Published online Oct 15, 2022. doi: 10.4239/wjd.v13.i10.861
Peer-review started: March 26, 2022
First decision: June 11, 2022
Revised: June 25, 2022
Accepted: August 24, 2022
Article in press: August 24, 2022
Published online: October 15, 2022
Processing time: 202 Days and 0.1 Hours
Gestational diabetes mellitus (GDM) places both the mother and offspring at high risk of complications. Increasing evidence suggests that the gut microbiota plays a role in the pathogenesis of GDM. However, it is still unclear whether the gut microbiota is related to blood biochemical traits, particularly glucagon-like peptide-1 (GLP-1), in GDM patients.
To explore the correlation between the gut microbiota and blood biochemical traits, particularly GLP-1, in GDM patients.
The V4 region of the 16S ribosomal ribonucleic acid (rRNA) gene was sequenced based on the fecal samples of 35 pregnant women with GDM and was compared to that of 25 pregnant women with normal glucose tolerance (NGT).
The results showed that Ruminococcaceae_UCG-002, Ruminococcaceae_UCG-005, Clostri-dium_sensu_stricto_1, and Streptococcus were more abundant in the NGT group than in the GDM group. Bacteroides and Lachnoclostridium were more abundant in the GDM group than in the NGT group. Spearman’s correlation analysis was performed to identify the relationships between microbiota genera and blood biochemical traits. Paraprevotella, Roseburia, Faecalibacterium, and Ruminococcaceae_UCG-002 were significantly negatively correlated with glucose. Ruminococcaceae_UCG-002 was significantly negatively correlated with hemoglobin A1c. Bacteroides was significantly positively correlated with glucose. Sutterella, Oscillibacter, and Bifidobacterium were significantly positively correlated with GLP-1. A random forest model showed that 20 specific genera plus glucose provided the best discriminatory power, as indicated by the area under the receiver operating characteristic curve (0.94).
The results of this study reveal novel relationships between the gut microbiome, blood bio-chemical traits, particularly GLP-1, and GDM status. These findings suggest that some genera are crucial for controlling blood glucose-related indices and may be beneficial for GDM treatment. Alteration in the microbial composition of the gut may potentially serve as a marker for identifying individuals at risk of GDM.
Core Tip: Increasing evidence suggests that the gut microbiota plays a role in the pathogenesis of gestational diabetes mellitus (GDM). However, it is still unclear whether the gut microbiota is related to blood biochemical traits, particularly glucagon-like peptide-1 (GLP-1), in GDM patients. To the best of our knowledge, this is the first study to analyze the relationship between GLP-1 and the gut microbiota in patients with GDM, and this is the first report on the relationship between Paraprevotella, Roseburia, and Faecalibacterium and glucose in GDM, and the first report on the associations between GLP-1 and genera including Sutterella, Oscillibacter, and Bifidobacterium in GDM.
- Citation: Liang YY, Liu LY, Jia Y, Li Y, Cai JN, Shu Y, Tan JY, Chen PY, Li HW, Cai HH, Cai XS. Correlation between gut microbiota and glucagon-like peptide-1 in patients with gestational diabetes mellitus. World J Diabetes 2022; 13(10): 861-876
- URL: https://www.wjgnet.com/1948-9358/full/v13/i10/861.htm
- DOI: https://dx.doi.org/10.4239/wjd.v13.i10.861
Gestational diabetes mellitus (GDM) is defined as abnormal glucose tolerance in pregnancy[1]. It is one of the most common complications of pregnancy. The incidence of GDM has increased due to lifestyle changes, increasing maternal age, and changes to the GDM diagnostic criteria. The incidence of GDM is reported to be 13.20%. GDM is closely related to the occurrence of perinatal maternal and neonatal complications, and also significantly increases the risk of long-term metabolic diseases in pregnant women and newborns. The pathogenesis of GDM is complex and has not yet been comprehensively elucidated. Timely diagnosis and intervention are of great significance to the long-term health of pregnant women and their fetuses.
In recent years, with increased research and understanding of gastrointestinal hormones, their roles in the occurrence and development of GDM have attracted growing attention. Research suggests that GDM patients exhibit insufficient glucagon-like peptide-1 (GLP-1) secretion during pregnancy and after delivery relative to their blood glucose level. Bonde et al[2] found that the postprandial GLP-1 level of pregnant women was decreased, especially in GDM patients. However, with the recovery of blood glucose homeostasis after delivery, postprandial GLP-1 secretion gradually returned to normal. Kosinski et al[3] confirmed that decreased GLP-1 in patients with GDM is reversible. Changes in GLP-1 levels may be related to insulin resistance (IR) as a result of high blood glucose levels. However, it cannot be ruled out that changes in GLP-1 levels may be involved in the occurrence and development of GDM.
Evidence indicates that the gut microbiota is closely related to GDM[4]. The mechanism underlying the effect of probiotics in diabetes has not yet been fully elucidated, but it may be related to reductions in oxidative stress, regulation of the immune response, reductions in inflammation, and regulation of the gut microbiota[5,6]. In addition, probiotics can also reduce postprandial blood lipid levels and improve the absorption of antioxidants, which are related to oxidative stress[7]. Numerous studies have demonstrated that GLP-1 has insulin tropic and antioxidant effects[8-10]. Since GLP-1 and the gut microbiota each play roles in GDM, is there a correlation between the two?
Both clinical and animal studies have reported correlations between changes in the GLP-1 level and changes in the gut microbiota after gastrointestinal bypass surgery in type 2 diabetes mellitus (T2DM) patients or mice[11]. Therefore, it is speculated that GLP-1 may regulate blood glucose by regulating the number and structure of gut microbiota. Several authors have argued that GLP-1 may play a role in regulating blood glucose by increasing the diversity of the gut microbiota[12] and increasing the proportion of probiotics. Together, the current literature provides a comprehensive explanation of the hypoglycemic mechanism of GLP-1 and a reliable experimental basis for the study of GDM therapeutic targets and therapeutic drugs based on GLP-1. Accordingly, one study found that bifidobacteria improved insulin sensitivity by increasing the production of GLP-1[13].
The rapid increase in the prevalence of GDM in recent years cannot be easily explained by genetic factors; thus, it has been hypothesized that environmental factors may play a more important role. The gut microbiota constitutes an important environmental factor. GLP-1, as the most important representative of gastrointestinal hormones, may also be involved in the pathogenesis of GDM. The gastrointestinal microbiota and gastrointestinal hormones share the same root, are inseparable, influence and restrict each other, and jointly participate in the occurrence, development, and prognosis of GDM. Thus, a comprehensive study of the correlations between changes in the gut microbiota and GLP-1 will help to further clarify the pathogenesis of GDM. This is of great significance for the prevention, treatment, and prognosis of GDM, and may provide a novel and sensitive index for the clinical evaluation of GDM.
Patients were screened for GDM in the obstetric outpatient department according to the GDM diagnostic criteria (2014). Thirty-five patients with GDM were randomly selected from the patients who met the diagnostic criteria for GDM; these patients formed the GDM group. A further 25 pregnant women with normal glucose tolerance (NGT) were selected as the NGT group. Each subject provided written informed consent before inclusion in the study. The study was approved and carried out in accordance with the guidelines of the Ethics Committee of Nanhai District People’s Hospital of Foshan.
The inclusion criteria were as follows: 18-45 years old, being female, and any education level. Based on the diagnostic criteria for gestational diabetes (2014), during the 24-28th week of gestation, the 75 g oral glucose tolerance test (OGTT) was used to measure each patient’s blood glucose levels before, 1 h after, and 2 h after consuming sugar. If the patient’s blood glucose level reached or exceeded 5.1, 10.0, or 8.5 mmol/L, respectively, they were deemed to have GDM and were eligible for inclusion in the GDM group.
The exclusion criteria were as follows: (1) History of chronic digestive system disorder; (2) history of treatment with GLP-1 analogues or GLP-1 receptor agonists; (3) history of cardiac, renal, or liver dysfunction; (4) multiple pregnancy; (5) pregnancy-induced hypertension syndrome, placental insufficiency, placenta previa, placental abruption, pelvic or soft birth canal abnormalities, or other pregnancy complications; (6) history of mental disorders; (7) exposure to a large amount of radiation, chemical poisons, or drugs that can affect the fetus during pregnancy; (8) tumor history or history of radiotherapy and chemotherapy within the past 6 mo; (9) participation in other research studies; (10) patients lost to follow-up due to various reasons, including the occurrence of other serious diseases during the study; and (11) consumption of antibiotics or probiotics within 1 mo prior to admission.
Fresh fecal samples were collected from the participants and immediately frozen in a refrigerator at -80 °C. After collection of all samples, they were sent to the Treat Gut company for 16S rDNA sequencing.
Blood samples were collected after fasting and then 1 h and 2 h following consumption of sugar. Plasma glucose (Glu), glycosylated serum protein (GSP), low-density lipoprotein (LDL), uric acid (UA), hemoglobin (HB), total cholesterol (TCH), triglyceride (TG), and high-density lipoprotein (HDL) were determined with a Beckman AU5800 fully automatic biochemical analyzer. Glycosylated hemoglobin A1c (HbA1c) was determined using an ADAMS™ A1c HA-8180 automatic glycosylated hemoglobin analyzer. Insulin (INS), thyroid-stimulating hormone (TSH), and free tetraiodothyronine (FT4) were measured with a Maglumi2000plus automatic chemiluminescence immunoanalyzer.
The active forms of GLP-1 in the plasma samples of patients with and without GDM were measured using a GLP-1 (active) ELISA kit (ELabscience, Wuhan, Hubei Province, China).
The total genomic DNA of each sample was extracted using a fecal genomic DNA extraction kit (Tiangen Company). Sixteen S rDNA sequencing was performed by PCR amplification of V4 variable regions (39 to 297 base pairs), and a purified product library was established. The library construction steps followed the library construction method of the Illumina sequencing platform. The sequencing analysis was as follows. First, the Illumina Miseq 2 × 300bp paired-end sequencing data were analyzed. According to the barcode information, the samples were distinguished. Then, the data were merged, spliced, and filtered, and quality control analysis was conducted, including Q20 and Q30 scores. The final clean data were analyzed by operational taxonomic unit (OTU) cluster analysis and species taxonomy.
The data were filtered using Mothur software and clustered into OTUs (species) at a similarity level of 97% using Quantitative Insights into Microbial Ecology (QIIME) software version 1.80[14]. Based on the OTU analysis, the Ace, Shannon, observed species, Simpson, Chao1, and J indices were calculated as alpha diversity metrics. To compare the microbial composition between the samples, beta diversity analysis was performed using principal component analysis (PCA) and principal coordinate analysis (PCoA). Analysis of similarities (ANOSIM) was applied to evaluate the statistical significance of differences between the groups. A linear discriminant analysis (LDA) effect size (LEfSe) method was employed to evaluate any differences in the gut microbe between the groups.
GraphPad Prism (version 7.0) and R version 3.0.2 (R Foundation for Statistical Computing) were using for statistical analyses. The measurement data are expressed as the mean ± SD. Differences between groups were analyzed using oneway ANOVA. The differences were considered statistically significant at P < 0.05. Random-forest classification was performed for discriminating the samples from different groups using the R package “random forest”. The model was employed for five-fold cross-validation of the relative species abundance profile. Case probabilities were calculated by drawing receiver operating characteristic (ROC) curves.
GDM was diagnosed in 35 women based on fasting or oral glucose-stimulated hyperglycemia, or a combination of the two. Markers of glucose and insulin homeostasis were higher in the GDM group compared with the NGT group (Table 1). Individuals with GDM also had higher hemoglobin A1C (P = 0.003) and fasting blood glucose levels (P < 0.001). There were no significant differences in pre-pregnancy body weight, BMI, UA, TCH, TG, HDL, LDL, TSH, or FT4 between the two groups.
| Variable | Control (25) | GDM (31) | P value |
| Age (yr) | 28.42 (3.11) | 30.18 (3.26) | 0.055 |
| Pre-body weight (kg) | 52.42 (7.68) | 55.18 (6.48) | 0.165 |
| Height (cm) | 160.13 (5.44) | 157.88 (4.10) | 0.063 |
| Pre-BMI | 20.73 (2.85) | 22.2 (2.37) | 0.054 |
| GLU (mmol/L) | 4.49 (0.38) | 5.77 (0.95) | 3.53 × 10-7 |
| GLP-1 0 h (ug/L) | 67.72 (22.89) | 75.45(23.23) | 0.223 |
| GLP-1 1 h (ug/L) | 75.33 (26.14) | 84.34 (19.84) | 0.099 |
| GLP-1 2 h (ug/L) | 71.75 (23.83) | 79.21 (24.20) | 0.312 |
| OGTT 0 h (mmol/L) | 4.47 (0.39) | 5.74 (0.99) | 3.53 × 10-7 |
| OGTT 1 h (mmol/L) | 7.77 (1.55) | 11.23 (2.95) | 3.00 × 10-6 |
| OGTT 2 h (mmol/L) | 6.26 (0.87) | 9.64 (3.12) | 3.96 × 10-6 |
| Insulin 0 h (uIU/mL) | 9.78 (3.41) | 14.03 (15.93) | 0.239 |
| Insulin 1 h (uIU/mL) | 85.85 (43.99) | 64.52 (39.67) | 0.061 |
| Insulin 2 h (uIU/mL ) | 57.51 (39.36) | 66.52 (45.21) | 0.675 |
| GSP (mmol/L) | 1.68 (0.35) | 1.97 (0.62) | 0.054 |
| HbA1c (%) | 5.04 (0.30) | 5.79 (1.12) | 0.003 |
| UA (umol/L) | 279.52(68.47) | 263.80(81.27) | 0.463 |
| TCH (mmol/L) | 5.40 (1.06) | 5.35 (1.08) | 0.993 |
| TG (mmol/L) | 1.82 (0.72) | 2.45 (1.54) | 0.051 |
| HDL (mmol/L) | 2.11 (0.52) | 2.08 (0.63) | 0.946 |
| LDL (mmol/L) | 2.41 (0.89) | 2.34 (0.82) | 0.739 |
| TSH (uIU/mL) | 1.82 (3.43) | 1.22 (1.24) | 0.337 |
| FT4 (pg/mL) | 11.39 (3.27) | 11.77 (1.96) | 0.598 |
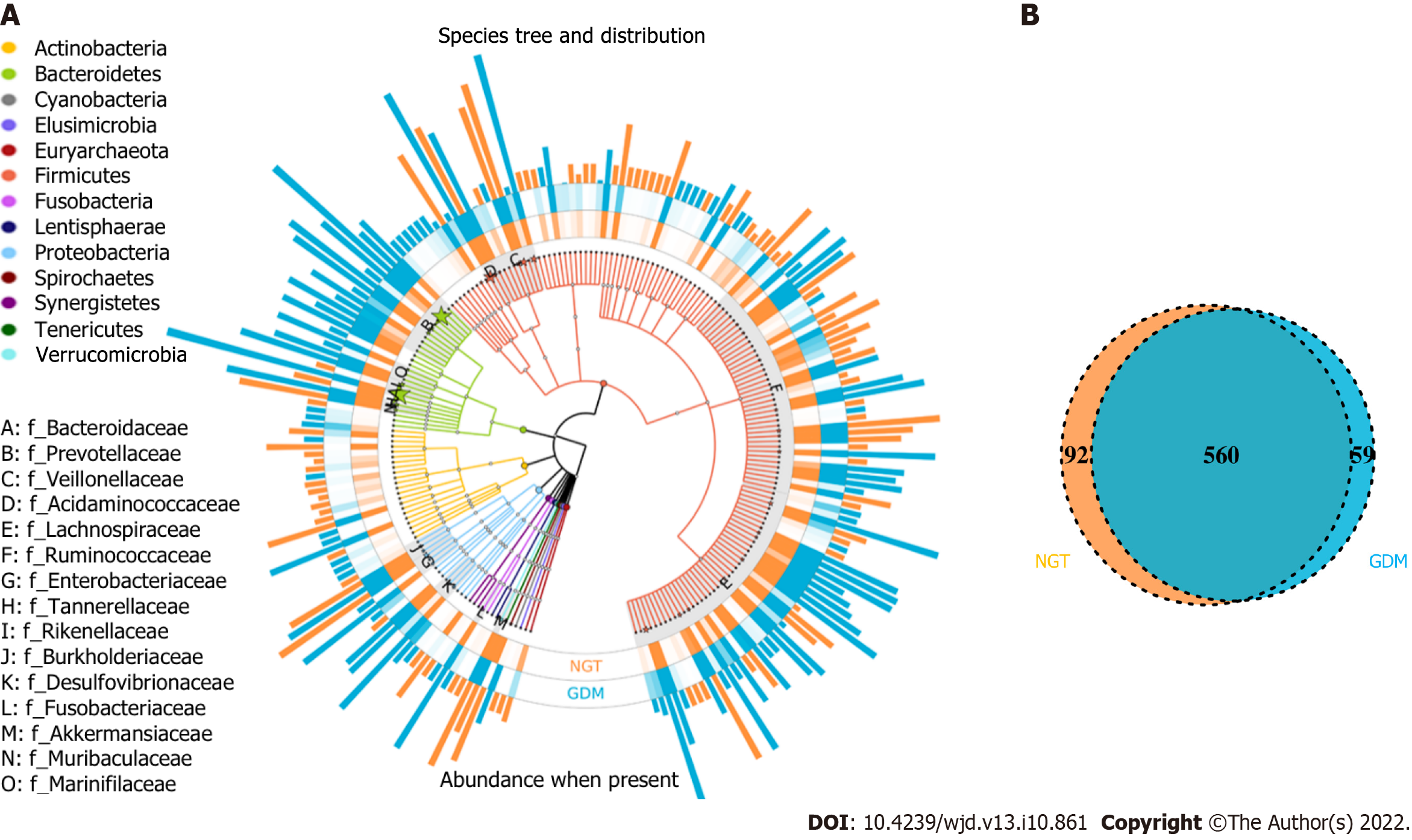
In this study, the OTUs annotated included 14 phyla, 62 families, and 214 genera of gut microbiota; the similarity among samples was 97% (Figure 1A). The total number of OTUs of the NGT group (at the 97% similarity level) was 652, and for the GDM group, it was 619. Venn diagram shows that 560 OTUs were shared by the NGT and GDM groups (Figure 1B).
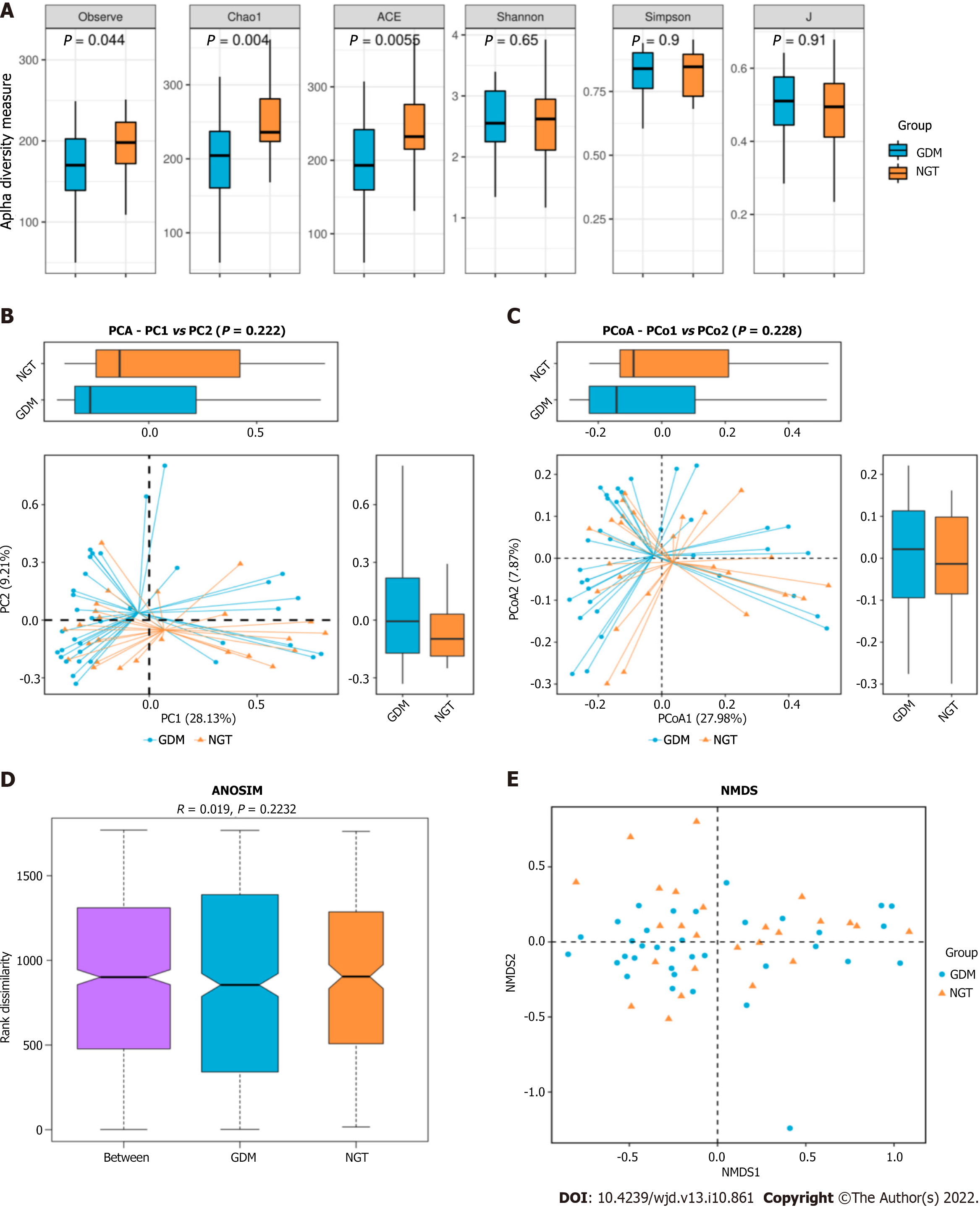
The observed species index of the GDM group was significantly difference from that of the NGT group (25; P = 0.044). The Chao1 richness index of the GDM group was significantly different from that of the NGT group (43; P = 0.004). The ACE index of the GDM group differed significantly from that of the NGT group (25; P = 0.0055) (Figure 2). There were no significant differences in the Shannon, Simpson, or J indices between the GDM group and NGT group (Shannon, P = 0.65; Simpson, P = 0.9; J, P = 0.91; Figure 2A). PCA and PCoA indicated that the gut microbiota in GDM patients differed significantly from that of the NGT subjects. There was no difference in the gut microbiota structure between the groups (ANOSIM, r = 0.019, P = 0.2232). NMDS cluster analysis indicated marked differences between the GDM patients and NGT subjects (Figure 2B).
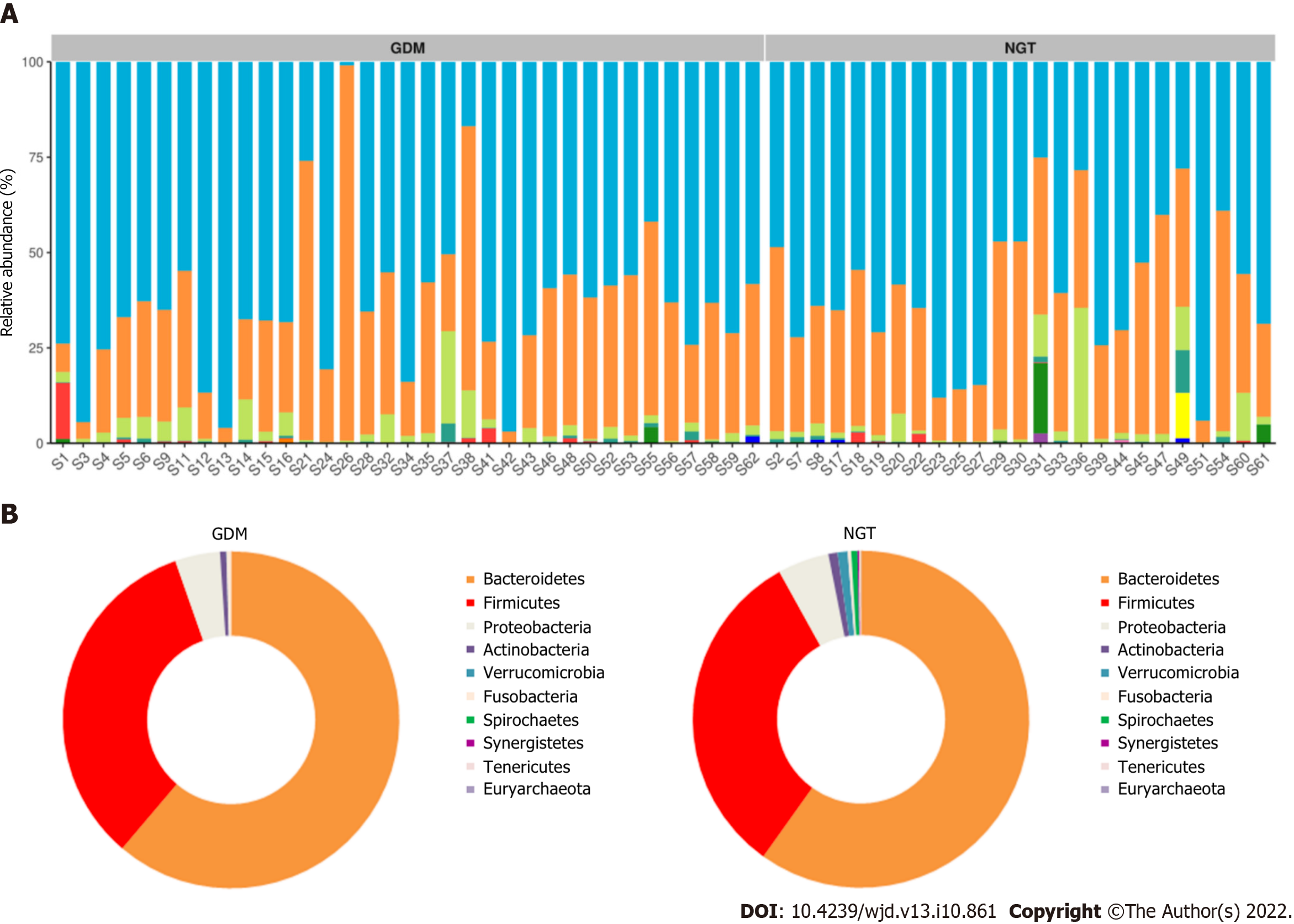
The composition of gut microbiota was different between the groups at the phylum, family, and genus levels. At the phylum level, Bacteroidetes, Proteobacteria, Firmicutes, Verrucomicrobia, Actinobacteria, Fusobacteria, Synergistetes, and Tenericutes were common phyla in both the GDM group and NGT group, accounting for 98.81% and 98.58% of the gut bacteria of each group, respectively (Figure 3). The GDM group had a lower abundance of Firmicutes (31.4% vs 33.2%), Verrucomicrobia (0.18% vs 1.01%, P < 0.05), Synergistetes (0.003% vs 0.110%, P < 0.01), and Tenericutes (0.05% vs 0.08%), and a higher abundance of Bacteroidetes (63.50% vs 60.81%), Proteobacteria (3.03% vs 2.56%), and Fusobacteria (0.33% vs 0.29%), compared to the NGT group. The ratio of Firmicutes to Bacteroidetes was decreased in the GDM group compared to the NGT group (0.49 vs 0.54).
At the family level, a greater number of different families were identified between the two groups (Figure 4). Fifty-five and 54 of the dominant families were detected in the GDM group and NGT group, respectively. Bacteroidaceae (phylum Bacteroidetes), Prevotellaceae (phylum Bacteroidetes), Acidaminococcaceae (phylum Firmicutes), Veillonellaceae (phylum Firmicutes), Lachnospiraceae (phylum Firmicutes), Ruminococcaceae (phylum Firmicutes), Enterobacteriaceae (phylum Proteobacteria), and Tannerellaceae (phylum Bacteroidetes) had the highest relative abundance in the GDM group, while Bacteroidaceae, Prevotellaceae, Acidaminococcaceae, Veillonellaceae, Lachnospiraceae, Enterobacteriaceae, Ruminococcaceae, and Rikenellaceae were the eight most abundant families in the NGT group.
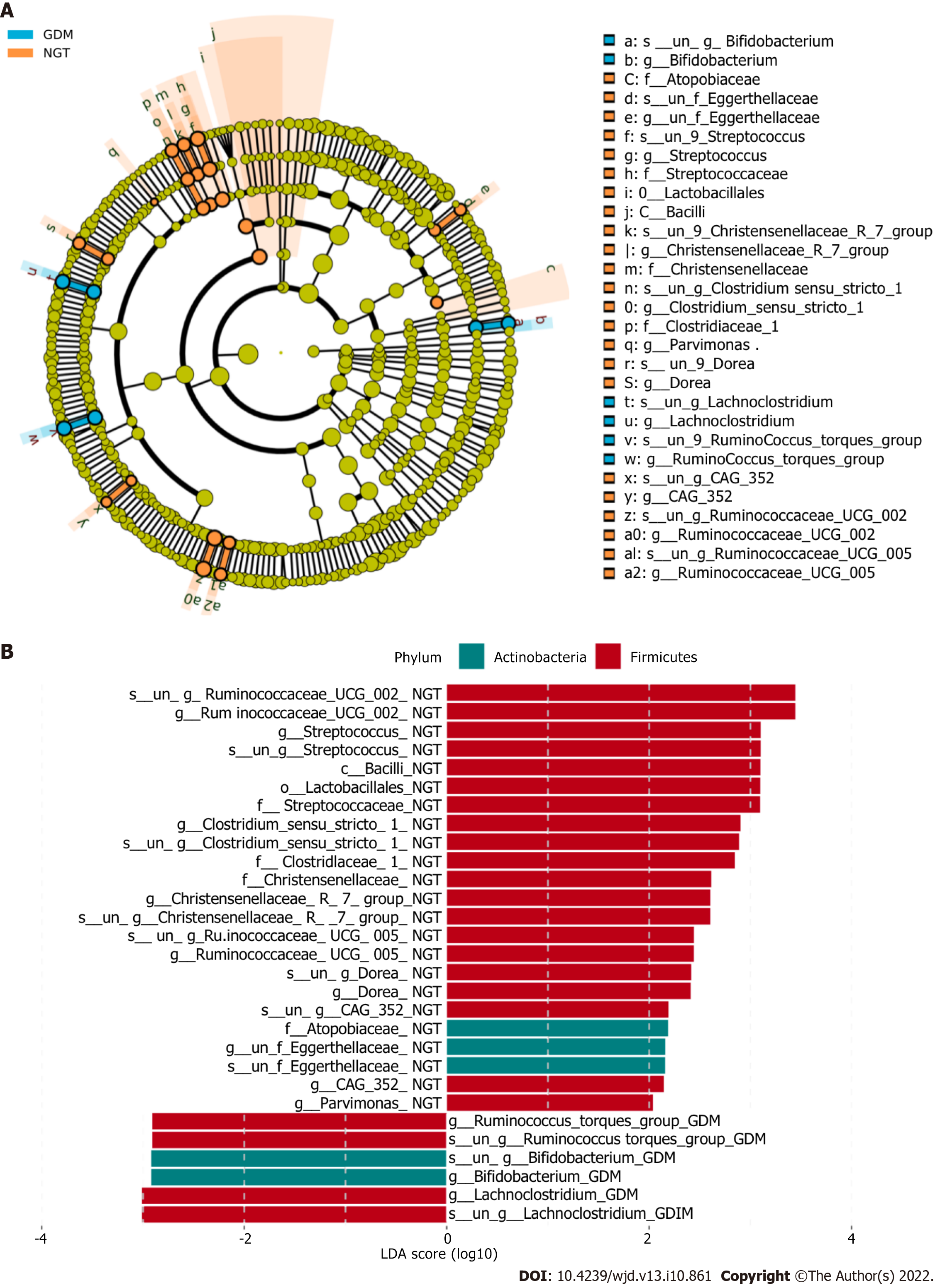
The bacterial taxa whose levels differed significantly between the two groups were identified by LEfSE analysis (Figure 4). At the family level, Atopobiaceae, Eggerthellaceae, Streptococcaceae, Christensenellaceae, Clostridiaceae, Bifidobacteriaceae, Lachnospiraceae, and Ruminococcaceae were significantly more abundant in the NGT group than in the GDM group.
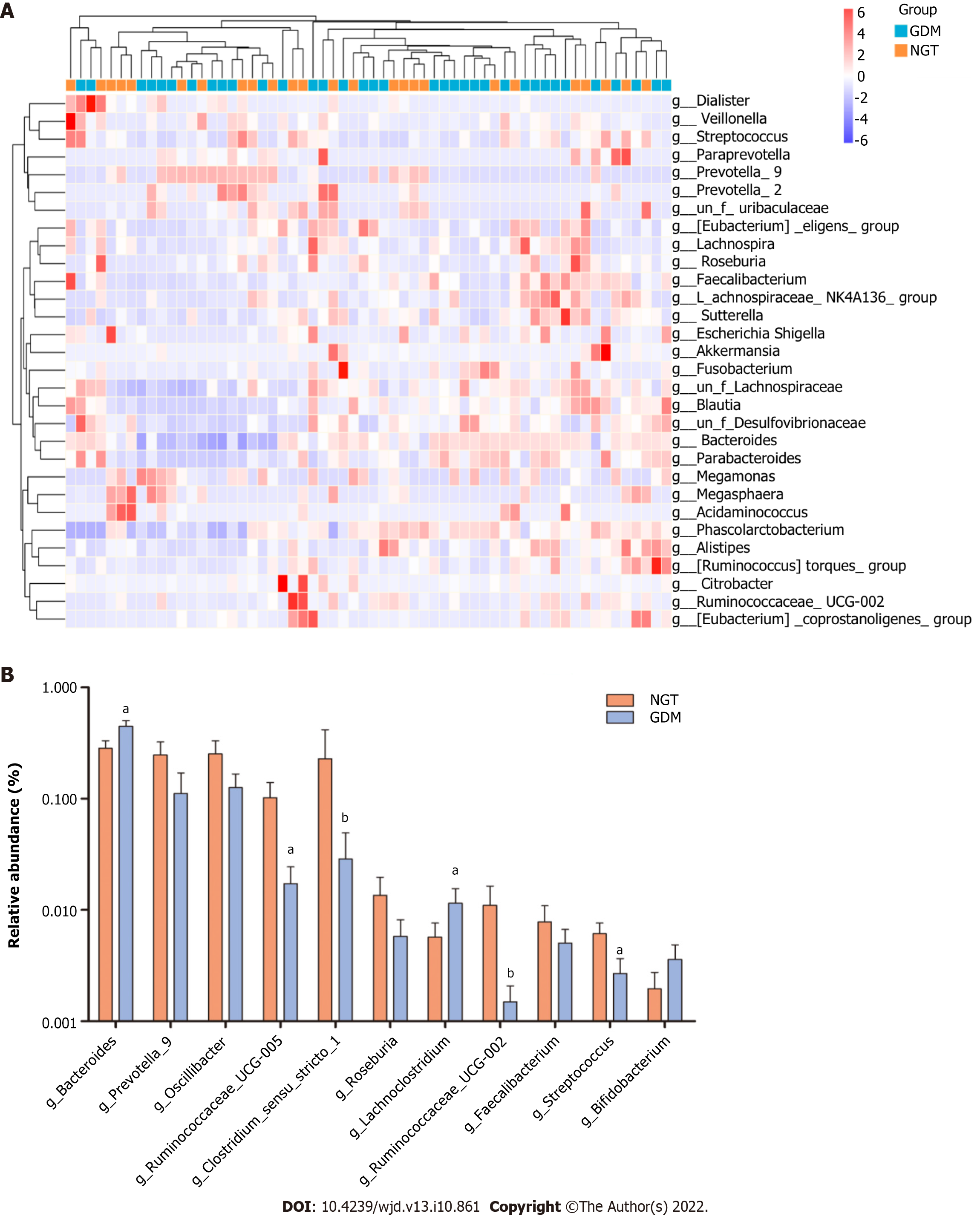
At the genus level, bacterial genera exhibited significant differences between the two groups (Figure 5). In the NGT group, Bacteroides (phylum Bacteroidetes), Prevotella_9 (phylum Bacteroidetes), Phascolarctobacterium (phylum Firmicutes), Megasphaera (phylum Firmicutes), Megamonas (phylum Firmicutes), Lachnospiraceae (phylum Firmicutes), Escherichia-Shigella (phylum Proteobacteria), and Prevotella_2 (phylum Bacteroidetes) were the eight most dominant genera. The eight most dominant genera in the GDM group were Bacteroides (phylum Bacteroidetes), Prevotella_9 (phylum Bacteroidetes), Megamonas (phylum Firmicutes), Phascolarctobacterium (phylum Firmicutes), Lachnospiraceae (phylum Firmicutes), Megasphaera (phylum Firmicutes), Prevotella_2 (phylum Bacteroidetes), and Parabacteroides (phylum Bacteroidetes).
Ruminococcaceae_UCG-002, Ruminococcaceae_UCG-005, Clostridium_sensu_stricto_1, and Streptococcus were more abundant in the NGT group than in the GDM group (P < 0.05). Bacteroides and Lachnoclostridium were more abundant in the GDM group than in the NGT group (P < 0.05). Prevotella_9, Oscillibacter, Roseburia, and Faecalibacterium were slightly more abundant in the NGT group than in the GDM group.
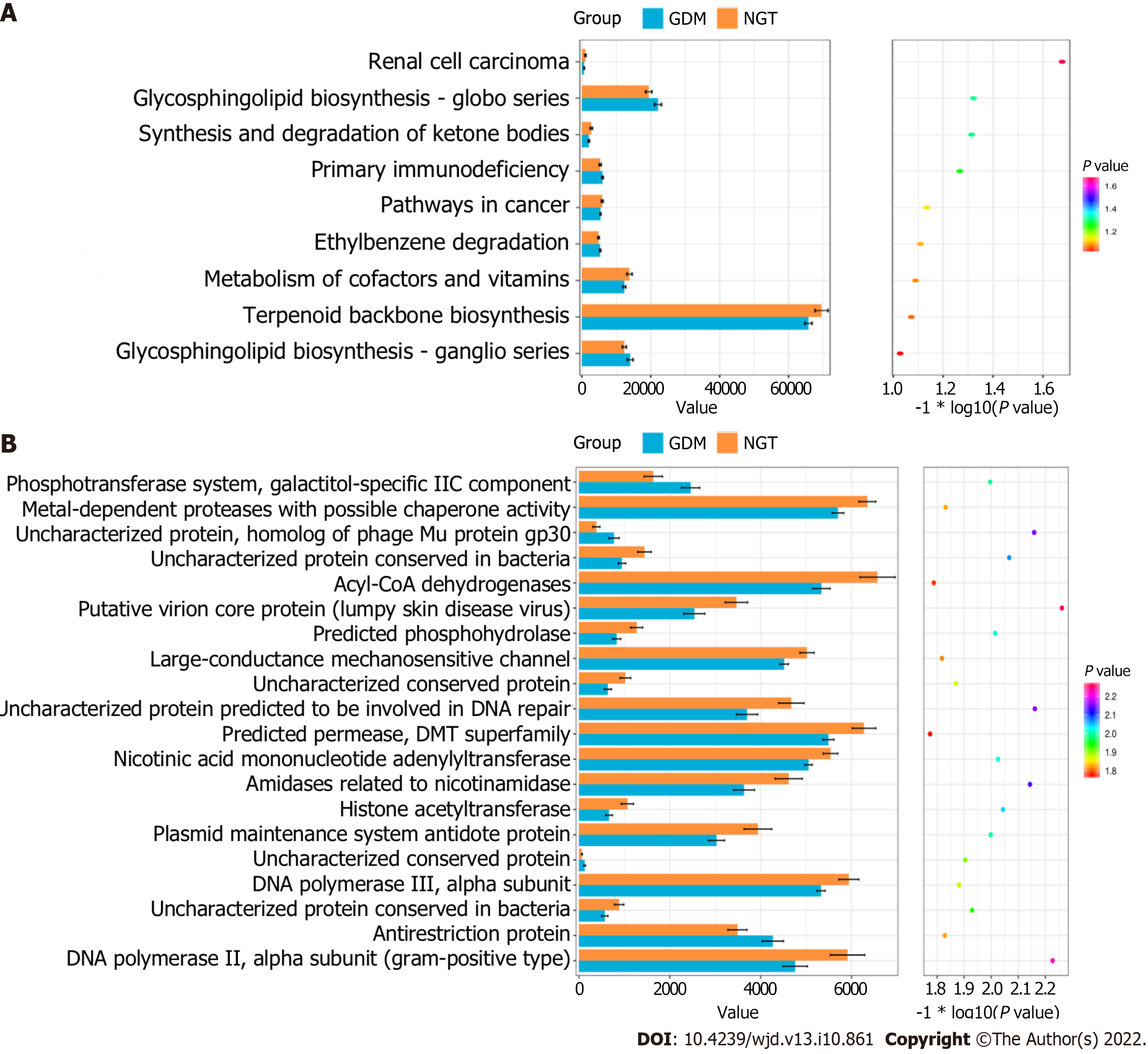
The cluster of orthologous groups (COG) categories and KEGG pathways were compared between the GDM and NGT groups. Figure 6A shows that three functional KEGG pathways differed between the GDM group and NGT group, including the glycosphingolipid biosynthesis-globo series, synthesis and degradation of ketone bodies, and renal cell carcinoma pathways. Figure 6B shows that 20 COG categories differed between the GDM group and NGT group, including the phosphotransferase system, galactitol-specific IIC component, metal-dependent proteases with possible chaperone activity, uncharacterized protein, homolog of phage Mu protein gp30, uncharacterized protein conserved in bacteria, Acyl-CoA dehydrogenases, putative virion core protein (lumpy skin disease virus), predicted phosphohydrolase, large-conductance mechanosensitive channel, uncharacterized conserved protein, uncharacterized protein predicted to be involved in DNA repair, predicted permease, DMT superfamily, nicotinic acid mononucleotide adenylyltransferase, amidases related to nicotinamidase, histone acetyltransferase, plasmid maintenance system antidote protein, uncharacterized conserved protein, DNA polymerase III, alpha subunit, uncharacterized protein conserved in bacteria, antirestriction protein, NA polymerase III, and alpha subunit (Gram-positive type).
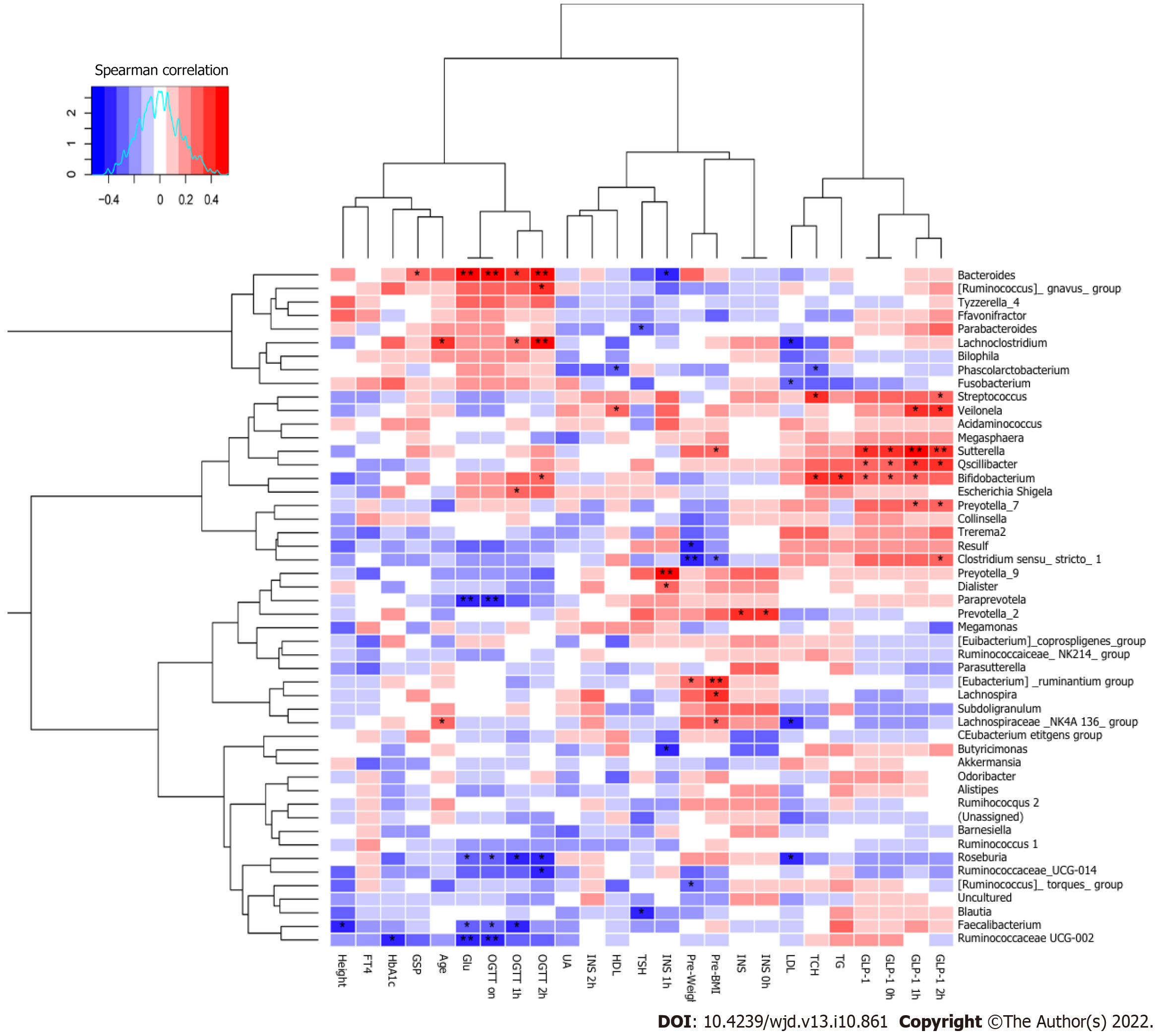
Spearman’s correlation analysis was performed to identify whether the different dominant genera were associated with blood biochemical traits in the second trimester of pregnancy (Figure 7). Paraprevotella, Roseburia, Faecalibacterium, and Ruminococcaceae_UCG-002 were negatively correlated with Glu (P < 0.05). Ruminococcaceae_UCG-002 was negatively correlated with HbA1c (P < 0.05). Clostridium_sensu_stricto_1, Desulfovibrio, and (Ruminococcus)_torques_group were negatively correlated with pre-pregnancy body weight (P < 0.05). Phascolarctobacterium was negatively correlated with HDL (P < 0.05). Ruminococcaceae_UCG-003 and Faecalibacterium were negatively correlated with height (P < 0.05). Lachnoclostridium and Lachnospiraceae_NK4A136_group were positively correlated with age (P < 0.05). Bacteroides was significantly positively correlated with Glu (P < 0.01). Sutterella, Oscillibacter, and Bifidobacterium were positively correlated with GLP-1 (P < 0.05).
Roseburia was negatively correlated with OGTT (0 h), OGTT (1 h), and OGTT (2 h) (P < 0.05). Faecalibacterium was negatively correlated with OGTT (0 h) and OGTT (1 h) (P < 0.05). Bacteroides was positively correlated with OGTT (0 h), OGTT (1 h), OGTT (2 h), and GSP (P < 0.05). Lachnoclostridium was positively correlated with OGTT (1 h) and OGTT (2 h) (P < 0.05). Sutterella was positively correlated with GLP-1(0 h), GLP-1(1 h), GLP-1(2 h), and pre-pregnancy BMI (P < 0.05). Oscillibacter was positively correlated with GLP-1(0 h), GLP-1(1 h), and GLP-1(2 h) (P < 0.05). Bifidobacterium was positively correlated with GLP-1(0 h), GLP-1(1 h), OGTT (2 h), TG, and TCH (P < 0.05).
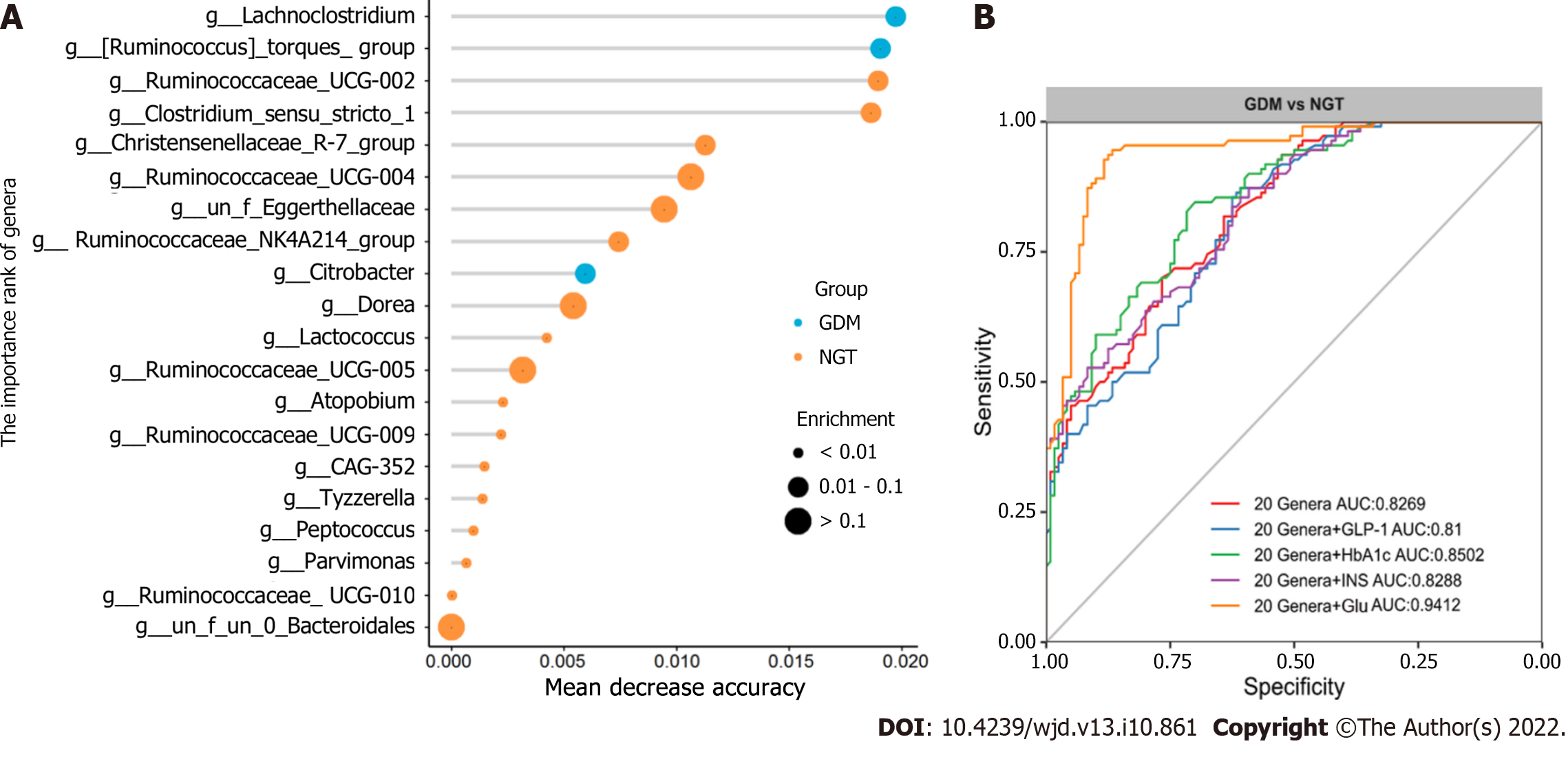
Finally, random forest models were used to assess the ability of the genera abundance profiles to predict GDM status (Figure 8A). Twenty genera plus Glu provided the best discriminatory power, as indicated by the area under the receiver operating characteristic (AUROCC) value of 0.94. The value was higher than that achieved with a model including just 20 genera (the best AUC was 0.828) (Figure 8B). Further, models with 20 genera plus GLP-1, INS, or HbA1c had lower AUROCC values than the model with 20 genera plus Glu. The AUROCC values were 0.81, 0.8288, and 0.8502, respectively.
In recent years, the relationships between the gut microbiota and diabetes as well as other endocrine diseases have become research hotspots. Similarly, the characteristics of the gut microbiota among pregnant women with GDM have received widespread research attention. To date, research on GDM has focused on the correlation between the gut microbiota and blood glucose or insulin, but there is still a lack of research on the relationship between the gut microbiota and GLP-1. Many studies have reported that GLP-1 is closely related to the gut microbiota and short-chain fatty acids (SCFAs)[15-17], and changes in the gut microbiota directly affect the secretion of GLP-1, which, in turn, affects insulin and blood glucose. These are closely related to the occurrence of GDM. Therefore, we focused on the relationship between GLP-1 and the gut microbiota in GDM patients. To the best of our knowledge, it is the first report on the relationship between GLP-1 and the gut microbiota in patients with GDM.
At the phylum level, the abundance of Firmicutes in the gut microbiota of the GDM group was lower than that in NGT group. Firmicutes are known to transform carbohydrates and undigested proteins into SCFAs, producing energy for the host organism. As a crucial SCFA, butyrate participates in the activation of multiple physiological signal pathways, including the proliferation and differentiation of regulatory T cells and anti-inflammatory activities[18,19]. Moreover, the GDM group exhibited reduced phylum levels of Verrucomicrobia, Synergistetes, and Tenericutes compared to the NGT group. Mucin-degrading bacteria Verrucomicrobia contribute to glucose homeostasis and intestinal health, and play a key role in the interaction between the host tissues and gut microbiome[20]. The gut microbiota in the GDM patients also exhibited a higher abundance of Bacteroidetes, Fusobacteria, and Proteobacteria compared to healthy subjects. Proteobacteria is an opportunistic pathogen that creates a major structural imbalance in the gut microbiota of GDM patients. The ratio of Firmicutes to Bacteroidetes in GDM patients is lower than that of NGT individuals.
At the family level, Atopobiaceae, Eggerthellaceae, Streptococcaceae, Christensenellaceae, Clostridiaceae, Bifidobacteriaceae, Lachnospiraceae, and Ruminococcaceae were more abundant in the NGT subjects than in the GDM patients. Zhang et al[21] reported that Ruminococcaceae, Bifidobacteriaceae, Christensenellaceae, Erysipelotrichaceae, Peptostreptococcaceae, and Eggerthellaceae were more abundant in the NGT subjects, which is consistent with the current study. In line with the study of Ma et al[22], the current results revealed that Ruminococcaceae were more abundant in the NGT group than in the GDM group. However, other studies have observed the opposite result[23,24]. The mechanisms remain unclear.
At the genus level, Ruminococcaceae_UCG-002, Ruminococcaceae_UCG-005, Clostridium_sensu_stricto_1, and Streptococcus were more abundant in the NGT group than in the GDM group. Bacteroides and Lachnoclostridium were more abundant in the GDM group than in the NGT group. Kuang et al[25] found that the proportion of Bifidobacterium in the gut microbiota of GDM pregnant women was significantly reduced, while the proportions of Bacteroides and Klebsiella were significantly increased. Liu et al[26] found that compared with normal pregnant women, the proportion of Faecalis in GDM patients was significantly lower, while the proportion of Prevotella was significantly higher. In the present study, there were no significant differences in Bifidobacterium, Klebsiella, or Prevotella between the GDM group and NGT group. Ma et al found that Ruminococcaceae_UCG-002 and Ruminococcaceae_UCG-005 were reduced in women with GDM[22].
One study found that supplementation with Lactobacillus rhamnosus in pregnant women may reduce the prevalence of GDM[27]. Another study showed that additional probiotic supplementation from pregnancy to 12 months post-delivery can reduce insulin levels and improve insulin sensitivity[28]. In this study, to identify beneficial bacteria for pregnant women, Spearman’s correlation analysis was performed to identify the relationships between bacterial genera and blood biochemical traits. Paraprevotella, Roseburia, Faecalibacterium, and Ruminococcaceae_UCG-002 were negatively correlated with Glu. Ruminococcaceae_UCG-002 was negatively correlated with HbA1c. Clostridium_sensu_stricto_1, Desulfovibrio, and (Ruminococcus)_torques_group were negatively correlated with pre-pregnancy body weight. Phascolarctobacterium was negatively correlated with HDL. Ruminococcaceae_UCG-003 and Faecalibacterium were negatively correlated with height. Lachnoclostridium and Lachnospiraceae_NK4A136_group were positively correlated with age. Zhang et al[21] found that Ruminococcaceae_UCG-002 was negatively correlated with fasting blood glucose levels. In the study of Crusell, Clostridium (sensu stricto) was positively correlated with gestational weight[29]. To the best of our knowledge, no studies have yet reported on the relationships between Paraprevotella, Roseburia, and Faecalibacterium and Glu in GDM. The current findings suggest that these genera are crucial for controlling blood glucose-related indices and may be beneficial for GDM treatment.
GLP-1 and its receptor agonist can promote insulin secretion only when the blood glucose level is elevated[30]. This safety feature makes GLP-1 and its agonist suitable for the treatment of GDM, which requires strict maintenance of blood glucose levels and stable, safe blood glucose regulation. Thus, the current study aimed to examine the correlations between the gut microbiota and GLP-1 levels, and identify beneficial bacteria that can improve the expression of GLP-1 in patients with GDM, so as to more safely control blood glucose. In the present study, Sutterella was significantly positively correlated with GLP-1 (0 h), GLP-1 (1 h), GLP-1 (2 h), and pre-pregnancy BMI. Wang et al[31] reported that subjects taking metformin exhibited a significantly increased relative abundance of Sutterella, whereas liraglutide dosing was associated with a significant increase in the genus Akkermansia. Another study showed that Sutterella was associated with C-reactive protein levels[32]. In the current study, Oscillibacter was significantly positively correlated with GLP-1 (0 h), GLP-1 (1 h), and GLP-1 (2 h). One study reported that Cyclocarya paliurus polysaccharides alleviated type 2 diabetic symptoms by increasing eleven SCFA-producing species, including Oscillibacter_valericigenes and Oscillibacter_ruminantium[33]. Oscillibacter belongs to the Clostridia class of Firmicutes, and in the human gut microbiota, this bacterium grow fermentatively, predominantly producing valerate when grown using glucose as a carbon source[34]. In the current study, Bifidobacterium was significantly positively correlated with GLP-1 (0 h), GLP-1 (1 h), TG, TCH, and OGTT (2 h). In the study of Zhao et al[35], Bifidobacterium longum DD98 improved the serum and intestinal cell GLP-1 levels, which protected pancreatic β-islet cells from damage induced by type 2 diabetes. To the best of our knowledge, this is the first report on the associations between GLP-1 and genera such as Sutterella, Oscillibacter, and Bifidobacterium in GDM.
In summary, this study contributes to a better understanding of the relationships between the gut microbiota and blood biochemical traits, particularly GLP-1, in individuals with GDM. The current findings suggest that some genera are crucial for controlling blood glucose-related indices and may be beneficial for GDM treatment. Alteration in the microbial composition of the gut may potentially serve as a marker for identifying individuals at risk of GDM. Future studies combining metagenomics and metabonomics would be of value for improving our understanding of the roles of specific strains and metabolites in patients with GDM and supporting precise prevention and intervention strategies for GDM.
Gestational diabetes mellitus (GDM) places both the mother and offspring at high risk of complications. Increasing evidence suggests that the gut microbiota plays a role in the pathogenesis of GDM.
To confirm whether the gut microbiota is related to blood biochemical traits, particularly glucagon-like peptide-1 (GLP-1), in GDM patients.
To explore the correlation between the gut microbiota and blood biochemical traits.
The V4 region of the 16S ribosomal ribonucleic acid (rRNA) gene was sequenced based on the fecal samples of 35 pregnant women with GDM and was compared to that of 25 pregnant women with normal glucose tolerance (NGT).
The results showed that Ruminococcaceae_UCG-002, Ruminococcaceae_UCG-005, Clostridium_sen-su_stricto_1, and Streptococcus were more abundant in the NGT group than in the GDM group. Bacteroides and Lachnoclostridium were more abundant in the GDM group than in the NGT group. Spearman’s correlation analysis was performed to identify the relationships between bacterial genera and blood biochemical traits. Paraprevotella, Roseburia, Faecalibacterium, and Ruminococcaceae_UCG-002 were significantly negatively correlated with glucose. Ruminococcaceae_UCG-002 was significantly negatively correlated with HbA1c. Bacteroides was significantly positively correlated with glucose. Sutterella, Oscillibacter, and Bifidobacterium were significantly positively correlated with GLP-1. A random forest model showed that 20 specific genera plus glucose provided the best discriminatory power, as indicated by the area under the receiver operating characteristic curve (0.94).
The results of this study reveal novel relationships between the gut microbiome, blood biochemical traits, particularly GLP-1, and GDM status.
These findings suggest some genera are crucial for controlling blood glucose-related indices and may be beneficial for GDM treatment. Alteration in the microbial composition of the gut may potentially serve as a marker for identifying individuals at risk of GDM.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Microbiology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Houri H, Iran; Janvilisri T, Thailand S-Editor: Chen YL L-Editor: Wang TQ P-Editor: Chen YX
| 1. | Feig DS, Zinman B, Wang X, Hux JE. Risk of development of diabetes mellitus after diagnosis of gestational diabetes. CMAJ. 2008;179:229-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 310] [Cited by in RCA: 311] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 2. | Bonde L, Vilsbøll T, Nielsen T, Bagger JI, Svare JA, Holst JJ, Larsen S, Knop FK. Reduced postprandial GLP-1 responses in women with gestational diabetes mellitus. Diabetes Obes Metab. 2013;15:713-720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 3. | Kosinski M, Knop FK, Vedtofte L, Grycewiczv J, Swierzewska P, Cypryk K, Vilsbøll T. Postpartum reversibility of impaired incretin effect in gestational diabetes mellitus. Regul Pept. 2013;186:104-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 4. | Huang L, Thonusin C, Chattipakorn N, Chattipakorn SC. Impacts of gut microbiota on gestational diabetes mellitus: a comprehensive review. Eur J Nutr. 2021;60:2343-2360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 5. | Gomes AC, Bueno AA, de Souza RG, Mota JF. Gut microbiota, probiotics and diabetes. Nutr J. 2014;13:60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 251] [Cited by in RCA: 224] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 6. | Pitocco D, Di Leo M, Tartaglione L, De Leva F, Petruzziello C, Saviano A, Pontecorvi A, Ojetti V. The role of gut microbiota in mediating obesity and diabetes mellitus. Eur Rev Med Pharmacol Sci. 2020;24:1548-1562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 32] [Reference Citation Analysis (0)] |
| 7. | Dang F, Jiang Y, Pan R, Zhou Y, Wu S, Wang R, Zhuang K, Zhang W, Li T, Man C. Administration of Lactobacillus paracasei ameliorates type 2 diabetes in mice. Food Funct. 2018;9:3630-3639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 110] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 8. | Nyström T. The potential beneficial role of glucagon-like peptide-1 in endothelial dysfunction and heart failure associated with insulin resistance. Horm Metab Res. 2008;40:593-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 54] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 9. | Aaboe K, Krarup T, Madsbad S, Holst JJ. GLP-1: physiological effects and potential therapeutic applications. Diabetes Obes Metab. 2008;10:994-1003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 63] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 10. | Cai X, She M, Xu M, Chen H, Li J, Chen X, Zheng D, Liu J, Chen S, Zhu J, Xu X, Li R, Yang X, Li H. GLP-1 treatment protects endothelial cells from oxidative stress-induced autophagy and endothelial dysfunction. Int J Biol Sci. 2018;14:1696-1708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 68] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 11. | Kaska L, Sledzinski T, Chomiczewska A, Dettlaff-Pokora A, Swierczynski J. Improved glucose metabolism following bariatric surgery is associated with increased circulating bile acid concentrations and remodeling of the gut microbiome. World J Gastroenterol. 2016;22:8698-8719. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 76] [Cited by in RCA: 78] [Article Influence: 8.7] [Reference Citation Analysis (1)] |
| 12. | Ren M, Zhang H, Qi J, Hu A, Jiang Q, Hou Y, Feng Q, Ojo O, Wang X. An Almond-Based Low Carbohydrate Diet Improves Depression and Glycometabolism in Patients with Type 2 Diabetes through Modulating Gut Microbiota and GLP-1: A Randomized Controlled Trial. Nutrients. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 80] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 13. | Stenman LK, Waget A, Garret C, Briand F, Burcelin R, Sulpice T, Lahtinen S. Probiotic B420 and prebiotic polydextrose improve efficacy of antidiabetic drugs in mice. Diabetol Metab Syndr. 2015;7:75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 14. | Houlden A, Goldrick M, Brough D, Vizi ES, Lénárt N, Martinecz B, Roberts IS, Denes A. Brain injury induces specific changes in the caecal microbiota of mice via altered autonomic activity and mucoprotein production. Brain Behav Immun. 2016;57:10-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 267] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 15. | Bliss ES, Whiteside E. The Gut-Brain Axis, the Human Gut Microbiota and Their Integration in the Development of Obesity. Front Physiol. 2018;9:900. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 123] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 16. | Jackson A, Forsyth CB, Shaikh M, Voigt RM, Engen PA, Ramirez V, Keshavarzian A. Diet in Parkinson's Disease: Critical Role for the Microbiome. Front Neurol. 2019;10:1245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 96] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 17. | Hernández MAG, Canfora EE, Jocken JWE, Blaak EE. The Short-Chain Fatty Acid Acetate in Body Weight Control and Insulin Sensitivity. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 371] [Article Influence: 61.8] [Reference Citation Analysis (1)] |
| 18. | Vuik F, Dicksved J, Lam SY, Fuhler GM, van der Laan L, van de Winkel A, Konstantinov SR, Spaander M, Peppelenbosch MP, Engstrand L, Kuipers EJ. Composition of the mucosa-associated microbiota along the entire gastrointestinal tract of human individuals. United European Gastroenterol J. 2019;7:897-907. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 19. | Nagpal R, Neth BJ, Wang S, Craft S, Yadav H. Modified Mediterranean-ketogenic diet modulates gut microbiome and short-chain fatty acids in association with Alzheimer's disease markers in subjects with mild cognitive impairment. EBioMedicine. 2019;47:529-542. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 424] [Article Influence: 70.7] [Reference Citation Analysis (0)] |
| 20. | Anderson JR, Carroll I, Azcarate-Peril MA, Rochette AD, Heinberg LJ, Peat C, Steffen K, Manderino LM, Mitchell J, Gunstad J. A preliminary examination of gut microbiota, sleep, and cognitive flexibility in healthy older adults. Sleep Med. 2017;38:104-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 116] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 21. | Zhang Y, Chen T, Zhang Y, Hu Q, Wang X, Chang H, Mao JH, Snijders AM, Xia Y. Contribution of trace element exposure to gestational diabetes mellitus through disturbing the gut microbiome. Environ Int. 2021;153:106520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 42] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 22. | Ma S, You Y, Huang L, Long S, Zhang J, Guo C, Zhang N, Wu X, Xiao Y, Tan H. Alterations in Gut Microbiota of Gestational Diabetes Patients During the First Trimester of Pregnancy. Front Cell Infect Microbiol. 2020;10:58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 78] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 23. | Su Y, Wang HK, Gan XP, Chen L, Cao YN, Cheng DC, Zhang DY, Liu WY, Li FF, Xu XM. Alterations of gut microbiota in gestational diabetes patients during the second trimester of pregnancy in the Shanghai Han population. J Transl Med. 2021;19:366. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 24. | Mokkala K, Houttu N, Vahlberg T, Munukka E, Rönnemaa T, Laitinen K. Gut microbiota aberrations precede diagnosis of gestational diabetes mellitus. Acta Diabetol. 2017;54:1147-1149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 65] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 25. | Kuang YS, Lu JH, Li SH, Li JH, Yuan MY, He JR, Chen NN, Xiao WQ, Shen SY, Qiu L, Wu YF, Hu CY, Wu YY, Li WD, Chen QZ, Deng HW, Papasian CJ, Xia HM, Qiu X. Connections between the human gut microbiome and gestational diabetes mellitus. Gigascience. 2017;6:1-12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 223] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 26. | Liu H, Pan LL, Lv S, Yang Q, Zhang H, Chen W, Lv Z, Sun J. Alterations of Gut Microbiota and Blood Lipidome in Gestational Diabetes Mellitus With Hyperlipidemia. Front Physiol. 2019;10:1015. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 76] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 27. | Wickens KL, Barthow CA, Murphy R, Abels PR, Maude RM, Stone PR, Mitchell EA, Stanley TV, Purdie GL, Kang JM, Hood FE, Rowden JL, Barnes PK, Fitzharris PF, Crane J. Early pregnancy probiotic supplementation with Lactobacillus rhamnosus HN001 may reduce the prevalence of gestational diabetes mellitus: a randomised controlled trial. Br J Nutr. 2017;117:804-813. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 128] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 28. | Laitinen K, Poussa T, Isolauri E. Probiotics and dietary counselling contribute to glucose regulation during and after pregnancy: A randomised controlled trial. Br J Nutr. 2009;101:1679-1687. [RCA] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 237] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 29. | Crusell MKW, Hansen TH, Nielsen T, Allin KH, Rühlemann MC, Damm P, Vestergaard H, Rørbye C, Jørgensen NR, Christiansen OB, Heinsen FA, Franke A, Hansen T, Lauenborg J, Pedersen O. Gestational diabetes is associated with change in the gut microbiota composition in third trimester of pregnancy and postpartum. Microbiome. 2018;6:89. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 219] [Cited by in RCA: 302] [Article Influence: 43.1] [Reference Citation Analysis (0)] |
| 30. | Huthmacher JA, Meier JJ, Nauck MA. Efficacy and Safety of Short- and Long-Acting Glucagon-Like Peptide 1 Receptor Agonists on a Background of Basal Insulin in Type 2 Diabetes: A Meta-analysis. Diabetes Care. 2020;43:2303-2312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 57] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 31. | Wang Z, Saha S, Van Horn S, Thomas E, Traini C, Sathe G, Rajpal DK, Brown JR. Gut microbiome differences between metformin- and liraglutide-treated T2DM subjects. Endocrinol Diabetes Metab. 2018;1:e00009. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 32. | Ferrocino I, Ponzo V, Gambino R, Zarovska A, Leone F, Monzeglio C, Goitre I, Rosato R, Romano A, Grassi G, Broglio F, Cassader M, Cocolin L, Bo S. Changes in the gut microbiota composition during pregnancy in patients with gestational diabetes mellitus (GDM). Sci Rep. 2018;8:12216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 171] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 33. | Yao Y, Yan L, Chen H, Wu N, Wang W, Wang D. Cyclocarya paliurus polysaccharides alleviate type 2 diabetic symptoms by modulating gut microbiota and short-chain fatty acids. Phytomedicine. 2020;77:153268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 153] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 34. | Fouré M, Dugardin C, Foligné B, Hance P, Cadalen T, Delcourt A, Taminiau B, Daube G, Ravallec R, Cudennec B, Hilbert JL, Lucau-Danila A. Chicory Roots for Prebiotics and Appetite Regulation: A Pilot Study in Mice. J Agric Food Chem. 2018;66:6439-6449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 35. | Zhao D, Zhu H, Gao F, Qian Z, Mao W, Yin Y, Tan J, Chen D. Antidiabetic effects of selenium-enriched Bifidobacterium longum DD98 in type 2 diabetes model of mice. Food Funct. 2020;11:6528-6541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |