Published online Apr 15, 2021. doi: 10.4239/wjd.v12.i4.366
Peer-review started: January 16, 2021
First decision: January 24, 2021
Revised: January 30, 2021
Accepted: February 18, 2021
Article in press: February 18, 2021
Published online: April 15, 2021
Processing time: 82 Days and 16.8 Hours
According to the developmental origin of health and disease concept, the risk of many age-related diseases is not only determined by genetic and adult lifestyle factors but also by factors acting during early development. In particular, maternal obesity and neonatal accelerated growth predispose offspring to overweight and type 2 diabetes (T2D) in adulthood. This concept mainly relies on the developmental plasticity of adipose tissue and pancreatic β-cell programming in response to suboptimal milieu during the perinatal period. These changes result in unhealthy hypertrophic adipocytes with decreased capacity to store fat, low-grade inflammation and loss of insulin-producing pancreatic β-cells. Over the past years, many efforts have been made to understand how maternal obesity induces long-lasting adipose tissue and pancreatic β-cell dysfunction in offspring and what are the molecular basis of the transgenerational inheritance of T2D. In particular, rodent studies have shed light on the role of epigenetic mechanisms in linking maternal nutritional manipulations to the risk for T2D in adulthood. In this review, we discuss epigenetic adipocyte and β-cell remodeling during development in the progeny of obese mothers and the persistence of these marks as a basis of obesity and T2D predisposition.
Core Tip: According to the developmental origin of health and disease concept, maternal obesity and neonatal accelerated growth predispose offspring to metabolic diseases. White adipose tissue and pancreatic β-cells are key targets of developmental programming, although the underlying mechanisms remain elusive. Human and rodent studies have contributed to decipher the role of epigenetic mechanisms in the transgenerational inheritance of obesity and type 2 diabetes (T2D). In this review, we discuss the current understanding of the link between obesogenic maternal nutritional environment, developmental epigenetic adipocyte and β-cell remodeling and predisposition to obesity and T2D later in life.
- Citation: Lecoutre S, Maqdasy S, Breton C. Maternal obesity as a risk factor for developing diabetes in offspring: An epigenetic point of view. World J Diabetes 2021; 12(4): 366-382
- URL: https://www.wjgnet.com/1948-9358/full/v12/i4/366.htm
- DOI: https://dx.doi.org/10.4239/wjd.v12.i4.366
Obesity and related metabolic diseases have doubled since 1980 and reached epidemic proportions over the past decades[1]. Obesity and expansion and dysfunction of white adipose tissue (WAT) are major drivers of type 2 diabetes (T2D), through induction of insulin resistance[2]. Insulin resistance is observed locally in hypertrophic adipocytes long before glucose intolerance develops[3]. The inability of further WAT expansion accelerates fat spillover, leading to ectopic fat deposition in skeletal muscle and liver and insulin resistance in those tissues[4,5]. Chronic inflammation in WAT is also considered a crucial causal factor for the development of insulin resistance and T2D in obese individuals[6,7]. Indeed, obesity-induced WAT remodeling provides intrinsic signals capable of initiating a local inflammatory response that may spread into the circulation, resulting in systemic insulin resistance and T2D[8]. An obesogenic environment and lipotoxic conditions also result in dysfunction and loss of insulin-producing pancreatic β-cells due to dedifferentiation, transdifferentiation, or death. These phenotypic alterations ultimately hamper insulin secretion[9].
Although genetics account for the variation in body weight and T2D, the dramatic rise in their incidence cannot be solely explained by genetic predisposition. Hence, environmental factors such as overnutrition, sedentary lifestyle, xenobiotics, and chemical exposure appear to be major contributors to the rapidly increasing prevalence[1]. In particular, studies in both humans and animal models suggest that excess nutrient supply in the fetal or neonatal period result in long-term programming of body weight set-point and predispose individuals to obesity and T2D[10-12]. Thus, the developmental origin of obesity and T2D perpetuate the vicious cycle of metabolic diseases across generations[12]. Among different mechanisms of transmission, epigenetics has emerged as a very important determinant[13]. In this review, we will present data on how maternal obesity programs T2D risk via epigenetic mechanisms by focusing on changes in WAT and β-cell physiology.
The developmental origin of health and disease concept (DOHaD) proposes a link between environmental challenges during early stages of growth and predisposition to metabolic disorders later in life. In particular, this concept states that suboptimal nutritional environment (i.e., under or overnutrition) during the perinatal period can program or imprint the development of key tissues that play a central role in regulating energy homeostasis. Later in life, it might permanently determine physiological responses and ultimately produce energy balance dysfunction and metabolic diseases, such as obesity and T2D[10].
Originally called the Barker hypothesis or fetal programming, this concept arises from epidemiological studies. Indeed, David Barker was the first to report that intrauterine growth retardation (IUGR) and low birth weight were associated with increased risk of metabolic syndrome-related diseases during adulthood[10]. As illustrated by the Dutch famine of 1944-45, offspring of mothers exposed to the famine presented with low birth weight associated with an increase in the incidence of dyslipidemia, obesity, and T2D later in life[13]. More recently, adults born during the Chinese famine of 1959-61 were also predisposed to overweight and T2D, constituting a major contributor to China’s current T2D epidemic[14]. David Barker proposed the notion of a “thrifty phenotype,” which placed an emphasis on development, arguing that nutritionally inadequate conditions in pregnancy not only affected fetal growth but also induced permanent changes in insulin secretory capacity and in glucose metabolism[15]. In humans with low birth weights, postnatal hypercaloric nutrition, and more specifically rapid catch-up growth, are also important accelerators in the etiology of adult-onset diseases[16]. As stated by the predictive adaptive response concept, the degree of mismatch between the pre-and postnatal environments is the key paradigm in developmental metabolic programming[17].
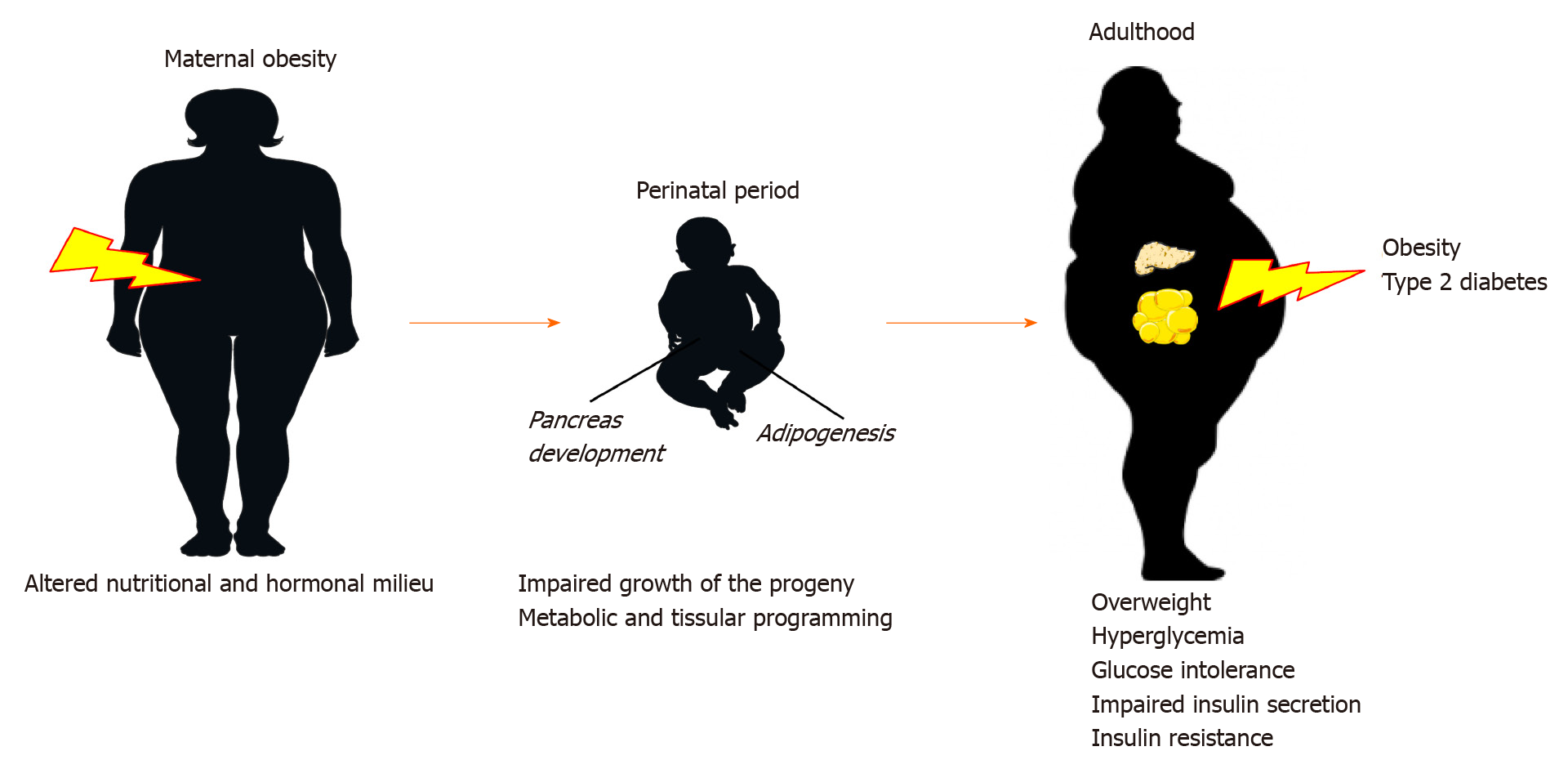
This concept has evolved from undernutrition to overnutrition. As shown in Figure 1, epidemiological and clinical studies have reported that individuals exposed to maternal overnutrition and/or obesity during pregnancy and lactation are also predisposed to increased risk of metabolic syndrome-related diseases later in life[18]. Subsequent meta-analyses have highlighted birth weight as a predictor of obesity and T2D. A U-shaped curve was proposed to explain the relationship between birth weight (a marker of fetal nutritional exposure) and the propensity to develop obesity in adulthood. Hence, it is currently well accepted that individuals born small or large (i.e., low or high body fat percentage) have similar increased risks of obesity and related diseases later in life[19]. Over the past decades, animal studies have confirmed that maternal obesity during gestation and lactation, gestational diabetes, and accelerated growth of neonates predispose offspring to obesity and T2D[20,21].
Two main questions arise about the DOHaD concept. First, what is the basis of the persistent cellular memory of a developmental event, even when the initial stimulus has disappeared and despite continuous cellular turnover? Second, how two opposite maternal nutritional manipulations (under- vs overnutrition) may result in similar outcomes in adult offspring. Little is known about the cellular and molecular mechanisms underlying the phenomenon known as developmental programming. Among them, epigenetic modifications are likely to play a key role in the heritability of obesity and T2D[12,22,23].
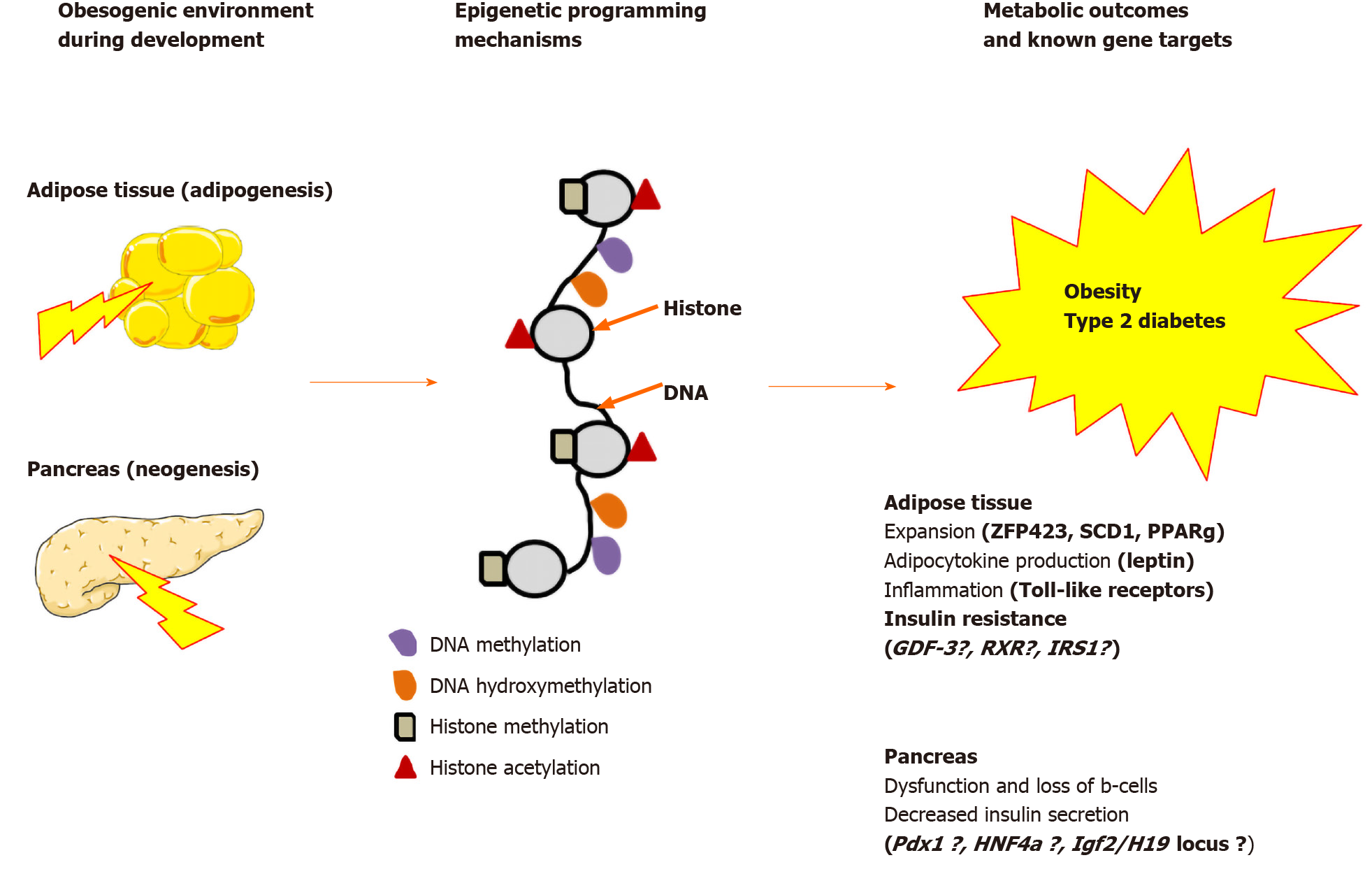
Epigenetics can be defined as somatically heritable states of gene expression resulting from changes in chromatin structure without alterations in the DNA sequence[24]. Epigenetic modifications are transmitted from one cell generation to the next (mitotic inheritance) and can also be transmitted across generations (meiotic inheritance)[25]. These processes include DNA (hydroxy) methylation, histone post-translational modifications (PTMs) such as acetylation, phosphorylation, ubiquitination, and sumoylation, and noncoding RNA that regulates gene expression at both transcriptional and post-transcriptional levels[26]. DNA methylation, which results from the transfer of a methyl group, by DNA methyltransferase (DNMT), takes place at cytosines, mainly in the CpG islands, to form 5-methylcytosine (5mC). It serves to establish long-term gene silencing[27]. 5-hydroxymethylcytosine (5hmC) is another important cytosine modification catalyzed by the enzymes of the ten-eleven translocation methylcytosine dioxygenase (TET) family[28]. It serves as an intermediate for demethylation of 5hmC and is enriched in active transcriptional regulatory regions. Based on the histone code hypothesis, PTMs play crucial roles in controlling gene expression by adapting the local chromatin architecture and accessibility, allowing the recruitment of partners that modulate the transcriptional machinery. For example, acetylation of histone H3 lysine residues (H3Kac) and methylation of H3K4 (H3K4me1/3) are associated with active transcription while methylation of H3K9 (H3K9me3) generally indicates silenced chromatin. These histone PTMs are catalyzed by various enzymes including histone acetyltransferase (HAT), histone deacetylase (HDAC), histone methyltransferases (MTs)/demethylases[29,30]. Thus, the term epigenome refers to the combination of all chromatin modifications (i.e., DNA methylation and PTMs) of a given cell type in an individual.
Histone-modifying enzyme activity is sensitive to cellular energy status and hormonal response. In particular, it is highly dependent on intermediary metabolites that act as enzyme cofactors[6,31]. For example, HATs use acetyl-coenzyme A (CoA), histone MTs use S-adenosyl methionine (SAM), HDACs can use nicotinamide adenine dinucleotide, and histone demethylases can use flavin adenine dinucleotide or symbol-ketoglutarate as coenzymes[29,30]. Interestingly, sirtuin 1 is a nutrient-sensing HDAC and is associated with the risk of metabolic syndrome including T2D[32]. Hence, modifications of nutritional and hormonal milieu in offspring from obese dams may affect the developmental program of adipocyte and β-cells by impairing chromatin remodeling activities and DNA methylation[30]. Several studies have also reported the role of noncoding RNAs in WAT and pancreas development as well as in adipocyte and β-cell differentiation and function[33,34].
However, here we emphasize DNA methylation and chromatin modifications in progenitor cells, whose inappropriate editing during gestation and lactation may serve as a deleterious memory of exposure to a maternal obesogenic environment. The persistence of these marks throughout life and across generations may program permanent changes in gene expression and may account for inheritance of metabolic diseases.
Based on the DOHaD concept, there are two main reasons why adipocyte and β-cells are key targets of perinatal programming. First, numerous studies have shown that manipulation of epigenetic machinery can alter cell fate and identity as well as cell-type-specific gene expression during both adipogenesis[33] and β-cell neogenesis (i.e., adipocyte and β-cell formation, respectively)[35-41]. Epigenetic mechanisms are also known to play a crucial role in the control of β-cell identity and plasticity (i.e., dedifferentiation and transdifferentiation) in adulthood[42-45]. Second, adipogenesis and β-cell neogenesis occur primarily during the second part of gestation in rodents, accelerate during early postnatal life, and remain active after weaning. The deleterious effects of maternal obesity operate during periods of development in which precursor cells are plastic (i.e., possess the flexibility to adapt to the changing microenvironment) and where epigenetic remodeling is particularly dynamic and sensitive to the nutritional and hormonal milieu[12,19,46,47]. Hence, disturbance of the developmental program might result in severe dysfunctions in fetal pancreatic β-cells and adipose tissue and a profound perturbation of systemic glucose homeostasis in adulthood.
WAT exists in multiple locations in the body and has two major subtypes, visceral and subcutaneous[12,48]. Unlike visceral WAT (vWAT), the metabolic plasticity of subcu-taneous WAT (sWAT) is associated with increased insulin sensitivity and decreased occurrence of T2D[49]. In rodents, adipogenesis is particularly active during the perinatal period. These processes occur primarily during the last week of gestation (the first fat cells appear between the fourteenth and the eighteenth days of gestation)[50-52] and accelerate during early postnatal life until pups are weaned. However, sWAT develops during late gestation and lactation whereas vWAT formation is mainly initiated after birth[50]. The developmental origin of adipocytes remains elusive. Cell lineage tracing, adipocyte precursor fate studies and subsequent molecular analysis demonstrated a heterogeneity of the adipose lineage between fat depots, but also within individual adipose depots, in terms of cell origin, spatio
Adipocytes are derived from multipotent mesenchymal stem cells (MSCs), which are first committed to the adipogenic lineage and then transformed into preadipocytes. This phase is then followed by terminal differentiation during which preadipocytes become mature adipocytes that develop the ability to store lipids in a large monolocular lipid droplet and display endocrine properties. The process of adipocyte differentiation involves three defined steps. The first step is the commitment of MSCs to the adipocyte lineage. The second step is mitotic clonal expansion involving DNA replication and duplication of cells. The third step is terminal differentiation, which involves transcriptional factors such as CCAAT-enhancer-binding proteins (C/EBP) , β and δ, peroxisome proliferator-activated receptor γ (PPARγ), and significant expression of adipocyte-specific genes such as adiponectin and leptin[55-58].
Our understanding of adipogenesis comes mainly from studies using preadipocyte cells such as 3T3-L1[33,59]. Adipogenesis involves a dynamic reorganization of the chromatin landscape at specific developmental stages that is associated with the recruitment of multiple transcription factors controlling the expression of adipocyte-specific genes. This process is regulated through the remodeling of cell-specific histone marks and DNA (hydroxy)methylation[60,61]. Thus, in undifferentiated adipocytes, the master adipogenic transcription factors, zinc finger protein 423 (ZFP423), C/EBPβ, and PPARγ, are in a poised state owing to the bivalent presence of the active H3K4me3 and repressive H3K9me3 marks in their promoters[59]. ZFP423 was identified as crucial for adipogenesis by promoting PPARγ expression[62]. During the early phase of adipocyte differentiation, the reorganization of the chromatin structure and chromatin opening along with the recruitment of several early transcription factors, such as C/EBPβ/δ, coincide with the removal of repressive histone marks (i.e., H3K9me3) and the enrichment of active chromatin marks, including H3K27ac, and H3K4me3, as well as DNA hydroxymethylation, in their promoters[60]. During differentiation, chromatin remodeling primes genomic regions to allow the expression of CEBPα and PPARγ, their specific binding on the chromatin as well as their target genes[60]. For instance, H3K4 MT mixed-lineage leukemia (MLL) 3 and MLL4 are recruited by C/EBPβ to activate the enhancers of C/EBPα and PPARγ and induce C/EBPα and PPARγ expression during adipogenesis. Once induced, C/EBPα and PPARγ recruit MLL3/MLL4 to further activate enhancers of downstream target genes[63].
The pancreas is derived from the ventral and dorsal endoderm, which stepwise differentiate into exocrine and endocrine lineages. Most of our current understanding of neogenesis is derived from rodent studies. In the developing pancreas, cell fate commitment towards specific endocrine subtypes is controlled by a complex orchestrated expression of specific transcription factors. In rodents, pancreas development undergoes two transitionary periods, a first wave transition (from embryonic days E9.5-E12.5) during which the endocrine cells that are formed are primarily -cells or multi-hormonal cells and a second wave transition (from E12.5 to birth), which is the main period of endocrine cell formation including β-cells[34,64-66].
Pancreatic endocrine cells arise from multipotent endocrine progenitor-precursors (EPs) that express PDX1. The master regulator of EP formation and differentiation is Ngn3, which is indispensable for endocrine cell formation. Higher expression levels of Arx and Pax4 favor formation of - and β/δ-cells, respectively. Differentiation toward β-cell fate depends on the expression of several other transcription factors such as FOXA2, NKX6-1, NEUROD1, NKX2-2 and MAFA which are important for the establishment and maintenance of β-cell identity. In rodents, embryonic β-cells appear during the perinatal period, are immature, highly proliferative, and plastic but respond poorly to glucose stimulation[67,68]. After birth, β-cells follow a biphasic pattern of maturation[69]. The first 2 wk of life define a first maturation wave characterized by active proliferation that results in an increase in the β-cell mass, a characteristic that is progressively lost. The second wave of maturation coincides with the third week of life and the weaning period[70]. During that time, β-cells differentially regulate metabolic pathways and acquire physiological functions, such as glucose-stimulated insulin secretion in response to extracellular glucose[71,72]. In rodents, the postnatal maturation of β-cells is driven by weaning (dietary change from high-fat milk to high-carbohydrate chow), and studies have suggested that microRNAs (miRNAs) have a central role in regulating postnatal β-cell maturation[70,73].
Several studies support the notion that the development and heterogeneity of EPs are regulated at the chromatin level. Our understanding of epigenetic mechanisms involved in β-cell formation and maintenance of identity comes from in vitro differentiation protocols of pluripotent stem cells[74] and studies using inhibitors of epigenetic enzymes or employing mice deficient models for different classes of epigenetic modifiers. First, the use of HDAC inhibitors on embryonic pancreas explants resulted in increased numbers of endocrine progenitors and β-cells whereas embryonic pancreatic overexpression of HDAC reduced the β-cell mass[35]. Generation of HDAC5- or -9-deficient mice increased the insulin-secreting β-cell mass[36,44]. Second, deletion of Jmjd3, a histone demethylase for the repressive H3K27me3 mark at the pancreatic progenitor stage, impaired the efficiency of endocrine cell fate transition and subsequent islet formation in mice[38]. Third, numerous genes critical for β-cell function that are bivalently marked in -cells by both active H3K4me3 and repressive H3K27me3 histone modifications are monovalently marked by active H3K4me3 in β-cells. This bivalency suggests a plastic epigenetic state for key β-cell genes in α-cells indicating a paused state with potential for activation[45]. To maintain β-cell identity, genes important for -cell function have to be actively repressed by the DNA methy
Clinical observations among human and animal studies have determined two major determinants for metabolic health and insulin sensitivity. On one hand, the ability of sWAT to store excess fat (i.e., storage capacity) rather than allowing it to accumulate in ectopic depots such as liver, muscle and vWAT, is of prime importance. On the other hand, the ability to recruit and differentiate new adipocytes (i.e., activation of adipogenesis resulting in increased storage capacity) in sWAT also reduces risk of metabolic diseases including T2D in overweight individuals[78]. In obesity, WAT expands either by hyperplasia (increase in adipocyte number) or hypertrophy (increase in adipocyte size), where the latter is associated with insulin resistance and inflammation.
Numerous studies have shown that maternal obesity modifies the expansion capacity of WAT in offspring throughout life. In particular, the activity of key adipogenic transcription factors was impacted by maternal obesity during development, leading to impaired expandability of WAT in offspring[46,47,79]. One of the most well-studied mechanisms is the modulation of Zfp423 gene expression and activity in rodents. Zfp423 expression defines committed preadipocytes, and its expression persists throughout adipocyte differentiation[62,80]. Inactivation of Zfp423 during WAT development results in arrested differentiation, specifically of sWAT[81]. As a key developmental gene, Zfp423 promoter has a bivalent region with enrichment of both H3K27me3 and H3K4me3 histone marks[82]. Offspring of obese mice were overweight, with an increased fat mass that was correlated with persistently elevated Zfp423 activity in WAT[83]. Developmental epigenomic remodeling of these marks in the Zfp423 promoter accounts for persistent higher Zfp423 expression later in life. During the second part of gestation (E14.5) in which adipogenic activity was elevated, the H3K27me3 histone mark and DNA methylation were lower in the Zfp423 promoter, whereas the H3K4me3 histone mark was higher in the fetuses of obese dams[83]. At weaning, WAT neonates still showed elevated Zfp423 activity and exacerbated adipogenesis resulting in increased numbers of adipocytes and adiposity[83-85]. Overweight adult mice showed persistent increased gene expression with DNA hypomethylation in the Zfp423 promoter despite impaired hyperplasia when fed a high-fat (HF) diet[84].
A possible interpretation is that individuals from obese mothers with dysfunctional WAT (i.e., hypertrophic adipocytes), inflammation, and insulin resistance displayed a failure of WAT plasticity and inappropriate expansion of the adipose progenitors in sWAT. This might be due to maternal obesogenic environment and premature exhaustion of the stock of resident adipocyte progenitors during development that favors hypertrophy vs hyperplasia to store excess energy later in life[12]. In line with these findings, human MSC from the umbilical cords of infants born to obese mothers exhibit a greater potential for adipogenesis[86]. Godfrey et al[87], also reported a strong association between methylation of the retinoid-X-receptor α (RXRα) promoter region from DNA extracted from the umbilical cord of infants born to mothers with low carbohydrate intake during pregnancy and the degree of adiposity 6-9 years later. High methylation in the RXRα promoter reduces gene expression and alters insulin sensitivity and glucose metabolism in differentiated adipocytes[88].
Another well-studied epigenetic mechanism is the modulation of Pparγ expression and activity in offspring of obese dams. The regulation of Pparγ expression in WAT via DNA methylation and histone modification at its promoter region is well illustrated[89-91]. Decreased Pparγ2 expression in the WAT of adult rat offspring is generally associated with tissue dysfunction[92]. Several studies suggest that the reduction in Pparγ2 expression may be due to epigenomic remodeling occurring during WAT development. In weanling rats from obese dams, reduced Pparγ2 mRNA levels were observed together with DNA hypermethylation and decreased enrichment of H3ac and H3K4me3 active marks in the Pparγ2 promoter region. In adulthood, DNA hypermethylation of the Pparγ2 promoter and the reduction of Pparγ2 mRNA expression levels were still observable[92]. It is tempting to speculate that the decreased expression of the master regulator of adipogenesis and lipid storage is an adaptative mechanism to avoid further deleterious adipocyte hypertrophy[93]. Interestingly, adult offspring from dams fed an HF diet only during lactation were predisposed to obesity, with increased expression of stearoyl-CoA desaturase 1 (SCD1), a key enzyme of lipid storage. Higher Scd1 gene expression was associated with reduced DNA methylation in the Scd1 promoter surrounding a Pparγ-binding region[94].
Maternal obesity also predisposes to the development of a chronic low-grade proinflammatory state associated with insulin resistance in WAT offspring[12,95,96]. Several studies showed that elevated proinflammatory adipocytokine production such as leptin, WAT inflammation, and macrophage infiltration can be transmitted across generations via epigenetic mechanisms. For instance, obesity-prone offspring from obese rats displayed elevated leptin gene expression, hyperleptinemia, and adipocyte hypertrophy in WAT[79,97]. During lactation, increased leptin gene expression arises from higher DNA hydroxymethylation and active histone H3K4me1/H3K27ac marks in an enhancer region[97]. These histone marks persisted in the WAT of adult offspring in hypertrophic adipocytes[97]. Maternal obesity in mice also results in persistent hypermethylation of the. H4K20 histone mark in the promoter region in offspring that persists across generations[98-100]. Interestingly, multigenerational HF diet feeding in female mice resulted in gradually increased WAT weight, proinflammatory markers, and immune cell infiltration associated with a gradual decrease in DNA methylation of inflammation-associated genes (i.e., Toll-like receptors) in WAT across generations (up to F2)[12]. However, the effects of a maternal HF diet vs maternal obesity on offspring WAT inflammation and glucose homeostasis remain to be determined[12,101].
It is interesting to note that programmed changes in miRNAs may also account for both adipose tissue expandability and insulin resistance in offspring from malnourished dams. On the one hand, an increase in miR-483 and parallel reduction in growth differentiation factor 3 have been reported in WAT from the offspring of dams fed a low protein (LP) diet[102]. That may lead to a reduction in the expandability of WAT, and therefore, increased ectopic fat deposition. Similar observations were also described in WAT from individuals with low birthweights, showing conservation of this programmed mechanism. On the other hand, maternal obesity resulted in higher miR-126 levels in WAT of mice offspring which led to reduced expression of key insulin signaling proteins, including insulin receptor substrate-1[103,104].
The pancreas is an organ that is particularly sensitive to nutritional imbalance during intrauterine organogenesis[34]. Rodent models have been mainly used to investigate the effects of maternal obesity on islet development and function and to decipher underlying programming mechanisms. In rodent models, maternal obesity results in decreased β-cell mass and insulin secretion at birth[105]. Islets from offspring born to obese or HF diet-fed mothers have decreased pancreatic insulin content, Pdx1 expression in adult islets[106,107] with remodeling of the architecture of the islets, characterized by an increase of α-cells in the centers of pancreatic islets[108,109]. Interestingly, pancreatic β-cell dysfunctions occur in offspring from obese dams in a sex-dependent manner. Very few data are available regarding the effects of maternal obesity on the β-cell programming in offspring in terms of epigenetics. However, consistent with the DOHaD concept, several studies highly suggest that maternal nutritional manipulations have a transgenerational influence on β-cell development and function through long-lasting effects on the offspring epigenome, predisposing to T2D later in life.
The first evidence comes from a rat model of IUGR caused by bilateral uterine artery ligation leading to a lower body weight at birth[110]. This deficient intrauterine environment affects fetal development through progressive and permanent dysregulation of gene expression and function of β-cells resulting in the development of T2D. For instance, adult IUGR rats exhibited a persistent reduction of Pdx1 expression levels in β-cells associated with chromatin remodeling throughout development. In the fetus, prior to the onset of T2D, deacetylation of histones H3 and H4 and recruitment of Hdac1 in the promoter of Pdx1 were associated with decreased mRNA expression levels. Loss of acetylation was accompanied by loss of binding of the key transcription factor Usf-1. In neonates, the active histone H3K4me3 mark was lower, and the repressive histone H3K9me2 mark was higher, at the hypomethylated Pdx1 promoter of IUGR islets. Once T2D occurs in adulthood, the promoter was hypermethylated, resulting in permanent silencing of the Pdx1 gene[111]. To achieve a more complete picture of DNA methylation changes, Thompson et al[112] have generated a whole DNA methylation map of the rat genome in IUGR pancreatic islet cells. They showed that IUGR changes cytosine methylation at approximately 1400 loci in IUGR rats before the onset of diabetes. Interestingly, epigenetic dysregulation occurred preferentially at intergenic sequences close to genes regulating cellular processes that were impaired in IUGR islets, including β-cell proliferation, insulin secretion, and apoptosis. The modifications of the DNA methylation were associated with changes in mRNA expression levels[112]. Consistent with this notion, early postnatal overnutrition (newborns suckled in a small litter) accelerates aging-associated epigenetic DNA hypermethylation in dysfunctional pancreatic islets of weaned and adult offspring[113].
Data obtained from a rat model of maternal protein restriction (LP) during pregnancy and lactation that resulted in IUGR reinforce this hypothesis. Maternal LP altered the pancreatic structure, islet areas and quantities and resulted in abnormal morphological changes during pancreatic development[114]. LP offspring had normal glucose tolerance in young adult life but suffered from an age-dependent loss of glucose tolerance and developed T2D in adulthood[115]. The authors showed that the Hnf4a gene encoding a transcription factor required for β-cell differentiation, and which has been implicated in the etiology of T2D, is epigenetically regulated by maternal LP diet and aging in rat islets[116]. IUGR offspring have progressive epigenetic silencing at the promoter-enhancer regions (i.e., decreased active H3ac and H3K4me1/3 and increased repressive H3K9me2 and H327me3 histone marks), which weakens their interaction and results in a permanent reduction in Hnf4a expression[116]. It is interesting to note that changes in DNA methylation also take place in the pancreatic islets of mice born to mothers with gestational diabetes mellitus. In the offspring, hypermethylation of the imprinted Igf2/H19 (insulin-like growth factor-2) locus in pancreatic islets may account, at least in part, for impaired islet ultrastructure and function that has been shown to be transmitted to subsequent generations[117]. Although precise mechanisms linking offspring β-cell programming and maternal obesity are lacking, it is tempting to speculate that the developmental pathways of pancreatic endocrine lineages could be epigenetically reprogrammed through similar mechanisms, ultimately resulting in impaired β-cell number and plasticity.
Growing evidence suggests that epigenetic dysregulation of key metabolic genes implicated in adipocyte and β-cell development and function in offspring contribute to developmental programming of T2D[118]. As summarized in Figure 2, chromatin remodeling and changes in DNA methylation may account, at least in part, for the molecular basis of intergenerational effects. The establishment of epigenetic marks on somatic stem cells will give rise to mature adipocytes and β-cells of the fetus (F1) as well as the germline of the fetus (the future F2). The persistence of these marks into adulthood may program obesity-associated insulin resistance up to F2. Consistent with this notion, maternal obesity before and throughout pregnancy and lactation results in altered development of the pancreas in F1 and F2 mouse offspring[119]. However, it is less clear so far how developmentally induced epigenetic modifications may persist beyond the F2. Only persistent phenotypes in the F3 and subsequent generations represent true transgenerational epigenetic inheritance, as they are stably transmitted through the F2 germline, which is not directly exposed to the initial maternal nutritional insult[120,121]. While the mitotic heritability of epigenetic marks is widely accepted, the existence and role of transgenerational epigenetic inheritance in mammals remain controversial. Most of our knowledge concerning germ cell formation in mammals comes from mouse models. In mice, primordial germ cells undergo a global epigenetic remodeling during germline development and following fertilization resulting in a complete resetting of the epigenetic memory arising from the parents and the establishment of sex-specific gamete identity. This event limits the stable transmission of epigenetic marks acquired during development or imposed by the environment from one generation to the next. However, In rodents, it has been clearly demonstrated that the epigenetic states are not entirely reprogrammed. Some imprinted genes escape demethylation processes resulting in intergenerational inheritance[122].
Increasing evidence suggests that epigenetic modifications of the sperm and the spermatozoa are key players in transgenerational epigenetic inheritance in subsequent F2 male generations and beyond[98,123-127]. Indeed, a significant increase in body size, adiposity and reduced insulin sensitivity were reported in F1 and F2 after maternal obesity through both maternal and paternal lineages[128]. However, in the F3 generation, those metabolic alterations were only displayed by females and only via paternal lineage in the absence of any further nutritional stimulus[129,130]. Although the implication of DNA methylation and histone modifications cannot be totally excluded, noncoding RNAs have emerged as an alternative mode of transgenerational epigenetic inheritance from the male germline[129-131].
To our knowledge, there are very few data providing evidence for transgenerational epigenetic inheritance in humans. Human epidemiological studies suggest that grandparental overnutrition increases the rates of diabetes and cardiovascular diseases risk in F2[132]. Increased risk for obesity and related metabolic diseases was observed in children whose parents were of normal weight but whose grandparents were obese[133].
Consistent with the DOHaD concept, epigenetic research conducted on inter- and transgenerational inheritance of obesity and T2D has shed light on new molecular mechanisms. An important challenge for the scientific community is that the solutions to the transmission of obesity and T2D beyond the scope of health system prevention programs. In this context, a better understanding of the underlying mechanisms involved in the epigenetic regulation of adipocyte and β-cells programming becomes a necessity. Of high interest, deciphering the epigenetic mechanisms leading to enhanced β-cell mass, β-cell proliferation, and function defects during T2D should guide toward the identification of novel therapeutic targets.
The reversible nature of epigenetic modifications, together with recent advances in epigenome-targeting methods, provide a new opportunity for alternative epigenetic therapies[134,135]. Indeed, targeting epigenetic machinery during early development is an attractive way to reduce adverse outcomes of maternal obesity. On one hand, it is crucial to better understand when and how an obesogenic environment may affect the fate of stem cells during adipocyte and β-cell development via epigenetic mechanisms. In the near future, determining the nature and kinetics of recruitment of enzymes controlling the PTMs of histones and DNA methylation involved in the complex transcriptional program will be needed. High-throughput DNA sequencing approaches in epigenomics for genome-wide profiling of global DNA methylation and histone modifications should allow determining changes in chromatin landscape throughout development. As a follow-up, high-throughput CRISPR-Cas9 technologies for epigenome editing might allow efficient targeting of key epigenetic marks as therapeutic option[136].
On the other hand, the use of natural compounds or pharmacological agents leading to DNA methylation and histone modifications, such as DNMT inhibitors and HDAC inhibitors have been already validated as an innovative approach[132]. Dietary supplementation of methyl donors during perinatal period was found to alleviate the adverse consequences of maternal malnutrition[137,138]. For instance, pharmacological modulation of epigenetic enzymatic machineries via drugs to improve β-cell functionality has already been recognized as promising new avenue for future therapeutic purposes[135,139-141]. Most important, targeting transient and reversible epigenetic modifications during early stages of life, either by genetic or pharma-cological means, provides a promising therapeutic way to counteract adverse programming effects on maternal obesity in the progeny.
Conflict of interest statement: The authors declare that they have no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Manuscript source: Invited manuscript
Specialty type: Endocrinology and metabolism
Country/Territory of origin: France
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Athyros VG S-Editor: Gao CC L-Editor: Filipodia P-Editor: Ma YJ
| 1. | González-Muniesa P, Mártinez-González MA, Hu FB, Després JP, Matsuzawa Y, Loos RJF, Moreno LA, Bray GA, Martinez JA. Obesity. Nat Rev Dis Primers. 2017;3:17034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 593] [Cited by in RCA: 802] [Article Influence: 100.3] [Reference Citation Analysis (0)] |
| 2. | Mejhert N, Rydén M. Novel aspects on the role of white adipose tissue in type 2 diabetes. Curr Opin Pharmacol. 2020;55:47-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 3. | Czech MP. Mechanisms of insulin resistance related to white, beige, and brown adipocytes. Mol Metab. 2020;34:27-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 152] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 4. | Pellegrinelli V, Carobbio S, Vidal-Puig A. Adipose tissue plasticity: how fat depots respond differently to pathophysiological cues. Diabetologia. 2016;59:1075-1088. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 243] [Cited by in RCA: 298] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 5. | Virtue S, Vidal-Puig A. Adipose tissue expandability, lipotoxicity and the Metabolic Syndrome--an allostatic perspective. Biochim Biophys Acta. 2010;1801:338-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 616] [Cited by in RCA: 707] [Article Influence: 47.1] [Reference Citation Analysis (0)] |
| 6. | Lecoutre S, Maqdasy S, Petrus P, Ludzki A, Couchet M, Mejhert N, Rydén M. Glutamine metabolism in adipocytes: a bona fide epigenetic modulator of inflammation. Adipocyte. 2020;9:620-625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 7. | Zatterale F, Longo M, Naderi J, Raciti GA, Desiderio A, Miele C, Beguinot F. Chronic Adipose Tissue Inflammation Linking Obesity to Insulin Resistance and Type 2 Diabetes. Front Physiol. 2019;10:1607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 265] [Cited by in RCA: 637] [Article Influence: 127.4] [Reference Citation Analysis (0)] |
| 8. | Reilly SM, Saltiel AR. Adapting to obesity with adipose tissue inflammation. Nat Rev Endocrinol. 2017;13:633-643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 681] [Cited by in RCA: 860] [Article Influence: 107.5] [Reference Citation Analysis (0)] |
| 9. | Hudish LI, Reusch JE, Sussel L. β Cell dysfunction during progression of metabolic syndrome to type 2 diabetes. J Clin Invest. 2019;129:4001-4008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 219] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 10. | Barker DJ. Developmental origins of adult health and disease. J Epidemiol Community Health. 2004;58:114-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 300] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 11. | Hanson MA, Gluckman PD. Early developmental conditioning of later health and disease: physiology or pathophysiology? Physiol Rev. 2014;94:1027-1076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 822] [Cited by in RCA: 775] [Article Influence: 70.5] [Reference Citation Analysis (0)] |
| 12. | Lecoutre S, Petrus P, Rydén M, Breton C. Transgenerational Epigenetic Mechanisms in Adipose Tissue Development. Trends Endocrinol Metab. 2018;29:675-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 13. | Painter RC, Osmond C, Gluckman P, Hanson M, Phillips DI, Roseboom TJ. Transgenerational effects of prenatal exposure to the Dutch famine on neonatal adiposity and health in later life. BJOG. 2008;115:1243-1249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 381] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 14. | Li C, Lumey LH. Exposure to the Chinese famine of 1959-61 in early life and long-term health conditions: a systematic review and meta-analysis. Int J Epidemiol. 2017;46:1157-1170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 157] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 15. | Safi-Stibler S, Gabory A. Epigenetics and the Developmental Origins of Health and Disease: Parental environment signalling to the epigenome, critical time windows and sculpting the adult phenotype. Semin Cell Dev Biol. 2020;97:172-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 85] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 16. | Godfrey KM, Gluckman PD, Hanson MA. Developmental origins of metabolic disease: life course and intergenerational perspectives. Trends Endocrinol Metab. 2010;21:199-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 338] [Cited by in RCA: 320] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 17. | Lumey LH, Stein AD, Susser E. Prenatal famine and adult health. Annu Rev Public Health. 2011;32:237-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 259] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 18. | Harder T, Rodekamp E, Schellong K, Dudenhausen JW, Plagemann A. Birth weight and subsequent risk of type 2 diabetes: a meta-analysis. Am J Epidemiol. 2007;165:849-857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 443] [Cited by in RCA: 437] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 19. | Parlee SD, MacDougald OA. Maternal nutrition and risk of obesity in offspring: the Trojan horse of developmental plasticity. Biochim Biophys Acta. 2014;1842:495-506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 121] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 20. | Williams L, Seki Y, Vuguin PM, Charron MJ. Animal models of in utero exposure to a high fat diet: a review. Biochim Biophys Acta. 2014;1842:507-519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 168] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 21. | Samuelsson AM, Matthews PA, Argenton M, Christie MR, McConnell JM, Jansen EH, Piersma AH, Ozanne SE, Twinn DF, Remacle C, Rowlerson A, Poston L, Taylor PD. Diet-induced obesity in female mice leads to offspring hyperphagia, adiposity, hypertension, and insulin resistance: a novel murine model of developmental programming. Hypertension. 2008;51:383-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 665] [Cited by in RCA: 679] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 22. | Desai M, Jellyman JK, Han G, Lane RH, Ross MG. Programmed regulation of rat offspring adipogenic transcription factor (PPARγ) by maternal nutrition. J Dev Orig Health Dis. 2015;6:530-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 23. | Hanson M, Godfrey KM, Lillycrop KA, Burdge GC, Gluckman PD. Developmental plasticity and developmental origins of non-communicable disease: theoretical considerations and epigenetic mechanisms. Prog Biophys Mol Biol. 2011;106:272-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 194] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 24. | Bird A. Perceptions of epigenetics. Nature. 2007;447:396-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1909] [Cited by in RCA: 1830] [Article Influence: 101.7] [Reference Citation Analysis (0)] |
| 25. | Tuscher JJ, Day JJ. Multigenerational epigenetic inheritance: One step forward, two generations back. Neurobiol Dis. 2019;132:104591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 26. | Skvortsova K, Iovino N, Bogdanović O. Functions and mechanisms of epigenetic inheritance in animals. Nat Rev Mol Cell Biol. 2018;19:774-790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 323] [Article Influence: 53.8] [Reference Citation Analysis (0)] |
| 27. | Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057-1068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1865] [Cited by in RCA: 1974] [Article Influence: 141.0] [Reference Citation Analysis (0)] |
| 28. | Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930-935. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4860] [Cited by in RCA: 4375] [Article Influence: 273.4] [Reference Citation Analysis (0)] |
| 29. | Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16:9-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 472] [Cited by in RCA: 510] [Article Influence: 39.2] [Reference Citation Analysis (0)] |
| 30. | Berger SL, Sassone-Corsi P. Metabolic Signaling to Chromatin. Cold Spring Harb Perspect Biol. 2016;8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 110] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 31. | Öst A, Pospisilik JA. Epigenetic modulation of metabolic decisions. Curr Opin Cell Biol. 2015;33:88-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 32. | Kitada M, Koya D. SIRT1 in Type 2 Diabetes: Mechanisms and Therapeutic Potential. Diabetes Metab J. 2013;37:315-325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 205] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 33. | Lee JE, Schmidt H, Lai B, Ge K. Transcriptional and Epigenomic Regulation of Adipogenesis. Mol Cell Biol. 2019;39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 194] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 34. | Yu XX, Xu CR. Understanding generation and regeneration of pancreatic β cells from a single-cell perspective. Development. 2020;147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 35. | Haumaitre C, Lenoir O, Scharfmann R. Histone deacetylase inhibitors modify pancreatic cell fate determination and amplify endocrine progenitors. Mol Cell Biol. 2008;28:6373-6383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 148] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 36. | Lenoir O, Flosseau K, Ma FX, Blondeau B, Mai A, Bassel-Duby R, Ravassard P, Olson EN, Haumaitre C, Scharfmann R. Specific control of pancreatic endocrine β- and δ-cell mass by class IIa histone deacetylases HDAC4, HDAC5, and HDAC9. Diabetes. 2011;60:2861-2871. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 116] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 37. | Scavuzzo MA, Hill MC, Chmielowiec J, Yang D, Teaw J, Sheng K, Kong Y, Bettini M, Zong C, Martin JF, Borowiak M. Endocrine lineage biases arise in temporally distinct endocrine progenitors during pancreatic morphogenesis. Nat Commun. 2018;9:3356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 68] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 38. | Yu XX, Qiu WL, Yang L, Li LC, Zhang YW, Xu CR. Dynamics of chromatin marks and the role of JMJD3 during pancreatic endocrine cell fate commitment. Development. 2018;145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 39. | Liu J, Banerjee A, Herring CA, Attalla J, Hu R, Xu Y, Shao Q, Simmons AJ, Dadi PK, Wang S, Jacobson DA, Liu B, Hodges E, Lau KS, Gu G. Neurog3-Independent Methylation Is the Earliest Detectable Mark Distinguishing Pancreatic Progenitor Identity. Dev Cell 2019; 48: 49-63. e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 40. | Dhawan S, Georgia S, Tschen SI, Fan G, Bhushan A. Pancreatic β cell identity is maintained by DNA methylation-mediated repression of Arx. Dev Cell. 2011;20:419-429. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 228] [Cited by in RCA: 211] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 41. | Papizan JB, Singer RA, Tschen SI, Dhawan S, Friel JM, Hipkens SB, Magnuson MA, Bhushan A, Sussel L. Nkx2.2 repressor complex regulates islet β-cell specification and prevents β-to-α-cell reprogramming. Genes Dev. 2011;25:2291-2305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 162] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 42. | Arda HE, Li L, Tsai J, Torre EA, Rosli Y, Peiris H, Spitale RC, Dai C, Gu X, Qu K, Wang P, Wang J, Grompe M, Scharfmann R, Snyder MS, Bottino R, Powers AC, Chang HY, Kim SK. Age-Dependent Pancreatic Gene Regulation Reveals Mechanisms Governing Human β Cell Function. Cell Metab. 2016;23:909-920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 185] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 43. | Avrahami D, Li C, Zhang J, Schug J, Avrahami R, Rao S, Stadler MB, Burger L, Schübeler D, Glaser B, Kaestner KH. Aging-Dependent Demethylation of Regulatory Elements Correlates with Chromatin State and Improved β Cell Function. Cell Metab. 2015;22:619-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 165] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 44. | Campbell SA, Hoffman BG. Chromatin Regulators in Pancreas Development and Diabetes. Trends Endocrinol Metab. 2016;27:142-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 45. | Bramswig NC, Everett LJ, Schug J, Dorrell C, Liu C, Luo Y, Streeter PR, Naji A, Grompe M, Kaestner KH. Epigenomic plasticity enables human pancreatic α to β cell reprogramming. J Clin Invest. 2013;123:1275-1284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 318] [Cited by in RCA: 319] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 46. | Lecoutre S, Breton C. Maternal nutritional manipulations program adipose tissue dysfunction in offspring. Front Physiol. 2015;6:158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 47. | Lecoutre S, Breton C. The cellularity of offspring's adipose tissue is programmed by maternal nutritional manipulations. Adipocyte. 2014;3:256-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 48. | Berry DC, Stenesen D, Zeve D, Graff JM. The developmental origins of adipose tissue. Development. 2013;140:3939-3949. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 232] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 49. | Goodpaster BH, Sparks LM. Metabolic Flexibility in Health and Disease. Cell Metab. 2017;25:1027-1036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 478] [Cited by in RCA: 641] [Article Influence: 80.1] [Reference Citation Analysis (0)] |
| 50. | Berry DC, Jiang Y, Graff JM. Emerging Roles of Adipose Progenitor Cells in Tissue Development, Homeostasis, Expansion and Thermogenesis. Trends Endocrinol Metab. 2016;27:574-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 73] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 51. | Ailhaud G, Grimaldi P, Négrel R. Cellular and molecular aspects of adipose tissue development. Annu Rev Nutr. 1992;12:207-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 478] [Cited by in RCA: 465] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 52. | Holtrup B, Church CD, Berry R, Colman L, Jeffery E, Bober J, Rodeheffer MS. Puberty is an important developmental period for the establishment of adipose tissue mass and metabolic homeostasis. Adipocyte. 2017;6:224-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 53. | Hepler C, Gupta RK. The expanding problem of adipose depot remodeling and postnatal adipocyte progenitor recruitment. Mol Cell Endocrinol. 2017;445:95-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 67] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 54. | Sanchez-Gurmaches J, Guertin DA. Adipocyte lineages: tracing back the origins of fat. Biochim Biophys Acta. 2014;1842:340-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 189] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 55. | Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14:1293-1307. [PubMed] |
| 56. | Siersbæk R, Nielsen R, Mandrup S. Transcriptional networks and chromatin remodeling controlling adipogenesis. Trends Endocrinol Metab. 2012;23:56-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 233] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 57. | Farmer SR. Regulation of PPARgamma activity during adipogenesis. Int J Obes (Lond). 2005;29 Suppl 1:S13-S16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 340] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 58. | Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell. 2014;156:20-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1414] [Cited by in RCA: 1776] [Article Influence: 161.5] [Reference Citation Analysis (0)] |
| 59. | Matsumura Y, Nakaki R, Inagaki T, Yoshida A, Kano Y, Kimura H, Tanaka T, Tsutsumi S, Nakao M, Doi T, Fukami K, Osborne TF, Kodama T, Aburatani H, Sakai J. H3K4/H3K9me3 Bivalent Chromatin Domains Targeted by Lineage-Specific DNA Methylation Pauses Adipocyte Differentiation. Mol Cell. 2015;60:584-596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 163] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 60. | Siersbæk R, Nielsen R, John S, Sung MH, Baek S, Loft A, Hager GL, Mandrup S. Extensive chromatin remodelling and establishment of transcription factor 'hotspots' during early adipogenesis. EMBO J. 2011;30:1459-1472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 294] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 61. | Oger F, Dubois-Chevalier J, Gheeraert C, Avner S, Durand E, Froguel P, Salbert G, Staels B, Lefebvre P, Eeckhoute J. Peroxisome proliferator-activated receptor γ regulates genes involved in insulin/insulin-like growth factor signaling and lipid metabolism during adipogenesis through functionally distinct enhancer classes. J Biol Chem. 2014;289:708-722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 62. | Gupta RK, Arany Z, Seale P, Mepani RJ, Ye L, Conroe HM, Roby YA, Kulaga H, Reed RR, Spiegelman BM. Transcriptional control of preadipocyte determination by Zfp423. Nature. 2010;464:619-623. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 435] [Cited by in RCA: 404] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 63. | Lee JE, Wang C, Xu S, Cho YW, Wang L, Feng X, Baldridge A, Sartorelli V, Zhuang L, Peng W, Ge K. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife. 2013;2:e01503. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 300] [Cited by in RCA: 382] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 64. | Pan FC, Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn. 2011;240:530-565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 425] [Cited by in RCA: 450] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 65. | Shih HP, Wang A, Sander M. Pancreas organogenesis: from lineage determination to morphogenesis. Annu Rev Cell Dev Biol. 2013;29:81-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 226] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 66. | Bastidas-Ponce A, Scheibner K, Lickert H, Bakhti M. Cellular and molecular mechanisms coordinating pancreas development. Development. 2017;144:2873-2888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 110] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 67. | Blum B, Hrvatin S, Schuetz C, Bonal C, Rezania A, Melton DA. Functional beta-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nat Biotechnol. 2012;30:261-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 299] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 68. | Henquin JC, Nenquin M. Immaturity of insulin secretion by pancreatic islets isolated from one human neonate. J Diabetes Investig. 2018;9:270-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 69. | Dore BA, Grogan WM, Madge GE, Webb SR. Biphasic development of the postnatal mouse pancreas. Biol Neonate. 1981;40:209-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 70. | Stolovich-Rain M, Enk J, Vikesa J, Nielsen FC, Saada A, Glaser B, Dor Y. Weaning triggers a maturation step of pancreatic β cells. Dev Cell. 2015;32:535-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 122] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 71. | Jermendy A, Toschi E, Aye T, Koh A, Aguayo-Mazzucato C, Sharma A, Weir GC, Sgroi D, Bonner-Weir S. Rat neonatal beta cells lack the specialised metabolic phenotype of mature beta cells. Diabetologia. 2011;54:594-604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 104] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 72. | Wortham M, Benthuysen JR, Wallace M, Savas JN, Mulas F, Divakaruni AS, Liu F, Albert V, Taylor BL, Sui Y, Saez E, Murphy AN, Yates JR 3rd, Metallo CM, Sander M. Integrated In Vivo Quantitative Proteomics and Nutrient Tracing Reveals Age-Related Metabolic Rewiring of Pancreatic β Cell Function. Cell Rep 2018; 25: 2904-2918. e8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 73. | Jacovetti C, Matkovich SJ, Rodriguez-Trejo A, Guay C, Regazzi R. Postnatal β-cell maturation is associated with islet-specific microRNA changes induced by nutrient shifts at weaning. Nat Commun. 2015;6:8084. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 84] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 74. | Astro V, Adamo A. Epigenetic Control of Endocrine Pancreas Differentiation in vitro: Current Knowledge and Future Perspectives. Front Cell Dev Biol. 2018;6:141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 75. | Gilbert ER, Liu D. Epigenetics: the missing link to understanding β-cell dysfunction in the pathogenesis of type 2 diabetes. Epigenetics. 2012;7:841-852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 90] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 76. | Lu TT, Heyne S, Dror E, Casas E, Leonhardt L, Boenke T, Yang CH, Sagar, Arrigoni L, Dalgaard K, Teperino R, Enders L, Selvaraj M, Ruf M, Raja SJ, Xie H, Boenisch U, Orkin SH, Lynn FC, Hoffman BG, Grün D, Vavouri T, Lempradl AM, Pospisilik JA. The Polycomb-Dependent Epigenome Controls β Cell Dysfunction, Dedifferentiation, and Diabetes. Cell Metab 2018; 27: 1294-1308. e7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 77. | Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343-349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2760] [Cited by in RCA: 2517] [Article Influence: 179.8] [Reference Citation Analysis (0)] |
| 78. | Ghaben AL, Scherer PE. Adipogenesis and metabolic health. Nat Rev Mol Cell Biol. 2019;20:242-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 513] [Cited by in RCA: 941] [Article Influence: 156.8] [Reference Citation Analysis (0)] |
| 79. | Lecoutre S, Deracinois B, Laborie C, Eberlé D, Guinez C, Panchenko PE, Lesage J, Vieau D, Junien C, Gabory A, Breton C. Depot- and sex-specific effects of maternal obesity in offspring's adipose tissue. J Endocrinol. 2016;230:39-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 75] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 80. | Vishvanath L, MacPherson KA, Hepler C, Wang QA, Shao M, Spurgin SB, Wang MY, Kusminski CM, Morley TS, Gupta RK. Pdgfrβ+ Mural Preadipocytes Contribute to Adipocyte Hyperplasia Induced by High-Fat-Diet Feeding and Prolonged Cold Exposure in Adult Mice. Cell Metab. 2016;23:350-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 268] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 81. | Shao M, Hepler C, Vishvanath L, MacPherson KA, Busbuso NC, Gupta RK. Fetal development of subcutaneous white adipose tissue is dependent on Zfp423. Mol Metab. 2017;6:111-124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 82. | Wang B, Yang Q, Harris CL, Nelson ML, Busboom JR, Zhu MJ, Du M. Nutrigenomic regulation of adipose tissue development - role of retinoic acid: A review. Meat Sci. 2016;120:100-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 83. | Yang QY, Liang JF, Rogers CJ, Zhao JX, Zhu MJ, Du M. Maternal obesity induces epigenetic modifications to facilitate Zfp423 expression and enhance adipogenic differentiation in fetal mice. Diabetes. 2013;62:3727-3735. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 108] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 84. | Liang X, Yang Q, Fu X, Rogers CJ, Wang B, Pan H, Zhu MJ, Nathanielsz PW, Du M. Maternal obesity epigenetically alters visceral fat progenitor cell properties in male offspring mice. J Physiol. 2016;594:4453-4466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 68] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 85. | Borengasser SJ, Zhong Y, Kang P, Lindsey F, Ronis MJ, Badger TM, Gomez-Acevedo H, Shankar K. Maternal obesity enhances white adipose tissue differentiation and alters genome-scale DNA methylation in male rat offspring. Endocrinology. 2013;154:4113-4125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 136] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 86. | Boyle KE, Patinkin ZW, Shapiro AL, Baker PR 2nd, Dabelea D, Friedman JE. Mesenchymal Stem Cells From Infants Born to Obese Mothers Exhibit Greater Potential for Adipogenesis: The Healthy Start BabyBUMP Project. Diabetes. 2016;65:647-659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 89] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 87. | Godfrey KM, Sheppard A, Gluckman PD, Lillycrop KA, Burdge GC, McLean C, Rodford J, Slater-Jefferies JL, Garratt E, Crozier SR, Emerald BS, Gale CR, Inskip HM, Cooper C, Hanson MA. Epigenetic gene promoter methylation at birth is associated with child's later adiposity. Diabetes. 2011;60:1528-1534. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 616] [Cited by in RCA: 496] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 88. | Ngo S, Li X, O'Neill R, Bhoothpur C, Gluckman P, Sheppard A. Elevated S-adenosylhomocysteine alters adipocyte functionality with corresponding changes in gene expression and associated epigenetic marks. Diabetes. 2014;63:2273-2283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 89. | Fujiki K, Kano F, Shiota K, Murata M. Expression of the peroxisome proliferator activated receptor gamma gene is repressed by DNA methylation in visceral adipose tissue of mouse models of diabetes. BMC Biol. 2009;7:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 192] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 90. | Zwamborn RA, Slieker RC, Mulder PC, Zoetemelk I, Verschuren L, Suchiman HE, Toet KH, Droog S, Slagboom PE, Kooistra T, Kleemann R, Heijmans BT. Prolonged high-fat diet induces gradual and fat depot-specific DNA methylation changes in adult mice. Sci Rep. 2017;7:43261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 91. | Salma N, Xiao H, Mueller E, Imbalzano AN. Temporal recruitment of transcription factors and SWI/SNF chromatin-remodeling enzymes during adipogenic induction of the peroxisome proliferator-activated receptor gamma nuclear hormone receptor. Mol Cell Biol. 2004;24:4651-4663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 155] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 92. | Lecoutre S, Pourpe C, Butruille L, Marousez L, Laborie C, Guinez C, Lesage J, Vieau D, Eeckhoute J, Gabory A, Oger F, Eberlé D, Breton C. Reduced PPARγ2 expression in adipose tissue of male rat offspring from obese dams is associated with epigenetic modifications. FASEB J. 2018;32:2768-2778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 93. | Lukaszewski MA, Eberlé D, Vieau D, Breton C. Nutritional manipulations in the perinatal period program adipose tissue in offspring. Am J Physiol Endocrinol Metab. 2013;305:E1195-E1207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 90] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 94. | Butruille L, Marousez L, Pourpe C, Oger F, Lecoutre S, Catheline D, Görs S, Metges CC, Guinez C, Laborie C, Deruelle P, Eeckhoute J, Breton C, Legrand P, Lesage J, Eberlé D. Maternal high-fat diet during suckling programs visceral adiposity and epigenetic regulation of adipose tissue stearoyl-CoA desaturase-1 in offspring. Int J Obes (Lond). 2019;43:2381-2393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 95. | Murabayashi N, Sugiyama T, Zhang L, Kamimoto Y, Umekawa T, Ma N, Sagawa N. Maternal high-fat diets cause insulin resistance through inflammatory changes in fetal adipose tissue. Eur J Obstet Gynecol Reprod Biol. 2013;169:39-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 94] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 96. | del Bas JM, Crescenti A, Arola-Arnal A, Oms-Oliu G, Arola L, Caimari A. Grape seed procyanidin supplementation to rats fed a high-fat diet during pregnancy and lactation increases the body fat content and modulates the inflammatory response and the adipose tissue metabolism of the male offspring in youth. Int J Obes (Lond). 2015;39:7-15. [PubMed] [DOI] [Full Text] |
| 97. | Lecoutre S, Oger F, Pourpe C, Butruille L, Marousez L, Dickes-Coopman A, Laborie C, Guinez C, Lesage J, Vieau D, Junien C, Eberlé D, Gabory A, Eeckhoute J, Breton C. Maternal obesity programs increased leptin gene expression in rat male offspring via epigenetic modifications in a depot-specific manner. Mol Metab. 2017;6:922-930. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 98. | Masuyama H, Mitsui T, Eguchi T, Tamada S, Hiramatsu Y. The effects of paternal high-fat diet exposure on offspring metabolism with epigenetic changes in the mouse adiponectin and leptin gene promoters. Am J Physiol Endocrinol Metab. 2016;311:E236-E245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 81] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 99. | Masuyama H, Mitsui T, Nobumoto E, Hiramatsu Y. The Effects of High-Fat Diet Exposure In Utero on the Obesogenic and Diabetogenic Traits Through Epigenetic Changes in Adiponectin and Leptin Gene Expression for Multiple Generations in Female Mice. Endocrinology. 2015;156:2482-2491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 69] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 100. | Masuyama H, Hiramatsu Y. Additive effects of maternal high fat diet during lactation on mouse offspring. PLoS One. 2014;9:e92805. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 101. | Umekawa T, Sugiyama T, Du Q, Murabayashi N, Zhang L, Kamimoto Y, Yoshida T, Sagawa N, Ikeda T. A maternal mouse diet with moderately high-fat levels does not lead to maternal obesity but causes mesenteric adipose tissue dysfunction in male offspring. J Nutr Biochem. 2015;26:259-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 102. | Ferland-McCollough D, Fernandez-Twinn DS, Cannell IG, David H, Warner M, Vaag AA, Bork-Jensen J, Brøns C, Gant TW, Willis AE, Siddle K, Bushell M, Ozanne SE. Programming of adipose tissue miR-483-3p and GDF-3 expression by maternal diet in type 2 diabetes. Cell Death Differ. 2012;19:1003-1012. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 111] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 103. | de Almeida Faria J, Duque-Guimarães D, Carpenter AA, Loche E, Ozanne SE. A post-weaning obesogenic diet exacerbates the detrimental effects of maternal obesity on offspring insulin signaling in adipose tissue. Sci Rep. 2017;7:44949. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 104. | Fernandez-Twinn DS, Alfaradhi MZ, Martin-Gronert MS, Duque-Guimaraes DE, Piekarz A, Ferland-McCollough D, Bushell M, Ozanne SE. Downregulation of IRS-1 in adipose tissue of offspring of obese mice is programmed cell-autonomously through post-transcriptional mechanisms. Mol Metab. 2014;3:325-333. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 91] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 105. | Bringhenti I, Ornellas F, Martins MA, Mandarim-de-Lacerda CA, Aguila MB. Early hepatic insult in the offspring of obese maternal mice. Nutr Res. 2015;35:136-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 106. | Yokomizo H, Inoguchi T, Sonoda N, Sakaki Y, Maeda Y, Inoue T, Hirata E, Takei R, Ikeda N, Fujii M, Fukuda K, Sasaki H, Takayanagi R. Maternal high-fat diet induces insulin resistance and deterioration of pancreatic β-cell function in adult offspring with sex differences in mice. Am J Physiol Endocrinol Metab. 2014;306:E1163-E1175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 91] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 107. | Han J, Xu J, Epstein PN, Liu YQ. Long-term effect of maternal obesity on pancreatic beta cells of offspring: reduced beta cell adaptation to high glucose and high-fat diet challenges in adult female mouse offspring. Diabetologia. 2005;48:1810-1818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 54] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 108. | Gregorio BM, Souza-Mello V, Mandarim-de-Lacerda CA, Aguila MB. Maternal high-fat diet is associated with altered pancreatic remodelling in mice offspring. Eur J Nutr. 2013;52:759-769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 109. | Bringhenti I, Ornellas F, Mandarim-de-Lacerda CA, Aguila MB. The insulin-signaling pathway of the pancreatic islet is impaired in adult mice offspring of mothers fed a high-fat diet. Nutrition. 2016;32:1138-1143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 110. | Simmons RA, Templeton LJ, Gertz SJ. Intrauterine growth retardation leads to the development of type 2 diabetes in the rat. Diabetes. 2001;50:2279-2286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 452] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 111. | Park JH, Stoffers DA, Nicholls RD, Simmons RA. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest. 2008;118:2316-2324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 273] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 112. | Thompson RF, Fazzari MJ, Niu H, Barzilai N, Simmons RA, Greally JM. Experimental intrauterine growth restriction induces alterations in DNA methylation and gene expression in pancreatic islets of rats. J Biol Chem. 2010;285:15111-15118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 112] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 113. | Li G, Petkova TD, Laritsky E, Kessler N, Baker MS, Zhu S, Waterland RA. Early postnatal overnutrition accelerates aging-associated epigenetic drift in pancreatic islets. Environ Epigenet. 2019;5:dvz015. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 114. | Zhang L, Chen W, Dai Y, Zhu Z, Liu Q. Detection of expressional changes induced by intrauterine growth restriction in the developing rat pancreas. Exp Biol Med (Maywood). 2016;241:1446-1456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 115. | Petry CJ, Dorling MW, Pawlak DB, Ozanne SE, Hales CN. Diabetes in old male offspring of rat dams fed a reduced protein diet. Int J Exp Diabetes Res. 2001;2:139-143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 196] [Cited by in RCA: 186] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 116. | Sandovici I, Smith NH, Nitert MD, Ackers-Johnson M, Uribe-Lewis S, Ito Y, Jones RH, Marquez VE, Cairns W, Tadayyon M, O'Neill LP, Murrell A, Ling C, Constância M, Ozanne SE. Maternal diet and aging alter the epigenetic control of a promoter-enhancer interaction at the Hnf4a gene in rat pancreatic islets. Proc Natl Acad Sci USA. 2011;108:5449-5454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 280] [Cited by in RCA: 239] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 117. | Ding GL, Wang FF, Shu J, Tian S, Jiang Y, Zhang D, Wang N, Luo Q, Zhang Y, Jin F, Leung PC, Sheng JZ, Huang HF. Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes. 2012;61:1133-1142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 195] [Cited by in RCA: 209] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 118. | Vaiserman A, Lushchak O. Developmental origins of type 2 diabetes: Focus on epigenetics. Ageing Res Rev. 2019;55:100957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 119. | Graus-Nunes F, Dalla Corte Frantz E, Lannes WR, da Silva Menezes MC, Mandarim-de-Lacerda CA, Souza-Mello V. Pregestational maternal obesity impairs endocrine pancreas in male F1 and F2 progeny. Nutrition. 2015;31:380-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 120. | Heard E, Martienssen RA. Transgenerational epigenetic inheritance: myths and mechanisms. Cell. 2014;157:95-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1080] [Cited by in RCA: 1078] [Article Influence: 98.0] [Reference Citation Analysis (0)] |
| 121. | Grossniklaus U, Kelly WG, Kelly B, Ferguson-Smith AC, Pembrey M, Lindquist S. Transgenerational epigenetic inheritance: how important is it? Nat Rev Genet. 2013;14:228-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 179] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 122. | Daxinger L, Whitelaw E. Transgenerational epigenetic inheritance: more questions than answers. Genome Res. 2010;20:1623-1628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 162] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 123. | Fullston T, McPherson NO, Owens JA, Kang WX, Sandeman LY, Lane M. Paternal obesity induces metabolic and sperm disturbances in male offspring that are exacerbated by their exposure to an "obesogenic" diet. Physiol Rep. 2015;3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 84] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 124. | de Castro Barbosa T, Ingerslev LR, Alm PS, Versteyhe S, Massart J, Rasmussen M, Donkin I, Sjögren R, Mudry JM, Vetterli L, Gupta S, Krook A, Zierath JR, Barrès R. High-fat diet reprograms the epigenome of rat spermatozoa and transgenerationally affects metabolism of the offspring. Mol Metab. 2016;5:184-197. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 230] [Cited by in RCA: 271] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 125. | Wei Y, Yang CR, Wei YP, Zhao ZA, Hou Y, Schatten H, Sun QY. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc Natl Acad Sci USA. 2014;111:1873-1878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 315] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 126. | Donkin I, Barrès R. Sperm epigenetics and influence of environmental factors. Mol Metab. 2018;14:1-11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 210] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 127. | Champroux A, Cocquet J, Henry-Berger J, Drevet JR, Kocer A. A Decade of Exploring the Mammalian Sperm Epigenome: Paternal Epigenetic and Transgenerational Inheritance. Front Cell Dev Biol. 2018;6:50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 118] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 128. | Dunn GA, Bale TL. Maternal high-fat diet effects on third-generation female body size via the paternal lineage. Endocrinology. 2011;152:2228-2236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 268] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 129. | Sarker G, Berrens R, von Arx J, Pelczar P, Reik W, Wolfrum C, Peleg-Raibstein D. Transgenerational transmission of hedonic behaviors and metabolic phenotypes induced by maternal overnutrition. Transl Psychiatry. 2018;8:195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 130. | Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16:71-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 678] [Cited by in RCA: 710] [Article Influence: 71.0] [Reference Citation Analysis (0)] |
| 131. | Hur SSJ, Cropley JE, Suter CM. Paternal epigenetic programming: evolving metabolic disease risk. J Mol Endocrinol. 2017;58:R159-R168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 132. | Kaati G, Bygren LO, Pembrey M, Sjöström M. Transgenerational response to nutrition, early life circumstances and longevity. Eur J Hum Genet. 2007;15:784-790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 342] [Cited by in RCA: 282] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 133. | Davis MM, McGonagle K, Schoeni RF, Stafford F. Grandparental and parental obesity influences on childhood overweight: implications for primary care practice. J Am Board Fam Med. 2008;21:549-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 55] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 134. | Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. Editing DNA Methylation in the Mammalian Genome. Cell 2016; 167: 233-247. e17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 729] [Cited by in RCA: 847] [Article Influence: 94.1] [Reference Citation Analysis (0)] |
| 135. | Ling C, Rönn T. Epigenetics in Human Obesity and Type 2 Diabetes. Cell Metab. 2019;29:1028-1044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 561] [Article Influence: 93.5] [Reference Citation Analysis (0)] |
| 136. | Thakore PI, Black JB, Hilton IB, Gersbach CA. Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat Methods. 2016;13:127-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 318] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 137. | Burdge GC, Lillycrop KA, Phillips ES, Slater-Jefferies JL, Jackson AA, Hanson MA. Folic acid supplementation during the juvenile-pubertal period in rats modifies the phenotype and epigenotype induced by prenatal nutrition. J Nutr. 2009;139:1054-1060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 152] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 138. | Cordero P, Milagro FI, Campion J, Martinez JA. Maternal methyl donors supplementation during lactation prevents the hyperhomocysteinemia induced by a high-fat-sucrose intake by dams. Int J Mol Sci. 2013;14:24422-24437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 139. | Daneshpajooh M, Eliasson L, Bacos K, Ling C. MC1568 improves insulin secretion in islets from type 2 diabetes patients and rescues β-cell dysfunction caused by Hdac7 upregulation. Acta Diabetol. 2018;55:1231-1235. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 140. | Daneshpajooh M, Bacos K, Bysani M, Bagge A, Ottosson Laakso E, Vikman P, Eliasson L, Mulder H, Ling C. HDAC7 is overexpressed in human diabetic islets and impairs insulin secretion in rat islets and clonal beta cells. Diabetologia. 2017;60:116-125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 64] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 141. | Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1025] [Cited by in RCA: 1058] [Article Influence: 81.4] [Reference Citation Analysis (0)] |