | Hamidi-Asl E, Palchetti I, Hasheminejad E, Mascini M. A review on the electrochemical biosensors for determination of microRNAs. Talanta. 2013;115:74-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 88] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 144. | Arata H, Komatsu H, Hosokawa K, Maeda M. Rapid and sensitive microRNA detection with laminar flow-assisted dendritic amplification on power-free microfluidic chip. PLoS One. 2012;7:e48329. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 145. | Salim B, Athira MV, Kandaswamy A, Vijayakumar M, Saravanan T, Sairam T. Microfluidic device for novel breast cancer screening by blood test using miRNA beacon probe. Biomed Microdevices. 2017;19:89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 146. | Lässer C. Exosomal RNA as biomarkers and the therapeutic potential of exosome vectors. Expert Opin Biol Ther. 2012;12 Suppl 1:S189-S197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 98] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 147. | Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513-10518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5636] [Cited by in RCA: 6315] [Article Influence: 371.5] [Reference Citation Analysis (0)] |
| 148. | Subra C, Grand D, Laulagnier K, Stella A, Lambeau G, Paillasse M, De Medina P, Monsarrat B, Perret B, Silvente-Poirot S, Poirot M, Record M. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J Lipid Res. 2010;51:2105-2120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 425] [Cited by in RCA: 506] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 149. | Nik Mohamed Kamal NNSB, Shahidan WNS. Non-Exosomal and Exosomal Circulatory MicroRNAs: Which Are More Valid as Biomarkers? Front Pharmacol. 2019;10:1500. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 139] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 150. | Fu F, Jiang W, Zhou L, Chen Z. Circulating Exosomal miR-17-5p and miR-92a-3p Predict Pathologic Stage and Grade of Colorectal Cancer. Transl Oncol. 2018;11:221-232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 163] [Article Influence: 23.3] [Reference Citation Analysis (1)] |
| 151. | Goto T, Fujiya M, Konishi H, Sasajima J, Fujibayashi S, Hayashi A, Utsumi T, Sato H, Iwama T, Ijiri M, Sakatani A, Tanaka K, Nomura Y, Ueno N, Kashima S, Moriichi K, Mizukami Y, Kohgo Y, Okumura T. An elevated expression of serum exosomal microRNA-191, - 21, -451a of pancreatic neoplasm is considered to be efficient diagnostic marker. BMC Cancer. 2018;18:116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 187] [Cited by in RCA: 191] [Article Influence: 27.3] [Reference Citation Analysis (1)] |
| 152. | Jackson R, Eade S, Zehbe I. An epithelial organoid model with Langerhans cells for assessing virus-host interactions. Philos Trans R Soc Lond B Biol Sci. 2019;374:20180288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 153. | Driehuis E, Kolders S, Spelier S, Lõhmussaar K, Willems SM, Devriese LA, de Bree R, de Ruiter EJ, Korving J, Begthel H, van Es JH, Geurts V, He GW, van Jaarsveld RH, Oka R, Muraro MJ, Vivié J, Zandvliet MMJM, Hendrickx APA, Iakobachvili N, Sridevi P, Kranenburg O, van Boxtel R, Kops GJPL, Tuveson DA, Peters PJ, van Oudenaarden A, Clevers H. Oral Mucosal Organoids as a Potential Platform for Personalized Cancer Therapy. Cancer Discov. 2019;9:852-871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 260] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 154. | Fiorini E, Veghini L, Corbo V. Modeling Cell Communication in Cancer With Organoids: Making the Complex Simple. Front Cell Dev Biol. 2020;8:166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 72] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 155. | Lõhmussaar K, Oka R, Espejo Valle-Inclan J, Smits MHH, Wardak H, Korving J, Begthel H, Proost N, van de Ven M, Kranenburg OW, Jonges TGN, Zweemer RP, Veersema S, van Boxtel R, Clevers H. Patient-derived organoids model cervical tissue dynamics and viral oncogenesis in cervical cancer. Cell Stem Cell. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 122] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 156. | Liu X. Up-regulation of miR-20a by HPV16 E6 exerts growth-promoting effects by targeting PDCD6 in cervical carcinoma cells. Biomed Pharmacother. 2018;102:996-1002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 157. | Cheng Y, Geng L, Zhao L, Zuo P, Wang J. Human papillomavirus E6-regulated microRNA-20b promotes invasion in cervical cancer by targeting tissue inhibitor of metalloproteinase 2. Mol Med Rep. 2017;16:5464-5470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 158. | Au Yeung CL, Tsang TY, Yau PL, Kwok TT. Human papillomavirus type 16 E6 induces cervical cancer cell migration through the p53/microRNA-23b/urokinase-type plasminogen activator pathway. Oncogene. 2011;30:2401-2410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 114] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 159. | Wu YL, Hsu NY, Cheau-Feng Lin F, Lee H, Cheng YW. MiR-30c-2* negative regulated MTA-1 expression involved in metastasis and drug resistance of HPV-infected non-small cell lung cancer. Surgery. 2016;160:1591-1598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 160. | Wang X, Wang HK, McCoy JP, Banerjee NS, Rader JS, Broker TR, Meyers C, Chow LT, Zheng ZM. Oncogenic HPV infection interrupts the expression of tumor-suppressive miR-34a through viral oncoprotein E6. RNA. 2009;15:637-647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 180] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 161. | Zhang R, Su J, Xue SL, Yang H, Ju LL, Ji Y, Wu KH, Zhang YW, Zhang YX, Hu JF, Yu MM. HPV E6/p53 mediated down-regulation of miR-34a inhibits Warburg effect through targeting LDHA in cervical cancer. Am J Cancer Res. 2016;6:312-320. [PubMed] |
| 162. | Shi M, Du L, Liu D, Qian L, Hu M, Yu M, Yang Z, Zhao M, Chen C, Guo L, Wang L, Song L, Ma Y, Guo N. Glucocorticoid regulation of a novel HPV-E6-p53-miR-145 pathway modulates invasion and therapy resistance of cervical cancer cells. J Pathol. 2012;228:148-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 70] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 163. | Zhong J, Yuan H, Xu X, Kong S. MicroRNA195 inhibits cell proliferation, migration and invasion by targeting defective in cullin neddylation 1 domain containing 1 in cervical cancer. Int J Mol Med. 2018;42:779-788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 164. | Martinez I, Gardiner AS, Board KF, Monzon FA, Edwards RP, Khan SA. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene. 2008;27:2575-2582. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 270] [Cited by in RCA: 276] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 165. | Jiang Z, Song Q, Zeng R, Li J, Lin X, Chen X, Zhang J, Zheng Y. MicroRNA-218 inhibits EMT, migration and invasion by targeting SFMBT1 and DCUN1D1 in cervical cancer. Oncotarget. 2016;7:45622-45636. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 166. | Díaz-González Sdel M, Deas J, Benítez-Boijseauneau O, Gómez-Cerón C, Bermúdez-Morales VH, Rodríguez-Dorantes M, Pérez-Plasencia C, Peralta-Zaragoza O. Utility of microRNAs and siRNAs in cervical carcinogenesis. Biomed Res Int. 2015;2015:374924. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 167. | Liu S, Song L, Yao H, Zhang L, Xu D, Gao F, Li Q. MiR-375 Is Epigenetically Downregulated by HPV-16 E6 Mediated DNMT1 Upregulation and Modulates EMT of Cervical Cancer Cells by Suppressing lncRNA MALAT1. PLoS One. 2016;11:e0163460. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 168. | Xu J, Wan X, Chen X, Fang Y, Cheng X, Xie X, Lu W. miR-2861 acts as a tumor suppressor via targeting EGFR/AKT2/CCND1 pathway in cervical cancer induced by human papillomavirus virus 16 E6. Sci Rep. 2016;6:28968. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 169. | Hu J, Ge W, Xu J. HPV 16 E7 inhibits OSCC cell proliferation, invasion, and metastasis by upregulating the expression of miR-20a. Tumour Biol. 2016;37:9433-9440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 170. | Kong Q, Wang W, Li P. Regulator role of HPV E7 protein on miR-21 expression in cervical carcinoma cells and its functional implication. Int J Clin Exp Pathol. 2015;8:15808-15813. [PubMed] |
| 171. | Bumrungthai S, Ekalaksananan T, Evans MF, Chopjitt P, Tangsiriwatthana T, Patarapadungkit N, Kleebkaow P, Luanratanakorn S, Kongyingyoes B, Worawichawong S, Pientong C. Up-Regulation of miR-21 Is Associated with Cervicitis and Human Papillomavirus Infection in Cervical Tissues. PLoS One. 2015;10:e0127109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 48] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 172. | Liu F, Zhang S, Zhao Z, Mao X, Huang J, Wu Z, Zheng L, Wang Q. MicroRNA-27b up-regulated by human papillomavirus 16 E7 promotes proliferation and suppresses apoptosis by targeting polo-like kinase2 in cervical cancer. Oncotarget. 2016;7:19666-19679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 173. | Yao J, Deng B, Zheng L, Dou L, Guo Y, Guo K. miR-27b is upregulated in cervical carcinogenesis and promotes cell growth and invasion by regulating CDH11 and epithelial-mesenchymal transition. Oncol Rep. 2016;35:1645-1651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 174. | McKenna DJ, Patel D, McCance DJ. miR-24 and miR-205 expression is dependent on HPV onco-protein expression in keratinocytes. Virology. 2014;448:210-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 175. | Mandal P, Saha SS, Sen S, Bhattacharya A, Bhattacharya NP, Bucha S, Sinha M, Chowdhury RR, Mondal NR, Chakravarty B, Chatterjee T, Roy S, Chattapadhyay A, Sengupta S. Cervical cancer subtypes harbouring integrated and/or episomal HPV16 portray distinct molecular phenotypes based on transcriptome profiling of mRNAs and miRNAs. Cell Death Discov. 2019;5:81. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 176. | Xin M, Qiao Z, Li J, Liu J, Song S, Zhao X, Miao P, Tang T, Wang L, Liu W, Yang X, Dai K, Huang G. miR-22 inhibits tumor growth and metastasis by targeting ATP citrate lyase: evidence in osteosarcoma, prostate cancer, cervical cancer and lung cancer. Oncotarget. 2016;7:44252-44265. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 142] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 177. | Fan Z, Cui H, Xu X, Lin Z, Zhang X, Kang L, Han B, Meng J, Yan Z, Yan X, Jiao S. MiR-125a suppresses tumor growth, invasion and metastasis in cervical cancer by targeting STAT3. Oncotarget. 2015;6:25266-25280. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 127] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 178. | Peta E, Sinigaglia A, Masi G, Di Camillo B, Grassi A, Trevisan M, Messa L, Loregian A, Manfrin E, Brunelli M, Martignoni G, Palù G, Barzon L. HPV16 E6 and E7 upregulate the histone lysine demethylase KDM2B through the c-MYC/miR-146a-5p axys. Oncogene. 2018;37:1654-1668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 179. | Melar-New M, Laimins LA. Human papillomaviruses modulate expression of microRNA 203 upon epithelial differentiation to control levels of p63 proteins. J Virol. 2010;84:5212-5221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 141] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 180. | McKenna DJ, McDade SS, Patel D, McCance DJ. MicroRNA 203 expression in keratinocytes is dependent on regulation of p53 Levels by E6. J Virol. 2010;84:10644-10652. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 66] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 181. | Witwer KW, Watson AK, Blankson JN, Clements JE. Relationships of PBMC microRNA expression, plasma viral load, and CD4+ T-cell count in HIV-1-infected elite suppressors and viremic patients. Retrovirology. 2012;9:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 131] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 182. | Houzet L, Yeung ML, de Lame V, Desai D, Smith SM, Jeang KT. MicroRNA profile changes in human immunodeficiency virus type 1 (HIV-1) seropositive individuals. Retrovirology. 2008;5:118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 178] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 183. | Sun G, Li H, Wu X, Covarrubias M, Scherer L, Meinking K, Luk B, Chomchan P, Alluin J, Gombart AF, Rossi JJ. Interplay between HIV-1 infection and host microRNAs. Nucleic Acids Res. 2012;40:2181-2196. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 114] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 184. | Balasubramaniam M, Pandhare J, Dash C. Are microRNAs Important Players in HIV-1 Infection? Viruses. 2018;10:110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 185. | Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, Reynes J, Corbeau P, Jeang KT, Benkirane M. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579-1582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 489] [Cited by in RCA: 508] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 186. | Wang P, Qu X, Zhou X, Shen Y, Ji H, Fu Z, Deng J, Lu P, Yu W, Lu H, Zhu H. Two cellular microRNAs, miR-196b and miR-1290, contribute to HIV-1 Latency. Virology. 2015;486:228-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 187. | Duskova K, Nagilla P, Le HS, Iyer P, Thalamuthu A, Martinson J, Bar-Joseph Z, Buchanan W, Rinaldo C, Ayyavoo V. MicroRNA regulation and its effects on cellular transcriptome in human immunodeficiency virus-1 (HIV-1) infected individuals with distinct viral load and CD4 cell counts. BMC Infect Dis. 2013;13:250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 188. | Huang J, Lai J, Liang B, Jiang J, Ning C, Liao Y, Zang N, Wang M, Qin F, Yu J, Wei W, Ye L, Liang H. mircoRNA-3162-3p is a potential biomarker to identify new infections in HIV-1-infected patients. Gene. 2018;662:21-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 189. | Swaminathan S, Suzuki K, Seddiki N, Kaplan W, Cowley MJ, Hood CL, Clancy JL, Murray DD, Méndez C, Gelgor L, Anderson B, Roth N, Cooper DA, Kelleher AD. Differential regulation of the Let-7 family of microRNAs in CD4+ T cells alters IL-10 expression. J Immunol. 2012;188:6238-6246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 124] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 190. | Biswas S, Haleyurgirisetty M, Lee S, Hewlett I, Devadas K. Development and validation of plasma miRNA biomarker signature panel for the detection of early HIV-1 infection. EBioMedicine. 2019;43:307-316. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 191. | Guha D, Mancini A, Sparks J, Ayyavoo V. HIV-1 Infection Dysregulates Cell Cycle Regulatory Protein p21 in CD4+ T Cells Through miR-20a and miR-106b Regulation. J Cell Biochem. 2016;117:1902-1912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 192. | Sánchez-Del Cojo M, López-Huertas MR, Díez-Fuertes F, Rodríguez-Mora S, Bermejo M, López-Campos G, Mateos E, Jiménez-Tormo L, Gómez-Esquer F, Díaz-Gil G, Alcamí J, Coiras M. Changes in the cellular microRNA profile by the intracellular expression of HIV-1 Tat regulator: A potential mechanism for resistance to apoptosis and impaired proliferation in HIV-1 infected CD4+ T cells. PLoS One. 2017;12:e0185677. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 193. | Rahimian P, He JJ. HIV-1 Tat-shortened neurite outgrowth through regulation of microRNA-132 and its target gene expression. J Neuroinflammation. 2016;13:247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 194. | Sardo L, Vakil PR, Elbezanti W, El-Sayed A, Klase Z. The inhibition of microRNAs by HIV-1 Tat suppresses beta catenin activity in astrocytes. Retrovirology. 2016;13:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 195. | Mishra R, Singh SK. HIV-1 Tat C modulates expression of miRNA-101 to suppress VE-cadherin in human brain microvascular endothelial cells. J Neurosci. 2013;33:5992-6000. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 62] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 196. | Hu G, Liao K, Yang L, Pendyala G, Kook Y, Fox HS, Buch S. Tat-Mediated Induction of miRs-34a & -138 Promotes Astrocytic Activation via Downregulation of SIRT1: Implications for Aging in HAND. J Neuroimmune Pharmacol. 2017;12:420-432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 197. | Aqil M, Naqvi AR, Mallik S, Bandyopadhyay S, Maulik U, Jameel S. The HIV Nef protein modulates cellular and exosomal miRNA profiles in human monocytic cells. J Extracell Vesicles. 2014;3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 198. | Yan Q, Zhao R, Shen C, Wang F, Li W, Gao SJ, Lu C. Upregulation of MicroRNA 711 Mediates HIV-1 Vpr Promotion of Kaposi's Sarcoma-Associated Herpesvirus Latency and Induction of Pro-proliferation and Pro-survival Cytokines by Targeting the Notch/NF-κB-Signaling Axis. J Virol. 2018;92:e00580-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |