Published online Oct 15, 2021. doi: 10.4239/wjd.v12.i10.1655
Peer-review started: January 28, 2021
First decision: May 3, 2021
Revised: May 22, 2021
Accepted: July 5, 2021
Article in press: July 5, 2021
Published online: October 15, 2021
Processing time: 257 Days and 22 Hours
During infections, nucleic acids of pathogens are also engaged in recognition via several exogenous and cytosolic pattern recognition receptors, such as the toll-like receptors, retinoic acid inducible gene-I-like receptors, and nucleotide-binding and oligomerization domain-like receptors. The binding of the pathogen-derived nucleic acids to their corresponding sensors initiates certain downstream signaling cascades culminating in the release of type-I interferons (IFNs), especially IFN-α and other cytokines to induce proinflammatory responses towards invading pathogens leading to their clearance from the host. Although these sensors are hardwired to recognize pathogen associated molecular patterns, like viral and bacterial nucleic acids, under unusual physiological conditions, such as excessive cellular stress and increased apoptosis, endogenous self-nucleic acids like DNA, RNA, and mitochondrial DNA are also released. The presence of these self-nucleic acids in extranuclear compartments or extracellular spaces or their association with certain proteins sometimes leads to the failure of discriminating mechanisms of nucleic acid sensors leading to proinflammatory responses as seen in autoimmune disorders, like systemic lupus erythematosus, psoriasis and to some extent in type 1 diabetes (T1D). This review discusses the involvement of various nucleic acid sensors in autoimmunity and discusses how aberrant recognition of self-nucleic acids by their sensors activates the innate immune responses during the pathogenesis of T1D.
Core Tip: Under abnormal physiological conditions, such as excessive cellular stress or apoptosis, endogenous self-nucleic acids like DNA, RNA or mitochondrial DNA accumulate in extranuclear compartments or extracellular spaces or form complexes with host proteins. Such situations sometimes lead to the failure of discriminating mechanisms of nucleic acid sensors leading to proinflammatory responses as seen in autoimmune diseases like systemic lupus erythematosus, psoriasis and to some extent in type 1 diabetes (T1D). The understanding of the role of nucleic acid-sensing and their downstream signaling pathways is gradually evolving and provides another avenue in exploring therapeutic options for treating autoimmune diseases like T1D.
- Citation: Badal D, Sachdeva N, Maheshwari D, Basak P. Role of nucleic acid sensing in the pathogenesis of type 1 diabetes. World J Diabetes 2021; 12(10): 1655-1673
- URL: https://www.wjgnet.com/1948-9358/full/v12/i10/1655.htm
- DOI: https://dx.doi.org/10.4239/wjd.v12.i10.1655
Type 1 diabetes (T1D) is a complex autoimmune disorder that involves infiltration of innate and adaptive immune cells culminating in the killing of insulin producing beta (β)-cells, mainly through T-cell dependent mechanisms. Pathogenesis of T1D involves an initial infiltration of mononuclear cells consisting of neutrophils, dendritic cells (DCs) and macrophages[1] in the pancreatic islets[2] followed by lymphocytic infiltration[3]. Beta-cell death is mainly mediated by autoreactive CD8+ T cells that release cytolytic granules, perforins facilitating the entry of granzymes in target β-cells[4,5]. The innate immune cells carry a variety of specialized receptors known as pattern-recognition receptors (PRRs) whose main function is to detect well-conserved structural motifs that are indispensable to pathogen survival and are known as pathogen-associated molecular patterns (PAMPs)[6]. In addition to recognizing PAMPs, these receptors under certain circumstances can also recognize damage associated molecular patterns (DAMPs) released by dying autologous cells, including β-cells, and can activate signaling cascade in a fashion similar to PAMPs recognition[7]. This recognition initiates a canonical immune signaling cascade driven by type 1 interferons (IFNs), mainly IFN-α to induce IFN-stimulated genes (ISGs) which activate inflammatory mediators, release cytokines responsible for instituting an inflammatory state in the pancreatic islets, and overexpression of HLA class-1 molecules on β-cells that enhances uptake of autoantigens by antigen-presenting cells (APCs)[8-10]. Nucleic acids, like other PAMPs, are vital for the survival and propagation of pathogens, and hence, the PRRs of the human innate immune system were evolved to recognize and mount an appropriate response against the pathogens bearing them. In various autoimmune conditions, like systemic lupus erythematosus (SLE), psoriasis, etc. and to some extent in T1D, the nucleic acids released by self-cells under certain physiological conditions, such as inflammation, stress, apoptosis, necrosis, pyroptosis, necroptosis, and NETosis act as ligands of PRRs, leading to either initiation of these autoimmune conditions or worsening of their pathogenesis[1,11,12]. In this review, we have summarized the recent advances in understanding the role of self-nucleic acids, their sensors, and downstream signaling pathways involved in the pathogenesis of T1D and discussed the novel therapeutic approaches targeting autoimmune diseases, including T1D.
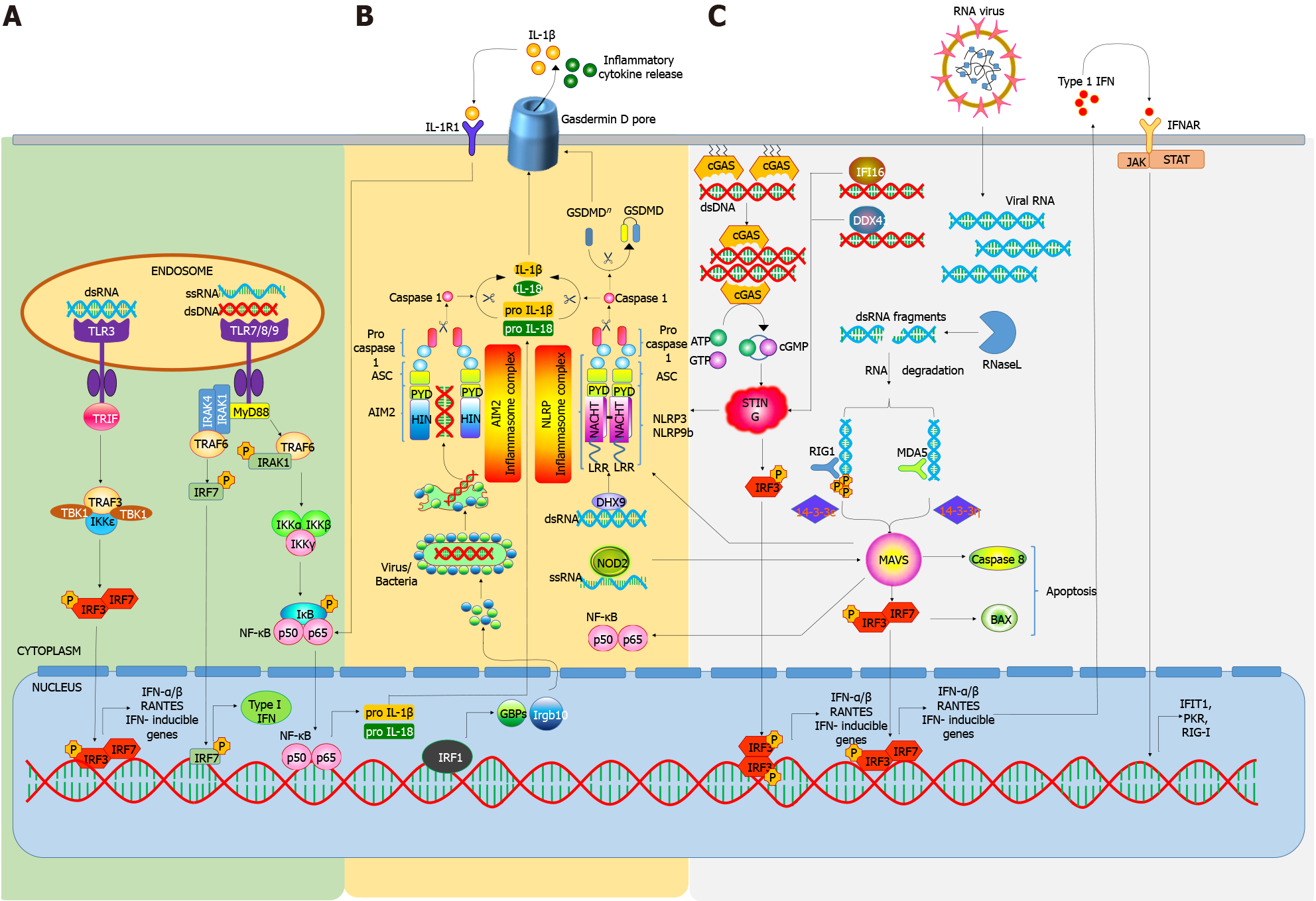
As a part of the innate immune system, PRRs are the primary sentinels against the microbes, and initiation of immune responses through PRR recognition is crucial for the host defenses. PAMPs, such as viral or bacterial nucleic acids, in addition to other bacterial or fungal cellular components, are commonly recognized by the host PRRs. Recognition of PAMPs by PRRs initiates a downstream signaling cascade resulting in the innate immune responses by promoting the expression of pro-inflammatory cytokines, IFNs, etc.[13]. These cytokines signal the adjacent cells to promote the expression of various ISG to impair replication of pathogens. Besides microbial infection, PRRs activation by nucleic acids can also be initiated by the host cells. Stress or cell-death induced release of self-nucleic acids, such as genomic DNA, mRNA, tRNA and mitochondrial DNA (mtDNA) can also be recognized by PRRs to trigger inflammatory cytokines and type-I IFN, leading to chronic inflammation. Inappropriate or prolonged detection of these nucleic acids has been shown to be associated with many autoimmune diseases[11]. Presently, PRRs are classified into 4 main categories as follows: Toll-like receptors (TLRs), retinoic acid inducible gene-I (RIG-I)-like receptors (RLRs), absent-in-melanoma (AIM)-Like Receptors (ALRs), nucleotide-binding and oligomerization domain (NOD)-like receptors (NLRs), and C-type lectins (CTLs). CTLs and most TLRs are located in the plasma membrane, while the NLRs, RLRs, ALRs and a few TLRs are located intracellularly[13].
TLRs are a conserved class of PRRs belonging to the family of type-I transmembrane receptor proteins consisting of an extracellular Leucine-Rich Repeat (LRR) domain and an intracellular C-terminal toll/IL-1 receptor (TIR) domain[14]. This domain is required for the interaction and recruitment of various adaptor molecules to activate downstream signaling pathways involving the transcription factors Activator Protein-1 (AP-1), Nuclear Factor Kappa-Light-Chain-Enhancer of Activated B Cells (NF-κB), and Interferon Regulatory Factor (IRF)[15]. To date, 13 different types of TLRs (TLR 1-13) have been identified. TLRs 1-9 are expressed by both humans and mice; whereas only humans, express TLR10, while mice are known to express TLR11-13[16]. TLRs are broadly expressed in both immune and non-immune cells in two distinct cellular compartments, extracellular and intracellular (mainly in endosomes)[17]. In T1D, upon recognition of pathogenic and/or foreign material, TLRs influence many immunologic mechanisms, including activation and maturation of APCs, antibody production, down regulating regulatory T cell (Treg) responses, and facilitating a pro-inflammatory environment through the secretion of a plethora of cytokines and chemokines[18].
TLR-TLR ligation and interaction transduces signals through MyD88 (Myeloid differentiation primary response 88)-dependent or independent pathways. Upon activation, MyD88 recruits Interleukin 1 Receptor Associated Kinase (IRAK-1), IRAK-4, and Tumor Necrosis Factor receptor (TNFR)-Associated Factor 6 (TRAF-6), which then activate c-Jun N-terminal Kinase (JNK), Ikβ Kinase (IKK), AP-1, and NF-κB. The MyD88-independent pathway is mediated by TIR-domain-containing adapter-inducing IFN-β (TRIF) and TRIF Related Adaptor Molecule (TRAM), leading to the activation of NF-κB, AP-1, or IRFs[19], while the TLR3 signaling is mediated through TRIF, TLR7, TLR8, and TLR9 signals through MyD88. It has also been demonstrated that TLR signaling can efficiently promote the uptake of autoantigens by APCs[8-10]. Under normal physiological conditions apoptotic cell derived antigens are not presented efficiently by MHC class II molecules. However, TLR ligand co-administration not only enhances antigen presentation but also promotes antigen specific responses by CD4+ T cells[8]. Thus, it means that TLRs not only acts as danger signal sensors but also regulators of self-and non-self-antigen discrimination[20,21]. In support of this fact, it has been demonstrated that stimulation of TLRs enhances antigen processing by up-regulating scavenger receptors via the MyD88-dependent pathway[22].
The role of TLRs especially those involved in the recognition of nucleic acids is also being recognized in autoimmune diabetes. TLRs can recognize various forms of endogenous DNA or RNA produced during virus infection induced cell death[23]. However, TLR3, TLR7, TLR8, and TLR9 specifically recognize viral-associated nucleic acids with comparatively higher affinity and have been implicated in the pathogenesis of T1D. TLR3-/- NOD mice have shown high mortality from Coxsackie B4 virus (CVB4) infections and the few that survived develop T1D[24]. Certain polymorphisms in the TLR3 gene (rs3775291 and rs13126816) have also been shown to be related with a higher risk of T1D and a more aggressive pathology[25]. A double stranded RNA (dsRNA) mimetic polyinosinic: polycytidylic (poly I: C) has been reported to be recognized by TLR3, leading to induction and increase in the severity of T1D in mice, depending on dose and administration[25].
Stimulation of TLR7 (in addition to CD40 activation of DCs) can induce diabetogenic cytotoxic CD8+ T cells in the pancreatic lymph nodes of NOD mice to promote the onset of autoimmunity[26]. Repeated topical administration of a TLR7 agonist, imiquimod, is sufficient to promote T1D development while inhibition using IRS661 can significantly lower disease onset[26]. Similarly, TLR7 signaling in plasmacytoid DCs (pDCs) triggers B and T cell activation via IFN-I secretion in rotavirus infections, on the other hand, inhibition of TLR7 can block this process and prevent the acceleration of T1D following infection[27]. Zhang et al[28] have shown that TLR9 blockade can impede the activation of diabetogenic CD8+ T cells and, delay autoimmune diabetes in NOD mice. Liu et al[29] generated TLR9 knockout NOD mice and observed improvements in insulin secretion, glucose tolerance, and β-cell function. These improvements were partially mediated by the upregulation of CD140a on β-cells. Similar results have been observed by the use of TLR9 antagonists or by genetic targeting on ontogenesis and function of β-cells to protect NOD mice from T1D.
Hence, these and other reports further necessitate more research to understand and improve defects associated with self-nucleic acid recognition by TLRs associated with T1D pathology.
RLRs are a group of intracellular receptors that recognize viral dsRNA and are comprised of 3 proteins: (1) RIG-1; (2) Melanoma differentiation-associated gene 5 (MDA5); and (3) Laboratory of genetics and physiology 2 (LGP2), which is composed of a DExD/H box RNA helicase domain and a C-terminal domain[30]. Both RIG-1 and MDA5 contain additional N-terminal caspase activation and recruitment domains (CARDs) that transmit downstream signaling. RIG-I and MDA5 have similar functions and they initiate antiviral signals to induce IFN gene activation, while LGP2 acts as a regulator of MDA5 and RIG-1[31]. Upon recognition of RNA, an ATP-dependent conformational change occurs in RLR[32] resulting in the activation of CARD and further activation of an adaptor molecule, mitochondrial antiviral signaling (MAVS) protein[33]. Activation of MAVS, in turn, triggers signaling cascades involving TRAF3/6, caspase 8/10, RIP-1, fas-associated death domain, and TNF receptor-associated death domain ultimately activating TANK binding kinase 1 (TBK1)/IKK-ε and IKKα/IKKβ to induce transcription of type-I IFNs and proinflammatory cytokines by activating IRF-3 and NF-κB.
When challenged with pathogenic stress, various single nucleotide polymorphism (SNP) in the interferon induced with helicase C domain 1 (IFIH1) gene have been found to cause greater or reduced susceptibility in the pathogenesis of T1D via altering MDA5 activation and expression[34]. The IFIH1 mutation A946T (rs1990760) has been involved in the pathogenesis and development of various autoimmune diseases like T1D, SLE, and multiple sclerosis (MS)[35,36]. Two independent studies conducted on subjects with diabetes showed that subjects with heterozygous A946T SNP have a more prominent immune response and ISG expression to Coxsackie virus challenge in comparison to healthy controls, suggesting greater IFNs and ISGs expression during infection[37,38]. In another study, Cinek et al[39] demonstrated a positive correlation between IFIH1 polymorphism (rs1990760), which is known to be strongly associated with T1D, and enteroviral RNA frequency in the blood of T1D subjects. The authors further suggested that rs1990760 can modify enteroviral frequency in the blood of healthy children harboring IFIH1 polymorphism, predisposing them towards T1D[39]. Gain-of-function mutations in IFIH1 have been also found to be associated with overexpression of type 1 and type 3 IFN[40]. A study by Gorman et al[41] observed mice that were homozygous for IFIH1 SNP (946T) or exhibiting IFIH1 risk alleles (843R and 946T) simultaneously, had enhanced expression of IFIH1-related genes, increased rate of autoimmunity development, and ability to recognize self-RNA. Such mutations may alter the expression of inflammatory molecules and the dynamics of target binding, and activation may also be altered, resulting in more potent/enhanced IFN response leading to the risk of T1D. For example, MDA5 mutation E627 causes loss of a portion of C-terminal region, resulting in loss of dsRNA ligand and binding[42]. Overall, these reports provide us with enough knowledge about the role of RLRs in the pathogenesis of T1D.
A few PRRs also include some members of the family of proteins containing pyrin and hematopoietic interferon-inducible nuclear (HIN) domain[43]. The Pyrin and HIN domain (PHYIN) family of proteins comprises of ALR, which contains an N-terminal Pyrin domain and one or two C-terminal hematopoietic IFN-inducible nuclear proteins with 200 amino acids (HIN-200) domains, containing an oligonucleo
AIM2 is a cytosolic dsDNA receptor that oligomerizes on recognizing cytosolic foreign dsDNA and promotes the polymerization of the adaptor protein, Apoptosis-associated Speck-like (ASC) protein and eventually forming a caspase-1 activating inflammasome[44]. AIM2 binds to small DNA fragments up to 20bp; however, in order to initiate immune responses against longer DNA fragments, oligomerization of AIM2 is required. ALRs can sense self-DNA through leakage from nuclear envelope and exosomes engulfed by phagocytes; however, the ability of ALRs to elicit type 1 IFN responses is questionable, as mice deficient in ALRs can mount effective type 1 IFN responses to DNA viruses and lentiviruses[45].
NLRs are comprised of various cytosolic PRRs, which are characterized by the presence of a conserved NOD[46]. NLRs consist of an N-terminal effector binding region, which is further comprised of: (1) Protein-protein interaction domain such as the: (a) CARD; (b) Pyrin domain (PYD); and (c) Baculovirus inhibitor repeat domain; (2) NOD domain, which is needed for self-oligomerization and nucleotide binding; and (3) Array of C-terminal LRR motifs to recognize the pathogenic pattern and regulate NLR activity.
Upon recognition of nucleic acids by the C-terminal LRR motifs, the downstream signaling gets initiated, involving conformational changes that result in oligomerization of NLR via the NOD domain. NLR exposes the effector domains to initiate CARD and PYD recruitment and activation by enhancing their oligomerization[47]. NLRs interact with receptor interacting serine/threonine protein kinase 2 to trigger mitogen-activated protein kinase (MAPK) and NF-κB[48]. The NLRs have a proven role in antiviral immunity; however, their role in sensing self-nucleic acids is gradually emerging[49]. NLRs also recognize oxidized forms of mitochondrial DNA, which could have important implications in inflammation and cancers[50].
Inflammasomes are a diverse class of cytosolic multiprotein complexes consisting of an adaptor protein containing CARD, a sensor protein and caspase-1 which is highly proinflammatory. Their assembly can be triggered by a variety of stimuli, ultimately leading to caspase-1 activation and synthesis of proinflammatory cytokines. Inflammasomes play a crucial role in the mobilization and activation of various immune cells in maintaining tissue homeostasis by initiating acute immune responses. Inflammasomes can also initiate chronic immune response leading to uncontrolled inflammation which eventually causes cell death via pyroptosis[51]. Among them, NLRP3 and NLRP1 inflammasomes are the most common subtypes[52]. ALRs and NLRs initiate the immune response by forming inflammasomes, thereby alleviating IL-1β and IL-18 maturation and release[53,54]. Activated caspase-1 then cleaves pro-IL-1β or pro-IL-18 ,enabling the release of the mature active cytokines IL-1β and IL-18[53,55].
NLRP3 inflammasomes have been reported to play crucial roles in the pathogenesis of various autoimmune disorders, including T1D[56,57]. In 2019, Sun et al[58], showed the association of SNPs with T1D pathogenesis and diabetes onset in the NLRP1 gene of T1D patients of Chinese Han origin. Increased susceptibility to T1D and celiac disease have been reported to be associated with SNPs within the NLRP3 gene. A study by Hu et al[59] showed an important role of NLRP3 in the pathogenesis of T1D in NOD mice. Elimination of NLRP3 altered T cell maturation via regulation of CCR5 and CXCR3 expression, as well as pathogenic T cell mobilization to the pancreatic islets, which is a crucial process leading to β-cell death and disease progression. Also, knockout of NLRP3 downregulated C-C motif chemokine ligand 5 (CCL-5) and C-X-C motif chemokine ligand 10 (CXCL10) expression in the pancreatic islets via IRF-1 signaling[59]. Furthermore, in STZ induced diabetic mice model, NLRP3 activation via mtDNA initiated IL-1β production in caspase-1 dependent manner, suggesting a direct role of NLRP3-caspase1 signaling in T1D[60]. Pereira et al[61] recently highlighted the role of mtDNA in the involvement of vascular endothelial dysfunction in human subjects with T1D and asserted on the connection between NLRP3 inflammasomes and T1D complications. In this study, mtDNA isolated from diabetic mice promoted NLRP3 inflammasome activation via mechanisms involving mitochondrial ROS and Ca2+ influx, which was abrogated in NLRP3 knockout mice.
The cyclic GMP-AMP synthase-stimulator of IFN genes (cGAS-STING) is a DNA sensing receptor present in the cytoplasm that recognizes host/pathogenic DNA[62]. When DNA binds on the active site of cGAS, its C-terminal containing the catalytic unit undergoes a variety of conformational changes, resulting in cyclic guanosine monophosphate–adenosine monophosphate (cGAMP) formation from ATP and GTP[63,64]. cGAMP formation results in STING activation by inducing conformational changes upon binding to its active site and also facilitates STING transportation from the endoplasmic reticulum to the Golgi apparatus[65,66]. Upon activation, STING further forms a complex with TBK1, which further phosphorylates IRF3 in endoly
The role of cGAS-STING in various autoimmune disorders is being widely explored, while its role in T1D has not been reported earlier. Lemos et al[72] reported that the activation of STING resulted in suppression of T1D onset and progression when NOD mice were administered with DNA nanoparticles, which promoted indoleamine 2,3 dioxygenase (IDO) activity, thus modulating T cell immunity in pancreatic lymph nodes and pancreas.
Overall, many studies have yielded important information on how the nucleic acid sensors lead to the activation of downstream signaling pathways (Figure 1). These sensors and their signaling mediators have been implicated in different autoimmune diseases including T1D (Table 1)[73-84].
| No. | Nucleic acid sensor | Downstream signaling molecule | Autoimmune disease | Ref. |
| 1 | TLR9 | Myd88/ IRF3/7 | SLE | [73,74] |
| 2 | TLR7 | Myd88 | T1D | [27] |
| 3 | TLR3 | TRIF | T1D | [25,75] |
| 4 | RLR | IRF3 | Singleton-Merton Syndrome, | [76,77] |
| AGS and T1D | [34,78] | |||
| 5 | cGAS-STING | cGMP | SLE and AGS | [79,80] |
| 6 | NLR | Inflammasome activation | T1D and SLE | [58,81] |
| 7 | AIM | SLE | [81] | |
| 8 | IFI16 | Inflammasome | Primary Sjogren’s Syndrome | [82] |
| Activation | Rheumatoid Arthritis | [83] | ||
| 9 | CTL | Bcl10/CARD9 | Multiple Sclerosis | [84] |
Most of the nucleic acid recognition pathways culminate in the release of type 1 IFN, especially IFN-α, via mediators like IRFs, which makes them one of the most crucial part of the nucleic acid sensing pathway. IFN-α has multiple roles, including upregulation of human leukocyte antigen (HLA) class I and HLA class II to enhance antigenic presentation, increase in immunoproteasome activity, induce ER stress and cellular inflammation through TYK2 activation, induction of transcription factors, and signal transducer and activator of transcription 2 and IRF9. It also acts synergistically with IL-1β and induces β-cell apoptosis[85]. Heightened IFN-α secretion in peripheral blood mononuclear cells of T1D subjects by stimulation with influenza viruses has been attributed to the recognition of viral nucleic acids by endosomal TLRs of pDCs. Additionally, in vitro studies have demonstrated that pDCs secreted IFN-α enhances Th1 responses[86]. Another study observed higher levels of secreted IFN-α by pDCs obtained from the relatives of T1D subjects following their stimulation with CpG 2216[87]. The transition of prediabetic stage to full-blown diabetes is also found to be controlled by IFN-α signaling. The study demonstrated that the infiltration of autoreactive T cells and β-cell killing can be prevented by blocking IFN-α signaling by sphingosine-1 receptor agonist prior to the clinical onset of disease[88]. Rodrigues et al[89] in a recent study revealed IFN-1 hyper-responsiveness in T1D after innate immune stimulation of whole blood cells with CpG DNA. They observed higher induced IFN-1-associated gene expression in monocytes from NOD mice. Similarly, in human participants, ex vivo whole blood stimulation showed higher induced IFN-1 responses in participants with T1D compared with healthy controls. In our recent study, we, too, observed increased secretion of IFN-α by the peripheral pDCs from T1D subjects compared to non-diabetic controls. Enhanced IFN-α secretion was also observed after stimulation with DNA-LL37 complexes indicating the inflammatory nature of pDCs derived from T1D subjects. Collectively, these data support the notion that IFN-α mediated effects play an important role in the early pathogenic events during initiation of autoimmune diabetes, and the presence of early type 1 IFN signature in susceptible individuals and animal models suggests the role of viral nucleic acids, and to some extent, the self-nucleic acids in T1D pathogenesis.
During the initial phase of T1D, innate immune cells, like DCs, neutrophils and macrophages, infiltrate the islets much before the infiltration of T and B cells[2,90-92]. This buildup of innate immune cells is persistent during the later β-cell destructive insulitis as well[93]. Therefore, the entry of DCs and macrophages/monocytes can be considered an initial sign of the autoimmune process during the pathogenesis of T1D[1,20,94].
Although there is ambiguity regarding the exact role of innate immune cells and other initial triggers involved in the loss of β-cell tolerance, certain factors, like viral infection and ER stress are known to provoke an immune response in β-cells leading to the activation of pro-inflammatory pathways. Additionally, β-cells themselves might also participate in their demise by invoking apoptosis rather than being an innocent victim of autoimmune attack as previously thought [95]. One of the outcomes of β-cell destruction is the release of self-nucleic acids along with other cellular debris. Among the nucleic acids, the role of self-DNA in the development of T1D is highlighted by few studies, Diana et al[1] demonstrated that neutrophils, B-1a cells, and plasmacytoid dendritic cells are recruited to islets during physiological periods of β-cell death. Activated B-1a cells secrete dsDNA specific IgGs, which activate neutrophils to release DNA-binding cathelicidin-related antimicrobial peptide (CRAMP), which binds self-DNA, and along with DNA-specific IgG, activating pDCs through the TLR9–MyD88 pathway, leading to IFN-α production in pancreatic islets and initiation autoimmune diabetes in NOD mice. Mollah et al[96] observed increased incidence of diabetes associated with increased accumulation of ssDNA in the immune cells of granzyme A (protease degrading intracellular DNA) deficient NOD mouse due to induction of IFN response in pancreatic islets. The study identified DNA as a novel endogenous trigger of autoimmune diabetes and an in vivo role for granzyme A in maintaining immune tolerance. Earlier, Zentsova et al[97] had also observed that monocytes contribute to DNA sensing in patients with T1D via the TBK1 and STING pathways by recognizing CpG-DNA leading to the release of IFN-α and proinflammatory cytokines. These studies highlight the importance of investigating the interaction of DNA sensors of innate immune cells during the early pathogenesis of T1D. However, limitations in obtaining pancreatic tissues pose a big challenge in assessing such interactions.
Besides DNA, the role of self-RNA in the progression of T1D is also being speculated. A study by Kocic et al[98] demonstrated that accumulation of circulating self-RNA can lead to the progression of autoimmune or inflammatory conditions in subjects with juvenile T1D. Recently, studies from several groups suggested that adenosine deaminase acting on RNA (Adar1) deficiency leads to the accumulation of retroelements, such as Alu:Alu hybrids, in the cytoplasm, which are then recognized by MDA5, resulting in excessive proinflammatory response[99,100]. Furthermore, mouse models deficient in Adar1 established that dysregulated RNA editing caused MDA5-driven autoimmunity[101,102]. The role of mtDNA acting as a ligand for nucleic acid sensors is also being observed by various research groups. When mtDNA is released into extracellular space and cytoplasm, it activates a variety of innate immune responses. West et al[103] showed that the mitochondrial transcription factor A (TFAM) deficiency leads to mis-packaged mtDNA, resulting into its cytoplasmic release where it bound and activated cGAS initiating a type-I IFN response. mtDNA has also been involved in the activation of inflammasome[104]. Carlos et al[105] shown that mtDNA activates NLRP3 to trigger IL-1β secretion via caspase-1-dependent pathway to precipitate the onset of streptozotocin (STZ) induced T1D in C57BL/6 mice. In 2020, Pereira et al[61] observed that mtDNA promoted NLRP3 inflammasome activation that contributed to inflammation and endothelial dysfunction in patients with T1D.
The role of nucleic acids and their signaling has been explored by several studies in many autoimmune diseases, yet there is very little data on the aberrations in nucleic acid sensing mechanisms in autoimmune vs non-autoimmune conditions. Parallels are drawn from those autoimmune diseases, like psoriasis and SLE, where nucleic acids are targeted by the immune cells. During the pathogenesis of SLE, the pDCs get activated due to facilitated recognition of autoantibodies against nucleic acids by TLR7 and TLR9 leading to increased secretion of type 1 IFNs[106,107]. A similar role of self-DNA complexes and specific antibodies was also suggested by Diana et al[1] during the initial stages of T1D in the activation of TLR9 in pancreatic pDCs, which release IFN-α in NOD mice, as explained earlier.
An important study by Revelo et al[108], explored the possible role of different types of nucleic acids contributing to glucose intolerance during diet induced obesity (DIO). The study concluded that oxidized mtDNA derived from abnormal formation of extracellular traps (ETs) can promote inflammation of metabolic tissues via TLR7 and TLR9 in pDCs. The same study has also explored the possible role of exogenous sources of nucleic acids like CpG-ODN, which worsened glucose tolerance in lean mice, possibly by the recognition of CpG DNA by TLR9. A similar study has also shown that increased levels of circulating cell free DNA are involved in the activation of macrophages via TLR9 during DIO[109]. A recent study by Zentsova et al[97] demonstrated altered DNA sensing in subjects with T1D in response to microbial DNA. Prominent proinflammatory responses were observed in pDCs and monocytes of T1D patients compared to healthy controls. Furthermore, monocytes isolated from T1D subjects were shown to bind and internalize DNA and responded by releasing higher levels of proinflammatory cytokines as compared to control subjects. Surprisingly, this cytokine production was independent of the TLR9 signaling pathway but dependent on other intracellular receptors like, TBK1 and STING for recognition of CpG-DNA and NETs, which were used to mimic self-DNA in the study. During our study on the role of self-DNA in T1D, we have also observed that the pDCs and monocytes of T1D subjects behave differently from those of healthy subjects. We observed that the pDCs and monocytes of T1D subjects were more prompt on acquiring an inflammatory phenotype upon stimulation with molecules like DNA-LL37 complexes by initiating inflammation through IFN-α and augmenting autoimmunity by activating CD4+ T cells[110]. Therefore, it appears that either altered forms of nucleic acids or alterations in their sensors underlie the dysregulations in nucleic acid sensing in autoimmunity.
In normal circumstances, the self-nucleic acids are considered non-immunogenic in nature and in the extracellular environment, they undergo rapid degradation by various extracellular nucleases[111]. However, their binding to peptides like, LL37 and HMGB1 (released by neutrophils and monocytes, respectively)[112,113] can lead to the formation of complexes that are resistant to nuclease degradation. These complexes are transported to endosomal compartments of pDCs and monocytes, which are recognized by TLR9[114]. In the case of NOD mice, CRAMP (mouse equivalent of LL37) is known to form complexes with self-DNA and DNA-specific IgG to induce IFN-α production via the TLR9 and MyD88 pathways. In T1D, we have also observed that LL37 forms stable complexes with self-DNA to protect it from DNase degradation and, at the same time, it increases the efficiency by which pDCs and monocytes engulf DNA complexes in their cytosol[110]. Moreover, delayed clearance of apoptotic cells and other cellular debris by the macrophages also causes their accumulation, which in turn results in increased uptake of nucleic acids by innate immune cells, like pDCs and DCs that express abundant nucleic acid sensors. Apart from self-DNA, self-RNA is also capable of forming stable immune complexes with LL37, which was first observed by some researchers where they observed stable formation of complexes that readily enter endosomes of both pDCs and mDCs to induce TLR7 activation that finally triggers IFN-α secretion. Taking cue from these aforementioned studies it can be concluded that self-nucleic acids, like RNA, DNA and mtDNA, that are released from the dying β cells can form complexes with certain peptides and activate innate immune cells like pDCs, DCs and macrophages, and tilting the local immune homeostasis towards proinflammation.
However, the main unanswered question that remains is how does the uptake of self-nucleic acids or their complexes with proteins confer a proinflammatory phenotype to innate immune cells like the uptake of nucleic acids of viral and bacterial origin. Comparative studies done in past have shed some light and indicated that self-nucleic acids can induce similar if not heightened immune responses during the progression of autoimmune diseases, including T1D, although this hypothesis is still in its nascent stages and require some solid comparative studies, especially in T1D pathogenesis. The role of molecular mimicry by self-nucleic acids cannot be denied as they share similar motifs to pathogenic genomes like that of viruses and bacteria, a very good example of which is the presence of CpG islands in mtDNA. The role of nucleic acid induced innate immune inflammation also becomes particularly important, especially when viral infections alone cannot explain the initial infiltration and activation of innate immune cells, like pDCs, DCs, and monocytes, during the initial stages of T1D.
With the increasing understanding of their roles and the signaling cascades in initiating inflammatory responses, novel therapies involving PRRs, have been attempted to target autoimmune diseases (Table 2).
| No. | Inhibitor | Disease | Target | Phase (Preclinical/Clinical-trial ID) | Ref. |
| 1 | Hydroxychloroquine | Rheumatoid arthritis and SLE | TLR7, TLR9, cGAS-STING | NCT0380218 (Ongoing Trial) | [139] |
| 2 | SM934 | SLE | TLR7 and TLR9 | NCT03951259 (Phase II) | [125] |
| 3 | Amlexanox | T2D | TBK1 and IKKε | NCT01975935 (Phase II) | [140] |
| 4 | TJ-M2010-6 | T1D | Myd88 | Preclinical | [129] |
| 5 | ST-2825 | SLE | IRAK1 and IRAK4 | Preclinical | [126] |
| 6 | Aspirin | AGS | cGAS | Preclinical | [141] |
| 7 | ODN-1411 | Rheumatoid Arthritis | TLR8 | Preclinical | [142] |
| 8 | INH-ODNs | SLE | TLR3 and TLR9 | Preclinical | [143] |
| 9 | X6 | Autoimmune myocarditis | cGAS | Preclinical | [144] |
| 10 | PF-06650833 | Rheumatoid Arthritis | IRAK4 | NCT02996500 (Phase II) | [127] |
| 11 | Compound II | SLE and AGS | TBK1 | Preclinical | [128] |
| 12 | Sifalimumab (MEDI-545) | SLE | IFN-α | NCT00979654 (Phase II) | [145] |
| 13 | AGS-009 | SLE and Rheumatoid Arthritis | IFN-α | NCT00960362 (Phase I) | [132] |
| 14 | IMO-8400 | Plaque Psoriasis | TLR-7, 8, and 9 | NCT01899729 (Phase IIa) | [122] |
| 15 | CpG-52364 | SLE | TLR-7, 8, and 9 | NCT00547014 (Phase I) | [116] |
Historically, targeting of downstream TLR signaling pathways using antimalarial drugs like chloroquine, quinacrine, and hydroxyl-chloroquine (HCQ) have been used in the treatment of autoimmune diseases since the 1940s, suggesting the effectiveness and importance of blocking endosomal TLR signaling rather than blocking TLR ligand themselves[115]. Compared to HCQ, CpG-52364, a quinacrine derivative and small-molecule antagonist of TLR7/8/9 is therapeutically more effective and has fewer side effects in animal studies. A phase I clinical trial for treatment of SLE (NCT00547014) showed inhibition of disease development without causing general immunosuppression[116]. Next, the idea of reducing exogenous DNA and RNA associated DAMPs has also been tried as an alternative and broader approach to suppress non-TLR dependent pathways of IFN production for the treatment of autoimmune diseases. Pulmozyme, a recombinant human DNase, has been in use since 1994 for the treatment of cystic fibrosis[117]. Additionally, Macanovic et al[118] showed that murine DNase can improve renal histology in NZB/NZW F1 Lupus-prone mice. A bovine DNase preparation also had initial success in improving clinical outcomes in a patient trial of SLE, but further studies were precluded due to the development of antibodies to the bovine DNase[119].
Oligodeoxynucleotides (ODNs) were first designed for direct binding and for antagonizing endosomal TLRs as an alternate strategy to treat SLE, which despite showing initial success the therapy, failed to garner support due to several reports of adverse effects like thrombocytopenia and neutropenia. Although greater promise was shown by ODNs, like immunomodulatory oligonucleotides (IMO)-8400 in psoriasis that target TLR7, TLR8, and TLR9 to reduce the expression of IL-17 signaling associated genes[120,121]. A phase 2a clinical trial, sponsored by Idera Pharmaceuticals, involving use of IMO-8400 for the treatment of plaque psoriasis exhibited reduced psoriasis severity with good tolerance in the recruited subjects (NCT01899729)[122]. A preclinical study on INH-ODN-24888, a guanine modified oligonucleotide was initiated for the treatment of lupus patients based on its activity as a TLR7 and TLR9 antagonist, and it was observed to be more efficient than the unmodified oligonucleotide (INH-ODN-2088)[123,124].
Other peptide compounds designed to inhibit TLR signaling pathways in autoimmune diseases include SM934 (b-aminoarteether maleate). It targets TLR7 and TLR9 signaling cascades, thereby promoting their downregulation along with regulation of MyD88 expression and NF-kB activation through an unknown mechanism. Finally, it inhibits TLR-induced activation of B cells leading to a decrease in proliferation and antibody secretion in MRL/Lpr mice (animal model of SLE)[125]. Another peptide ST-2825 that blocks the dimerization of MyD88[126] by interfering with the recruitment of IRAK1 and IRAK4 to TLR7- and TLR9-MyD88 complexes was found to be of therapeutic importance in inhibiting TLR-mediated inflammatory responses. Recently, PF-06650833, a small molecule inhibitor of IRAK4 has been reported to be effective in ameliorating some symptoms in patients with moderate to severe rheumatoid disease[127]. Another molecule, reported as “Compound II” in the study by Hasan et al[128], was shown to inhibit TBK1 and consequently douse the hyper-inflammatory responses in Trex-/- mice. Another novel inhibitor, TJ-M2010-6, has also shown the ability to suppress homo-dimerization of MyD88 by interacting with amino acid residues of its TIR domain, thereby preventing and treating T1D in NOD mice. Upon deducing the mechanistic pathways, it was observed that TJ-M2010-6 treatment prevents insulitis in vivo, whereas in vitro experiments showed inhibition of DCs maturation, leading to suppression of T cell activation and production of inflammatory cytokines[129]. To directly target the interaction of TLRs with their corresponding ligands, several antibodies have been designed, including Sifalimumab (NCT00979654, NCT01283139) and AGS-009 (NCT00960362). Both of the antibodies showed significant reduction of the IFN-α signature in the clinical trials aimed at SLE treatment[130-132]. However, despite the indispensable role of endosomal TLRs in the pathology of several type 1 IFN-driven autoimmune diseases, the therapeutic strategies against TLR7, TLR8, and TLR9 have yet to see appreciable success in various clinical trials.
Recent data on the involvement of molecular pathways leading to NETosis, and the components of NETs, like myeloperoxidase MPO, neutrophil elastase NE, and nucleic acids, have made them an attractive target for therapeutic strategies in autoimmune diseases, including T1D[133]. The best studied and the viable target is PAD4, which is a nuclear enzyme mediating NET formation by chromatin de-condensation[134], several inhibitors against NETs have been tried, of which GSK484 has shown persistent activity in animal models of inflammatory disease[135]. Additionally, an enzyme, staphylococcal nuclease, has shown some promise by degrading intestinal NETs and ameliorating both intestine and pancreatic islet inflammation to effectively regulate the blood glucose homeostasis in NOD mice[136]. Keeping in view the important roles played by nucleic acid sensing in shaping immune responses, specifically via modulation of innate immunity, researchers are actively exploring the nucleic acid-based nanoparticles that can be designed and functionalized with known therapeutic immunomodulatory domains and motifs, for the treatment of various nucleic acid centered autoimmune diseases[137,138]. Collectively, these studies emphasize the scope of further exploration of novel approaches to targeting key checkpoints in nucleic acid recognition and their downstream signaling pathways.
There are ample studies on T1D pathogenesis in both humans and animal models, and significant progress has been made in understanding the role of various cellular mechanisms involved in the initiation of the disease. Emerging data on the contribution of nucleic acids and their receptors on innate immune cells is challenging the current dogmatic and historical view of T1D as being a T cell driven disease.
The evolving view, that we have tried to support in this review, is that the initiation of autoimmune diabetes and its etiopathogenesis is much more complex and might involve aberrant recognition of self-nucleic acids at a very early stage. Recent findings from several groups have suggested the role of self-nucleic acids in elevating IFN induced responses by involving several PRRs in various autoimmune disorders including T1D. We would further like to propose that recognition of these self-nucleic acids by various innate immune cell subsets may have a similar outcome as in other autoimmune diseases, like SLE and psoriasis, where DAMPs like self-nucleic acids play a crucial role in the precipitation of the disease. However, despite this growing knowledge, further insights are required on the role of various nucleic acids and their sensors particularly in the context of the regulation of their downstream signaling mediators during the pathogenesis of T1D. Thus, it becomes necessary to search for novel inhibitors or receptor antagonists as a way of modulating dysregulated nucleic acid sensing, which might be useful in preventing or delaying the progression of T1D and similar autoimmune diseases.
Manuscript source: Invited manuscript
Corresponding Author's Membership in Professional Societies: Immunology of Diabetes Society; Indian Immunology Society; and The Cytometry Society (India).
Specialty type: Immunology
Country/Territory of origin: India
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Feng L S-Editor: Ma YJ L-Editor: Filipodia P-Editor: Guo X
| 1. | Diana J, Simoni Y, Furio L, Beaudoin L, Agerberth B, Barrat F, Lehuen A. Crosstalk between neutrophils, B-1a cells and plasmacytoid dendritic cells initiates autoimmune diabetes. Nat Med. 2013;19:65-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 311] [Cited by in RCA: 347] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 2. | Pietropaolo M, Barinas-Mitchell E, Kuller LH. The heterogeneity of diabetes: unraveling a dispute: is systemic inflammation related to islet autoimmunity? Diabetes. 2007;56:1189-1197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 53] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 3. | Nakayama M. Insulin as a key autoantigen in the development of type 1 diabetes. Diabetes Metab Res Rev. 2011;27:773-777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 4. | Knight RR, Kronenberg D, Zhao M, Huang GC, Eichmann M, Bulek A, Wooldridge L, Cole DK, Sewell AK, Peakman M, Skowera A. Human β-cell killing by autoreactive preproinsulin-specific CD8 T cells is predominantly granule-mediated with the potency dependent upon T-cell receptor avidity. Diabetes. 2013;62:205-213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 5. | van Belle TL, Coppieters KT, von Herrath MG. Type 1 diabetes: etiology, immunology, and therapeutic strategies. Physiol Rev. 2011;91:79-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 634] [Cited by in RCA: 684] [Article Influence: 48.9] [Reference Citation Analysis (0)] |
| 6. | Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22:240-273, Table of Contents. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1862] [Cited by in RCA: 2191] [Article Influence: 136.9] [Reference Citation Analysis (0)] |
| 7. | Newton K, Dixit VM. Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol. 2012;4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1055] [Cited by in RCA: 1174] [Article Influence: 90.3] [Reference Citation Analysis (0)] |
| 8. | West AP, Koblansky AA, Ghosh S. Recognition and signaling by toll-like receptors. Annu Rev Cell Dev Biol. 2006;22:409-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 490] [Cited by in RCA: 534] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 9. | Doyle SE, O'Connell RM, Miranda GA, Vaidya SA, Chow EK, Liu PT, Suzuki S, Suzuki N, Modlin RL, Yeh WC, Lane TF, Cheng G. Toll-like receptors induce a phagocytic gene program through p38. J Exp Med. 2004;199:81-90. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 299] [Cited by in RCA: 322] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 10. | Blander JM, Medzhitov R. Regulation of phagosome maturation by signals from toll-like receptors. Science. 2004;304:1014-1018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 756] [Cited by in RCA: 765] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 11. | Fischer S. Pattern Recognition Receptors and Control of Innate Immunity: Role of Nucleic Acids. Curr Pharm Biotechnol. 2018;19:1203-1209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 12. | Barrat FJ, Elkon KB, Fitzgerald KA. Importance of Nucleic Acid Recognition in Inflammation and Autoimmunity. Annu Rev Med. 2016;67:323-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 113] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 13. | Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30:16-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1380] [Cited by in RCA: 1545] [Article Influence: 110.4] [Reference Citation Analysis (0)] |
| 14. | Matsushima N, Tanaka T, Enkhbayar P, Mikami T, Taga M, Yamada K, Kuroki Y. Comparative sequence analysis of leucine-rich repeats (LRRs) within vertebrate toll-like receptors. BMC Genomics. 2007;8:124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 271] [Cited by in RCA: 275] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 15. | Takeuchi O, Akira S. Signaling pathways activated by microorganisms. Curr Opin Cell Biol. 2007;19:185-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 62] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 16. | McGettrick AF, O'Neill LA. Toll-like receptors: key activators of leucocytes and regulator of haematopoiesis. Br J Haematol. 2007;139:185-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 90] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 17. | Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Front Immunol. 2014;5:461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1579] [Cited by in RCA: 2310] [Article Influence: 210.0] [Reference Citation Analysis (0)] |
| 18. | Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5843] [Cited by in RCA: 6747] [Article Influence: 449.8] [Reference Citation Analysis (0)] |
| 19. | Zhong JX, Xu JF, Yang P, Liang Y, Wang CY. Innate Immunity in the Recognition of β-Cell Antigens in Type 1 Diabetes. Wagner DD, editor. InTech Open. 2011;. [DOI] [Full Text] |
| 20. | Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2255] [Cited by in RCA: 2378] [Article Influence: 118.9] [Reference Citation Analysis (0)] |
| 21. | Takeda K, Akira S. Roles of Toll-like receptors in innate immune responses. Genes Cells. 2001;6:733-742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 198] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 22. | van Kooyk Y, Geijtenbeek TB. Toll-like receptors keep antigen sorting on the right track. Immunity. 2006;25:525-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 23. | Rifkin IR, Leadbetter EA, Busconi L, Viglianti G, Marshak-Rothstein A. Toll-like receptors, endogenous ligands, and systemic autoimmune disease. Immunol Rev. 2005;204:27-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 305] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 24. | Richer MJ, Lavallée DJ, Shanina I, Horwitz MS. Toll-like receptor 3 signaling on macrophages is required for survival following coxsackievirus B4 infection. PLoS One. 2009;4:e4127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 110] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 25. | Assmann TS, Brondani Lde A, Bouças AP, Canani LH, Crispim D. Toll-like receptor 3 (TLR3) and the development of type 1 diabetes mellitus. Arch Endocrinol Metab. 2015;59:4-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 26. | Lee AS, Ghoreishi M, Cheng WK, Chang TY, Zhang YQ, Dutz JP. Toll-like receptor 7 stimulation promotes autoimmune diabetes in the NOD mouse. Diabetologia. 2011;54:1407-1416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 27. | Pane JA, Webster NL, Coulson BS. Rotavirus activates lymphocytes from non-obese diabetic mice by triggering toll-like receptor 7 signaling and interferon production in plasmacytoid dendritic cells. PLoS Pathog. 2014;10:e1003998. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 28. | Zhang Y, Lee AS, Shameli A, Geng X, Finegood D, Santamaria P, Dutz JP. TLR9 blockade inhibits activation of diabetogenic CD8+ T cells and delays autoimmune diabetes. J Immunol. 2010;184:5645-5653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 60] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 29. | Liu M, Peng J, Tai N, Pearson JA, Hu C, Guo J, Hou L, Zhao H, Wong FS, Wen L. Toll-like receptor 9 negatively regulates pancreatic islet beta cell growth and function in a mouse model of type 1 diabetes. Diabetologia. 2018;61:2333-2343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 30. | Yoneyama M, Fujita T. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity. 2008;29:178-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 207] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 31. | Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M Jr, Akira S, Yonehara S, Kato A, Fujita T. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851-2858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1264] [Cited by in RCA: 1281] [Article Influence: 64.1] [Reference Citation Analysis (0)] |
| 32. | Chiang C, Gack MU. Post-translational Control of Intracellular Pathogen Sensing Pathways. Trends Immunol. 2017;38:39-52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 122] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 33. | Nakhaei P, Genin P, Civas A, Hiscott J. RIG-I-like receptors: sensing and responding to RNA virus infection. Semin Immunol. 2009;21:215-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 172] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 34. | Smyth DJ, Cooper JD, Bailey R, Field S, Burren O, Smink LJ, Guja C, Ionescu-Tirgoviste C, Widmer B, Dunger DB, Savage DA, Walker NM, Clayton DG, Todd JA. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet. 2006;38:617-619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 533] [Cited by in RCA: 490] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 35. | Cen H, Wang W, Leng RX, Wang TY, Pan HF, Fan YG, Wang B, Ye DQ. Association of IFIH1 rs1990760 polymorphism with susceptibility to autoimmune diseases: a meta-analysis. Autoimmunity. 2013;46:455-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 72] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 36. | Nejentsev S, Howson JM, Walker NM, Szeszko J, Field SF, Stevens HE, Reynolds P, Hardy M, King E, Masters J, Hulme J, Maier LM, Smyth D, Bailey R, Cooper JD, Ribas G, Campbell RD, Clayton DG, Todd JA; Wellcome Trust Case Control Consortium. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450:887-892. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 470] [Cited by in RCA: 434] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 37. | Downes K, Pekalski M, Angus KL, Hardy M, Nutland S, Smyth DJ, Walker NM, Wallace C, Todd JA. Reduced expression of IFIH1 is protective for type 1 diabetes. PLoS One. 2010;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 76] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 38. | Schulte BM, Gielen PR, Kers-Rebel ED, Prosser AC, Lind K, Flodström-Tullberg M, Tack CJ, Elving LD, Adema GJ. Enterovirus Exposure Uniquely Discriminates Type 1 Diabetes Patients with a Homozygous from a Heterozygous Melanoma Differentiation-Associated Protein 5/Interferon Induced with Helicase C Domain 1 A946T Genotype. Viral Immunol. 2016;29:389-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 39. | Cinek O, Tapia G, Witsø E, Kramna L, Holkova K, Rasmussen T, Stene LC, Rønningen KS. Enterovirus RNA in peripheral blood may be associated with the variants of rs1990760, a common type 1 diabetes associated polymorphism in IFIH1. PLoS One. 2012;7:e48409. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 40. | Rice GI, Del Toro Duany Y, Jenkinson EM, Forte GM, Anderson BH, Ariaudo G, Bader-Meunier B, Baildam EM, Battini R, Beresford MW, Casarano M, Chouchane M, Cimaz R, Collins AE, Cordeiro NJ, Dale RC, Davidson JE, De Waele L, Desguerre I, Faivre L, Fazzi E, Isidor B, Lagae L, Latchman AR, Lebon P, Li C, Livingston JH, Lourenço CM, Mancardi MM, Masurel-Paulet A, McInnes IB, Menezes MP, Mignot C, O'Sullivan J, Orcesi S, Picco PP, Riva E, Robinson RA, Rodriguez D, Salvatici E, Scott C, Szybowska M, Tolmie JL, Vanderver A, Vanhulle C, Vieira JP, Webb K, Whitney RN, Williams SG, Wolfe LA, Zuberi SM, Hur S, Crow YJ. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat Genet. 2014;46:503-509. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 401] [Cited by in RCA: 478] [Article Influence: 43.5] [Reference Citation Analysis (0)] |
| 41. | Gorman JA, Hundhausen C, Errett JS, Stone AE, Allenspach EJ, Ge Y, Arkatkar T, Clough C, Dai X, Khim S, Pestal K, Liggitt D, Cerosaletti K, Stetson DB, James RG, Oukka M, Concannon P, Gale M Jr, Buckner JH, Rawlings DJ. The A946T variant of the RNA sensor IFIH1 mediates an interferon program that limits viral infection but increases the risk for autoimmunity. Nat Immunol. 2017;18:744-752. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 110] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 42. | Shigemoto T, Kageyama M, Hirai R, Zheng J, Yoneyama M, Fujita T. Identification of loss of function mutations in human genes encoding RIG-I and MDA5: implications for resistance to type I diabetes. J Biol Chem. 2009;284:13348-13354. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 129] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 43. | Connolly DJ, Bowie AG. The emerging role of human PYHIN proteins in innate immunity: implications for health and disease. Biochem Pharmacol. 2014;92:405-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 44. | Kawasaki T, Kawai T. Discrimination Between Self and Non-Self-Nucleic Acids by the Innate Immune System. Int Rev Cell Mol Biol. 2019;344:1-30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 45. | Benmerzoug S, Ryffel B, Togbe D, Quesniaux VFJ. Self-DNA Sensing in Lung Inflammatory Diseases. Trends Immunol. 2019;40:719-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 46. | Inohara N, Nuñez G. The NOD: a signaling module that regulates apoptosis and host defense against pathogens. Oncogene. 2001;20:6473-6481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 179] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 47. | Inohara N, Nuñez G. NODs: intracellular proteins involved in inflammation and apoptosis. Nat Rev Immunol. 2003;3:371-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 757] [Cited by in RCA: 738] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 48. | Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem. 2001;276:4812-4818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1034] [Cited by in RCA: 1041] [Article Influence: 43.4] [Reference Citation Analysis (0)] |
| 49. | Zheng C. The emerging roles of NOD-like receptors in antiviral innate immune signaling pathways. Int J Biol Macromol. 2021;169:407-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 57] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 50. | Saxena M, Yeretssian G. NOD-Like Receptors: Master Regulators of Inflammation and Cancer. Front Immunol. 2014;5:327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 196] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 51. | Franklin BS, Bossaller L, De Nardo D, Ratter JM, Stutz A, Engels G, Brenker C, Nordhoff M, Mirandola SR, Al-Amoudi A, Mangan MS, Zimmer S, Monks BG, Fricke M, Schmidt RE, Espevik T, Jones B, Jarnicki AG, Hansbro PM, Busto P, Marshak-Rothstein A, Hornemann S, Aguzzi A, Kastenmüller W, Latz E. The adaptor ASC has extracellular and 'prionoid' activities that propagate inflammation. Nat Immunol. 2014;15:727-737. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 600] [Cited by in RCA: 618] [Article Influence: 56.2] [Reference Citation Analysis (0)] |
| 52. | Grishman EK, White PC, Savani RC. Toll-like receptors, the NLRP3 inflammasome, and interleukin-1β in the development and progression of type 1 diabetes. Pediatr Res. 2012;71:626-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 77] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 53. | Bauernfeind FG, Horvath G, Stutz A, Alnemri ES, MacDonald K, Speert D, Fernandes-Alnemri T, Wu J, Monks BG, Fitzgerald KA, Hornung V, Latz E. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol. 2009;183:787-791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2409] [Cited by in RCA: 2342] [Article Influence: 146.4] [Reference Citation Analysis (1)] |
| 54. | Krishnan SM, Sobey CG, Latz E, Mansell A, Drummond GR. IL-1β and IL-18: inflammatory markers or mediators of hypertension? Br J Pharmacol. 2014;171:5589-5602. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 175] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 55. | Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514-518. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2094] [Cited by in RCA: 2025] [Article Influence: 126.6] [Reference Citation Analysis (0)] |
| 56. | Tourkochristou E, Aggeletopoulou I, Konstantakis C, Triantos C. Role of NLRP3 inflammasome in inflammatory bowel diseases. World J Gastroenterol. 2019;25:4796-4804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 43] [Cited by in RCA: 77] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 57. | Soares JL, Oliveira EM, Pontillo A. Variants in NLRP3 and NLRC4 inflammasome associate with susceptibility and severity of multiple sclerosis. Mult Scler Relat Disord. 2019;29:26-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 58. | Sun X, Xia Y, Liu Y, Wang Y, Luo S, Lin J, Huang G, Li X, Xie Z, Zhou Z. Polymorphisms in NLRP1 Gene Are Associated with Type 1 Diabetes. J Diabetes Res. 2019;2019:7405120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 59. | Hu C, Ding H, Li Y, Pearson JA, Zhang X, Flavell RA, Wong FS, Wen L. NLRP3 deficiency protects from type 1 diabetes through the regulation of chemotaxis into the pancreatic islets. Proc Natl Acad Sci U S A. 2015;112:11318-11323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 105] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 60. | Carlos D, Costa F, Leite J, André C, Tostes R. NLRP3 Inflammasome: From Pathogenesis to Therapeutic Strategies in Type 1 Diabetes. J Autoimmu Disord. 2017;3:30. |
| 61. | Pereira CA, Carlos D, Ferreira NS, Silva JF, Zanotto CZ, Zamboni DS, Garcia VD, Ventura DF, Silva JS, Tostes RC. Mitochondrial DNA Promotes NLRP3 Inflammasome Activation and Contributes to Endothelial Dysfunction and Inflammation in Type 1 Diabetes. Front Physiol. 2019;10:1557. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 59] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 62. | Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390-1394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 705] [Cited by in RCA: 897] [Article Influence: 74.8] [Reference Citation Analysis (0)] |
| 63. | Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51:226-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 615] [Cited by in RCA: 836] [Article Influence: 69.7] [Reference Citation Analysis (0)] |
| 64. | Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3162] [Cited by in RCA: 3576] [Article Influence: 298.0] [Reference Citation Analysis (0)] |
| 65. | Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674-678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2812] [Cited by in RCA: 2682] [Article Influence: 157.8] [Reference Citation Analysis (0)] |
| 66. | Dobbs N, Burnaevskiy N, Chen D, Gonugunta VK, Alto NM, Yan N. STING Activation by Translocation from the ER Is Associated with Infection and Autoinflammatory Disease. Cell Host Microbe. 2015;18:157-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 473] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 67. | Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, Yamamoto N, Kawai T, Ishii K, Takeuchi O, Yoshimori T, Akira S. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:20842-20846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 563] [Cited by in RCA: 677] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 68. | Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788-792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1620] [Cited by in RCA: 2100] [Article Influence: 131.3] [Reference Citation Analysis (0)] |
| 69. | Fitzgerald KA. The interferon inducible gene: Viperin. J Interferon Cytokine Res. 2011;31:131-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 132] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 70. | Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5:ra20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1034] [Cited by in RCA: 993] [Article Influence: 76.4] [Reference Citation Analysis (0)] |
| 71. | Barber GN. STING: infection, inflammation and cancer. Nat Rev Immunol. 2015;15:760-770. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 645] [Cited by in RCA: 1006] [Article Influence: 111.8] [Reference Citation Analysis (0)] |
| 72. | Lemos H, Mohamed E, Huang L, Chandler PR, Ou R, Pacholczyk R, Mellor AL. Stimulator of interferon genes agonists attenuate type I diabetes progression in NOD mice. Immunology. 2019;158:353-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 73. | Christensen SR, Shupe J, Nickerson K, Kashgarian M, Flavell RA, Shlomchik MJ. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25:417-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 782] [Cited by in RCA: 837] [Article Influence: 44.1] [Reference Citation Analysis (0)] |
| 74. | Devarapu SK, Anders HJ. Toll-like receptors in lupus nephritis. J Biomed Sci. 2018;25:35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 66] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 75. | McCall KD, Thuma JR, Courreges MC, Benencia F, James CB, Malgor R, Kantake N, Mudd W, Denlinger N, Nolan B, Wen L, Schwartz FL. Toll-like receptor 3 is critical for coxsackievirus B4-induced type 1 diabetes in female NOD mice. Endocrinology. 2015;156:453-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 76. | Jang MA, Kim EK, Now H, Nguyen NT, Kim WJ, Yoo JY, Lee J, Jeong YM, Kim CH, Kim OH, Sohn S, Nam SH, Hong Y, Lee YS, Chang SA, Jang SY, Kim JW, Lee MS, Lim SY, Sung KS, Park KT, Kim BJ, Lee JH, Kim DK, Kee C, Ki CS. Mutations in DDX58, which encodes RIG-I, cause atypical Singleton-Merten syndrome. Am J Hum Genet. 2015;96:266-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 173] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 77. | Oda H, Nakagawa K, Abe J, Awaya T, Funabiki M, Hijikata A, Nishikomori R, Funatsuka M, Ohshima Y, Sugawara Y, Yasumi T, Kato H, Shirai T, Ohara O, Fujita T, Heike T. Aicardi-Goutières syndrome is caused by IFIH1 mutations. Am J Hum Genet. 2014;95:121-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 171] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 78. | Lincez PJ. MDA5 and a type 1 interferon signature in the development of type 1 diabetes. Univers British Columbia. 2015;. [DOI] [Full Text] |
| 79. | An J, Durcan L, Karr RM, Briggs TA, Rice GI, Teal TH, Woodward JJ, Elkon KB. Expression of Cyclic GMP-AMP Synthase in Patients With Systemic Lupus Erythematosus. Arthritis Rheumatol. 2017;69:800-807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 146] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 80. | Crowl JT, Gray EE, Pestal K, Volkman HE, Stetson DB. Intracellular Nucleic Acid Detection in Autoimmunity. Annu Rev Immunol. 2017;35:313-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 178] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 81. | Yang CA, Huang ST, Chiang BL. Sex-dependent differential activation of NLRP3 and AIM2 inflammasomes in SLE macrophages. Rheumatology (Oxford). 2015;54:324-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 105] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 82. | Baer AN, Petri M, Sohn J, Rosen A, Casciola-Rosen L. Association of Antibodies to Interferon-Inducible Protein-16 With Markers of More Severe Disease in Primary Sjögren's Syndrome. Arthritis Care Res (Hoboken). 2016;68:254-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 83. | Alunno A, Caneparo V, Bistoni O, Caterbi S, Terenzi R, Gariglio M, Bartoloni E, Manzo A, Landolfo S, Gerli R. Circulating Interferon-Inducible Protein IFI16 Correlates With Clinical and Serological Features in Rheumatoid Arthritis. Arthritis Care Res (Hoboken). 2016;68:440-445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 84. | N'diaye M, Brauner S, Flytzani S, Kular L, Warnecke A, Adzemovic MZ, Piket E, Min JH, Edwards W, Mela F, Choi HY, Magg V, James T, Linden M, Reichardt HM, Daws MR, van Horssen J, Kockum I, Harris RA, Olsson T, Guerreiro-Cacais AO, Jagodic M. C-type lectin receptors Mcl and Mincle control development of multiple sclerosis-like neuroinflammation. J Clin Invest. 2020;130:838-852. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 85. | Marroqui L, Dos Santos RS, Op de Beeck A, Coomans de Brachène A, Marselli L, Marchetti P, Eizirik DL. Interferon-α mediates human beta cell HLA class I overexpression, endoplasmic reticulum stress and apoptosis, three hallmarks of early human type 1 diabetes. Diabetologia. 2017;60:656-667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 149] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 86. | Xia CQ, Peng R, Chernatynskaya AV, Yuan L, Carter C, Valentine J, Sobel E, Atkinson MA, Clare-Salzler MJ. Increased IFN-α-producing plasmacytoid dendritic cells (pDCs) in human Th1-mediated type 1 diabetes: pDCs augment Th1 responses through IFN-α production. J Immunol. 2014;193:1024-1034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 87. | Kayserova J, Vcelakova J, Stechova K, Dudkova E, Hromadkova H, Sumnik Z, Kolouskova S, Spisek R, Sediva A. Decreased dendritic cell numbers but increased TLR9-mediated interferon-alpha production in first degree relatives of type 1 diabetes patients. Clin Immunol. 2014;153:49-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 88. | Marro BS, Ware BC, Zak J, de la Torre JC, Rosen H, Oldstone MB. Progression of type 1 diabetes from the prediabetic stage is controlled by interferon-α signaling. Proc Natl Acad Sci U S A. 2017;114:3708-3713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 89. | Rodrigues KB, Dufort MJ, Llibre A, Speake C, Rahman MJ, Bondet V, Quiel J, Linsley PS, Greenbaum CJ, Duffy D, Tarbell KV. Innate immune stimulation of whole blood reveals IFN-1 hyper-responsiveness in type 1 diabetes. Diabetologia. 2020;63:1576-1587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 90. | Delovitch TL, Singh B. The nonobese diabetic mouse as a model of autoimmune diabetes: immune dysregulation gets the NOD. Immunity. 1997;7:727-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 511] [Cited by in RCA: 508] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 91. | Rosmalen JG, Martin T, Dobbs C, Voerman JS, Drexhage HA, Haskins K, Leenen PJ. Subsets of macrophages and dendritic cells in nonobese diabetic mouse pancreatic inflammatory infiltrates: correlation with the development of diabetes. Lab Invest. 2000;80:23-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 45] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 92. | Charré S, Rosmalen JG, Pelegri C, Alves V, Leenen PJ, Drexhage HA, Homo-Delarche F. Abnormalities in dendritic cell and macrophage accumulation in the pancreas of nonobese diabetic (NOD) mice during the early neonatal period. Histol Histopathol. 2002;17:393-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 93. | Jansen A, Homo-Delarche F, Hooijkaas H, Leenen PJ, Dardenne M, Drexhage HA. Immunohistochemical characterization of monocytes-macrophages and dendritic cells involved in the initiation of the insulitis and beta-cell destruction in NOD mice. Diabetes. 1994;43:667-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 216] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 94. | Pearson AM. Scavenger receptors in innate immunity. Curr Opin Immunol. 1996;8:20-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 204] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 95. | Eizirik DL, Colli ML, Ortis F. The role of inflammation in insulitis and beta-cell loss in type 1 diabetes. Nat Rev Endocrinol. 2009;5:219-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 722] [Cited by in RCA: 785] [Article Influence: 49.1] [Reference Citation Analysis (0)] |
| 96. | Mollah ZUA, Quah HS, Graham KL, Jhala G, Krishnamurthy B, Dharma JFM, Chee J, Trivedi PM, Pappas EG, Mackin L, Chu EPF, Akazawa S, Fynch S, Hodson C, Deans AJ, Trapani JA, Chong MMW, Bird PI, Brodnicki TC, Thomas HE, Kay TWH. Granzyme A Deficiency Breaks Immune Tolerance and Promotes Autoimmune Diabetes Through a Type I Interferon-Dependent Pathway. Diabetes. 2017;66:3041-3050. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 97. | Zentsova I, Parackova Z, Kayserova J, Palova-Jelinkova L, Vrabcova P, Volfova N, Sumnik Z, Pruhova S, Petruzelkova L, Sediva A. Monocytes contribute to DNA sensing through the TBK1 signaling pathway in type 1 diabetes patients. J Autoimmun. 2019;105:102294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 98. | Kocic G, Pavlovic R, Najman S, Nikolic G, Sokolovic D, Jevtovic-Stoimenov T, Musovic D, Veljkovic A, Kocic R, Djindjic N. Circulating ribonucleic acids and metabolic stress parameters may reflect progression of autoimmune or inflammatory conditions in juvenile type 1 diabetes. ScientificWorldJournal. 2011;11:1496-1508. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 99. | Chung H, Calis JJA, Wu X, Sun T, Yu Y, Sarbanes SL, Dao Thi VL, Shilvock AR, Hoffmann HH, Rosenberg BR, Rice CM. Human ADAR1 Prevents Endogenous RNA from Triggering Translational Shutdown. Cell. 2018;172:811-824.e14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 260] [Cited by in RCA: 396] [Article Influence: 56.6] [Reference Citation Analysis (0)] |
| 100. | Ahmad S, Mu X, Yang F, Greenwald E, Park JW, Jacob E, Zhang CZ, Hur S. Breaching Self-Tolerance to Alu Duplex RNA Underlies MDA5-Mediated Inflammation. Cell. 2018;172:797-810.e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 301] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 101. | Mannion NM, Greenwood SM, Young R, Cox S, Brindle J, Read D, Nellåker C, Vesely C, Ponting CP, McLaughlin PJ, Jantsch MF, Dorin J, Adams IR, Scadden AD, Ohman M, Keegan LP, O'Connell MA. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014;9:1482-1494. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 391] [Cited by in RCA: 505] [Article Influence: 45.9] [Reference Citation Analysis (0)] |
| 102. | Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH, Walkley CR. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015;349:1115-1120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 657] [Cited by in RCA: 686] [Article Influence: 68.6] [Reference Citation Analysis (0)] |
| 103. | West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, Kaech SM, Smiley JR, Means RE, Iwasaki A, Shadel GS. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520:553-557. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 911] [Cited by in RCA: 1356] [Article Influence: 135.6] [Reference Citation Analysis (0)] |
| 104. | Zhong Z, Liang S, Sanchez-Lopez E, He F, Shalapour S, Lin XJ, Wong J, Ding S, Seki E, Schnabl B, Hevener AL, Greenberg HB, Kisseleva T, Karin M. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature. 2018;560:198-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 740] [Cited by in RCA: 860] [Article Influence: 122.9] [Reference Citation Analysis (0)] |
| 105. | Carlos D, Costa FR, Pereira CA, Rocha FA, Yaochite JN, Oliveira GG, Carneiro FS, Tostes RC, Ramos SG, Zamboni DS, Camara NO, Ryffel B, Silva JS. Mitochondrial DNA Activates the NLRP3 Inflammasome and Predisposes to Type 1 Diabetes in Murine Model. Front Immunol. 2017;8:164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 83] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 106. | Colonna M, Trinchieri G, Liu YJ. Plasmacytoid dendritic cells in immunity. Nat Immunol. 2004;5:1219-1226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1216] [Cited by in RCA: 1244] [Article Influence: 59.2] [Reference Citation Analysis (0)] |
| 107. | Liu Z, Davidson A. Taming lupus-a new understanding of pathogenesis is leading to clinical advances. Nat Med. 2012;18:871-882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 331] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 108. | Revelo XS, Ghazarian M, Chng MH, Luck H, Kim JH, Zeng K, Shi SY, Tsai S, Lei H, Kenkel J, Liu CL, Tangsombatvisit S, Tsui H, Sima C, Xiao C, Shen L, Li X, Jin T, Lewis GF, Woo M, Utz PJ, Glogauer M, Engleman E, Winer S, Winer DA. Nucleic Acid-Targeting Pathways Promote Inflammation in Obesity-Related Insulin Resistance. Cell Rep. 2016;16:717-730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 59] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 109. | Nishimoto S, Fukuda D, Higashikuni Y, Tanaka K, Hirata Y, Murata C, Kim-Kaneyama JR, Sato F, Bando M, Yagi S, Soeki T, Hayashi T, Imoto I, Sakaue H, Shimabukuro M, Sata M. Obesity-induced DNA released from adipocytes stimulates chronic adipose tissue inflammation and insulin resistance. Sci Adv. 2016;2:e1501332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 196] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 110. | Badal D, Dayal D, Singh G, Sachdeva N. Role of DNA-LL37 complexes in the activation of plasmacytoid dendritic cells and monocytes in subjects with type 1 diabetes. Sci Rep. 2020;10:8896. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 111. | Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783-801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8061] [Cited by in RCA: 8810] [Article Influence: 463.7] [Reference Citation Analysis (0)] |
| 112. | Agerberth B, Charo J, Werr J, Olsson B, Idali F, Lindbom L, Kiessling R, Jörnvall H, Wigzell H, Gudmundsson GH. The human antimicrobial and chemotactic peptides LL-37 and alpha-defensins are expressed by specific lymphocyte and monocyte populations. Blood. 2000;96:3086-3093. [PubMed] |
| 113. | Edfeldt K, Agerberth B, Rottenberg ME, Gudmundsson GH, Wang XB, Mandal K, Xu Q, Yan ZQ. Involvement of the antimicrobial peptide LL-37 in human atherosclerosis. Arterioscler Thromb Vasc Biol. 2006;26:1551-1557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 109] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 114. | Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang YH, Homey B, Cao W, Su B, Nestle FO, Zal T, Mellman I, Schröder JM, Liu YJ, Gilliet M. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564-569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1298] [Cited by in RCA: 1372] [Article Influence: 76.2] [Reference Citation Analysis (0)] |
| 115. | Ruiz-Irastorza G, Ramos-Casals M, Brito-Zeron P, Khamashta MA. Clinical efficacy and side effects of antimalarials in systemic lupus erythematosus: a systematic review. Ann Rheum Dis. 2010;69:20-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 771] [Cited by in RCA: 680] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 116. | Anwar MA, Shah M, Kim J, Choi S. Recent clinical trends in Toll-like receptor targeting therapeutics. Med Res Rev. 2019;39:1053-1090. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 126] [Cited by in RCA: 212] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 117. | Fuchs HJ, Borowitz DS, Christiansen DH, Morris EM, Nash ML, Ramsey BW, Rosenstein BJ, Smith AL, Wohl ME. Effect of aerosolized recombinant human DNase on exacerbations of respiratory symptoms and on pulmonary function in patients with cystic fibrosis. The Pulmozyme Study Group. N Engl J Med. 1994;331:637-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1067] [Cited by in RCA: 1071] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 118. | Macanovic M, Sinicropi D, Shak S, Baughman S, Thiru S, Lachmann PJ. The treatment of systemic lupus erythematosus (SLE) in NZB/W F1 hybrid mice; studies with recombinant murine DNase and with dexamethasone. Clin Exp Immunol. 1996;106:243-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 119] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 119. | Lachmann PJ. Lupus and desoxyribonuclease. Lupus. 2003;12:202-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 120. | Cline A, Felix KH, Oussedik E, Cardwell LA, Feldman SR. Future Therapeutics in Psoriasis. Evidence-Based Psoriasis: Springer, 2018: 93-112. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 121. | Zhu FG, Jiang WW, Dong Y, Kandimalla E, La Monica N, Agrawal S. IMO-8400, a novel TLR7, TLR8 and TLR9 antagonist, inhibits disease development in mouse models of psoriasis (119.8). J Immunol. 2012;188. |
| 122. | Balak DM, van Doorn MB, Arbeit RD, Rijneveld R, Klaassen E, Sullivan T, Brevard J, Thio HB, Prens EP, Burggraaf J, Rissmann R. IMO-8400, a toll-like receptor 7, 8, and 9 antagonist, demonstrates clinical activity in a phase 2a, randomized, placebo-controlled trial in patients with moderate-to-severe plaque psoriasis. Clin Immunol. 2017;174:63-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 123. | Römmler F, Jurk M, Uhlmann E, Hammel M, Waldhuber A, Pfeiffer L, Wagner H, Vollmer J, Miethke T. Guanine modification of inhibitory oligonucleotides potentiates their suppressive function. J Immunol. 2013;191:3240-3253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 124. | Römmler F, Hammel M, Waldhuber A, Müller T, Jurk M, Uhlmann E, Wagner H, Vollmer J, Miethke T. Guanine-modified inhibitory oligonucleotides efficiently impair TLR7- and TLR9-mediated immune responses of human immune cells. PLoS One. 2015;10:e0116703. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 125. | Wu Y, He S, Bai B, Zhang L, Xue L, Lin Z, Yang X, Zhu F, He P, Tang W, Zuo J. Therapeutic effects of the artemisinin analog SM934 on lupus-prone MRL/Lpr mice via inhibition of TLR-triggered B-cell activation and plasma cell formation. Cell Mol Immunol. 2016;13:379-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 90] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 126. | Loiarro M, Capolunghi F, Fantò N, Gallo G, Campo S, Arseni B, Carsetti R, Carminati P, De Santis R, Ruggiero V, Sette C. Pivotal Advance: Inhibition of MyD88 dimerization and recruitment of IRAK1 and IRAK4 by a novel peptidomimetic compound. J Leukoc Biol. 2007;82:801-810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 143] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 127. | Danto SI, Shojaee N, Singh RSP, Li C, Gilbert SA, Manukyan Z, Kilty I. Safety, tolerability, pharmacokinetics, and pharmacodynamics of PF-06650833, a selective interleukin-1 receptor-associated kinase 4 (IRAK4) inhibitor, in single and multiple ascending dose randomized phase 1 studies in healthy subjects. Arthritis Res Ther. 2019;21:269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 128. | Hasan M, Dobbs N, Khan S, White MA, Wakeland EK, Li QZ, Yan N. Cutting Edge: Inhibiting TBK1 by Compound II Ameliorates Autoimmune Disease in Mice. J Immunol. 2015;195:4573-4577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 64] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 129. | Zhang X, Xing S, Li M, Zhang L, Xie L, He W, Liu J, Chang S, Jiang F, Zhou P. Beyond knockout: A novel homodimerization-targeting MyD88 inhibitor prevents and cures type 1 diabetes in NOD mice. Metabolism. 2016;65:1267-1277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 130. | Merrill JT, Wallace DJ, Petri M, Kirou KA, Yao Y, White WI, Robbie G, Levin R, Berney SM, Chindalore V, Olsen N, Richman L, Le C, Jallal B, White B; Lupus Interferon Skin Activity (LISA) Study Investigators. Safety profile and clinical activity of sifalimumab, a fully human anti-interferon α monoclonal antibody, in systemic lupus erythematosus: a phase I, multicentre, double-blind randomised study. Ann Rheum Dis. 2011;70:1905-1913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 191] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 131. | Petri M, Wallace DJ, Spindler A, Chindalore V, Kalunian K, Mysler E, Neuwelt CM, Robbie G, White WI, Higgs BW, Yao Y, Wang L, Ethgen D, Greth W. Sifalimumab, a human anti-interferon-α monoclonal antibody, in systemic lupus erythematosus: a phase I randomized, controlled, dose-escalation study. Arthritis Rheum. 2013;65:1011-1021. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 201] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 132. | Yao Y, Richman L, Higgs BW, Morehouse CA, de los Reyes M, Brohawn P, Zhang J, White B, Coyle AJ, Kiener PA, Jallal B. Neutralization of interferon-alpha/beta-inducible genes and downstream effect in a phase I trial of an anti-interferon-alpha monoclonal antibody in systemic lupus erythematosus. Arthritis Rheum. 2009;60:1785-1796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 225] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 133. | Parackova Z, Zentsova I, Vrabcova P, Klocperk A, Sumnik Z, Pruhova S, Petruzelkova L, Hasler R, Sediva A. Neutrophil Extracellular Trap Induced Dendritic Cell Activation Leads to Th1 Polarization in Type 1 Diabetes. Front Immunol. 2020;11:661. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 134. | Wang Y, Li M, Stadler S, Correll S, Li P, Wang D, Hayama R, Leonelli L, Han H, Grigoryev SA, Allis CD, Coonrod SA. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J Cell Biol. 2009;184:205-213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 872] [Cited by in RCA: 1164] [Article Influence: 72.8] [Reference Citation Analysis (0)] |
| 135. | Lewis HD, Liddle J, Coote JE, Atkinson SJ, Barker MD, Bax BD, Bicker KL, Bingham RP, Campbell M, Chen YH, Chung CW, Craggs PD, Davis RP, Eberhard D, Joberty G, Lind KE, Locke K, Maller C, Martinod K, Patten C, Polyakova O, Rise CE, Rüdiger M, Sheppard RJ, Slade DJ, Thomas P, Thorpe J, Yao G, Drewes G, Wagner DD, Thompson PR, Prinjha RK, Wilson DM. Inhibition of PAD4 activity is sufficient to disrupt mouse and human NET formation. Nat Chem Biol. 2015;11:189-191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 368] [Cited by in RCA: 547] [Article Influence: 54.7] [Reference Citation Analysis (0)] |
| 136. | Liang Y, Wang X, He D, You Q, Zhang T, Dong W, Fei J, Xing Y, Wu J. Ameliorating gut microenvironment through staphylococcal nuclease-mediated intestinal NETs degradation for prevention of type 1 diabetes in NOD mice. Life Sci. 2019;221:301-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 137. | Chandler M, Afonin KA. Smart-Responsive Nucleic Acid Nanoparticles (NANPs) with the Potential to Modulate Immune Behavior. Nanomaterials (Basel). 2019;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 138. | Chandler M, Johnson MB, Panigaj M, Afonin KA. Innate immune responses triggered by nucleic acids inspire the design of immunomodulatory nucleic acid nanoparticles (NANPs). Curr Opin Biotechnol. 2020;63:8-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 139. | Lamphier M, Zheng W, Latz E, Spyvee M, Hansen H, Rose J, Genest M, Yang H, Shaffer C, Zhao Y, Shen Y, Liu C, Liu D, Mempel TR, Rowbottom C, Chow J, Twine NC, Yu M, Gusovsky F, Ishizaka ST. Novel small molecule inhibitors of TLR7 and TLR9: mechanism of action and efficacy in vivo. Mol Pharmacol. 2014;85:429-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 112] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 140. | Beyett TS, Gan X, Reilly SM, Chang L, Gomez AV, Saltiel AR, Showalter HD, Tesmer JJG. Carboxylic Acid Derivatives of Amlexanox Display Enhanced Potency toward TBK1 and IKKε and Reveal Mechanisms for Selective Inhibition. Mol Pharmacol. 2018;94:1210-1219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 141. | Dai J, Huang YJ, He X, Zhao M, Wang X, Liu ZS, Xue W, Cai H, Zhan XY, Huang SY, He K, Wang H, Wang N, Sang Z, Li T, Han QY, Mao J, Diao X, Song N, Chen Y, Li WH, Man JH, Li AL, Zhou T, Liu ZG, Zhang XM. Acetylation Blocks cGAS Activity and Inhibits Self-DNA-Induced Autoimmunity. Cell. 2019;176:1447-1460.e14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 264] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 142. | Duffy L, O'Reilly SC. Toll-like receptors in the pathogenesis of autoimmune diseases: recent and emerging translational developments. Immunotargets Ther. 2016;5:69-80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 79] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 143. | Lenert P, Yasuda K, Busconi L, Nelson P, Fleenor C, Ratnabalasuriar RS, Nagy PL, Ashman RF, Rifkin IR, Marshak-Rothstein A. DNA-like class R inhibitory oligonucleotides (INH-ODNs) preferentially block autoantigen-induced B-cell and dendritic cell activation in vitro and autoantibody production in lupus-prone MRL-Fas(lpr/Lpr) mice in vivo. Arthritis Res Ther. 2009;11:R79. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 144. | An J, Woodward JJ, Lai W, Minie M, Sun X, Tanaka L, Snyder JM, Sasaki T, Elkon KB. Inhibition of Cyclic GMP-AMP Synthase Using a Novel Antimalarial Drug Derivative in Trex1-Deficient Mice. Arthritis Rheumatol. 2018;70:1807-1819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 93] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 145. | Morehouse C, Chang L, Wang L, Brohawn P, Ueda S, Illei G, Greth W, Yoo S, Roskos L, Yao Y. Target Modulation of a Type I Interferon (IFN) Gene Signature with Sifalimumab or Anifrolumab in Systemic Lupus Erythematosus (SLE) Patients in Two Open Label Phase 2 Japanese Trials.: 719. Arthrit Rheumatol. 2014;. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |