Published online Oct 15, 2023. doi: 10.4239/wjd.v14.i10.1463
Peer-review started: June 29, 2023
First decision: July 18, 2023
Revised: August 1, 2023
Accepted: September 1, 2023
Article in press: September 1, 2023
Published online: October 15, 2023
Processing time: 101 Days and 23.5 Hours
There is a recent increase in the worldwide prevalence of both obesity and diabetes. In this review we assessed insulin signaling, genetics, environment, lipid metabolism dysfunction and mitochondria as the major determinants in diabetes and to identify the potential mechanism of gut microbiota in diabetes diseases. We searched relevant articles, which have key information from laboratory experiments, epidemiological evidence, clinical trials, experimental models, meta-analysis and review articles, in PubMed, MEDLINE, EMBASE, Google scholars and Cochrane Controlled Trial Database. We selected 144 full-length articles that met our inclusion and exclusion criteria for complete assessment. We have briefly discussed these associations, challenges, and the need for further research to manage and treat diabetes more efficiently. Diabetes involves the complex network of physiological dysfunction that can be attributed to insulin signaling, genetics, environment, obesity, mitochondria and stress. In recent years, there are intriguing findings regarding gut microbiome as the important regulator of diabetes. Valid approaches are necessary for speeding medical advances but we should find a solution sooner given the burden of the metabolic disorder ― What we need is a collaborative venture that may involve laboratories both in academia and industries for the scientific progress and its application for the diabetes control.
Core Tip: We have read through the references, gathered information and then summarized the literature focusing on the complex physiological networks that play important roles in diabetes. This review highlight that how impairment of insulin signaling, mitochondrial dysfunction can bring about changes in energy balance resulting in diabetes epidemic. We have covered studies from laboratory experiments, clinical trials, epidemiological, and several review articles making this review is a good reference point for further understanding and control of diabetes epidemic in human population.
- Citation: Wang H, Akbari-Alavijeh S, Parhar RS, Gaugler R, Hashmi S. Partners in diabetes epidemic: A global perspective. World J Diabetes 2023; 14(10): 1463-1477
- URL: https://www.wjgnet.com/1948-9358/full/v14/i10/1463.htm
- DOI: https://dx.doi.org/10.4239/wjd.v14.i10.1463
Diabetes mellitus is a widespread endocrine disorder. A dysfunctional carbohydrate, lipid, and protein metabolism lead to diabetes mellitus which is identified by prolong hyperglycemia, resulting from insufficient insulin secretion, insulin action or both. Prolonged hyperglycemia in partner with other metabolic abnormalities in patients with diabetes mellitus can cause a significant negative impact on many organs, leading to a life-threatening health problem, including retinopathy, nephropathy, and neuropathy and can also lead to an increased risk of cardiovascular diseases. Karamanou et al[1] gathered information from published research and review articles and presented a notable story of Diabetes mellitus in a review article in 2016.
Diabetes mellitus is largely classified into insulin dependent Type 1 Diabetes (T1D) and non-insulin-dependent, Type 2 Diabetes (T2D). In addition, there is also Gestational diabetes, a common medical complication that arises in women during pregnancy[2,3]. Several lines of evidence support the view that both genetic and the environmental risk factors act cooperatively in the pathogenesis of diabetes[4].
There is a recent increase in the worldwide prevalence of both obesity and diabetes [the International Diabetes Federation (IDF) Diabetes Atlas 9th edition 2019]. According to IDF report the diabetes prevalence in 2019 was 463 million people, will rise to 578 million by 2030 and 700 million by 2045. The global incidence of impaired glucose tolerance was around 374 million in 2019 and projected to reach 454 million by 2030 and 548 million by 2045. According to a World Health Organization (WHO) report, diabetes will become one of the most significant diseases or major diseases in the future[5]. A relatively recent WHO global reports (2016) stated that the number of diabetic adults ages between 40 and 59 escalated to 422 million in 2014. Although most countries are experiencing dramatic increase in diabetes, it appears to be more prevalent in middle- and low-income countries. Diabetes is not transmissible however risk factors including impaired glucose tolerance, insulin resistance, genetics, environment, and stress can cause the disease. Mitochondria are also important in many phases of diabetes disorder; however, their role in the pathophysiology of the disease is much dispersed involving both insulin sensitivity and secretion. The human microbiome including both the oral and gut microbiota are linked with diabetes and therefore, in recent years the world scientific communities and medical professionals are beginning to focus attention on the relationship between human microbiome and diabetes. The recent microbiome studies have linked gut microbiome to diabetes. For instance, Li et al[6] have assessed auspicious studies which allow a better understanding of the probable mechanism of microbiota in diabetes epidemic in 2020.
To protect the population from diabetes a number of approaches can be adopted by which it can be treated and its effects eluded with balanced diet, physical activity, and medication. Recent technological advancement has offered unique opportunities for the development of strategies to minimize or control the spread of diabetes. What we need is a collaborative venture that may involve laboratories both in academia and industries to understand the mechanism and control of diabetes. This review aims to assess literatures providing insights into factors associated with diabetes.
The initial search was performed in September 2019, the search was restricted to articles published in English focusing on the factors associated with diabetes. Then an updated search was performed in July 2023. In both searches we used the medical and biological databases (PubMed, MEDLINE, EMBASE, Google scholars and Cochrane Controlled Trial Database) using the search terms (diabetes, insulin resistance, insulin secretion, obesity, genetics-diabetes, environment-diabetes, mitochondria-diabetes and gut microbe-diabetes).
The authors of this review were consulted for the inclusion of appropriate articles. EndNote was used to manage references. We examined each article according to the following inclusion and exclusion criteria: The study: (1) Described the factors linked with diabetes; (2) be an original study; and (3) have key information from laboratory experiments, epidemiological evidence, clinical trials, experimental models, meta-analysis and review articles. We included a wide range of study designs used in laboratory studies, cross-sectional, prospective studies and clinical trials. The following studies were excluded: Irrelevant to our main objective and low-quality articles. All abstracts and full-text articles were assessed independently and in duplicate according to pre-defined inclusion/ exclusion criteria. Articles that met all criteria were selected for data extraction. In this review, we followed The Preferred Reporting Items for Systematic reviews and Meta-Analyses guidelines. The studies used in this review were published between 1981 and 2021.
We identified studies that discussed diabetes including T1D, T2D and gestational diabetes, and then the data were extracted by two authors who focused on first author name, year of publication, title of study, study design, study location and duration and the journals in which articles were published. The selected articles were discussed and then final decision was made for the inclusion in this systematic review. The quality of articles was assessed by three authors on the basis of relevance to the topic. Two authors independently evaluated the characteristics of the study population, as well as the quality of the methods, results and the discussion used in the selected studies.
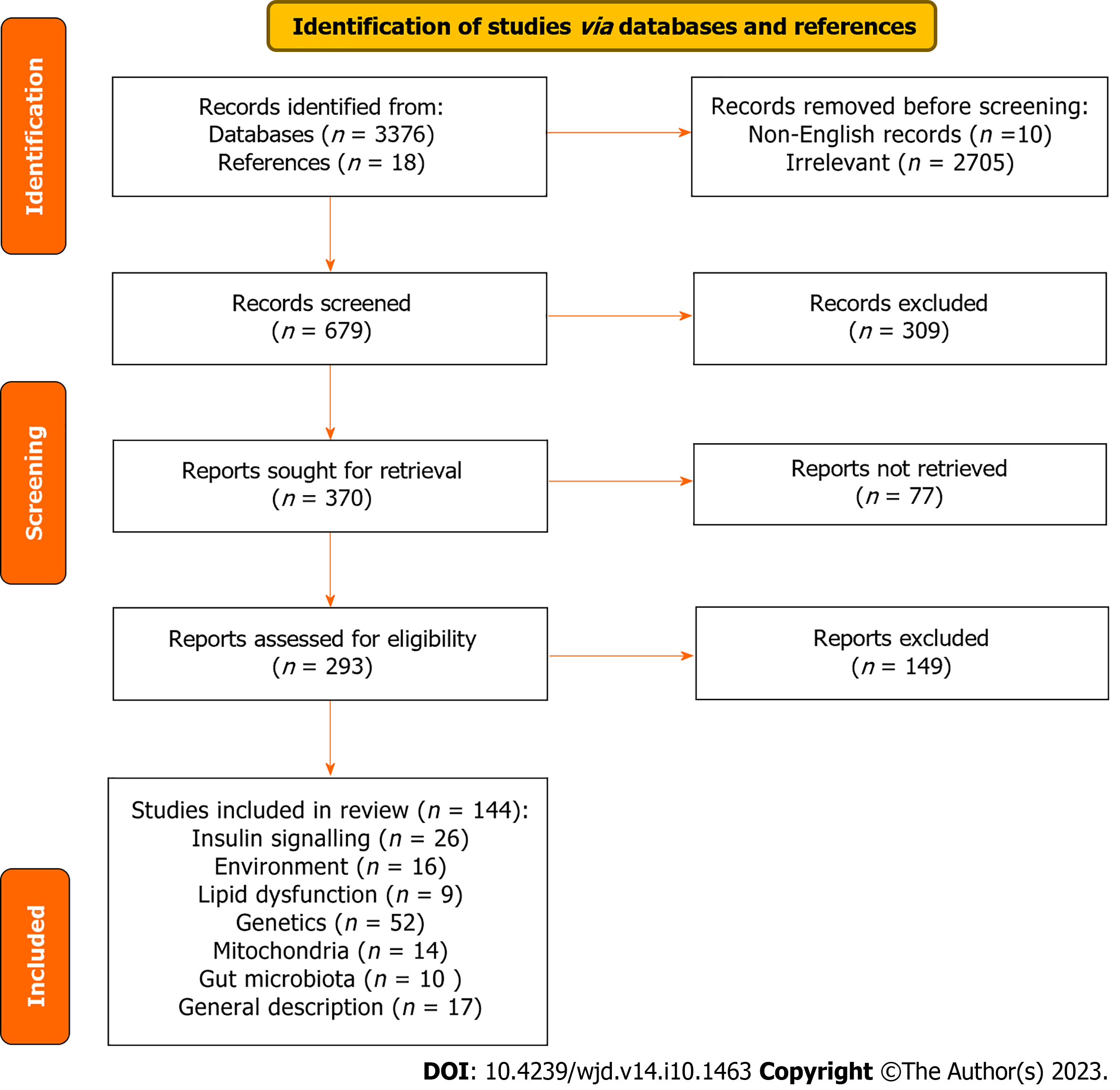
We recognized 3376 possibly pertinent papers in our initial search as well as 18 published articles from reference lists, assessed the title and abstracts of all 3376 articles and then selected 144 full-length articles that met our inclusion and exclusion criteria for complete assessment (Figure 1). Studies have shown a possible association of genetics, environment, mitochondria, obesity and insulin resistance with diabetes. As identified in 144 publications; 26 articles evaluated the relationship between diabetes and insulin signaling; 16 articles evaluated the link between diabetes and environmental factors; 9 articles linked diabetes with lipid dysfunction; 52 articles assessed the link between diabetes and genetics; 14 articles evaluated the relation between diabetes and mitochondria; 10 articles evaluated the relation between diabetes and gut microbiota; 17 articles presented general description of diabetes (Figure 1).
Diabetes can cause long-term damage to individuals suffering from this disease. It may cause impairment of heart, damages to kidneys. Adults with diabetes confers greater risk of cardiovascular complications including heart attack and strokes[7]. The elevated blood glucose levels can result in fat deposits in blood vessel walls, causing obstruction in blood flow and may increase the possibility of developing atherosclerosis. Diabetes complication can lead to diabetic retinopathy; an estimated 2.6% of blindness reported from around the world is related to diabetes[8]. Apparently, severe diabetic condition can also result in diabetic nephropathy[9]. T1D is the result of pancreatic beta cell damage, by autoimmune mechanisms which may lead to poor or no insulin production and hence the individuals need exogenous insulin to regulate blood glucose levels[10]. T2D results from the body’s resistance to insulin as well as inefficient secretion of insulin involving muscle, adipocytes, hepatocytes and may also involve the central nervous system. T2D is usually the result of excess body weight. Ordinarily, diabetes starts at or around the age of 40, but now there are reports of T2D in many children[11]. Gestational diabetes develops during pregnancy; it causes high blood glucose that can affect pregnancy and baby' s health[12].
There is a strong link between human microbiome and diabetes and therefore, the world scientific communities are beginning to focus attention on the relationship between human microbiome and diabetes. It is critical to understand that how microbes interact with the fundamental mechanisms of diabetes in humans and how much close is the relationship? We have discussed this topic in the preceding paragraphs.
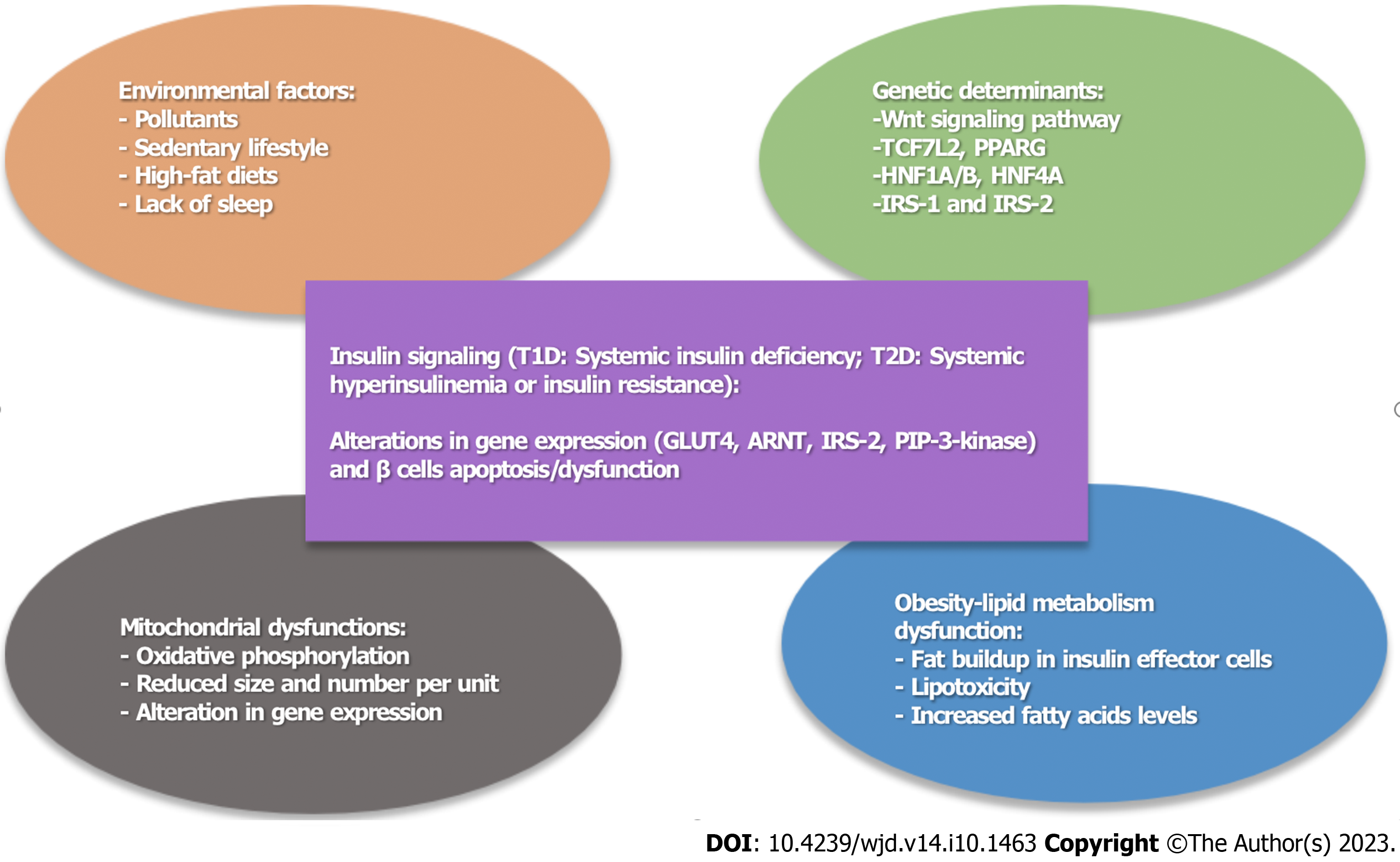
Most diabetes-related problems can be minimized by managing glucose, triglycerides and cholesterol levels within normal range. Although the molecular mechanisms of diabetes are not fully understood, it may result from defects in diverse molecular pathways or from genetic defects that cause both insulin resistance and insulin deficiency (Figure 2). The multidimensional interventions involving organizational changes including a change in the structure of health care system which is one step forward that can provide a positive effect on patient’s care. Such as develop strategies to improve treatment of diabetes by managing hyperglycemia and hyperlipidemia in patient as well as physician’s faithfulness to ensure a monitoring system.
Insulin is the key hormone frequently produced by pancreatic β cells regulates the fat storage from absorbed nutrients while acting as adiposity signal to the brain for regulation of energy balance[13], affecting skeletal muscle, liver, and adipose tissue. Insulin secretion from β-cells is fueled by high glucose levels, maintains the normal levels of blood glucose[14]. The presence of insulin receptors on muscle and adipose tissues allows insulin-dependent uptake of glucose into these tissues and thus lowers blood glucose levels by taking away the excess glucose from the blood[15-17]. A fall in blood glucose results in lowering insulin release from β-cells and augmenting glucagon release from α-cells, thereby stimulating the glycogen to glucose conversion.
A lack of insulin and hyperglycemia intensify insulin resistance and affects insulin secretion. In insulin resistance state, high insulin level creates a reduced biological response; weekend sensitivity to insulin mediated glucose removal[18]. Most diabetic patients are obese, which is believed to be an important causal factor in the development of insulin resistance. During the disease development there seems to be a gradual injury to beta cells and finally, the insulin resistant becomes evident in liver resulting in hyperglycemia. A high blood glucose levels that may arise due to dysfunctional insulin action and/or insulin secretion[19] is a prime factor causing diabetes (Table 1).
| Defect | Phenomenon | Ref. |
| β cells failure | Insulin resistance, hyperglycaemia | Kahn[20] |
| ↓ GLUT 4 translocation in adipocytes | ↓ IRS-1 expression, ↓ PIP-3-kinase and PKB/Akt activities | Wilcox[23], Smith[24] |
| ↑ Plasma free fatty acid | Insulin resistance, ↓ lipoprotein lipase activity, hypertriglyceridaemia | Reaven et al[26], Frayne[27] |
| Suppression of GLUT4 activity (muscle and liver) | Insulin dysfunction | Leroith et al[33] |
| Suppression of insulin receptor activity (LIRKO) (liver) | Hyperinsulinemia, hepatic and peripheral insulin resistance, glucose intolerance, insulin dysfunction | Michael et al[37] |
| Nuclear receptor translocator (ARNT) | ↓ Insulin secretion, alterations in gene expression | Gunton et al[47], Kewley et al[48] |
| Suppression of IRS-2 | β-cell apoptosis, insulin resistance | Rhodes[53] |
The progressive failure of β cells in making up for insulin resistance results in reduced glucose tolerance and diabetes[20] and with a rise in glucose levels and a further decline in β cell function leads to low glucose sensitivity. Although there is genetic predisposition to insulin resistance, physical inactivity, fatty foods, stress[21] and sleep deficiency[22] are other risk factors. The detailed mechanism of insulin resistance is not clear but the general belief is that insulin resistance begins in the adipose tissue playing a critical part in initiating insulin resistance in the muscles and the liver. Adipocytes from T2D patients have been reported to do poor GLUT 4 translocation, reduced insulin's intracellular signaling activities including low insulin receptor substrate (IRS)-1 expression, as well as reduced insulin-stimulated PIP-3-kinase and PKB/Akt activities[23,24]. Insulin stimulates glucose and free fatty acids uptake, suppresses lipolysis, and perhaps stimulates de novo fatty acid synthesis[25]. An elevated plasma free fatty acid is generally associated with insulin-resistant states, and T2D[26,27]. Perseghin et al[28] performed a cross-sectional study of young, normal-weight offspring of T2D patients and found an inverse relationship between fasting plasma fatty acid levels and insulin sensitivity, consistent with the premise that changes in fatty acid metabolism add to insulin resistance in T2D patients in 1997[29]. Later, Perseghin et al[30] studied 18 patients with T1D in 2003, 7 older and overweight/obese patients with T2D, and 15 nondiabetic, and insulin-resistant offspring of T2D parents. They reported an increased adiponectin levels in insulin-resistant patients with T1D, and a reduced levels in patients with T2D. The increased adiponectin levels in insulin-resistant patients with T1DM, in contrast to the reduced levels found in patients with T2DM showed an undefined relationship of adiponectin to insulin resistance in humans[30].
Acting as a neuropeptide, insulin also functions in satiety, appetite and olfaction[31]. Whereas angiotensinogen and leptin increase insulin resistance, adiponectin reduces insulin resistance suggesting that both leptin and insulin are possibly a part of a common signaling system in the hypothalamus.
It is generally believed that impairment of glucose transporter GLUT4 in adipose tissue is responsible for insulin resistant, obesity, and diabetes[32] and its key sites of expression are white and brown adipocytes, skeletal muscle, and cardiac muscle, but it is also present in some isolated areas of brain and kidney[33]. Study with tissue-specific targets have identified the specific insulin responsive organs to glucose homeostasis[34]. In 2003, Minokoshi et al[35] reported data from tissue-conditional knock-out mice showing that while suppression of muscle-specific insulin receptor activity did not affect glucose tolerance regardless of insulin resistance, the suppression of muscle specific GLUT4 activity caused insulin resistance and T2D. In addition, suppression of GLUT4 expression in white adipose showed insulin resistance, glucose intolerance, T2D and a deficiency in glucose uptake[35]. Surprisingly, knockout of insulin receptor in adipocyte did improve insulin sensitivity suggesting a fundamental role of adipocytes in diabetes. Many groups have shown that the suppression of insulin receptor activity in the liver resulted in hyperinsulinemia along with peripheral insulin resistance[34,36-38]. In 2003, Fisher and Kahn[36] performed high-dose hyperinsulinemia-euglycemic clamps using [3-(3) H]-glucose in liver-specific insulin receptor knockout (LIRKO) mice, and LIRKO mice treated with streptozotocin (STZ) (LIRKO + STZ) and found that in LIRKO mice, both direct and indirect effects of insulin required an intact insulin-signaling pathway in the liver, primary hepatic insulin resistance led to hyperinsulinemia and secondary extrahepatic insulin resistance.
Adipocyte-targeted GLUT4 knockout mice developed insulin resistance comparable to that shown by muscle-specific GLUT4 knockout mice would suggest that GLUT4 deficient adipocyte may release molecules involved in organ cross-talk[39,40]. Later, Yang et al[41] (2005) noted that retinol binding protein-4 (RBP4) could be involved in the organ cross-talk in the adipose tissue of adipose-specific Glut4 deficient mice. Not only, they found elevated serum RBP4 protein levels in insulin resistant mice but also found this protein in obese and diabetic individuals. Mice injected with recombinant RBP4 protein showed the sign of insulin resistance, whereas Rbp4 knockout mice increased insulin sensitivity[41]. Both visceral and peripheral adipocytes secrete multiple cytokines and hormone-like molecules such as adiponectin, leptin, cytokines interleukin-6 and tumor necrosis factor-α, visfatin, RBP4, and free fatty acids which may produce significant effect on insulin action and hepatic glucose production[42-44]. High fat deposits and increased levels of cytokine secretion give rise to inflammatory response that leads to insulin resistance[44-46].
While gene expression profiling of pancreatic islets obtained from T2D individuals, Gunton et al[47] (2005) observed major reduction in the expression of hepatocyte nuclear factor 4 alpha, insulin receptor, IRS-2, Akt2, and several glucose-metabolic-pathway genes. They also found a very high reduction in the transcription factor, aryl hydrocarbon nuclear receptor translocator (ARNT) in T2D islets compared with nondiabetic individuals. Basic helix-loop-helix Per/AhR/ARNT/Sim family ARNT and its partner proteins form heterodimers acting as transcription factors[48] and in association with other transcription factors show hypoxic stress response, may bring about the negative effects of both genetics and environment in T2D pathology[49], chronic hyperglycemia[50], hyperlipidemia[51] and oxidative stress[52]. Rhodes[53] (2005) argued that the failure of β-cell mass to compensate for insulin resistance is caused by a significant increase in β-cell apoptosis, stimulated by chronic hyperglycemia, hyperlipidemia, or specific cytokines affecting pathways responsible for maintaining healthy β-cell. Insulin receptor substrate, IRS-2 is fundamental and necessary for maintaining the adult β-cell in its normal state, and is a key factor in keeping the balance between β-cell and insulin resistance. The mechanisms pertinent to T2D pathogenies is possibly boost IRS-2 serine/threonine phosphorylation that leads to IRS-2 ubiquitination and proteasomal loss[54,55].
Consistent evidence has shown that the wide spread incidence of T2D is in part due to obesity, none or reduced physical activity and aging. However, many individuals exposed to these risk factors do not develop diabetes suggest that genetics may be involved in diabetes pathology. Obesity prompts T2D in individuals with susceptibility alleles in T2D associated genes acting at several points on the diabetes pathway[56].
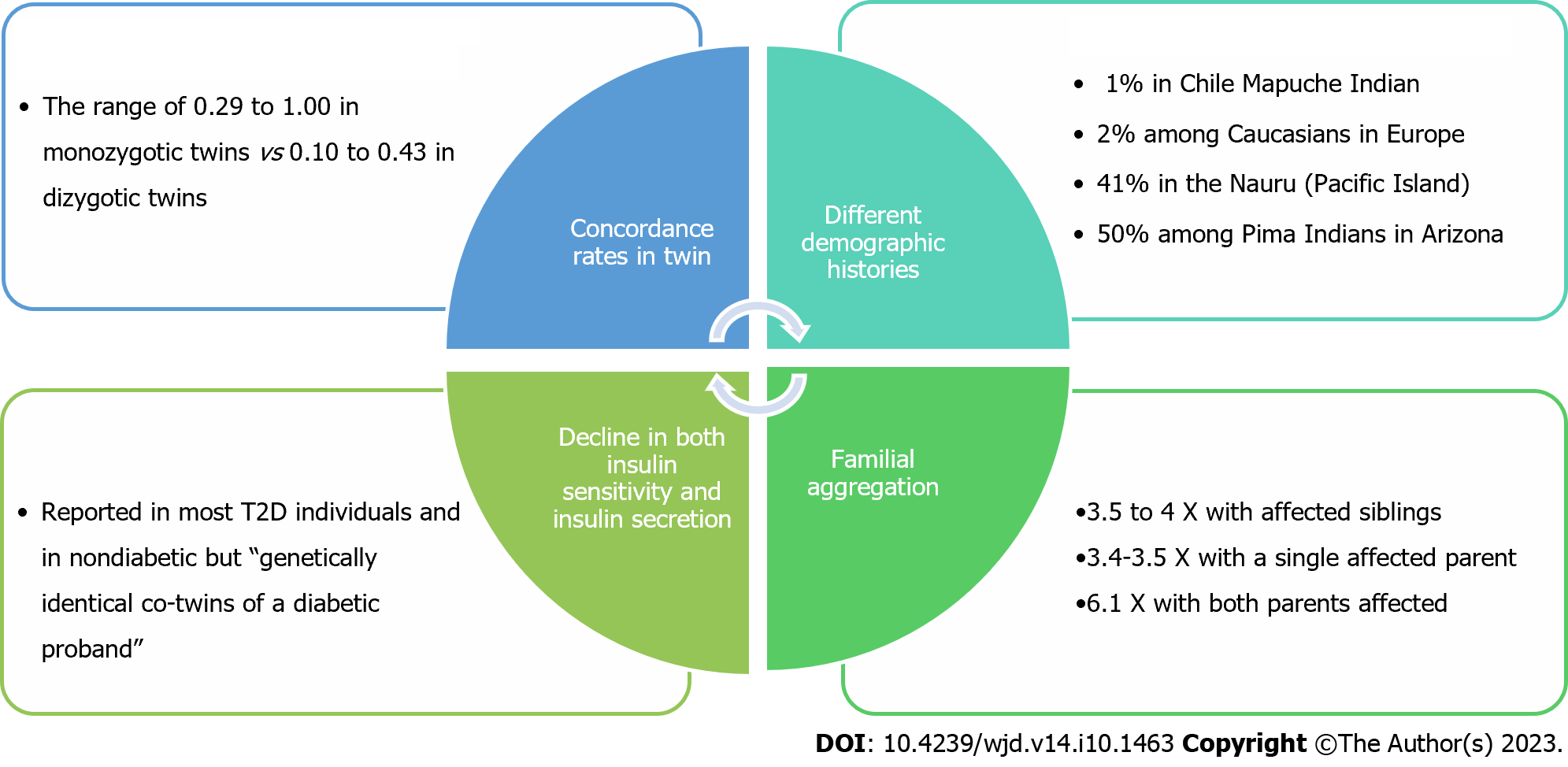
Das and Elbein[57] (2006) presented some visible scenario suggesting the role of genetics in diabetes pathology (Figure 3). First, incidence of T2D varies among populations with different demographic histories[58]. Second, approximately a 4X higher risk of T2D was found in siblings of a diabetic over the normal population with a single diabetic parent, and 6.1 when both parents were affected[59]. Third, in twin studies this rate has been found ranging 0.29 to 1.00 in monozygotic twins, and 0.10-0.43 in dizygotic twins[60-63] with a consistent decline in both insulin sensitivity and insulin secretion in most T2D individuals[64].
T1D and T2D are in part, genetically controlled[65], a key element is located within major histocompatibility complex (MHC) on chromosome 6p21 that add to the ancestral clustering of T1D[66]. The data from United States, United Kingdom and Scandinavian countries along with recent data from T1D Genetics Consortium (http://www.t1dgc.org), 1435 multiplex families suggested a link of T1D “to the MHC (IDDM1), insulin (INS, IDDM2)” region containing many genes including “CTLA4 (2q31-q33 [IDDM12 and IDDM7]) and seven other chromosome regions”[67]. The genome-wide studies have identified multiple T2D risk genes including TCF7L2, KCNQ1 and KCNJ11[68]. Ali[68] (2013) has presented possible explanations for missing heritability including the role of rare variants, gene-environment interactions and epigenetics. The susceptibility variants within CAPN10 gene[69] has been identified because of an association between T2D and chromosome 2q37 in Mexican Americans[70]. Peroxisome proliferator-activated receptor-γ[71] is common variants but variants affecting IRS-1 pathway[72] and glucose homeostasis PTPN1[73] are not very common indicating that mixtures of sporadic and conjoint modification may enhance T2D risk in diverse communities. Applying a linkage analysis, Hanis et al[70] (1996) identified CAPN10 cysteine protease linked to T2D but as shown in a meta-analysis’ variations in CAPN10 is likely[74] but not always linked to T2D[75].
The parents, siblings and children of T2D individuals have 3X greater chances to acquire diabetes than those who do not have a T2D family history[73]. The ancestral risk is greater in parents in the range of 35-60 years of age suggesting that environmental factors play a role in older population[76]. However, epigenetic factors can also yield congenital risk for subsequent generations. The genetic risk factor for T1D is very much intense in human leucocyte antigen region but this risk is not concerted in single region for T2D. It is because of possible interaction of many genes that are dispersed throughout the genome. A number of single-nucleotide polymorphisms in the transcription factor TCF7L2 and a member of Wnt signaling pathway has been linked to T2D in many ethnic groups[77]. TCF7L2 is known to function in beta cells, was identified through a linkage signal on chromosome 10q in a Mexican-American population[78]. Later, the region was identified in the population of other three countries including the United States[79]. TCF7L2 was also identified in a large-scale genome-wide association study that was performed in a French population[80]. Studies conducted in multiple ethnic groups indicated that the risk allele in intron 3 of the TCF7L2 gene increased the level of its protein in beta cells, impaired insulin secretion, and elevate hepatic glucose production[81]. Ali[68] (2013) lists PPARG, IRS-1 and IRS-2, potassium inwardly-rectifying channel, subfamily member 11 (KCNJ11), Wolfram syndrome 1 (wolframin-WFS1), HNF1 homeobox A, HNF1 homeobox B and HNF4A that are associated with T2D[68]. IRS-1 and IRS-2 the two-insulin receptor substrate play an important role in insulin signal transduction. Polymorphisms in both irs-1 and irs-2 results in reduce insulin sensitivity in some populations[82,83].
While genotyping 2000 T2D individuals, Wellcome Trust Case Control Consortium[84] has identified TCF7L2 as the most robust T2D signal but mutation in TCF7L2 shows no effect in beta cells[85]. In their meta-analysis, Fu et al[86] (2013) pooled 24 articles involving 88229 cases and 210239 controls and identified -30G>A polymorphism of glucokinase as a risk factor associated with increased T2D susceptibility, however, those associations vary in different ethnic populations.
Although genome studies, twin studies and linkage analysis have identified few T2D risk genes, their global impact on the perceived heritability of T2D remained low[87]. Phenotypes may depend on the nature of genetic variation within and across different ethnic group. We believe that T2D develop as a result of interaction between environmental factors and hereditary factors.
A series of epidemiological and clinical papers have shown serious effects of behavioral and environmental changes on the occurrence of diabetes. The changes in the environment ranges from endocrine disruption, sleep deprivation, physical inactivity, over eating and pollutants[88,89]. In addition, the sedentary lifestyles and the high-fat diets are interactive factors that are associated with high incidence of T2D (Figure 2). There are some excellent publications on these topics[88,90,91]. Numerous environmental factors including refined carbohydrates, stress, and exposure to chemical pollutants produce gradual weight gains increasing the risk of T2D, heart diseases and some cancers[92] but the relative contributions of these factors influencing T2D are not fully understood. Environment does play a critical role in diabetes development; it does not affect all individuals in a similar manner. Even living under the same environment some individuals are more vulnerable to diabetes risk because of some inherited factors suggesting that T2D occurs because of intense interactions between many genes and the environment[93]. Cells use several mechanisms in regulating gene expression in response to environmental cues not only remain in individual’s lifespan but can also pass on to few generations[94]. Changes in maternal environment in early childhood have been implicated in long-lasting diseases[95]. This may also explain that some heritability of T2D can occur because of epigenetic changes that happen in intra-uterine which may be influenced by maternal environment. As our knowledge of the epigenetics changes and the detailed mechanisms of epigenetic become widely available, we may be able to understand clearly the effect of these changes on diabetes pathology. The molecular basis of genetic risk factors in T2D is not yet clear and it is certainly an area of intensive investigation.
The gene mutations in mitochondrial DNA also cause mitochondrial diabetes[96]. The role of mitochondria in the pathophysiology of diabetes is very imprecise involving both insulin sensitivity and secretion (Table 2). Our knowledge about the connection between mitochondrial dysfunction and defective insulin sensitivity and secretion is, however, sketchy[97,98]. According to Lu et al[97] (2010) both mitochondrial oxidative phosphorylation dysfunction and its morphology play an essential part in the pathology of insulin resistance-induced β-cell failure. As a result of oxidation, mitochondria create large amount of reactive oxygen species which are important in the pathophysiology of diabetes and its complications. Both clinical and rodent data demonstrate decreased oxidative phosphorylation in muscle mitochondria in insulin-resistant states. Kelley et al[99] (2002) investigated mitochondria obtained from vastus lateralis muscle by percutaneous biopsy during fasting from T2D, obese, and lean individuals to examine the effect of perturbation of mitochondrial function. They noted a reduction in both nicotinamide adenine dinucleotide oxidoreductase and citrate synthase activity in their mitochondria. They also found mitochondria of smaller size and numbers per unit volume compared with those in lean controls[100]. A reduced skeletal muscle oxidative phosphorylation was also noticed in insulin-resistant offspring of T2D individuals linked to elevated levels of fat droplets in muscle cells[101]. Petersen et al[101] (2003) investigated healthy, lean, elderly and young volunteers corresponded for lean body mass and fat mass and noted that elderly or aged individuals were insulin-resistant compared to young controls and the changes were linked to increased fat deposits in muscle and liver tissue and an approximately 40% reduction in mitochondrial oxidative and phosphorylation activity suggesting age-linked deterioration in mitochondrial function add to insulin resistance in the older population. Boushel et al[102] (2007) conducted individual based study where they investigated mitochondrial function in skeletal muscle obtained from 11 individuals with T2D and found reduced oxygen use in diabetic patients which could be linked to reduced mitochondrial content in muscles. The mitochondrial dysfunction and/or reduced mitochondria can cause insulin resistance. Dysfunctional mitochondria can result in reduced oxidation leading to increased fatty acyl-CoA, diacylglycerol and activates protein kinase C[103,104].
| Defect | Consequence | Ref. |
| ↑ Mitochondrial oxidative phosphorylation in pancreatic β-cells | ↑ROS, β-cell failure | American Diabetes Association[96], Marroqui et al[98] |
| ↓ Mitochondrial oxidative phosphorylation in skeletal muscle | ↓ NADH oxidoreductase activity, ↓ citrate synthase activity, ↑ fat droplets in muscle cells, insulin resistance | Petersen et al[100], Petersen et al[101], Zhang et al[106] |
| ↓ Size and number of mitochondria per unit | ↑ Fatty acyl-CoA and diacylglycerol, ↑ protein kinase C activities, insulin resistance | Abdul-Ghani and DeFronzo[103], Lowell and Shulman[104] |
| ↓ Expression of NRF-dependent genes | Insulin resistance | Lu et al[97], Patti et al[105] |
| UCP2 deficiency in β-cells | ↑ Islet ATP levels, ↑ glucose-stimulated insulin secretion, obesity, β-cell failure | Zhang et al[106] |
Patti et al[105] (2003) have demonstrated that insulin resistance and T2D link with low level of various nuclear respiratory factor-1 (NRF-1)-dependent genes encoding key enzymes in mitochondrial function. The authors noted low levels of proliferator-activated receptor gamma coactivator (PGC)-1α and PGC-1β, coactivators of NRF-1 and PPARγ-dependent transcription involved in oxidative phosphorylation in both diabetic subjects and family history-positive nondiabetic subjects. Their conclusion was that the low PGC1 expression led to reduced NRF-dependent genes expression, thereby metabolic instabilities known for insulin resistance. Liver and skeletal muscle are involved in fatty acids oxidation but their failure to efficiently oxidize fatty acids leads to insulin resistance.
Mitochondria are known to play a key role in regulating insulin secretion. Beta cells detect glucose amid its metabolism and then subsequent increase in adenosine triphosphate (ATP) promotes insulin secretion. Zhang et al[106] (2001) found that uncoupling protein 2 (UCP2)-deficient mice had higher islet ATP levels and increased glucose-stimulated insulin secretion, suggesting that UCP2 negatively regulates insulin secretion. The UCP2 deficient ob/ob mice had restored first-phase insulin secretion, elevated serum insulin levels, and reduced levels of glycaemia suggesting UCP2 as a key component of beta cell glucose sensing, and as a vital link between obesity, beta cell failure, and T2D[106]. Bugger et al[107] (2008) suggested that mechanisms for mitochondrial dysfunction differ between insulin-deficient type 1 and insulin-resistant T2D hearts. Sivitz and Yorek[108] (2010) observed liver mitochondria of the STZ-diabetic rats and noted a significant tendency of reduced respiration. Karakelides et al[109] (2007) found that depriving diabetes patients from insulin did reduce muscle mitochondrial ATP production and expression of oxidative phosphorylation genes in T1D patients despite an increase in whole-body oxygen consumption. Although there are inconsistencies in the result outcome, most studies appear to imply that respiration and/or ATP production in muscle and heart mitochondria are lower when insulin level is low at least in isolated mitochondria. Friederich et al[110] (2008) in their decade old immune-histochemical studies on isolated mitochondria from kidneys showed an elevated proximal tubular UCP2 expression in STZ diabetic rats resulting in mitochondrial uncoupling and increased O2 consumption. The successive low O2 presence may add to diabetes-induced continuing kidney damage. However, other study exhibiting elevated mitochondrial membrane potential in mitochondria of STZ diabetic rat kidney[111].
Lipids play an important function in the pathogenesis of diabetes but the mechanistic links between lipids and diabetes is not very clear. Lipids metabolism involves a large number of enzymes catalyzed metabolic reactions engaging brain, adipose tissue, muscles, liver, and gut and its dysfunction results in fat build up that may also lead to diabetes. These organs are part of complex homeostatic system, communicating through hormones, neurons and metabolites. Just a small shift in the regulation of lipid metabolism can lead to a large change in energy homeostasis; it can result in diabetes, obesity, atherosclerosis, and accelerated aging. In its full-blown state T2D manifests two hallmarks in clinical patients, insulin resistance and β-cell failure. Fat buildup in insulin effector cells (liver cells, muscle cells, and adipocytes) can decrease their sensitivity to insulin and ultimately lead to insulin resistance. Conversely, fat buildup in non-adipose tissues may promote lipotoxicity and this toxicity can diminish or impair β-cell function to disrupt insulin supply by affecting insulin biosynthesis, processing, and secretion[112]. Abnormal fat buildup may also trigger inflammatory response, which in turn impairs both effector and source cells of insulin. High-fat diet negatively affect insulin resistance may result fatty acids overloads in mitochondria. Sparks et al[113] (2005) found that high-fat diets downregulated genes linked to oxidative phosphorylation and mitochondrial biogenesis, and those changes were interpreted as seen in diabetes. The adipose mitochondrial dysfunction causes increase in fatty acids levels which in turn can contribute to the insulin resistance. Wolfrum et al[114] (2004) noted that inactivating Foxa2 transcription factor in insulin-resistant mice led to lipid deposits in the liver, and promoted fat as well as glucose export. The adenoviral expression of Foxa2T156A, a nuclear, constitutively active Foxa2 in insulin resistant mice reduced hepatic triglyceride content, increased hepatic insulin sensitivity, reduced glucose production, and reduced plasma insulin. An et al[115] (2004) found that rats fed with high fat diets, the degradation of malonyl CoA in liver did encourage fat oxidation and decreased circulating free fatty acids, increased insulin sensitivity in both muscle and liver. Mice deficient in acetyl-CoA carboxylase 2 showed reduce malonyl-CoA, improve fatty acid oxidation but withstood diet-induced obesity and diabetes (Table 3)[116].
| Defect | Consequence | Ref. |
| Fat buildup in insulin effector cells (liver cells, muscle cells, and adipocytes) | Insulin resistance, lipotoxicity, β-cell disruption | Muoio and Newgard [112] |
| Adipose mitochondrial dysfunction | ↑ Fatty acids levels, insulin resistance | Sparks et al[113] |
| Inactivated Foxa2 | Lipid deposits in the liver, insulin resistance | Wolfrum et al[114] |
| ↓ Malonyl Co-A in liver | ↑ Fat oxidation, ↓ circulating free fatty acids, ↑ insulin sensitivity in both muscle and liver | An et al[115] |
What about the presence of gut microbes and their possible mechanism in diabetes? The findings from recent microbiome studies have indicated significant association of gut microbiome with diabetes. While the clinical significance of gut microbes in diabetes can be measured the many variables related to these microbes remains to be fully understood. In a fascinating review, Li et al[6] (2020) have assessed auspicious studies which allow a better understanding of the probable mechanism of microbiota in diabetes epidemic. The human microbiome including both the oral and gut microbiota are linked with diabetes and therefore, in recent years the world scientific communities and medical professionals are beginning to focus attention on the relationship between human microbiome and diabetes. It is critical to understand that how microbes interact with the fundamental mechanisms of diabetes in humans and how much close is the relationship? The human gut is a complex network involving microbiome, host cells and nutrients[117]. Diet induced-obesity promotes insulin resistance by mechanisms involving self-regulation and dependent on gut microbiota. Saad et al[118] (2016) have deliberated that the lipopolysaccharide from gut bacteria can prompt a chronic inflammatory process, inducing insulin resistance via TLR4 activation. Han and Lin[119] suggest that gut microbiota can impact on body weight, bile-acid metabolism, proinflammatory activity and insulin resistance. A defect in short-chain fatty acids synthesis is a common feature across studies that suggest a relationship between gut microbiota with T1D[120]. Both T1D and T2D are linked with multifaceted immune system and gut microbiome interactions. Thus, gut microbiota disarrays can lead to T1D, which is allied to the interaction between gut microbiota and the innate immunity. Hänninen and colleague profiled intestinal microbiota investigated the incidence of T1D between two non-obese diabetic mouse groups with different gut microbiota. They found that a single symbiont, Akkermansia muciniphila with favorable metabolic and immune signaling may be able to minimize diabetes incidence when given as a probiotic[121]. In many studies Akkermansia muciniphila has been reported to reduce insulin resistance and also reduces damage of the intestinal wall[6]. The oral cavity and gut are the two uninterrupted regions linked via gastrointestinal tract serving as microbial environments, have a key function in microbiome-linked diseases. The oral and gut microbiome stay apart because of the presence of oral–gut obstacle. Yet, the transmission of the oral microbiota can take place to the intestinal mucosa in the event the oral–gut wall does not function properly. The oral and gut microbiomes have been found interdependently regulating human physiological functions and disease pathology[122]. The intestinal colonization of oral microbiota and fecal–oral transmission occurs regularly, which can affect the microbial ecosystem in both habitats, to modulate pathophysiology[123-125]. Research on gut microbes seems reasonably important because it could provide valuable insights for evaluating gut microbiome for the diagnosis and treatment of diabetes. In addition, it certainly ensures the future discovery of the microbiota-related underlying mechanisms of diabetes.
A key issue for diabetes research is to develop a eukaryotic preclinical model, such as Caenorhabditis elegans that enables researchers to understand mechanistic insight into the biology and genetics of diabetes.
We are using a Caenorhabditis elegans (C. elegans) pre-clinical model to study Krüppel-like transcription factor (KLF) for their functional roles in obesity and diabetics. C. elegans encodes 3 members: klf-1[126], klf-2[127] and klf-3 all contain three highly conserved C-terminal C2H2 zinc fingers. KLF extensively expressed throughout larval development and during adulthood with a predominant expression in intestine, a major endocrine system positioned close to sexual organs and engaged in nutrient sensing and energy metabolism[128-130]. Mutation or RNA interference in klf-1, klf-2, or klf-3 leads to excess deposit of large fat droplets in the intestine of the mutant worm. Our detailed study on klf-3 mutant (ok1975) suggest that mutation in klf-3 also dysregulate insulin signaling. Most likely, the excessive fat buildup and defects in insulin signaling associated with mutation in klf-3 result from damage that gradually takes place during development. We have also shown that KLF-3 is an important regulator of fatty acid biosynthesis, lipid absorption and secretion, mitochondrial proliferation, β-oxidation and physically interacts with genes essential in lipid metabolism[131-133]. These regulatory functions of KLF-3 provide an important lead aimed at studying the mechanism of human diabetes.
The worm KLFs share the highest identity with members of several mammalian KLFs, including KLF-2, 3, 4, 5 and 6 in terms of their C-terminal C2H2 zinc fingers. Some members of mammalian KLFs (KLF 2-7 and 15) have been recently identified as one of the major transcription factors controlling adipogenesis, lipogenesis, obesity and diabetes[134-137]. The insulin signaling pathway that controls aging and metabolism was built on experiments in C. elegans and the identification of genes underlying daf phenotypes[138]. Several KLFs have been implicated in lipogenesis via their residence and action in adipose tissue and non-adipose tissues (pancreas, liver or muscle): They regulate adipocyte differentiation[139-141] or promote lipogenesis[142,143] or tune glucose and lipid homeostasis[134,135]. However, no direct evidence is shown that the KLF circuit intersects the insulin system. Small et al[144] (2011) have shown that the maternally expressed KLF14 which is associated with T2D and the cis-acting expression quantitative trait locus of high-density lipoprotein act as a master trans-regulator of adipose gene expression. Thus, klf14 acts as a major regulator of events in fat tissue, with these alterations in the levels of klf14 leading, through as yet unspecified mechanisms, to peripheral insulin resistance and T2D. Despite these advances a direct role for these KLFs in fat buildup and insulin resistance at an organism level remains to be established.
Due to the heterogeneity of the studies cited, the duration of the study and the complexity of interactions of the molecules, genetic factors, environmental factors and gut microbes limited the ability to make direct comparisons between diabetes epidemic, and genetic, biological, environmental factors and the gut microbiota. However, we acknowledge the risk of developing diabetes or alleviating diabetes. We performed an arduous literature search, but we might have missed some studies published as an abstract. To sum up, bias is expected since articles published in languages other than English were not included in this review but we think this is the general limitations for many review articles published in English or vice versa.
Diabetes is a pressing health issue with a disturbing epidemic forecast, it could be attributed to diets, stress, absence of physical activities, insulin resistance, genetic and environmental factors. Diabetes involves the complex network of physiological dysfunction that can be attributed to insulin signaling, genetics, environment, obesity, and mitochondria. Although clinical severity of the disease can be measured the many variables’ interactions in the incidence of diabetes remain to be fully understood.
The simple reason is that more or less severe clinical involvement of a specific factor has been difficult to identify. The emerging picture of genetics continues to support the general conclusion that there are a large number of risk susceptibility genes, each of them with relatively small effect. Yet, we lack understanding that how genes interact with each other and with the known environmental and other influencers that predispose to develop diabetes. There are intriguing findings regarding gut microbiome as the important regulator of diabetes and it is now known that oral and gut microbiomes interdependently regulate physiological functions and disease pathology. Although there are many models available to study the physiology, biology and genetics of diabetes, the difference between cellular, and animal models and human’s biology restrain the applicability of these models in the mechanistic investigations and therapeutic intervention of diabetics. We are yet to be able to overcome the problems in the genetic manipulation to produce a reliable model to understand human diabetes. Although C. elegans enables us to understand the fundamental mechanistic insight into the physiology and genetics of diabetes it does not present a true picture of human diabetes. Our work in model system may not always translate to human disease problem. We still need to focus our basic research efforts toward methods that are more directly relevant to human physiology to understand the mechanism for diabetes treatment. In recent years. there are intriguing findings regarding gut microbiome as the important regulator of diabetes. Valid approaches are necessary for speeding medical advances but we should find a solution sooner given the burden of the metabolic disorder- What we need is a collaborative venture that may involve laboratories both in academia and industries for the scientific progress and its application for the diabetes control.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Endocrinology and metabolism
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Nawab M, India; Sun H, China S-Editor: Lin C L-Editor: A P-Editor: Chen YX
| 1. | Karamanou M, Protogerou A, Tsoucalas G, Androutsos G, Poulakou-Rebelakou E. Milestones in the history of diabetes mellitus: The main contributors. World J Diabetes. 2016;7:1-7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 98] [Cited by in RCA: 101] [Article Influence: 11.2] [Reference Citation Analysis (24)] |
| 2. | Picke AK, Campbell G, Napoli N, Hofbauer LC, Rauner M. Update on the impact of type 2 diabetes mellitus on bone metabolism and material properties. Endocr Connect. 2019;8:R55-R70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 97] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 3. | Carrillo-Larco RM, Barengo NC, Albitres-Flores L, Bernabe-Ortiz A. The risk of mortality among people with type 2 diabetes in Latin America: A systematic review and meta-analysis of population-based cohort studies. Diabetes Metab Res Rev. 2019;35:e3139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 4. | Tremblay J, Hamet P. Environmental and genetic contributions to diabetes. Metabolism. 2019;100S:153952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 67] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 5. | Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3:e442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6517] [Cited by in RCA: 6898] [Article Influence: 363.1] [Reference Citation Analysis (0)] |
| 6. | Li WZ, Stirling K, Yang JJ, Zhang L. Gut microbiota and diabetes: From correlation to causality and mechanism. World J Diabetes. 2020;11:293-308. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 64] [Cited by in RCA: 91] [Article Influence: 18.2] [Reference Citation Analysis (5)] |
| 7. | Emerging Risk Factors Collaboration, Sarwar N, Gao P, Seshasai SR, Gobin R, Kaptoge S, Di Angelantonio E, Ingelsson E, Lawlor DA, Selvin E, Stampfer M, Stehouwer CD, Lewington S, Pennells L, Thompson A, Sattar N, White IR, Ray KK, Danesh J. Diabetes mellitus, fasting blood glucose concentration, and risk of vascular disease: a collaborative meta-analysis of 102 prospective studies. Lancet. 2010;375:2215-2222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3857] [Cited by in RCA: 3460] [Article Influence: 230.7] [Reference Citation Analysis (0)] |
| 8. | Bourne RR, Stevens GA, White RA, Smith JL, Flaxman SR, Price H, Jonas JB, Keeffe J, Leasher J, Naidoo K, Pesudovs K, Resnikoff S, Taylor HR; Vision Loss Expert Group. Causes of vision loss worldwide, 1990-2010: a systematic analysis. Lancet Glob Health. 2013;1:e339-e349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 955] [Cited by in RCA: 1132] [Article Influence: 94.3] [Reference Citation Analysis (0)] |
| 9. | Erratum Regarding "US Renal Data System 2014 Annual Data Report: Epidemiology of Kidney Disease in the United States". Am J Kidney Dis. 2015;66:545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 10. | Genuth S, Alberti KG, Bennett P, Buse J, Defronzo R, Kahn R, Kitzmiller J, Knowler WC, Lebovitz H, Lernmark A, Nathan D, Palmer J, Rizza R, Saudek C, Shaw J, Steffes M, Stern M, Tuomilehto J, Zimmet P; Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. Follow-up report on the diagnosis of diabetes mellitus. Diabetes Care. 2003;26:3160-3167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2379] [Cited by in RCA: 2592] [Article Influence: 117.8] [Reference Citation Analysis (0)] |
| 11. | Pulgaron ER, Delamater AM. Obesity and type 2 diabetes in children: epidemiology and treatment. Curr Diab Rep. 2014;14:508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 270] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 12. | Szmuilowicz ED, Josefson JL, Metzger BE. Gestational Diabetes Mellitus. Endocrinol Metab Clin North Am. 2019;48:479-493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 200] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 13. | Benoit SC, Clegg DJ, Seeley RJ, Woods SC. Insulin and leptin as adiposity signals. Recent Prog Horm Res. 2004;59:267-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 178] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 14. | Komatsu M, Takei M, Ishii H, Sato Y. Glucose-stimulated insulin secretion: A newer perspective. J Diabetes Investig. 2013;4:511-516. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 172] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 15. | Khan AH, Pessin JE. Insulin regulation of glucose uptake: a complex interplay of intracellular signalling pathways. Diabetologia. 2002;45:1475-1483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 285] [Cited by in RCA: 276] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 16. | Kohn AD, Summers SA, Birnbaum MJ, Roth RA. Expression of a constitutively active Akt Ser/Thr kinase in 3T3-L1 adipocytes stimulates glucose uptake and glucose transporter 4 translocation. J Biol Chem. 1996;271:31372-31378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 943] [Cited by in RCA: 974] [Article Influence: 33.6] [Reference Citation Analysis (0)] |
| 17. | Bellou V, Belbasis L, Tzoulaki I, Evangelou E. Risk factors for type 2 diabetes mellitus: An exposure-wide umbrella review of meta-analyses. PLoS One. 2018;13:e0194127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 418] [Cited by in RCA: 421] [Article Influence: 60.1] [Reference Citation Analysis (0)] |
| 18. | Reaven G. The metabolic syndrome or the insulin resistance syndrome? Different names, different concepts, and different goals. Endocrinol Metab Clin North Am. 2004;33:283-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 321] [Cited by in RCA: 301] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 19. | Philp I, Ghosh U. Community care services: views of patients attending a geriatric day hospital. Health Bull (Edinb). 1992;50:296-301. [PubMed] |
| 20. | Kahn SE. The relative contributions of insulin resistance and beta-cell dysfunction to the pathophysiology of Type 2 diabetes. Diabetologia. 2003;46:3-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1276] [Cited by in RCA: 1369] [Article Influence: 62.2] [Reference Citation Analysis (0)] |
| 21. | Björntorp P. Do stress reactions cause abdominal obesity and comorbidities? Obes Rev. 2001;2:73-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 772] [Cited by in RCA: 743] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 22. | Bass J, Turek FW. Sleepless in America: a pathway to obesity and the metabolic syndrome? Arch Intern Med. 2005;165:15-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 60] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 23. | Wilcox G. Insulin and insulin resistance. Clin Biochem Rev. 2005;26:19-39. [PubMed] |
| 24. | Smith U. Impaired ('diabetic') insulin signaling and action occur in fat cells long before glucose intolerance--is insulin resistance initiated in the adipose tissue? Int J Obes Relat Metab Disord. 2002;26:897-904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 142] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 25. | Giorgino F, Laviola L, Eriksson JW. Regional differences of insulin action in adipose tissue: insights from in vivo and in vitro studies. Acta Physiol Scand. 2005;183:13-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 141] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 26. | Reaven GM, Hollenbeck C, Jeng CY, Wu MS, Chen YD. Measurement of plasma glucose, free fatty acid, lactate, and insulin for 24 h in patients with NIDDM. Diabetes. 1988;37:1020-1024. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 422] [Cited by in RCA: 462] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 27. | Frayne KN. Insulin resistance and lipid metabolism. Curr Opin Lipidol. 1993;4:197-204. [DOI] [Full Text] |
| 28. | Perseghin G, Ghosh S, Gerow K, Shulman GI. Metabolic defects in lean nondiabetic offspring of NIDDM parents: a cross-sectional study. Diabetes. 1997;46:1001-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 172] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 29. | Shulman GI. Cellular mechanisms of insulin resistance. J Clin Invest. 2000;106:171-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1893] [Cited by in RCA: 1870] [Article Influence: 74.8] [Reference Citation Analysis (0)] |
| 30. | Perseghin G, Lattuada G, Danna M, Sereni LP, Maffi P, De Cobelli F, Battezzati A, Secchi A, Del Maschio A, Luzi L. Insulin resistance, intramyocellular lipid content, and plasma adiponectin in patients with type 1 diabetes. Am J Physiol Endocrinol Metab. 2003;285:E1174-E1181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 130] [Article Influence: 5.9] [Reference Citation Analysis (1)] |
| 31. | Gerozissis K. Brain insulin and feeding: a bi-directional communication. Eur J Pharmacol. 2004;490:59-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 74] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 32. | Shepherd PR, Kahn BB. Glucose transporters and insulin action--implications for insulin resistance and diabetes mellitus. N Engl J Med. 1999;341:248-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 906] [Cited by in RCA: 868] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 33. | Leroith D, Taylor SI, Olefsky JM. Diabetes mellitus: a fundamental and clinical text. 3th ed. Philadelphia: Lippincott Williams Co., 2004: 627-641. |
| 34. | Mauvais-Jarvis F, Kulkarni RN, Kahn CR. Knockout models are useful tools to dissect the pathophysiology and genetics of insulin resistance. Clin Endocrinol (Oxf). 2002;57:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 57] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 35. | Minokoshi Y, Kahn CR, Kahn BB. Tissue-specific ablation of the GLUT4 glucose transporter or the insulin receptor challenges assumptions about insulin action and glucose homeostasis. J Biol Chem. 2003;278:33609-33612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 174] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 36. | Fisher SJ, Kahn CR. Insulin signaling is required for insulin's direct and indirect action on hepatic glucose production. J Clin Invest. 2003;111:463-468. [PubMed] [DOI] [Full Text] |
| 37. | Michael MD, Kulkarni RN, Postic C, Previs SF, Shulman GI, Magnuson MA, Kahn CR. Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol Cell. 2000;6:87-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 805] [Cited by in RCA: 805] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 38. | Baudry A, Leroux L, Jackerott M, Joshi RL. Genetic manipulation of insulin signaling, action and secretion in mice. Insights into glucose homeostasis and pathogenesis of type 2 diabetes. EMBO Rep. 2002;3:323-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 39. | Abel ED, Peroni O, Kim JK, Kim YB, Boss O, Hadro E, Minnemann T, Shulman GI, Kahn BB. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409:729-733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 895] [Cited by in RCA: 894] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 40. | Zisman A, Peroni OD, Abel ED, Michael MD, Mauvais-Jarvis F, Lowell BB, Wojtaszewski JF, Hirshman MF, Virkamaki A, Goodyear LJ, Kahn CR, Kahn BB. Targeted disruption of the glucose transporter 4 selectively in muscle causes insulin resistance and glucose intolerance. Nat Med. 2000;6:924-928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 533] [Cited by in RCA: 575] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 41. | Yang Q, Graham TE, Mody N, Preitner F, Peroni OD, Zabolotny JM, Kotani K, Quadro L, Kahn BB. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature. 2005;436:356-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1461] [Cited by in RCA: 1540] [Article Influence: 77.0] [Reference Citation Analysis (0)] |
| 42. | Gimeno RE, Klaman LD. Adipose tissue as an active endocrine organ: recent advances. Curr Opin Pharmacol. 2005;5:122-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 127] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 43. | Lazar MA. How obesity causes diabetes: not a tall tale. Science. 2005;307:373-375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 399] [Cited by in RCA: 401] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 44. | Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J Clin Invest. 2005;115:1111-1119. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1714] [Cited by in RCA: 2347] [Article Influence: 117.4] [Reference Citation Analysis (0)] |
| 45. | de Luca C, Olefsky JM. Stressed out about obesity and insulin resistance. Nat Med. 2006;12:41-2; discussion 42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 64] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 46. | Lazar MA. The humoral side of insulin resistance. Nat Med. 2006;12:43-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 59] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 47. | Gunton JE, Kulkarni RN, Yim S, Okada T, Hawthorne WJ, Tseng YH, Roberson RS, Ricordi C, O'Connell PJ, Gonzalez FJ, Kahn CR. Loss of ARNT/HIF1beta mediates altered gene expression and pancreatic-islet dysfunction in human type 2 diabetes. Cell. 2005;122:337-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 380] [Cited by in RCA: 389] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 48. | Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int J Biochem Cell Biol. 2004;36:189-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 415] [Cited by in RCA: 448] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 49. | Czech MP. ARNT misbehavin' in diabetic beta cells. Nat Med. 2006;12:39-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 50. | Donath MY, Halban PA. Decreased beta-cell mass in diabetes: significance, mechanisms and therapeutic implications. Diabetologia. 2004;47:581-589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 284] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 51. | Poitout V, Robertson RP. Minireview: Secondary beta-cell failure in type 2 diabetes--a convergence of glucotoxicity and lipotoxicity. Endocrinology. 2002;143:339-342. [PubMed] [DOI] [Full Text] |
| 52. | Kaneto H, Nakatani Y, Kawamori D, Miyatsuka T, Matsuoka TA, Matsuhisa M, Yamasaki Y. Role of oxidative stress, endoplasmic reticulum stress, and c-Jun N-terminal kinase in pancreatic beta-cell dysfunction and insulin resistance. Int J Biochem Cell Biol. 2006;38:782-793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 89] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 53. | Rhodes CJ. Type 2 diabetes-a matter of beta-cell life and death? Science. 2005;307:380-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 683] [Cited by in RCA: 676] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 54. | Werner ED, Lee J, Hansen L, Yuan M, Shoelson SE. Insulin resistance due to phosphorylation of insulin receptor substrate-1 at serine 302. J Biol Chem. 2004;279:35298-35305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 183] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 55. | White MF. IRS proteins and the common path to diabetes. Am J Physiol Endocrinol Metab. 2002;283:E413-E422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 620] [Cited by in RCA: 627] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 56. | O'Rahilly S, Barroso I, Wareham NJ. Genetic factors in type 2 diabetes: the end of the beginning? Science. 2005;307:370-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 191] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 57. | Das SK, Elbein SC. The Genetic Basis of Type 2 Diabetes. Cellscience. 2006;2:100-131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 50] [Reference Citation Analysis (0)] |
| 58. | Diamond J. The double puzzle of diabetes. Nature. 2003;423:599-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 244] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 59. | Meigs JB, Cupples LA, Wilson PW. Parental transmission of type 2 diabetes: the Framingham Offspring Study. Diabetes. 2000;49:2201-2207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 416] [Cited by in RCA: 426] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 60. | Barnett AH, Eff C, Leslie RD, Pyke DA. Diabetes in identical twins. A study of 200 pairs. Diabetologia. 1981;20:87-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 633] [Cited by in RCA: 586] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 61. | Newman B, Selby JV, King MC, Slemenda C, Fabsitz R, Friedman GD. Concordance for type 2 (non-insulin-dependent) diabetes mellitus in male twins. Diabetologia. 1987;30:763-768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 281] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 62. | Poulsen P, Kyvik KO, Vaag A, Beck-Nielsen H. Heritability of type II (non-insulin-dependent) diabetes mellitus and abnormal glucose tolerance--a population-based twin study. Diabetologia. 1999;42:139-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 390] [Cited by in RCA: 393] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 63. | Medici F, Hawa M, Ianari A, Pyke DA, Leslie RD. Concordance rate for type II diabetes mellitus in monozygotic twins: actuarial analysis. Diabetologia. 1999;42:146-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 102] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 64. | Vaag A, Henriksen JE, Madsbad S, Holm N, Beck-Nielsen H. Insulin secretion, insulin action, and hepatic glucose production in identical twins discordant for non-insulin-dependent diabetes mellitus. J Clin Invest. 1995;95:690-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 110] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 65. | Pociot F, McDermott MF. Genetics of type 1 diabetes mellitus. Genes Immun. 2002;3:235-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 208] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 66. | Rich SS. Mapping genes in diabetes. Genetic epidemiological perspective. Diabetes. 1990;39:1315-1319. [RCA] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 124] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 67. | Rich SS. Mapping genes in diabetes. Genetic epidemiological perspective. Diabetes. 1990;39:1315-1319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 173] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 68. | Ali O. Genetics of type 2 diabetes. World J Diabetes. 2013;4:114-123. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 188] [Cited by in RCA: 206] [Article Influence: 17.2] [Reference Citation Analysis (23)] |
| 69. | Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, Hinokio Y, Lindner TH, Mashima H, Schwarz PE, del Bosque-Plata L, Horikawa Y, Oda Y, Yoshiuchi I, Colilla S, Polonsky KS, Wei S, Concannon P, Iwasaki N, Schulze J, Baier LJ, Bogardus C, Groop L, Boerwinkle E, Hanis CL, Bell GI. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet. 2000;26:163-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1007] [Cited by in RCA: 933] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 70. | Hanis CL, Boerwinkle E, Chakraborty R, Ellsworth DL, Concannon P, Stirling B, Morrison VA, Wapelhorst B, Spielman RS, Gogolin-Ewens KJ, Shepard JM, Williams SR, Risch N, Hinds D, Iwasaki N, Ogata M, Omori Y, Petzold C, Rietzch H, Schröder HE, Schulze J, Cox NJ, Menzel S, Boriraj VV, Chen X, Lim LR, Lindner T, Mereu LE, Wang YQ, Xiang K, Yamagata K, Yang Y, Bell GI. A genome-wide search for human non-insulin-dependent (type 2) diabetes genes reveals a major susceptibility locus on chromosome 2. Nat Genet. 1996;13:161-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 408] [Cited by in RCA: 367] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 71. | Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, Tuomi T, Gaudet D, Hudson TJ, Daly M, Groop L, Lander ES. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1044] [Cited by in RCA: 1038] [Article Influence: 41.5] [Reference Citation Analysis (0)] |
| 72. | Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Görgün C, Glimcher LH, Hotamisligil GS. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2756] [Cited by in RCA: 2869] [Article Influence: 136.6] [Reference Citation Analysis (0)] |
| 73. | Florez JC, Hirschhorn J, Altshuler D. The inherited basis of diabetes mellitus: implications for the genetic analysis of complex traits. Annu Rev Genomics Hum Genet. 2003;4:257-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 207] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 74. | Song Y, Niu T, Manson JE, Kwiatkowski DJ, Liu S. Are variants in the CAPN10 gene related to risk of type 2 diabetes? A quantitative assessment of population and family-based association studies. Am J Hum Genet. 2004;74:208-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 82] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 75. | Bodhini D, Radha V, Ghosh S, Sanapala KR, Majumder PP, Rao MR, Mohan V. Association of calpain 10 gene polymorphisms with type 2 diabetes mellitus in Southern Indians. Metabolism. 2011;60:681-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 76. | Almgren P, Lehtovirta M, Isomaa B, Sarelin L, Taskinen MR, Lyssenko V, Tuomi T, Groop L; Botnia Study Group. Heritability and familiality of type 2 diabetes and related quantitative traits in the Botnia Study. Diabetologia. 2011;54:2811-2819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 197] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 77. | Tong Y, Lin Y, Zhang Y, Yang J, Liu H, Zhang B. Association between TCF7L2 gene polymorphisms and susceptibility to type 2 diabetes mellitus: a large Human Genome Epidemiology (HuGE) review and meta-analysis. BMC Med Genet. 2009;10:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 176] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 78. | Duggirala R, Blangero J, Almasy L, Dyer TD, Williams KL, Leach RJ, O'Connell P, Stern MP. Linkage of type 2 diabetes mellitus and of age at onset to a genetic location on chromosome 10q in Mexican Americans. Am J Hum Genet. 1999;64:1127-1140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 243] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 79. | Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, Magnusson KP, Walters GB, Palsdottir E, Jonsdottir T, Gudmundsdottir T, Gylfason A, Saemundsdottir J, Wilensky RL, Reilly MP, Rader DJ, Bagger Y, Christiansen C, Gudnason V, Sigurdsson G, Thorsteinsdottir U, Gulcher JR, Kong A, Stefansson K. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1578] [Cited by in RCA: 1580] [Article Influence: 83.2] [Reference Citation Analysis (0)] |
| 80. | Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881-885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2162] [Cited by in RCA: 2103] [Article Influence: 116.8] [Reference Citation Analysis (1)] |
| 81. | Lyssenko V, Lupi R, Marchetti P, Del Guerra S, Orho-Melander M, Almgren P, Sjögren M, Ling C, Eriksson KF, Lethagen AL, Mancarella R, Berglund G, Tuomi T, Nilsson P, Del Prato S, Groop L. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest. 2007;117:2155-2163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 570] [Cited by in RCA: 571] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 82. | Clausen JO, Hansen T, Bjørbaek C, Echwald SM, Urhammer SA, Rasmussen S, Andersen CB, Hansen L, Almind K, Winther K. Insulin resistance: interactions between obesity and a common variant of insulin receptor substrate-1. Lancet. 1995;346:397-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 141] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 83. | Le Fur S, Le Stunff C, Bougnères P. Increased insulin resistance in obese children who have both 972 IRS-1 and 1057 IRS-2 polymorphisms. Diabetes. 2002;51 Suppl 3:S304-S307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 84. | Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661-678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7741] [Cited by in RCA: 7160] [Article Influence: 397.8] [Reference Citation Analysis (0)] |
| 85. | Boj SF, van Es JH, Huch M, Li VS, José A, Hatzis P, Mokry M, Haegebarth A, van den Born M, Chambon P, Voshol P, Dor Y, Cuppen E, Fillat C, Clevers H. Diabetes risk gene and Wnt effector Tcf7 L2/TCF4 controls hepatic response to perinatal and adult metabolic demand. Cell. 2012;151:1595-1607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 194] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 86. | Fu D, Cong X, Ma Y, Cai H, Cai M, Li D, Lv M, Yuan X, Huang Y, Lv Z. Genetic polymorphism of glucokinase on the risk of type 2 diabetes and impaired glucose regulation: evidence based on 298,468 subjects. PLoS One. 2013;8:e55727. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 87. | Withers DJ, White M. Perspective: The insulin signaling system--a common link in the pathogenesis of type 2 diabetes. Endocrinology. 2000;141:1917-1921. [PubMed] [DOI] [Full Text] |
| 88. | Ershow AG. Environmental influences on development of type 2 diabetes and obesity: challenges in personalizing prevention and management. J Diabetes Sci Technol. 2009;3:727-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 89. | den Braver NR, Lakerveld J, Rutters F, Schoonmade LJ, Brug J, Beulens JWJ. Built environmental characteristics and diabetes: a systematic review and meta-analysis. BMC Med. 2018;16:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 153] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 90. | Murea M, Ma L, Freedman BI. Genetic and environmental factors associated with type 2 diabetes and diabetic vascular complications. Rev Diabet Stud. 2012;9:6-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 233] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 91. | Dendup T, Feng X, Clingan S, Astell-Burt T. Environmental Risk Factors for Developing Type 2 Diabetes Mellitus: A Systematic Review. Int J Environ Res Public Health. 2018;15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 255] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 92. | Hashmi S, Wang Y, Suman DS, Parhar RS, Collison K, Conca W, Al-Mohanna F, Gaugler R. Human cancer: is it linked to dysfunctional lipid metabolism? Biochim Biophys Acta. 2015;1850:352-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 93. | Franks PW. Gene × environment interactions in type 2 diabetes. Curr Diab Rep. 2011;11:552-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 59] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 94. | Skinner MK. Environmental epigenetic transgenerational inheritance and somatic epigenetic mitotic stability. Epigenetics. 2011;6:838-842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 241] [Cited by in RCA: 234] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 95. | Seki Y, Williams L, Vuguin PM, Charron MJ. Minireview: Epigenetic programming of diabetes and obesity: animal models. Endocrinology. 2012;153:1031-1038. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 122] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 96. | American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2014;37 Suppl 1:S81-S90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2986] [Cited by in RCA: 3449] [Article Influence: 313.5] [Reference Citation Analysis (16)] |
| 97. | Lu H, Koshkin V, Allister EM, Gyulkhandanyan AV, Wheeler MB. Molecular and metabolic evidence for mitochondrial defects associated with beta-cell dysfunction in a mouse model of type 2 diabetes. Diabetes. 2010;59:448-459. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 152] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 98. | Marroqui L, Tudurí E, Alonso-Magdalena P, Quesada I, Nadal Á, Dos Santos RS. Mitochondria as target of endocrine-disrupting chemicals: implications for type 2 diabetes. J Endocrinol. 2018;239:R27-R45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 99. | Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes. 2002;51:2944-2950. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1699] [Cited by in RCA: 1737] [Article Influence: 75.5] [Reference Citation Analysis (0)] |
| 100. | Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med. 2004;350:664-671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1619] [Cited by in RCA: 1593] [Article Influence: 75.9] [Reference Citation Analysis (0)] |
| 101. | Petersen KF, Befroy D, Dufour S, Dziura J, Ariyan C, Rothman DL, DiPietro L, Cline GW, Shulman GI. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science. 2003;300:1140-1142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1633] [Cited by in RCA: 1566] [Article Influence: 71.2] [Reference Citation Analysis (0)] |
| 102. | Boushel R, Gnaiger E, Schjerling P, Skovbro M, Kraunsøe R, Dela F. Patients with type 2 diabetes have normal mitochondrial function in skeletal muscle. Diabetologia. 2007;50:790-796. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 387] [Cited by in RCA: 406] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 103. | Abdul-Ghani MA, DeFronzo RA. Mitochondrial dysfunction, insulin resistance, and type 2 diabetes mellitus. Curr Diab Rep. 2008;8:173-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 105] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 104. | Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307:384-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1461] [Cited by in RCA: 1517] [Article Influence: 75.9] [Reference Citation Analysis (0)] |
| 105. | Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, Landaker EJ, Goldfine AB, Mun E, DeFronzo R, Finlayson J, Kahn CR, Mandarino LJ. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A. 2003;100:8466-8471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1507] [Cited by in RCA: 1543] [Article Influence: 70.1] [Reference Citation Analysis (0)] |
| 106. | Zhang CY, Baffy G, Perret P, Krauss S, Peroni O, Grujic D, Hagen T, Vidal-Puig AJ, Boss O, Kim YB, Zheng XX, Wheeler MB, Shulman GI, Chan CB, Lowell BB. Uncoupling protein-2 negatively regulates insulin secretion and is a major link between obesity, beta cell dysfunction, and type 2 diabetes. Cell. 2001;105:745-755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 698] [Cited by in RCA: 704] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 107. | Bugger H, Boudina S, Hu XX, Tuinei J, Zaha VG, Theobald HA, Yun UJ, McQueen AP, Wayment B, Litwin SE, Abel ED. Type 1 diabetic akita mouse hearts are insulin sensitive but manifest structurally abnormal mitochondria that remain coupled despite increased uncoupling protein 3. Diabetes. 2008;57:2924-2932. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 147] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 108. | Sivitz WI, Yorek MA. Mitochondrial dysfunction in diabetes: from molecular mechanisms to functional significance and therapeutic opportunities. Antioxid Redox Signal. 2010;12:537-577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 478] [Cited by in RCA: 540] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 109. | Karakelides H, Asmann YW, Bigelow ML, Short KR, Dhatariya K, Coenen-Schimke J, Kahl J, Mukhopadhyay D, Nair KS. Effect of insulin deprivation on muscle mitochondrial ATP production and gene transcript levels in type 1 diabetic subjects. Diabetes. 2007;56:2683-2689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 96] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 110. | Friederich M, Fasching A, Hansell P, Nordquist L, Palm F. Diabetes-induced up-regulation of uncoupling protein-2 results in increased mitochondrial uncoupling in kidney proximal tubular cells. Biochim Biophys Acta. 2008;1777:935-940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 87] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 111. | de Cavanagh EM, Ferder L, Toblli JE, Piotrkowski B, Stella I, Fraga CG, Inserra F. Renal mitochondrial impairment is attenuated by AT1 blockade in experimental Type I diabetes. Am J Physiol Heart Circ Physiol. 2008;294:H456-H465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 112. | Muoio DM, Newgard CB. Mechanisms of disease:Molecular and metabolic mechanisms of insulin resistance and beta-cell failure in type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9:193-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 822] [Cited by in RCA: 879] [Article Influence: 51.7] [Reference Citation Analysis (0)] |
| 113. | Sparks LM, Xie H, Koza RA, Mynatt R, Hulver MW, Bray GA, Smith SR. A high-fat diet coordinately downregulates genes required for mitochondrial oxidative phosphorylation in skeletal muscle. Diabetes. 2005;54:1926-1933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 467] [Cited by in RCA: 467] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 114. | Wolfrum C, Asilmaz E, Luca E, Friedman JM, Stoffel M. Foxa2 regulates lipid metabolism and ketogenesis in the liver during fasting and in diabetes. Nature. 2004;432:1027-1032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 326] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 115. | An J, Muoio DM, Shiota M, Fujimoto Y, Cline GW, Shulman GI, Koves TR, Stevens R, Millington D, Newgard CB. Hepatic expression of malonyl-CoA decarboxylase reverses muscle, liver and whole-animal insulin resistance. Nat Med. 2004;10:268-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 352] [Cited by in RCA: 367] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 116. | Abu-Elheiga L, Oh W, Kordari P, Wakil SJ. Acetyl-CoA carboxylase 2 mutant mice are protected against obesity and diabetes induced by high-fat/high-carbohydrate diets. Proc Natl Acad Sci U S A. 2003;100:10207-10212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 310] [Cited by in RCA: 294] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 117. | Azad MAK, Sarker M, Li T, Yin J. Probiotic Species in the Modulation of Gut Microbiota: An Overview. Biomed Res Int. 2018;2018:9478630. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 394] [Cited by in RCA: 454] [Article Influence: 64.9] [Reference Citation Analysis (0)] |
| 118. | Saad MJ, Santos A, Prada PO. Linking Gut Microbiota and Inflammation to Obesity and Insulin Resistance. Physiology (Bethesda). 2016;31:283-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 471] [Article Influence: 58.9] [Reference Citation Analysis (0)] |
| 119. | Han JL, Lin HL. Intestinal microbiota and type 2 diabetes: from mechanism insights to therapeutic perspective. World J Gastroenterol. 2014;20:17737-17745. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 111] [Cited by in RCA: 117] [Article Influence: 10.6] [Reference Citation Analysis (2)] |
| 120. | Gavin PG, Hamilton-Williams EE. The gut microbiota in type 1 diabetes: friend or foe? Curr Opin Endocrinol Diabetes Obes. 2019;26:207-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 121. | Hänninen A, Toivonen R, Pöysti S, Belzer C, Plovier H, Ouwerkerk JP, Emani R, Cani PD, De Vos WM. Akkermansia muciniphila induces gut microbiota remodelling and controls islet autoimmunity in NOD mice. Gut. 2018;67:1445-1453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 265] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 122. | Park SY, Hwang BO, Lim M, Ok SH, Lee SK, Chun KS, Park KK, Hu Y, Chung WY, Song NY. Oral-Gut Microbiome Axis in Gastrointestinal Disease and Cancer. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 131] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 123. | Olsen I, Yamazaki K. Can oral bacteria affect the microbiome of the gut? J Oral Microbiol. 2019;11:1586422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 127] [Cited by in RCA: 214] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 124. | Shaffer M, Lozupone C. Prevalence and Source of Fecal and Oral Bacteria on Infant, Child, and Adult Hands. mSystems. 2018;3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 125. | Schmidt TS, Hayward MR, Coelho LP, Li SS, Costea PI, Voigt AY, Wirbel J, Maistrenko OM, Alves RJ, Bergsten E, de Beaufort C, Sobhani I, Heintz-Buschart A, Sunagawa S, Zeller G, Wilmes P, Bork P. Extensive transmission of microbes along the gastrointestinal tract. Elife. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 331] [Cited by in RCA: 367] [Article Influence: 61.2] [Reference Citation Analysis (0)] |
| 126. | Hashmi S, Ji Q, Zhang J, Parhar RS, Huang CH, Brey C, Gaugler R. A Krüppel-like factor in Caenorhabditis elegans with essential roles in fat regulation, cell death, and phagocytosis. DNA Cell Biol. 2008;27:545-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 127. | Ling J, Brey C, Schilling M, Lateef F, Lopez-Dee ZP, Fernandes K, Thiruchelvam K, Wang Y, Chandel K, Rau K, Parhar R, Al-Mohanna F, Gaugler R, Hashmi S. Defective lipid metabolism associated with mutation in klf-2 and klf-3: important roles of essential dietary salts in fat storage. Nutr Metab (Lond). 2017;14:22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 128. | Zhang J, Yang C, Brey C, Rodriguez M, Oksov Y, Gaugler R, Dickstein E, Huang CH, Hashmi S. Mutation in Caenorhabditis elegans Krüppel-like factor, KLF-3 results in fat accumulation and alters fatty acid composition. Exp Cell Res. 2009;315:2568-2580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 129. | Zhang J, Bakheet R, Parhar RS, Huang CH, Hussain MM, Pan X, Siddiqui SS, Hashmi S. Regulation of fat storage and reproduction by Krüppel-like transcription factor KLF3 and fat-associated genes in Caenorhabditis elegans. J Mol Biol. 2011;411:537-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 130. | Hashmi S, Zhang J, Siddiqui SS, Parhar RS, Bakheet R, Al-Mohanna F. Partner in fat metabolism: role of KLFs in fat burning and reproductive behavior. 3 Biotech. 2011;1:59-72. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 131. | Brey CW, Nelder MP, Hailemariam T, Gaugler R, Hashmi S. Krüppel-like family of transcription factors: an emerging new frontier in fat biology. Int J Biol Sci. 2009;5:622-636. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 69] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 132. | Hashmi S, Wang Y, Parhar RS, Collison KS, Conca W, Al-Mohanna F, Gaugler R. A C. elegans model to study human metabolic regulation. Nutr Metab (Lond). 2013;10:31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 133. | Zhang J, Hashmi S, Cheema F, Al-Nasser N, Bakheet R, Parhar RS, Al-Mohanna F, Gaugler R, Hussain MM. Regulation of lipoprotein assembly, secretion and fatty acid β-oxidation by Krüppel-like transcription factor, klf-3. J Mol Biol. 2013;425:2641-2655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 134. | Oishi Y, Manabe I, Tobe K, Tsushima K, Shindo T, Fujiu K, Nishimura G, Maemura K, Yamauchi T, Kubota N, Suzuki R, Kitamura T, Akira S, Kadowaki T, Nagai R. Krüppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 2005;1:27-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 345] [Cited by in RCA: 354] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 135. | Wu J, Srinivasan SV, Neumann JC, Lingrel JB. The KLF2 transcription factor does not affect the formation of preadipocytes but inhibits their differentiation into adipocytes. Biochemistry. 2005;44:11098-11105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 82] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 136. | Li D, Yea S, Li S, Chen Z, Narla G, Banck M, Laborda J, Tan S, Friedman JM, Friedman SL, Walsh MJ. Krüppel-like factor-6 promotes preadipocyte differentiation through histone deacetylase 3-dependent repression of DLK1. J Biol Chem. 2005;280:26941-26952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 144] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 137. | Birsoy K, Chen Z, Friedman J. Transcriptional regulation of adipogenesis by KLF4. Cell Metab. 2008;7:339-347. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 293] [Cited by in RCA: 277] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 138. | Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1636] [Cited by in RCA: 1658] [Article Influence: 59.2] [Reference Citation Analysis (0)] |
| 139. | Bieker JJ. Krüppel-like factors: three fingers in many pies. J Biol Chem. 2001;276:34355-34358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 471] [Cited by in RCA: 494] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 140. | Kaczynski J, Cook T, Urrutia R. Sp1- and Krüppel-like transcription factors. Genome Biol. 2003;4:206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 664] [Cited by in RCA: 768] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 141. | van Vliet J, Crofts LA, Quinlan KG, Czolij R, Perkins AC, Crossley M. Human KLF17 is a new member of the Sp/KLF family of transcription factors. Genomics. 2006;87:474-482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 97] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 142. | Gray S, Feinberg MW, Hull S, Kuo CT, Watanabe M, Sen-Banerjee S, DePina A, Haspel R, Jain MK. The Krüppel-like factor KLF15 regulates the insulin-sensitive glucose transporter GLUT4. J Biol Chem. 2002;277:34322-34328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 192] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 143. | Wei D, Kanai M, Huang S, Xie K. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis. 2006;27:23-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 177] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 144. | Small KS, Hedman AK, Grundberg E, Nica AC, Thorleifsson G, Kong A, Thorsteindottir U, Shin SY, Richards HB; GIANT Consortium; MAGIC Investigators; DIAGRAM Consortium, Soranzo N, Ahmadi KR, Lindgren CM, Stefansson K, Dermitzakis ET, Deloukas P, Spector TD, McCarthy MI; MuTHER Consortium. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat Genet. 2011;43:561-564. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 221] [Article Influence: 15.8] [Reference Citation Analysis (0)] |