Published online Jul 15, 2023. doi: 10.4239/wjd.v14.i7.1077
Peer-review started: March 6, 2023
First decision: March 14, 2023
Revised: March 27, 2023
Accepted: April 27, 2023
Article in press: April 27, 2023
Published online: July 15, 2023
Processing time: 129 Days and 0.2 Hours
Cataracts remain a prime reason for visual disturbance and blindness all over the world, despite the capacity for successful surgical replacement with artificial lenses. Diabetic cataract (DC), a metabolic complication, usually occurs at an earlier age and progresses faster than age-related cataracts. Evidence has linked N6-methyladenosine (m6A) to DC progression. However, there exists a lack of understanding regarding RNA m6A modifications and the role of m6A in DC pathogenesis.
To elucidate the role played by altered m6A and differentially expressed mRNAs (DEmRNAs) in DC.
Anterior lens capsules were collected from the control subjects and patients with DC. M6A epitranscriptomic microarray was performed to investigate the altered m6A modifications and determine the DEmRNAs. Through Gene Ontology and pathway enrichment (Kyoto Encyclopedia of Genes and Genomes) analyses, the potential role played by dysregulated m6A modification was predicted. Real-time polymerase chain reaction was further carried out to identify the dysregulated expression of RNA methyltransferases, demethylases, and readers.
Increased m6A abundance levels were found in the total mRNA of DC samples. Bioinformatics analysis predicted that ferroptosis pathways could be associated with m6A-modified mRNAs. The levels of five methylation-related genes-RBM15, WTAP, ALKBH5, FTO, and YTHDF1-were upregulated in DC samples. Upregulation of RBM15 expression was verified in SRA01/04 cells with high-glucose medium and in samples from DC patients.
M6a mRNA modifications may be involved in DC progression via the ferroptosis pathway, rendering novel insights into therapeutic strategies for DC.
Core Tip: Diabetic cataracts (DCs) are associated with elevated blood sugar levels and usually occur at an earlier age with more rapid progression than age-related cataracts. However, the specific molecular mechanisms underlying DC progression remain to be elucidated. As environmental factors are essential in the pathogenesis of diabetes mellitus, epigenetic changes may be particularly important. Recently, N6-methyladenosine (m6A) has been suggested to play a part in DC progression. The present study elucidated the m6A landscape in DC and simultaneously analyzed the methylation and expression of related mRNA. These analyses indicate that m6A mRNA modifications in lens epithelial cells might be involved in DC progression.
- Citation: Cai L, Han XY, Li D, Ma DM, Shi YM, Lu Y, Yang J. Analysis of N6-methyladenosine-modified mRNAs in diabetic cataract. World J Diabetes 2023; 14(7): 1077-1090
- URL: https://www.wjgnet.com/1948-9358/full/v14/i7/1077.htm
- DOI: https://dx.doi.org/10.4239/wjd.v14.i7.1077
Over the last several decades, the prevalence of diabetes mellitus (DM) in adults has increased globally. There were approximately 110 million DM cases in China in 2015, and the number is estimated to be 150 million by 2040, as indicated by the International Diabetes Federation[1]. DM, a systemic condition, affects various organs and thus can induce several complications, including cataracts[2]. Despite the increasing maturity of modern cataract surgery technology, cataracts remain a prime reason for vision loss and blindness globally[3,4]. Diabetic cataracts (DCs) usually develop at an earlier age and pro-gresses more rapidly than age-related cataracts do[5]. Evidence has linked DC to polyol pathway, nonenzymatic glycation, and oxidative stress (OS)[4]. Yet, the molecular mechanism underlying DC progression remains largely unknown.
As environmental factors play critical roles in the pathogenesis of DM, epigenetic changes may be particularly important[6]. N6-methyladenosine (m6A), one of the most prevalent epigenetic modifications in mammals[7], is increasingly shown to be crucial in several pathological processes (e.g., tumorigenesis, angiogenesis, tissue degeneration, and inflammatory responses)[8,9]. A study on DC patho-genesis based on m6A-RNA immunoprecipitation (MeRIP)-sequencing reported that the level of the methyltransferase protein complex, methyltransferase-like 3 (METTL3), is upregulated in high glucose-induced human lens epithelial cells (LECs) and that METTL3 mediates a higher methylation level[10]. However, the RNA m6A modification landscape in DC and the role of m6A in DC pathogenesis are still largely undetermined.
Herein, we performed an m6A epitranscriptomic microarray analysis to identify differentially methylated mRNAs and determined their potential roles using bioinformatics analyses, rendering novel insights into the pathogenic mechanisms of DC as well as clues for future biological interventions.
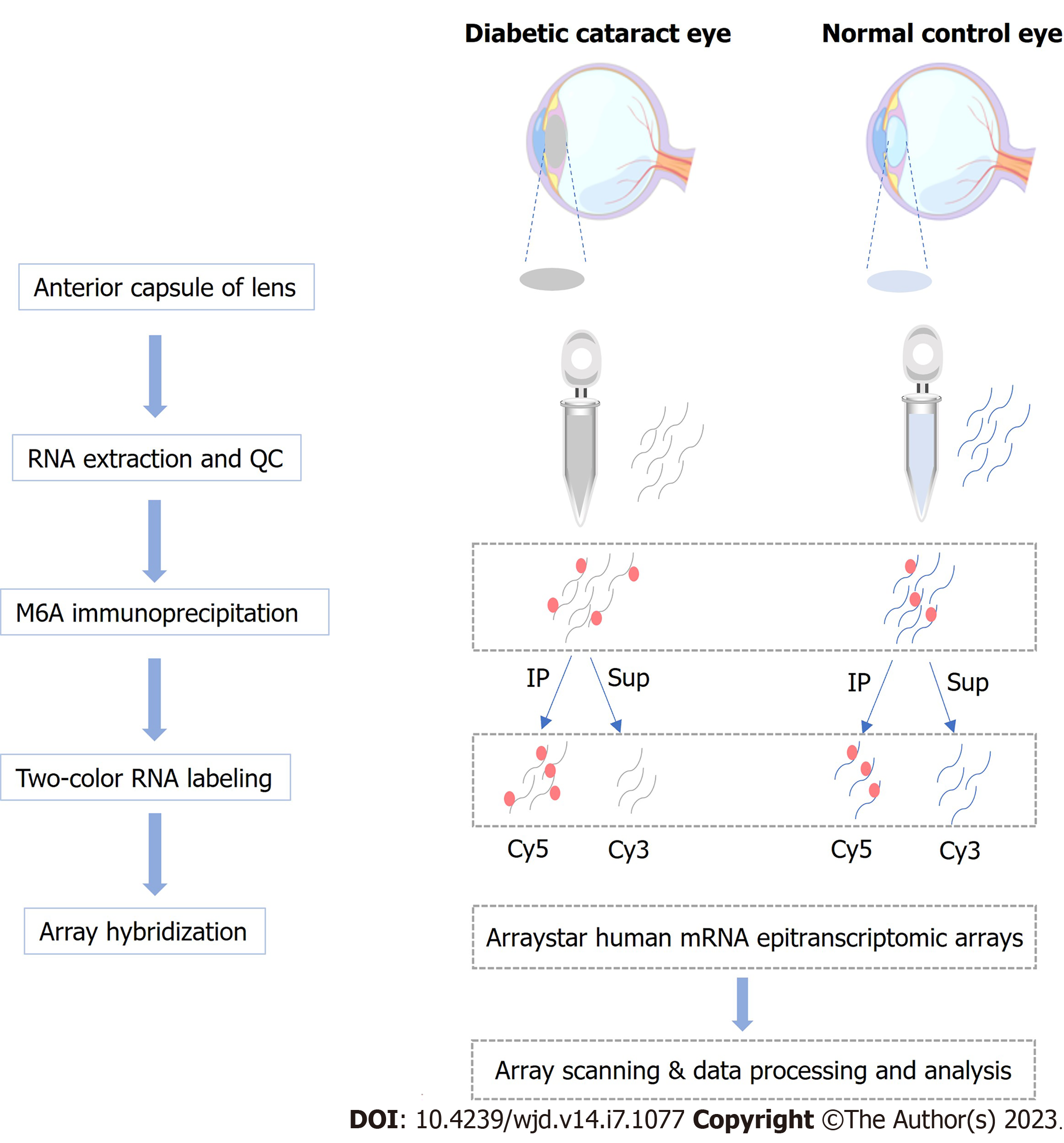
The anterior lens capsule (ALC) tissue of three DC patients had been living with diabetes for more than 5 years was collected, and the cataract severity was graded using the Lens Opacities Classification System III[11]. In addition, ALCs collected from age-matched transparent crystals of cadaveric eyes were used as normal controls (NC). Patients with other eye diseases, such as high myopia, trauma, uveitis, or glaucoma were excluded from the study. Patients’ information is presented in Table 1. The workflow of sample collection and processing is shown in Figure 1. This study has obtained approval from the Ethics Committee of the Eye and ENT Hospital of Fudan University and written informed consent from all participants, and the principles of the Declaration of Helsinki were strictly follows throughout the research period. This study was registered with ClinicalTrials.gov, number NCT05682001.
| No. | Gender | Age (yr) | AL (mm) | Lens opacity grading | Duration of DM (year) | FBG (mmol/L) | HbA1c (%) |
| 1 | Male | 64 | 22.09 | C4N3P3 | 10 | 7.89 | 7.50 |
| 2 | Female | 68 | 21.77 | C3N4P4 | 7 | 8.30 | 7.80 |
| 3 | Female | 63 | 23.43 | C4N4P5 | 7 | 8.30 | 7.90 |
The human LEC line SRA01/04, obtained from Genechem, was immersed in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific, United States) where 5.5 mmol/L glucose, 10% fetal bovine serum (Invitrogen, Carlsbad, CA, United States), 100 IU/mL penicillin (Thermo Fisher Scientific), and 100 mg/mL streptomycin (Thermo Fisher Scientific) were added, for cultivation in a 5% CO2 humidified atmosphere with the temperature maintained at 37 °C. Confluent cells (75%-80%) were then randomly grouped as a normal- (NG group; 5.5 mmol/L glucose-supplemented medium) and a high-glucose group (HG group; 25.0 mmol/L glucose-supplemented medium), and cultured for 24 h for subsequent examinations.
Using TRIzol (Invitrogen) and following kit recommendations, total RNA was isolated from the LECs of the included DC patients and controls, as well as SRA01/04 cells, followed by RNA quantification and purity evaluation with a NanoDrop ND-1000 spectrophotometer purchased from Thermo Fisher Scientific. This was followed by immunoprecipitation (IP) of the extracted total RNA from the NC (n = 3) and DC samples (n = 3) with an anti-m6A antibody by referring to the manufacturer’s recommendations. In brief, we placed 2 μg total RNA and m6A spike-in control mixture into a 300 μL IP buffer supplemented with 2 μg anti-m6A rabbit polyclonal antibody (Synaptic Systems, Goettingen, Germany), and let the reaction mixture rotate head-over-tail for 2 h at 4 °C. A DynabeadsTM M-280 sheep anti-rabbit immunoglobulin G (IgG) suspension (20 μL) was blocked with freshly prepared 0.5% bovine serum albumin at 4 °C for 2 h, followed by three rinses with IP buffer (300 μL) and resuspension in the total RNA-antibody mixture prepared. The RNA was then allowed to bind to the m6A-antibody beads for 2 h at 4 °C via head-over-tail rotation. After washing the beads thrice with 500 μL 1 × IP buffer and twice with 500 μL wash buffer, and incubation with 200 μL elution buffer (50 °C, 1 h), the enriched RNA was eluted and extracted using acid phenol-chloroform for ethanol precipitation.
The immunoprecipitated m6A-enriched RNAs were eluted from the magnetic beads as “IP”, while the unmodified RNAs were collected from the supernatant as “Sup”, which were then labeled with Cy5 and Cy3 (cRNAs), respectively, using an Arraystar Super RNA Labeling Kit (Arraystar, AL-SE-005). Purification of the synthesized cRNAs employed a RNeasy Mini Kit (QIAGEN, 74105), and the determination of concentrations and specific activities used the NanoDrop ND-1000. Following Arraystar’s standard protocol, microarray hybridization was performed. We combined and hybridized Cy3 and Cy5 Labeled cRNAs to an Arraystar Human mRNA Epitranscriptomic Microarray (4 × 44 K, Arraystar, China), after which the slices were washed for array scanning using an Agilent Scanner G2505C (Agilent, Beijing, China).
Analyses of the acquired array images were carried out using Agilent’s Feature Extraction software v11.0.1.1. Cy5-labeled IP and Cy3-labeled Sup raw intensities were normalized to the mean of log2-scaled spike-in RNA intensities. The m6A methylation level was counted as a percentage of modified RNA (% modified) from total RNA, based on IP and Sup normalized intensities. The m6A quantity of each transcript was calculated according to normalized IP (Cy5-labeled) intensities. RNA expression was determined from the total IP and Sup normalized RNA intensities.
The online gene ontology (GO) (URL: http://www.geneontology.org) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (URL: http://www.genome.jp/kegg) were utilized for determining the enriched GO terms and pathways in the mRNAs with significantly different m6A expression levels.
To validate microarray data quality, MeRIP-quantitative polymerase chain reaction (qPCR) was performed on four randomly selected mRNAs. In brief, the IP RNAs from ALC tissues of patients with DC and NCs were analyzed for microarray data validation, with primers used presented in Supplementary Table 1.
Reverse transcription of total RNA to cDNA was performed as per the instruction of the PrimeScript RT Reagent Kit (Takara, Dalian, Liaoning province, China). Reverse transcription-qPCR (qRT-PCR) primers, designed with the use of Primer 5.0, were blasted for specificity in NCBI (Supplementary Table 1). An Applied Biosystems ViiA 7 Real-Time thermal cycler (Thermo Fisher Scientific) and SYBR Green PCR Master Mix (Arraystar) were then utilized to perform the qRT-PCR. The expression of target mRNAs were normalized against Actin, and fold changes were determined by the comparative CT (2-ΔΔCT) method.
The significance threshold was P < 0.05 in this study. For the microarray analysis, statistical significance in methylation levels between DC cases and NCs was identified using an unpaired two-sided t-test. For GO and KEGG analyses, GO terms and KEGG pathway identifiers with significant differences were identified using the Fisher’s exact test p-value and -log10(p) transformed as the enrichment score. While the relative genes’ expression in MeRIP-qPCR and qRT-PCR was worked out by 2-ΔΔCT.
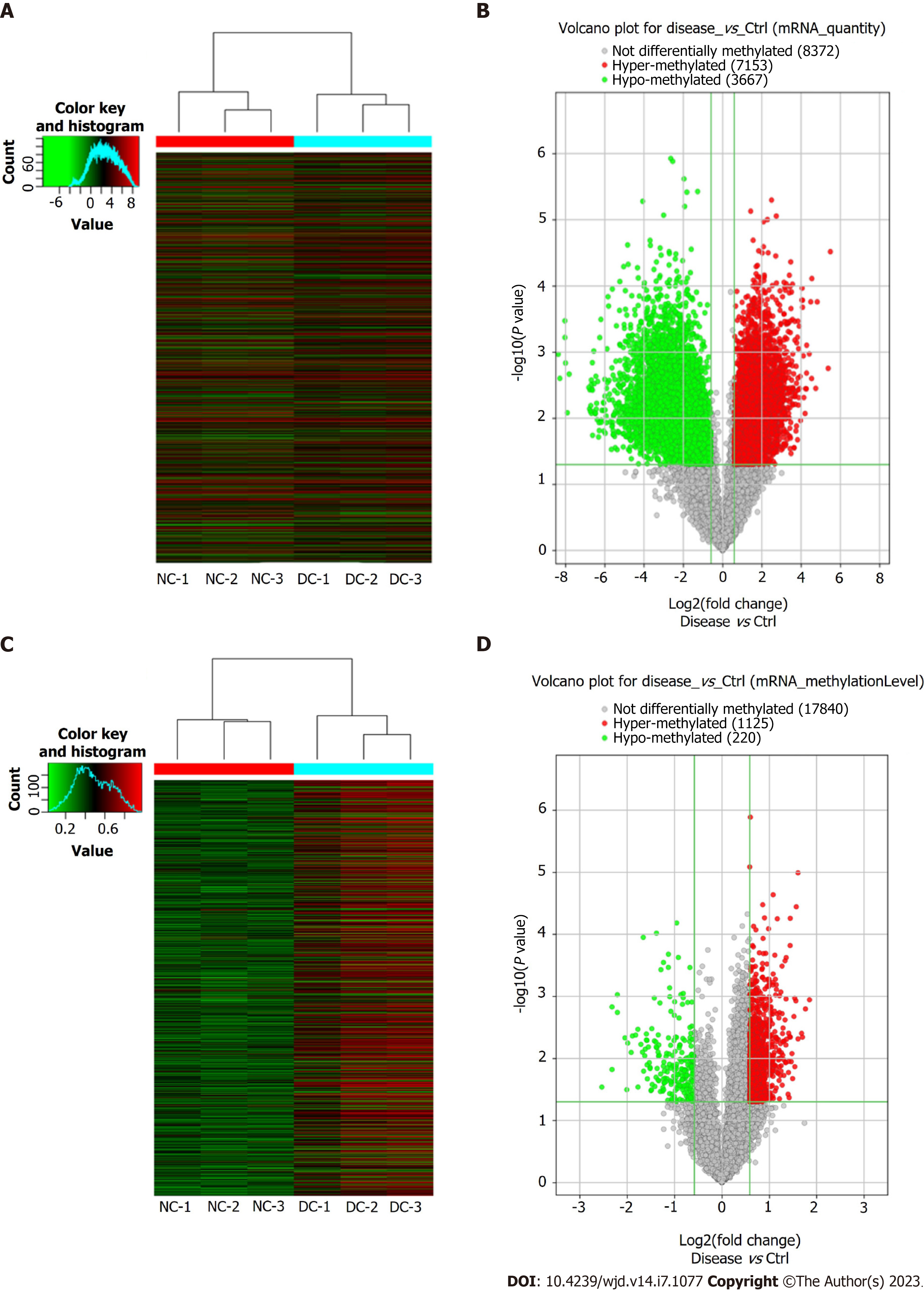
Microarray analyses of the mRNAs extracted from the lens anterior capsule tissues of the DC and NC samples showed differential m6A-methylated mRNAs, as identified by the “m6A-mRNA quantity and m6A-mRNA methylation level”. The results have been presented as heatmaps (Figure 2A and C) and volcano plots (Figure 2B and D). According to the m6A quantity results, there were 7153 hypermethylated mRNAs and 3667 hypomethylated mRNAs. As per the m6A methylation level results, 1125 mRNAs had higher m6A methylation levels, whereas 220 mRNAs had lower levels. See Table 2 for the top 20 mRNAs with the most significant hyper- and hypomethylation levels between DCs and NCs.
| Gene symbol | P value | Fold change | Regulation | Chromosome |
| AQP2 | 0.001135123 | 3.60 | Hyper | chr12 |
| RPL10 | 0.001594064 | 3.40 | Hyper | chrX |
| ACP5 | 0.004544885 | 3.25 | Hyper | chr19 |
| ACAP1 | 0.003944032 | 3.21 | Hyper | chr17 |
| CALML6 | 0.00484061 | 3.05 | Hyper | chr1 |
| DDX31 | 1.02006E-05 | 3.04 | Hyper | chr9 |
| KRTAP29-1 | 3.59728E-05 | 2.97 | Hyper | chr17 |
| TRIM39-RPP21 | 0.001179594 | 2.95 | Hyper | chr6 |
| DGCR8 | 0.013600714 | 2.90 | Hyper | chr22 |
| LRAT | 0.001067196 | 2.90 | Hyper | chr4 |
| NR1H3 | 0.028959935 | 5.78 | Hypo | chr11 |
| PNPT1 | 0.001479448 | 4.99 | Hypo | chr2 |
| TSEN2 | 0.015094744 | 4.99 | Hypo | chr3 |
| C3orf80 | 0.000939962 | 4.61 | Hypo | chr3 |
| TBCD | 0.001813039 | 4.61 | Hypo | chr17 |
| RPS19 | 0.004668423 | 4.12 | Hypo | chr19 |
| TEAD2 | 0.031922173 | 4.02 | Hypo | chr19 |
| RAB3IP | 0.005656491 | 3.96 | Hypo | chr12 |
| HN1L | 0.008004647 | 3.77 | Hypo | chr16 |
| TFEB | 0.004349132 | 3.70 | Hypo | chr6 |
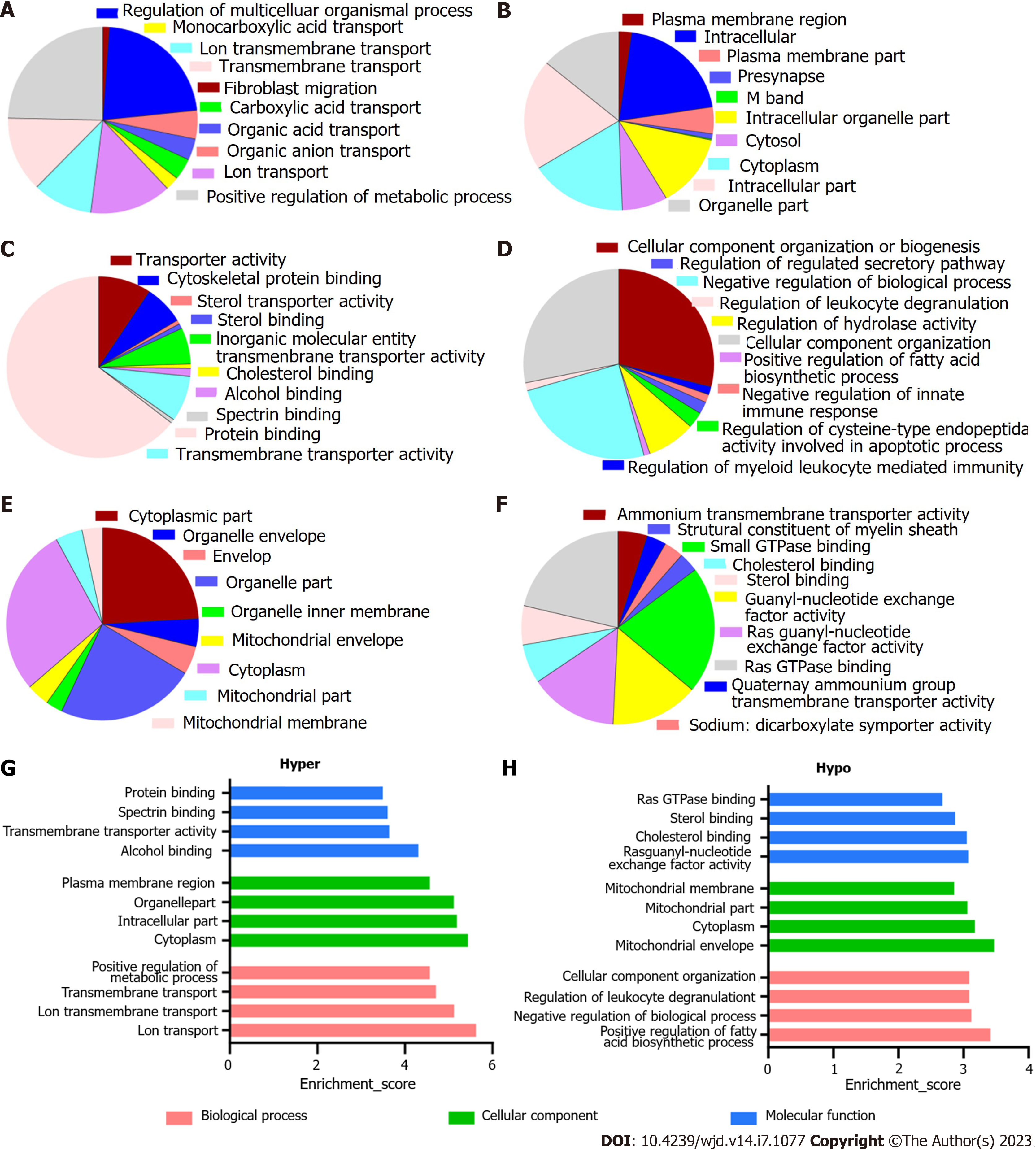
The enriched GO annotations can be fall into biological process (BP), cellular component (CC), or molecular function (MF). For hypermethylated mRNAs, 580 BPs, 110 CCs, and 100 MFs were enriched. The quantity of differentially methylated mRNAs related to the listed GO ID was recorded; of them, the top 10 most significantly enriched terms are presented as pie charts (Figure 3A-C, G). In addition, the top four terms with the highest enrichment score are shown in Figure 3G. For the hypomethylated mRNAs, 288 BPs, 47 CCs, and 67 MFs were enriched. See Figures 3D-F and H for the top 10 most significantly enriched terms and the top 4 terms with the highest enrichment scores.
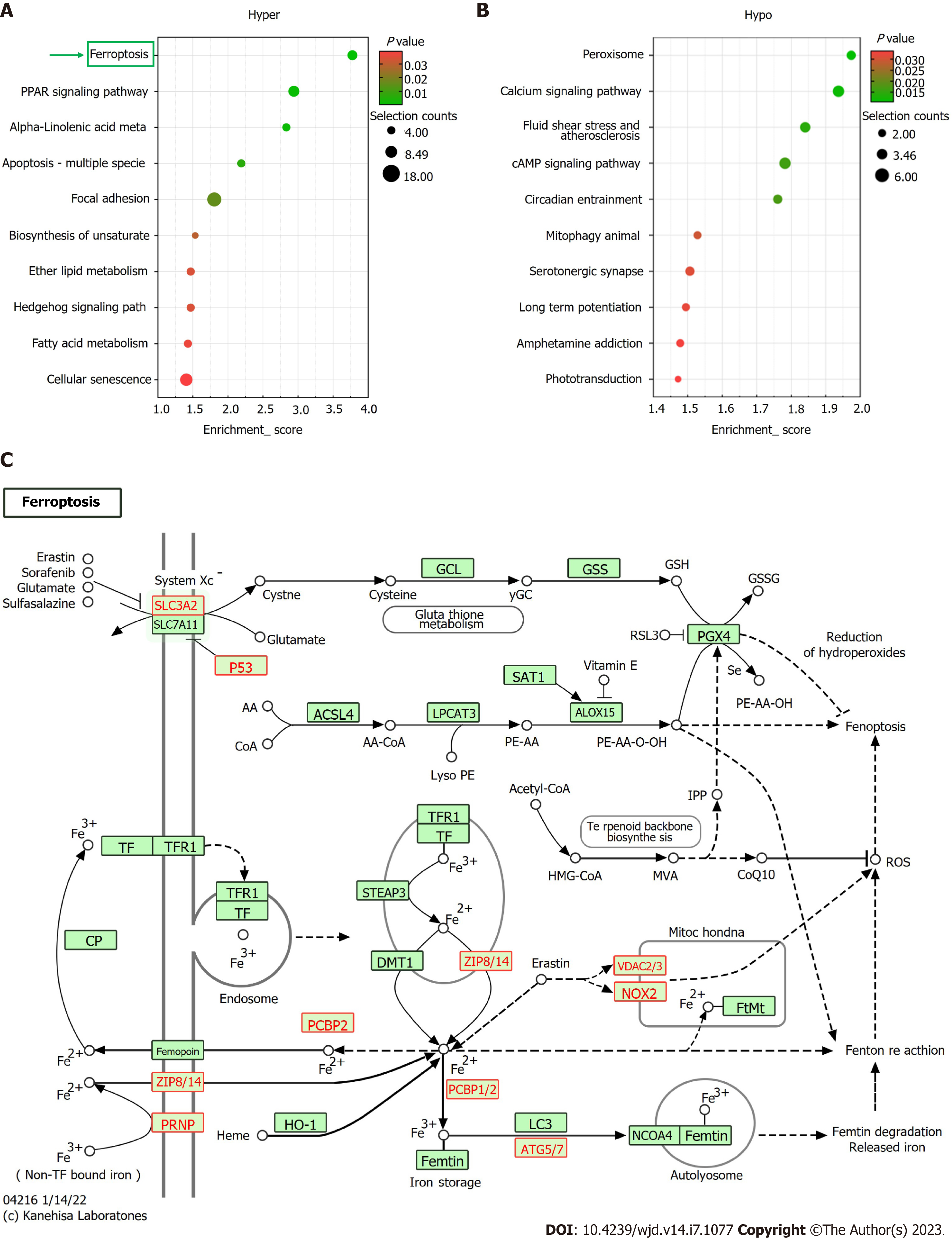
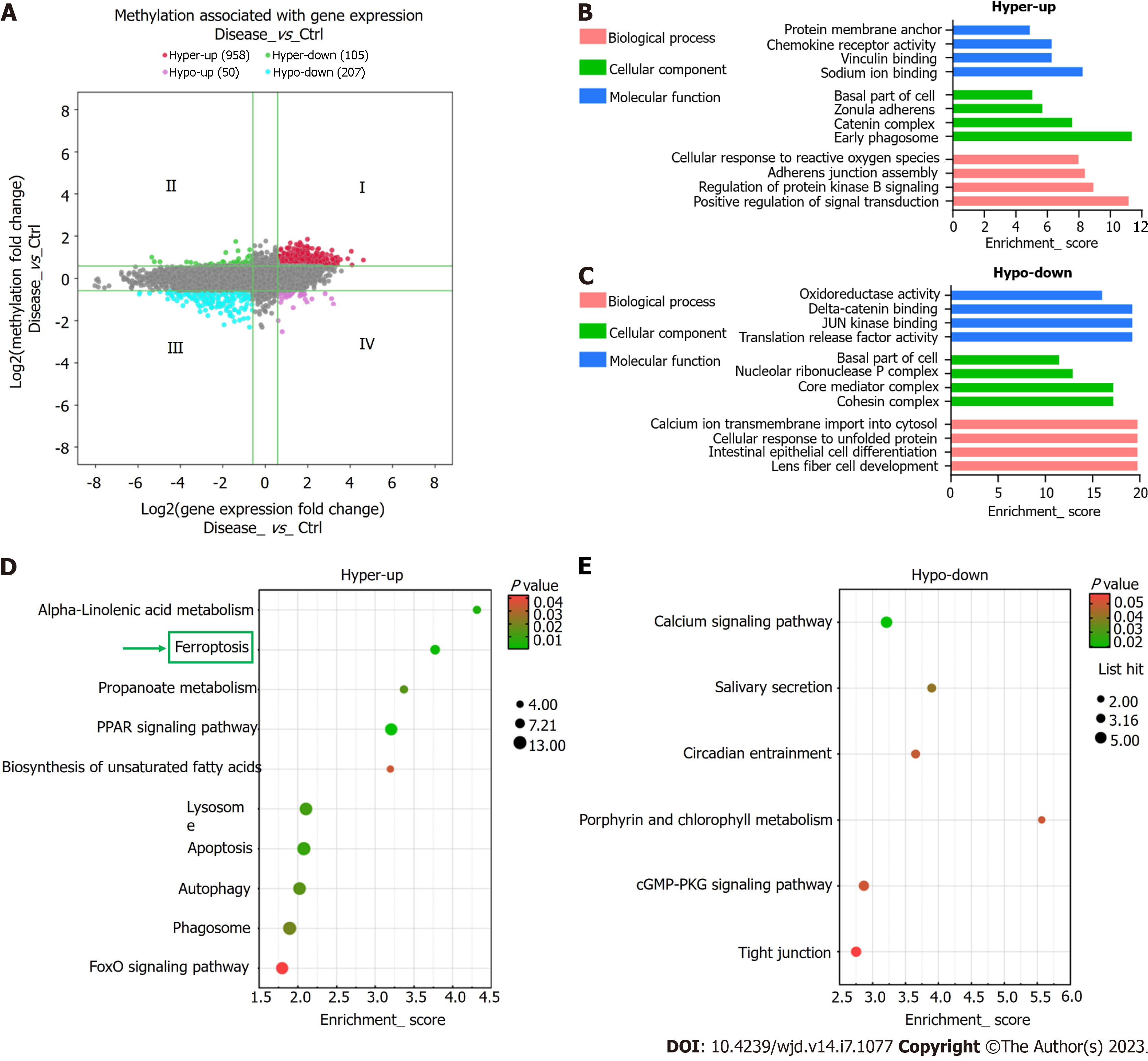
Based on KEGG pathway analysis, the mRNAs differentially methylated by m6A participated in 27 pathways (Figure 4A and B). Most of the hypermethylated mRNAs were primarily enriched in “ferroptosis”, “PPAR axis”, and “alpha-linolenic acid metabolism”. The ferroptosis pathway map is illustrated in Figure 4C.
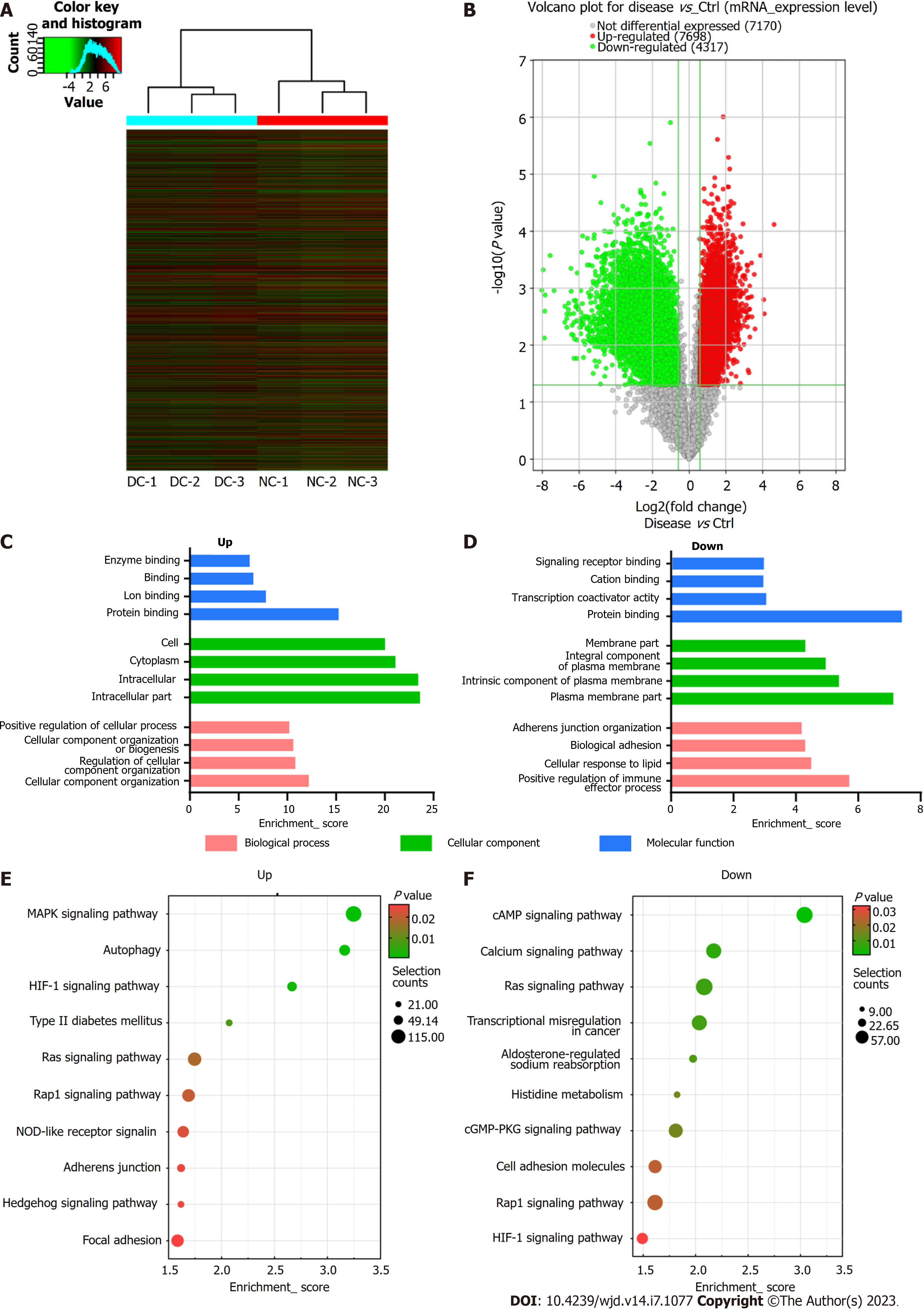
Besides m6A modification levels, the m6A microarray analyses provided data for mRNA expression (Figure 5A and B). A total of 12015 mRNAs in the DC and NC groups showed significantly different expression [P ≤ 0.05, fold change (FC) ≥ 1.5], 7698 of which were upregulated, whereas 4317 were downregulated. The functions of the top 20 differentially expressed mRNAs (DEmRNAs) (Supplementary Table 2) were analyzed using GO and KEGG pathway analyses. Among the enriched GO terms, 780 BPs, 137 CCs, and 101 MFs were associated with downregulated mRNA expression, with the top 10 displayed in Figure 5C. Moreover, 1199 BPs, 119 CCs, and 190 MFs were identified to be linked to upregulated mRNA expression, with the top 10 presented in Figure 5D. Among the upregulated mRNAs of the BP category, “cellular component organization” had the highest GO term enrichment score, whereas for the downregulated mRNAs, the highest score belonged to “positive regulation of immune effector process”. For the CC category, “intracellular” and “plasma membrane” were the most prominent GO terms for up- and down-regulated mRNA expression, respectively. In the MF category, “protein binding” was the most significant term for both up- and downregulated mRNAs.
According to KEGG pathway analysis, DEmRNAs participated in 55 pathways, most of which were primarily enriched in the “MAPK axis”, “Type II DM”, and “cAMP axis” (Figure 5E and F).
Using the thresholds FC ≥ 1.5 and P ≤ 0.05, the combined analysis revealed significantly altered m6A methylation and mRNA expression levels in 1,320 mRNAs. Conjoint analysis of these 1320 mRNAs resulted in the formation of four mRNA groups: Group I, 958 hypermethylated and upregulated mRNAs; Group II, 105 hypermethylated and downregulated mRNAs; Group III, 207 hypomethylated and downregulated mRNAs; Group IV, 50 hypomethylated and upregulated mRNAs (Figure 6A). Several key genes of ferroptosis (PRNP, SLC39A8, VDAC2, P53, CYBB, ATG7, and SLC3A2) were found in Group I.
Hypermethylated-upregulated (hyper-up) and -downregulated (hypo-down) mRNAs were further identified using GO and KEGG pathway analyses. For Group I mRNAs, the most enriched GO terms in BP, CC, and MF categories were found to be “protein membrane anchor”, “early phagosome”, and “sodium ion binding”, respectively. For Group III mRNAs, the terms were “lens fiber cell development”, “cohesin complex”, and “translation release factor activity binding” (Figure 6B). KEGG pathway analysis showed that DEmRNAs participated in 26 pathways. Most mRNAs in Group I were mainly enriched in “alpha-linolenic acid metabolism,” “ferroptosis”, and “apoptosis”, whereas Group III mRNAs were primarily enriched in “calcium axis”, “cGMP-PKG axis”, and “tight junction” (Figure 6C and D).
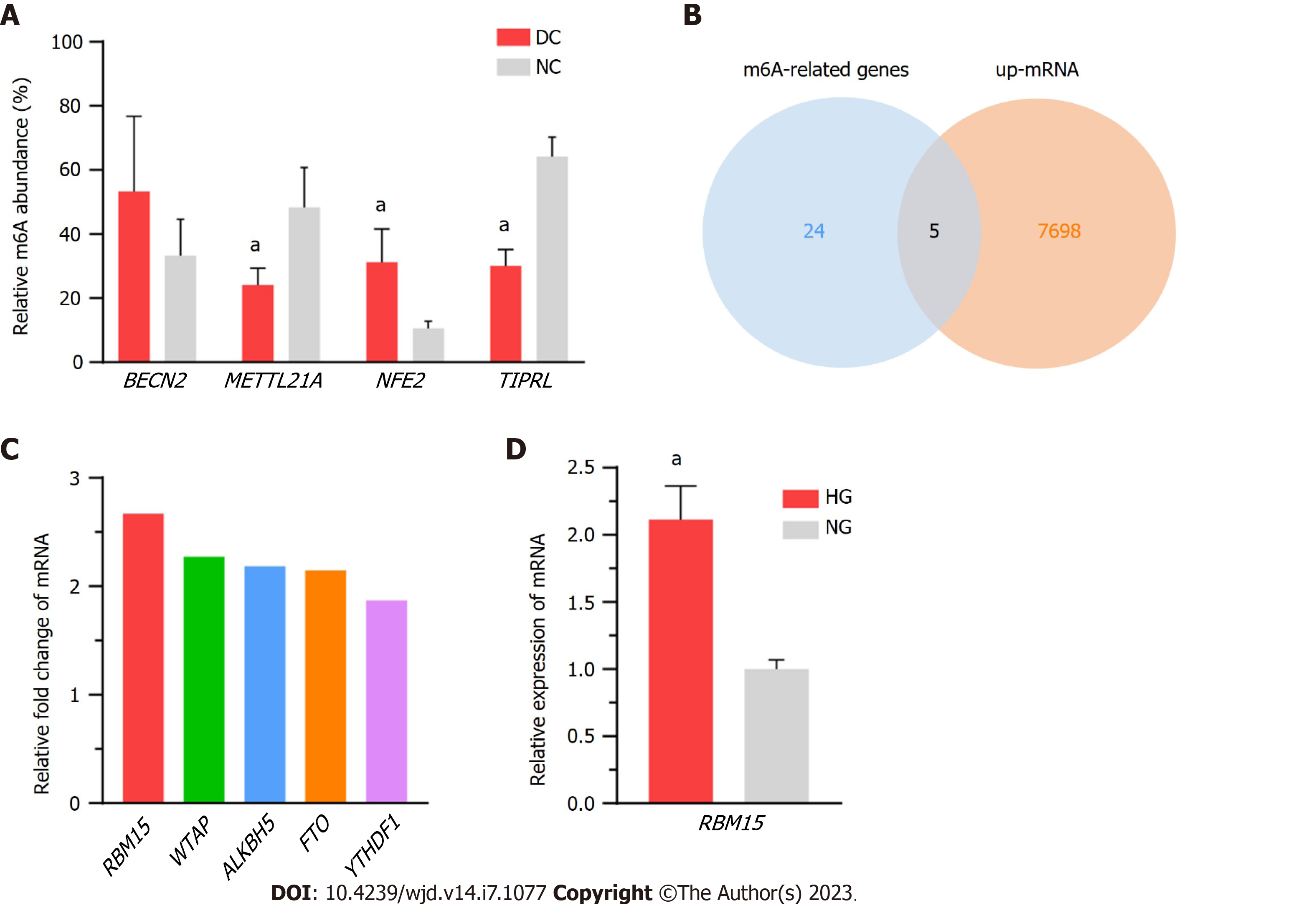
We randomly selected four mRNAs (BECN2, METTL21A, NFE2, and TIPRL) for MeRIP-qPCR to validate the microarray data quality; specifically, we screened the differentially methylated mRNAs under the criteria of P value ≤ 0.05 and FC ≥ 1.5, and then we selected genes with multiple expression folds for verification with the primers can be designed for those mRNAs. Finally, we selected 4 methylated mRNAs with different fold changes for verification to ensure the reliability of the results to a certain extent. Details of the selected genes are listed in Supplementary Table 3, and the results accorded with the microarray data (Figure 7A). Furthermore, to explore the possible genes participating in m6A modification, we compared the expression of the DEmRNAs in our epitranscriptomic micro-array with that of 24 known methylation-related genes (METTL3, METTL14, WTAP, VIRMA, KIAA1429, RNA binding motif protein 15 (RBM15), RBM15B, ALKBH5, FTO, AlkB-H9, HNRNPA2, HNRNPB1, HNRNPC1, HNRNPC2, YTHDC1, YTHDC2, YTHDF1, YTHDF2, YTHDF3, EIF3A, EIF3B, IGF2BPs, DGCR8, and ELAVL1). The expression of five genes (RBM15, WTAP, ALKBH5, FTO, and YTHDF1) was found to be upregulated in the microarray results (Figure 7B; the FC values of these genes are shown in Figure 7C); and of these, RBM15 exhibited the highest change in expression level. Additionally, the upregulation of RBM15 in SRA01/04 cells cultured in HG medium verified its expression in vitro, supporting the qRT-PCR results in DC specimens (Figure 7D).
The present study elucidated the m6A landscape in DC using an epitranscriptomic microarray, which simultaneously analyzed the methylation and expression of related mRNA. According to the microarray results, a total of 1345 mRNAs exhibiting significantly different m6A modification levels between DC cases and NCs were identified. Most of these mRNAs (1125/1345) had higher m6A methylation levels in the DC samples. First identified in the 1970s, abundant m6A modifications in polyadenylated RNA were accidentally discovered by some research groups when they were characterizing the 5’ structures of mRNA in mammalian cells[12]. In multiple human pathophysiological processes, m6A extensively modifies RNA transcription and protein generation[13]. Modification by m6A modulates gene expression by affecting mRNA splicing, localisation, stability, and translation. Over the past few years, the development of techniques such as MeRIP-sequencing and epitranscriptomic microarrays has made the high-throughput measurement of m6A modification sites possible[14-16]. These approaches allow simultaneous screening of modified transcript types and modification changes under different con-ditions, as well as detection of modification proportions per transcript. The development of microarray method has allowed for a more subtle mapping of the m6A modification, providing better insights into its importance in gene regulation.
In this study, RBM15 was found most upregulated in the DC group, which was verified in DC samples and HG-cultured LECs. Methylation through m6A is a reversible process, dynamically regulated by three different types of protein complexes: methyltransferases, demethylases, and readers[17]. RBM15 and its paralog, RBM15B, are additional components of the methyltransferase complex[18]. RBM15, a split-end protein family member, modulates m6A methylation for RNA modification[19]. As part of the methyltransferase complex, it participates in hematopoietic cell homeostasis and alternative mRNA splicing[20]. The main role of RBM15 in m6A methylation catalysis is recruiting the m6A methyltransferase complex to U-rich regions adjacent to m6A sites[18,21]. Pollreisz et al[22] reported markedly increased global mRNA m6A methylation level and RBM15 expression in laryngeal squa-mous cell cancer patients; however, inhibiting RBM15 led to a notable reduction in the m6A methylation level. But the potential roles played by RBM15 in DC pathogenicity need further research. It could be suggested that RBM15-mediated m6A modification of LECs may promote DC progression. Further studies are warranted to clarify the mechanisms underlying m6A modification in DC.
Further, based on the KEGG pathway analysis, ferroptosis was identified as one of the most enriched pathways in the m6A-hypermethylated and upregulated mRNAs in DC samples (Figure 4C). In human lens development, LECs play a key role in transport, metabolism, and detoxification[23]. The integrity and survival of LECs are critical for lens transparency[24]. LEC death due to apoptosis and autophagy plays pathophysiological roles in DC progression[25]. Ferroptosis is a newly defined programmed death mode that is implicated in various reactive oxygen species (ROS)-related pathophysiological states, such as age-related macular degeneration and cardiovascular diseases[26,27]. OS is vital in DC pathogenesis[28]. The process of ferroptosis is characterized by glutathione (GSH) depletion, lipid peroxidation, and intracellular ROS accumulation with iron overload as well as accelerated cell death[29-31]. GSH levels are markedly lower in DC patients than in non-diabetic senile cataract patients and non-diabetic type 2 DM patients as well as in healthy individuals[32].
The subsequent combined analysis of m6A methylation and mRNA expression levels showed several ferroptosis-associated key genes (PRNP, SLC39A8, VDAC2, P53, CYBB, ATG7, and SLC3A2) to be hypermethylated and upregulated in the DC group, suggesting enhanced ferroptosis in LECs of patients with DC. P53 can potentiate ferroptosis by inhibiting the transcription of system xc-subunit SLC7A11[33]. Reportedly, its expression was upregulated in the LECs of patients with DC[34]. Therefore, we speculate that m6A mRNA modifications of LECs are involved in DC progression via the ferroptosis pathway. In future, more comprehensive research is warranted to elucidate ferroptosis-associated mechanisms in DC pathogenesis.
Collectively, the m6A abundance level in total mRNA increased in patients with DC. Conjoint analysis indicated that m6A mRNA modifications of LECs might be involved in DC progression via the ferroptosis pathway. The expression level of RBM15 increased, which provided a better understanding of the mechanisms underlying upregulated m6A demethylation levels.
Cataract remains a prime reason for visual disturbance and blindness all over the world, despite successful surgical replacement with artificial lenses. Diabetic cataract (DC) usually occurs at an earlier age with more rapid progression than age-related cataracts. The polyol pathway, oxidative stress, and nonenzymatic glycation have been shown to be linked to the pathogenesis of DC. But the exact molecular mechanisms underlying DC progression remains largely unknown. As environmental factors play critical roles in the pathogenesis of diabetes mellitus, epigenetic changes may be particularly important.
Despite successful surgical replacement with artificial lenses, cataract remains a prime reason for visual disturbance and blindness globally. It has been recently suggested that N6-methyladenosine (m6A) plays a role in DC progression. However, there exists a lack of understanding regarding RNA m6A modifications and the role of m6A in DC pathogenesis.
To investigate the roles played by altered m6A and differentially expressed mRNAs (DEmRNAs) in DC.
M6A epitranscriptomic microarray was used to investigate altered m6A modifications and determine DEmRNAs. The possible roles played by dysregulated m6A modification was predicted through Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses. Real-time polymerase chain reaction was carried out to identify dysregulated expression patterns of RNA methyltransferases, demethylases, and readers.
Increased m6A abundance levels were found in the total mRNA of DC samples. Bioinformatics analysis predicted that ferroptosis pathways could be associated with m6A-modified mRNAs. The levels of five methylation-related genes-RBM15, WTAP, ALKBH5, FTO, and YTHDF1-were upregulated in DC samples. Upregulation of RBM15 expression was verified in SRA01/04 cells with high-glucose medium and in samples from patients with DC.
M6A abundance level in total mRNA increased in patients with DC. Ferroptosis pathways could be associated with m6A-modified mRNAs.
M6A mRNA modifications may be involved in DC progression via the ferroptosis pathway.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Ophthalmology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Godfrey KM, United Kingdom; Sathish T, Canada S-Editor: Chen YL L-Editor: A P-Editor: Chen YX
| 1. | Andley UP. The lens epithelium: focus on the expression and function of the alpha-crystallin chaperones. Int J Biochem Cell Biol. 2008;40:317-323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 69] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 2. | Bertrand RL. Iron accumulation, glutathione depletion, and lipid peroxidation must occur simultaneously during ferroptosis and are mutually amplifying events. Med Hypotheses. 2017;101:69-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 89] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 3. | Cao JY, Dixon SJ. Mechanisms of ferroptosis. Cell Mol Life Sci. 2016;73:2195-2209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1001] [Cited by in RCA: 1117] [Article Influence: 124.1] [Reference Citation Analysis (0)] |
| 4. | Chen L, Chen Y, Ding W, Zhan T, Zhu J, Zhang L, Wang H, Shen B, Wang Y. Oxidative Stress-Induced TRPV2 Expression Increase Is Involved in Diabetic Cataracts and Apoptosis of Lens Epithelial Cells in a High-Glucose Environment. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 5. | Chokkalla AK, Mehta SL, Kim T, Chelluboina B, Kim J, Vemuganti R. Transient Focal Ischemia Significantly Alters the m(6)A Epitranscriptomic Tagging of RNAs in the Brain. Stroke. 2019;50:2912-2921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 123] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 6. | Chylack LT Jr, Wolfe JK, Singer DM, Leske MC, Bullimore MA, Bailey IL, Friend J, McCarthy D, Wu SY. The Lens Opacities Classification System III. The Longitudinal Study of Cataract Study Group. Arch Ophthalmol. 1993;111:831-836. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1693] [Cited by in RCA: 1959] [Article Influence: 61.2] [Reference Citation Analysis (0)] |
| 7. | Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A. 1974;71:3971-3975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 974] [Cited by in RCA: 1351] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 8. | Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, Morrison B 3rd, Stockwell BR. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060-1072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4711] [Cited by in RCA: 11702] [Article Influence: 900.2] [Reference Citation Analysis (1)] |
| 9. | Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8:176-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 390] [Cited by in RCA: 522] [Article Influence: 43.5] [Reference Citation Analysis (0)] |
| 10. | Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m⁶A RNA methylation. Nat Rev Genet. 2014;15:293-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 956] [Cited by in RCA: 1426] [Article Influence: 129.6] [Reference Citation Analysis (0)] |
| 11. | Hiriart E, Gruffat H, Buisson M, Mikaelian I, Keppler S, Meresse P, Mercher T, Bernard OA, Sergeant A, Manet E. Interaction of the Epstein-Barr virus mRNA export factor EB2 with human Spen proteins SHARP, OTT1, and a novel member of the family, OTT3, links Spen proteins with splicing regulation and mRNA export. J Biol Chem. 2005;280:36935-36945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 70] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 12. | Jain AK, Lim G, Langford M, Jain SK. Effect of high-glucose levels on protein oxidation in cultured lens cells, and in crystalline and albumin solution and its inhibition by vitamin B6 and N-acetylcysteine: its possible relevance to cataract formation in diabetes. Free Radic Biol Med. 2002;33:1615-1621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 13. | Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, Baer R, Gu W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1173] [Cited by in RCA: 2399] [Article Influence: 239.9] [Reference Citation Analysis (0)] |
| 14. | Kaliaperumal R, Venkatachalam R, Nagarajan P, Sabapathy SK. Association of Serum Magnesium with Oxidative Stress in the Pathogenesis of Diabetic Cataract. Biol Trace Elem Res. 2021;199:2869-2873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 15. | Klungland A, Dahl JA. Dynamic RNA modifications in disease. Curr Opin Genet Dev. 2014;26:47-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 79] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 16. | Lim SA, Joo CK, Kim MS, Chung SK. Expression of p53 and caspase-8 in lens epithelial cells of diabetic cataract. J Cataract Refract Surg. 2014;40:1102-1108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 17. | McCarty CA, Taylor HR. Recent developments in vision research: light damage in cataract. Invest Ophthalmol Vis Sci. 1996;37:1720-1723. [PubMed] |
| 18. | Meyer KD, Jaffrey SR. Rethinking m(6)A Readers, Writers, and Erasers. Annu Rev Cell Dev Biol. 2017;33:319-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 689] [Cited by in RCA: 841] [Article Influence: 105.1] [Reference Citation Analysis (0)] |
| 19. | Ogurtsova K, da Rocha Fernandes JD, Huang Y, Linnenkamp U, Guariguata L, Cho NH, Cavan D, Shaw JE, Makaroff LE. IDF Diabetes Atlas: Global estimates for the prevalence of diabetes for 2015 and 2040. Diabetes Res Clin Pract. 2017;128:40-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2306] [Cited by in RCA: 2509] [Article Influence: 313.6] [Reference Citation Analysis (0)] |
| 20. | Pascolini D, Mariotti SP. Global estimates of visual impairment: 2010. Br J Ophthalmol. 2012;96:614-618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2555] [Cited by in RCA: 2155] [Article Influence: 165.8] [Reference Citation Analysis (0)] |
| 21. | Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 852] [Cited by in RCA: 1321] [Article Influence: 146.8] [Reference Citation Analysis (0)] |
| 22. | Pollreisz A, Schmidt-Erfurth U. Diabetic cataract-pathogenesis, epidemiology and treatment. J Ophthalmol. 2010;2010:608751. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 193] [Cited by in RCA: 245] [Article Influence: 16.3] [Reference Citation Analysis (1)] |
| 23. | Qin Y, Liu Q, Tian S, Xie W, Cui J, Wang RF. TRIM9 short isoform preferentially promotes DNA and RNA virus-induced production of type I interferon by recruiting GSK3β to TBK1. Cell Res. 2016;26:613-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 69] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 24. | Robman L, Taylor H. External factors in the development of cataract. Eye (Lond). 2005;19:1074-1082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 138] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 25. | Ru W, Zhang X, Yue B, Qi A, Shen X, Huang Y, Lan X, Lei C, Chen H. Insight into m(6)A methylation from occurrence to functions. Open Biol. 2020;10:200091. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 26. | Sharma KK, Santhoshkumar P. Lens aging: effects of crystallins. Biochim Biophys Acta. 2009;1790:1095-1108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 265] [Cited by in RCA: 250] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 27. | Totsuka K, Ueta T, Uchida T, Roggia MF, Nakagawa S, Vavvas DG, Honjo M, Aihara M. Oxidative stress induces ferroptotic cell death in retinal pigment epithelial cells. Exp Eye Res. 2019;181:316-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 128] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 28. | Wang X, Tian L, Li Y, Wang J, Yan B, Yang L, Li Q, Zhao R, Liu M, Wang P, Sun Y. RBM15 facilitates laryngeal squamous cell carcinoma progression by regulating TMBIM6 stability through IGF2BP3 dependent. J Exp Clin Cancer Res. 2021;40:80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 169] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 29. | Yan G, Yuan Y, He M, Gong R, Lei H, Zhou H, Wang W, Du W, Ma T, Liu S, Xu Z, Gao M, Yu M, Bian Y, Pang P, Li X, Yu S, Yang F, Cai B, Yang L. m(6)A Methylation of Precursor-miR-320/RUNX2 Controls Osteogenic Potential of Bone Marrow-Derived Mesenchymal Stem Cells. Mol Ther Nucleic Acids. 2020;19:421-436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 117] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 30. | Yang J, Liu J, Zhao S, Tian F. N(6)-Methyladenosine METTL3 Modulates the Proliferation and Apoptosis of Lens Epithelial Cells in Diabetic Cataract. Mol Ther Nucleic Acids. 2020;20:111-116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 77] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 31. | Yu Y, Yan Y, Niu F, Wang Y, Chen X, Su G, Liu Y, Zhao X, Qian L, Liu P, Xiong Y. Ferroptosis: a cell death connecting oxidative stress, inflammation and cardiovascular diseases. Cell Death Discov. 2021;7:193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 444] [Article Influence: 111.0] [Reference Citation Analysis (0)] |
| 32. | Zhang C, Fu J, Zhou Y. A Review in Research Progress Concerning m6A Methylation and Immunoregulation. Front Immunol. 2019;10:922. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 200] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 33. | Zhang L, Tran NT, Su H, Wang R, Lu Y, Tang H, Aoyagi S, Guo A, Khodadadi-Jamayran A, Zhou D, Qian K, Hricik T, Côté J, Han X, Zhou W, Laha S, Abdel-Wahab O, Levine RL, Raffel G, Liu Y, Chen D, Li H, Townes T, Wang H, Deng H, Zheng YG, Leslie C, Luo M, Zhao X. Cross-talk between PRMT1-mediated methylation and ubiquitylation on RBM15 controls RNA splicing. Elife. 2015;4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 135] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 34. | Zhu S, Wang JZ, Chen D, He YT, Meng N, Chen M, Lu RX, Chen XH, Zhang XL, Yan GR. An oncopeptide regulates m(6)A recognition by the m(6)A reader IGF2BP1 and tumorigenesis. Nat Commun. 2020;11:1685. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 181] [Article Influence: 36.2] [Reference Citation Analysis (0)] |