Published online Mar 15, 2023. doi: 10.4239/wjd.v14.i3.255
Peer-review started: December 2, 2022
First decision: December 19, 2022
Revised: December 31, 2022
Accepted: February 7, 2023
Article in press: February 7, 2023
Published online: March 15, 2023
Processing time: 103 Days and 7.8 Hours
In recent years, the incidence of type 2 diabetes (T2DM) has shown a rapid growth trend. Goto Kakizaki (GK) rats are a valuable model for the study of T2DM and share common glucose metabolism features with human T2DM patients. A series of studies have indicated that T2DM is associated with the gut microbiota composition and gut metabolites. We aimed to systematically characterize the faecal gut microbes and metabolites of GK rats and analyse the relationship between glucose and insulin resistance.
To evaluate the gut microbial and metabolite alterations in GK rat faeces based on metagenomics and untargeted metabolomics.
Ten GK rats (model group) and Wistar rats (control group) were observed for 10 wk, and various glucose-related indexes, mainly including weight, fasting blood glucose (FBG) and insulin levels, homeostasis model assessment of insulin resistance (HOMA-IR) and homeostasis model assessment of β cell (HOMA-β) were assessed. The faecal gut microbiota was sequenced by metagenomics, and faecal metabolites were analysed by untargeted metabolomics. Multiple metabolic pathways were evaluated based on the differential metabolites identified, and the correlations between blood glucose and the gut microbiota and metabolites were analysed.
The model group displayed significant differences in weight, FBG and insulin levels, HOMA-IR and HOMA-β indexes (P < 0.05, P < 0.01) and a shift in the gut microbiota structure compared with the control group. The results demonstrated significantly decreased abundances of Prevotella sp. CAG:604 and Lactobacillus murinus (P < 0.05) and a significantly increased abundance of Allobaculum stercoricanis (P < 0.01) in the model group. A correlation analysis indicated that FBG and HOMA-IR were positively correlated with Allobaculum stercoricanis and negatively correlated with Lactobacillus murinus. An orthogonal partial least squares discriminant analysis suggested that the faecal metabolic profiles differed between the model and control groups. Fourteen potential metabolic biomarkers, including glycochenodeoxycholic acid, uric acid, 13(S)-hydroxyoctadecadienoic acid (HODE), N-acetylaspartate, β-sitostenone, sphinganine, 4-pyridoxic acid, and linoleic acid, were identified. Moreover, FBG and HOMA-IR were found to be positively correlated with glutathione, 13(S)-HODE, uric acid, 4-pyridoxic acid and allantoic acid and ne-gatively correlated with 3-α, 7-α, chenodeoxycholic acid glycine conjugate and 26-trihydroxy-5-β-cholestane (P < 0.05, P < 0.01). Allobaculum stercoricanis was positively correlated with linoleic acid and sphinganine (P < 0.01), and 2-methyl-3-hydroxy-5-formylpyridine-4-carboxylate was negatively associated with Prevotella sp. CAG:604 (P < 0.01). The metabolic pathways showing the largest differences were arginine biosynthesis; primary bile acid biosynthesis; purine metabolism; linoleic acid metabolism; alanine, aspartate and glutamate metabolism; and nitrogen metabolism.
Metagenomics and untargeted metabolomics indicated that disordered compositions of gut microbes and metabolites may be common defects in GK rats.
Core Tip: Studies have suggested that the gut microbial and metabolites play an essential role in Goto Kakizaki rats. The results revealed evidence of a decrease in Prevotella sp. CAG:604 and increases in Lactobacillus_murinus and Allobaculum_stercoricanis. Fourteen potential metabolism biomarkers included glycochenodeoxycholic acid, uric acid, N-acetylaspartate, β-sitostenone, sphinganine, 4-pyridoxic acid, 13(S)-hydroxyoctadecadienoic acid, and linoleic acid, etc.
- Citation: Zhao JD, Sun M, Li Y, Yu CJ, Cheng RD, Wang SH, Du X, Fang ZH. Characterization of gut microbial and metabolite alterations in faeces of Goto Kakizaki rats using metagenomic and untargeted metabolomic approach. World J Diabetes 2023; 14(3): 255-270
- URL: https://www.wjgnet.com/1948-9358/full/v14/i3/255.htm
- DOI: https://dx.doi.org/10.4239/wjd.v14.i3.255
In recent years, the incidence of type 2 diabetes (T2DM) has shown a rapid growth trend in China, where its current prevalence is 12.8%[1]. T2DM is a metabolic disorder whose major pathological hallmark is insulin resistance[2]. Goto-Kakizaki (GK) noninsulin-dependent rats are a valuable animal model for studies of T2DM[3]. At 4 wk of age, GK rats exhibit insulin resistance, basal hyperglycaemia development, increased hepatic glucose production, and impaired insulin secretion[4,5]. GK rats share common features with human T2DM. Therefore, the results obtained using GK rats as the research subjects will be more similar to those of from patients with T2DM than the results obtained using other systems and will thus provide more useful evidence for analysing the pathogenesis and treatment of T2DM.
The gut microbiota of the host constitutes a massive, complex microecosystem. Exploring the composition of the gut microbiota will improve our understanding of the relationship with the host as one of the key factors in host health or disease. Recent studies suggest that both humans and animal models of T2DM and its complications, including GK rats, exhibit dysbiosis of the gut microbiota and further indicate that this characteristic microbiota imbalance, which may involve decreased bacterial diversity, contributes to the development of T2DM[6-8]. In addition, studies have shown that un-desirable shifts in metagenomic- and microbiota-associated metabolite production may negatively impact glucose tolerance and insulin resistance[9].
Because the human dietary structure is complex and diverse, the changes in gut microbes and metabolites observed in different studies vary, even if the type and amount of the diets administered to patients with T2DM (e.g., dietary fibre contents or low-carbohydrate diets) and nutritional advice are controlled[10-12]. In this study, we selected a fixed diet with balanced nutrient proportions for rats to avoid deviations in the results due to dietary differences. In animal models of diabetes, streptozotocin or alloxan is usually selected as an agent for damaging pancreatic function. This type of model is more similar to type 1 diabetes. In the classification of diabetes, T2DM accounts for approximately 90% of cases. Therefore, the further exploration of gut the microbes and metabolites in a T2DM model is desired. Moreover, a joint analysis of the gut microbiota and metabolite structure has not previously been conducted[13]. Therefore, the faeces of GK rats were collected as the study object, and this study aimed to systematically characterize the faecal gut microbes and metabolites of GK rats using metagenomic and untargeted metabolomic approaches and to analyse the relationship of gut microbes and metabolites with glucose and insulin resistance.
Ten male GK rats (aged 5-6 wk) were procured from Changzhou Cavens Model Animal Co., Ltd. (Changzhou, China; certificate No. 202145537), and ten male Wistar rats (aged 5-6 wk) were purchased from Sipeifu (Beijing) Biotechnology Co., Ltd. (Beijing, China; certificate No. 110324210106676238). The rats were acclimated in a controlled laboratory (25 ± 2 °C temperature, 60% ± 5% humidity) with free access to breeding feed and water. The Animal Committee of Anhui University of Chinese Medicine (Hefei, China; Approval AHUCM-rats-2021133) approved the experiments.
The rats were fed growth and reproduction feed during the 1-wk acclimatization period and were observed for 10 wk. The fasting blood glucose (FBG) level was measured, and nine rats in the model group showed levels exceeding 11.1 mmol/L, whereas the FBG level of the other rat was not as high. Additionally, one rat in the control group escaped due to poor management, and the analysis was thus performed using nine rats from each group. The body weight and FBG were measured every two weeks. The FBG levels in blood sampled from the tail vein were measured with a glucose metre (ACCU-CHEK Performa, ROCHE, Basel, Switzerland) after food deprivation for 12 h overnight.
Eighteen rats were anaesthetized via the intraperitoneal administration of pentobarbital sodium (30 mg/kg, Merck, United States). Blood samples were collected in nonheparinized tubes and centrifuged to obtain serum. The indexes FBG, insulin, homeostasis model assessment of insulin resistance (HOMA-IR) and homeostasis model assessment of β cell (HOMA-β) were assessed. The faecal were placed in Eppendorf tubes, and the serum and faecal were stored at -80 °C[14].
Total DNA was extracted from 1 g of faeces using a kit (Omega Bio-Tek, Norcross, GA, United States), and the concentration, purity and quality of the DNA was determined[15].
The extracted DNA (average length of 400 bp) was fragmented using a Covaris M220 ultrasonicator (Gene Company Limited, China). A paired-end library was constructed using NEXTFLEX Rapid DNA-Seq (Bioo Scientific, Austin, TX, United States). Sequencing was performed with an Illumina NovaSeq system (Illumina Inc., San Diego, CA, United States) using NovaSeq Reagent Kits according to the manufacturer’s instructions[16]. The paired-end Illumina reads were trimmed of adaptors, and reads with low quality (length < 50 bp, quality value < 20, containing N bases) were removed by fastp[17].
Metagenomic data were collected using MEGAHIT. Contigs with a length ≥ 300 bp were selected as the final assembly output and used for further gene annotation. Amino acid sequences from the predicted open reading frames with a length ≥ 100 bp were retrieved and translated from the NCBI database[18]. The nonredundant gene catalogue was constructed using CD-HIT and aligned to high-quality reads using SOAP aligner.
The supernatant was extracted from 200 mg of faeces and transferred to sample vials[19]. Two microlitres of a sample was separated with an HSS T3 column and used for LC-MS/MS analysis. Mass spectrometric data were collected using a UHPLC-Q Exactive system (Thermo Fisher Scientific, Waltham, MA, United States) with an electrospray ionization source operating in the positive- and negative-ion modes. Data acquisition was performed in the data-dependent acquisition mode.
The raw LC-MS/MS data were preprocessed using Progenesis QI (Waters Corporation, Milford, MA, United States) software. Internal standard peaks and false-positive peaks were removed from the data matrix, redundant signals were removed, and the peaks were pooled. In addition, the metabolites were searched and identified in the HMDB, KEGG and Metlin databases.
Metabolites detected in at least 80% of any set of samples were retained[20]. After filtering, the metabolite response intensity of the mass spectrum peaks was normalized using the sum-normalization method. Moreover, variables with a relative standard deviation > 30% relative to the quality control samples were removed, and log10 logarithmization was performed to obtain the final data matrix for subsequent analysis.
Statistical and graphical analyses of the data were performed by Student’s t tests and fold difference analysis using SPSS 23.0 (International Business Machines Corporation, NY, United States) and GraphPad Prism 9.0 (San Diego, CA, United States) software. The heatmap data were normalized via the z score method [z = (x-μ)/σ] and graphically visualized using the pheatmap package.
Variance analysis was performed with the matrix file after data preprocessing. The R package ropls (Version 1.6.2) was used to perform orthogonal partial least squares discriminant analysis (OPLS-DA). The stability of the model was assessed by 7-cycle interactive validation. The significantly different metabolites were selected based on the following criteria: variable importance in the projection score (> 1) of obtained by OPLS-DA and the P value < 0.05 obtained by Student’s t test[21].
The differential metabolites were screened and mapped to their biochemical pathways through metabolic enrichment and pathway analysis based on the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/). These metabolites were classified according to their functions. Significantly enriched pathways were identified using MetaboAnalyst based on degree centrality and Fisher’s exact test (https://www.metaboanalyst.ca/). The molecular weights of potential biomarkers were obtained from biochemical databases (https://www.genome.jp/kegg/compound/). The data were analysed using the Majorbio Cloud Platform (https://www.majorbio.com) and Personalbio Cloud Platform (https://www.genescloud.cn/).
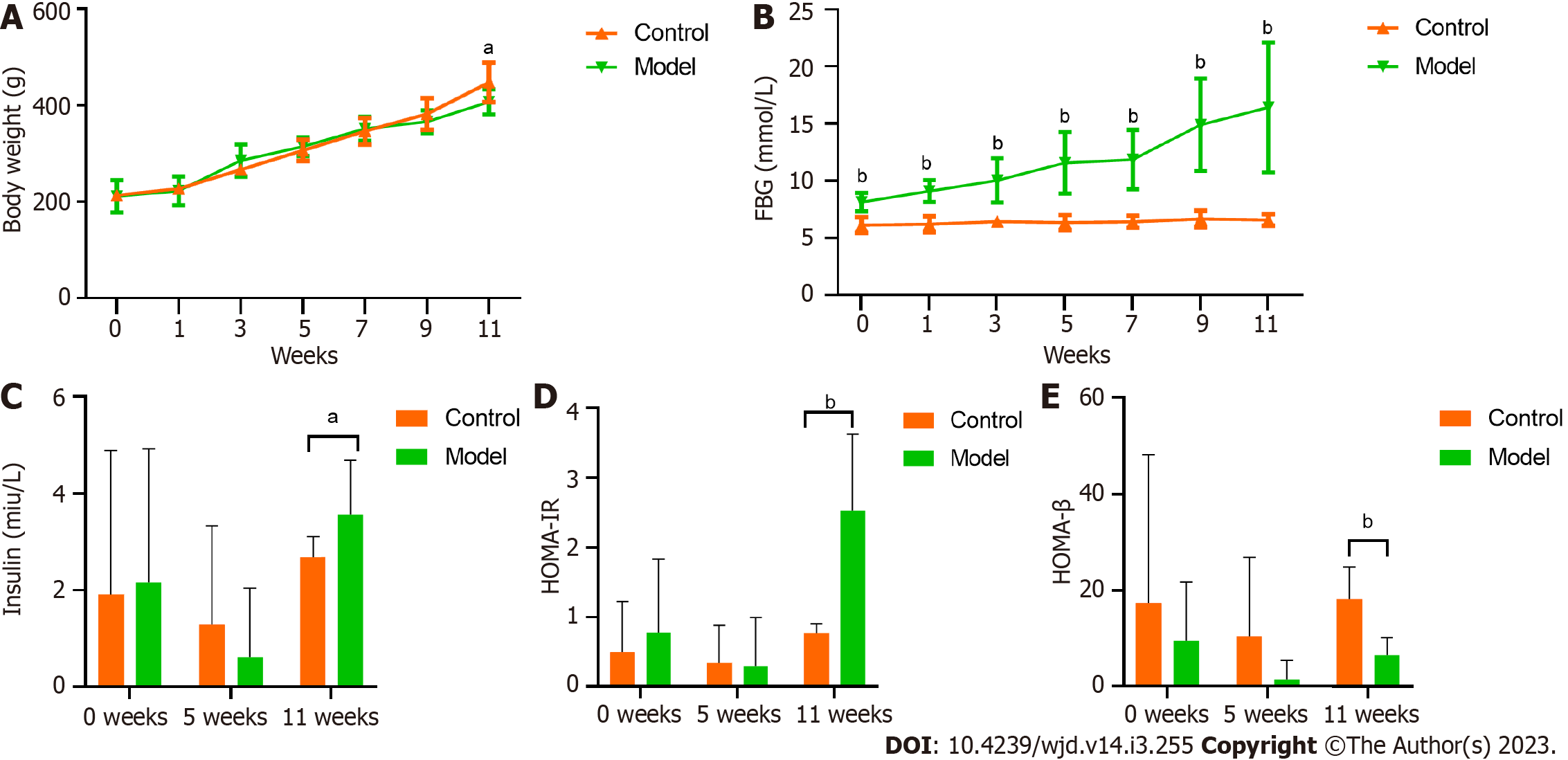
The initial body weights did not significantly differ between the groups (P > 0.05). In the final weeks of the experimental period, the model group had a significantly lower weight than the control group (P < 0.05; Figure 1A).
The FBG levels showed significantly differences from weeks 0 to 11, and over this time period, the significant difference became increasingly pronounced (Figure 1B). The mean FBG of the model group exceeded 11.1 mmol/L at week 5 and was close to 16.7 mmol/L at week 11.
Starting from week 11, the fasting insulin level of the model group was significantly higher than that of the control group (Figure 1C). Additionally, the HOMA-IR index of the model group was markedly higher than that of the control group from week 11 onwards (Figure 1D). Moreover, the HOMA-β index of the model group was significantly lower than that of the control group (Figure 1E).
In total, 4.15 × 1010 raw bases (bp) and 4.33 × 108 raw reads were obtained. Subsequently, 1.59 × 108 nonredundant genes were predicted from the reads after quality control and the removal of host sequences. In total, five domains, 14 kingdoms, 181 phylum, 316 classes, 551 orders, 940 families, 2764 genus and 12302 species were identified. At the domain level, the gut microbiota comprised bacteria, archaea, eukaryotes, viruses and unclassified bacteria, and bacteria accounted for more than 99.67% ± 0.05% of the organisms in both groups.
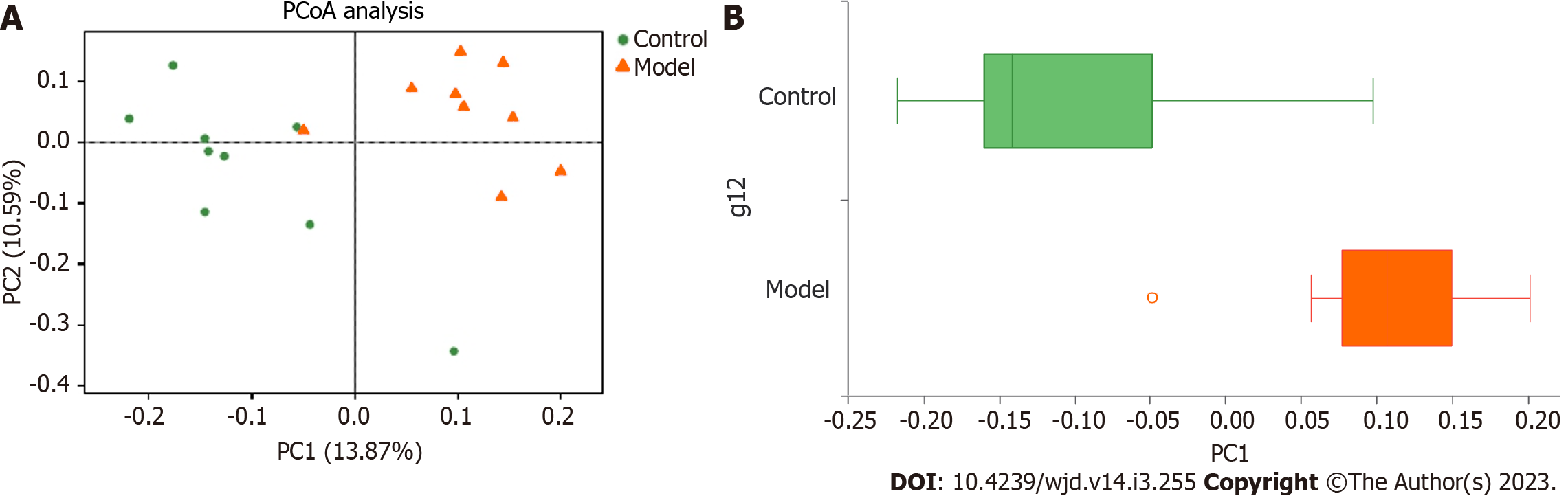
A principal coordinate analysis of the distance matrix showed that the first principal component (PC1) and the second principal components accounted for 13.87% and 10.59% of the observed variation, respectively (Figure 2A). The boxplots illustrate the distribution of different groups on the PC1 axis and reveal that the greatest difference existed between the control and model groups (Figure 2B).
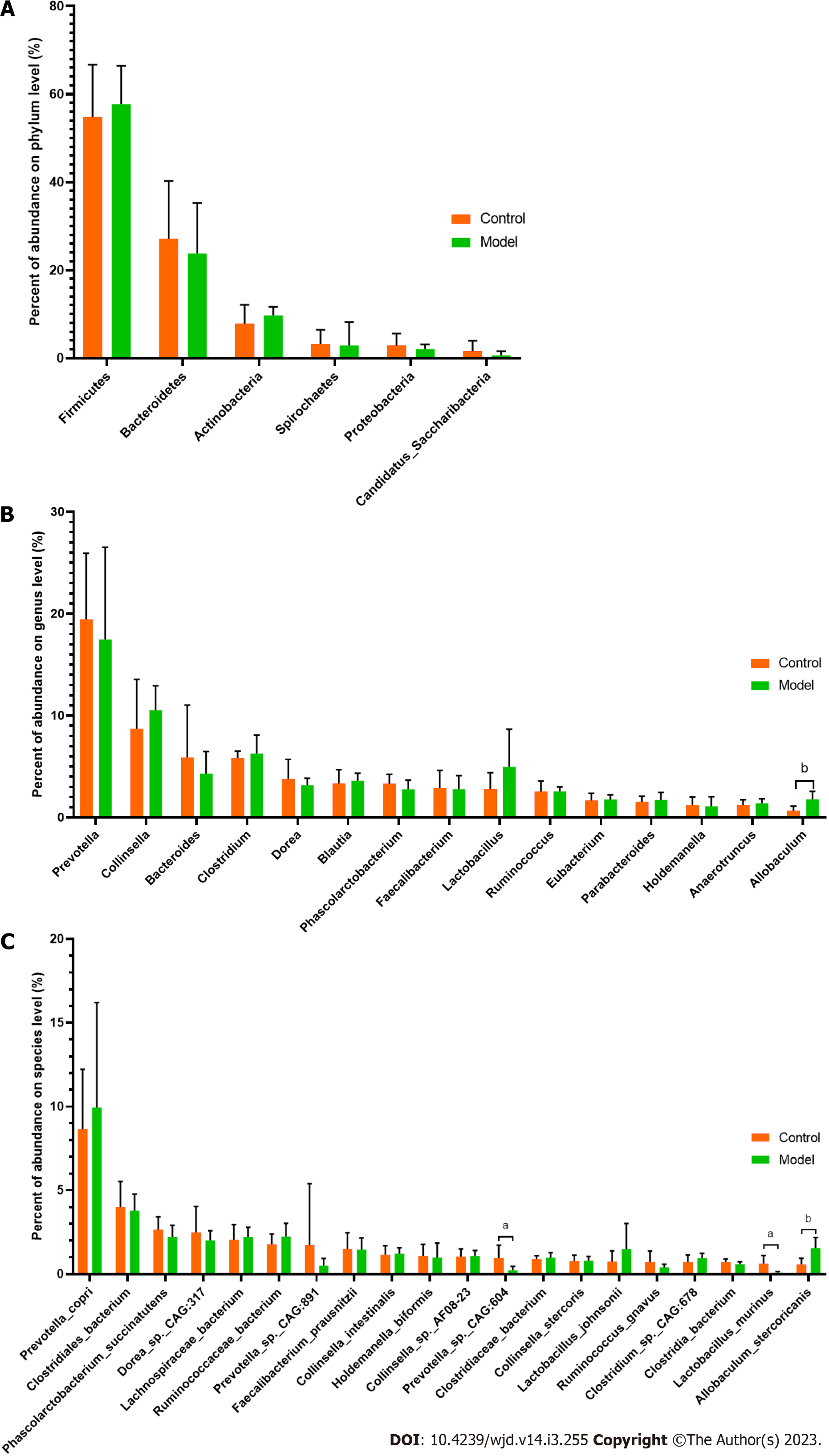
At the phylum level, Firmicutes, Bacteroidetes, Actinobacteria, Spirochaetes, Proteobacteria and Candidatus Saccharibacteria were identified as the main phylum (Figure 3A). The relative abundances of Firmicutes and Bacteroidetes were 54.85% and 27.16% in the control group and 57.75% and 23.82% in the model group, respectively. Although no significant differences were detected, a greater abundance of Firmicutes and a lower abundance of Bacteroidetes were found in the model group than in the control group.
In addition to Allobaculum, which was the only genus showing a significant difference, Prevotella, Bacteroides, Dorea, Phascolarctobacterium, Faecalibacterium and Ruminococcus were enriched in the control group, and Collinsella, Clostridium, Blautia and Lactobacillus were the main components found in the model group (Figure 3B). Among the 30 species with the highest abundance, 3 species showed significant differences between the control and model groups (P < 0.05). A higher abundance of Allobaculum stercoricanis and lower abundances of Prevotella sp. CAG:604 and Lactobacillus murinus were found in the model group compared with the control group (Figure 3C).
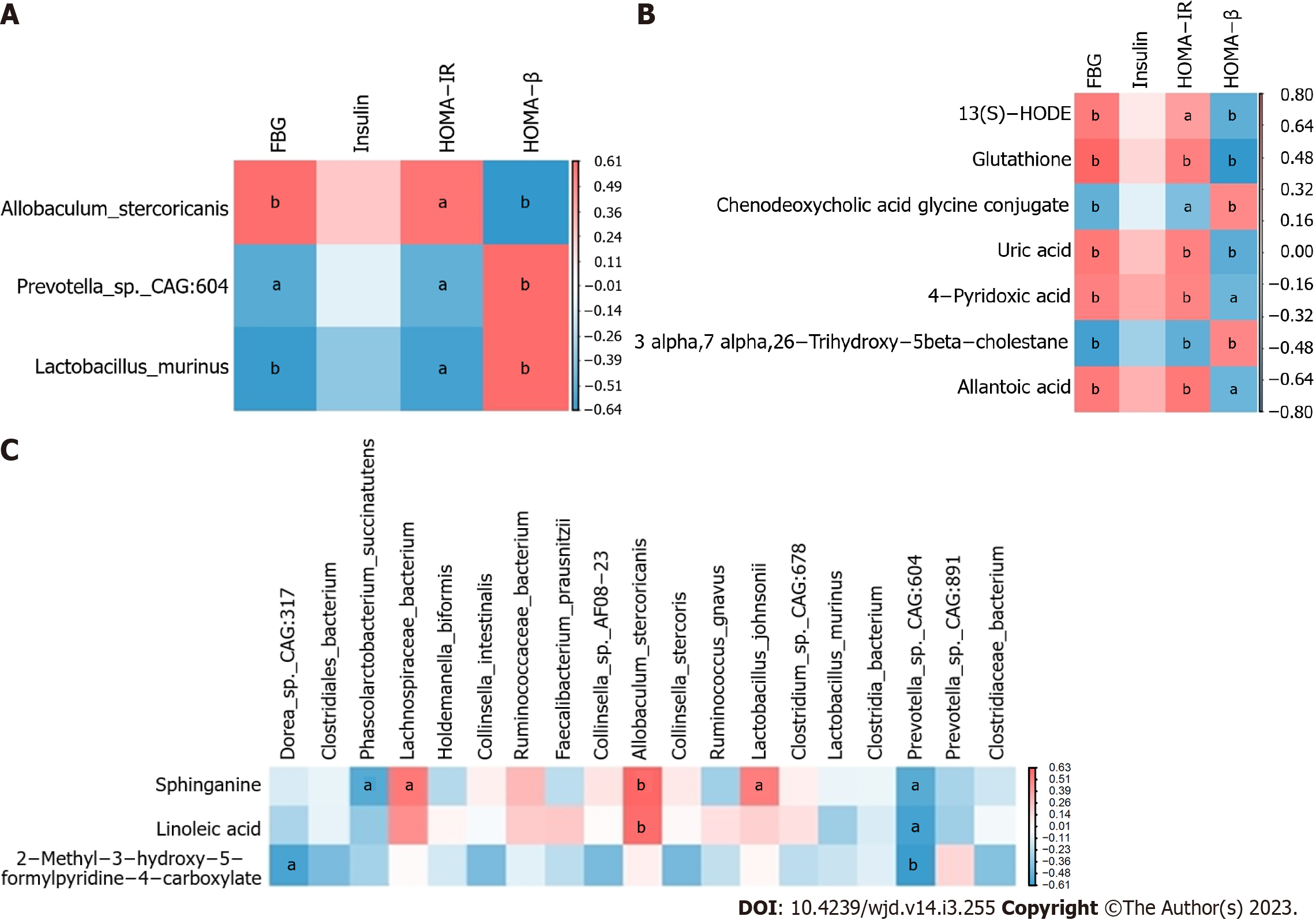
The constructed heatmap revealed that FBG and HOMA-IR were moderately positively correlated with Allobaculum stercoricanis and moderately negatively correlated with Lactobacillus murinus (P < 0.05, P < 0.01; Figure 4A).
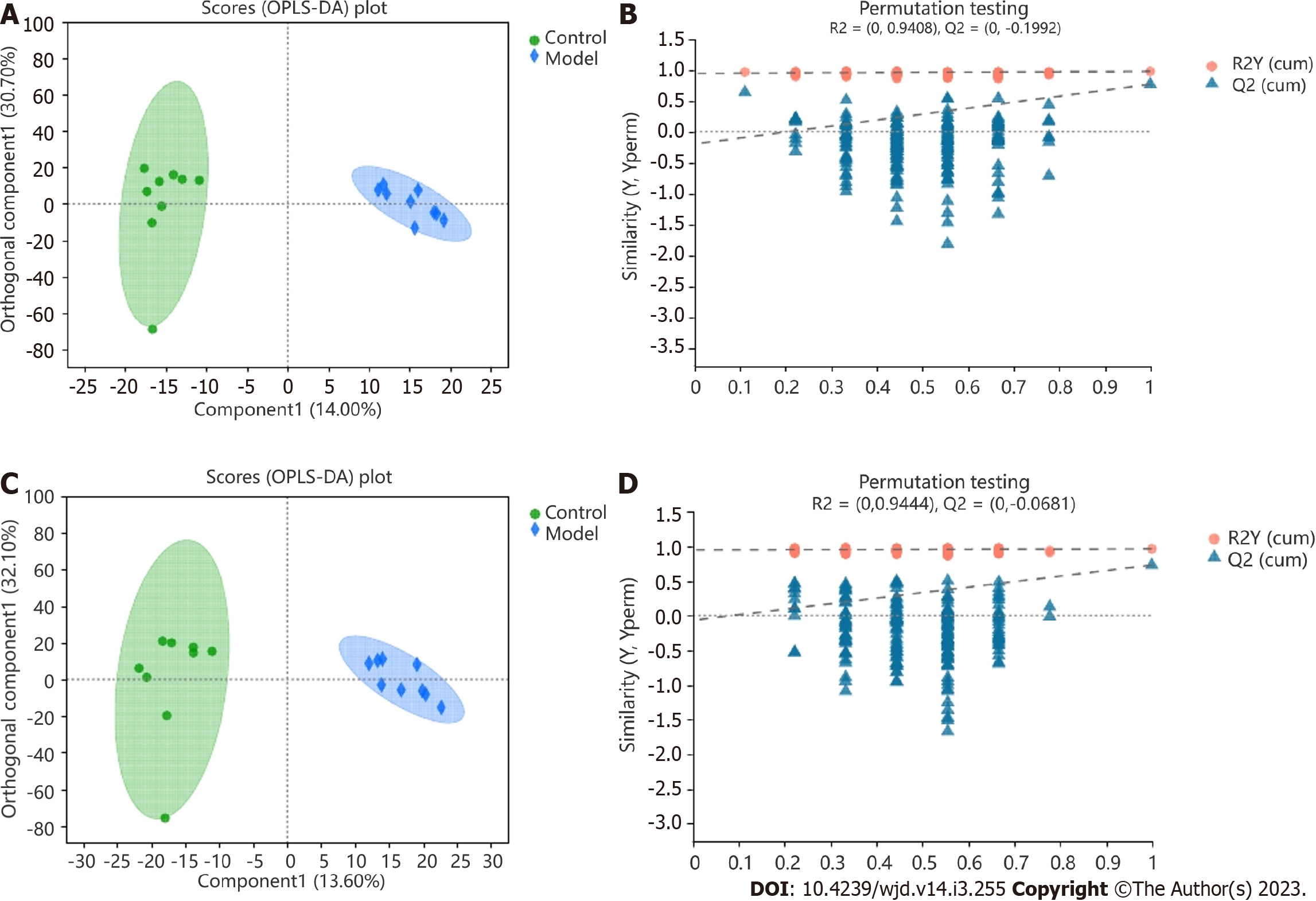
The faecal metabolic profile data of the control and model groups were separated by OPLS-DA model, which indicated that the positive- and negative-ion metabolic profiles of the samples differed between the two groups. The evaluation parameters of the OPLS-DA models from the positive- and negative-ion profiles showed values of Q2 = -0.199 and Q2 = -0.068, respectively, demonstrating good explanation and prediction with 200× permutation testing. For the positive-ion profiles, the component 1 showed a value of 14.00%, and the orthogonal component 1 showed a value of 30.70%. For the negative-ion profiles, the component 1 showed a value of 13.60%, and the orthogonal component 1 showed a value of 32.10% (Figure 5A-D).
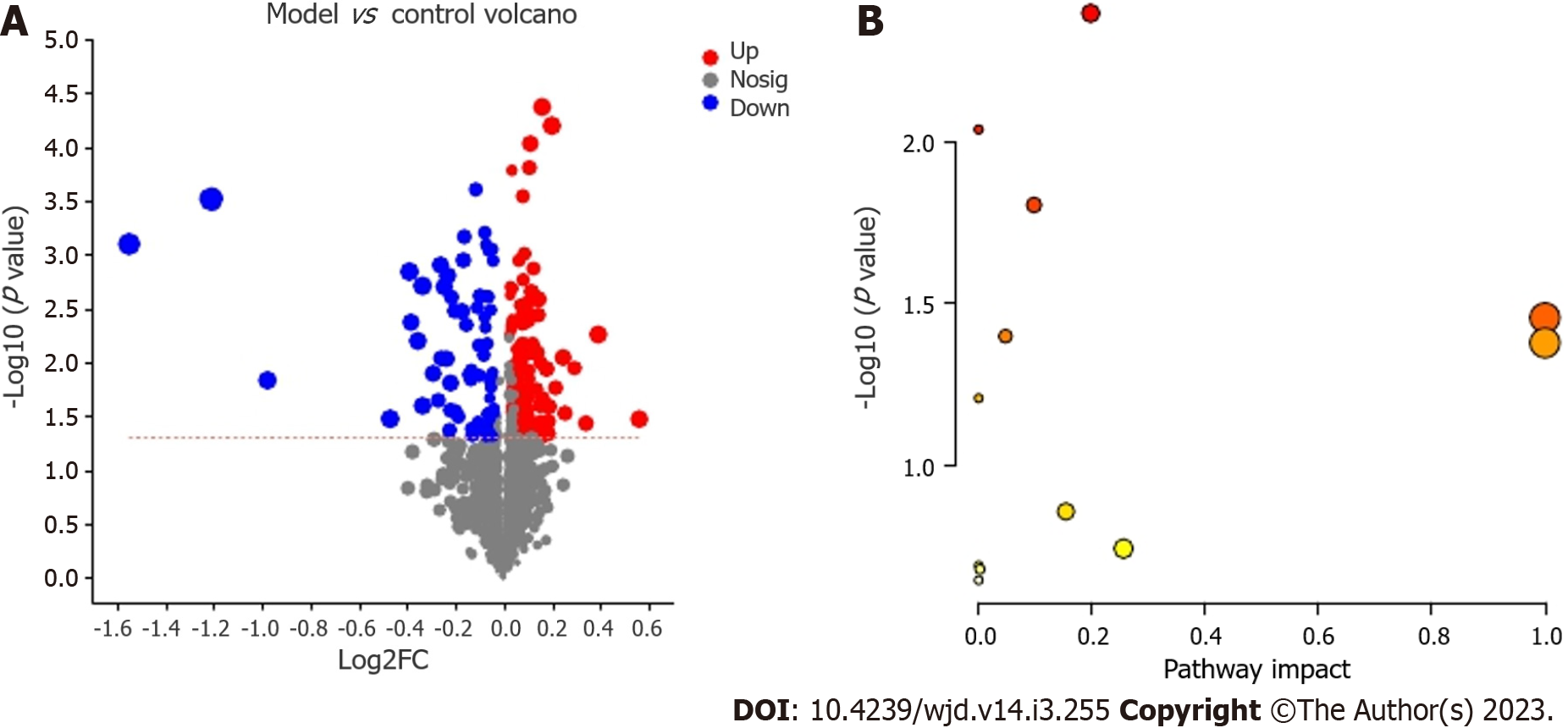
We identified 815 positive-ion and 678 negative-ion metabolites, and 596 metabolites were found in the KEGG compound database. The differential metabolites (including 145 positive- and 106 negative-ion metabolites or 180 upregulated and 71 downregulated metabolites) were filtered in volcano plots (Figure 6A). The most reasonable molecular formula was obtained from a search of the KEGG Compound database. According to the abovementioned principle, 14 potential metabolic biomarkers were identified in GK rats (Table 1).
| Metabolite | Class | KEGG compound ID | Mass M/Z | Mode | Formula | Retention time (min) | Change trend (model/control) |
| Glycochenodeoxycholic acid | Bile acids | C05466 | 414.2993 | Pos | C26H43NO5 | 5.333 | Down |
| Uric acid | Benzenoids | C00366 | 169.0355 | Pos | C5H4N4O3 | 1.3882 | Up |
| Glutathione | Amino acids | C00051 | 371.1005 | Pos | C10H17N3O6S | 1.0579 | Up |
| Glycocholic acid | Bile acids | C01921 | 446.2898 | Neg | C26H43NO6 | 5.3482 | Down |
| Β-sitostenone | Steroid | C00014 | 413.3772 | Pos | C29H48O | 9.9336 | Up |
| Allantoic acid | Amino acids | C00499 | 175.0461 | Neg | C4H8N4O4 | 0.6403 | Up |
| N-acetylaspartate | Amino acids | C01042 | 174.0396 | Neg | C6H9NO5 | 1.1621 | Up |
| N-acetyl-l-glutamic acid | Amino acids | C00624 | 188.0553 | Neg | C7H11NO5 | 1.4766 | Up |
| Sphinganine | Amines | C00836 | 302.3045 | Pos | C18H39NO2 | 6.9062 | Up |
| 4-pyridoxic acid | Pyridine carboxylic acids and derivatives | C00847 | 184.0604 | Pos | C8H9NO4 | 1.6417 | Up |
| 2-methyl-3-hydroxy-5-formylpyridine-4-carboxylate | Pyridine carboxylic acids and derivatives | C06050 | 180.029 | Neg | C8H7NO4 | 2.2041 | Up |
| 13(s)-HODE | Fatty acids | C14762 | 279.2311 | Pos | C18H32O3 | 7.4255 | Up |
| Linoleic acid | Linoleic acids and derivatives | C01595 | 281.2469 | Pos | C18H32O2 | 8.1846 | Up |
| 3-α, 7-α, 26-trihydroxy-5-β-cholestane | Bile acids | C05444 | 419.3521 | Neg | C27H48O3 | 8.5577 | Down |
The heatmap revealed that FBG and HOMA-IR were positively correlated with 13(S)-hydroxyoctadecadienoic acid (HODE), glutathione, uric acid, 4-pyridoxic acid and allantoic acid and negatively associated with 3-α, 7-α, 26-trihydroxy-5-β-cholestane and chenodeoxycholic acid glycine conjugate (P < 0.05, P < 0.01; Figure 4B).
The heatmap also showed that sphinganine and linoleic acid were positively correlated with Allobaculum stercoricanis (P < 0.01) and that 2-methyl-3-hydroxy-5-formylpyridine-4-carboxylate was negatively associated with Prevotella sp. CAG:604 (P < 0.01; Figure 4C).
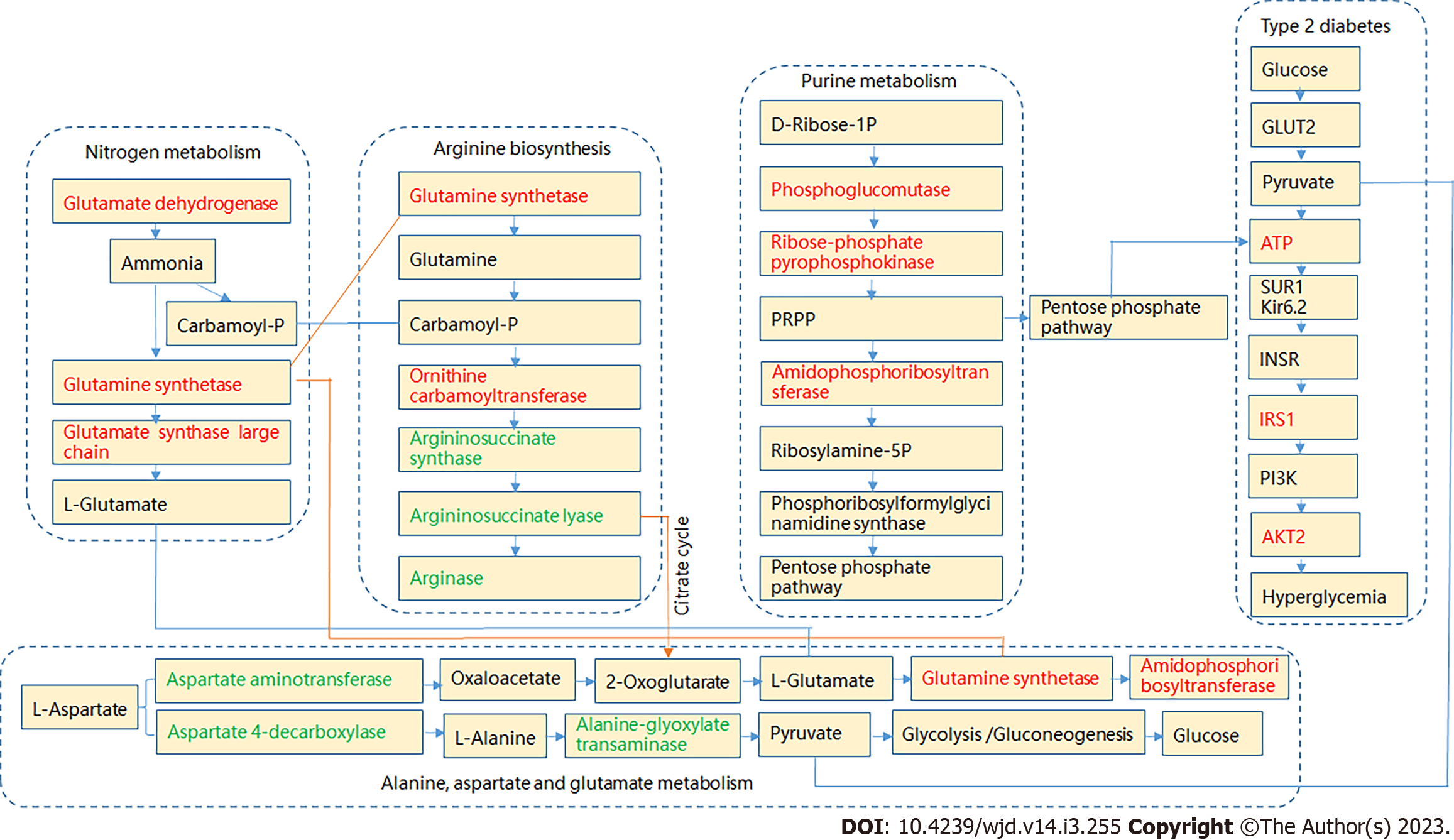
The KEGG compound IDs and compound names of endogenous metabolites were then introduced into the MetaboAnalyst 5.0 system for pathway and visual analyses. Based on the faecal biomarker data, the enriched pathways were found to include vitamin B6 metabolism; primary bile acid biosynthesis; arginine biosynthesis; purine metabolism; alanine, aspartate and glutamate metabolism; linoleic acid metabolism; and nitrogen metabolism. Among these pathways, six pathways were found to show differences (Figure 6B; Table 2). Figure 7 illustrates the key potential proteases of the metabolic pathways related to hyperglycaemia levels based on the KEGG database.
| Pathway name | Match status | P value | -log (p) | Impact | Pathway level |
| Arginine biosynthesis | 2/14 | 0.0055854 | 2.2529 | 0.19797 | Amino acid metabolism |
| Primary bile acid biosynthesis | 3/46 | 0.0056824 | 2.2455 | 0.05721 | Lipid metabolism |
| Purine metabolism | 3/65 | 0.014896 | 1.8269 | 0.0 | Nucleotide metabolism |
| Alanine, aspartate and glutamate metabolism | 2/28 | 0.021712 | 1.6633 | 0.09776 | Amino acid metabolism |
| Linoleic acid metabolism | 1/5 | 0.04129 | 1.3842 | 1.0 | Lipid metabolism |
| Nitrogen metabolism | 1/6 | 0.049357 | 1.3066 | 1.0 | Energy metabolism |
| Vitamin B6 metabolism | 1/9 | 0.073183 | 1.1356 | 0.0 | Metabolism of cofactors and vitamins |
| Glutathione metabolism | 1/28 | 0.21173 | 0.67421 | 0.25596 | Metabolism of other amino acids |
| Glyoxylate and dicarboxylate metabolism | 1/32 | 0.23835 | 0.62279 | 0.0 | Carbohydrate metabolism |
| Glycine, serine and threonine metabolism | 1/33 | 0.24487 | 0.61107 | 0.00245 | Amino acid metabolism |
| Biosynthesis of unsaturated fatty acids | 1/36 | 0.26413 | 0.57818 | 0.0 | Lipid metabolism |
The aetiology and pathogenesis of T2DM are complex, and most experts believe that this disorder is a clinical syndrome caused by genetic and environmental factors. Because GK rats develop spontaneous diabetes and show insulin resistance, these rats constitute one of the best models for the study of T2DM. In clinical practice, we have found that some patients who suffer from T2DM do not consume a high-sugar and high-fat diet. Therefore, this study did not use a high-sugar and high-fat diet, which reduced the impacts of excess nutrition on glucose. Studies have shown that the glucose levels of 14- to 16-wk-old GK rats meet the diagnostic criteria for T2DM[3]. In this study, at 14 wk, the GK rats appeared to drink water and urinate more than the control rats, and their hair colour turned yellow. Before 12 wk, the GK rats were heavier than the control Wistar rats. However, at 14 wk, the body weight of the GK rats was lower than that of the control rats, and the FBG level of the former exceeded 11.1 mmol/L. By the 16th week, the body weight of the GK rats was further reduced, and the glucose level was further increased to levels exceeding 16 mmol/L. Furthermore, the increase in glucose was related an increase in the HOMA-IR index and a decrease in the HOMA-β index. This finding may be related to the observed decrease in the numbers of pancreatic β-cell islets, the irregular shape of the islets, and the presence of amyloid degeneration. Some researchers believe that weight loss is related to a reduction in the Firmicutes/Bacteroidetes ratio, but this phenomenon is not observed universally[22,23] and was not found in our research. Muñoz-Garach et al[24] believe that weight loss is related to a reduction in the level of bile acid, and the results of the present study are consistent with this view.
A clear separation among the communities was observed between the control and model groups, which suggested that the gut microbiota shows substantial differences between the diabetes and nondiabetes groups. At the phylum level, Bacteroidetes and Firmicutes were dominant in GK and Wistar rats, with proportions greater than 80%. Although no significant differences were detected, a higher relative abundance of Firmicutes and a lower relative abundance of Bacteroidetes were found in the model group than in the control group. These findings may be related to the fact that the rats were fed breeding feed after weaning. These results may also be related to the shorter disease course in the T2DM rats because the gut microbiota is seriously disturbed, and the development of characteristic hyperglycaemia lags behind.
At the species level, a higher abundance of Allobaculum stercoricanis and lower abundances of Prevotella sp. CAG:604 and Lactobacillus murinus were found in the model group compared with the control group. Allobaculum stercoricanis belongs to the Allobaculum genus (Firmicutes), for which a significant difference was found at the genus level. A previous study showed that Allobaculum presented a 10.95-fold increase in HF diabetic mice compared with ND-fed mice[25], and the corresponding increase found in this study was 2.75-fold. In a previous study, Allobaculum was identified as an important biomarker of metabolic disorders, particularly diabetes[26,27]. This study also showed that Allobaculum stercoricanis was closely related to FBG, HOMA-IR index and a decrease in HOMA-β index. Different individual animals, even within the same species, show different feed conversion efficiencies. Animals with a high feed conversion efficiency show higher mRNA expression levels of insulin-like growth factor[28], which can further regulate glucose. The abundance of Prevotella sp. CAG:604 is related to the feed conversion efficiency[29]. This study revealed a decrease in the abundance of Prevotella sp. CAG:604, which may be one of the reasons for the diabetic phenotype of GK rats, and a direct negative correlation was found between these factors. Prevotella sp. CAG:604 could be considered a potential biomarker of diabetes. Lactobacillus murinus was decreased in GK rats, and this finding is consistent with the results reported by Cui et al[30] and Yue et al[31]. With increases in the abundance of Lactobacillus murinus, the glucose level and HOMA-IR index showed a downwards trend. Lactobacillus murinus is considered a beneficial gut microbe[32]. In a previous study, a probiotic group whose members drank Lactobacillus casei strain milk showed a significant increase in the faecal count of Lactobacillus. The condition of patients with T2DM has also been controlled[33]. We speculate that drinking milk containing Lactobacillus murinus may also control the glucose levels.
The OPLS-DA model suggested that the positive- and negative-ion metabolic profiles differed between the GK and Wistar rat faeces. Fourteen potential metabolic biomarkers were assigned to bile acids, benzenoids, amino acids, steroids, amines, pyridine carboxylic acids and derivatives, and fatty acids. The topological analysis of the metabolic pathways showed that the difference between GK and Wistar rats was mainly due to arginine biosynthesis; primary bile acid biosynthesis; alanine, aspartate and glutamate metabolism; purine metabolism; linoleic acid metabolism; and nitrogen metabolism. These results were consistent with those found in previous studies[34,35]. The metabolic pathway analysis showed that the differences were mainly due to amino acid metabolism, lipid metabolism, nucleotide metabolism, and energy metabolism. These pathways provide ideas for future research, and the results show that in addition to abnormal carbohydrate pathways, fat and protein are also metabolized in a disorderly manner leading to dysfunctions of the eyes, kidneys, blood vessels, and nerves, among other organs.
We observed marked decreases in the excretion of glycochenodeoxycholic acid and glycocholic acid in rat faeces, as reported by Sun et al[36]. The levels of glycochenodeoxycholic acid are increased in the liver, blood and ileum[36-38]. Lu et al[39] showed that increased plasma glycochenodeoxycholic acid levels are associated with an increased risk of T2DM. The relationship among 3-α, 7-α, 26-trihydroxy-5-β-cholestane and T2DM has been less well studied, and we found that 3-α, 7-α, 26-trihydroxy-5-β-cholestane was positively related to FBG and HOMA-IR, which may be a research topic that will be addressed in the future. Hyperuricaemia is particularly common in patients with T2DM[40]. Some clinical studies have observed that ursodeoxycholic acid, as the representative drug regulating bile acid metabolism, exerts a certain effect on reducing glucose, glycosylated haemoglobin A1c, and weight and increasing GLP-1 secretion[41,42]. Although ursodeoxycholic acid is not widely used in clinical practice, it may be used as a potential drug for the treatment of T2DM. In addition to excretion in the kidneys, uric acid can also be secreted into the intestine and metabolized by microorganisms[43]. This interaction could potentially modulate the serum uric acid levels[44]. This study revealed an increase in the intestinal excretion of uric acid, indicating that uric acid in the body is likely to be a risk factor for T2DM and could aggravate this condition. In our study, we also found that uric acid was positively related to FBG and HOMA-IR.
In the presence of glutathione, a series of physiological reactions result in a low antioxidative defence capacity. β cells gradually die, and progression to T2DM occurs[45]. This study showed that the glutathione level was increased in the GK group and was positively related to the FBG and HOMA-IR. Hyperglycaemia is a potential risk factor for the N-acetylaspartate network[46]. The present study also identified this phenomenon. The N-acetylaspartate level was higher in GK rats. A high level of N-acetyl-L-glutamic acid may result in increased blood pressure[47]. Uncontrolled blood pressure can significantly increase the risk of macroangiopathy in diabetes. The relationship of allantoic acid with diabetes has not been studied, but the present study revealed that allantoic acid was positively related to FBG and HOMA-IR. At baseline, a previous study showed that amino acids are upregulated in T2DM patients. Dysregulation of amino acid metabolism may occur earlier than glucose metabolism in T2DM[48]. This finding suggests that patients with T2DM should not consume a diet with a high protein level.
We found that the concentration of sphinganine was increased in GK rats. Sphinganine is positively related to Lachnospiraceae bacteria, Allobaculum stercoricanis, and Lactobacillus johnsonii and is negatively related to Prevotella sp. CAG:604 and Phascolarctobacterium succinatutens. Studies have shown that when obese people lose weight, they exhibit an increase in the abundance of Phascolarctobacterium succinatutens[49]. Weight loss is conducive to glucose control. Sphinganine is formed from ceramide and is a risk factor for β-cell dysfunction in T2DM[50]. Under these conditions, insulin secretion was observed to be reduced, and the inhibition of ceramide metabolism was weakened. An increase in 13(S)-HODE has been observed in individuals with nonalcoholic fatty liver disease[51]. Because the common pa-thogenesis of nonalcoholic fatty liver and diabetes is insulin resistance, diabetes may also be accompanied by elevated levels of 13(S)-HODE, and our research confirms this hypothesis. 13(S)-HODE was found to be positively related to FBG and HOMA-IR, and Phascolarctobacterium succinatutens was enriched in the model group.
We observed that the linoleic acid level was elevated in GK rats. Linoleic acid levels are markedly higher among populations with a higher prevalence of T2DM[52]. Linoleic acid is positively related to Allobaculum stercoricanis and is negatively related to Prevotella sp. CAG:604. 4-Pyridoxic acid is a catabolite of vitamin B6 metabolism, and our study showed that the 4-pyridoxic acid levels were increased in the model group and positively related to FBG and HOMA-IR. A higher level of vitamin B6 is associated with a higher HOMA-IR values and T2DM[53]. Furthermore, the risk of all-cause mortality in patients with T2DM is higher among those with 4-pyridoxic acid levels in the highest quartile[54]. 2-Methyl-3-hydroxy-5-formylpyridine-4-carboxylate is also a catabolite of vitamin B6 metabolism, and a reduction in its content can help regulate the glucose levels[55,56]. This compound was was detected at to be negatively related to Prevotella sp. CAG:604 and Dorea sp. CAG:317 and showed increased levels in the model group in this study. The results indicated that the glucose level was elevated in GK rats and that chenodeoxycholic acid glycine conjugate was negatively correlated with FBG and HOMA-IR, which was consistent with the results reported by Zhang et al[57].
Feng et al[58] found that glutamine synthetase activity was significantly increased after the induction of diabetes by streptozotocin in rats. The serum levels of ornithine carbamoyltransferase are increased in KK-Ay diabetic mice[59]. The liver is involved in glucose metabolism. In a state of hyperglycaemia, the liver is damaged to varying degrees, resulting in the abnormal expression of some proteases. Phosphoglucomutase is a key protease in carbohydrate metabolism[60]. In particular, this protease is closely related to obesity. Endothelial cell function damage mostly occurs in T2DM and is related to arg-ininosuccinate synthase and argininosuccinate lyase. In particular, TNF-α downregulates argininosuccinate synthase expression[61,62]. The activity of amidophosphoribosyltransferase is higher in the kidneys of diabetic rats[63]. The results indicate that amidophosphoribosyltransferase is related to the accretion of nucleic acids. A low level of aspartate aminotransferase is an independent risk factor for frail T2DM patients[64]. Glycogen is synthesized in the liver, and changes in a series of liver enzymes affect the synthesis of glycogen. For example, aspartate 4-decarboxylase and alanine-glyoxylate transaminase were detected in this study. The occurrence of T2DM is related to many signalling pathways. Pyruvate, which is the end product of glycolysis, is transported into mitochondria and drives ATP production[65]. This study showed that ATP imbalance was the main reason for insulin resistance and thus mainly affected the IRS1 and AKT2 potential in the insulin resistance signalling pathway.
We identified different gut microbes and metabolites in GK rats, and the results provide new research directions related to faecal bacteria transplantation, supplementary metabolites, and drug screening, among other topics. This study also has some limitations, such as a lack of drug interventions for verification and a lack of long-term observations of the dynamic changes in gut microbes and metabolites. In future research, we will further improve the design scheme for studying metabolic disorders through the influence of nitrogen metabolism, arginine biosynthesis, primary bile acid biosynthesis, purine metabolism, alanine, aspartate and glutamate metabolism and insulin resistance, which can lead to the emergence of T2DM. In addition to the characteristics of hyperglycaemia, disordered fat and protein levels are also observed in patients with T2DM. T2DM is a clinical syndrome with multiple causes, and we thus believe that the treatment strategy for T2DM should be comprehensive and focus on glucose, lipids, protein, weight and other factors. This finding may provide ideas for the future management of patients with T2DM.
The present study revealed that disordered compositions of gut microbes and metabolites are putative common defects observed in GK rats by metagenomics and untargeted metabolomics. T2DM-related changes in gut microbes and metabolites may contribute to hyperglycaemia. However, further experiments are needed to summarize the different stages of T2DM.
Goto Kakizaki (GK) rats share common features with human type 2 diabetes (T2DM). Therefore, the results obtained from analyses of the gut microbiota and metabolites of GK rats will be more similar to those of patients with T2DM.
The gut microbiota and metabolites are critical in T2DM. Therefore, alterations in different gut microbiota and metabolites may provide useful evidence for analysing the pathogenesis and treatment of T2DM.
To investigate the alterations in gut microbiota and metabolites in the faeces of T2DM rats.
Systematic characterization of the faecal gut microbes and metabolites of GK rats using metagenomic and untargeted metabolomic approaches and analysis of the relationship between gut microbes and metabolites under conditions of glucose and insulin resistance.
The GK rats displayed significant differences in the gut microbiota structure compared with the control group. The results demonstrated that the GK rats presented significantly decreased abundances of Prevotella sp. CAG:604 and Lactobacillus murinus (P < 0.05) and a significantly higher abundance of Allobaculum stercoricanis (P < 0.01). Orthogonal partial least squares discriminant analysis suggested that the faecal metabolic profiles differed between the GK and control groups. Fourteen potential metabolic biomarkers, including glycochenodeoxycholic acid, uric acid, 13(S)-hydroxyoctadecadienoic acid, N-acetylaspartate, β-sitostenone, sphinganine, 4-pyridoxic acid, and linoleic acid, were identified. The metabolic pathways showing the main differences were arginine biosynthesis; primary bile acid biosynthesis; purine metabolism; linoleic acid metabolism; alanine, aspartate and glutamate metabolism; and nitrogen metabolism.
The present study revealed that disordered compositions of gut microbes and metabolites are putative common defects observed in GK rats by metagenomics and untargeted metabolomics.
Gut microbes and metabolites play a key role in carbohydrate metabolic pathways. Therefore, an evaluation of the involvement of dynamic changes in gut microbes and metabolites may be important in the future.
We thank Majorbio Li-Fen Huang and Ji-Long Feng (Majorbio Biotech Co., Ltd., Shanghai, China) for their excellent technical assistance.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Endocrinology and metabolism
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Kotlyarov S, Russia; Millman JF, Japan S-Editor: Zhang H L-Editor: A P-Editor: Zhao S
| 1. | Li Y, Teng D, Shi X, Qin G, Qin Y, Quan H, Shi B, Sun H, Ba J, Chen B, Du J, He L, Lai X, Li Y, Chi H, Liao E, Liu C, Liu L, Tang X, Tong N, Wang G, Zhang JA, Wang Y, Xue Y, Yan L, Yang J, Yang L, Yao Y, Ye Z, Zhang Q, Zhang L, Zhu J, Zhu M, Ning G, Mu Y, Zhao J, Teng W, Shan Z. Prevalence of diabetes recorded in mainland China using 2018 diagnostic criteria from the American Diabetes Association: national cross sectional study. BMJ. 2020;369:m997. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1030] [Cited by in RCA: 1002] [Article Influence: 200.4] [Reference Citation Analysis (1)] |
| 2. | Kahn SE. The relative contributions of insulin resistance and beta-cell dysfunction to the pathophysiology of Type 2 diabetes. Diabetologia. 2003;46:3-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1276] [Cited by in RCA: 1369] [Article Influence: 62.2] [Reference Citation Analysis (0)] |
| 3. | Nagao M, Esguerra JLS, Wendt A, Asai A, Sugihara H, Oikawa S, Eliasson L. Selectively Bred Diabetes Models: GK Rats, NSY Mice, and ON Mice. Methods Mol Biol. 2020;2128:25-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 4. | Zhang Q, Hong Z, Zhu J, Zeng C, Tang Z, Wang W, Huang H. Biliopancreatic Limb Length of Small Intestinal Bypass in Non-obese Goto-Kakizaki (GK) Rats Correlates with Gastrointestinal Hormones, Adipokines, and Improvement in Type 2 Diabetes. Obes Surg. 2021;31:4419-4426. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 5. | Portha B. Programmed disorders of beta-cell development and function as one cause for type 2 diabetes? Diabetes Metab Res Rev. 2005;21:495-504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 94] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 6. | Scheithauer TPM, Rampanelli E, Nieuwdorp M, Vallance BA, Verchere CB, van Raalte DH, Herrema H. Gut Microbiota as a Trigger for Metabolic Inflammation in Obesity and Type 2 Diabetes. Front Immunol. 2020;11:571731. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 379] [Article Influence: 75.8] [Reference Citation Analysis (0)] |
| 7. | Yang G, Wei J, Liu P, Zhang Q, Tian Y, Hou G, Meng L, Xin Y, Jiang X. Role of the gut microbiota in type 2 diabetes and related diseases. Metabolism. 2021;117:154712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 226] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 8. | Vangipurapu J, Fernandes Silva L, Kuulasmaa T, Smith U, Laakso M. Microbiota-Related Metabolites and the Risk of Type 2 Diabetes. Diabetes Care. 2020;43:1319-1325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 102] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 9. | Iatcu CO, Steen A, Covasa M. Gut Microbiota and Complications of Type-2 Diabetes. Nutrients. 2021;14:166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 208] [Article Influence: 52.0] [Reference Citation Analysis (0)] |
| 10. | Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, Fu H, Xue X, Lu C, Ma J, Yu L, Xu C, Ren Z, Xu Y, Xu S, Shen H, Zhu X, Shi Y, Shen Q, Dong W, Liu R, Ling Y, Zeng Y, Zhang Q, Wang J, Wang L, Wu Y, Zeng B, Wei H, Zhang M, Peng Y, Zhang C. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018;359:1151-1156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1058] [Cited by in RCA: 1542] [Article Influence: 220.3] [Reference Citation Analysis (68)] |
| 11. | Ren M, Zhang H, Qi J, Hu A, Jiang Q, Hou Y, Feng Q, Ojo O, Wang X. An Almond-Based Low Carbohydrate Diet Improves Depression and Glycometabolism in Patients with Type 2 Diabetes through Modulating Gut Microbiota and GLP-1: A Randomized Controlled Trial. Nutrients. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 80] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 12. | Palacios T, Vitetta L, Coulson S, Madigan CD, Denyer GS, Caterson ID. The effect of a novel probiotic on metabolic biomarkers in adults with prediabetes and recently diagnosed type 2 diabetes mellitus: study protocol for a randomized controlled trial. Trials. 2017;18:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 13. | Koh A, Bäckhed F. From Association to Causality: the Role of the Gut Microbiota and Its Functional Products on Host Metabolism. Mol Cell. 2020;78:584-596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 195] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 14. | Zhao JD, Li Y, Sun M, Yu CJ, Li JY, Wang SH, Yang D, Guo CL, Du X, Zhang WJ, Cheng RD, Diao XC, Fang ZH. Effect of berberine on hyperglycaemia and gut microbiota composition in type 2 diabetic Goto-Kakizaki rats. World J Gastroenterol. 2021;27:708-724. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 32] [Cited by in RCA: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 15. | Chen J, Li F, Zhao X, Wang Y, Zhang L, Yan L, Yu L. Change in composition and potential functional genes of microbial communities on carbonatite rinds with different weathering times. Front Microbiol. 2022;13:1024672. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 16. | Hu R, Wang XP, Xu JS, Zhang YF, Pan YX, Su X. The mechanism of soil nitrogen transformation under different biocrusts to warming and reduced precipitation: From microbial functional genes to enzyme activity. Sci Total Environ. 2020;722:137849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 17. | Wang B, Song L, Li W, Hou L, Li J, Xu X, Sheng G. Distribution and migration of antibiotic resistance genes, as well as their correlation with microbial communities in swine farm and its surrounding environments. Environ Pollut. 2023;316:120618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 12] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 18. | Li F, Jiang L, Pan S, Jiang S, Fan Y, Jiang C, Gao C, Leng Y. Multi-omic Profiling Reveals that Intra-abdominal-Hypertension-Induced Intestinal Damage Can Be Prevented by Microbiome and Metabolic Modulations with 5-Hydroxyindoleacetic Acid as a Diagnostic Marker. mSystems. 2022;7:e0120421. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 19. | Wang Z, Yu Y, Liao J, Hu W, Bian X, Wu J, Zhu YZ. S-Propargyl-Cysteine Remodels the Gut Microbiota to Alleviate Rheumatoid Arthritis by Regulating Bile Acid Metabolism. Front Cell Infect Microbiol. 2021;11:670593. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 20. | Kaleem MM, Nawaz MA, Ding X, Wen S, Shireen F, Cheng J, Bie Z. Comparative analysis of pumpkin rootstocks mediated impact on melon sensory fruit quality through integration of non-targeted metabolomics and sensory evaluation. Plant Physiol Biochem. 2022;192:320-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 21. | Liu T, Wang D, Zhou X, Song J, Yang Z, Shi C, Li R, Zhang Y, Zhang J, Yan J, Zhu X, Li Y, Gong M, Wang C, Yuan C, Cui Y, Wu X. Study on the mechanism of American ginseng extract for treating type 2 diabetes mellitus based on metabolomics. Front Pharmacol. 2022;13:960050. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 22. | Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022-1023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7222] [Cited by in RCA: 6409] [Article Influence: 337.3] [Reference Citation Analysis (0)] |
| 23. | Schwiertz A, Taras D, Schäfer K, Beijer S, Bos NA, Donus C, Hardt PD. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring). 2010;18:190-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1670] [Cited by in RCA: 1787] [Article Influence: 119.1] [Reference Citation Analysis (0)] |
| 24. | Muñoz-Garach A, Diaz-Perdigones C, Tinahones FJ. Gut microbiota and type 2 diabetes mellitus. Endocrinol Nutr. 2016;63:560-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 100] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 25. | Jia L, Li D, Feng N, Shamoon M, Sun Z, Ding L, Zhang H, Chen W, Sun J, Chen YQ. Anti-diabetic Effects of Clostridium butyricum CGMCC0313.1 through Promoting the Growth of Gut Butyrate-producing Bacteria in Type 2 Diabetic Mice. Sci Rep. 2017;7:7046. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 122] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 26. | Nobel YR, Cox LM, Kirigin FF, Bokulich NA, Yamanishi S, Teitler I, Chung J, Sohn J, Barber CM, Goldfarb DS, Raju K, Abubucker S, Zhou Y, Ruiz VE, Li H, Mitreva M, Alekseyenko AV, Weinstock GM, Sodergren E, Blaser MJ. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat Commun. 2015;6:7486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 252] [Cited by in RCA: 286] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 27. | Van Hul M, Geurts L, Plovier H, Druart C, Everard A, Ståhlman M, Rhimi M, Chira K, Teissedre PL, Delzenne NM, Maguin E, Guilbot A, Brochot A, Gérard P, Bäckhed F, Cani PD. Reduced obesity, diabetes, and steatosis upon cinnamon and grape pomace are associated with changes in gut microbiota and markers of gut barrier. Am J Physiol Endocrinol Metab. 2018;314:E334-E352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 116] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 28. | He B, Li T, Wang W, Gao H, Bai Y, Zhang S, Zang J, Li D, Wang J. Metabolic characteristics and nutrient utilization in high-feed-efficiency pigs selected using different feed conversion ratio models. Sci China Life Sci. 2019;62:959-970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 29. | Tan Z, Yang T, Wang Y, Xing K, Zhang F, Zhao X, Ao H, Chen S, Liu J, Wang C. Metagenomic Analysis of Cecal Microbiome Identified Microbiota and Functional Capacities Associated with Feed Efficiency in Landrace Finishing Pigs. Front Microbiol. 2017;8:1546. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 30. | Cui X, Chen J, Yang Y. Administration of selenomethionine in combination with serine benefits diabetes via gut microbiota. Front Microbiol. 2022;13:1007814. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 31. | Yue S, Shan B, Peng C, Tan C, Wang Q, Gong J. Theabrownin-targeted regulation of intestinal microorganisms to improve glucose and lipid metabolism in Goto-Kakizaki rats. Food Funct. 2022;13:1921-1940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 30] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 32. | Goldstein EJ, Tyrrell KL, Citron DM. Lactobacillus species: taxonomic complexity and controversial susceptibilities. Clin Infect Dis. 2015;60 Suppl 2:S98-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 224] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 33. | Sato J, Kanazawa A, Azuma K, Ikeda F, Goto H, Komiya K, Kanno R, Tamura Y, Asahara T, Takahashi T, Nomoto K, Yamashiro Y, Watada H. Probiotic reduces bacterial translocation in type 2 diabetes mellitus: A randomised controlled study. Sci Rep. 2017;7:12115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 99] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 34. | Guo Q, Niu W, Li X, Guo H, Zhang N, Wang X, Wu L. Study on Hypoglycemic Effect of the Drug Pair of Astragalus Radix and Dioscoreae Rhizoma in T2DM Rats by Network Pharmacology and Metabonomics. Molecules. 2019;24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 35. | Ma Q, Li Y, Wang M, Tang Z, Wang T, Liu C, Wang C, Zhao B. Progress in Metabonomics of Type 2 Diabetes Mellitus. Molecules. 2018;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 36. | Sun L, Pang Y, Wang X, Wu Q, Liu H, Liu B, Liu G, Ye M, Kong W, Jiang C. Ablation of gut microbiota alleviates obesity-induced hepatic steatosis and glucose intolerance by modulating bile acid metabolism in hamsters. Acta Pharm Sin B. 2019;9:702-710. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 125] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 37. | Mantovani A, Dalbeni A, Peserico D, Cattazzo F, Bevilacqua M, Salvagno GL, Lippi G, Targher G, Danese E, Fava C. Plasma Bile Acid Profile in Patients with and without Type 2 Diabetes. Metabolites. 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 42] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 38. | Osuna-Prieto FJ, Rubio-Lopez J, Di X, Yang W, Kohler I, Rensen PCN, Ruiz JR, Martinez-Tellez B. Plasma Levels of Bile Acids Are Related to Cardiometabolic Risk Factors in Young Adults. J Clin Endocrinol Metab. 2022;107:715-723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 39. | Lu J, Wang S, Li M, Gao Z, Xu Y, Zhao X, Hu C, Zhang Y, Liu R, Hu R, Shi L, Zheng R, Du R, Su Q, Wang J, Chen Y, Yu X, Yan L, Wang T, Zhao Z, Wang X, Li Q, Qin G, Wan Q, Chen G, Xu M, Dai M, Zhang D, Tang X, Wang G, Shen F, Luo Z, Qin Y, Chen L, Huo Y, Ye Z, Liu C, Wang Y, Wu S, Yang T, Deng H, Li D, Lai S, Mu Y, Zhao J, Xu G, Ning G, Bi Y, Wang W; 4C Study Group. Association of Serum Bile Acids Profile and Pathway Dysregulation With the Risk of Developing Diabetes Among Normoglycemic Chinese Adults: Findings From the 4C Study. Diabetes Care. 2021;44:499-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 54] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 40. | Katsiki N, Papanas N, Fonseca VA, Maltezos E, Mikhailidis DP. Uric acid and diabetes: Is there a link? Curr Pharm Des. 2013;19:4930-4937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 41. | Harrison SA, Gunn N, Neff GW, Kohli A, Liu L, Flyer A, Goldkind L, Di Bisceglie AM. A phase 2, proof of concept, randomised controlled trial of berberine ursodeoxycholate in patients with presumed non-alcoholic steatohepatitis and type 2 diabetes. Nat Commun. 2021;12:5503. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 69] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 42. | Shima KR, Ota T, Kato KI, Takeshita Y, Misu H, Kaneko S, Takamura T. Ursodeoxycholic acid potentiates dipeptidyl peptidase-4 inhibitor sitagliptin by enhancing glucagon-like peptide-1 secretion in patients with type 2 diabetes and chronic liver disease: a pilot randomized controlled and add-on study. BMJ Open Diabetes Res Care. 2018;6:e000469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 43. | Sorensen LB. Role of the intestinal tract in the elimination of uric acid. Arthritis Rheum. 1965;8:694-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 156] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 44. | Ichida K, Matsuo H, Takada T, Nakayama A, Murakami K, Shimizu T, Yamanashi Y, Kasuga H, Nakashima H, Nakamura T, Takada Y, Kawamura Y, Inoue H, Okada C, Utsumi Y, Ikebuchi Y, Ito K, Nakamura M, Shinohara Y, Hosoyamada M, Sakurai Y, Shinomiya N, Hosoya T, Suzuki H. Decreased extra-renal urate excretion is a common cause of hyperuricemia. Nat Commun. 2012;3:764. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 468] [Cited by in RCA: 488] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 45. | Lenzen S. The mechanisms of alloxan- and streptozotocin-induced diabetes. Diabetologia. 2008;51:216-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1173] [Cited by in RCA: 1222] [Article Influence: 71.9] [Reference Citation Analysis (0)] |
| 46. | Zyśk M, Pikul P, Kowalski R, Lewandowski K, Sakowicz-Burkiewicz M, Pawełczyk T. Neither Excessive Nitric Oxide Accumulation nor Acute Hyperglycemia Affects the N-Acetylaspartate Network in Wistar Rat Brain Cells. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 47. | Mar Rodríguez M, Pérez D, Javier Chaves F, Esteve E, Marin-Garcia P, Xifra G, Vendrell J, Jové M, Pamplona R, Ricart W, Portero-Otin M, Chacón MR, Fernández Real JM. Obesity changes the human gut mycobiome. Sci Rep. 2015;5:14600. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 194] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 48. | Lai M, Liu Y, Ronnett GV, Wu A, Cox BJ, Dai FF, Röst HL, Gunderson EP, Wheeler MB. Amino acid and lipid metabolism in post-gestational diabetes and progression to type 2 diabetes: A metabolic profiling study. PLoS Med. 2020;17:e1003112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 49. | Cuevas-Sierra A, Romo-Hualde A, Aranaz P, Goni L, Cuervo M, Martínez JA, Milagro FI, Riezu-Boj JI. Diet- and sex-related changes of gut microbiota composition and functional profiles after 4 months of weight loss intervention. Eur J Nutr. 2021;60:3279-3301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 50. | Górska M, Dobrzyń A, Baranowski M. Concentrations of sphingosine and sphinganine in plasma of patients with type 2 diabetes. Med Sci Monit. 2005;11:CR35-CR38. [PubMed] |
| 51. | Sun T, Zhang B, Ru QJ, Chen XM, Lv BD. Tocopheryl quinone improves non-alcoholic steatohepatitis (NASH) associated dysmetabolism of glucose and lipids by upregulating the expression of glucagon-like peptide 1 (GLP-1) via restoring the balance of intestinal flora in rats. Pharm Biol. 2021;59:723-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 52. | Henderson G, Crofts C, Schofield G. Linoleic acid and diabetes prevention. Lancet Diabetes Endocrinol. 2018;6:12-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 53. | Spratlen MJ, Grau-Perez M, Umans JG, Yracheta J, Best LG, Francesconi K, Goessler W, Balakrishnan P, Cole SA, Gamble MV, Howard BV, Navas-Acien A. Arsenic, one carbon metabolism and diabetes-related outcomes in the Strong Heart Family Study. Environ Int. 2018;121:728-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 54. | Zhang D, Li Y, Lang X, Zhang Y. Associations of Serum Vitamin B6 Status and Catabolism With All-Cause Mortality in Patients With T2DM. J Clin Endocrinol Metab. 2022;107:2822-2832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 55. | Gu X, Al Dubayee M, Alshahrani A, Masood A, Benabdelkamel H, Zahra M, Li L, Abdel Rahman AM, Aljada A. Distinctive Metabolomics Patterns Associated With Insulin Resistance and Type 2 Diabetes Mellitus. Front Mol Biosci. 2020;7:609806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 56. | Liu Z, Li P, Zhao ZH, Zhang Y, Ma ZM, Wang SX. Vitamin B6 Prevents Endothelial Dysfunction, Insulin Resistance, and Hepatic Lipid Accumulation in Apoe (-/-) Mice Fed with High-Fat Diet. J Diabetes Res. 2016;2016:1748065. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 57. | Zhang J, Li H, Zhou H, Fang L, Xu J, Yan H, Chen S, Song Q, Zhang Y, Xu A, Fang Q, Ye Y, Jia W. Lowered fasting chenodeoxycholic acid correlated with the decrease of fibroblast growth factor 19 in Chinese subjects with impaired fasting glucose. Sci Rep. 2017;7:6042. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 58. | Feng B, Banner C, Max SR. Effect of diabetes on glutamine synthetase expression in rat skeletal muscles. Am J Physiol. 1990;258:E762-E766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 59. | Murayama H, Ikemoto M, Hamaoki M. Serum ornithine carbamyltransferase reflects hepatic damage in diabetic obese mice. J Gastroenterol Hepatol. 2010;25:413-419. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 60. | Gloria-Bottini F, Magrini A, Antonacci E, La Torre M, Di Renzo L, De Lorenzo A, Bergamaschi A, Bottini E. Phosphoglucomutase genetic polymorphism and body mass. Am J Med Sci. 2007;334:421-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 61. | Goodwin BL, Pendleton LC, Levy MM, Solomonson LP, Eichler DC. Tumor necrosis factor-alpha reduces argininosuccinate synthase expression and nitric oxide production in aortic endothelial cells. Am J Physiol Heart Circ Physiol. 2007;293:H1115-H1121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 87] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 62. | Oyadomari S, Gotoh T, Aoyagi K, Araki E, Shichiri M, Mori M. Coinduction of endothelial nitric oxide synthase and arginine recycling enzymes in aorta of diabetic rats. Nitric Oxide. 2001;5:252-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 46] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 63. | Beardsley SJ, Kunjara S, Sochor M, Greenbaum AL. Enzymes of the de novo and salvage pathways of purine synthesis in renal hypertrophy. Contrasting effects of early diabetes and unilateral nephrectomy. Enzyme. 1988;40:231-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 64. | Yanagita I, Fujihara Y, Iwaya C, Kitajima Y, Tajima M, Honda M, Teruya Y, Asakawa H, Ito T, Eda T, Yamaguchi N, Kayashima Y, Yoshimoto M, Harada M, Yoshimoto S, Aida E, Yanase T, Nawata H, Muta K. Low serum albumin, aspartate aminotransferase, and body mass are risk factors for frailty in elderly people with diabetes-a cross-sectional study. BMC Geriatr. 2020;20:200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 65. | Gray LR, Tompkins SC, Taylor EB. Regulation of pyruvate metabolism and human disease. Cell Mol Life Sci. 2014;71:2577-2604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 386] [Cited by in RCA: 633] [Article Influence: 52.8] [Reference Citation Analysis (0)] |